Abstract
The retinoblastoma-related pocket proteins pRb, p107, and p130 are implicated in the control of cell proliferation, differentiation, and transformation. The function of pocket proteins is in part mediated by their ability to inhibit specific E2F transcription factors. The transcriptional activity of E2Fs is controlled by alteration of their nucleocytoplasmic localization during the cell cycle. p130 was observed to shuttle between the nucleus and cytoplasm in a heterokaryon fusion assay, suggesting the presence of nuclear and cytoplasmic localization signals. Two independent nuclear localization signals (NLS) that could target reporter proteins to the nucleus in transient transfection and microinjection experiments were identified in the C terminus of p130. In addition to the C-terminal NLS, the intact pocket domain of p130 itself was sufficient for nuclear translocation. Moreover, an additional functional NLS was mapped within the unique Loop region of p130. An N-terminal domain that conferred cytoplasmic localization was identified. Removal of the entire N terminus did not affect the ability of p130 to interact with E2F and to induce growth arrest. A model suggesting that the activity of pRb family members can be regulated by intracellular trafficking of the proteins is proposed.
The retinoblastoma family of proteins is comprised of pRb and the related proteins p107 and p130 (also known as pRb2 and RBL2) (24, 28, 32, 47, 54). Rb-1 has been identified as a tumor suppressor gene deleted or mutated in childhood retinoblastoma and in a wide variety of adult cancers (28, 80). Although the role of p107 and p130 in tumor suppression is less clear than that of pRb, there are several reports of p130 inactivating mutations identified in human cancers (10, 13, 14, 37). As is the case for pRb, overexpression of p107 and p130 can induce growth arrest in certain cell types (12, 91) and can bind to and inhibit transcriptional activity of E2F transcription factors. All three members of the pRb family undergo cell cycle-dependent phosphorylation (6, 19, 51, 52, 85). It has been proposed that phosphorylation during the transition from G0/G1 to S phase inactivates the growth-suppressive properties of pRb family members (11, 40, 91) by dissociation from E2F transcription factors.
All three pRb family members share a high degree of sequence homology. The characteristic pocket domain is comprised of A and B boxes separated by a spacer domain. The A and B boxes define the minimal region essential for binding to the LXCXE-containing proteins (41, 44) including adenovirus E1A, simian virus 40 (SV40) large T antigen (T-Ag), and human papillomavirus E7 (18, 23, 81), as well as cellular proteins such as histone deacetylase 1 and 2 (5, 50), BRG1, BRM (22, 73, 76), and others (20). The ability of the LXCXE motif of viral oncoproteins to interact with pRb family members is indispensable for virus-mediated transformation (9, 74, 83, 88, 89). However, pRb mutants defective in binding to LXCXE-containing proteins were capable of inducing growth arrest and remained resistant to viral-mediated transformation (8, 15, 20). The N- and C-terminal regions, as well as the spacer domain of the pocket proteins, share less sequence homology, suggesting that these fragments may endow functions specific for each family member.
The ability of pocket proteins to bind to E2F and inhibit E2F-dependent transcription is an important element of their cell cycle control. E2F regulates the transcription of genes coding for cell cycle regulatory factors (4, 17, 62, 67, 75, 90), proto-oncogenes (68), and enzymes required for DNA synthesis (21, 43, 55, 61, 69). The E2F family consists of six members: E2F-1, E2F-2, E2F-3, E2F-4, E2F-5, and E2F-6. E2Fs heterodimerize with DP proteins DP-1 and DP-2, and this association is essential for high-affinity sequence-specific DNA binding, transcriptional activity, and interaction with pRb family members (1, 38, 45, 65, 84). pRb family members associate with different subsets of E2F transcription factors both in vivo and in vitro. p130 and p107 specifically bind to E2F-4 and E2F-5, while pRb binds to E2F-1, E2F-2, E2F-3, and E2F-4 (2, 30, 39, 65). Typically, E2F-1, -2, -3, and -5 levels are low in most cell types. In contrast, E2F-4 accounts for the majority of E2F complexes at every stage of the cell cycle (58). E2F-4 is found associated with p130 at the G0/G1 stage of the cell cycle but then apparently switches to p107 upon progression into the S phase (58).
The cellular compartmentalization of E2F transcription factors is cell cycle dependent. Several laboratories demonstrated that upon progression from G0 to S phase E2F-4 is translocated from the nucleus to the cytoplasm (48, 59, 78). This finding suggests that cytoplasmic sequestration or nuclear export of E2Fs may provide an additional level of control of their transcriptional activity. While E2F-1, E2F-2, and E2F-3 contain a nuclear localization signal (NLS), E2F-4 and E2F-5 do not (48, 59). Coexpression of certain pocket and DP proteins may promote the nuclear localization of E2F-4 and -5 (48, 49). Furthermore, the nuclear translocation of E2F-4 may require interaction with p107 and p130 (48).
It remains unclear whether the pocket proteins themselves can shuttle between the nucleus and cytoplasm. There are several reports demonstrating that p130 may be found in the cytoplasm in early S phase of the cell cycle (10, 13, 14, 29). The dependence of cellular localization of E2F-4 on pocket proteins suggests the possibility that p130 may be involved in nucleocytoplasmic trafficking. In the present study, p130 was observed to be capable of shuttling between the nucleus and cytoplasm in an interspecies heterokaryon formation assay. Several independent NLSs were identified, including the C terminus, the pocket domain, and the unique Loop region of p130. In addition, an N-terminal region of p130 conferred cytoplasmic localization. Taken together, these results suggest that shuttling of p130 between the nucleus and the cytoplasm may take place under physiological conditions and may contribute to the regulation of p130 activity.
MATERIALS AND METHODS
Cells.
The human osteosarcoma U-2 OS and Saos-2, human glioblastoma T98G, and mouse NIH 3T3 cell lines were obtained from the American Type Culture Collection. The p107−/−;p130−/− double-knockout mouse embryo fibroblasts (MEFs) were obtained from N. Dyson (42). U-2 OS-R HA-p130 cells were generated from U-2 OS cells by retroviral transfer of HA-130 subcloned in pBabe-Puro shuttle vector (obtained from H. Land). After infection the cells were selected in puromycin-containing medium (1 μg/ml). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Cellgro) supplemented with 10% Fetal Clone-I serum (HyClone), 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Cells were transfected using FuGene 6 transfection reagent (Roche Biochemicals) according to the manufacturer’s protocol. Fifteen micrograms of total plasmid DNA was used per 100-mm-diameter dish transfected.
For pulse-chase, U-2 OS-R HA-p130 cells were incubated in methionine-free DMEM for 2 h prior to labeling. The cells were incubated in DMEM containing 2 mCi of [35S]methionine per ml for 20 min and then chased with complete DMEM supplemented with 10% Fetal Clone-I serum (HyClone).
For immunostaining, cells were cultured and transfected on glass coverslips. Twenty-four to 72 h posttransfection the coverslips were washed three times with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS for 30 min. The cells were washed with PBS three times, blocked with PBS-buffered 0.5% Triton X-100 containing 3% normal goat serum (Gibco BRL) for 20 min, and exposed to primary antibodies for 1.5 to 2 h. After being washed three times with PBS, cells were treated with a mixture of fluorescent-labeled secondary antibodies and 5 μg of 4′,6-diamidino-2-phenylindole (DAPI) per ml for 25 min. Fluorescent images were obtained by using Nikon Eclipse E800 and a Spot digital camera (Diagnostic Instruments, Inc.).
Immunoprecipitation and Western blotting were performed according to a previously described method (35). For immunoprecipitation, whole-cell extracts of U-2 OS and T98G cells were prepared in EBC buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 0.5% Nonidet P-40) supplemented with protease inhibitors Set I (Calbiochem). Fractionation of T98G cells was performed using NE-PER (Nuclear and Cytoplasmic Extraction Reagent; Pierce) according to the manufacturer’s protocol. Growth arrest assay in Saos-2 cells was performed as described previously (7).
Plasmids.
p130 mutants were generated by cloning p130 fragments in frame with the influenza virus hemagglutinin (HA) epitope (YPYDVPDYA [25]) in the pcDNA 3.1/Zeo expression vector (Invitrogen). Point mutations were introduced by PCR using two overlapping mutagenesis primers (64). The SV40 NLS (PKKKRKVED) and the human immunodeficiency virus type 1 (HIV-1) Rev nuclear export signal (NES) (LPPLERLTLD [26]) were inserted in frame. Fusion proteins of p130 with enhanced green fluorescent protein (EGFP) were prepared with a pEGFP-C1 expression vector (Clontech).
Antibodies.
The following antibodies were used for Western blot analysis: anti-p130 polyclonal (C-20), anti-E2F-1 polyclonal (C-20), anti-E2F-4 polyclonal (C-108), anti-p107 polyclonal (C-18), and anti-p53 DO5 monoclonal antibodies from Santa Cruz; anti-pRb and anti-Ki-67 rabbit polyclonal antibodies from PharMingen; and anti-α-tubulin monoclonal antibodies from Sigma. The following antibodies were used for immunofluorescence experiments: anti-HA monoclonal antibodies (HA-11) from Babco or polyclonal antibodies (Y-11) from Santa Cruz; anti-T-Ag monoclonal PAb 419 (34), anti-p130 monoclonal (211.6) from Santa Cruz, and anti-cyclin B monoclonal antibodies CB169 from UBI (16).
Heterokaryon formation assay.
