Abstract
Depletion of any of the five essential proteins Lsm2p to Lsm5p and Lsm8p leads to strong accumulation of all tested unspliced pre-tRNA species, as well as accumulation of 5′ and 3′ unprocessed species. Aberrant 3′-extended pre-tRNAs were detected, presumably due to stabilization of transcripts that fail to undergo correct transcription termination, and the accumulation of truncated tRNA fragments was also observed. Tandem affinity purification-tagged Lsm3p was associated with pre-tRNA primary transcripts and, less efficiently, with other unspliced pre-tRNA intermediates but not mature tRNAs. Association of the Saccharomyces cerevisiae La homologue Lhp1p with pre-tRNAs was reduced approximately threefold on depletion of Lsm3p or Lsm5p. The association of Lhp1p with larger RNA polymerase III transcripts, pre-RNase P RNA and the signal recognition particle RNA (scR1), was more drastically reduced. The impaired pre-tRNA processing seen on Lsm depletion is not, however, due solely to reduced Lhp1p association as evidenced by analysis of lhp1-Δ strains depleted of Lsm3p or Lsm5p. These data are consistent with roles for an Lsm complex as a chaperone that facilitates the efficient association of pre-tRNA processing factors with their substrates.
The U1, U2, U4, and U5 snRNAs associate with the seven Sm proteins (20), which form a closed ring structure (23). In contrast, U6 snRNA associates with seven related proteins, Lsm2p (Lsm stands for “like Sm”) to Lsm8p (8, 17, 29, 34, 40, 41, 44), which also form a heptameric ring structure (1, 2, 7). The Lsm2p-Lsm8p complex is involved in pre-mRNA splicing, probably by facilitating biogenesis of the U6 snRNP and its structural rearrangement during spliceosome assembly. The related Lsm1p-Lsm7p complex associates with cytoplasmic mRNA decay factors and functions in mRNA decapping and degradation (4, 5, 19, 45). The Lsm2p-Lsm8p complex binds U6 snRNA via the 3′ poly(U) tract (1, 2). Newly synthesized U6 also binds Lhp1p (La-homologous protein), the Saccharomyces cerevisiae homologue of the human La phosphoprotein, which protects the RNA against degradation (34).
Recent analyses have identified Sm-like proteins in both the domains Archaea and Bacteria (2, 7, 30, 31, 47, 52). Since these organisms lack both the U6 snRNA and capped mRNAs, this suggested that the ancestral Sm-like proteins had different functions in RNA metabolism. Indeed, evidence has been presented for their association with the RNA component of RNase P in Archaea and with small regulatory RNAs in Escherichia coli, where the Sm-like Hfq protein was proposed previously to act as a chaperone facilitating RNA-RNA interactions (30, 47, 52). We have therefore investigated possible additional roles for the eukaryotic Lsm proteins.
Poly(U) tracts are present in all RNA polymerase III (PolIII) transcribed RNAs, including pre-tRNAs, pre-P RNA (the RNA component of RNase P), pre-5S rRNA, and scR1 (the RNA component of signal recognition particle), all of which also bind Lhp1p/La (6, 37, 38, 43, 51). Here we show that Lsm proteins associate with some of these RNA precursors and are required for their normal association with Lhp1p, particularly in the case of complex ribonucleoprotein (RNP) substrates.
All tRNA species undergo posttranscriptional 5′ and 3′ maturation. In addition, many but not all pre-tRNAs contain intervening sequences that are removed by cleavage and ligation. Analysis of pre-tRNA processing in Drosophila melanogaster and in the budding yeast S. cerevisiae showed that processing of the 3′ end involved an endonucleolytic cleavage and occurred after 5′-end maturation (11, 12, 14, 32). In contrast, there is no obligatory order of pre-tRNA end maturation and splicing (32), and either activity will continue when the other is inhibited. In most cases end maturation is more rapid, however, and the unspliced but end-matured pre-tRNAs are readily visible in wild-type cells for most pre-tRNA species. Lhp1p stimulates endonucleolytic cleavage of tRNA 3′ ends (by an as-yet-unidentified enzyme) while suppressing maturation by exonucleases (51).
