Abstract
Transcriptional corepressors of the Groucho/transducin-like Enhancer of split (Gro/TLE) family regulate a number of developmental pathways in both invertebrates and vertebrates. They form transcription repression complexes with members of several DNA-binding protein families and participate in the regulation of the expression of numerous genes. Despite their pleiotropic roles, little is known about the mechanisms that regulate the functions of Gro/TLE proteins. It is shown here that Gro/TLEs become hyperphosphorylated in response to neural cell differentiation and interaction with the DNA-binding cofactor Hairy/Enhancer of split 1 (Hes1). Hyperphosphorylation of Gro/TLEs is correlated with a tight association with the nuclear compartment through interaction with chromatin, suggesting that hyperphosphorylated Gro/TLEs may mediate transcriptional repression via chromatin remodeling mechanisms. Pharmacological inhibition of protein kinase CK2 reduces the Hes1-induced hyperphosphorylation of Gro/TLEs and causes a decrease in the chromatin association of the latter. Moreover, the transcription repression activity of Gro/TLEs is reduced by protein kinase CK2 inhibition. Consistent with these observations, Gro/TLEs are phosphorylated in vitro by purified protein kinase CK2. Taken together, these results implicate protein kinase CK2 in Gro/TLE functions. They suggest further that this kinase is involved in a hyperphosphorylation mechanism activated by Hes1 that promotes the transcription repression functions of Hes1-Gro/TLE protein complexes.
Drosophila Groucho (Gro) is the founding member of a family of evolutionarily conserved transcriptional corepressors that are involved in the regulation of a variety of cell differentiation events (7). Groucho and its vertebrate counterparts, the transducin-like Enhancer of split/Groucho-related gene products 1 to 4 (TLEs 1 to 4) (27, 43, 45), lack DNA-binding ability but can be targeted to a variety of specific gene regulatory regions through interactions with numerous DNA-binding proteins. Gro/TLE-binding partners include members of several important groups of developmental regulators including, among others, basic helix-loop-helix (bHLH) proteins of the Hairy/Enhancer of split (Hes) family (16, 19, 32, 38, 42), Runt-homology domain proteins of the Runt/RUNX family (2, 22, 28, 31), homeodomain proteins containing engrailed-homology region 1 motifs (14, 23, 35, 47, 54), and winged-helix transcription factors (51, 54).
As a result of their ability to functionally associate with such a wide array of transcriptional regulators, Gro/TLEs are involved in numerous developmental processes. For example, Drosophila Gro is required for sex determination, segmentation, dorsoventral pattern formation, eye development, and neurogenesis (7). Vertebrate Gro/TLE proteins have been directly or indirectly implicated in such events as neuronal differentiation (53), dorsoventral patterning of the neural tube (35), skeletogenesis (11, 22, 46), and hematopoiesis (14, 28, 41). Thus, Gro/TLE proteins define a class of general transcriptional regulators whose multiple gene regulatory functions depend on their context-dependent association with different transcriptional partners and consequent recruitment to specific target genes.
The fundamental role played by protein-protein interactions in Gro/TLE activities is further underscored by their proposed mechanism of transcriptional repression. Previous studies have shown that Gro/TLEs interact with each other to form oligomeric structures (5, 37) that can interact with both histones (15, 37) and histone deacetylases (4, 6, 8, 54). These observations suggest that specific transcriptional partners target oligomers of Gro/TLEs to DNA. These oligomers may interact with adjacent nucleosomes, resulting in the formation of heterochromatic complexes that spread along the chromosome. The presence of histone deacetylase proteins in these complexes may then establish and/or maintain a transcriptionally silenced chromatin structure. These events may be facilitated by dynamic interactions between chromatin and the filamentous ribonucleoprotein network of the nucleus (operationally defined as the nuclear matrix), where Gro/TLEs (22), as well as several Gro/TLE-binding proteins (20, 32, 34), were shown to localize.
These combined observations suggest that the ability of Gro/TLEs to interact with different transcriptional cofactors, as well as specific nuclear structures, must be precisely controlled and coordinated. Virtually nothing is known, however, about the mechanisms involved in these regulatory events. In this regard, previous studies have suggested that phosphorylation may be involved in the modulation of Gro/TLE function. These proteins are basally phosphorylated in both insect and mammalian cells (21) and can associate with at least one protein kinase, the homeodomain interacting protein kinase 2 (8). Moreover, induction of neural and chondrocytic cell differentiation in vitro is correlated with hyperphosphorylation of the mammalian family member Gro/TLE1, suggesting that an increase in the phosphorylation state of these proteins underlies their functions during differentiation (21, 52). Finally, interaction between Gro/TLEs and the paired domain-containing protein Pax5 results in increased phosphorylation of Gro/TLE4 (14), suggesting that transcriptional cofactors may be involved in the regulation of Gro/TLE phosphorylation. Thus, regulation of the phosphorylation state of Gro/TLEs may represent an important mechanism in the control of the functions of these proteins.
Here we describe experiments showing that neural cell differentiation and interaction with both the bHLH protein Hes1 and the Runt-domain protein RUNX1 result in hyperphosphorylation of Gro/TLEs. Hyperphosphorylation is correlated with a strong association of Gro/TLEs with the nuclear compartment through interaction with chromatin. We also provide evidence that the pleiotropic and ubiquitous kinase, protein kinase CK2 (formerly referred to as casein kinase II) (1, 33, 39), is involved in the hyperphosphorylation of Gro/TLEs induced by Hes1. Moreover, inhibition of protein kinase CK2 activity inhibits the transcription repression ability of Gro/TLEs. Taken together, these findings suggest a model in which the interaction of Gro/TLEs with Hes1 initiates a phosphorylation mechanism involving protein kinase CK2 that results in a functional association of Gro/TLEs with chromatin and transcriptional repression.
MATERIALS AND METHODS
Plasmids.
Plasmids pCMV2-FLAG-Hes1(1-281), pCMV2-FLAG-Hes1(Δ276-281), pEBG-Hes1(3-281), pEBG-Hes1(Δ276-281), pCMV2-FLAG-RUNX1b, and pcDNA3-GAL4bd-Gro/TLE1 were described previously (19, 31, 32). Construct pCMV2-FLAG-RUNX1a was generated by PCR amplification and subcloning into the EcoRV site of pCMV2-FLAG. To express Drosophila Gro in mammalian cells, construct pCMX-gro was obtained by first digesting the gro cDNA with NheI and NotI and subcloning it into pCaSper4 digested with XbaI and NotI. The ligated plasmid was digested with XhoI and NotI, and the gro insert was subcloned into pCMX digested with the same enzymes. Plasmids pEF-BOS-RUNX1b and pEF-BOS-RUNX1b(R80C) (encoding a DNA-binding defective form of RUNX1b) have been described previously (36).
Cell culture, preparation of cell lysates, Western blotting analysis, and phosphatase treatment.
Primary neural progenitor cell cultures were established from telencephalic cortices dissected from embryonic day 13.0 (E13.0) mouse embryos and then allowed to differentiate in vitro exactly as described previously (18, 44). For time course experiments, dissected telencephalic tissue that was dissociated but not plated was considered “day 0.” After 24 h in vitro (“day 1”), ∼90% of the cultured cells were mitotic (as indicated by bromodeoxyuridine incorporation studies (44; data not shown). Neuronal differentiation was monitored by immunocytochemistry and Western blotting by using antibodies against microtubule-associated protein 2 (MAP2), and significant numbers of neurons were detected by day 3. Mouse P19 embryonic carcinoma cells were cultured and induced to undergo neural differentiation with retinoic acid as described previously (21). Induction was normally terminated 10 days after the addition of retinoic acid (“day 10”). Drosophila S2, rat ROS17/2.8, and human HeLa, Jurkat, and 293 cells were cultured and, when appropriate, transfected by using the SuperFect reagent (Qiagen) as described previously (21, 31, 32). Transfected cells were treated with the following protein kinase inhibitors as indicated in the figure legends: chrysin (Sigma), 5,6-dichloro-1-β-𝒹-ribofuranosylbenzimidazole (DRB), and LY294002 (Calbiochem). Cell lysates were prepared as described elsewhere (21), and Western blotting analysis with panTLE (32, 37, 45), anti-Gro/TLE1 (21, 52, 53), anti-Gro (12, 21, 37), anti-MAP2 (Sigma), anti-FLAG (Sigma), anti-histone deacetylase 1 (HDAC1) (Santa Cruz Biotechnology), or anti-histone H3 (32) antibodies was as described previously. Incubation of cell extracts with calf intestinal phosphatase was exactly as described previously (21).
