Abstract
A variety of transcription factors are targets for conjugation to the ubiquitin-like protein Smt3 (also called SUMO). While many such factors exhibit enhanced activity under conditions that favor conjugation, the mechanisms behind this enhancement are largely unknown. We previously showed that the Drosophila melanogaster rel family factor, Dorsal, is a substrate for Smt3 conjugation. The conjugation machinery was found to enhance Dorsal activity at least in part by counteracting the Cactus-mediated inhibition of Dorsal nuclear localization. In this report, we show that Smt3 conjugation occurs at a single site in Dorsal (lysine 382), requires just the Smt3-activating and -conjugating enzymes, and is reversed by the deconjugating enzyme Ulp1. Mutagenesis of the acceptor lysine eliminates the response of Dorsal to the conjugation machinery and results in enhanced levels of synergistic transcriptional activation. Thus, in addition to controlling Dorsal localization, Smt3 also appears to regulate Dorsal-mediated activation, perhaps by modulating an interaction with a negatively acting nuclear factor. Finally, since Dorsal contributes to innate immunity, we examined the role of Smt3 conjugation in the immune response. We find that the conjugation machinery is required for lipopolysaccharide-induced expression of antimicrobial peptides in cultured cells and larvae, suggesting that Smt3 regulates Dorsal function in vivo.
Ubiquitin is the prototype for a family of polypeptides that become covalently attached to other proteins via an isopeptide bond between the C-terminal carboxyl group of the ubiquitin-like polypeptide and a lysine side chain of the target protein. In addition to ubiquitin, members of this family include Rub1, Smt3 (also called SUMO), Apg12, and Urm1 (54). While the covalent conjugation of ubiquitin to a target protein almost always leads to proteasomal degradation of the target, the roles of the other ubiquitin-related modifiers are more diverse.
All ubiquitin-like modifiers appear to attach to their targets via a conserved series of enzymatic reactions. For example, Smt3 is initially activated in an ATP-dependent reaction that results in the formation of a thioester bond between the C terminus of Smt3 and a cysteine side chain in the heterodimeric Smt3-activating enzyme SAE1/SAE2 (10, 27, 41). Smt3 is then handed off to a cysteine side chain in the Smt3-conjugating enzyme, Ubc9, before being transferred to the ɛ-amino group of a target protein (11, 25). SAE1/SAE2 and Ubc9 show significant homology to the E1 (ubiquitin-activating) and E2 (ubiquitin-conjugating) enzymes that play analogous roles in the ubiquitin conjugation pathway. In ubiquitin conjugation, the selection of target proteins is the responsibility of a diverse set of E3 enzymes (ubiquitin ligases), which catalyze the transfer of ubiquitin from an E2 enzyme to the target protein (24). Although a recent report strongly suggests the existence of analogous Smt3 ligases (26), such enzymes are not essential for Smt3 conjugation or for the selection of conjugation targets, which appears to be a responsibility of the conjugating enzyme Ubc9. Smt3 conjugation is believed to occur, at least in part, in the nucleus, and many Smt3 conjugation targets are found in the nucleus or at the nuclear periphery. These targets include components of the nuclear pore complex, proteins associated with the nuclear bodies (NBs), and numerous regulatory transcription factors (54).
The Smt3 conjugation pathway appears to play many functional roles. For example, conjugation of Smt3 to RanGAP1 is required to target it to the nuclear pore complex, where it associates with the Ran-GTP-binding protein RanBP2 (33, 37). Within the nucleus, Smt3 conjugation may also be essential for the integrity of NBs, as viruses that alter Smt3 modification of the NB-associated factors PML and Sp100 also cause disassembly of the NBs (40, 56). Many transcription factors, including c-jun, p53, and the androgen receptor, have also been identified as targets for Smt3 conjugation (17, 32, 42, 43, 46).
Unlike ubiquitin conjugation, in which the target lysine residues do not need to be in a particular sequence context, Smt3 is almost always conjugated to sites that match the consensus sequence (I/L/V)KXE, in which the K is the acceptor lysine and X is any amino acid (45). Intriguingly, a so-called synergy control (SC) motif found in the glucocorticoid receptor (GR) matches this consensus very closely (22). The SC motif appears to down-regulate the synergistic activation that occurs when multiple molecules of the GR bind to multiple closely spaced binding sites in a compound hormone response element (HRE). Thus, mutagenesis of the SC motif in the GR results in significantly increased activation of promoters containing compound HREs, but there is no increased activation of promoters linked to a single binding site for the GR. It was therefore hypothesized that the presence of multiple SC motifs at compound HREs allows for the recruitment of a putative SC factor (SCF) that attenuates synergy. The similarity between the SC motif and the Smt3 conjugation site suggests mechanistic or regulatory connections between Smt3 conjugation and transcriptional synergy.
It was recently discovered that the Drosophila melanogaster rel family transcription factor Dorsal is a substrate for Smt3 conjugation (3). Dorsal controls cell fate along the dorsoventral axis of the embryo by activating and repressing an array of target genes (12, 44). Dorsal and its vertebrate homolog NF-κB also contribute to the innate immune response (14, 18, 21). In both insects and vertebrates, Toll family receptors mediate an immune response by triggering the degradation of IκB or its Drosophila homolog Cactus, inhibitory factors that bind rel family proteins and block their nuclear translocation. After degradation of IκB or Cactus, rel family proteins are free to enter the nucleus, where they then activate the promoters of genes encoding antimicrobial peptides.
Previously, it was found that Dorsal binds to Ubc9 in vivo and in vitro (3). In cultured Drosophila S2 cells, Ubc9 enhanced the nuclear localization of a Dorsal-green fluorescent protein (GFP) fusion protein in the presence of Cactus. This result was corroborated by the finding that Ubc9, Smt3, and SAE1/SAE2 all work to oppose inhibition of Dorsal-dependent reporter activity by Cactus. Lastly, we showed that Ubc9 catalyzes the conjugation of Smt3 to Dorsal in vivo. These findings suggest that conjugation of Smt3 to Dorsal enhances its activity, at least in part by enhancing either its nuclear uptake or its nuclear retention in the presence of Cactus.
In the present report, we present experiments that tighten the experimental link between Smt3 conjugation and Dorsal function. First, we show that the Smt3 conjugation machinery stimulates Dorsal activity even under conditions in which Dorsal is constitutively nuclear, suggesting that conjugation may directly influence the ability of Dorsal to activate transcription. Second, we map the lysine in Dorsal that serves as the acceptor for Smt3 and show that mutagenesis of this lysine abolishes the ability of Dorsal to respond to the Smt3 conjugation machinery. Furthermore, we show that this lysine resides within a motif that controls transcriptional synergy. Finally, we demonstrate that the Dorsal-dependent activation of genes encoding certain antimicrobial peptides is compromised in the absence of a functional Smt3 conjugation system, thereby showing that Smt3 conjugation is likely to regulate Dorsal function in the intact organism.
MATERIALS AND METHODS
Stable transfections.
To generate the stably transformed 529SU cell line, the full-length ubc9 open reading frame (ORF) was amplified using PCR primers containing SmaI sites and the full-length smt3 ORF was amplified with PCR primers containing SacI sites. The 5′ ubc9 primer encoded a hemagglutinin (HA) epitope tag, while the 5′ smt3 primer encoded a FLAG epitope tag. HA-ubc9 or FLAG-smt3 PCR products were then inserted into the pRM vector (3) digested with SmaI or SacI, respectively. These vectors were cotransfected into Drosophila S2 cells along with pHYGRO (Invitrogen) and pRM-GFP (3) by calcium phosphate precipitation. Beginning 5 days posttransfection, the cells were cultured in Schneider’s insect medium (Sigma) containing 300 μg of hygromycin (Invitrogen)/ml for 1 month, replacing the medium every 5 days. Hygromycin-resistant cells were then cultured as usual in the presence of 300 μg of hygromycin/ml.
Site-directed mutagenesis.
The DorsalK382R point mutation was generated within pBS-dl (49) with primers designed to change the AAA codon corresponding to lysine 382 to AGA (which encodes arginine) or GCA (which encodes alanine). The Ubc9* point mutation was generated within pGEM-Ubc9 (3) with primers designed to change the TGC codon corresponding to cysteine 93 to CGC (which encodes arginine). All mutagenesis was carried out using the QuickChange mutagenesis kit (Stratagene) according to the manufacturer’s protocol.
In vitro Smt3 conjugation and deconjugation assays.