U-2 OS and NIH 3T3 heterokaryons were prepared by a modification of a previously described procedure (63). Briefly, U-2 OS cells were cultured on coverslips and transfected with 2 μg of total DNA in a six-well plate (Falcon). Eighteen to 20 h after transfection, cells were washed three times with serum-free DMEM containing 100 μM cycloheximide (Fluka). NIH 3T3 cells were trypsinized, washed with DMEM containing 10% fetal calf serum, washed, resuspended in serum-free DMEM supplemented with 100 μM cycloheximide, and added to the six-well plate containing transfected U-2 OS cells. Alternatively, adherent T98G cells were fused with trypsinized double-knockout p107−/−;p130−/− MEFs. The plate was centrifuged at 300 × g for 5 min at room temperature. Each coverslip was inverted onto a 100-μl drop of 50% polyethylene glycol (PEG) (Roche Molecular Biochemicals) and incubated for 2 min at room temperature. The coverslips were placed into a new six-well plate filled with serum-free DMEM containing 100 μM cycloheximide, rinsed three times, and incubated in DMEM containing 10% fetal calf serum and 100 μM cycloheximide for 3 h at 37°C under 10% CO2. Cells were fixed and immunostained as described above.
Microinjection.
EGFP-1081P-S1110 was subcloned in frame to the C terminus of glutathione S-transferase (GST) in pFastBac-GST-2T baculovirus and used to infect SF-21 insect cells. Seventy-two hours postinfection the cells were harvested and lysed in EBC buffer containing protease inhibitors Set I (Calbiochem). The cell extract was centrifuged at 15,000 × g for 15 min to remove insoluble particles, and the resulting supernatant was mixed with glutathione-Sepharose (Pharmacia). The resin was extensively washed with EBC buffer and then equilibrated with GST elution buffer (100 mM Tris-HCl [pH 8.0] and 125 mM NaCl). The bound material was eluted with 15 mM glutathione dissolved in the GST elution buffer. The eluted GST-EGFP-1081P-S1110 protein was dialyzed against PBS, and the concentration was adjusted to 5 mg/ml. This purified protein was microinjected into U-2 OS cells using Femtotips attached to Micromanipulator 5171 and Transjector 5246 (Eppendorf). Fluorescent and light images were taken with an Eclipse TE300 inverted fluorescent microscope (Nikon) equipped with a Spot digital camera (Diagnostic Instruments, Inc.).
RESULTS
Nuclear and cytoplasmic localization of pRb proteins.
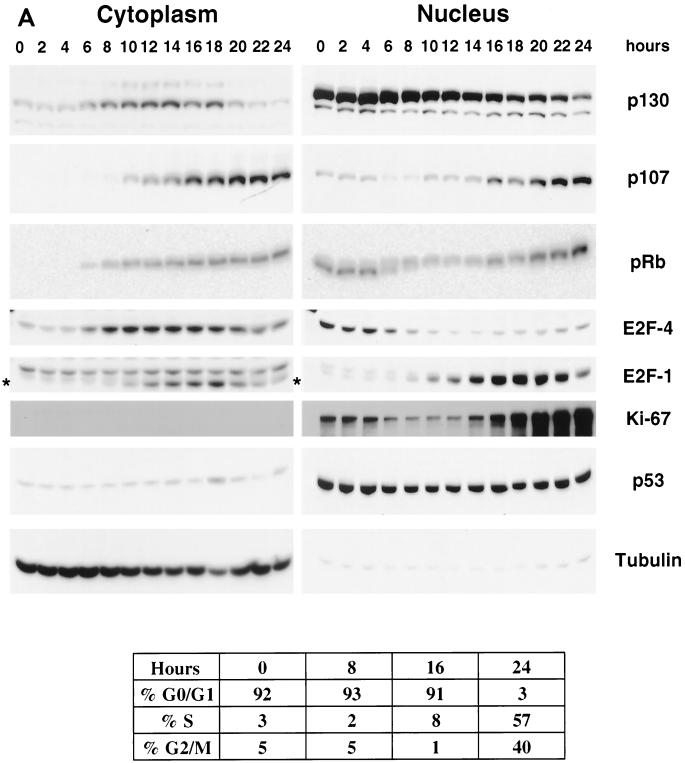
Cell fractionation experiments were performed to analyze cellular distribution of p130 throughout the cell cycle. T98G cells were growth arrested in G0/early G1 by serum deprivation for 72 h and then stimulated to reenter the cell cycle by addition of medium containing 20% serum. Cells were harvested at 2-h time intervals after the stimulation and subjected to a fractionation protocol. The efficiency and accuracy of the fractionation was verified by Western blot analysis of α-tubulin as a cytoplasmic marker and Ki-67 as markers for the nuclear fraction (Fig. 1). Ki-67 is a nuclear antigen that is expressed only in proliferating cells (36, 66, 72) and is a marker for exit from G0. As shown in Fig. 1, Ki-67 expression was reduced in the G0-arrested cells and its expression increased dramatically 16 to 18 h after serum stimulation. p53 was Western blotted as an additional marker for nuclear fraction and as a tumor suppresser protein that is regulated by nucleocytoplasmic shuttling (56, 79). We found that p53 was localized predominantly in the nucleus and its level did not fluctuate under these conditions.
FIG. 1.
Appearance of p130 in nuclear and cytoplasmic fractions. (A) Fractionation of T98G cells upon serum stimulation. Serum-starved (72 h) T98G cells were stimulated with serum for the indicated time (in hours) and subjected to fractionation and cell cycle analysis. Equal-volume fractions of the obtained cytoplasmic (80 μg) and nuclear (16 μg) extracts were separated by SDS-PAGE and Western blotted with indicated antibodies. Asterisks indicate bands specific for E2F-1 signal in the cytoplasmic fraction. The fluorescence-activated cell sorter analysis of cellular DNA content was performed at the indicated time points (shown at the bottom). (B) Fractionation and immunoprecipitation of [35S]methionine-labeled HA-p130. Retroviral transduced U-2 OS-R HA-p130 cells were labeled with [35S]methionine for 20 min, chased with complete medium for the indicated times, and extracted with NE-PER reagent. HA-p130 was immunoprecipitated from the resulting nuclear and cytoplasmic fractions using anti-HA antibodies, separated by SDS-PAGE, and transferred onto nitrocellulose. Autoradiography (top panel) for 35S was performed first, followed by Western blotting (bottom panel) with anti-HA antibody. Arrows indicate HA-p130, and asterisks indicate hyperphosphorylated forms of HA-p130.
The distributions of p130, p107, and pRb in the nuclear and cytoplasmic fractions were analyzed by Western blotting. In G0 and during the first 6 h after serum stimulation, the relative amount of all pocket proteins in the nuclear fraction was elevated compared to that in the corresponding cytoplasmic fractions. At 6 to 8 h poststimulation, the cytoplasmic signal for each of the three pocket proteins started to increase. After 18 h of serum stimulation, both the cytoplasmic and the nuclear signals for p130 became significantly reduced, while the signals for pRb and p107 were maintained at a steady level. The overall reduction in the p130 levels during late S and G2 phases is in agreement with data reported previously (51, 70). Thus, all three pocket proteins appear to accumulate in the cytoplasmic fraction several hours after serum stimulation.
Analysis of the cellular distribution of E2F-4 revealed that its nuclear fraction predominated in growth-arrested cells and for 6 h after serum stimulation. In contrast, cytoplasmic E2F-4 was more evident after 8 h of serum stimulation. Notably, the increased fraction of E2F-4 in the cytoplasm coincided with the increase of cytoplasmic p130. Furthermore, the cytoplasmic pool of E2F-4 peaked between 10 and 16 h after serum stimulation and was reduced afterwards. This reduction was in parallel to the declining cytoplasmic signal for p130. In contrast, both the cytoplasmic and the nuclear signals for E2F-1 increased after 10 h of serum stimulation and were elevated up to late S phase (20 to 24 h time points).
To address the possibility that the appearance of p130 in the cytoplasmic fraction could be attributed to de novo synthesis, we carried out fractionation of the cells labeled with [35S]methionine. A stable line of U-2 OS cells was infected by a retroviral vector carrying HA-p130 (U-2 OS-R HA-p130) that constitutively expressed HA-p130 at a level twofold greater than did endogenous p130. Unsynchronized U-2 OS-R HA-p130 cells were labeled with [35S]methionine for a 20-min pulse and chased with cold methionine. Cells were extracted to prepare nuclear and cytoplasmic fractions, and HA-p130 was immunoprecipitated with anti-HA antibodies. The immunoprecipitated material was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto a nitrocellulose membrane, and exposed to autoradiography film to detect the 35S signal (Fig. 1B). The [35S]-labeled HA-p130 was detected in the nuclear fraction in the initial pulse and in all chase points, although it appeared to decrease with prolonged chase (nucleus, 120-min chase time). In contrast, hyperphosphorylated p130 was not observed in the cytoplasmic fraction until 50 min into the chase and the cytoplasmic HA-p130 signal appeared to increase with increasing chase time. The combination of decreasing nuclear signal and increasing cytoplasmic fractions is consistent with export from the nucleus into the cytoplasm.
p130 shuttles between the nucleus and cytoplasm.
To evaluate whether the appearance of p130 in the cytoplasm could be attributed to nuclear export, an in vivo shuttling assay was performed. The migration of p130 between cellular compartments was studied in a human-mouse interspecies heterokaryon formation assay. HA-p130 and the K1 mutant of SV40 large T-Ag were transiently cotransfected into the U-2 OS human cell line. K1 T-Ag contains a mutated LXCXE motif and is unable to bind to p130 or other pRb family members (88). K1 contains the strong SV40 T-Ag NLS but no export signal and is not known to shuttle. Twenty-four hours posttransfection, NIH 3T3 mouse fibroblasts were fused to the transfected U-2 OS cells in the presence of cycloheximide to block synthesis of new proteins. The hybrid cells were double stained with HA and T-Ag antibodies to detect expression of HA-p130 and K1 T-Ag, respectively.