Preparation of subcellular fractions.
Postnuclear supernatant and nuclear fractions were prepared as described earlier (21), except in the studies depicted in Fig. 4D, wherein 0.5% Triton X-100 was added to the homogenization buffer. Chromatin fractions were obtained by subjecting cells to sequential extraction and solubilization steps exactly as described by Merriman et al. (34).
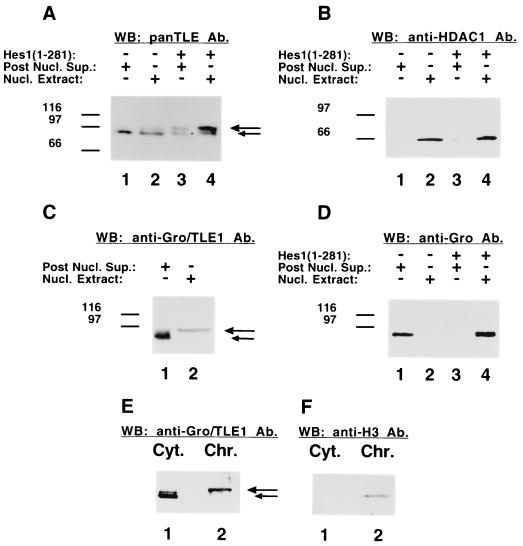
FIG. 4.
Preferential nuclear association of hyperphosphorylated Gro/TLEs. (A and B) ROS17/2.8 cells were either not transfected (lanes 1 and 2) or transfected with Hes1 (lanes 3 and 4), followed by a biochemical fractionation yielding postnuclear supernatant (Post Nucl. Sup., lanes 1 and 3) or nuclear (Nucl. Extract, lanes 2 and 4) fractions. Equivalent amounts of protein from these fractions were subjected to Western blotting with either panTLE (A) or anti-HDAC1 (B) antibodies (Ab). The hyperphosphorylated species induced by Hes1 associated preferentially with the nuclear compartment (long arrow). Hes1 had no effect on the mobility or nuclear interaction of HDAC1. (C) Analysis of the nuclear association of Gro/TLE1 in Jurkat cells. Postnuclear supernatant (lane 1) or nuclear (lane 2) fractions were subjected to Western blotting with anti-Gro/TLE1 antibodies. Hyperphosphorylated Gro/TLE1 was found preferentially in the nuclear fraction. (D) Effect of Hes1 expression on the nuclear association of Drosophila Gro. ROS17/2.8 cells were transfected with Drosophila Gro in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of Hes1. Cells were homogenized in the presence of 0.5% Triton X-100 to yield a postnuclear supernatant fraction (lanes 1 and 3) and a nuclear extract (lanes 2 and 4). Western blotting analysis of these fractions with anti-Gro antibodies showed that, while Gro was recovered in the non-nuclear fraction in the absence of Hes1, it was found bound to the nuclear compartment when Hes1 was present. (E and F) Analysis of the chromatin association of Gro/TLE1. Cytoplasmic (Cyt., lane 1, ∼30 μg of protein/lane) and nuclease treated, chromatin-enriched (Chr., lane 2, ∼10 μg of protein/lane) fractions were subjected to Western blotting with either anti-Gro/TLE1 (E) or anti-histone H3 (F) antibodies. Hypophosphorylated Gro/TLE1 (short arrow) was found in the soluble faction (lane 1), while hyperphosphorylated Gro/TLE1 (long arrow) was preferentially associated with chromatin (lane 2).
In vitro phosphorylation of immunoprecipitated Gro/TLE proteins.
Immunoprecipitation of either Gro from Drosophila S2 cell extracts or Gro/TLE1 from human Jurkat cells was performed as described previously (21). Immunoprecipitated proteins were incubated as described previously (10) in the presence of 25 μM [γ-32P]ATP and 0.5 U of purified protein kinase CK2 (New England Biolabs)/μl for 15 min at 30°C. Reactions were terminated by the addition of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and incubation at 65°C for 5 min. After gel electrophoresis, proteins were transferred to nitrocellulose and exposed to film. After autoradiography, membranes were subjected to Western blotting with either anti-Gro or panTLE antibodies.
Transcription assays.
HeLa cells were transfected by using the SuperFect reagent (Qiagen) as described previously (31, 32, 54). The amount of DNA transfected was adjusted by using plasmid pcDNA3 so that the total amount of DNA used in each transfection was the same (2.0 μg). Transcription assays were performed by using 0.5 μg of reporter plasmid p5xGAL4UAS-tk-luciferase in the absence or presence of plasmids pcDNA3-GAL4bd and pcDNA3-GAL4bd-Gro/TLE1 (1.0 μg) as described previously (19, 32). In each case, 0.5 μg of pRSV-β-galactosidase DNA was cotransfected to provide a means of normalizing the assays for transfection efficiency. At 24 h after transfection, cells were either not treated or treated with DRB (25, 50, or 75 μM) and then cultured for an additional 20 h. Results were expressed as mean values ± the standard deviation.
RESULTS
Neural cell differentiation is correlated with Gro/TLE hyperphosphorylation.
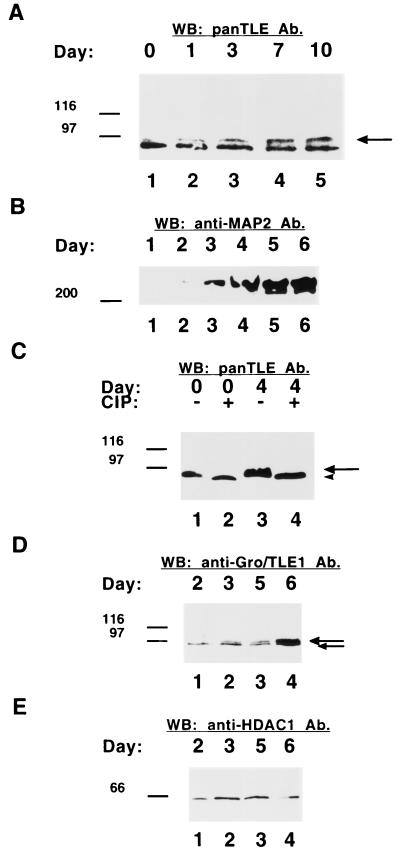
Gro/TLE proteins participate in the regulation of a variety of cell differentiation events, including neuronal development (7). Previous studies (21) have shown that Gro/TLEs become increasingly phosphorylated during the retinoic acid-induced neural differentiation of P19 embryonic carcinoma cells, resulting in a gradual increase of more slowly migrating forms (Fig. 1A, see long arrow). To determine whether Gro/TLEs are also phosphorylated during the differentiation of primary neural progenitor cells, the dorsal telencephalon was dissected from E13.0 mouse embryos, and cultures of telencephalic neural progenitor cells were established and allowed to undergo neuronal differentiation in vitro as described previously (18, 44). At the time of dissection (day 0), the vast majority of the cells present in the dorsal telencephalon were mitotically active and did not express neuronal marker proteins, like MAP2 (Fig. 1B). However, neuronal differentiation occurred after as little as 3 days in vitro (day 3) and was quite robust by day 5 (Fig. 1B). Examination of Gro/TLE expression with monoclonal antibodies that recognize all members of this family (11, 37, 45) revealed expression of these proteins both at day 0 (Fig. 1C, lane 1) and in cultures that exhibited extensive neuronal differentiation (Fig. 1C, lane 3). The electrophoretic mobility of Gro/TLEs appeared to become slower during differentiation, suggesting that these proteins are phosphorylated during the differentiation of neural progenitor cells (Fig. 1C, lane 3, see long arrow). To examine this possibility, the same samples were subjected to SDS-PAGE after incubation in the presence of calf intestinal phosphatase. This treatment resulted in the appearance of a single species that migrated faster than any Gro/TLE bands from untreated samples (Fig. 1C, lanes 2 and 4, see arrowhead). These observations show that Gro/TLEs exist as phosphorylated proteins in differentiating telencephalic neural progenitor cells. In agreement with this, analysis of the expression of Gro/TLE1, a specific member of this family that was shown to be involved in the regulation of telencephalic neurogenesis in vivo (53), revealed that this protein became progressively more abundant during in vitro differentiation (Fig. 1D). Moreover, we observed a gradual increase in the amount of a more slowly migrating Gro/TLE1 species (Fig. 1D, see long arrow). Treatment with calf intestinal phosphatase eliminated this slower species and resulted in only one band of faster mobility (not shown). No similar changes were observed when the same samples were examined using antibodies against the protein HDAC1 (Fig. 1E). Taken together, these results show an increase in both the amount and the phosphorylation of Gro/TLEs during neural cell differentiation and suggest that the functions of these proteins during neurogenesis are regulated by phosphorylation mechanisms.
