Abstract
We have previously shown that the pilL, pilN, pilQ, pilS, pilU, and pilV genes of plasmid R64 encode outer membrane lipoprotein, secretin, cytoplasmic ATPase, major pilin, prepilin peptidase, and minor pilin, respectively, which are required for thin-pilus formation. In this work, we characterized the products of the remaining essential genes, pilK, pilM, pilO, pilP, pilR, and pilT, with regard to their localization and processing. Overexpression systems containing pilM, pilO, and pilP genes fused with N-terminal glutathione S-transferase (GST) or a His tag were constructed. Overproduced proteins were purified and used to raise specific antibodies. Localization of PilM, PilO, and PilP proteins was performed by Western blot analysis with anti-GST-PilM, anti-PilO, and anti-PilP antibodies, respectively. The pilK, pilR, and pilT products were produced with a C-terminal His tag and then detected by anti-His tag antibody. Subcellular fractionation experiments with Escherichia coli cells producing R64 thin pili revealed that PilK, PilM, and PilR are inner membrane proteins, and PilP and PilT are periplasmic proteins. PilO protein was localized to the outer membrane in the presence of other Pil proteins, whereas it was localized to the cytoplasm in the absence of these proteins. Furthermore, the cleavage site of PilP protein was determined by N-terminal amino acid sequencing of purified mature PilP protein. We predict that PilK, PilM, PilO, PilP, and PilT proteins function as the components of the pilin transport apparatus and thin-pilus basal body.
Type IV pili are flexible, filamentous structures protruding from the cell surface of gram-negative bacteria (15, 33). In many pathogenic bacteria, including Pseudomonas aeruginosa, Neisseria gonorrhoeae, Vibrio cholerae, and enteropathogenic Escherichia coli, type IV pili promote the attachment of bacterial pathogens to the specific receptors of host cells during colonization (5). Such attachment of bacterial pathogens to host cells by type IV pili is an essential event for the initiation of infection. Many laboratories have investigated type IV pilus biogenesis to reveal the mechanism of bacterial infection. Type IV pilus is composed of small pilin subunits, which are derived from type IV prepilin through cleavage of the N-terminal prepeptide. Cleavage of prepilin occurs at the cytoplasmic side of the inner membrane by cognate prepilin peptidase (19). In many cases, the N-terminal amino acid of mature pilin is methylated. Both cleavage and methylation of pilin are catalyzed by a single bifunctional prepilin peptidase (34). After modification, mature pilins are transported across the inner membrane to form the pilus structure. Many additional gene products are required for the biogenesis of type IV pili (9, 15, 21, 32).
The type IV pilus biogenesis system is closely related to type II secretion pathways (also called general secretion pathways) and DNA uptake systems. Many type II secretion systems have been identified in gram-negative bacteria (17, 23, 27). To form type II secretion machinery, usually 10 to 14 gsp genes are required. Several common genes have been identified among type IV pilus biogenesis systems and type II secretion systems, including prepilin or pseudopilin, NTP-binding proteins, secretins, prepilin peptidases, and integral membrane proteins. In many type II secretion systems, most of the general secretion pathway (Gsp) proteins have been localized to the membrane fraction (24, 27).
The self-transmissible IncI1 plasmids, including R64 and ColIb-P9, produce thick and thin pili (12, 39). The thick pilus is generally involved in DNA transfer, while the thin pilus is involved in recognition of and binding to recipient cells in liquid matings. One-third of the 54-kb R64 transfer region is required for the formation of thin pili (Fig. 1A). This region contains 14 pil genes, 12 of which are essential for thin-pilus biogenesis (9, 11, 40). Several pil gene products share amino acid sequence similarity with proteins involved in type IV pilus biogenesis, indicating that R64 thin pilus belongs to the type IV pilus family. R64 pilS, pilV, and pilU genes encode the major pilin, minor pilin, and prepilin peptidase, respectively (9, 40). The C-terminal segments of the pilV genes are under the control of multiple DNA inversion of shufflon and determine the recipient specificity in liquid matings (10). The pilL and pilN gene products are outer membrane lipoproteins, and the pilQ gene product is a cytoplasmic ATPase (28, 29). The remaining pil genes are likely to encode structural proteins that function in the establishment of the pilin transport apparatus and thin-pilus basal body. Most of these pil gene products contain signal sequences or transmembrane domains (9), suggesting that they are transported to the periplasmic space, inner membrane, and outer membrane.
FIG. 1.
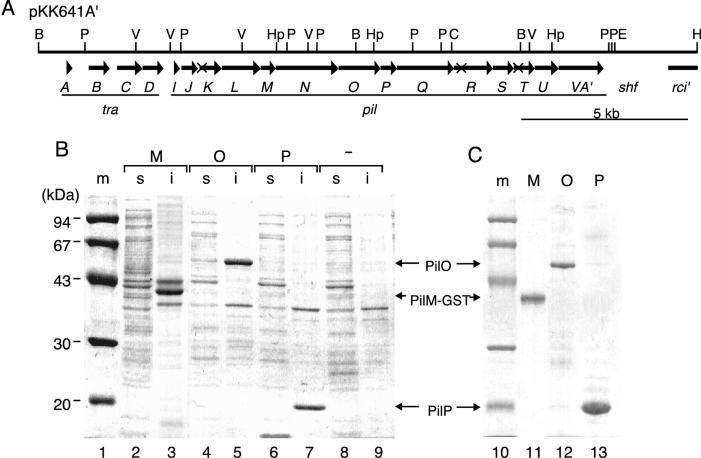
(A) Gene organization of the traA to -D and pilI to -V region of pKK641A′. The top horizontal line represents a restriction map: B, BglII; C, ClaI; E, EcoRI; H, HindIII; Hp, HpaI; P, PstI; and V, PvuII. Below the map, coding sequences of various genes are represented by arrows: tra, transfer; pil, formation of thin pilus; shf, shufflon; and rci, shufflon-specific recombinase. The locations of pilK1, pilR2, and pilT1 mutations are indicated by X. (B) Overexpression of PilM-GST and His-tagged PilO and PilP proteins. E. coli BL21(DE3) cells harboring pEM-GST (lanes 2 and 3), pEO28 (lanes 4 and 5), and pEP28 (lanes 6 and 7) and control cells without plasmid (lanes 8 and 9) were treated with 1 mM IPTG for 3 h. Soluble (lanes 2, 4, 6, and 8) and insoluble (lanes 3, 5, 7, and 9) fractions were prepared from the induced cells. After SDS-PAGE, proteins were stained with Coomassie brilliant blue. Molecular size markers (lanes 1 and 10 [m]), in kilodaltons, are indicated on the left side. (C) Purification of PilM-GST and His-tagged PilO and PilP proteins. His-tagged PilO and PilP proteins in the insoluble fraction were dissolved in 6 M guanidine hydrochloride. Solubilized proteins were applied to Co2+ affinity columns. Bound protein was eluted with 500 mM imidazole. PilM-GST fusion protein in the insoluble fraction was purified by washing in detergent. After SDS-PAGE, proteins were stained with Coomassie brilliant blue. Lanes: 11, purified PilM-GST; 12, purified His-tagged PilO; and 13, purified His-tagged PilP.