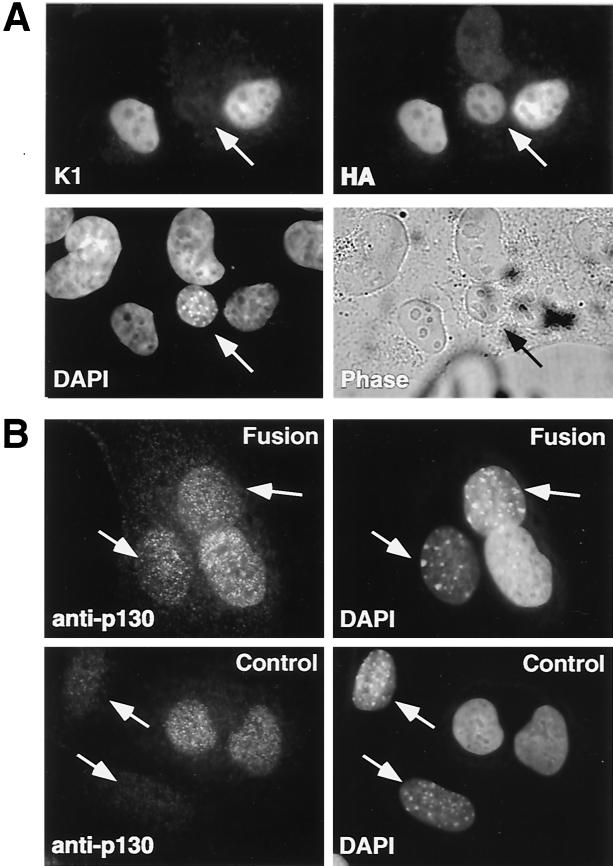
Figure 2A shows a representative hybrid of mouse and human cells fused together without fusion of the nuclear membranes. As revealed by DAPI staining, mouse nuclei were smaller and rounder than human nuclei and filled with characteristic small bright dots (Fig. 2, DAPI panels). Both HA-p130 and K1 were efficiently expressed and localized in the nuclei of human cells. However, HA-p130 was also detected in the mouse nucleus 3 h after the formation of the heterokaryon, while K1 remained in the nucleus of the human cell. The presence of HA-p130 in the nucleus of the mouse cell was not due to de novo protein synthesis, given the presence of cycloheximide but suggested the possibility of a combination of export from the human nucleus and import into the mouse nucleus. Similar results were observed when HA-p130 was transfected in the absence of the K1 mutant, demonstrating that the shuttling was independent of T-Ag (data not shown).
FIG. 2.
Transiently expressed and endogenous p130 shuttles between nuclei in interspecies heterokaryons. (A) U-2 OS cells were cotransfected with HA-p130 and the K1 mutant of SV40 large T-Ag. Twenty-four hours after transfection, NIH 3T3 cells were fused to the transfected U-2 OS cells in the presence of PEG and cycloheximide. Heterokaryons were incubated for 3 h and then fixed and immunostained with T-Ag-specific (K1) and anti-HA-specific (HA) antibodies. Nuclei of U-2 OS and NIH 3T3 cells were distinguished by DAPI staining (DAPI). The phase panel shows the corresponding field visualized by phase-contrast microscopy. Arrows point to mouse nuclei. (B) MEFs with homozygously deleted p107 and p130 were plated on human T98G cells in the presence (Fusion) or absence (Control) of PEG and incubated for 3 h in the presence of cycloheximide. Endogenous p130 was detected with specific monoclonal antibodies (anti-p130). Arrows point to mouse nuclei.
To address whether nucleocytoplasmic shuttling of the transfected p130 could be attributed to nonphysiological overexpression conditions, we performed a similar assay monitoring the translocation of the endogenous protein. In this experiment we employed the human T98G glioblastoma cell line because of the relatively higher expression level of p130. The adherent T98G cells were fused with MEFs which had homozygous deletions of both p107 and p130. Double-knockout p107−/−;p130−/− MEFs were chosen to avoid cross-reactivity of anti-p130 antibodies with p107 present in p130−/− MEFs (data not shown). We found that endogenous p130 was translocated from human to mouse nuclei (Fig. 2B). A specific signal for p130 in mouse nuclei was detected only in the presence of PEG, which caused fusion of the cell membranes. In the control mixed cell population (not treated with PEG), p130 was undetectable in murine nuclei (Fig. 2B). The fact that the nucleocytoplasmic shuttling of p130 occurred with physiological levels of expression suggested that this trafficking occurs in vivo and may play a role in the regulation of p130 activity.
Cellular localization of p130 deletion mutants.
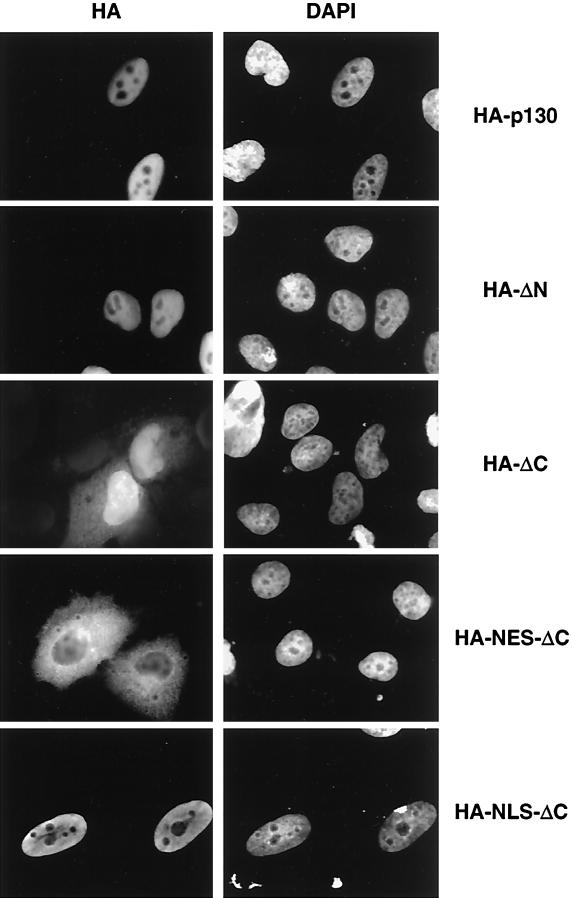
The fact that p130 was able to shuttle between nuclei of multinucleated cells suggests that p130 may contain signals for nuclear as well as cytoplasmic localization. To identify these signals, a series of HA-tagged deletion mutants of p130 were prepared (Fig. 3). HA-tagged p130 constructs were transiently expressed in U-2 OS, immunostained with anti-HA monoclonal antibodies, and counterstained with DAPI. Full-length HA-p130 and HA-ΔN, deleting the N-terminal 410 amino acids, were detected only in the nucleus (Fig. 4). In contrast, deletion of the C-terminal 112 residues of p130 (HA-ΔC) resulted in increased cytoplasmic localization compared to the wild-type p130 distribution, although the overall localization of the mutant remained predominantly nuclear. To clarify the ambiguous cellular distribution of HA-ΔC, well-characterized nuclear localization or nuclear export signals were added. The fusion of HIV-1 Rev protein NES to the mutant resulted in a prominent cytoplasmic localization of HA-NES-ΔC. The fusion of NLS from SV40 large T-Ag resulted in exclusively nuclear localization of HA-NLS-ΔC.
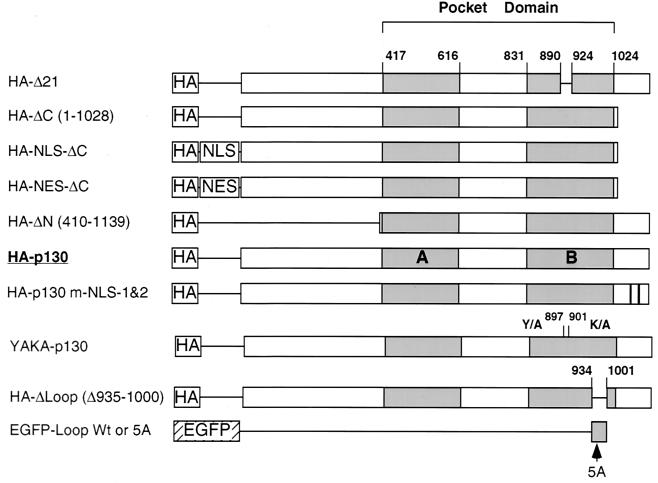
FIG. 3.
Design of p130 mutants. Fragments of human p130 were ligated in frame with various epitope tags. The numbers indicate the positions of the amino acid residues in the primary structure of human p130. HA, hemagglutinin epitope; NLS, SV40 large T-Ag NLS; NES, nuclear export signal of HIV-1 Rev protein; EGFP, enhanced green fluorescent protein. HA-p130-m-NLS-1&2 results from the combination of alanine substitutions at C-terminal clusters of basic amino acid residues 1083KRLR1086 and 1100KKR1102 (relative positions are shown by black bars). YAKA (Y897A, K901A) is a double point substitution of residues in the B pocket domain that disrupt binding to LXCXE-containing proteins. HA-Δ Loop is the deletion of the fragment 935S-E1000 in the context of full-length HA-epitope-tagged p130. EGFP-Loop and EGFP-Loop-5A represent fusion proteins of EGFP with either the intact 935S-E1000 fragment of p130 or the same fragment with substitution of 935KRKRR939 with five alanines, respectively. The arrow indicates the approximate position of the 5A substitution.
FIG. 4.
Cellular localization of p130 mutants. U-2 OS cells were cultured on coverslips and transiently transfected with the indicated constructs. Forty-eight hours posttransfection, the cells were fixed and the transfected proteins were immunostained with HA antibodies. Nuclei were visualized with DAPI.
The C terminus of p130 contains NLSs.