FIG. 1.
Induction of Gro/TLE phosphorylation during neural differentiation. (A) Western blotting analysis of Gro/TLE expression in differentiating P19 cells. P19 cells were induced to undergo neural differentiation by treatment with retinoic acid. Whole-cell lysates were collected either before (day 0, lane 1) or 1, 3, 7, or 10 days after the addition of retinoic acid (lanes 2 to 5). Equal amounts of proteins were subjected to SDS-PAGE, transfer to nitrocellulose, and Western blotting with panTLE monoclonal antibodies. Neural differentiation was correlated with a progressive increase in the amount of more slowly migrating forms of Gro/TLE (long arrow). (B to E) Western blotting analysis of extracts from differentiating telencephalic progenitor cells. The dorsal telencephalon was dissected from E13 mouse embryos, and primary cultures of neural progenitor cells were established. Cells were allowed to differentiate in vitro and whole-cell lysates were prepared 1, 2, 3, 4, 5, or 6 days after plating. Equal amounts of proteins were subjected to SDS-PAGE, transfer to nitrocellulose, and Western blotting with either anti-MAP2 (B), panTLE (C), anti-Gro/TLE1 (D), or anti-HDAC1 (E) antibodies (Ab). (B) The expression of the neuronal protein MAP2 increased during in vitro differentiation. (C) Protein extract obtained at the time of plating (day 0, lanes 1 and 2) or 4 days after plating (day 4, lanes 3 and 4) were incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of calf intestinal phosphatase (CIP), followed by Western blotting analysis. Phosphatase treatment abolished the slower Gro/TLE forms (long arrow), resulting in a single band of faster mobility (arrowhead). (D) The expression of both slower (long arrow) and faster (short arrow) forms of Gro/TLE1 increased during in vitro differentiation. (E) Neither the electrophoretic mobility nor the expression of HDAC1 changed during in vitro differentiation. Here and in succeeding figures, the positions of size standards are indicated in kilodaltons.
Induction of Gro/TLE hyperphosphorylation by Hes1.
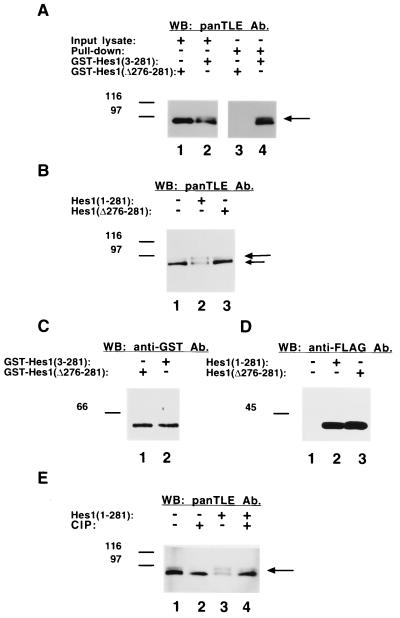
Gro/TLEs can interact with a variety of DNA-binding proteins that target them to selected DNA sites (7). One of these cofactors is the bHLH protein Hes1, which interacts with Gro/TLEs in a manner dependent on its C-terminal WRPW motif (32) and is coexpressed with the former during in vivo and in vitro neural development (11, 21, 42, 49). We noticed that when rat ROS17/2.8 osteoblastic cells were transfected with a fusion protein of glutathione S-transferase (GST) and full-length Hes1 [Hes1(3-281)], the electrophoretic mobility of endogenous Gro/TLEs appeared to be retarded compared to cells transfected with a fusion of GST and a C-terminally truncated form of Hes1 [Hes1(Δ276-281)] lacking the last 6 amino acids necessary for Gro/TLE binding (Fig. 2A, cf. lanes 1 and 2, see long arrow). GST-Hes1 interacted with both faster and slower forms of Gro/TLEs, while GST-Hes1(Δ276-281) did not interact with either one (Fig. 2A, lanes 3 and 4). Both GST-Hes1(3-281) and GST-Hes1(Δ276-281) were expressed at equivalent levels (Fig. 2C). These observations suggested that Hes1 might be involved in the regulation of Gro/TLE phosphorylation. To examine this possibility further, the electrophoretic mobility of Gro/TLE was compared in the absence or presence of FLAG epitope-tagged Hes1(1-281) or Hes1(Δ276-281). Western blotting analysis of lysates from nontransfected cells revealed the presence of a major Gro/TLE band of faster mobility (Fig. 2B and E, lane 1, see short arrow) and a less abundant component of slower mobility (Fig. 2B and E, lane 1, see long arrow). The slower band became more prominent after transfection of a plasmid encoding full-length Hes1 (Fig. 2B, lane 2, see long arrow), while the faster component became concurrently less abundant (Fig. 2B, lane 2, see short arrow). In contrast, expression of Hes1(Δ276-281) did not affect the Gro/TLE electrophoretic profile (Fig. 2B, lane 3). Reprobing of the nitrocellulose replicas showed that both Hes1 and Hes1(Δ276-281) were expressed at equal levels (Fig. 2D). The retardation in the electrophoretic mobility of Gro/TLEs induced by Hes1 was abolished by incubation with calf intestinal phosphatase prior to electrophoresis, indicating that it was due to an increase in phosphorylation (Fig. 2E, cf. lanes 3 and 4). Taken together, these findings show that Hes1 induces Gro/TLE hyperphosphorylation upon physical interaction with the latter.
FIG. 2.
Induction of Gro/TLE phosphorylation by Hes1. (A) Coprecipitation of Hes1 and Gro/TLEs. ROS17/2.8 cells were transfected with plasmids encoding fusion proteins of GST and either near full-length Hes1 [Hes1(3-281], lane 2) or a truncated form of Hes1 that cannot bind Gro/TLEs [Hes1(Δ276-281], lane 1), followed by isolation of the fusion proteins on glutathione-Sepharose beads. The isolated material was subjected to SDS-PAGE (lanes 3 and 4) together with one-tenth of each input lysate (lanes 1 and 2), followed by Western blotting with panTLE antibodies (Ab). Expression of full-length Hes1, but not Hes1(Δ276-281), resulted in the induction of a more slowly migrating Gro/TLE form (long arrow). Both faster and slower species interacted with Hes1. (B) ROS17/2.8 cells were either not transfected (lane 1) or were transfected with FLAG-tagged Hes1(1-281) (lane 2) or Hes1(Δ276-281) (lane 3), followed by Western blotting analysis with panTLE antibodies. Expression of full-length Hes1 but not Hes1(Δ276-281) resulted in the induction of a more slowly migrating Gro/TLE form (long arrow). (C) Both GST-Hes1(3-281) and GST-Hes1(Δ276-281) were expressed at equal levels. (D) Both FLAG-Hes1(1-281) and FLAG-Hes1(Δ276-281) were expressed at equivalent levels. (E) Effect of alkaline phosphatase treatment of lysates from cells transfected with Hes1. Equivalent amounts of protein extract from cells not transfected (lanes 1 and 2) or transfected (lanes 3 and 4) with full-length Hes1 were incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of calf intestinal phosphatase (CIP), followed by Western blotting analysis with panTLE antibodies. Phosphatase treatment reduced the slower Gro/TLE form induced by Hes1 (long arrow) resulting in a single band of faster mobility.