For the Smt3 conjugation assay, the glutathione S-transferase (GST)-tagged expression vectors pGex-HA-Smt3FL and pGex-HA-Smt3GG were generated by using primers containing a 5′ BamHI site and a 3′ EcoRI site to amplify either the full-length ORF or the ORF without the final two amino acids, respectively. Vectors pVL-FLAG-SAE1 and pVL-FLAG-SAE2 for the expression of FLAG-tagged SAE1 and SAE2 were generated by using primers containing XbaI sites to amplify FLAG-tagged versions of each full-length ORF from the pPAC-FLAG-SAE1 and pPAC-FLAG-SAE2 templates, respectively (3). GST fusion proteins (Smt3GG, Smt3FL, and Ubc9) were expressed in Escherichia coli and purified on glutathione-agarose beads. The full-length, precursor GST-Smt3FL and the processed GST-Smt3GG were cleaved by thrombin to yield Smt3FL and Smt3GG. FLAG-SAE1, FLAG-SAE2, and FLAG-Dorsal were expressed in baculovirus-infected Sf9 cells and purified on anti-FLAG (M2) beads (Sigma). The in vitro Smt3 conjugation assay included 50 mM Tris (pH 7.5), 5 mM MgCl2, and an ATP regeneration system (3.5 U of creatine kinase/ml, 0.6 U of inorganic pyrophosphatase/ml, 10 mM creatine phosphate, and 2 mM ATP) in addition to the following (as indicated): 600 ng of FLAG-Dorsal, 90 ng of FLAG-SAE1, 90 ng of FLAG-SAE2, 600 ng of GST-Ubc9, and/or 1.2 μg of Smt3FL or Smt3GG. Reaction mixtures were incubated at 37°C for 2 h, and reactions were terminated by boiling in sodium dodecyl sulfate (SDS) loading buffer.
A GST-Ulp1 expression construct was generated by amplifying the Ulp1 cDNA from the expressed sequence tag clone GH02751 by using PCR primers containing BamHI sites and inserting it into the BamHI site in the pGex-4T1 vector. The GST-HA-Smt3-GFP construct was generated by first amplifying GFP from pRM-GFP (3) by using primers containing EcoRI (5′) and XhoI (3′) and amplifying HA-Smt3 from pPAC-HA-Smt3 (3) by using primers containing BamHI (5′) and EcoRI (3′). These fragments were ligated into pGex-4T1 using its BamHI and XhoI sites. GST-HA-Smt3 and GST-HA-Smt3-GFP fusion proteins were expressed in E. coli, purified on glutathione-agarose beads, and treated with thrombin to remove the GST tag. The Smt3 processing assays were performed at 30°C and included 150 ng of HA-Smt3-GFP (or HA-Smt3) and 3 μg of crude GST-Ulp1 (or GST). Reaction mixtures contained 150 mM NaCl, 1 mM dithiothreitol, 10 mM Tris (pH 8.0), and 0.2% Triton X-100. The plus N-ethylmaleimide (+NEM) reaction mixture included GST-Ulp1 preincubated with 20 mM NEM (Sigma) for 15 min at room temperature. Reactions were stopped at different time points (from 5 min to 2 h) by boiling in SDS loading buffer.
Cotransfection assays.
A pPAC-FLAG-Ulp1 vector was created by amplifying the ulp1 cDNA by PCR with primers containing BamHI sites and inserting it into a BamHI-digested pPAC-FLAG vector (3). To generate pPAC-FLAG-DorsalK382R, PCR primers containing KpnI sites were used to amplify a fragment encoding DorsalK382R. The resulting fragment was then inserted into a KpnI-digested pPAC-FLAG vector. The DE1 vector was generated by ligating a KpnI-digested, double-stranded oligonucleotide (top strand, 5′ CGG GGT ACC AAA CAT ATG CTC TCG GGT TGG GAA AAT CCC CCA GGT ACC CCG 3′) consisting of one Dorsal binding site and one Twist binding into the KpnI-digested −37tkluc vector (Promega). The construction of the pPAC-FLAG-Dorsal, pPAC-Twist, pPAC-HA-Ubc9, and pPAC-HA-Smt3 expression vectors and the DE5 (same as pDE5-37tkluc) and p-37Rluc reporter plasmids has been previously described (3, 7, 49).
Calcium phosphate cotransfections into Drosophila S2 cells were carried out as described previously (9). For reporter assays, each transfection consisted of 5 μg of DE5 or DE1, 0.5 μg of p-37Rluc (internal control), 20 ng of pPAC-Twist, 50 ng of pPAC-dl, and the indicated amounts of pPAC-HA-Ubc9, pPAC-HA-Smt3, and/or pPAC-FLAG-Ulp1. The luciferase reporter activity was determined with the Dual-luciferase reporter assay system (Promega).
For immunoblot experiments, the indicated combinations of pPAC-FLAG-Dorsal, pPAC-HA-Ubc9, pPAC-HA-Smt3, and/or pPAC-FLAG-Ulp1 were brought to 20 μg total with empty vector and cotransfected into S2 cells by the calcium phosphate method as described previously (9). 529SU cells were induced with 500 μM CuSO4, approximately 18 to 24 h after transfection. After approximately 48 h, cells were washed once in phosphate-buffered saline (PBS) and lysed directly in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (5% SDS, 0.15 M Tris-HCl [pH 6.7], 30% glycerol). SDS-PAGE sample buffer lysates were fractionated by SDS-8% PAGE, and proteins were transferred to polyvinylidene difluoride membranes and detected by enhanced chemiluminescence according to the manufacturer’s recommendations (Roche Molecular Biochemicals). The primary antibodies for the Western blots in this study were anti-FLAG M2 (Sigma), anti-HA 12CA5 (Roche Molecular Biochemicals), and rabbit anti-Dorsal (13).
Fly stocks and manipulations.
The P-element insertion lines l(2)05486, inserted in the 5′ untranslated region of the ubc9 locus (2, 15), and l(2)04493, inserted approximately 250 bp upstream of the smt3 transcriptional start site (http://flybase.bio.indiana.edu/.bin/fbidq.html), were procured from the Bloomington Stock Center. The null dorsal allele dl1 has been described previously (23). These lines were balanced over either the CyO;Act-GFP or the CyO;Gal4-Kr;UAS-GFP chromosome, and homozygous mutant larvae were sorted by fluorescence microscopy.
The lipopolysaccharide (LPS) challenge was carried out by collecting 100 first instar larvae in a 1.5-ml microcentrifuge tube containing 50 μl of 10 μg of LPS (E. coli 026:B6; Sigma)/ml or 50 μl of PBS (negative control). The larvae were incubated at room temperature for 6 h and vortexed briefly every hour. At the end of the incubation, the larvae were frozen in liquid nitrogen, and total RNA was extracted as described below.
RNAi, RNA isolation, and RT-PCR.
RNA interference (RNAi) was carried out essentially as described previously (8). Briefly, double-stranded RNA (dsRNA) was generated by in vitro transcription (Megascript kit; Ambion) from pGEM vectors (Promega) containing a 400- to 600-bp region of the indicated ORF. Single-stranded RNA transcribed in the sense direction (with the T7 promoter) was annealed to its antisense complement (transcribed with the SP6 promoter) by heating mixtures of equal amounts of each species to 94°C and slowly cooling them to room temperature. Ten micrograms of each dsRNA was then added to S2 cells grown to approximately 20 to 30% confluence in 100-mm-diameter cell culture dishes. After 36 h, cells were challenged with 10 μg of LPS/ml for 6 h. The cells were then washed once with PBS, and total RNA was isolated using Trizol reagent according to the manufacturer’s protocol (Life Technologies). After degrading residual genomic DNA with RNase-free DNase (Promega), the RNA was reprecipitated and subjected to reverse transcription-PCR (RT-PCR) (Access RT-PCR kit; Promega) with primers specific for CecA1, Drs, dorsal, ubc9, smt3, or β-actin mRNA. The optimal number of cycles was determined according to the manufacturer’s protocol, and the appropriate amount of template was determined empirically for each target by titration of the starting material and comparison with β-actin control primers.
RESULTS
The Smt3-activating and -conjugating enzymes are sufficient for covalent attachment of Smt3 to Dorsal.
It was previously demonstrated that Dorsal is conjugated to Smt3 in Drosophila S2 cells when expressed with Ubc9 and Smt3 (3). In that study, coexpression of FLAG-Dorsal with HA-Smt3 resulted in the appearance of a species with an apparent molecular mass that was ∼20 kDa larger than that of FLAG-Dorsal. This 20-kDa shift is in accord with the apparent molecular mass of Smt3 (as determined by SDS-PAGE) and with the shift in apparent mass reported for RanGAP1 and other Smt3-conjugated proteins (33). Moreover, additional expression of Ubc9 with HA-Smt3 and FLAG-Dorsal greatly increased the intensity of the slower migrating species. This novel species was recognized by both the anti-FLAG and anti-HA antibodies, proving that it is an Smt3-Dorsal conjugate.