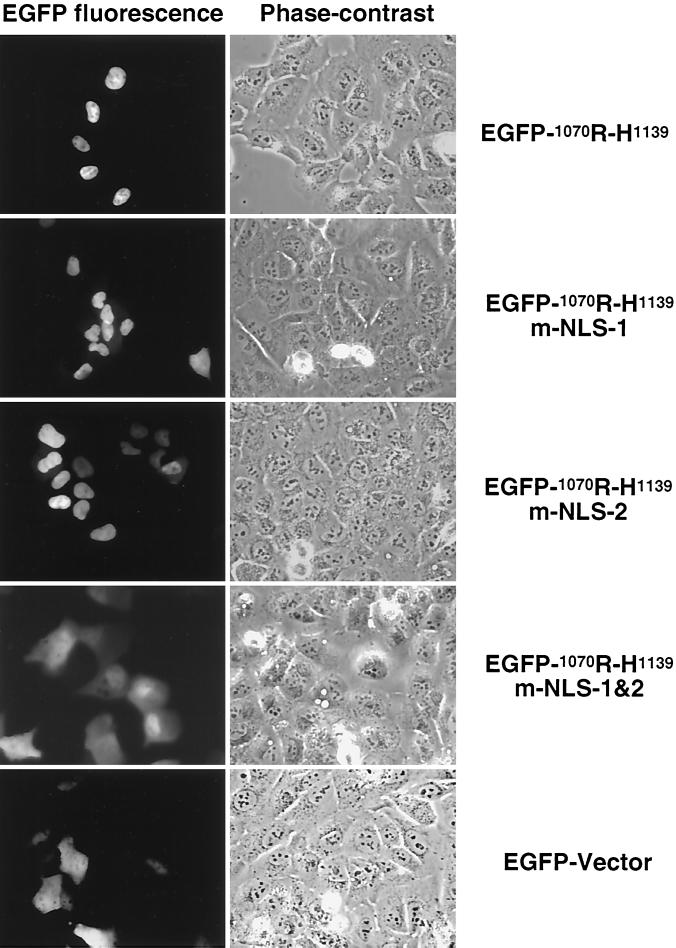
Since removal of the C terminus of p130 resulted in an increase in the cytoplasmic signal for p130 (Fig. 4), it was thought that the C terminus might contain an NLS. The C terminus of p130 shares high homology with pRb, for which a bipartite NLS has been previously reported (86). Analysis of p130 by PSORT software identified two putative NLS in the C terminus, 1081PSKRLRE1087 and 1098PTKKRGI1104, separated by 11 residues. To evaluate whether these sequences could function independently or form a bipartite NLS, the last 69 amino acid residues of p130 were fused to EGFP. The construct was transiently expressed in U-2 OS cells, and the chimeric proteins were visualized by direct fluorescence. The experiment revealed that the EGFP-tagged wild-type C-terminal fragment was localized in the nucleus (Fig. 5, panel EGFP-1070R-H1139). To evaluate the contribution of each potential NLS sequence, all positively charged amino acid residues were replaced with alanines either separately or together, as follows: 1081PSKRLRE1087 to 1081PSAALAE1087 (m-NLS-1) and 1098PTKKRGI1104 to 1098PTAAAGI1104 (m-NLS-2); the combination of the two substitutions was designated m-NLS-1&2. Separate alanine substitutions of the basic clusters (Fig. 5, EGFP-1070R-H1139-m-NLS-1 and EGFP-1070R-H1139-m-NLS-2) were observed to be exclusively nuclear. However, the combination of alanine substitutions in both basic regions resulted in reduced nuclear localization of the protein (Fig. 5, panel EGFP-1070R-H1139-m-NLS-1&2), with a distribution pattern very similar to that of the EGFP-vector alone. Given that the 1081PSKRLRE1087 and the 1098PTKKRGI1104 fragments functioned independently suggested that the C terminus of p130 contains two independent NLSs and not a bipartite one as was previously described for pRb (86, 87).
FIG. 5.
C terminus of p130 contains two NLSs. Fusion proteins of EGFP and the C-terminal 69 residues of p130 were transiently expressed in U-2 OS cells. The EGFP-1070R-H1139 contained either wild-type or mutant C-terminal basic clusters (1083KRLR1086 and 1100KKR1102) fused to EGFP (see text for details). Autofluorescence of the chimeric proteins was detected in living cells with an inverted fluorescent microscope. The phase-contrast column represents light images of the corresponding fields.
Cytoplasmic microinjection of the C-terminal NLS of p130 results in nuclear translocation.
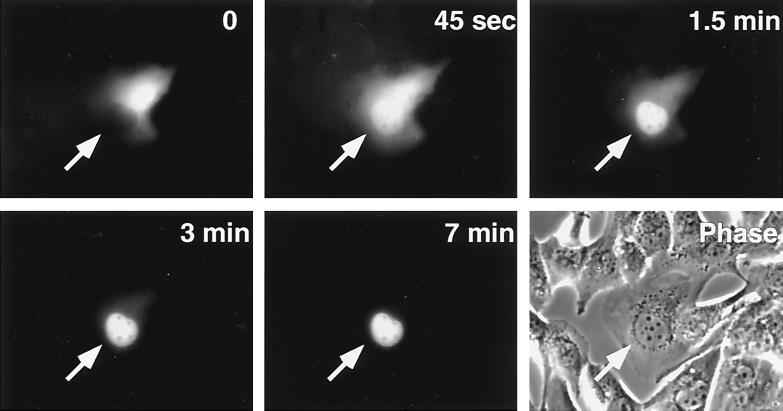
To prove unequivocally that the C-terminal region contains an authentic NLS we performed microinjection studies. A fusion construct comprised of GST, EGFP, and the 1081P-S1110 fragment of p130 containing both C-terminal NLSs was microinjected into U-2 OS cells. At the time of injection the green fluorescence of the GST-EGFP-1081P-S1110 protein was detected exclusively in the cytoplasm (Fig. 6, time 0). As early as 45 s after microinjection, a substantial fraction of the protein entered the nucleus. The nuclear translocation of the chimeric protein increased with time, reaching its maximum (exclusively nuclear staining) within 5 to 7 min (Fig. 6). A control protein, rabbit immunoglobulin G conjugated to red fluorescent dye, remained in the cytoplasm 20 min after cytoplasmic microinjection (data not shown). Unaltered GST-EGFP protein was also microinjected into U-2 OS cells, and it was found that its green fluorescence did not change localization with time and remained in the same compartment where it had been microinjected, i.e., either in the cytoplasm or in the nucleus (data not shown). This experiment demonstrates that the 1081P-S1110 p130 fragment contains an authentic NLS.
FIG. 6.
Microinjection of the C terminus of p130 results in nuclear import. A chimeric protein containing an in-frame fusion of GST, EGFP, and the 1081P–S1110 fragment of p130 was expressed in insect cells, purified, and microinjected into the cytoplasm of U-2 OS cells. Detection of autofluorescence of the injected protein was performed on living cells with an inverted fluorescent microscope. Microphotographs were taken at the indicated times after microinjection. The phase panel represents the phase-contrast image of the corresponding field. Arrows point to the microinjected cell.
p130 contains multiple NLSs.
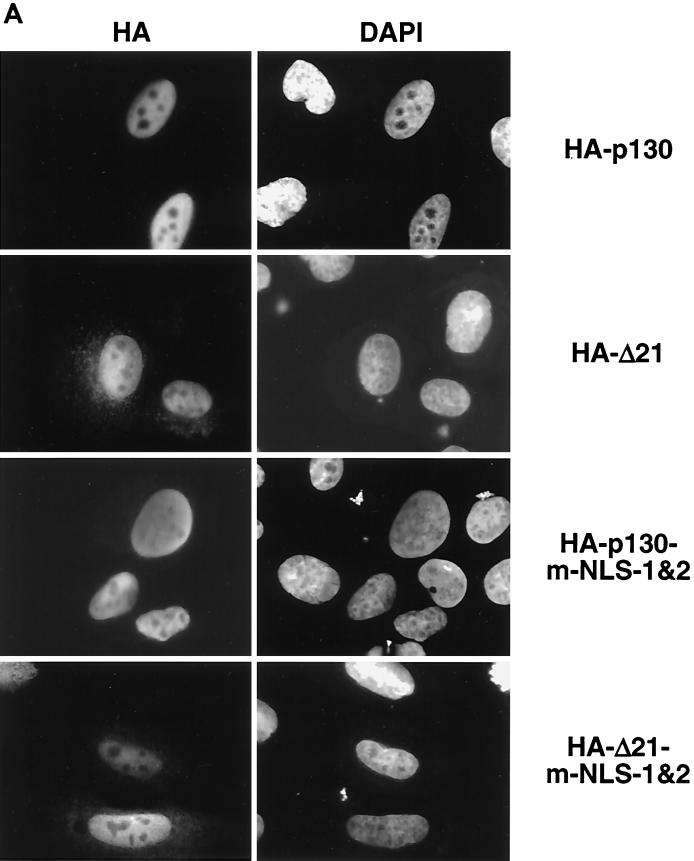
The results described above unambiguously demonstrate the presence of two functional NLSs in the C terminus of p130. However, deletion of the entire C terminus did not completely abolish nuclear localization of the HA-ΔC mutant (Fig. 4), suggesting the presence of additional NLS within p130. Full-length p130 containing alanine substitutions in both C-terminal NLSs (HA-p130-m-NLS-1&2) was also located exclusively in the nucleus when transiently expressed in U-2 OS cells (Fig. 7A). Neither deletion (HA-ΔC) nor mutation (HA-p130-m-NLS-1&2) of the C-terminal NLS sequences resulted in complete abolishment of nuclear localization of p130, suggesting the possibility that p130 contains additional NLSs.
FIG. 7.