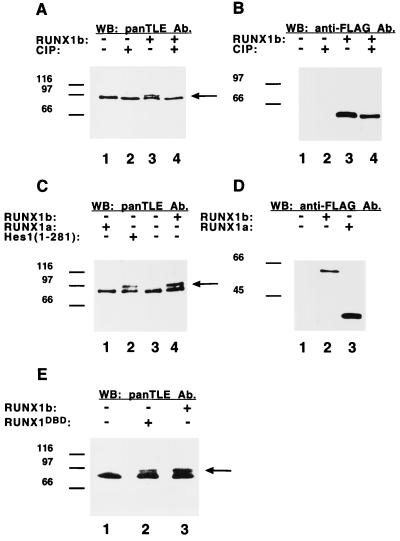
To determine whether other known Gro/TLE-binding transcription factors might also influence the phosphorylation of these proteins, we tested RUNX1, which is coexpressed with Gro/TLEs in a number of differentiating cells (11, 21, 28, 30). Similar to Hes1, full-length RUNX1, referred to hereafter as RUNX1b (30), induced an increase in Gro/TLE phosphorylation (Fig. 3A, cf. lanes 1 and 3, see long arrow). This effect could be abolished by phosphatase treatment (Fig. 3A, cf. lanes 3 and 4), without affecting the stability or expression of RUNX1b (Fig. 3B). In contrast to RUNX1b and Hes1 (Fig. 3C, lanes 2 and 4), a naturally occurring truncated form of RUNX1, RUNX1a (30), which cannot bind Gro/TLEs (28), had no effect on the phosphorylation of the latter (Fig. 3C, lane 1). RUNX1b and RUNX1a were both expressed in the transfected cells (Fig. 3D). Expression of a previously described (36) DNA-binding defective form of RUNX1 (RUNX1DBD) also resulted in Gro/TLE hyperphosphorylation compared to nontransfected cells (Fig. 3E, cf. lanes 1 and 2). We noticed that when equivalent amounts of RUNX1b and RUNX1DBD were expressed, the Gro/TLE hyperphosphorylation induced by the former was more robust, raising the possibility that DNA recruitment might contribute in part to Gro/TLE hyperphosphorylation (cf. lanes 2 and 3). Other proteins containing an amino-terminal FLAG epitope but lacking Gro/TLE-binding ability had no effect on Gro/TLE phosphorylation (not shown). Taken together, these findings demonstrate that both Hes1 and RUNX1 can promote Gro/TLE phosphorylation and that these effects depend on their ability to interact with the latter.
FIG. 3.
Induction of Gro/TLE phosphorylation by RUNX1. (A and B) ROS17/2.8 cells were either not transfected (lanes 1 and 2) or transfected with full-length RUNX1 (RUNX1b, lanes 3 and 4). Equivalent amounts of protein extracts were then incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of calf intestinal phosphatase (CIP), followed by Western blotting analysis with panTLE (A) or anti-FLAG (B) antibodies (Ab). Expression of RUNX1b resulted in the induction of a more slowly migrating Gro/TLE form (A, lane 3, long arrow). Phosphatase treatment abolished this slower form resulting in a single band of faster mobility (A, lane 4). (C and D) Both RUNX1b (lane 4) and Hes1 (lane 2), but not a truncated form of RUNX1 that does not interact with Gro/TLE (RUNX1a, lane 1), induced Gro/TLE hyperphosphorylation, as revealed by Western blotting with panTLE antibodies (C). (D) Expression of transfected RUNX1a and RUNX1b proteins was detected with anti-FLAG antibodies. (E) Expression of a DNA-binding defective form of RUNX1b, RUNX1DBD, also resulted in the induction of a more slowly migrating Gro/TLE form (lane 2, long arrow). RUNX1DBD and RUNX1b were expressed at equivalent levels (not shown).
Association of hyperphosphorylated Gro/TLEs with chromatin.
Previous immunocytochemical studies have shown that Gro/TLEs are localized to the nucleus in intact cells (11, 12, 45). However, biochemical fractionation studies showed that a certain amount of Gro/TLEs remained in a soluble nuclear form that was easily dissociated from the nuclei by exposure to low-ionic-strength conditions (Fig. 4A, lane 1). A second pool was more tightly bound to the nuclear compartment and was not released by these conditions (Fig. 4A, lane 2). In particular, more slowly migrating Gro/TLE forms (present in low amounts compared to faster forms) exhibited a preferential association with the nuclear compartment (Fig. 4A, lane 2, see long arrow), whereas faster-migrating forms were observed in both fractions (Fig. 4A, lanes 1 and 2, see short arrow). This suggested that hyperphosphorylated Gro/TLEs are preferentially associated with the nucleus. To examine this possibility further, ROS17/2.8 cells were transfected with Hes1 to induce Gro/TLE hyperphosphorylation and then subjected to a biochemical fractionation. These studies showed that hyperphosphorylated Gro/TLE species accumulated preferentially in the nuclear fraction (Fig. 4A, cf. lanes 3 and 4, see long arrow). Reprobing of the same nitrocellulose replicas showed that neither the mobility nor the nuclear association of HDAC1 was affected by the expression of Hes1 (Fig. 4B). We found that hyperphosphorylated Gro/TLEs are also preferentially associated with the nuclear compartment in human Jurkat cells, which endogenously express Gro/TLE-binding proteins such as RUNX1 and Hes1 (Fig. 4C, lane 2, see long arrow), suggesting that this behavior is neither a cell type-specific event nor an effect of Hes1 overexpression. Together, these findings show that hyperphosphorylated Gro/TLE species are preferentially associated with the nuclear compartment and that this process is promoted by Hes1. Consistent with these findings, when Drosophila Gro was expressed in mammalian cells in the absence of Hes1, Gro immunoreactivity (detected with a monoclonal antibody that does not cross-react with endogenous TLEs [12, 21]) was found in the non-nuclear fraction (Fig. 4D, lane 1). In contrast, expression of Hes1 resulted in the association of Gro with the nuclear fraction (Fig. 4D, lane 4).
To further our investigations, we tested whether the preferential nuclear association of hyperphosphorylated Gro/TLEs was due to interaction with chromatin, a subnuclear compartment where these proteins were shown to localize (37). ROS17/2.8 cells were subjected to a more rigorous subcellular fractionation procedure (34) to yield a soluble fraction and a nuclease-treated nuclear fraction enriched in chromatin components but not in components of the nuclear matrix. Western blotting analysis with Gro/TLE1 antibodies revealed two main species: an underphosphorylated form that was found in the nonchromatin fraction (Fig. 4E, lane 1, see short arrow) and a hyperphosphorylated form that was found in the chromatin fraction (Fig. 4E, lane 2, see long arrow). The chromatin protein histone H3 was only detected in the chromatin fraction (Fig. 4F, lane 2). Taken together, these findings show that hyperphosphorylated Gro/TLEs interact with chromatin, while underphosphorylated forms either do not interact or interact weakly with this compartment, and suggest further that Hes1 promotes the chromatin association of Gro/TLEs.
Involvement of protein kinase CK2 in Gro/TLE hyperphosphorylation.
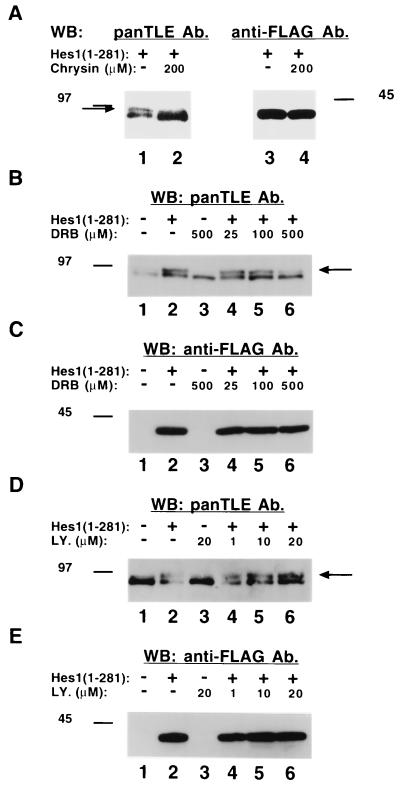
All Gro/TLE family members share conserved sequences that resemble possible sites for phosphorylation by protein kinase CK2 (45). We therefore tested whether this kinase was involved in the Hes1-induced hyperphosphorylation of Gro/TLEs by determining whether this process might be affected by the selective protein kinase CK2 inhibitors, chrysin and DRB (9). Incubation in the presence of chrysin significantly reduced the hyperphosphorylation of Gro/TLE induced by Hes1, with a concomitant increase in the abundance of faster-migrating species (Fig. 5A, cf. lanes 1 and 2, see long arrow). Neither the stability nor the electrophoretic mobility of Hes1 appeared to be affected by chrysin (Fig. 5A, lanes 3 and 4). Similarly, DRB reduced the Gro/TLE hyperphosphorylation induced by Hes1 in a dose-dependent manner (Fig. 5B, see long arrow) but had no effect on the expression or mobility of Hes1 (Fig. 5C). In contrast, the selective phosphatidylinositol 3-kinase (PI 3-kinase) inhibitor LY294002 had no detectable effect on the Hes1-induced hyperphosphorylation of Gro/TLEs (Fig. 5D and E). Together, these observations implicate protein kinase CK2 in the hyperphosphorylation of Gro/TLE induced by Hes1.
FIG. 5.