The present work was performed to identify and localize the products of the R64 pilK, pilM, pilO, pilP, pilR, and pilT genes. Overexpression systems for the pilM, pilO, and pilP genes were constructed, and antisera against the purified proteins were generated. The products of the pilK, pilR, and pilT genes were produced with a C-terminal His tag. Localization of these proteins was performed by immunological detection. Furthermore, the cleavage site of mature PilP protein was determined.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| JM83 | ara Δ(lac-proAB) rpsL thi φ80 dlacZΔM15 | 38 |
| NF83 | recA56 ara Δ(lac-proAB) rpsL thi φ80 dlacZΔM15 | 29 |
| BL21(DE3) | dcm ompT hsdS gal/λ(DE3) | 35 |
| TN102 | = W3110 Nalr | 11 |
| Plasmids | ||
| pUC119 | Apr, pMB1 ori, lacZ′ | 38 |
| pUH23a | Apr, pMB1 ori, lac promoter, 6-His tag | This study |
| pET11a | Apr, pMB1 ori, T7 promoter | 35 |
| pET28b | Kmr, pMB1 ori, T7 promoter, 6-His tag | 35 |
| pET11 km-GST | Kmr, pMB1 ori, T7 promoter, GST fusion | 20 |
| pKK641A′ | Kmr, R64drd-11 derivative carrying 18.5-kb rep and pil segment | 11 |
| pKK641A′ pilK1 | pKK641A′ carrying the pilK1 frameshift mutation | 40 |
| pKK641A′ pilR2 | pKK641A′ carrying the pilR2 frameshift mutation | 40 |
| pKK641A′ pilT1 | pKK641A′ carrying the pilT1 frameshift mutation | 40 |
| pKK661 | Cmr, pHSG576 (pSC101 rep) derivative carrying 35.6-kb tra segment | 11 |
| pKK698a | 0.8-kb pilM fragment in pUC119 | 40 |
| pKK700a | 2.0-kb pilO fragment in pUC119 | 40 |
| pKK701a | 1.0-kb pilP fragment in pUC119 | 40 |
| pEO28 | 1.3-kb pilO fragment in pET28b | This study |
| pEP28 | 0.5-kb pilP fragment in pET28b | This study |
| pEM-GST | 0.5-kb pilM fragment in pET11 km-GST | This study |
| pEP11 | 0.5-kb pilP fragment in pET11a | This study |
| pUK23 | 0.6-kb pilK fragment in pUH23a | This study |
| pUR23 | 1.1-kb pilR fragment in pUH23a | This study |
| pUT23 | 0.5-kb pilT fragment in pUH23a | This study |
Luria-Bertani (LB) and M9 glucose media were prepared as previously described (30). The solid medium contained 1.5% agar. Antibiotics were added to the liquid and solid media at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml; and nalidixic acid, 20 μ/ml.
Construction of plasmids.
Recombinant DNA techniques were performed as previously described (30).
To construct the pilO, pilP, and pilM overexpression plasmids, an NdeI site was introduced at the translation initiation site of the pilO, pilP, and pilM genes, respectively, by PCR with synthetic oligonucleotides. The NdeI-BamHI fragments of PCR products were inserted into the NdeI-BamHI site of pET28b to give pEO28 (containing the pilO gene) and pEP28 (containing the pilP gene), or into the NdeI-BamHI site of the glutathione S-transferase (GST) fusion plasmid pET11 km-GST to give pEM-GST (containing the pilM gene). The NdeI-BamHI fragment containing the pilP gene was inserted into the NdeI-BamHI site of pET11a to give pEP11.
To express the pilK, pilR, and pilT genes with a C-terminal His tag, a pUC119-based vector, pUH23a, was first constructed as follows. The DNA segment between the T7 promoter and terminator of pET23a was amplified by PCR with T7 promoter and terminator primers. The PCR product was ligated with pUC119 DNA digested with EcoRI and HindIII and treated with Klenow enzyme, to give pUH23a. The NdeI and XhoI sites were introduced at the translation initiation and termination sites, respectively, of pilK, pilR, and pilT genes by PCR with synthetic oligonucleotides. PCR products digested with NdeI and XhoI were inserted into the NdeI-XhoI site of pUH23a to give pUK23, pUR23, and pUT23, respectively.
Overexpression and purification of His-tagged PilO and PilP proteins.