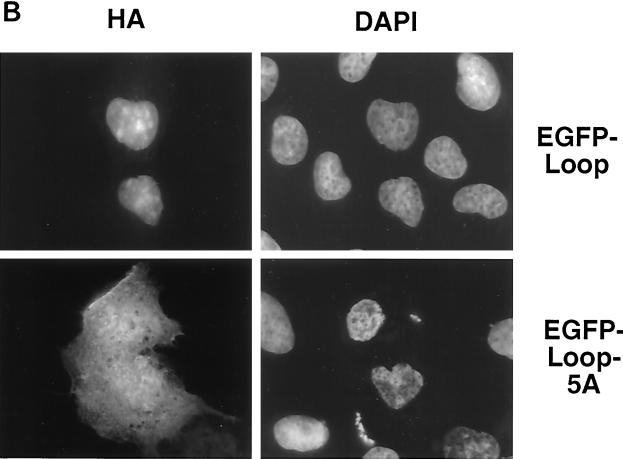
p130 contains multiple NLSs. (A) The indicated constructs were transiently transfected into U-2 OS cells and visualized by staining with anti-HA antibody 24 h posttransfection. Corresponding fields were stained with DAPI. (B) The Loop fragment of p130 (region 935S-E1000) was fused to EGFP and transiently expressed in U-2 OS cells (EGFP-Loop). Similarly, in the EGFP-Loop-5A construct the entire basic cluster 935KRKRR939 was replaced by alanines. The cells were fixed, and the chimeric proteins were visualized by autofluorescence.
It has been reported that pRb containing mutations both in the pocket domain and in the C-terminal NLS showed cytoplasmic localization (86, 87). By analogy with pRb, we expected a similar double mutant of p130 to be cytoplasmic. To address this, the C-terminal NLS sequences were mutated in the context of p130 with an inactivated pocket domain HA-Δ21 (3). Unexpectedly, HA-Δ21-m-NLS-1&2 was localized to the nucleus (Fig. 7A). This finding suggested that p130 may contain an NLS in addition to the C-terminal NLS and the pocket domain.
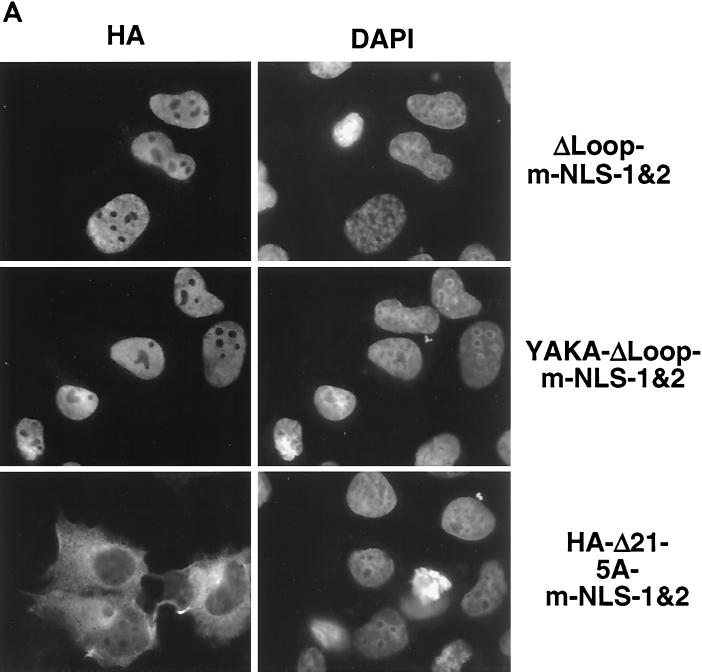
Analysis of the primary structure of p130 revealed that the 928R-E999 fragment of domain B of the pocket contained a potential NLS, 935KRKRR939. The 928R-E999 region, referred to as the Loop domain of p130, is multiply phosphorylated in G0 growth-arrested cells (7). The combination of a deleted Loop region and mutation of both C-terminal NLSs (HA-p130-ΔLoop-m-NLS-1&2) resulted in nuclear localization of the protein (Fig. 7 A). This finding supported the involvement of the pocket domain in nuclear localization of p130. To rule out the possibility of any additional NLS, we prepared a mutant with inactivating mutations in the pocket domain, the Loop region, and C-terminal NLSs. All five basic residues in the 935KRKRR939 Loop cluster were replaced by alanines (designated 5A mutation) in the context of p130 with mutated pocket domain (Δ21) and both C-terminal NLSs (m-NLS-1&2). This mutant (HA-p130-Δ21-5A-m-NLS-1&2) was found exclusively in the cytoplasm when transiently expressed in U-2 OS cells (Fig. 7A).
To confirm the nuclear targeting properties of the 935KRKRR939 region, we prepared a fusion protein of EGFP and the Loop region of p130, 929S-E1000 (Fig. 3), and a similar construct containing 5 alanine substitutions at residues 935 to 939. Transient expression of the construct in U-2 OS cells revealed that the wild-type Loop was able to target the chimeric protein to the nucleus (Fig. 7), while the EGFP-Loop-5A construct displayed a pancellular localization pattern similar to nonmodified EGFP (Fig. 5). We conclude that 935KRKRR939 is a functional NLS in the context of both full-length p130 and the Loop fragment. This result strongly suggested that p130 contains at least four independent signals for nuclear translocation: three short basic regions and one that requires an intact pocket domain.
We considered the possibility that the nuclear localization activity of the pocket domain of p130 was dependent on binding to LXCXE motif-containing proteins. A mutant p130 was prepared by analogy to a human pRb mutant that lacked the ability to bind to LXCXE motif-containing proteins but retained the ability to bind and inactivate E2F (15). This p130 mutation, designated YAKA, consisted of a double substitution from 897Y to A and from 901K to A, corresponding to the 709Y and 713K positions in human pRb. To verify that the YAKA-p130 mutant was defective in binding to the LXCXE motif, we tested its ability to associate with SV40 large T-Ag. We found that YAKA-p130 failed to bind to T-Ag in a coimmunoprecipitation assay (data not shown). Expression of YAKA-ΔLoop-m-NLS-1&2 mutant was localized in the nuclei of the transfected cells (Fig. 7A). This finding suggested that binding to cellular LXCXE-containing proteins did not contribute to the nuclear localization of the pocket domain.
A cytoplasm-targeting activity of the N terminus of p130.
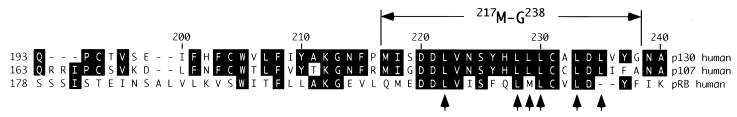
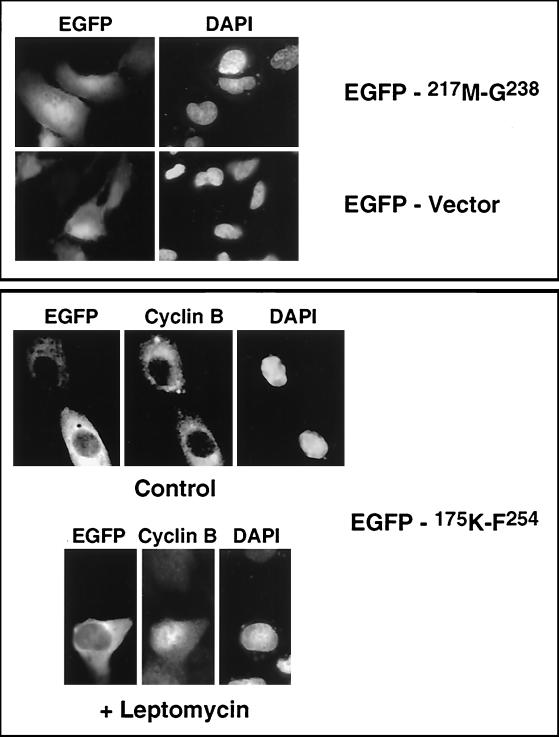
It has been reported that the nuclear export by the Crm1/exportin 1 transporter is dependent on interaction with a leucine-rich consensus sequence (71, 77). A short colinear sequence within the N terminus of p130 contains a leucine-rich sequence that resembles the consensus sequence (L-X2-3-L/V/M/I/F-X2-3-LXL) recognized by Crm1/exportin 1 (Fig. 8) (46). The Clustal algorithm alignment of human members of pRb family revealed a high degree of homology within this fragment (Fig. 8). To test whether this leucine-rich region (217M-G238) would act as an NES, an EGFP fusion construct was transiently expressed in U-2 OS cells. We found that the EGFP-217M-G238 fusion protein was present in both nucleus and cytoplasm (Fig. 9) and that the overall distribution pattern was not distinguishable from the distribution of the EGFP vector (Fig. 5).
FIG. 8.
Alignment of the N-terminal region of human p130, p107 and pRb. p130, p107, and pRb were aligned using the Clustal alignment algorithm. Identical amino acid residues are highlighted in black. 217M-G238 contains the leucine-rich motif (arrows indicate positions of leucine residues) that resembles an NES consensus sequence, L-X2-3-L/V/M/I/F-X2-3-LXL, recognized by Crm1/exportin 1 (46).
FIG. 9.
A cytoplasm localization sequence in the N terminus of p130. EGFP-175K-F254 and EGFP-217M-G238 chimeras were transiently expressed in U-2 OS cells and visualized by autofluorescence (EGFP). Endogenous cyclin B was detected with anti-cyclin B antibodies (Cyclin B), and nuclei were stained with DAPI (DAPI). The EGFP-175K-F254 panel shows the cellular localization of the proteins in cells treated with dimethyl sulfoxide (Control) and typical immunostaining of the cells treated with leptomycin B (10 ng/ml for 4 h) (+ Leptomycin).
To exclude the possibility that the 217M-G238 fragment did not encompass the entire putative NES, we prepared an EGFP fusion protein of a larger fragment of p130 (175K-F254 region). The predicted molecular mass of the fusion protein was about 35 kDa, a size presumably small enough to allow diffusion between nucleus and cytoplasm. Transient expression of the EGFP-175K-F254 construct in U-2 OS cells resulted in exclusive localization of the chimera in the cytoplasm (Fig. 9, EGFP-175K-F254). To test whether the cytoplasmic localization of EGFP-175K-F254 was dependent on nuclear export mediated by Crm1/exportin 1, cells were treated with leptomycin B, a specific inhibitor of this transporter (27, 77). Cyclin B was used as a positive control for inhibition of the Crm1/exportin 1 activity. Cyclin B has been reported to be exported from the nucleus in a Crm1/exportin 1-dependent manner (31). Treatment of transfected cells with leptomycin B resulted in the accumulation of cyclin B in the nucleus, while the EGFP-175K-F254 fusion protein remained in the cytoplasm (Fig. 9). These results suggested that either the potential NES in p130 was not sensitive to leptomycin or the cytoplasmic localization of the chimeric protein was not due to nuclear export.