Effect of selective protein kinase inhibitors on Gro/TLE hyperphosphorylation induced by Hes1. (A) ROS17/2.8 cells were transfected with Hes1 and cultured for 24 h. During the last 2 h, cells were cultured in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of the protein kinase CK2 inhibitor, chrysin, followed by cell lysis and Western blotting with either panTLE (lanes 1 and 2) or anti-FLAG (lanes 3 and 4) antibodies (Ab). The presence of chrysin had two effects: it abolished the hyperphosphorylated form induced by Hes1 (long arrow), and it increased the amount of Gro/TLE in the cell extract. (B to E) ROS17/2.8 cells were not transfected (lanes 1 and 3) or transfected (lanes 2 and 4 to 6) with Hes1 and then incubated for the last 2 h in the absence (lanes 1 and 2) or presence (lanes 3 to 6) of the indicated amounts of either the protein kinase CK2 inhibitor DRB (B and C) or the PI 3-kinase inhibitor LY294002 (LY., D and E). Lysates were then collected and analyzed by Western blotting with either panTLE (B and D) or anti-FLAG (C and E) antibodies. DRB effectively reduced Hes1-induced hyperphosphorylation of Gro/TLEs without affecting the expression or mobility of Hes1, while LY294002 had no effect on the Hes1-induced phosphorylation of Gro/TLEs.
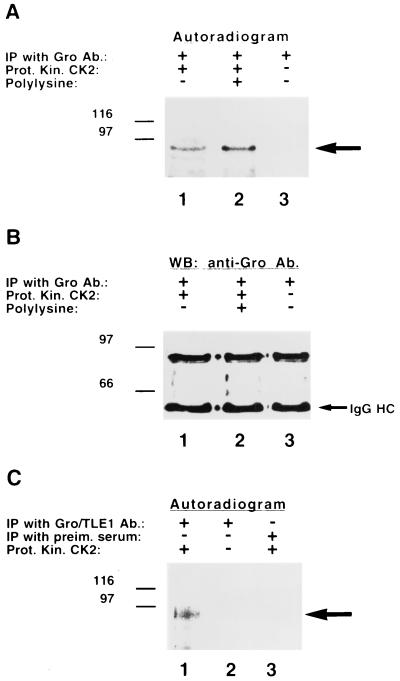
To further our investigations, we next tested whether protein kinase CK2 can phosphorylate Gro/TLE proteins. Drosophila S2 cell lysates were prepared, followed by immunoprecipitation of endogenous Gro and incubation in the presence or absence of purified protein kinase CK2. Addition of the kinase resulted in the phosphorylation of Gro (Fig. 6A, lane 1). This phosphorylation was enhanced by the presence of polylysine, which stimulates the phosphorylation of most substrates for protein kinase CK2 (3, 10) (Fig. 6A, lane 2). Gro was not phosphorylated in the absence of this kinase (Fig. 6A, lane 3), even though it was efficiently immunoprecipitated (Fig. 6B). Protein kinase CK2 also phosphorylated Gro/TLE proteins immunoprecipitated from mammalian cells (Fig. 6C, lane 1). Taken together with the results shown above, these findings strongly suggest that Gro/TLEs are phosphorylated by protein kinase CK2 and that this phosphorylation is promoted by Hes1 binding.
FIG. 6.
Phosphorylation of Gro/TLEs by protein kinase CK2. Lysates from either Drosophila S2 (A and B) or human Jurkat (C) cells were subjected to immunoprecipitation with anti-Gro (A and B, lanes 1 to 3), anti-Gro/TLE1 (C, lanes 1 and 2), or control (C, preim. serum, lane 3) antibodies. Immunoprecipitates were extensively washed, followed by incubation with [γ-32P]ATP in the presence or absence of purified protein kinase CK2, as indicated. Samples were then subjected to SDS-PAGE, transfer to nitrocellulose, and autoradiography. In panel B, autoradiography was followed by Western blotting with anti-Gro antibodies. Gro/TLEs were phosphorylated in vitro by purified protein kinase CK2.
Involvement of protein kinase CK2 in Gro/TLE transcriptional repression.
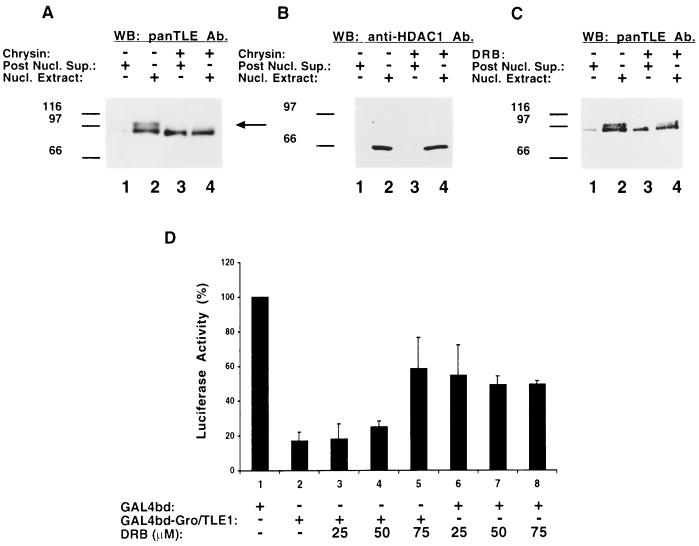
We next tested whether protein kinase CK2 is involved in the association of hyperphosphorylated Gro/TLEs with the nuclear compartment. ROS17/2.8 cells were transfected with Hes1 to induce Gro/TLE hyperphosphorylation and then cultured in the absence or presence of chrysin, followed by a biochemical fractionation. When compared to untreated cells, the presence of chrysin resulted in a significant decrease in the amount of hyperphosphorylated Gro/TLE present in the nuclear fraction (Fig. 7A, cf. lanes 2 and 4, see long arrow). This was correlated with an increase in the amount of underphosphorylated Gro/TLEs in the non-nuclear fraction (Fig. 7A, cf. lanes 1 and 3). Reprobing with anti-HDAC1 antibodies showed that chrysin had no effect on the subcellular distribution of this protein (Fig. 7B). Similar results were obtained in experiments with the DRB compound (Fig. 7C). These observations show that inhibition of protein kinase CK2 activity reduces the amount of hyperphosphorylated Gro/TLEs that can associate with chromatin.
FIG. 7.
Effect of protein kinase CK2 inhibition on Gro/TLE1-mediated transcriptional repression. (A to C) Analysis of the effect of chrysin and DRB on the nuclear association of hyperphosphorylated Gro/TLE. ROS17/2.8 cells were transfected with Hes1 and cultured for the last 4 h in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of either chrysin (A and B) or DRB (C). Non-nuclear (Post Nucl. Sup., lanes 1 and 3) and nuclear (Nucl. Extract, lanes 2 and 4) fractions were prepared and subjected to Western blotting analysis with either panTLE (A and C) or anti-HDAC1 (B) antibodies. Chrysin and DRB reduced the nuclear association of hyperphosphorylated Gro/TLE species (A and C; see long arrow in panel A). No effects on HDAC1 were observed. (D) Transient transfection-transcription assays. HeLa cells were transfected with the reporter plasmid p5xGAL4UAS-tk-luciferase (0.5 μg) alone or in the presence of GAL4bd (1.0 μg, lanes 1 and 6 to 8) or GAL4bd-Gro/TLE1 (1.0 μg, lanes 2 to 5). After 24 h, the cells were incubated in the absence or presence of the indicated amounts of DRB (lanes 3 to 5 and lanes 6 to 8) and then cultured for an additional 20 h. Luciferase activity measured in the presence of GAL4bd alone (approximately twofold higher than basal activity in the absence of GAL4bd) was considered 100%. Values represent the means ± the standard deviation of four experiments performed in duplicate. The ability of Gro/TLE1 to mediate transcriptional repression was reduced by DRB.
We then determined whether transcriptional repression mediated by Gro/TLEs might be reduced as a result of inhibition of protein kinase CK2. HeLa cells were transfected with a reporter plasmid containing the luciferase reporter gene under the control of the tk promoter linked to five tandem copies of the GAL4 upstream activation sequence. This 5xGAL4UAS-tk-luciferase reporter gene can form active chromatin after transfection into HeLa cells (8, 24). Cotransfection of this reporter with GAL4bd alone resulted in an approximately twofold activation of transcription above the basal level driven by the tk promoter. In contrast, expression of GAL4bd-Gro/TLE1 led to a significant repression of both activated and basal transcription (Fig. 7B, lane 2). The transcription repression ability of GAL4bd-Gro/TLE1 was reduced in the presence of increasing amounts of DRB (Fig. 7B, lanes 3 to 5). Importantly, this derepression was opposite to an inhibitory effect of DRB on transcription in the absence of GAL4bd-Gro/TLE1 (Fig. 7B, lanes 6 to 8), which was consistent with the previous demonstration that inhibition of protein kinase CK2 can reduce the activity of RNA polymerases (1). Thus, when this decrease in basal transcription is taken into account, these results show that inhibition of protein kinase CK2 activity results in a significant inhibition of the ability of Gro/TLE proteins to repress transcription.