A possible role for Lsm proteins in pre-tRNA processing was suggested by the two-hybrid interactions reported between Lsm8p and the putative RNA helicase Sen1p, which acts as a positive effector of the tRNA splicing endonuclease, and between Lsm2p and Tpt1p, the 2′-phosphotransferase that functions in tRNA splicing (9, 10, 13, 36, 48, 49). In addition, deletion of the LHP1 gene is synthetically lethal with mutation in several LSM genes (33).
We report here that pre-tRNAs are indeed associated with Lsm proteins, which are required for their normal processing.
MATERIALS AND METHODS
Strains.
The transformation procedure was as described previously (16). Yeast strains used in this work are listed in Table 1. Strain YJK20 was constructed by inserting the ADH1 terminator sequence into Lhp1p-ProtA in the strain YDL579 by one-step PCR with pFA6a-3HA-His3MX6 as a template (27). The Kluyveromyces lactis URA3 marker was replaced with the His3MX6 module containing the Schizosaccharomyces pombe his5+ gene. Expression of the Lhp1p-ProtA fusion was tested by Northern hybridization and by Western blotting with peroxidase-antiperoxidase antibodies (Sigma). Strains YJK21 and YJK22 were constructed by the same PCR strategy using the YJK20 strain. The LHP1::ProtA-TADH1-HIS3MX6 gene was amplified and transformed into strains AEMY31 and AEMY47. Transformants were tested by Western blotting with peroxidase-antiperoxidase antibodies. StrainYJK34 was constructed by a PCR strategy described previously (35); construction was confirmed by PCR analysis, and the expression of Lsm3p-tandem affinity purification (TAP) was tested by Western blotting. Strains YCA50 and YCA51 were generated by PCR-based gene disruption of LHP1 in strains AEMY31 and AEMY47 with plasmid pTL54 as PCR template (25). Disruption was confirmed by PCR analysis.
TABLE 1.
Yeast strains used in this work
| Strain | Genotype | Reference or source |
|---|---|---|
| AEMY19 | MATα ade2-1 his3Δ200 leu2-3,112 trp1Δ1 ura3-1 LSM6::HIS3 | 29 |
| AEMY22 | MATα ade2-1 his3Δ200 leu2-3,112 trp1Δ1 ura3-1 LSM7::HIS3 | 29 |
| AEMY24 | MATα ade2-1 his3-11,15 leu2-3,112 trp1Δ1 ura3-1 LSM1::TRP1 | 29 |
| AEMY29 | MATα ade2-1 his3-11,15 leu2-3,112 trp1Δ1 ura3-1 LSM5::TRP1 [pACTIIst-LSM5] | 29 |
| AEMY30 | MATα ade2-1 his3Δ200 leu2-3,112 trp1Δ1 ura3-1 LSM2::HIS3 [pACTIIst-LSM2] | 45 |
| AEMY31 | MATα ade2-1 his3-11,15 leu2-3,112 trp1Δ1 ura3-1 LSM3::TRP1 [pBM125-GAL1-HA-LSM3] | 29 |
| AEMY33 | MATα ade2-1 his3Δ200 leu2-3,112 trp1Δ1 ura3-1 LSM2::HIS3 [pBM125-GAL1-LSM2-HA] | 29 |
| AEMY46 | MATα ade2-1 his3-11,15 leu2-3,112 trp1Δ1 ura3-1 LSM8::TRP1 [pBM125-GAL1-HA-LSM8] | 29 |
| AEMY47 | MATα ade2-1 his3-11,15 leu2-3,112 trp1Δ1 ura3-1 LSM5::TRP1 [pBM125-GAL1-HA-LSM5] | 29 |
| MCY4 | MATaade1-101 his3-Δ1 trp1-289 ura3-52 LEU2-GAL1-LSM4 | 8 |
| YDL579 | MATahis3Δ200 leu2Δ1 trp1 ura3-52 gal2 galΔ108 LHP1::ProtA-URA3 | 24 |
| YJK20 | As YDL579 but LHP1::ProtA-TADH1-HIS3MX6 | This work |
| YJK21 | As AEMY31 but LHP1::ProtA-TADH1-HIS3MX6 | This work |
| YJK22 | As AEMY47 but LHP1::ProtA-TADH1-HIS3MX6 | This work |
| BMA64 | MATα ade2-1 his3-11,15 leu2-3,112 trp1Δura3-1 | F. Lacroute |
| YJV140 | MATaade2 his3 leu2 trp2 ura3 | 50 |
| YJK34 | As YJV140 but TAP-LSM3 | This work |
| YCA35 | MATahis3Δ1 trp1 ura3-52 gal2 galΔ108 LHP1::Kl URA | 24 |
| YCA50 | As AEMY31 but LHP1::HIS3 | This work |
| YCA51 | As AEMY47 but LHP1::HIS3 | This work |
RNA extraction and Northern hybridization.