E. coli BL21(DE3) cells harboring pEO28 or pEP28 were grown in LB medium until they had reached an A600 of 0.5. Then isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1.0 mM, and cultivation was continued for 3 h. IPTG-induced cells from 200-ml cultures were suspended in 50 ml of TS buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl), broken by a French pressure cell, and centrifuged at 100,000 × g for 30 min. The precipitate was dissolved in 50 ml of lysis buffer (TS buffer containing 6 M guanidine-HCl) and then centrifuged at 100,000 × g for 30 min. The supernatant was applied to a Talon Co2+ affinity column (Clontech) equilibrated with lysis buffer. After the column had been washed with lysis buffer, the bound proteins were eluted from the column with lysis buffer containing 500 mM imidazole. The His-tagged PilO and PilP proteins in the eluate fractions were precipitated by acetone.
Overexpression and purification of GST-PilM fusion protein.
E. coli BL21(DE3) cells harboring pLysS and pEM-GST were grown in LB medium until they reached an A600 of 0.5. IPTG was added to a final concentration of 1.0 mM, and cultivation was continued for 3 h. IPTG-induced cells from 200-ml cultures were suspended in 50 ml of TS buffer, broken by a French pressure cell, and centrifuged at 100,000 × g for 30 min. The precipitate was suspended in 8 ml of 50 mM Tris-HCl buffer (pH 8.0) containing 4% Triton X-100, shaken at room temperature for 30 min, and centrifuged 25,000 × g for 20 min. The precipitate was washed three times with 8 ml of 50 mM Tris-HCl buffer (pH 8.0) containing 4% Triton X-100 to remove contaminant proteins. To remove the detergent, the precipitate was suspended in 16 ml of distilled water, shaken at room temperature for 30 min, and centrifuged at 25,000 × g for 20 min. The pellet containing GST-PilM protein was stored at −20°C until use.
Preparation of anti-GST-PilM, anti-PilO, and anti-PilP antibodies.
Final purification of Pil proteins for the preparation of antibodies was carried out by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins in the pellets described above were dissolved in SDS sample buffer (2% SDS, 0.2 M 2-mercaptoethanol, 0.01% bromophenol blue, 10% glycerol, 80 mM Tris-HCl [pH 6.8]), boiled for 3 min, and separated by SDS-PAGE (12% polyacrylamide). The proteins in the gel were stained with ice-cold 1 M KCl; subsequently, the band containing the respective protein was cut out. Each protein in the gel was used to immunize rabbits. Anti-PilP antibody was purified by affinity chromatography with purified mature PilP protein-conjugated N-hydroxysuccinimide (NHS)-activated agarose beads (Amersham Pharmacia Biotech).
Purification of mature PilP protein.
E. coli BL21(DE3) cells harboring pEP11 were grown in LB medium at 37°C until they reached an A600 of 0.5. IPTG was added to a final concentration of 0.5 mM, and cultivation was continued for 3 h. The IPTG-induced cells from 3-liter cultures were suspended in 300 ml of OS buffer (100 mM Tris-HCl [pH 8.0], 10 mM EDTA, 20% sucrose). Lysozyme solution (0.5 ml [5 mg/ml in OS buffer]) was added, and then cells were incubated for 30 min at 0°C. After spheroplasts and cell debris were removed by centrifugation at 200,000 × g for 30 min, the supernatant (periplasmic fraction) was used for the purification of mature PilP protein. Ammonium sulfate was added to a final concentration of 40% (wt/vol) to the periplasmic fraction. After 1 h, samples were centrifuged at 20,000 × g for 30 min. The precipitate was dissolved in 20 mM Tris-HCl (pH 7.5) containing 20% ammonium sulfate and loaded onto a Phenyl 5-PW hydrophobic chromatography column (Tosoh) equilibrated with 20 mM Tris-HCl (pH 7.5) containing 20% ammonium sulfate. The column was washed with 20 mM Tris-HCl containing 20% ammonium sulfate, and then bound proteins were eluted with a linear gradient (from 20 to 0%) of ammonium sulfate. The PilP-enriched fractions were applied to a Superose 6 gel filtration column (Amersham Pharmacia Biotech) equilibrated with 20 mM Tris-HCl (pH 7.5). Peak fractions of PilP protein were concentrated and dialyzed against 20 mM Tris-HCl.
The N-terminal amino acid sequence of mature PilP protein was determined by Edman degradation in a model 477A protein sequencer (Applied Biosystems).
Subcellular fractionation.
E. coli cells harboring various plasmids were grown in LB medium at 37°C until they reached an A600 of 1.0. The cells harvested from 5-ml cultures were lysed by EDTA-lysozyme and fractionated to periplasmic, cytoplasmic, and crude membrane fractions as previously described (4). Crude membrane fraction was separated into inner and outer membranes by the three-step sucrose gradient procedure (25). In our hands, 67% of cellular succinate dehydrogenase activity was recovered in the inner membrane fraction, while 18% was recovered in the outer membrane fraction. By SDS-PAGE, OmpF protein was detected only in the outer membrane fraction. Approximately 85% of cellular alkaline phosphatase activity was recovered in the periplasmic fraction, while 15% was recovered in the cytoplasmic fraction.
Localization of PilM, PilO, and PilP proteins was determined by Western blot analysis with anti-GST-PilM, anti-PilO, and anti-PilP antibodies, respectively. To detect proteins containing His tag by Western blotting, anti-Penta-His antibody (Qiagen) was used.
Processing of pre-PilP protein.
E. coli BL21(DE3) cells harboring pEP11 were grown in M9 glucose medium until they reached an A600 of 0.5 at 37°C. IPTG was added to a final concentration of 1.0 mM, and cultivation was continued for 20 min. Rifampin was added to a final concentration of 10 μg/ml, after which the culture was shifted to 42°C and cultivation was continued for 30 min. To label PilP protein, 10 μCi of [35S]methionine (1,000 Ci/mmol; ICN) was added to the culture. After 2 min, incorporation was terminated by the addition of excess nonradioactive methionine (final concentration, 1 mM). For the detection of pre-PilP protein, sodium azide was added to a final concentration of 3 mM prior to the addition of [35S]methionine, and then cultivation was continued for 5 min at 42°C. The labeled cells were dissolved in SDS sample buffer and boiled for 3 min. The labeled proteins were separated by SDS-PAGE (15% polyacrylamide) and detected by fluorography.