Growth arrest activity of p130 mutants.
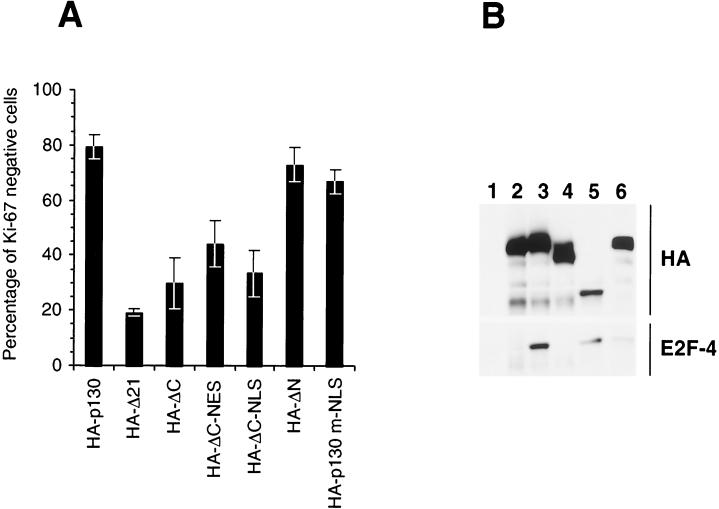
To evaluate the effects of mutations in the N and C termini on the activity of p130, a Ki-67 growth arrest assay was performed (7). Transient expression of wild-type, full-length HA-p130 resulted in a significant decrease in expression of Ki-67 in Saos-2 cells (Fig. 10A). In contrast, expression of the pocket mutant HA-Δ21 failed to reduce the expression of Ki-67 relative to untransfected cells. The HA-ΔN mutant retained at least 90% of the growth arrest activity seen with HA-p130 (Fig. 10A).
FIG. 10.
Growth arrest activity and E2F-4 binding of p130 mutants. (A) SaOS-2 cells, cultured on coverslips, were transfected with the indicated p130 mutants. Seventy-two hours after transfection, endogenous Ki-67 and transiently expressed proteins were immunostained with specific antibodies. The percent ratio of Ki-67-negative cells to the total number of counted transfected cells is shown. The results represent the mean values from at least three independent experiments. Error bars reflect standard deviations. (B) U-2OS cells were transiently transfected with vector (lane 1), HA-Δ21 (lane 2), HA-p130 (lane 3), HA-ΔC (lane 4), HA-ΔN (lane 5), or HA-p130 m-NLS-1&2 (lane 6) constructs. Forty-eight hours after transfection, immunoprecipitation with anti-HA antibodies was performed. The immune complexes were separated by SDS-PAGE, transferred to nitrocellulose, and Western blotted with anti-HA antibodies (HA) and anti-E2F-4 antibodies (E2F-4).
Although the HA-ΔC mutant was predominantly nuclear, it lacked the ability to induce growth arrest. Moreover, retargeting of the HA-ΔC mutant into the nucleus by exogenous NLS (HA-NLS-ΔC) did not restore its growth arrest activity. In addition, the HA-NES-ΔC mutant did not display a growth arrest activity significantly different from that of HA-ΔC or HA-NLS-ΔC (Fig. 10A). In contrast to the HA-ΔC mutant, the growth arrest activity of HA-p130-m-NLS-1&2 (Fig. 10A) protein was very similar to that of the wild-type p130. This finding indicates that nuclear localization activity of the C-terminal domain of p130 was not essential for the growth arrest activity.
To get an insight into the mechanisms critical for growth arrest, the association of p130 mutants with the E2F-4 transcription factor was evaluated. HA-tagged p130 and mutants were transfected into U-2 OS cells and were immunoprecipitated with anti-HA-tag-specific antibodies. The immunoprecipitated complexes were Western blotted to determine the ability to coprecipitate endogenous E2F-4. HA-p130, HA-ΔN, and HA-p130-m-NLS-1&2 retained E2F-4 binding activity (Fig. 10B, lanes 3, 5, and 6), while HA-ΔC failed to associate with E2F-4 (lane 4). This result demonstrated that E2F-4 binding is not dependent on the entire N terminus of p130, and although loss of the entire C terminus disrupted binding to E2F-4, specific mutations of the C-terminal NLS did not disrupt E2F-4 association or growth arrest activity.
DISCUSSION
Progression through the cell cycle requires the orchestrated action of the pRb family and E2F/DP transcription factors. In growth-arrested and differentiated cells, p130 is the major E2F partner (53). Under these conditions, the p130-E2F complexes are exclusively nuclear and transcription of E2F-dependent genes is repressed. Upon exit from the growth-arrested stage, several events occur. During mid-to-late G1, all pRb family members undergo a specific phosphorylation by cyclin-dependent kinases. This phosphorylation is thought to dissociate pRb family members from E2F, resulting in loss of repression of E2F-dependent transcription. In this paper, evidence is presented that in addition to phosphorylation, pRb family members also accumulate in the cytoplasm during the late G1 phase of the cell cycle. Furthermore, nucleocytoplasmic shuttling may be dependent on multiple nuclear and cytoplasmic localization sequences. Export to the cytoplasm may represent a novel mechanism providing a rapid and efficient way to relieve pocket protein-mediated repression of E2F-dependent transcription.
It has been reported that pRb contains a bipartite nuclear localization signal in the C-terminal part of the molecule (86, 87). Furthermore, nuclear localization of pRb could be achieved by two independent mechanisms: by the C-terminal bipartite NLS and by an intact pocket domain. While the present study was in progress, Cinti et al. reported that p130 also contains a C-terminal bipartite NLS (10). However, the experimental results presented here demonstrate that the C terminus of p130 apparently contains two independent NLS sequences and each of them can efficiently translocate reporter proteins into the nucleus. Mutation of the C-terminal p130 NLS separately or together did not disrupt the nuclear localization of p130 or its ability to bind to and repress E2F activity. In contrast, deletion of the C terminus of p130 resulted in loss of binding to E2F-4 and ability to induce growth arrest. This finding suggested that the functional importance of the p130 C terminus is in its contribution to the physical association of p130 with E2F-4, rather than in targeting p130 to the nucleus as previously suggested (10, 86). Furthermore, although addition of the Rev NES to p130 led to cytoplasmic localization, it was still capable of repressing E2F-dependent transcription.
As was the case for pRb, the nuclear localization of p130 was not entirely dependent on the C-terminal NLSs (86, 87). p130 and pRb use their pocket domains for nuclear targeting. It is plausible that the pocket domain does not contain a canonical NLS but is able to interact with another protein(s) that may target pocket proteins into the nucleus. We genetically separated the ability of p130 to bind LXCXE-containing proteins and nuclear targeting activity of the pocket domain. We also demonstrated the presence of an additional NLS (935KRKRR939) in the Loop region of p130. Given that the Loop region of p130 is not present in pRb, this may explain the difference in the nuclear localization of the double mutant of p130 and of pRb constructs.
Interestingly, it was reported that phosphorylation of pRb reduced the ability of pRb to bind to a nuclear anchor (57). In fact, all pocket proteins may share this decreased affinity for the nucleus when they become hyperphosphorylated during the G1/S phase transition. Phosphorylation of pocket proteins may affect the activity of the nuclear localization sequences. Both of the C-terminal NLSs (1081SPSKRLRE1087 and 1098TPTKKRGI1104) are preceded by serine or threonine/proline residues. The SP/TP sites may be phosphorylated by cyclin-dependent kinases and could disrupt their ability to target p130 to the nucleus. It was recently reported that the Loop region is also specifically phosphorylated at multiple sites (7, 33). This finding suggests that the NLS within the Loop may also be regulated by phosphorylation. More experimental data are required to prove such hypothesis.
In spite of the presence of multiple NLSs, fractionation experiments performed on cells released from growth arrest demonstrated that a substantial fraction of p130 was present in the cytoplasm. An interspecies heterokaryon formation assay also demonstrated that p130 was capable of shuttling between the nucleus and cytoplasm. These results suggest the existence of specific signals providing p130 with an ability to enter and leave the nucleus. Leucine-rich NES are demonstrated to export proteins by binding to Crm1/exportin 1. A leucine-rich fragment in the N terminus of p130 was identified (217M-G238) but was not sufficient for nuclear export of the protein. However, a slightly larger fragment containing the leucine-rich region localized to the cytoplasm. The cytoplasmic localization of the EGFP 175-254 protein containing the leucine-rich fragment of p130 was not changed upon treatment with leptomycin B, a specific inhibitor of Crm1/exportin 1 (27, 77). The failure of leptomycin B to affect the localization of the N terminus of p130 suggests that its cytoplasmic targeting might not be dependent on binding to Crm1/exportin 1. Alternatively, Crm1/exportin 1 may regulate the subcellular localization in a leptomycin-independent manner (60). It is also plausible that cytoplasmic translocation of p130 may be dependent on a different molecular mechanism, for example as was demonstrated for β-catenin (82). The N terminus of p130 may serve as a cytoplasmic retention signal rather than a nuclear export sequence. Alternatively, p130 may lack NES of its own but utilize the N-terminal domain to interact with another NES-containing protein for shuttling.