DISCUSSION
Induction of Gro/TLE hyperphosphorylation by cell differentiation and cofactor binding.
The transcription functions of a number of different DNA-binding proteins are regulated by interaction with Gro/TLE family members, which can either provide a corepressor activity to dedicated transcriptional repressors (16, 32, 38, 54) or convert transcriptional activators into repressors (8, 13, 28). By interacting with Gro/TLEs, these different DNA-binding proteins recruit general transcriptional corepressors to specific DNA sites, where the latter become involved in selected gene regulatory events. Our present studies were motivated by a lack of information about the mechanisms that regulate the functions of complexes of Gro/TLEs and their transcriptional partners. We have described new results that strongly suggest that one of the best-characterized cofactors of Gro/TLEs, the bHLH protein Hes1, plays a role in the regulation of Gro/TLE phosphorylation. More specifically, Hes1 induces Gro/TLE hyperphosphorylation, and this effect requires interaction between these proteins because it was not observed when a mutated form of Hes1 that cannot bind to Gro/TLE was tested. This conclusion is also consistent with the results of our protein-protein interaction experiments showing that Hes1 can be coprecipitated with both under- and hyperphosphorylated forms of Gro/TLEs, suggesting that hyperphosphorylation may follow the interaction between these proteins. Taken together with the demonstration that other Gro/TLE-binding factors, such as RUNX1 (this study), Pax5 (14), and BF-1 (H. Nuthall and S. Stifani, unpublished data), also induce Gro/TLE hyperphosphorylation, our findings suggest that changes in the phosphorylation state of Gro/TLEs induced by cofactor binding constitute an important event in the regulation of Gro/TLE functions. It is possible that different transcriptional cofactors share the ability to induce conformational changes in Groucho/TLEs, which in turn may expose the latter to the activity of specific kinases, including protein kinase CK2 (see below). It is tempting to speculate that these structural changes may be similar in response to interactions with DNA-binding proteins whose functions are modulated by Gro/TLEs in similar manners leading to the exposure of the same Gro/TLE phosphorylation sites and common functional effects. It is entirely possible, however, that conformational changes promoted by different cofactors are selective, resulting in hyperphosphorylation through different combinations of kinases that may lead to different regulatory effects. This latter possibility is consistent with the fact that Gro/TLEs contain many conserved sequences conforming to consensus phosphorylation sites for numerous kinases. In the future, it will therefore be important to determine whether or not different kinases mediate the Gro/TLE hyperphosphorylation induced by different transcriptional cofactors, particularly when Gro/TLEs are involved in interactions with either dedicated repressors or transactivators.
The hyperphosphorylation of Gro/TLE promoted by Hes1 resembles the increase in Gro/TLE phosphorylation observed during neural differentiation. In addition to Hes1 (42), Gro/TLEs are coexpressed in neural progenitor cells with several of their known binding partners, including BF-1 (54), Pax (50), SIX (17), and Nkx (35) proteins. It is possible that at least some of these factors are involved in promoting the hyperphosphorylation of Gro/TLEs observed during neural differentiation. This possibility is also consistent with the demonstration that the expression of a number of Gro/TLE-binding factors, including Hes1 (49) and Pax (40) proteins, is activated during the retinoic acid-induced neural differentiation of P19 cells, a process characterized by significant Gro/TLE hyperphosphorylation. Interactions leading to phosphorylation of Gro/TLEs may occur during periods of active differentiation to restrict the number of progenitor cells that will enter the neuronal fate. Alternatively, these proteins may become hyperphosphorylated in newly generated postmitotic neurons, perhaps to contribute to the establishment of the neuronal phenotype by silencing the expression of nonneuronal genes.
Regulation of the nuclear association of Gro/TLEs by phosphorylation.
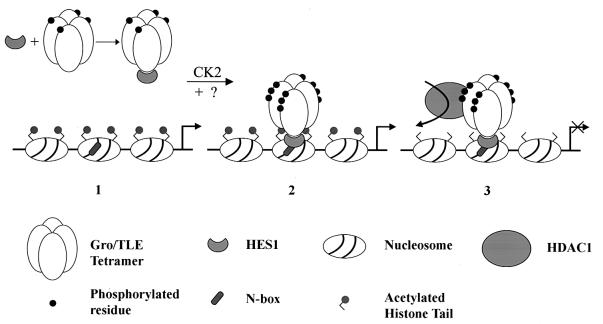
Our investigations have shown further that when Gro/TLE proteins become hyperphosphorylated in response to Hes1, they also become strongly bound to the nuclear compartment and cannot be readily eluted from the nucleus. The tight nuclear association of hyperphosphorylated Gro/TLEs appears to result from their interaction with chromatin because biochemical fractionation experiments show that the Gro/TLE population that is bound to chromatin and is resistant to extraction is hyperphosphorylated, while the fraction that is not bound to chromatin is underphosphorylated. These findings are in agreement with previous studies showing that Gro/TLEs interact with both core histones (15, 37) and histone deacetylases (4, 6, 8, 54). Taken together, these observations suggest that Hes1 both induces Gro/TLE hyperphosphorylation and targets hyperphosphorylated Gro/TLEs to specific promoter regions where the latter interact with chromatin components and mediate transcriptional repression (Fig. 8). In such a model, hyperphosphorylation induced by Hes1 (and possibly other Gro/TLE cofactors) is a crucial event contributing to the establishment of a strong interaction of Gro/TLE with chromatin. It does not appear that Gro/TLE hyperphosphorylation is solely the result of recruitment to DNA because a DNA-binding defective form of RUNX1 can induce Gro/TLE hyperphosphorylation, suggesting that this effect is not dependent on DNA binding. Moreover, Engrailed proteins, which can bind to both Gro/TLEs and DNA, do not induce Gro/TLE hyperphosphorylation (14, 23, 54), suggesting that the latter event is not simply the result of recruitment to DNA. However, our observation that wild-type RUNX1 induces a more robust Gro/TLE hyperphosphorylation than a DNA-binding defective form suggests that the interaction with DNA may also contribute to the hyperphosphorylation of Gro/TLEs. In any case, Hes1 appears to act as a crucial regulator of the ability of Gro/TLEs to mediate transcriptional repression by promoting the assembly of active repressor complexes at transcriptionally competent sites. As a result, selected chromatin domains become targeted by the chromatin remodeling activity of protein complexes containing Hes1 and hyperphosphorylated Gro/TLE, leading to transcriptional repression.
FIG. 8.
Proposed model for transcriptional repression mediated by Hes1 and Gro/TLEs. (Structures 1 and 2) The interaction of Hes1 with oligomeric Gro/TLE leads to hyperphosphorylation of the latter via a mechanism involving protein kinase CK2 and perhaps other as-yet-unidentified kinases. Hes1 then targets hyperphosphorylated Gro/TLE to specific DNA sequences (N boxes), where additional phosphorylation may occur. (Structure 3) Hyperphosphorylated Gro/TLE interacts with histones and recruits HDAC1 to the template, leading to histone deacetylation. The ability of Gro/TLE to oligomerize may result in the engagement of adjacent nucleosomes by the Gro/TLE polymer, resulting in chromatin modification over an extended domain and transcriptional repression.
Similar strategies may be used by other DNA-binding proteins that rely on the transcription repression activity provided by Gro/TLEs. In turn, this suggests that the functions of these repressor complexes may be regulated not only by the formation of stable complexes but also by mechanisms involving phosphorylation events following complex formation. For instance, conditions under which hyperphosphorylation of Gro/TLE does not occur due to inhibition of the kinase(s) involved may result in the formation of complexes that may interact with DNA but may be unable to engage chromatin components. Alternatively, controlled dephosphorylation mechanisms may be utilized to negatively regulate the functions of these complexes when their activities are no longer needed. Finally, it is important to stress that Gro/TLEs may utilize other mechanisms of transcriptional repression in addition to modification of the acetylation state of histones. This possibility would be consistent with our present observation that, in addition to the hyperphosphorylated Gro/TLE population that is bound to chromatin, there is a separate Gro/TLE population that also associates strongly with the nuclear compartment but is neither hyperphosphorylated nor bound to chromatin. This second pool may contain Gro/TLE proteins involved in different molecular mechanisms.