RESULTS AND DISCUSSION
Overexpression and purification of GST-PilM and His-tagged PilO and PilP proteins.
To determine the intracellular localization of the R64 pil products, pilM, pilO, and pilP genes were cloned into overexpression vectors carrying the T7 promoter. The respective coding sequences of these genes were amplified by PCR with primers containing an NdeI site at their translation initiation sites. The NdeI-BamHI fragments of the amplified DNAs were inserted into pET28b or pET11 km-GST, in which the cloned genes were fused with an N-terminal His tag or E. coli gst gene, respectively. Upon IPTG induction, E. coli BL21(DE3) cells harboring pEM-GST (pilM gene in pET11 km-GST), pEO28 (pilO gene in pET28b), and pEP28 (pilP gene in pET28b) overproduced proteins with molecular masses of approximately 40, 50, and 19 kDa, respectively (Fig. 1B, lanes 3, 5, and 7). Since these proteins were not produced in E. coli cells without plasmid (Fig. 1B, lanes 8 and 9) and the apparent molecular masses of these proteins are consistent with the calculated values of 39,312 Da (GST-PilM), 50,325 Da (His-tagged PilO), and 18,167 Da (His-tagged PilP), the overproduced 40-, 50-, and 19-kDa proteins are likely to be GST-PilM, His-tagged PilO, and His-tagged PilP proteins, respectively.
Overproduced His-tagged PilO and PilP proteins were recovered in the insoluble fractions (Fig. 1B, lanes 5 and 7). These proteins were dissolved in 6 M guanidine-HCl and then subjected to Co2+ affinity chromatography. The bound proteins were eluted with 500 mM imidazole, and the peak fractions of the His-tagged PilO and PilP proteins were pooled (Fig. 1C, lanes 12 and 13). Overproduced GST-PilM protein was also recovered in the insoluble fraction (Fig. 1B, lane 3). Since GST-PilM protein was insoluble in 4% Triton X-100 and most contaminant proteins were fairly soluble in this solution, GST-PilM protein was purified by washing the insoluble fraction with 4% Triton X-100 (Fig. 1C, lane 11).
To raise antibodies against GST-PilM and His-tagged PilO and PilP, the proteins were further purified by SDS-PAGE. The bands containing the respective proteins were excised out of the gel and used to immunize rabbits. PilM, PilO, and PilP proteins produced in E. coli cells harboring pKK641A′ were detected by Western blot analysis with anti-GST-PilM, anti-PilO, and anti-PilP antibodies, respectively (Fig. 2, left panel, lanes W). The apparent molecular masses of PilM and PilO proteins were approximately 16 and 48 kDa, respectively, which are consistent with the calculated values of 15,716 Da (PilM) and 48,162 Da (PilO), suggesting that PilM and PilO proteins are not processed. As will be described below, processing of PilP protein was demonstrated. Hence, the observed molecular mass, 16 kDa, of mature PilP protein is apparently higher than the calculated value of 13,646 Da. In a previous report (9), PilP protein was identified as 16-kDa protein by the maxicell procedure.
FIG. 2.
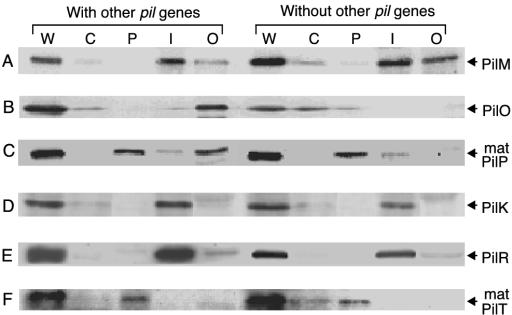
Subcellular localization of the PilM (A), PilO (B), PilP (C), PilK (D), PilR (E), and PilT (F) proteins. To determine the localization of the PilM, PilO, and PilP proteins, E. coli cells harboring pKK641A′ were grown in LB medium and fractionated to cytoplasmic, periplasmic, and inner and outer membrane fractions (with other pil genes). Cells harboring pKK698a, pKK700a, or pKK701a were also fractionated (without other pil genes). To determine the localization of His-tagged PilK, PilR, and PilT proteins, cells harboring pKK641A′ pilK1 and pUK23, pKK641A′ pilR1 and pUR23, or pKK641A′ pilT1 and pUT23 were fractionated (with other pil genes). Cells harboring only pUK23, pUR23, or pUT23 were also fractionated (without other pil genes). After SDS-PAGE, PilM, PilO, and PilP proteins were detected by Western blotting with anti-PilM-GST, anti-PilO, and anti-PilP antibodies, respectively. His-tagged PilK, PilR, and PilT proteins were detected by Western blotting with anti-Penta-His antibody. Only protein bands of interest are shown in this figure. The location of each protein is indicated on the right. mat, mature. Lanes: W, whole cell; C, cytoplasm; P, periplasm; I, inner membrane; and O, outer membrane.
Localization of R64 PilM and PilO proteins.
Localization of R64 PilM and PilO proteins within E. coli cells was performed by Western blot analysis with anti-GST-PilM and anti-PilO antibodies, respectively. Subcellular localization of various R64 Pil proteins was performed in E. coli cells with different backgrounds: (i) each Pil protein was expressed with other R64 Pil proteins, or (ii) each Pil protein was individually expressed (without other R64 Pil proteins). For the localization of PilM and PilO proteins with other R64 Pil proteins, E. coli cells harboring pKK641A′ (Fig. 1A) were grown in LB medium. For the localization of PilM or PilO proteins without other R64 Pil proteins, E. coli cells harboring pKK698a or pKK700a, respectively, were grown.