It has been reported that E2F-4 and E2F-5 do not contain intrinsic NLS but can be imported into the nucleus in complex with p130 or p107 (48, 49). It was also suggested that the nucleocytoplasmic shuttling of E2F-4 might regulate its transcriptional activity (29, 48, 58, 59, 78). Given that p130 has nuclear and cytoplasmic targeting signals, E2F-4 shuttling may be dependent upon its interaction with p130. Translocation of p130 with E2F-4 into the nucleus during G0 and early G1 could contribute to repression of E2F-dependent transcription, while shuttling out of the nucleus and into the cytoplasm as the cell enters the cell cycle would relieve repression. The nucleocytoplasmic shuttling of a p130 and E2F-4 complex may represent a novel mechanism for inhibition of E2F-dependent transcription that is utilized upon exit from G0.
Acknowledgments
We thank David Batt for pFastBac 2T GST fusion vector.
This work was supported in part by National Research Service Award 1 F32 CA84752-01 (A. Chestukhin). J. A. DeCaprio is a Scholar of the Leukemia and Lymphoma Society. This work was supported in part by Public Health Service grant RO1-CA63113.
REFERENCES
- 1.Bandara, L. R., V. M. Buck, M. Zamanian, L. H. Johnston, and N. B. La Thangue. 1993. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F. EMBO J. 12:4317–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beijersbergen, R. L., R. M. Kerkhoven, L. Zhu, L. Carlee, P. M. Voorhoeve, and R. Bernards. 1994. E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 8:2680–2690. [DOI] [PubMed] [Google Scholar]
- 3.Bookstein, R., J. Y. Shew, P. L. Chen, P. Scully, and W. H. Lee. 1990. Suppression of tumorigenicity of human prostate carcinoma cells by replacing a mutated RB gene. Science 247:712–715. [DOI] [PubMed] [Google Scholar]
- 4.Botz, J., K. Zerfass-Thome, D. Spitkovsky, H. Delius, B. Vogt, M. Eilers, A. Hatzigeorgiou, and P. Jansen-Durr. 1996. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol. Cell. Biol. 16:3401–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597–601. [DOI] [PubMed] [Google Scholar]
- 6.Buchkovich, K., L. A. Duffy, and E. Harlow. 1989. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell 58:1097–1105. [DOI] [PubMed] [Google Scholar]
- 7.Canhoto, A. J., A. Chestukhin, L. Litovchick, and J. A. DeCaprio. 2000. Phosphorylation of the retinoblastoma-related protein p130 in growth-arrested cells. Oncogene 19:5116–5122. [DOI] [PubMed] [Google Scholar]
- 8.Chen, T. T., and J. Y. Wang. 2000. Establishment of irreversible growth arrest in myogenic differentiation requires the RB LXCXE-binding function. Mol. Cell. Biol. 20:5571–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, J. B., and M. J. Imperiale. 1995. Inactivation of the retinoblastoma susceptibility protein is not sufficient for the transforming function of the conserved region 2-like domain of simian virus 40 large T antigen. J. Virol. 69:3945–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cinti, C., P. P. Claudio, C. M. Howard, L. M. Neri, Y. Fu, L. Leoncini, G. M. Tosi, N. M. Maraldi, and A. Giordano. 2000. Genetic alterations disrupting the nuclear localization of the retinoblastoma-related gene RB2/p130 in human tumor cell lines and primary tumors. Cancer Res. 60:383–389. [PubMed] [Google Scholar]
- 11.Claudio, P. P., A. De Luca, C. M. Howard, A. Baldi, E. J. Firpo, A. Koff, M. G. Paggi, and A. Giordano. 1996. Functional analysis of pRb2/p130 interaction with cyclins. Cancer Res. 56:2003–2008. [PubMed] [Google Scholar]
- 12.Claudio, P. P., C. M. Howard, A. Baldi, A. De Luca, Y. Fu, G. Condorelli, Y. Sun, N. Colburn, B. Calabretta, and A. Giordano. 1994. p130/pRb2 has growth suppressive properties similar to yet distinctive from those of retinoblastoma family members pRb and p107. Cancer Res. 54:5556–5560. [PubMed] [Google Scholar]
- 143.Claudio, P. P., C. M. Howard, Y. Fu, C. Cinti, L. Califano, P. Micheli, E. W. Mercer, M. Caputi, and A. Giordano. 2000. Mutations in the retinoblastoma-related gene RB2/p130 in primary nasopharyngeal carcinoma. Cancer Res. 60:8–12. [PubMed] [Google Scholar]
- 14.Claudio, P. P., C. M. Howard, C. Pacilio, C. Cinti, G. Romano, C. Minimo, N. M. Maraldi, J. D. Minna, L. Gelbert, L. Leoncini, G. M. Tosi, P. Hicheli, M. Caputi, G. G. Giordano, and A. Giordano. 2000. Mutations in the retinoblastoma-related gene RB2/p130 in lung tumors and suppression of tumor growth in vivo by retrovirus-mediated gene transfer. Cancer Res. 60:372–382. [PubMed] [Google Scholar]
- 15.Dahiya, A., M. R. Gavin, R. X. Luo, and D. C. Dean. 2000. Role of the LXCXE binding site in Rb function. Mol. Cell. Biol. 20:6799–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalal, S. N., C. M. Schweitzer, J. Gan, and J. A. DeCaprio. 1999. Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol. Cell. Biol. 19:4465–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalton, S. 1992. Cell cycle regulation of the human cdc2 gene. EMBO J. 11:1797–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeCaprio, J. A., J. W. Ludlow, J. Figge, J. Y. Shew, C. M. Huang, W. H. Lee, E. Marsilio, E. Paucha, and D. M. Livingston. 1988. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell 54:275–283. [DOI] [PubMed] [Google Scholar]
- 19.DeCaprio, J. A., J. W. Ludlow, D. Lynch, Y. Furukawa, J. Griffin, H. Piwnica-Worms, C. M. Huang, and D. M. Livingston. 1989. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell 58:1085–1095. [DOI] [PubMed] [Google Scholar]
- 20.Dick, F. A., E. Sailhamer, and N. J. Dyson. 2000. Mutagenesis of the pRB pocket reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol. Cell. Biol. 20:3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dou, Q. P., P. J. Markell, and A. B. Pardee. 1992. Thymidine kinase transcription is regulated at G1/S phase by a complex that contains retinoblastoma-like protein and a cdc2 kinase. Proc. Natl. Acad. Sci. USA 89:3256–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119–130. [DOI] [PubMed] [Google Scholar]
- 23.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934–937. [DOI] [PubMed] [Google Scholar]
- 24.Ewen, M. E., Y. G. Xing, J. B. Lawrence, and D. M. Livingston. 1991. Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein. Cell 66:1155–1164. [DOI] [PubMed] [Google Scholar]
- 25.Field, J., J. Nikawa, D. Broek, B. MacDonald, L. Rodgers, I. A. Wilson, R. A. Lerner, and M. Wigler. 1988. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 8:2159–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475–483. [DOI] [PubMed] [Google Scholar]
- 27.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051–1060. [DOI] [PubMed] [Google Scholar]
- 28.Friend, S. H., R. Bernards, S. Rogelj, R. A. Weinberg, J. M. Rapaport, D. M. Albert, and T. P. Dryja. 1986. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323:643–646. [DOI] [PubMed] [Google Scholar]
- 29.Gill, R. M., and P. A. Hamel. 2000. Subcellular compartmentalization of E2F family members is required for maintenance of the postmitotic state in terminally differentiated muscle. J. Cell Biol. 148:1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ginsberg, D., G. Vairo, T. Chittenden, Z. X. Xiao, G. Xu, K. L. Wydner, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 8:2665–2679. [DOI] [PubMed] [Google Scholar]
- 31.Hagting, A., C. Karlsson, P. Clute, M. Jackman, and J. Pines. 1998. MPF localization is controlled by nuclear export. EMBO J. 17:4127–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hannon, G. J., D. Demetrick, and D. Beach. 1993. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes Dev. 7:2378–2391. [DOI] [PubMed] [Google Scholar]
- 33.Hansen, K., T. Farkas, J. Lukas, K. Holm, L. Ronnstrand, and J. Bartek. 2001. Phosphorylation-dependent and -independent functions of p130 cooperate to evoke a sustained G1 block. EMBO J. 20:422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harlow, E., L. V. Crawford, D. C. Pim, and N. M. Williamson. 1981. Monoclonal antibodies specific for simian virus 40 tumor antigens. J. Virol. 39:861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratories, Cold Spring Harbor, N.Y.