For depletion of the essential Lsm proteins cells were harvested at intervals following the shift from RSG medium (2% galactose, 2% sucrose, 2% raffinose) or YPGal medium containing 2% galactose to YPD medium containing 2% glucose. Otherwise strains were grown in YPD medium. The lsm-Δ strains were pregrown at 23°C and transferred to 37°C. RNA extraction and Northern hybridization were performed as described previously (3, 46).
For RNA hybridization the following oligonucleotides were used: 032 (RNase P RNA), 5′-ATTTCTGATAACAACGGTCGG; 041 (5S), 5′-CTACTCGGTCAGGCTC; 250 (scR1), 5′-ATCCCGGCCGCCTCCATCAC; 261 (U6), 5′-AAAACGAAATAAATTCTTTGTAAAAC; 266 (5S-3′), 5′-AAAAAAAACAACTGCAGC; 301 ( ), 5′-GTCGAACCCATAATCTTC; 302 (tRNAAlaUGC), 5′-CTACCAACTGCGCCATG; 303 (tRNAGlyGCC), 5′-TACCACTAAACCACTTGC; 304 (tRNAProUGG), 5′-ACCCAGGGCCTCTCG; 305 (tRNATrp), 5′-AACCTGCAACCCTTCGA; 306 (
), 5′-GTCGAACCCATAATCTTC; 302 (tRNAAlaUGC), 5′-CTACCAACTGCGCCATG; 303 (tRNAGlyGCC), 5′-TACCACTAAACCACTTGC; 304 (tRNAProUGG), 5′-ACCCAGGGCCTCTCG; 305 (tRNATrp), 5′-AACCTGCAACCCTTCGA; 306 ( ), 5′-GCATCTTACGATACCTG; 307 (
), 5′-GCATCTTACGATACCTG; 307 ( -intron), 5′-CACAGTTAACTGCGGTC; 308 (tRNAPhe-intron), 5′-AACTTGACCGAAGTATTTC; 310 (tRNATyrGΨA-intron), 5′-AAGATTTCGTAGTGATAA; and 320 (
-intron), 5′-CACAGTTAACTGCGGTC; 308 (tRNAPhe-intron), 5′-AACTTGACCGAAGTATTTC; 310 (tRNATyrGΨA-intron), 5′-AAGATTTCGTAGTGATAA; and 320 ( ), 5′-ATCCTTGCTTAAGCAAATGCGC.
), 5′-ATCCTTGCTTAAGCAAATGCGC.
Immunoprecipitation.
Whole-cell extracts from strains GAL::HA-lsm3, Lhp1p-ProtA, Lhp1p-ProtA/GAL::HA-lsm3, and Lhp1p-ProtA/GAL::HA-lsm5 grown either in RSG medium or following the transfer to YPD medium for 8.5 or 24 h were prepared as described previously (42). Immunoprecipitation of ProtA-tagged strains was performed as described previously (28) at 150 mM KAc. Immunoprecipitation of TAP-tagged Lsm3p protein was performed as described previously (35) using extract equivalent to a cell optical density at 600 nm of 800. Copurified RNAs were recovered from the eluate of the immunoglobulin G (IgG) column by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation. Precursors and mature RNAs were identified by Northern hybridizations. An untagged isogenic strain (YJV140) was utilized as a control.