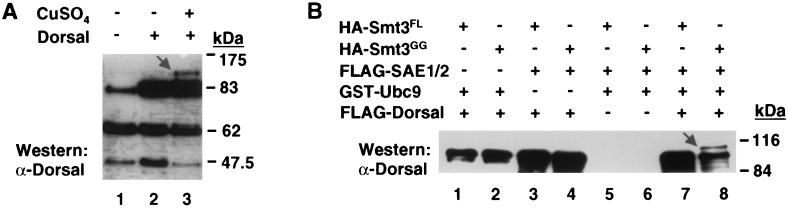
Because transient transfection efficiency varies, the level of conjugation can vary quite widely between experiments. To reduce this variability, we established a cell line stably transfected with expression vectors for FLAG-tagged Smt3 and HA-tagged Ubc9, both under the control of the Cu2+ inducible metallothionein promoter. These cells (referred to hereafter as 529SU cells) were then transiently transfected with an expression vector encoding Dorsal with or without Cu2+. Immunoblotting of the cell extracts with an anti-Dorsal antibody revealed the Smt3/Ubc9-dependent appearance of a species with an apparent molecular mass approximately 20 kDa larger than that of unmodified Dorsal (Fig. 1A), which, as explained above, represents Dorsal conjugated to a single molecule of Smt3.
FIG. 1.
Conjugation of Smt3 to Dorsal. (A) Detection of the Dorsal-Smt3 conjugate. 529SU cells (S2 cells stably transformed with expression vectors encoding HA-Ubc9 and FLAG-Smt3 under the control of a Cu2+ inducible promoter) were left untransfected (lane 1) or were transiently transfected with 10 μg of Dorsal expression vector with (lane 2) or without (lane 3) subsequent Cu2+ induction. Cells were lysed 48 h later, lysates were fractionated by SDS-10% PAGE and analyzed by immunoblotting with anti-Dorsal antibody. An arrow indicates Dorsal-Smt3 (lane 3); the bands with apparent molecular masses of approximately 63 and 46 kDa are unknown proteins that cross-react with the anti-Dorsal antibody nonspecifically. (B) Conjugation of Smt3 to Dorsal in vitro. Epitope-tagged forms of Dorsal and components of the Smt3 conjugation machinery were expressed in E. coli (Smt3FL, Smt3GG, Ubc9) or Sf9 cells (Dorsal, SAE1/2) and purified to homogeneity. These proteins were then used in an in vitro conjugation assay (described in Materials and Methods) in the indicated combinations. Reactions were fractionated by SDS-PAGE, blotted onto polyvinylidene difluoride membranes, and probed with anti-Dorsal antibodies. An arrow indicates Dorsal-Smt3 (lane 8).
To determine if the known components of the conjugation machinery were necessary and sufficient for conjugation of Smt3 to Dorsal, we established an in vitro Drosophila Smt3 conjugation system. Purified recombinant Smt3-activating enzyme (SAE1/SAE2), Ubc9, Smt3, and Dorsal were mixed in various combinations and incubated in the presence of an ATP regeneration system. Proteins were then subjected to SDS-PAGE and immunoblotting to detect modified forms of Dorsal. A Dorsal-Smt3 conjugate was detected only when all of the aforementioned recombinant proteins were included (Fig. 1B, lane 8). Previous studies indicated that Smt3 can be conjugated to target proteins only after endoproteolytic cleavage to remove the two C-terminal amino acids, thereby exposing a C-terminal glycine (10, 27, 41). In agreement with these studies, we find that only Smt3GG (equivalent to the processed form of Smt3), but not full-length Smt3 (Smt3FL), is competent for conjugation to Dorsal (Fig. 1B, compare lanes 7 and 8).
Smt3 conjugation stimulates Dorsal activity even when Dorsal is constitutively nuclear.
In a previous study (3), it was shown that the Smt3 conjugation machinery counteracts Cactus-mediated inhibition of Dorsal nuclear localization. In these S2 cell experiments, it was found that transiently expressed Dorsal protein was constitutively nuclear in the absence of transiently expressed Cactus, presumably because high levels of transiently expressed Dorsal overwhelm the endogenous Cactus. Transient expression of Cactus resulted in cytoplasmic localization of Dorsal, an effect that was largely reversed by additionally expressing Ubc9.
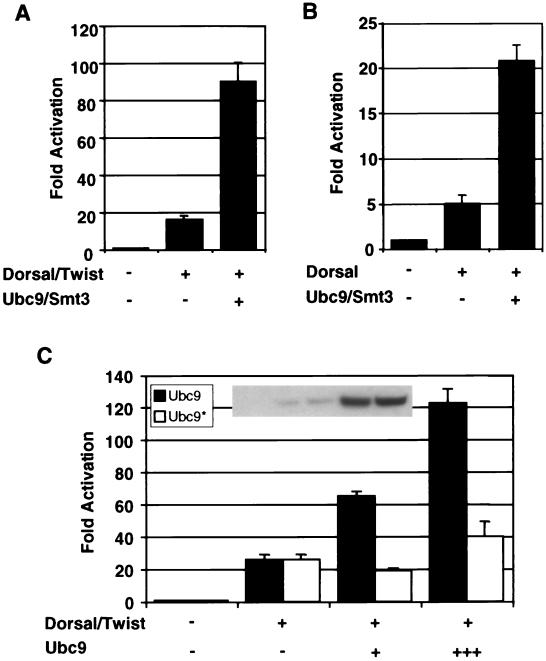
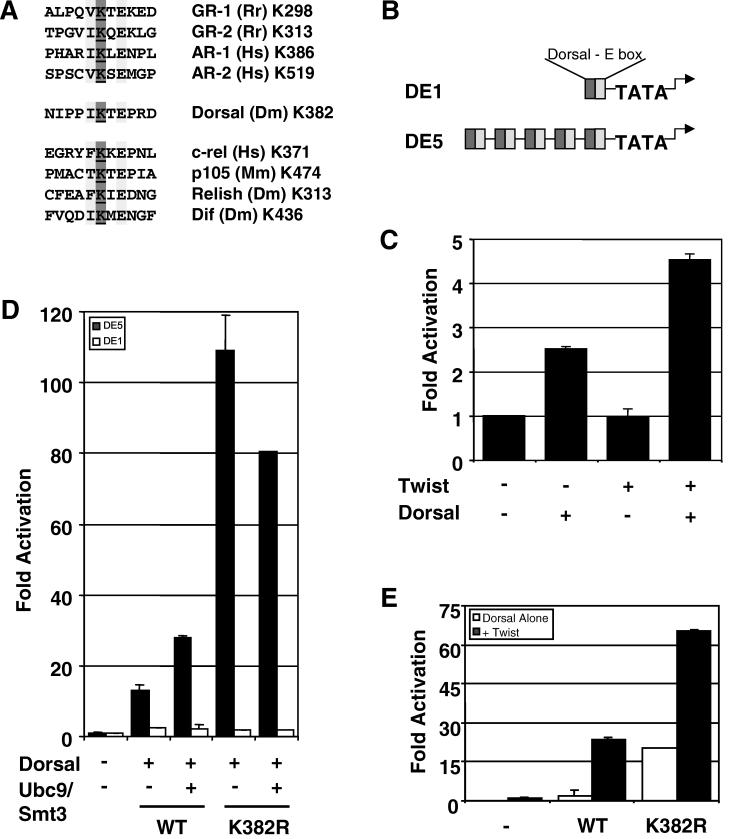
To determine whether or not conjugation of Smt3 to Dorsal can stimulate Dorsal activity even in the absence of Cactus (and therefore when Dorsal is constitutively nuclear), we used a reporter termed DE5 in which a regulatory module containing adjacent Twist and Dorsal binding sites is repeated five times upstream of a core promoter directing luciferase gene expression. Previous studies from our lab demonstrated that Dorsal and Twist interact to direct strong synergistic activation of this reporter, which therefore provides a sensitive means for measuring Dorsal activity (49). As expected, coexpression of Dorsal and Twist results in the activation of this reporter. Additionally, coexpressing Ubc9 and Smt3 in these cells results in an enhancement of Dorsal-dependent transcriptional activation (Fig. 2A). Dorsal is similarly stimulated by Ubc9 and Smt3 in the absence of Twist (Fig. 2B). This indicates that the observed effects of the Smt3 conjugation machinery on reporter activity are due to an effect on Dorsal and not to an effect on Twist. Thus, the Smt3 conjugation machinery is able to stimulate Dorsal activity even when Dorsal is constitutively nuclear, suggesting that Smt3 may be able to influence Dorsal-mediated transcriptional activation directly.
FIG. 2.
Stimulation of Dorsal-mediated activation by the Smt3 conjugation machinery in the absence of Cactus. (A) Transfection assays. S2 cells were transfected with a Dorsal-responsive luciferase gene reporter construct (DE5) alone or in combination with expression vectors encoding Dorsal and Twist, with or without Ubc9 and Smt3. A Renilla luciferase internal control reporter was also included in each transfection. (B) Smt3 and Ubc9 influence Dorsal in the absence of Twist. S2 cells were transfected with the DE5 reporter alone or in combination with expression vectors encoding Dorsal (without Twist) with or without Ubc9 and Smt3. (C) Catalytically defective Ubc9. The DE5 reporter was introduced into S2 cells with expression vectors for Dorsal and Twist and with (25 ng [+] or 500 ng [+++]) or without a mutant or wild-type expression vector for HA-tagged Ubc9. The inset shows equivalent expression levels of mutant and wild-type Ubc9, at both levels indicated in the bar graph, detected by anti-HA immunoblotting of whole-cell extracts. Each bar represents the average (+ standard deviation [error bar]) of duplicate experiments. Relative luminescence was first calculated by dividing the firefly luciferase activity (from the Dorsal-responsive reporter) by the Renilla luciferase activity (from the internal control). Fold activation values were then obtained by dividing each relative luminescence value by the relative luminescence value obtained for the reporters alone.