E. coli cells grown as described above were lysed open by EDTA-lysozyme and separated into cytoplasmic, periplasmic, and inner and outer membrane fractions. Each fraction was analyzed by SDS-PAGE followed by Western blot analysis with anti-GST-PilM or anti-PilO antibodies (Fig. 2). When the pilM gene was expressed without other pil genes in E. coli cells harboring pKK698a, the majority of PilM protein was found in the inner membrane fraction, while minor and residual amounts of PilM protein were detectable in the outer membrane and cytoplasmic fractions, respectively (Fig. 2A). When the pilM gene was expressed with other pil genes in E. coli cells harboring pKK641A′, the majority of PilM protein was found in the inner membrane fraction, while a small portion was found in the outer membrane fraction.
Since PilM protein has a putative transmembrane domain near its N terminus (9), it may be an integral membrane protein. Positively charged residues are present behind the putative transmembrane domain, suggesting that a large portion of PilM protein may exist in the cytoplasmic side of the inner membrane.
When the pilO gene was expressed without other pil genes, PilO protein was mainly found in the cytoplasmic fraction (Fig. 2B). In contrast, when the pilO gene was expressed with the other pil genes, the majority of PilO protein was found in the outer membrane fraction, while a small portion of PilO protein was found in the cytoplasmic fraction.
These results suggest that PilO protein is a cytoplasmic protein in the absence of other Pil proteins, but PilO protein is translocated to the outer membrane in the presence of other Pil proteins. Alternatively, PilO protein may form a complex with other Pil protein(s) that is recoverable in the outer membrane fraction, since PilO protein does not contain a signal peptide motif. Further investigations are required to reveal the translocation of PilO protein.
Processing and localization of PilP protein.
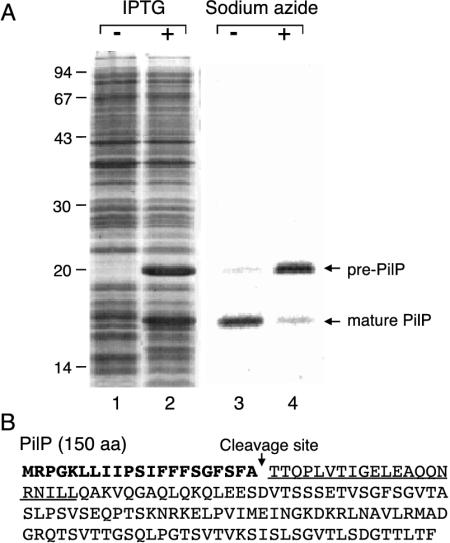
When the R64 pilP gene was overexpressed in E. coli cells harboring pEP11, two proteins with molecular masses of 20 and 16 kDa were overproduced (Fig. 3A, lane 2). The predicted amino acid sequence of PilP protein contains a signal peptide motif (Fig. 3B), suggesting that the 20- and 16-kDa proteins correspond to pre-PilP and mature PilP proteins, respectively. In fact, the 16-kDa mature PilP protein was detected mainly in the periplasmic fraction in the subcellular fractionation experiments as described below. Since SecA function has been demonstrated as being inactivated by the addition of sodium azide to the culture medium (22), inhibition of pre-PilP processing by sodium azide was expected. To analyze the processing of pre-PilP protein, labeling experiments were performed in the presence or absence of sodium azide. When the pilP product was labeled in the absence of sodium azide, the 16-kDa mature PilP protein was produced, indicating normal PilP processing (Fig. 3A, lane 3). In contrast, when the pilP product was labeled in the presence of 3 mM sodium azide, accumulation of the 20-kDa pre-PilP protein was observed (Fig. 3A, lane 4). These results indicate that the 20- and 16-kDa proteins are pre-PilP and mature PilP proteins, respectively.
FIG. 3.
Processing of the pilP gene product. (A) E. coli BL21(DE3) cells harboring pET11a (lane 1) and pEP11 (lane 2) were treated with 1 mM IPTG. The cell proteins were analyzed by SDS-PAGE, followed by Coomassie brilliant blue staining. E. coli cells harboring pEP11 were treated with 1 mM IPTG and then labeled with [35S]methionine in the absence (lane 3) or presence (lane 4) of 3 mM sodium azide. The cell proteins were analyzed by SDS-PAGE, followed by fluorography. The positions of pre-PilP and mature PilP proteins are indicated on the right. Numbers on the left indicate the sizes (in kilodaltons) of marker proteins. (B) Determination of the cleavage site of PilP protein. The amino acid sequence of PilP protein deduced from its nucleotide sequence (GenBank accession no. AB027308) is indicated. The signal peptide is indicated in boldface type. The N-terminal amino acid sequence determined for purified mature PilP protein is underlined. The signal peptidase cleavage site of pre-PilP protein is indicated by a downward arrow.
When the pilP gene was overexpressed with an N-terminal His tag, a portion of the His-tagged pre-PilP protein was also processed. After 3 h of IPTG induction of the cells harboring pEP28, approximately 20% of the overproduced PilP protein was processed and recovered in the soluble fraction (data not shown). Thus, the presence of the N-terminal His tag may reduce the efficiency of pre-PilP processing.
To define the cleavage site of pre-PilP protein, the N-terminal amino acid sequence of mature PilP protein was determined. First, mature PilP protein was purified from the periplasmic fraction of the IPTG-induced cells harboring pEP11. Induced cells overproduced the 16-kDa mature PilP protein in the periplasmic fraction. The mature PilP protein was purified by ammonium sulfate precipitation, followed by hydrophobic and gel filtration chromatography. Purified mature PilP protein ran as a single band by SDS-PAGE (data not shown). The N-terminal amino acid sequence of the mature PilP protein was determined to be TTQPLVTIGELEAQQNRNIL. This sequence corresponds to the 22nd to 41st residues of pre-PilP protein (Fig. 3B), indicating that pre-PilP protein is cleaved between Ala21 and Thr22. These results suggest that the R64 pilP product is processed and translocated to the periplasm by the Sec machinery. Features of the PilP signal peptide are similar to those of standard signal peptides (23).