RESULTS
Lsm proteins are required for normal tRNA processing.
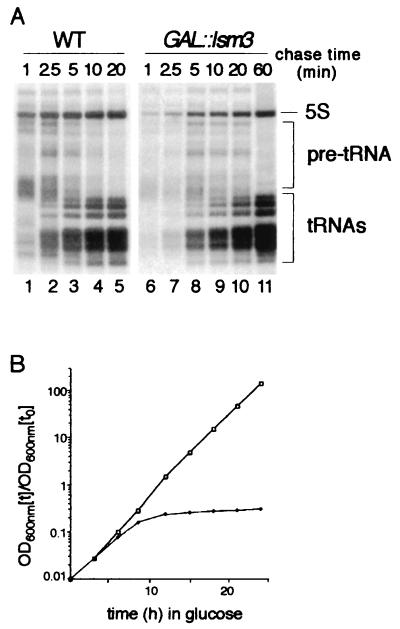
As an initial test of the effects of Lsm protein depletion on pre-tRNA processing, in vivo pulse-chase labeling was performed with [H3]uracil (Fig. 1A). The wild-type and GAL::HA-lsm3 (referred to throughout the text as GAL::lsm3) strains were pregrown in permissive RSG medium and then transferred to repressive glucose medium for 8.5 h. Cells were labeled with [3H]uracil for 1 min, followed by a chase with a large excess of unlabeled uracil. In the wild-type strain, pre-tRNAs could be seen at the 1-, 2.5-, and 5-min chase time-points. In the Lsm3p-depleted strain, pre-tRNAs were visible at the 1-min time point but were still readily detected after 20 min, strongly indicating that their processing was slowed. A delay in the accumulation of mature tRNAs was also evident. Incorporation of label into 5S was reduced in the Lsm3p-depleted strain, probably because this strain also shows defects in processing of pre-rRNAs, particularly the precursors to the 5.8S rRNA (data not shown; these results will be described in detail elsewhere, but see the Discussion). The 8.5-h time point corresponds to the earliest time at which any clear growth defect was seen in the GAL::lsm3 strain (Fig. 1B) (29), suggesting that the inhibition of tRNA processing may be a primary defect.
FIG. 1.
Processing of tRNAs is delayed in a strain depleted of Lsm3p. (A) Pulse-chase analysis of tRNA processing in GAL::HA-lsm3 strain. Strains were grown in permissive RSG medium and transferred to repressive glucose medium at 30°C for 8.5 h. [3H]uracil was added for 1 min, followed by a large excess of unlabeled uracil. RNA was extracted at the times indicated, separated on a 6% polyacrylamide gel, and visualized by fluorography. WT, wild type. (B) Growth curves of the wild-type (□) and GAL::HA-lsm3 (♦) strains pregrown in permissive RSG medium and transferred to repressive glucose medium for the times indicated. Strains were maintained in exponential growth by dilution with prewarmed medium. Cell densities measured by optical density at 600 nm are shown corrected for dilution. The GAL::lsm3 panel was exposed approximately 16-fold longer than the wild-type panel.
To confirm these observations, GAL-regulated hemagglutinin (HA)-tagged constructs were used to deplete the other essential Lsm proteins (strains GAL::lsm2, GAL::lsm4, GAL::lsm5, and GAL::lsm8). Deletion of the genes encoding nonessential Lsm proteins gives rise to temperature-sensitive (ts) strains (strains lsm6-Δ, lsm7-Δ, and lsm1-Δ) (29).