The ability of Ubc9 to catalyze Smt3 conjugation is critically dependent on a catalytic cysteine at position 93 (11). To ascertain whether Ubc9 is able to influence Dorsal in a manner independent of Smt3 conjugation, we generated a catalytically inactive version of Ubc9 (Ubc9*) by replacing the cysteine residue at position 93 with an arginine residue. Whereas wild-type Ubc9 enhances Dorsal-dependent reporter activity in a dose dependent manner, the mutant was severely compromised in its ability to influence Dorsal-dependent activation (Fig. 2C). The relatively low level of stimulation detected at the higher level of Ubc9* suggests that Ubc9 may have a minor effect on Dorsal that is independent of its enzymatic function. Nevertheless, our results strongly suggest that Ubc9 predominantly influences Dorsal activity via Smt3 conjugation, a finding that is consistent with our observation that the overexpression of Smt3 and the activating enzyme along with Ubc9 further stimulates Dorsal-mediated activation (3).
Conjugation of Smt3 to Dorsal is reversible.
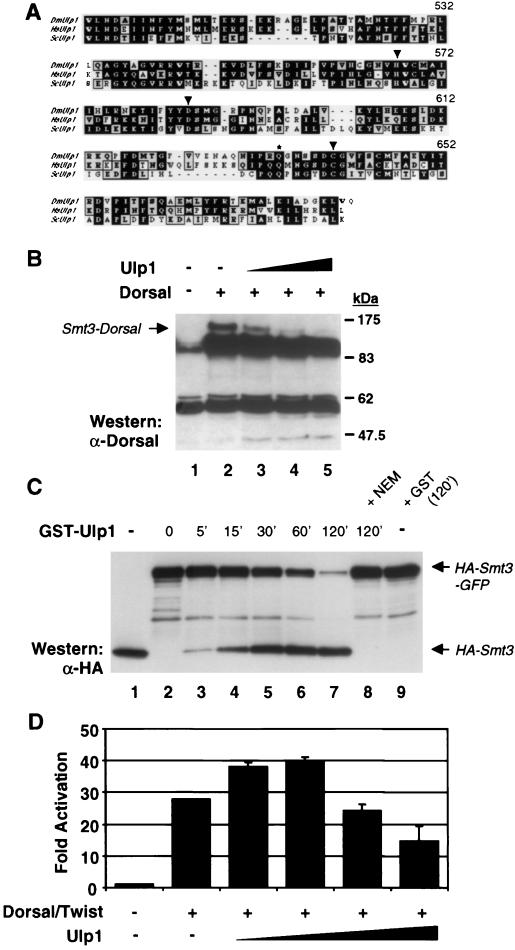
Studies with the yeast Saccharomyces cerevisiae suggest that Smt3 conjugation is a dynamic process and that Smt3 deconjugation can be catalyzed by the cysteine protease Ulp1 (30, 31, 39). This observation is consistent with our finding that only a small fraction of cellular Dorsal exists in the modified form at any given time, which suggests that modification may be transient. We identified a putative Drosophila ulp1 cDNA in the Drosophila Genome Project expressed sequence tag collection. Alignment of the C-terminal 181-amino-acid region of the encoded protein with the corresponding region in yeast Ulp1 reveals 34% identity and 48% similarity (Fig. 3A). The cysteine protease catalytic triad and a conserved glutamine residue that forms part of the oxyanion hole are fully conserved.
FIG. 3.
Conjugation of Smt3 to Dorsal is reversible. (A) Alignment of yeast and human Ulp1 with Drosophila Ulp1. Only the C-terminal 181 amino acids are shown. The conserved catalytic triad residues (arrowheads) and oxyanion hole Gln residue (asterisk) are indicated. (B) Deconjugation by Ulp1. 529SU cells were untransfected (lane 1) or transiently transfected with 10 μg of Dorsal and 0 (lane 2), 0.5 (lane 3), 1 (lane 4), or 2 (lane 5) μg of a Ulp1 expression vector. Whole-cell lysates were analyzed as described for Fig. 1A. (C) Processing of Smt3-GFP fusion by Ulp1. The contents of each lane were incubated in the presence of 150 mM NaCl, 1 mM dithiothreitol, 10 mM Tris (pH 8.0), and 0.2% Triton X-100. The reactions were stopped at the times indicated above the lanes (in minutes). Lane 1, HA-Smt3; lane 2, HA-Smt3-GFP; lanes 3 to 8, HA-Smt3-GFP and crude GST-Ulp1 (or GST). In lane 8, the GST-Ulp1 was preincubated with 20 mM NEM at room temperature for 15 min prior to its addition to the reaction mixture. Lane 9 contained the same ingredients as lane 7 except that GST-Ulp1 was replaced with GST. The reactions were stopped by boiling in SDS loading buffer, subjected to SDS-12% PAGE, and analyzed by anti-HA immunoblotting. A similar blot was also probed with an anti-GFP antibody, demonstrating that the appearance of HA-Smt3 was accompanied by the parallel appearance of GFP (data not shown). (D) Biphasic response of Dorsal to Ulp1. The DE5 reporter was cotransfected without Dorsal and Twist expression vectors or with Dorsal and Twist expression vectors plus 0, 1, 2, 5, or 10 μg of a Ulp1 expression vector. Data were analyzed as described for Fig. 2A.
The coding region of Drosophila ulp1 was inserted into an expression vector and transiently transfected into 529SU cells along with Dorsal. The cells were treated with Cu2+ to induce Ubc9 and Smt3 expression. Expression of increasing amounts of Ulp1 led to a dose-dependent decrease in Smt3-modified Dorsal (Fig. 3B). Thus, Drosophila Ulp1 is a true Smt3-deconjugating enzyme.
In addition to functioning as an Smt3-deconjugating enzyme, yeast Ulp1 functions as a site-specific endoprotease to process Smt3 into its mature form (30, 31, 39). As mentioned above, this processing normally involves the removal of two residues from the C terminus of Smt3, exposing a C-terminal glycine. Previous studies of yeast Ulp1 have shown that appending additional sequences to the C-terminal end of full-length Smt3 does not interfere with processing (48). Accordingly, we have found that Drosophila Ulp1 will cleave an Smt3-GFP fusion protein to generate polypeptides of the expected size for mature Smt3 and GFP (Fig. 3C).
Interestingly, in S2 cells, expression of low levels of Ulp1 augmented Dorsal-dependent reporter activity, whereas higher levels resulted in a reduction in reporter activity (Fig. 3D). These findings are consistent with the possibility that low levels of Ulp1 are required to process Smt3 into its mature form while high levels of Ulp1 result in deconjugation of Smt3 from Dorsal.
Mutagenesis of the Smt3 conjugation site in Dorsal.
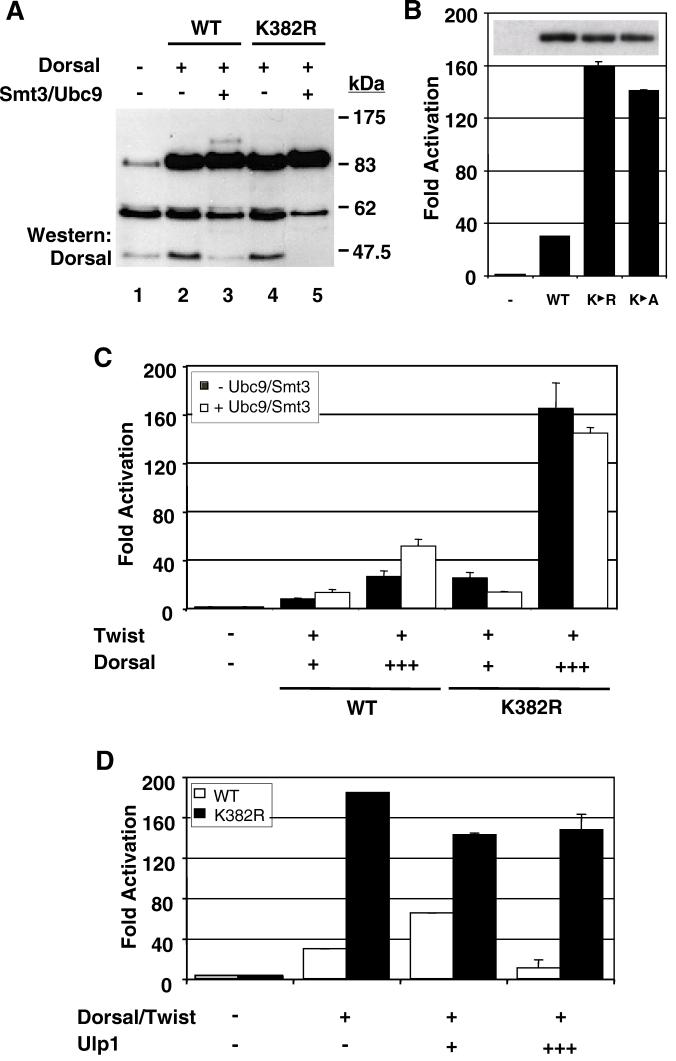
To further solidify the connection between Smt3 conjugation and Dorsal function, we identified and mutagenized the Smt3 conjugation site in Dorsal. As mentioned in the introduction, Smt3 is generally conjugated to a lysine that falls within a sequence of the type (I/L/V)KXE (45, 54). Inspection of the Dorsal sequence revealed a single lysine residue at position 382 that resides in such a context. To determine if K382 represents an Smt3 conjugation site, we generated a mutant form of Dorsal in which the lysine at position 382 is replaced with an arginine (DorsalK382R). When DorsalK382R was introduced into 529SU cells, Cu2+ induction no longer resulted in the appearance of the modified form of Dorsal, confirming that the lysine at position 382 is the only residue within Dorsal subject to Smt3 conjugation (Fig. 4A, compare lanes 3 and 5).