When the pilP gene was expressed without other pil genes, the majority of mature PilP protein was found in the periplasmic fraction, while a minor portion was found in the inner membrane fraction (Fig. 2C). Without overexpression, pre-PilP protein could not be detected. When the pilP gene was expressed with the other pil genes, equal amounts of mature PilP protein were found in the periplasmic and outer membrane fractions, and a minor portion of PilP protein was found in the inner membrane fraction.
TrbJ protein in the RP4 conjugation system has been localized to the periplasm when expressed in the absence of other Tra and Trb proteins. In contrast, TrbJ protein was loosely associated with the other Tra and Trb proteins in their presence (7). The mature PilP protein may interact with other Pil protein(s) in a similar manner.
Expression of R64 PilK, PilR, and PilT proteins with His tag.
Since overexpression of the pilK, pilR, and pilT genes in pET28b or pET11 km-GST was unsuccessful (data not shown), antibodies against the PilK, PilR, and PilT proteins could not be prepared. The pilK, pilR, and pilT genes were cloned into vector pUH23a, to give pUK23, pUR23, and pUT23, respectively, in which their products were produced with a C-terminal His tag. To assess the effects of C-terminal His tag on the function of each Pil protein, the ability of each Pil protein with His tag to complement the respective frameshift mutations of pKK641A′ was estimated by liquid matings (Table 2). E. coli NF83 donor cells harboring pKK661 and pKK641A′ transmitted pKK661 into the recipient cells at a frequency of 1.6%, while those harboring pKK661 and pKK641A′ pilK1, pKK641A′ pilR2, or pKK641A′ pilT1 did not. Transfer frequencies of E. coli cells harboring pKK661 and pKK641A′ pilK1 and those harboring pKK661 and pKK641A′ pilR2 were recovered to wild-type levels by the introduction of pUK119 and pUR119, respectively (Table 2). The transfer frequency of cells harboring pKK661, pKK641A′ pilT1, and pUT23 was 0.04%, indicating a low level of complementation. Failure of complementation of the pilT1 mutation by pUC118- and pUC119-derived plasmids carrying the pilT gene has been previously reported (40). These results suggest that PilK, PilR, and PilT proteins with a C-terminal His tag exhibit normal activity in R64 thin-pilus biogenesis and that their C-terminal His tag does not affect their intracellular localization.
TABLE 2.
Effects of C-terminal His tag fused to pilK, pilR, and pilT genes on the complementation of the respective pil mutations
| Plasmids | Transfer frequency (%)a |
|---|---|
| pKK661, pKK641A′ | 1.6 |
| pKK661, pKK641A′ pilK1 | <0.001 |
| pKK661, pKK641A′ pilK1, pUK23 | 1.4 |
| pKK661, pKK641A′ pilR2 | <0.001 |
| pKK661, pKK641A′ pilR2, pUR23 | 1.0 |
| pKK661, pKK641A′ pilT1 | <0.001 |
| pKK661, pKK641A′ pilT1, pUT23 | 0.04 |
Transfer frequency of pKK661 from E. coli NF83 donor cells harboring pKK661, pKK641A′ pilK1, pilR2 or pilT1 mutations with or without complementation plasmids to TN102 recipient cells in liquid matings was estimated as described previously (28). Transfer frequency is indicated as a percentage of transconjugants relative to donor cells.
Localization of R64 PilK, PilR, and PilT proteins.
To express the pilK, pilR, and pilT genes with the other R64 pil genes, E. coli NF83 cells harboring pKK641A′ pilK1 and pUK23, pKK641A′ pilR2 and pUR23, or pKK641A′ pilT1 and pUT23 were grown in LB medium. To express these genes without the other pil genes, E. coli cells harboring pUK23, pUR23, or pUT23 were grown. E. coli cells were fractionated to cytoplasmic, periplasmic, and inner and outer membrane fractions. Each fraction was subjected to SDS-PAGE followed by Western blot analysis with anti-Penta-His antibody.
When the pilK gene was expressed without other pil genes, PilK protein was found in the inner membrane fraction (Fig. 2D). When the pilK gene was expressed with other pil genes, localization of PilK protein was similar to that observed for individual expression. PilK protein has two putative transmembrane domains near the N terminus, which may span the inner membrane. The distribution of positively charged amino acid residues suggests that the C-terminal large part of PilK protein may be exposed to the cytoplasmic side of the inner membrane.
When the pilR gene was expressed without other pil genes, the majority of PilR protein was found in the inner membrane fraction, while a minor portion was detected in the outer membrane fraction (Fig. 2E). When the pilR gene was expressed with other pil genes, localization of PilR protein was similar to that observed for individual expression.
PilR protein, which contains three putative transmembrane domains, seems to be a polytopic integral membrane protein. R64 PilR protein shares amino acid sequence similarity with many bacterial proteins related to type IV pilus biogenesis, such as PilC of P. aeruginosa, BfpE of enteropathogenic E. coli, and TcpE of V. cholerae, as well as those related to type II secretion pathways, such as PulF of Klebsiella oxytoca and OutF of Erwinia chrysanthemi (2, 8, 14, 18, 23, 26, 32). In addition, ComGB of the B. subtilis DNA uptake system belongs to this group (1). These proteins carry three transmembrane domains located at similar positions within the molecule, implicating their inner membrane localization. Fusion of outF to blaM revealed that OutF consists of a large N-terminal cytoplasmic domain, a smaller periplasmic domain, and a large cytoplasmic loop (36), suggesting that R64 PilR also displays similar membrane topology.
When the pilT gene was expressed without or with other pil genes, the majority of PilT protein was found in the periplasmic fraction, while a minor portion was in the cytoplasmic fraction (Fig. 2F).