Involvement of protein kinase CK2 in Gro/TLE functions.
Examination of the sequence of Caenorhabditis elegans, Drosophila, and vertebrate Gro/TLE proteins reveals the presence of several evolutionarily conserved motifs containing the consensus protein kinase CK2 phosphorylation sequence S/T-X-X-D/E. This first suggested that protein kinase CK2 may be involved in Gro/TLE activities (45). Our present studies have provided new results that lend additional support to this notion. First, purified protein kinase CK2 phosphorylates Gro/TLEs in vitro. Second, pharmacological inhibition of protein kinase CK2 activity in cultured cells reduces the Hes1-induced hyperphosphorylation of Gro/TLEs, indicative of an involvement of this kinase in that response. Third, exposure to protein kinase CK2 inhibitors decreases the amount of hyperphosphorylated Gro/TLEs that are tightly associated with the nuclear compartment and inhibits Gro/TLE’s ability to mediate transcriptional repression. Taken together, these findings suggest that protein kinase CK2 (or a closely related kinase) is at least one of the kinases involved in Gro/TLE hyperphosphorylation and contributes to promoting the transcription repression functions of Gro/TLEs by increasing their interaction with chromatin.
Protein kinase CK2 is a multifunctional enzyme found in the nucleus and cytoplasm of all eukaryotic cells and can phosphorylate a multitude of substrates, including a variety of transcriptional regulators (reviewed in references 1, 33, and 39). In this regard, it is worth noting that a number of transcription factors that interact with Gro/TLEs are phosphorylated by protein kinase CK2. These include the Drosophila homeodomain protein Even-skipped (26, 29) and mammalian SIX (17) and Nkx (25) proteins. Although the functional significance of the phosphorylation of these proteins by protein kinase CK2 differs from case to case and remains to be further characterized, it is tempting to speculate that under appropriate physiological conditions these factors may interact with both protein kinase CK2 and Gro/TLE; in turn, this may facilitate and/or stimulate the phosphorylation of the latter by protein kinase CK2. A similar situation has been described in the case of the homeodomain interacting protein kinase 2, which can form complexes with both Gro/TLE and NK3 proteins (8). This possibility is also consistent with the recent demonstration that the Drosophila Hes family members, Enhancer of split m5, m7, and m8 (but not the closely related factors m3, mA, and mC) interact with, and are phosphorylated by, protein kinase CK2 (48). Since Enhancer of split proteins such as m7 and m8 physically bind to Gro (16, 38), it is conceivable that they may also facilitate the interaction between protein kinase CK2 and Gro proteins. It will be important to determine whether mammalian Hes1, which is related to the Enhancer of split proteins, may act in a similar manner, although it should be pointed out that Hes1 does not have a recognizable protein kinase CK2 phosphorylation site similar to that of m5, m7, and m8 and may not be phosphorylated by this enzyme (this is also suggested by our observation that pharmacological inhibition of protein kinase CK2 did not appear to change the electrophoretic mobility of Hes1).
In summary, our present findings provide the first evidence that protein kinase CK2 is involved in the regulation of the transcription repression functions of Gro/TLEs and point to a crucial role for Hes1 in the modulation of the phosphorylation of Gro/TLEs by this kinase. These observations provide new insight into the mechanisms that control the functional interaction between Gro/TLEs and Hes1 and raise the interesting possibility that similar regulatory events may underlie the interaction of Gro/TLEs with a number of other DNA-binding proteins.
Acknowledgments
We thank Elena Torban for providing neural progenitor cell extracts, Rita Lo for invaluable help during this work, K. Shigesada and M. Osato for pEF-BOS-RUNX1 plasmids, C. D. Allis for antibodies against histone H3, D. Kaplan for reagents, and Julie Nadeau for important observations.
This work was supported by grants from the Canadian Institutes of Health Research (GR-14971) (50%) and the Cancer Research Society, Inc. (50%) to S.S. S.S. is a Scholar of the Fonds de la Recherche en Santé du Québec and a Killam Scholar of the Montreal Neurological Institute.
REFERENCES
- 1.Allende, J. E., and C. C. Allende. 1995. Protein kinases 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 9:313–323. [DOI] [PubMed] [Google Scholar]
- 2.Aronson, B. D., A. L. Fisher, K. Blechman, M. Caudy, and J. P. Gergen. 1997. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol. Cell. Biol. 17:5581–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bidwai, A. P., J. C. Reed, and C. V. Glover. 1993. Phosphorylation of calmodulin by the catalytic subunit of casein kinase II is inhibited by the regulatory subunit. Arch. Biochem. Biophys. 300:265–270. [DOI] [PubMed] [Google Scholar]
- 4.Brantjes, H., J. Roose, M. van de Wetering, and H. Clevers. 2001. All Tcf HMG box transcription factors interact with Groucho-related corepressors. Nucleic Acids Res. 29:1410–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, G., P. H. Nguyen, and A. J. Courey. 1998. A role for Groucho tetramerization in transcriptional repression. Mol. Cell. Biol. 18:7259–7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, G., J. Fernandez, S. Mische, and A. J. Courey. 1999. A functional interaction between the histone deacetylase rpd3 and the corepressor Groucho in Drosophila development. Genes Dev. 13:2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, G., and A. J. Courey. 2000. Groucho/TLE family proteins and transcriptional repression. Gene 249:1–16. [DOI] [PubMed] [Google Scholar]
- 8.Choi, C. Y., Y. H. Kim, H. J. Kwon, and Y. Kim. 1999. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J. Biol. Chem. 274:33194–33197. [DOI] [PubMed] [Google Scholar]
- 9.Critchfield, J. W., J. E. Coligan, T. M. Folks, and S. T. Butera. 1997. Casein kinase II is a selective target of HIV-1 transcriptional inhibitors. Proc. Natl. Acad. Sci. USA 94:6110–6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davletov, B., J. M. Sontag, Y. Hata, A. G. Petrenko, E. M. Fykse, R. Jahn, and T. C. Sudhof. 1993. Phosphorylation of synaptotagmin I by casein kinase II. J. Biol. Chem. 268:6816–6822. [PubMed] [Google Scholar]
- 11.Dehni, G., Y. Liu, J. Husain, and S. Stifani. 1995. TLE expression correlates with mouse embryonic segmentation, neurogenesis, and epithelial determination. Mech. Dev. 53:369–381. [DOI] [PubMed] [Google Scholar]
- 12.Delidalis, C., A. Preiss, D. A. Hartley, and S. Artavanis-Tsakonas. 1991. Two genetically and molecularly distinct functions involved in early neurogenesis reside within the Enhancer of split locus of Drosophila melanogaster. Genetics 129:803–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubnicoff, T., S. A. Valentine, G. Chen, T. Shi, J. A. Lengyel, Z. Paroush, and A. J. Courey. 1997. Conversion of Dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev. 11:2952–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberhard, D., G. Jimenez, B. Heavy, and M. Busslinger. 2000. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 19:2292–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flores-Saaib, R. D., and A. J. Courey. 2000. Analysis of Groucho-histone interactions suggests mechanistic similarities between Groucho- and Tup1-mediated repression. Nucleic Acids Res. 28:4184–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher, A. L., S. Ohsako, and M. Caudy. 1996. The WRPW motif of the Hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 16:2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ford, H. L., E. Landesman-Bollag, C. S. Dacwag, P. T. Stukenberg, A. B. Pardee, and D. C. Seldin. 2000. Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J. Biol. Chem. 275:22245–22254. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh, A., and M. E. Greenberg. 1995. Distinct roles for bFGF and NT-3 in the regulation of cortical neurogenesis. Neuron 15:89–103. [DOI] [PubMed] [Google Scholar]
- 19.Grbavec, D., R. Lo, Y. Liu, and S. Stifani. 1998. Transducin-like Enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with Hairy/Enhancer of split proteins, and is expressed during neuronal development. Eur. J. Biochem. 258:339–349. [DOI] [PubMed] [Google Scholar]