FIG. 4.
Conjugation of Smt3 to Dorsal occurs at lysine 382. (A) DorsalK382R is resistant to Smt3 conjugation. 529SU cells were left untransfected (lane 1) or were transiently transfected with 10 μg of an expression vector for wild-type (WT) Dorsal (lanes 2 and 3) or DorsalK382R (lanes 4 and 5). Cells were either untreated (lanes 1, 2, and 4) or treated with Cu2+ to induce Ubc9 and Smt3 expression (lanes 3 and 5). (B) Disruption of the Smt3 conjugation site enhances transcriptional activation. S2 cells were transfected with the DE5 reporter and a Twist expression vector along with a Dorsal, DorsalK382R, or DorsalK382A expression vector. Data were analyzed as described in the legend to Fig. 2A. The inset displays the expression levels of the three Dorsal variants corresponding to the bar graph revealed by anti-Dorsal immunoblotting of whole-cell extracts. (C) DorsalK382R is unresponsive to Ubc9/Smt3. The DE5 reporter was introduced into S2 cells with expression vectors for mutant or wild-type Dorsal and Twist with or without Ubc9 and Smt3. Either 8 ng (+) or 64 ng (+++) of Dorsal expression vector was employed in this study. Data were analyzed as described in the legend to Fig. 2A. (D) DorsalK382R is not responsive to Ulp1. S2 cells were transfected with the DE5 reporter alone or in combination with expression vectors encoding Dorsal and Twist with or without either 1 or 10 μg of an expression vector for Ulp1.
To explore the influence of Smt3 conjugation on Dorsal activity further, we compared DorsalK382R with wild-type Dorsal in our reporter assay. Surprisingly, we found that this nonconjugable mutant was significantly more active than wild-type Dorsal. The mutant typically yielded about 5- to 10-fold-higher levels of reporter gene activity than did the wild-type protein (Fig. 4B). A similar result was obtained for another mutant, DorsalK382A, in which the lysine was changed to alanine rather than arginine. This difference was not due to variations in the level of protein, since each protein was expressed with similar efficiency (Fig. 4B, inset).
Assuming that stimulation of Dorsal by the Smt3 conjugation machinery is dependent upon the covalent attachment of Smt3 to Dorsal, mutagenesis of the target lysine should result in a transcription factor that is resistant to this stimulation. In accord with this expectation, the level of reporter gene activity driven by DorsalK382R does not increase upon coexpression of Ubc9 and Smt3 (Fig. 4C). To guard against the possibility that the lack of stimulation was due to the presence of a saturating dose of DorsalK382R, we examined the effect of the conjugation machinery at multiple concentrations of the wild-type and mutant forms of the factors. We consistently observed stimulation of wild-type Dorsal, but no stimulation of DorsalK382R, in accord with the results obtained with the catalytically inactive form of Ubc9 (Ubc9*). These results suggest that Ubc9 stimulates Dorsal primarily by catalyzing the covalent addition of Smt3 at lysine 382. We note that Ubc9 and Smt3 actually result in a small but reproducible decrease in the activity of DorsalK382R. The reason for this small decrease is unclear at this time.
The results presented earlier on Ulp1 (Fig. 3) showed a biphasic effect of Ulp1 on Dorsal activity. To test the possibility that this reflects the ability of Ulp1 to stimulate Smt3 conjugation at low concentrations (perhaps by catalyzing Smt3 maturation) and to mediate deconjugation at high concentrations, we coexpressed low or high levels of Ulp1 with Dorsal or DorsalK382R. Unlike wild-type Dorsal, which once again exhibited a biphasic response, the defective conjugation mutant was largely unresponsive to Ulp1 (Fig. 4D). These findings demonstrate that the effects of Ulp1 on Dorsal-mediated activation are specific and dependent upon an intact conjugation site in Dorsal.
The Dorsal Smt3 conjugation site lies within an SC motif.
The results described in the previous section are consistent with a model in which lysine 382 helps to mediate an interaction between Dorsal and a negative regulator. According to this model, both the covalent modification and mutagenesis of K382 would eliminate this interaction, resulting in enhanced transcriptional activity. Two known negative regulatory factors known to bind Dorsal are Cactus and Groucho (13, 19, 53). However, both of these factors bind DorsalWT (wild type) and DorsalK382R with equal affinity (data not shown), suggesting that neither one is the negative regulator responsible for the function of K382. Since Smt3 conjugation sites are very similar to SC motifs (Fig. 5A), and since SC motifs may be binding sites for a negatively acting factor (22), we decided to determine if the Smt3 conjugation site in Dorsal has the properties of an SC motif.
FIG. 5.
Lysine 382 is part of a potential SC motif. (A) Alignment of putative steroid receptor and rel family SC motifs. Many rel family transcription factors contain motifs that resemble possible Smt3 conjugation sites and/or SC motifs found in steroid receptors. The conserved residues in the (I/L/V)KXE consensus sequence are highlighted, and the target lysine is underlined. (B) Reporters used in this study. Both reporters used in this work contain a module consisting of tandem Dorsal and Twist binding sites driving expression of the firefly luciferase gene. The DE1 and DE5 reporters contain one or five tandem copies of this module, respectively, and are described in further detail in Materials and Methods. (C) The DE1 reporter is synergistically activated by Dorsal and Twist. S2 cells were transfected with the DE1 reporter alone or in combination with expression vectors encoding Dorsal and/or Twist. (D) DorsalK382R does not superactivate a reporter containing a single Dorsal binding site. S2 cells were transfected with the DE5 or DE1 reporter along with expression vectors for mutant or wild-type (WT) Dorsal, Twist, and/or Ubc9/Smt3 (2.5 μg), as indicated. (E) Superactivation by DorsalK382R does not require Twist. The DE5 reporters were transfected into S2 cells along with expression vectors for mutant or wild-type Dorsal with or without Twist, as indicated.
To determine whether the Smt3 conjugation site in Dorsal resides within an SC motif, we compared the ability of DorsalWT and DorsalK382R to activate a reporter containing five Dorsal binding sites (DE5) with the ability of these two Dorsal variants to activate a reporter containing just a single Dorsal binding site (DE1) (Fig. 5B). Whereas DorsalK382R was approximately eightfold-more potent than DorsalWT at the DE5 reporter, both forms of Dorsal yielded similar low levels of activation at the DE1 reporter (Fig. 5D). This is not due to a general need for multiple Dorsal binding sites for a Dorsal response, since Dorsal is able to stimulate the DE1 reporter about twofold in the absence of Twist and about fourfold in the presence of Twist (Fig. 5C). Importantly, synergy between Dorsal and Twist is not altered by the mutation at lysine 382 since both DorsalWT and DorsalK382R are strongly potentiated by Twist at the DE5 reporter (Fig. 5E). Taken together, these findings suggest that the K382R mutation specifically stimulates synergy between multiple molecules of Dorsal. Neither simple activation nor synergy between Dorsal and Twist is affected. The Smt3 conjugation site in Dorsal therefore lies within an SC motif.
Smt3 conjugation is required during the innate immune response.