PilT protein is likely to be cleaved between Ala25 and Ser26 to produce a periplasmic mature protein. The amino acid sequence of PilT protein has similarity to those of P19 proteins (gene X proteins) of F and R1 plasmids, as well as the C-terminal domain of E. coli Slt protein (9). Processing of P19 protein was demonstrated (3). Subcellular fractionation experiments indicated that mature P19 protein is a periplasmic protein that may be attached to the putative membrane-spanning DNA transport complex. X-ray crystallography revealed that Slt protein has a three-dimensional structure similar to those of hen egg and T4 muramidases (37). It is possible that R64 PilT carries muramidase activity and forms a pore in the peptidoglycan layer to allow passage of thin pilus.
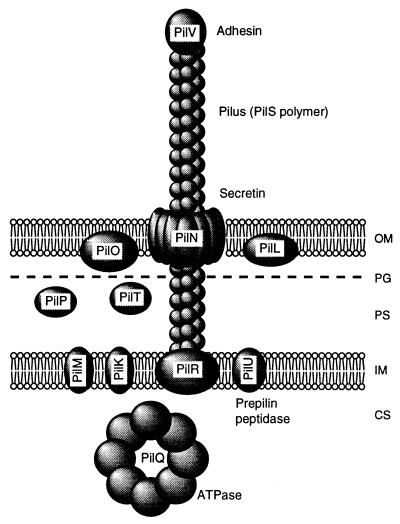
In the present work, we have identified and localized the products of the pilK, pilM, pilO, pilP, pilR, and pilT genes, which are essential for the formation of R64 thin pilus. The locations of these proteins are schematically illustrated in Fig. 4together with those of other Pil proteins previously described (28, 29, 39). We predict that PilK, PilM, PilO, PilP, and PilT proteins function as the components of the pilin transport apparatus and thin-pilus basal body, since they are localized to cell envelope. The features of the various pil gene products are summarized in Table 3.
FIG. 4.
Schematic representation of localization of R64 Pil proteins. The size and spatial arrangement of each Pil protein are hypothetical. OM, outer membrane; PG, peptidoglycan layer; PS, periplasmic space; IM, inner membrane; CS, cytoplasmic space.
TABLE 3.
Properties of R64 pil gene products
| Gene | No. of amino acids | Mol mass
|
Locationb
|
gsp homologue | Description and reference | ||
|---|---|---|---|---|---|---|---|
| Calculated (Da) | Found (kDa)a | Predicted | Found | ||||
| pilK | 196 | 23,184 | 30 [+His] | IM | IM | ||
| pilL | 355 | 38,603 | 38 | IM/OM | OM | Lipoprotein (28) | |
| pilM | 145 | 15,716 | 16 | P/OM | IM | ||
| pilN | 560 | 59,848 | 57 | M/OM | OM | gspD | Secretin (28) |
| pilO | 431 | 48,162 | 48 | IM | OM | ||
| pilP | 150 | 16,003 | 20 | IM | |||
| matc | 129 | 13,646 | 16 | P/OM | |||
| pilQ | 517 | 58,280 | 58 | C | C | gspE | ATPase (29) |
| pilR | 365 | 41,415 | 44 [+His] | IM | IM | gspF | Integral membrane protein |
| pilS | 204 | 21,265 | 22 | IM | gspGHIJ | Prepilin (39) | |
| mat | 181 | 18,715 | 19 | EC | Pilin (39) | ||
| pilT | 186 | 20,758 | IM | ||||
| mat | 161 | 18,140 | 24 [+His] | P | Putative muramidase | ||
| pilU | 218 | 24,600 | NTd | IM | NT | gspO | Prepilin peptidase (39) |
| pilVA′ | 430 | 45,758 | 44 | P/OM | EC | Adhesin (10, 39) | |
[+His], molecular mass was estimated for the protein containing a six-His tag.
C, cytoplasm; IM, inner membrane; OM, outer membrane; P, periplasm; EC, extracellular.
mat, mature form.
NT, not tested.
In type II secretion pathways, 10 to 14 proteins are thought to be involved. Two proteins, GspD and GspS, have been shown to be outer membrane proteins (16, 31): GspD is a secretin and forms a gated channel for protein secretion through the outer membrane. GspS lipoprotein binds to and stabilizes the GspD secretin channel. Other Gsp proteins, including GspGHIJ pseudopilins, are localized to the inner membrane (27). GspEFLM proteins were shown to constitute a complex in the inner membrane that may be used as a platform for assembling other parts of the secretion machinery (24). Since periplasmic proteins have not previously been found in the type II secretion pathways, the presence of two periplasmic proteins, PilP and PilT, in the R64 thin-pilus biogenesis system is of great interest. The presence of PilT in the periplasm is reasonable, since it may function as muramidase.
Components of the type I, III, and IV secretion machinery are thought to be organized in a supramolecular structure that spans both the inner and outer membranes (6, 7, 13). In these secretion pathways, protein transportation is independent of Sec machinery, while type II secretion occurs in two steps, including the Sec machinery. In type IV pilus, assembled pilin molecules themselves may span the inner and outer membranes, as described in Fig. 4. Further investigation is required for a better understanding of the structure and function of the type IV pilus biogenesis system.
Acknowledgments
We are grateful to K. Takayama for critical reading of the manuscript.