- 20.Hendzel, M. J., G. P. Delcuve, and J. R. Davie. 1991. Histone deacetylase is a component of the internal nuclear matrix. J. Biol. Chem. 266:21936–21942. [PubMed] [Google Scholar]
- 21.Husain, J., R. Lo, D. Grbavec, and S. Stifani. 1996. Affinity for the nuclear compartment and expression during cell differentiation implicate phosphorylated Groucho/TLE1 forms of higher molecular mass in nuclear functions. Biochem. J. 317:523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Javed, A., B. Guo, S. Hiebert, J. Y. Choi, J. Green, S. C. Zhao, M. A. Osborne, S. Stifani, J. L. Stein, J. B. Lian, A. J. van Wijnen, and G. S. Stein. 2000. Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBFα/AML/PEBP2α) dependent activation of tissue-specific gene transcription. J. Cell Sci. 113:2221–2231. [DOI] [PubMed] [Google Scholar]
- 23.Jimenez, G., Z. Paroush, and D. Ish-Horowicz. 1997. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev. 11:3072–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadosh, D., and K. Struhl. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18:5121–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasahara, H., and S. Izumo. 1999. Identification of the in vivo casein kinase II phosphorylation site within the homeodomain of the cardiac tissue-specific homeobox gene product Csx/Nkx2.5. Mol. Cell. Biol. 19:526–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi, M., R. E. Goldstein, M. Fujioka., Z. Paroush, and J. B. Jaynes. 2001. Groucho augments the repression of multiple Even skipped target genes in establishing parasegmental boundaries. Development 128:1805–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koop, K. E., L. M. MacDonald, and C. G. Lobe. 1996. Transcripts of Grg4, a murine groucho-related gene, are detected in adjacent tissues to other murine neurogenic gene homologues during embryonic development. Mech. Dev. 59:73–87. [DOI] [PubMed] [Google Scholar]
- 28.Levanon, D., R. E. Goldstein, Y. Bernstein, H. Tang, D. Goldenberg, S. Stifani, Z. Paroush, and Y. Groner. 1998. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. USA 95:11590–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, C., and J. L. Manley. 1999. Allosteric regulation of even-skipped repression activity by phosphorylation. Mol. Cell 3:77–86. [DOI] [PubMed] [Google Scholar]
- 30.Lutterbach, B., and S. W. Hiebert. 2000. Role of the transcription factor AML-1 in acute leukemia and hematopoietic differentiation. Gene 245:223–235. [DOI] [PubMed] [Google Scholar]
- 31.McLarren, K. W., R. Lo, D. Grbavec, K. Thirunavukkarasu, G. Karsenty, and S. Stifani. 2000. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the Runt-related factor Cbfa1. J. Biol. Chem. 275:530–538. [DOI] [PubMed] [Google Scholar]
- 32.McLarren, K. W., F. Theriault, and S. Stifani. 2001. Association with the nuclear matrix and interaction with Groucho and RUNX proteins regulate the transcription repression ability of the basic helix loop helix factor Hes1. J. Biol. Chem. 276:1578–1584. [DOI] [PubMed] [Google Scholar]
- 33.Meisner, H., and M. P. Czech. 1991. Phosphorylation of transcription factors and cell-cycle-dependent proteins by casein kinase II. Curr. Opin. Cell Biol. 3:474–483. [DOI] [PubMed] [Google Scholar]
- 34.Merriman, H. L., A. J. van Wijnen, S. Hiebert, J. P. Bidwell, E. Fey, J. Lian, J. Stein, and G. S. Stein. 1995. The tissue-specific nuclear matrix protein, NMP-2, is a family member of the AML/CBF/PEBP2/Runt domain transcription factor family: interactions with the osteocalcin gene promoter. Biochemistry 34:13125–13132. [DOI] [PubMed] [Google Scholar]
- 35.Muhr, J., E. Andersson, M. Persson, T. M. Jessell, and J. Ericson. 2001. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 104:861–873. [DOI] [PubMed] [Google Scholar]
- 36.Osato, M., N. Asou, E. Abdalla, K. Hoshino, H. Yamasaki, T. Okubo, H. Suzushima, K. Takatsuki, T. Kanno, K. Shigesada, and Y. Ito. 1999. Biallelic and heterozygous point mutations in the Runt domain of the AML1/PEBP2αB gene associated with myeloblastic leukemias. Blood 93:1817–1824. [PubMed] [Google Scholar]
- 37.Palaparti, A., A. Baratz, and S. Stifani. 1997. The Groucho/Transducin-like Enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J. Biol. Chem. 272:26604–26610. [DOI] [PubMed] [Google Scholar]
- 38.Paroush, Z., R. L. Finley, T. Kidd, S. M. Wainwright, P. W. Ingham, R. Brent, and D. Ish-Horowicz. 1994. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79:805–815. [DOI] [PubMed] [Google Scholar]
- 39.Pinna, L. A. 1990. Casein kinase 2: an “eminence grise” in cellular regulation? Biochim. Biophys. Acta 1054:267–284. [DOI] [PubMed] [Google Scholar]
- 40.Pruitt, S. C. 1992. Expression of Pax-3 and neuroectoderm-inducing activities during differentiation of P19 embryonal carcinoma cells. Development 116:573–583. [DOI] [PubMed] [Google Scholar]
- 41.Ren, B., K. J. Chee, T. H. Kim, and T. Maniatis. 1999. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 13:125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasai, Y., R. Kageyama, Y. Tagawa, R. Shigemoto, and S. Nakanishi. 1992. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 5:2620–2634. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt, C. J., and T. E. Sladek. 1993. A rat homolog of the Drosophila Enhancer of split (groucho) locus lacking WD-40 repeats. J. Biol. Chem. 268:25681–25686. [PubMed] [Google Scholar]
- 44.Slack, R. S., H. El-Bizri, J. Wong, D. J. Belliveau, and F. D. Miller. 1998. A critical temporal requirement for the retinoblastoma protein family during neuronal determination. J. Cell Biol. 140:1497–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stifani, S., C. M. Blaumueller, N. J. Redhead, R. E. Hill, and S. Artavanis-Tsakonas. 1992. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat. Genet. 2:119–127. [DOI] [PubMed] [Google Scholar]
- 46.Thirunavukkarasu, K., M. Mahajan, K. W. McLarren, S. Stifani, and G. Karsenty. 1998. Two domains unique to osteoblast-specific transcription factor Osf2/Cbf1 contribute to its transactivating function and its inability to heterodimerize with Cbfβ. Mol. Cell. Biol. 18:4197–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tolkunova, E. N., M. Fujioka, M. Kobayashi, D. Deka, and J. B. Jaynes. 1998. Two distinct types of repression domain in Engrailed: one interacts with the Groucho corepressor and is preferentially active on integrated target genes. Mol. Cell. Biol. 18:2804–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trott, R. L., K. Madhavi, Z. Paroush, and A. P. Bidwai. 2001. Drosophila melanogaster casein kinase II interacts with and phosphorylates the basic helix-loop-helix proteins m5, m7, and m8 derived from the Enhancer of split complex. J. Biol. Chem. 276:2159–2167. [DOI] [PubMed] [Google Scholar]
- 49.Wakabayashi, N., R. Kageyama, T. Habu, T. Doi, T. Morita, M. Nozaki, M. Yamamoto, and Y. Nishimune. 2000. A novel cis-acting element regulates HES-1 gene expression in P19 embryonal carcinoma cells treated with retinoic acid. J. Biochem. 128:1087–1095. [DOI] [PubMed] [Google Scholar]
- 50.Walther, C., and P. Gruss. 1991. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 113:1435–1449. [DOI] [PubMed] [Google Scholar]
- 51.Wang, J. C., M. Waltner-Law, K. Yamada, H. Osawa, S. Stifani, and D. K. Granner. 2000. Transducin-like Enhancer of split proteins, the human homologs of Drosophila Groucho, interact with hepatic nuclear factor 3β. J. Biol. Chem. 275:18418–18423. [DOI] [PubMed] [Google Scholar]
- 52.Yao, J., Y. Liu, J. Husain, R. Lo, A. Palaparti, J. Henderson, and S. Stifani. 1998. Combinatorial expression patterns of individual TLE proteins during cell determination and differentiation suggest non-redundant functions for mammalian homologs of Drosophila Groucho. Dev. Growth Differ. 40:133–146. [DOI] [PubMed] [Google Scholar]
- 53.Yao, J., Y. Liu, R. Lo, I. Tretjakoff, A. Peterson, and S. Stifani. 2000. Disrupted development of the cerebral hemispheres in transgenic mice expressing the mammalian Groucho homologue Transducin-like Enhancer of split 1 in postmitotic neurons. Mech. Dev. 93:105–115. [DOI] [PubMed] [Google Scholar]
- 54.Yao, J., E. Lai, and S. Stifani. 2001. The winged-helix protein brain factor 1 interacts with Groucho and Hes proteins to repress transcription. Mol. Cell. Biol. 21:1962–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]