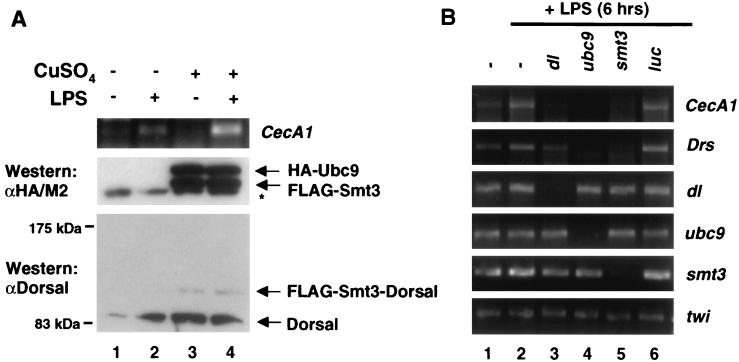
The foregoing analysis relied exclusively on transiently transfected reporter genes as a means of gauging the effects of Smt3 conjugation on Dorsal activity. We therefore sought to determine if the Smt3 conjugation machinery could influence the transcription of endogenous Dorsal target genes. Drosophila S2 cells can be induced to produce antimicrobial peptides by challenging them with LPS, a component of the bacterial cell wall (20, 28, 50). Of the numerous antimicrobial peptides induced in response to LPS, we have found that cecropinA1 (CecA1) is the most dependent upon Dorsal activity (data not shown). To determine whether the Smt3 conjugation machinery plays a role during the immune response, we treated 529SU cells with Cu2+ (to induce FLAG-Smt3 and HA-Ubc9 expression) and/or LPS. We then measured expression of the gene encoding CecA1 by quantitative RT-PCR (Fig. 6A). In 529SU cells, the LPS challenge results in an induction of CecA1 expression (Fig. 6A, lane 2), whereas treatment with Cu2+ alone results in potent expression of FLAG-Smt3 and HA-Ubc9 and the appearance of a conjugate between Smt3 and endogenous Dorsal, but it does not give rise to an appreciable increase in CecA1 mRNA (Fig. 6A, lane 3). Treatment with both LPS and Cu2+, however, results in a significantly greater rise in CecA1 mRNA than either LPS or Cu2+ induction alone (Fig. 6A, lane 4). These data suggest that the Smt3 conjugation pathway amplifies the induction of an endogenous Dorsal-dependent antimicrobial gene during the Drosophila immune response.
FIG. 6.
Ubc9 and Smt3 are required for the induction of antimicrobial peptides. (A) Conjugation of Smt3 to Dorsal correlates with increased LPS induction of CecA1. 529SU cells were induced with Cu2+ for 24 h (lanes 1 and 2) and/or challenged with LPS for 6 h (lanes 2 and 4). Total RNA was then isolated and subjected to quantitative RT-PCR with primers specific to the CecA1 mRNA (top panel). Whole-cell lysates from these cells were fractionated by SDS-PAGE (15 or 5% acrylamide) and analyzed by immunoblotting to confirm Ubc9 and Smt3 expression (middle panel, lanes 3 and 4) and Smt3-Dorsal production (bottom panel, lanes 3 and 4). (B) Smt3 and Ubc9 are required for the induction of CecA1 and Drs in S2 cells. mRNA corresponding to the genes indicated at the top were disrupted using dsRNAi (36 h). Total RNA was isolated following 6 h of LPS challenge. RT-PCR was then performed using primers specific to the genes indicated to the right of the gels.
In S2 cells, transcripts can be specifically targeted for degradation double-stranded RNA interference (dsRNAi) (5, 8). We decided to exploit this technique to determine whether components of the Smt3 conjugation pathway are required for the production of antimicrobial peptides upon LPS stimulation. S2 cells were challenged with LPS after treatment with dsRNA corresponding to the dorsal, ubc9, smt3, or luciferase gene (as a negative control) coding regions. RT-PCR was then employed to measure CecA1 mRNA (Fig. 6B). Depletion of dorsal, ubc9, or smt3 (Fig. 6B, lanes 3, 4, and 5, respectively) mRNA by dsRNAi abolished LPS-induced CecA1 expression, whereas dsRNAi against luciferase gene expression did not (Fig. 6B, lane 6), suggesting that the Smt3 conjugation pathway is required for the induction of this antimicrobial gene. The gene encoding another antimicrobial peptide, drosomycin (Drs), also fails to be induced upon interference with Smt3 conjugation (Fig. 6B). Interestingly, Drs induction was less affected by depletion of Dorsal than by depletion of Smt3 or Ubc9. A possible explanation for this result is considered in Discussion.
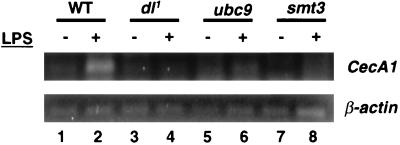
Having established that components of the Smt3 conjugation machinery are required for induction of CecA1 expression in cell culture, we decided to determine if the same is true in the intact organism. For these studies, we obtained previously described fly stocks bearing hypomorphic P-element disruptions within the ubc9 or smt3 transcription units (see Materials and Methods). The mutant chromosomes were each placed over a balancer chromosome marked with a reporter gene encoding the GFP. Larvae and adults were then examined from these stocks for possible homozygous mutant escapers (identified by lack of green fluorescence). No homozygous mutant flies survived to adulthood. However, we were able to isolate a small number of smt3−/− first instar larvae and a somewhat smaller number of ubc9−/− first instar larvae. After treating these larvae with LPS for 6 h, total larval RNA was extracted and subjected to quantitative RT-PCR with primers specific for the CecA1 or β-actin (control) mRNAs. While wild-type larvae showed significant induction of CecA1 expression in response to the LPS challenge (Fig. 7, lanes 1 and 2), larvae compromised in dl (Fig. 7, lanes 3 and 4), ubc9 (Fig. 7, lanes 5 and 6), or smt3 (Fig. 7, lanes 7 and 8) expression displayed no CecA1 induction.
FIG. 7.
Smt3 and Ubc9 are required for the induction of CecA1 in first instar larvae. Larvae homozygous for a null allele of dl (dl1) or for hypomorphic P-element disruptions of either ubc9 or smt3 were isolated and challenged with LPS for 6 h. Total RNA was isolated and subjected to quantitative RT-PCR using primers corresponding to either CecA1 or β-actin. WT, wild type.
DISCUSSION
Previous studies demonstrated that Dorsal is a target for Smt3 conjugation (3) and that the conjugation machinery helps Dorsal to overcome Cactus-mediated inhibition of nuclear localization. Here, we have expanded on the relationship between Dorsal and the Smt3 conjugation pathway in a number of ways. First, we have demonstrated that Smt3 is reversibly conjugated to lysine 382 of Dorsal. Second, we have found that the Smt3 conjugation machinery may modulate Dorsal activity in multiple ways, including the regulation of synergistic transcriptional activation. Finally, we have shown that the Smt3 conjugation machinery is required both in cultured cells and in larvae for rel protein-dependent induction by LPS of genes encoding antimicrobial peptides. Our studies on the effect of Smt3 conjugation on transcriptional synergy are among the first to address the mechanism by which Smt3 might regulate transcription factor activity. In addition, our studies looking at the role of Smt3 conjugation in the immune response are among the first to show a biological function for this process in an intact multicellular organism.
Reversible conjugation of Smt3 to Dorsal.
In vitro and in vivo conjugation assays demonstrate that, in the presence of ATP, Ubc9 and the Smt3-activating enzyme are sufficient to catalyze conjugation of mature Smt3 to lysine 382 of Dorsal. This finding supports the idea that Ubc9 is primarily responsible for the selection of substrates for conjugation, presumably via specific protein-protein interactions between Ubc9 and the substrates. Enzymes analogous to the ubiquitin ligases (E3 enzymes) are not absolutely required.
In S2 cells, only a small fraction of the cellular pool of Dorsal is conjugated to Smt3 at any given time, suggesting that modification of Dorsal is transient and that Smt3 may be actively removed from Dorsal after exerting its influence. Consistent with this idea, we have found that the Drosophila genome encodes multiple proteins with homology to the yeast ubiquitin-like protease Ulp1. Furthermore, we have found that one of these proteins (Drosophila Ulp1) can remove Smt3 from Dorsal. Moreover, it is apparently able to participate in the processing of Smt3 by removing the last two amino acids to yield the mature, conjugable form, Smt3GG. The bifunctional nature of Ulp1 may explain the biphasic response curve that we observe when we examine the effect of increasing concentrations of Ulp1 on Dorsal-dependent activation, but not DorsalK382R-dependent activation. At low concentrations, Ulp1 may primarily mediate Smt3 processing, thereby stimulating conjugation, while at high concentrations, deconjugation may be the dominant activity. While this theory is appealing given the aforementioned data, it is also possible that Ulp1 may modulate Smt3 conjugation in other ways. For example, a previous report suggests that yeast Ulp1 may localize to the nuclear pores where it could play a role in the nuclear targeting of the Smt3 conjugation machinery (51).