- 36.Heidebrecht, H. J., F. Buck, J. Steinmann, R. Sprenger, H. H. Wacker, and R. Parwaresch. 1997. p100: a novel proliferation-associated nuclear protein specifically restricted to cell cycle phases S, G2, and M. Blood 90:226–233. [PubMed] [Google Scholar]
- 37.Helin, K., K. Holm, A. Niebuhr, H. Eiberg, N. Tommerup, S. Hougaard, H. S. Poulsen, M. Spang-Thomsen, and P. Norgaard. 1997. Loss of the retinoblastoma protein-related p130 protein in small cell lung carcinoma. Proc. Natl. Acad. Sci. USA 94:6933–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helin, K., C. L. Wu, A. R. Fattaey, J. A. Lees, B. D. Dynlacht, C. Ngwu, and E. Harlow. 1993. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 7:1850–1861. [DOI] [PubMed] [Google Scholar]
- 39.Hijmans, E. M., P. M. Voorhoeve, R. L. Beijersbergen, L. J. van ’t Veer, and R. Bernards. 1995. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol. Cell. Biol. 15:3082–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinds, P. W., S. Mittnacht, V. Dulic, A. Arnold, S. I. Reed, and R. A. Weinberg. 1992. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell 70:993–1006. [DOI] [PubMed] [Google Scholar]
- 41.Hu, Q. J., N. Dyson, and E. Harlow. 1990. The regions of the retinoblastoma protein needed for binding to adenovirus E1A or SV40 large T antigen are common sites for mutations. EMBO J. 9:1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurford, R. K., D. Cobrinik, M. H. Lee, and N. Dyson. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447–1463. [DOI] [PubMed] [Google Scholar]
- 43.Johnson, L. F. 1992. G1 events and the regulation of genes for S-phase enzymes. Curr. Opin. Cell Biol. 4:149–154. [DOI] [PubMed] [Google Scholar]
- 44.Kaelin, W. G., Jr., M. E. Ewen, and D. M. Livingston. 1990. Definition of the minimal simian virus 40 large T antigen- and adenovirus E1A-binding domain in the retinoblastoma gene product. Mol. Cell. Biol. 10:3761–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krek, W., D. M. Livingston, and S. Shirodkar. 1993. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science 262:1557–1560. [DOI] [PubMed] [Google Scholar]
- 46.Kudo, N., H. Taoka, T. Toda, M. Yoshida, and S. Horinouchi. 1999. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J. Biol. Chem. 274:15151–15158. [DOI] [PubMed] [Google Scholar]
- 47.Li, Y., C. Graham, S. Lacy, A. M. Duncan, and P. Whyte. 1993. The adenovirus E1A-associated 130-kD protein is encoded by a member of the retinoblastoma gene family and physically interacts with cyclins A and E. Genes Dev. 7:2366–2377. [DOI] [PubMed] [Google Scholar]
- 48.Lindeman, G. J., S. Gaubatz, D. M. Livingston, and D. Ginsberg. 1997. The subcellular localization of E2F-4 is cell-cycle dependent. Proc. Natl. Acad. Sci. USA 94:5095–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magae, J., C. L. Wu, S. Illenye, E. Harlow, and N. H. Heintz. 1996. Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J. Cell Sci. 109:1717–1726. [DOI] [PubMed] [Google Scholar]
- 50.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601–605. [DOI] [PubMed] [Google Scholar]
- 51.Mayol, X., J. Garriga, and X. Grana. 1995. Cell cycle-dependent phosphorylation of the retinoblastoma-related protein p130. Oncogene 11:801–808. [PubMed] [Google Scholar]
- 52.Mayol, X., J. Garriga, and X. Grana. 1996. G1 cyclin/CDK-independent phosphorylation and accumulation of p130 during the transition from G1 to G0 lead to its association with E2F-4. Oncogene 13:237–246. [PubMed] [Google Scholar]
- 53.Mayol, X., and X. Grana. 1998. The p130 pocket protein: keeping order at cell cycle exit/re-entrance transitions. Front. Biosci. 3:D11–24. [DOI] [PubMed] [Google Scholar]
- 54.Mayol, X., X. Grana, A. Baldi, N. Sang, Q. Hu, and A. Giordano. 1993. Cloning of a new member of the retinoblastoma gene family (pRb2) which binds to the E1A transforming domain. Oncogene 8:2561–2566. [PubMed] [Google Scholar]
- 55.Means, A. L., J. E. Slansky, S. L. McMahon, M. W. Knuth, and P. J. Farnham. 1992. The HIP1 binding site is required for growth regulation of the dihydrofolate reductase gene promoter. Mol. Cell. Biol. 12:1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Middeler, G., K. Zerf, S. Jenovai, A. Thulig, M. Tschodrich-Rotter, U. Kubitscheck, and R. Peters. 1997. The tumor suppressor p53 is subject to both nuclear import and export, and both are fast, energy-dependent and lectin-inhibited. Oncogene 14:1407–1417. [DOI] [PubMed] [Google Scholar]
- 57.Mittnacht, S., and R. A. Weinberg. 1991. G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell 65:381–393. [DOI] [PubMed] [Google Scholar]
- 58.Moberg, K., M. A. Starz, and J. A. Lees. 1996. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol. Cell. Biol. 16:1436–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller, H., M. C. Moroni, E. Vigo, B. O. Petersen, J. Bartek, and K. Helin. 1997. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol. Cell. Biol. 17:5508–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neumann, G., M. T. Hughes, and Y. Kawaoka. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19:6751–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ogris, E., H. Rotheneder, I. Mudrak, A. Pichler, and E. Wintersberger. 1993. A binding site for transcription factor E2F is a target for trans activation of murine thymidine kinase by polyomavirus large T antigen and plays an important role in growth regulation of the gene. J. Virol. 67:1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohtani, K., J. DeGregori, and J. R. Nevins. 1995. Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl. Acad. Sci. USA 92:12146–12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pinol-Roma, S., and G. Dreyfuss. 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355:730–732. [DOI] [PubMed] [Google Scholar]
- 64.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning. a laboratory manual, 2nd ed., vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 65.Sardet, C., M. Vidal, D. Cobrinik, Y. Geng, C. Onufryk, A. Chen, and R. A. Weinberg. 1995. E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc. Natl. Acad. Sci. USA 92:2403–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schluter, C., M. Duchrow, C. Wohlenberg, M. H. Becker, G. Key, H. D. Flad, and J. Gerdes. 1993. The cell proliferation-associated antigen of antibody Ki-67: a very large, ubiquitous nuclear protein with numerous repeated elements, representing a new kind of cell cycle-maintaining proteins. J. Cell Biol. 123:513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schulze, A., K. Zerfass, D. Spitkovsky, S. Middendorp, J. Berges, K. Helin, P. Jansen-Durr, and B. Henglein. 1995. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc. Natl. Acad. Sci. USA 92:11264–11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slansky, J. E., and P. J. Farnham. 1996. Introduction to the E2F family: protein structure and gene regulation. Curr. Top. Microbiol. Immunol. 208:1–30. [DOI] [PubMed] [Google Scholar]
- 69.Slansky, J. E., Y. Li, W. G. Kaelin, and P. J. Farnham. 1993. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol. Cell. Biol. 13:1610–1618. (Erratum, 13: 7201.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith, E. J., G. Leone, and J. R. Nevins. 1998. Distinct mechanisms control the accumulation of the Rb-related p107 and p130 proteins during cell growth. Cell Growth Differ. 9:297–303. [PubMed] [Google Scholar]
- 71.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041–1050. [DOI] [PubMed] [Google Scholar]
- 72.Starborg, M., K. Gell, E. Brundell, and C. Hoog. 1996. The murine Ki-67 cell proliferation antigen accumulates in the nucleolar and heterochromatic regions of interphase cells and at the periphery of the mitotic chromosomes in a process essential for cell cycle progression. J. Cell Sci. 109:143–153. [DOI] [PubMed] [Google Scholar]
- 73.Strober, B. E., J. L. Dunaief, S. Guha, and S. P. Goff. 1996. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol. Cell. Biol. 16:1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stubdal, H., J. Zalvide, K. S. Campbell, C. Schweitzer, T. M. Roberts, and J. A. DeCaprio. 1997. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol. Cell. Biol. 17:4979–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tommasi, S., and G. P. Pfeifer. 1995. In vivo structure of the human cdc2 promoter: release of a p130–E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol. Cell. Biol. 15:6901–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trouche, D., C. Le Chalony, C. Muchardt, M. Yaniv, and T. Kouzarides. 1997. RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl. Acad. Sci. USA 94:11268–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turpin, P., B. Ossareh-Nazari, and C. Dargemont. 1999. Nuclear transport and transcriptional regulation. FEBS Lett. 452:82–86. [DOI] [PubMed] [Google Scholar]
- 78.Verona, R., K. Moberg, S. Estes, M. Starz, J. P. Vernon, and J. A. Lees. 1997. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol. Cell. Biol. 17:7268–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1:20–26. [DOI] [PubMed] [Google Scholar]
- 80.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323–330. [DOI] [PubMed] [Google Scholar]
- 81.Whyte, P., K. J. Buchkovich, J. M. Horowitz, S. H. Friend, M. Raybuck, R. A. Weinberg, and E. Harlow. 1988. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature 334:124–129. [DOI] [PubMed] [Google Scholar]
- 82.Wiechens, N., and F. Fagotto. 2001. CRM1- and Ran-independent nuclear export of beta-catenin. Curr. Biol. 11:18–27. [DOI] [PubMed] [Google Scholar]
- 83.Wolf, D. A., H. Hermeking, T. Albert, T. Herzinger, P. Kind, and D. Eick. 1995. A complex between E2F and the pRb-related protein p130 is specifically targeted by the simian virus 40 large T antigen during cell transformation. Oncogene 10:2067–2078. [PubMed] [Google Scholar]
- 84.Wu, C. L., L. R. Zukerberg, C. Ngwu, E. Harlow, and J. A. Lees. 1995. In vivo association of E2F and DP family proteins. Mol. Cell. Biol. 15:2536–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xiao, Z. X., D. Ginsberg, M. Ewen, and D. M. Livingston. 1996. Regulation of the retinoblastoma protein-related protein p107 by G1 cyclin-associated kinases. Proc. Natl. Acad. Sci. USA 93:4633–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zacksenhaus, E., R. Bremner, R. A. Phillips, and B. L. Gallie. 1993. A bipartite nuclear localization signal in the retinoblastoma gene product and its importance for biological activity. Mol. Cell. Biol. 13:4588–4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zacksenhaus, E., Z. Jiang, Y. J. Hei, R. A. Phillips, and B. L. Gallie. 1999. Nuclear localization conferred by the pocket domain of the retinoblastoma gene product. Biochim. Biophys. Acta 1451:288–296. [DOI] [PubMed] [Google Scholar]
- 88.Zalvide, J., and J. A. DeCaprio. 1995. Role of pRb-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol. Cell. Biol. 15:5800–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zalvide, J., H. Stubdal, and J. A. DeCaprio. 1998. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol. Cell. Biol. 18:1408–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zerfass, K., A. Schulze, D. Spitkovsky, V. Friedman, B. Henglein, and P. Jansen-Durr. 1995. Sequential activation of cyclin E and cyclin A gene expression by human papillomavirus type 16 E7 through sequences necessary for transformation. J. Virol. 69:6389–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu, L., S. van den Heuvel, K. Helin, A. Fattaey, M. Ewen, D. Livingston, N. Dyson, and E. Harlow. 1993. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 7:1111–1125. [DOI] [PubMed] [Google Scholar]