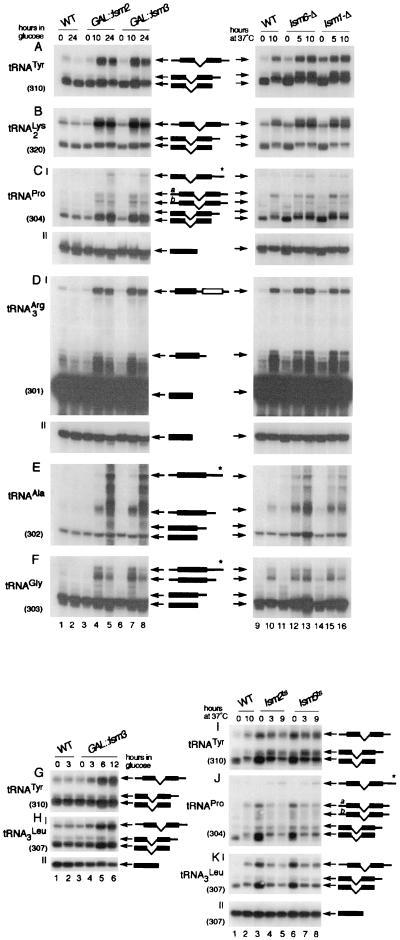
Processing of both intron-containing and intronless pre-tRNAs from both monocistronic and dicistronic transcripts was analyzed by Northern hybridization (Fig. 2). The identities of tRNA precursors were determined based on their relative electrophoretic mobilities, hybridization patterns, and comparison to previous reports (see, for example, references 32 and 51).
FIG. 2.
Processing of tRNAs is affected in lsm mutants. (A to F) Analysis of tRNA processing in GAL::lsm and lsm-Δ strains. (G and H) Analysis of processing of tRNATyr and  in GAL::lsm3 strain. (I to K) Analysis of tRNA processing in lsm2ts and lsm5ts strains. Strains carrying GAL-regulated HA-tagged constructs (GAL::lsm; A to F, lanes 3 to 8, and G and H, lanes 3 to 6) and the BMA64 wild-type strain (A to F, lanes 1 and 2, and G and H, lanes 1 and 2) were grown in permissive RSG medium (0 h) and transferred to repressive glucose medium at 30°C for the times indicated. Strains with deletions of Lsm6p or Lsm1p (A to F, lanes 11 to 16), the temperature-sensitive lsm2ts and lsm5ts strains (I to K, lanes 3 to 8), and the wild-type strain (A to F, lanes 9 and 10, and I to K, lanes 1 and 2) were pregrown at 23°C (0 h) and transferred to 37°C for the times indicated. RNA was separated on a 6% polyacrylamide gel and hybridized with oligonucleotide probes. Names of tRNAs are on the left; schematic representations of tRNA precursors, intermediates, and mature species are shown between the two columns (A to F) or on the right (G to K); and probe names are in parentheses. RNA precursors marked with asterisks represent species predicted to be 3′ extended beyond the wild-type transcripts. tRNAPro precursors designated by a and b represent two forms of PT (32). The minor size heterogeneity observed for pre-tRNAs and the mature tRNAs may result from the presence or absence of the 3′-end -CCAOH tail. WT, wild type.
in GAL::lsm3 strain. (I to K) Analysis of tRNA processing in lsm2ts and lsm5ts strains. Strains carrying GAL-regulated HA-tagged constructs (GAL::lsm; A to F, lanes 3 to 8, and G and H, lanes 3 to 6) and the BMA64 wild-type strain (A to F, lanes 1 and 2, and G and H, lanes 1 and 2) were grown in permissive RSG medium (0 h) and transferred to repressive glucose medium at 30°C for the times indicated. Strains with deletions of Lsm6p or Lsm1p (A to F, lanes 11 to 16), the temperature-sensitive lsm2ts and lsm5ts strains (I to K, lanes 3 to 8), and the wild-type strain (A to F, lanes 9 and 10, and I to K, lanes 1 and 2) were pregrown at 23°C (0 h) and transferred to 37°C for the times indicated. RNA was separated on a 6% polyacrylamide gel and hybridized with oligonucleotide probes. Names of tRNAs are on the left; schematic representations of tRNA precursors, intermediates, and mature species are shown between the two columns (A to F) or on the right (G to K); and probe names are in parentheses. RNA precursors marked with asterisks represent species predicted to be 3′ extended beyond the wild-type transcripts. tRNAPro precursors designated by a and b represent two forms of PT (32). The minor size heterogeneity observed for pre-tRNAs and the mature tRNAs may result from the presence or absence of the 3′-end -CCAOH tail. WT, wild type.