An alternative explanation for the low level of Smt3-conjugated Dorsal detected in our experiments is that another posttranslational modification may be a prerequisite for efficient Smt3 conjugation. For example, phosphorylation of Dorsal at multiple serine residues is known to modulate Dorsal activity, suggesting the possibility of a relationship between phosphorylation and Smt3 conjugation. However, we have not detected any difference between the ability of Smt3 and Ubc9 to stimulate phosphorylation-defective or wild-type versions of Dorsal (M. Smith, unpublished data), suggesting that there is no simple relationship between these two modifications.
Smt3 conjugation affects Dorsal activity in multiple ways.
In this report, we have used transient transfection assays to measure Dorsal activity in the absence of transiently expressed Cactus, that is, under conditions where transiently expressed Dorsal protein is constitutively nuclear. Under these conditions, we still observe stimulation of Dorsal activity when we coexpress Ubc9 and Smt3. This implies that Smt3 conjugation stimulates Dorsal activity by a mechanism not involving the modulation of nuclear localization. Experiments presented here suggest that this additional mechanism is the stimulation of transcriptional synergy via the modification of an SC motif.
SC motifs, which may mediate the recruitment of a putative negatively acting SCF, were initially characterized in GR (22). Further analysis revealed that this motif is present in a number of other transcription factors, including other steroid receptors, and its presence appears to correlate with the ability of these factors to synergize at compound regulatory modules containing multiple binding sites for the SC motif-containing factor.
In accord with the sequence similarity between SC motifs and Smt3 conjugation sites, we found that mutagenesis of the Smt3 acceptor lysine in Dorsal results in the increased activation of a reporter containing multiple Dorsal binding sites (DE5) but not of a reporter containing a single Dorsal binding site (DE1). Furthermore, under conditions where Dorsal is constitutively nuclear, the Smt3 conjugation machinery stimulates Dorsal-mediated activation of the DE5 reporter but not of the DE1 reporter. Thus, the Smt3 conjugation site in Dorsal fulfills the definition of an SC motif, and Smt3 may play a role in regulating transcription factor synergy.
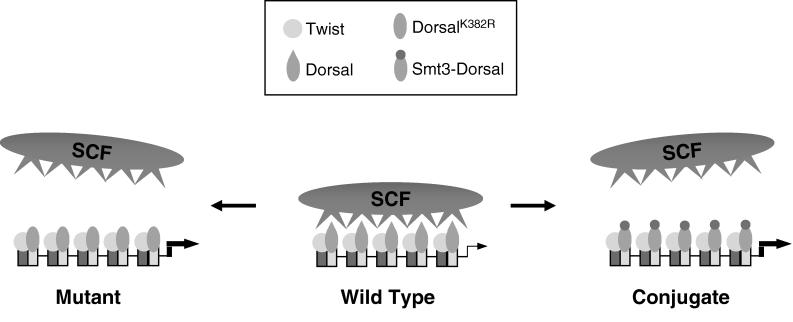
Considered in isolation, the finding that mutagenesis of the Smt3 conjugation site in Dorsal results in increased activation potential could be interpreted to suggest that Smt3 conjugation interferes with activation. However, this interpretation would be inconsistent with our finding that the overexpression of conjugation machinery components increases both Smt3 conjugation and Dorsal-mediated activation. It would also be inconsistent with our finding that reducing the intracellular concentration of conjugation machinery components (through the use of either dsRNAi or loss-of-function alleles) results in decreased transcription of endogenous Dorsal target genes. One model that is consistent with our observations and with previous studies of SC motifs in other factors (22) is that both mutagenesis of the Smt3 conjugation site and the conjugation of Smt3 to this site causes the displacement of the SCF, resulting in more potent transcriptional synergy at compound binding sites (Fig. 8). If this model is correct, then it suggests that Smt3 conjugation may influence Dorsal activity in at least two distinct ways, first by stimulating Dorsal nuclear localization and then by blocking the interaction between Dorsal and SCF.
FIG. 8.
Potential mechanism for synergy control via Smt3 conjugation. Twist and Dorsal bind to the multiple binding sites to bring about reporter activation. In its wild-type, unconjugated form, Dorsal recruits a putative SCF that attenuates Dorsal activity, giving rise to moderate levels of transcriptional activation. However, disruption of the SC motif by mutagenesis or Smt3 conjugation disallows binding of the SCF. As a result, Dorsal brings about the much higher levels of transcriptional activation observed experimentally.
Studies of the interactions between Ubc9 and the zebra fish protein Vsx-1, the androgen receptor, and Ets-1 have suggested that Ubc9 can influence nuclear localization and transcription factor activity independent of its ability to catalyze Smt3 conjugation (6, 29, 42). However, our finding that mutagenesis of the target lysine in Dorsal to arginine or alanine abolishes activation by Ubc9 suggests that, in the case of Dorsal, the effects of Ubc9 on activity depend, at least in part, upon Smt3 conjugation. Further support for this conclusion comes from our finding that a mutation in the catalytic cysteine of Ubc9 greatly reduces the ability of this protein to potentiate Dorsal-mediated activation.
The Smt3 conjugation machinery potentiates the immune response.
In addition to controlling dorsoventral pattern formation during embryogenesis, signaling through Toll family transmembrane receptor proteins mediates the innate immune response (1, 21, 55). As a transcription factor that controls gene expression in response to Toll signaling, Dorsal is a player in this response. In this study, we used overexpression experiments, dsRNAi, and loss-of-function alleles of smt3 and ubc9 to show that the conjugation machinery is required for the expression of the immune response genes CecA1 and Drs.
Our results indicate that in both S2 cells and first instar larvae, induction of CecA1 expression by LPS is strongly dependent upon Dorsal. This contrasts with previously published results (34, 38), which our own findings confirm (V. Bhaskar, unpublished data), suggesting that bacterial induction of CecA1 in third instar larvae and adults does not require Dorsal, perhaps due to redundant pathways for inducing CecA1 expression. By using first instar larvae in conjunction with LPS as the inducing agent, we have revealed a dependence of CecA1 induction upon Dorsal that had not been previously observed.
While LPS-induced CecA1 expression in S2 cells is strongly dependent upon both Dorsal and the Smt3 conjugation machinery, Drs expression is only weakly dependent on Dorsal, but it is strongly dependent on the Smt3 conjugation machinery. This suggests that Drs expression may be modulated by the conjugation of Smt3 to factors other than Dorsal. Prime candidates for such factors are Relish and Dif, two other rel family transcription factors in Drosophila, both of which contain sites that match the Smt3 conjugation site consensus sequence (Fig. 4A). The finding that the Smt3 conjugation pathway is an essential component of the immune response in insects suggests a possible functional role in the vertebrate immune response. Consistent with this idea, Ubc9 has been shown to potentiate c-rel activity in a mammalian cell line (52).
Yeast defective in the Smt3 conjugation machinery is either inviable or displays stress response defects (25, 27, 30, 31). Likewise, in vertebrate cell lines, a variety of cellular insults result in an increase in Smt3/SUMO-conjugated proteins. Stresses that have been shown to affect Smt3 conjugation include DNA damage, heat shock, and cytoskeletal perturbations (4, 17, 47, 48). Although these insults generally affect global Smt3 conjugation, a few specific stress response agents have been identified as substrates for Smt3 modification. These include p53 and topoisomerase, which are conjugated to Smt3/SUMO in response to DNA damage, and heat shock factor-2, which controls expression of certain heat shock proteins (16, 17, 35, 36, 46). Our identification of Dorsal as a substrate for Smt3 conjugation and our demonstration that components of the Smt3 conjugation machinery are required during the immune response indicate that Smt3 conjugation may be a general stress response mechanism.
Acknowledgments
This work was supported by grants to A.J.C. from the NIH (GM44522) and the March of Dimes (#1-FY01-299) and by an NIH training grant (GM07185) to V.B.