This work was supported in part by a grant from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Albano, M., R. Breitling, and D. A. Dubnau. 1989. Nucleotide sequence and genetic organization of the Bacillus subtilis comG operon. J. Bacteriol. 171:5386–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bally, M., A. Filloux, M. Akrim, G. Ball, A. Lazdunski, and J. Tommassen. 1992. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol. Microbiol. 6:1121–1131. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, M., K. Bischof, R. Noiges, and G. Koraimann. 2000. Subcellular localization and processing of the lytic transglycosylase of the conjugative plasmid R1. FEBS Lett. 466:389–393. [DOI] [PubMed] [Google Scholar]
- 4.Boeke, J. D., and P. Model. 1982. A prokaryotic membrane anchor sequence: carboxyl terminus of bacteriophage f1 gene III protein retains it in the membrane. Proc. Natl. Acad. Sci. USA 79:5200–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnenberg, M. S., J. A. Giron, J. P. Nataro, and J. B. Kaper. 1992. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol. Microbiol. 6:3427–3437. [DOI] [PubMed] [Google Scholar]
- 6.Fath, M. J., and R. Kolter. 1993. ABC transporters: bacterial exporters. Microbiol. Rev. 57:995–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grahn, A. M., J. Haase, D. H. Bamford, and E. Lanka. 2000. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J. Bacteriol. 182:1564–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaufman, M. R., C. E. Shaw, I. D. Jones, and R. K. Taylor. 1993. Biogenesis and regulation of the Vibrio cholerae toxin-coregulated pilus: analogies to other virulence factor secretory systems. Gene 126:43–49. [DOI] [PubMed] [Google Scholar]
- 9.Kim, S.-R., and T. Komano. 1997. The plasmid R64 thin pilus identified as a type IV pilus. J. Bacteriol. 179:3594–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komano, T. 1999. Shufflons: multiple inversion systems and integrons. Annu. Rev. Genet. 33:171–191. [DOI] [PubMed] [Google Scholar]
- 11.Komano, T., N. Funayama, S.-R. Kim, and T. Nisioka. 1990. Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J. Bacteriol. 172:2230–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komano, T., T. Yoshida, K. Narahara, and N. Furuya. 2000. The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol. Microbiol. 35:1348–1359. [DOI] [PubMed] [Google Scholar]
- 13.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S.-I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602–605. [DOI] [PubMed] [Google Scholar]
- 14.Lindeberg, M., and A. Collmer. 1992. Analysis of eight out genes required for pectic enzyme secretion by Erwinia chrysanthemi: sequence comparison with secretion genes from other gram-negative bacteria. J. Bacteriol. 174:7385–7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattick, J. S., C. B. Whitchurch, and R. A. Alm. 1996. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa—a review. Gene 179:147–155. [DOI] [PubMed] [Google Scholar]
- 16.Nouwen, N., N. Ranson, H. Saibil, B. Wolpensinger, A. Engel, A. Ghazi, and A. P. Pugsley. 1999. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl. Acad. Sci. USA 96:8173–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nunn, D. 1999. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 9:402–408. [DOI] [PubMed] [Google Scholar]
- 18.Nunn, D., S. Bergman, and S. Lory. 1990. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J. Bacteriol. 172:2911–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nunn, D. N., and S. Lory. 1991. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. USA 88:3281–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogawa, M., S. Fujitani, X. Mao, S. Inouye, and T. Komano. 1998. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol. Microbiol. 22:757–767. [DOI] [PubMed] [Google Scholar]
- 21.Ogierman, M. A., S. Zabihi, L. Mourtzios, and P. A. Manning. 1993. Genetic organization and sequence of the promoter-distal region of the tcp gene cluster of Vibrio cholerae. Gene 126:51–60. [DOI] [PubMed] [Google Scholar]
- 22.Oliver, D. B., R. J. Cabelli, K. M. Dolan, and G. P. Jarosik. 1990. Azide-resistant mutants of Escherichia coli alter the SecA protein, an azide-sensitive component of the protein export machinery. Proc. Natl. Acad. Sci. USA 87:8227–8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Py, B., L. Loiseau, and F. Barras. 2001. An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep. 2:244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramer, S. W., D. Bieber, and G. K. Schoolnik. 1996. BfpB, an outer membrane lipoprotein required for the biogenesis of bundle-forming pili in enteropathogenic Escherichia coli. J. Bacteriol. 178:6555–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reyss, I., and A. P. Pugsley. 1990. Five additional genes in the pulC-O of the gram-negative bacterium Klebsiella oxytoca UNF5023 which are required for pullulanase secretion. Mol. Gen. Genet. 222:176–184. [DOI] [PubMed] [Google Scholar]
- 27.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279:485–499. [DOI] [PubMed] [Google Scholar]
- 28.Sakai, D., T. Horiuchi, and T. Komano. 2001. ATPase activity and multimer formation of PilQ protein are required for thin pilus biogenesis in plasmid R64. J. Biol. Chem. 276:17968–17975. [DOI] [PubMed] [Google Scholar]
- 29.Sakai, D., and T. Komano. 2000. The pilL and pilN genes of IncI1 plasmids R64 and ColIb-P9 encode outer membrane lipoproteins responsible for thin pilus biogenesis. Plasmid 43:149–152. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Shevchik, V. E., J. Robert-Baudouy, and G. Condemine. 1997. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted protein. EMBO J. 16:3007–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sohel, I., J. L. Puente, S. W. Ramer, D. Bieber, C. Y. Wu, and G. K. Schoolnik. 1996. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J. Bacteriol. 178:2613–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565–596. [DOI] [PubMed] [Google Scholar]
- 34.Strom, M. S., D. N. Nunn, and S. Lory. 1993. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc. Natl. Acad. Sci. USA 90:2404–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned gene. J. Mol. Biol. 189:113. [DOI] [PubMed] [Google Scholar]
- 36.Thomas, J. D., P. J. Reeves, and G. P. C. Salmond. 1997. The general secretion pathway of Erwinia carotovora subsp. carotovora: analysis of the membrane topology of OutC and OutF. Microbiology 143:713–720. [DOI] [PubMed] [Google Scholar]
- 37.Thunnissen, A. M. D. H., A. J. Dijkstra, K. H. Kalk, H. J. Rozeboom, H. Engel, W. Keck, and B. W. Dijkstra. 1994. Doughnut-shaped structure of a bacterial muramidase revealed by X-ray crystallography. Nature 367:750–753. [DOI] [PubMed] [Google Scholar]
- 38.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida, T., N. Furuya, M. Ishikura, T. Isobe, K. Haino-Fukushima, T. Ogawa, and T. Komano. 1998. Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili. J. Bacteriol. 180:2842–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida, T., S.-R. Kim, and T. Komano. 1999. Twelve pil genes are required for biogenesis of the R64 thin pilus. J. Bacteriol. 181: FS2038–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]