REFERENCES
- 1.Aderem, A., and R. J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782–787 [DOI] [PubMed] [Google Scholar]
- 2.Apionishev, S., D. Malhotra, S. Raghavachari, S. Tanda, and R. S. Rasooly. 2001. The Drosophila UBC9 homologue lesswright mediates the disjunction of homologues in meiosis I. Genes Cells 6:215–224 [DOI] [PubMed] [Google Scholar]
- 3.Bhaskar, V., S. A. Valentine, and A. J. Courey. 2000. A functional interaction between dorsal and components of the Smt3 conjugation machinery. J. Biol. Chem. 275:4033–4040 [DOI] [PubMed] [Google Scholar]
- 4.Buschmann, T., D. Lerner, C. Lee, and Z. Ronai. 2001. Ubc-9 binding, which is required for SUMO-1 conjugation to Mdm2, is mapped to N-terminal region of Mdm2 and is reduced following UV-irradiation. J. Biol. Chem. 276:40389–40395 [DOI] [PubMed] [Google Scholar]
- 5.Caplen, N. J., J. Fleenor, A. Fire, and R. A. Morgan. 2000. dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene 252:95–105 [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarti, S. R., R. Sood, S. Ganguly, S. Bohlander, Z. Shen, and G. Nucifora. 1999. Modulation of TEL transcription activity by interaction with the ubiquitin-conjugating enzyme UBC9. Proc. Natl. Acad. Sci. USA 96:7467–7472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, G., P. H. Nguyen, and A. J. Courey. 1998. A role for Groucho tetramerization in transcriptional repression. Mol. Cell. Biol. 18:7259–7268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemens, J. C., C. A. Worby, N. Simonson-Leff, M. Muda, T. Maehama, B. A. Hemmings, and J. E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 97:6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courey, A. J., and R. Tjian. 1988. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55:887–898 [DOI] [PubMed] [Google Scholar]
- 10.Desterro, J. M., M. S. Rodriguez, G. D. Kemp, and R. T. Hay. 1999. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 274:10618–10624 [DOI] [PubMed] [Google Scholar]
- 11.Desterro, J. M., J. Thomson, and R. T. Hay. 1997. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417:297–300 [DOI] [PubMed] [Google Scholar]
- 12.Drier, E. A., and R. Steward. 1997. The dorsoventral signal transduction pathway and the Rel-like transcription factors in Drosophila. Semin. Cancer Biol. 8:83–92 [DOI] [PubMed] [Google Scholar]
- 13.Dubnicoff, T., S. A. Valentine, G. Chen, T. Shi, J. A. Lengyel, Z. Paroush, and A. J. Courey. 1997. Conversion of dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev. 11:2952–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dushay, M. S., and E. D. Eldon. 1998. Drosophila immune responses as models for human immunity. Am. J. Hum. Genet. 62:10–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epps, J. L., and S. Tanda. 1998. The Drosophila semushi mutation blocks nuclear import of bicoid during embryogenesis. Curr. Biol. 8:1277–1280 [DOI] [PubMed] [Google Scholar]
- 16.Goodson, M. L., Y. Hong, R. Rogers, M. J. Matunis, O. K. Park-Sarge, and K. D. Sarge. 2001. Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J. Biol. Chem. 276:18513–18518 [DOI] [PubMed] [Google Scholar]
- 17.Gostissa, M., A. Hengstermann, V. Fogal, P. Sandy, S. E. Schwarz, M. Scheffner, and G. Del Sal. 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18:6462–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govind, S. 1999. Control of development and immunity by rel transcription factors in Drosophila. Oncogene 18:6875–6887 [DOI] [PubMed] [Google Scholar]
- 19.Govind, S., E. Drier, L. H. Huang, and R. Steward. 1996. Regulated nuclear import of the Drosophila Rel protein Dorsal: structure-function analysis. Mol. Cell. Biol. 16:1103–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, Z. S., and Y. T. Ip. 1999. Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. J. Biol. Chem. 274:21355–21361 [DOI] [PubMed] [Google Scholar]
- 21.Imler, J. L., and J. A. Hoffmann. 2000. Signaling mechanisms in the antimicrobial host defense of Drosophila. Curr. Opin. Microbiol. 3:16–22 [DOI] [PubMed] [Google Scholar]
- 22.Iniguez-Lluhi, J. A., and D. Pearce. 2000. A common motif within the negative regulatory regions of multiple factors inhibits their transcriptional synergy. Mol. Cell. Biol. 20:6040–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isoda, K., S. Roth, and C. Nusslein-Volhard. 1992. The functional domains of the Drosophila morphogen dorsal: evidence from the analysis of mutants. Genes Dev. 6:619–630 [DOI] [PubMed] [Google Scholar]
- 24.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549–552 [DOI] [PubMed] [Google Scholar]
- 25.Johnson, E. S., and G. Blobel. 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272:26799–26802 [DOI] [PubMed] [Google Scholar]
- 26.Johnson, E. S., and A. A. Gupta. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106:735–744 [DOI] [PubMed] [Google Scholar]
- 27.Johnson, E. S., I. Schwienhorst, R. J. Dohmen, and G. Blobel. 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16:5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, Y. S., S. J. Han, J. H. Ryu, K. H. Choi, Y. S. Hong, Y. H. Chung, S. Perrot, A. Raibaud, P. T. Brey, and W. J. Lee. 2000. Lipopolysaccharide-activated kinase, an essential component for the induction of the antimicrobial peptide genes in Drosophila melanogaster cells. J. Biol. Chem. 275:2071–2079 [DOI] [PubMed] [Google Scholar]
- 29.Kurtzman, A. L., and N. Schechter. 2001. Ubc9 interacts with a nuclear localization signal and mediates nuclear localization of the paired-like homeobox protein Vsx-1 independent of SUMO-1 modification. Proc. Natl. Acad. Sci. USA 98:5602–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246–251 [DOI] [PubMed] [Google Scholar]
- 31.Li, S. J., and M. Hochstrasser. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20:2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, Q., C. Jin, X. Liao, Z. Shen, D. J. Chen, and Y. Chen. 1999. The binding interface between an E2 (UBC9) and a ubiquitin homologue (UBL1). J. Biol. Chem. 274:16979–16987 [DOI] [PubMed] [Google Scholar]
- 33.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97–107 [DOI] [PubMed] [Google Scholar]
- 34.Manfruelli, P., J. M. Reichhart, R. Steward, J. A. Hoffmann, and B. Lemaitre. 1999. A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. EMBO J. 18:3380–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mao, Y., S. D. Desai, and L. F. Liu. 2000. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J. Biol. Chem. 275:26066–26073 [DOI] [PubMed] [Google Scholar]
- 36.Mao, Y., M. Sun, S. D. Desai, and L. F. Liu. 2000. SUMO-1 conjugation to topoisomerase I: a possible repair response to topoisomerase-mediated DNA damage. Proc. Natl. Acad. Sci. USA 97:4046–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matunis, M. J., J. Wu, and G. Blobel. 1998. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140:499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng, X., B. S. Khanuja, and Y. T. Ip. 1999. Toll receptor-mediated Drosophila immune response requires Dif, an NF-kappaB factor. Genes Dev. 13:792–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mossessova, E., and C. D. Lima. 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5:865–876 [DOI] [PubMed] [Google Scholar]
- 40.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okuma, T., R. Honda, G. Ichikawa, N. Tsumagari, and H. Yasuda. 1999. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem. Biophys. Res. Commun. 254:693–698 [DOI] [PubMed] [Google Scholar]
- 42.Poukka, H., P. Aarnisalo, U. Karvonen, J. J. Palvimo, and O. A. Janne. 1999. Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J. Biol. Chem. 274:19441–19446 [DOI] [PubMed] [Google Scholar]
- 43.Poukka, H., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2000. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl. Acad. Sci. USA 97:14145–14150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray, R. P., K. Arora, C. Nusslein-Volhard, and W. M. Gelbart. 1991. The control of cell fate along the dorsal-ventral axis of the Drosophila embryo. Development 113:35–54 [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez, M. S., C. Dargemont, and R. T. Hay. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276:12654–12659 [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez, M. S., J. M. Desterro, S. Lain, C. A. Midgley, D. P. Lane, and R. T. Hay. 1999. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18:6455–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252–6258 [DOI] [PubMed] [Google Scholar]
- 48.Schwienhorst, I., E. S. Johnson, and R. J. Dohmen. 2000. SUMO conjugation and deconjugation. Mol. Gen. Genet. 263:771–786 [DOI] [PubMed] [Google Scholar]
- 49.Shirokawa, J. M., and A. J. Courey. 1997. A direct contact between the Dorsal rel homology domain and Twist may mediate transcriptional synergy. Mol. Cell. Biol. 17:3345–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silverman, N., R. Zhou, S. Stoven, N. Pandey, D. Hultmark, and T. Maniatis. 2000. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 14:2461–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi, Y., J. Mizoi, E. A. Toh, and Y. Kikuchi. 2000. Yeast Ulp1, an Smt3-specific protease, associates with nucleoporins. J. Biochem. (Tokyo) 128:723–725 [DOI] [PubMed] [Google Scholar]
- 52.Tashiro, K., M. P. Pando, Y. Kanegae, P. M. Wamsley, S. Inoue, and I. M. Verma. 1997. Direct involvement of the ubiquitin-conjugating enzyme Ubc9/Hus5 in the degradation of IkappaBalpha. Proc. Natl. Acad. Sci. USA 94:7862–7867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valentine, S. A., G. Chen, T. Shandala, J. Fernandez, S. Mische, R. Saint, and A. J. Courey. 1998. Dorsal-mediated repression requires the formation of a multiprotein repression complex at the ventral silencer. Mol. Cell. Biol. 18:6584–6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeh, E. T., L. Gong, and T. Kamitani. 2000. Ubiquitin-like proteins: new wines in new bottles. Gene 248:1–14 [DOI] [PubMed] [Google Scholar]
- 55.Zhang, G., and S. Ghosh. 2001. Toll-like receptor-mediated NF-kappaB activation: a phylogenetically conserved paradigm in innate immunity. J. Clin. Investig. 107:13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748–2752 [PubMed] [Google Scholar]