Substantial accumulation of all intron-containing precursors, particularly the primary transcripts (PT), was seen in strains depleted of Lsm2p to Lsm5p or Lsm8p. Data are shown for pre-tRNATyr,  , and pre-tRNAPro in GAL::lsm2 and GAL::lsm3 strains in Fig. 2A to C, lanes 3 to 8, and for pre-tRNATyr and pre-
, and pre-tRNAPro in GAL::lsm2 and GAL::lsm3 strains in Fig. 2A to C, lanes 3 to 8, and for pre-tRNATyr and pre- in GAL::lsm3 in Fig. 2G and H, lanes 3 to 6. Similar results were obtained for GAL::lsm4, GAL::lsm5, and GAL::lsm8 strains and for other intron-containing tRNAs, pre-tRNAPhe and pre-tRNASer (data not shown). Pre-tRNATrp was also accumulated in the lsm mutants (data not shown), but the processing of pre-tRNATrp was found to follow an unusual pathway, which will be reported elsewhere. Precursors to tRNAPro and tRNAPhe with mature 5′ and 3′ ends and 5′ processed pre-tRNAPhe were also elevated (Fig. 2C and data not shown). Defects in the processing of the intronless, dicistronic
in GAL::lsm3 in Fig. 2G and H, lanes 3 to 6. Similar results were obtained for GAL::lsm4, GAL::lsm5, and GAL::lsm8 strains and for other intron-containing tRNAs, pre-tRNAPhe and pre-tRNASer (data not shown). Pre-tRNATrp was also accumulated in the lsm mutants (data not shown), but the processing of pre-tRNATrp was found to follow an unusual pathway, which will be reported elsewhere. Precursors to tRNAPro and tRNAPhe with mature 5′ and 3′ ends and 5′ processed pre-tRNAPhe were also elevated (Fig. 2C and data not shown). Defects in the processing of the intronless, dicistronic  -tRNAAsp and monocistronic tRNAAla and tRNAGly were also observed in the GAL::lsm strains, showing that processing defects are not restricted to splicing. PT and 5′ processed, 3′ unprocessed precursors accumulated on depletion of Lsm2p to Lsm5p or Lsm8p (Fig. 2D to F, lanes 3 to 8, and data not shown); however, little depletion of the mature tRNAs was seen. In the experiment shown in Fig. 2A to F, pre-tRNA accumulation was strong 10 h after transfer to glucose medium. To confirm that this tRNA processing phenotype is not due to slowed growth, pre-tRNA processing was tested at early times after transfer to glucose medium (Fig. 2G and H). Mild accumulation of the PT of
-tRNAAsp and monocistronic tRNAAla and tRNAGly were also observed in the GAL::lsm strains, showing that processing defects are not restricted to splicing. PT and 5′ processed, 3′ unprocessed precursors accumulated on depletion of Lsm2p to Lsm5p or Lsm8p (Fig. 2D to F, lanes 3 to 8, and data not shown); however, little depletion of the mature tRNAs was seen. In the experiment shown in Fig. 2A to F, pre-tRNA accumulation was strong 10 h after transfer to glucose medium. To confirm that this tRNA processing phenotype is not due to slowed growth, pre-tRNA processing was tested at early times after transfer to glucose medium (Fig. 2G and H). Mild accumulation of the PT of 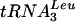 and tRNATyr was observed in the GAL::lsm3 strain 3 h after transfer, with marked pre-tRNA accumulation after 6 h. This is prior to growth inhibition and would be consistent with a direct role for the Lsm proteins.
and tRNATyr was observed in the GAL::lsm3 strain 3 h after transfer, with marked pre-tRNA accumulation after 6 h. This is prior to growth inhibition and would be consistent with a direct role for the Lsm proteins.
The GAL::lsm strains were analyzed at 30°C. Transfer of wild-type cells to 37°C resulted in some pre-tRNA accumulation (Fig. 2A to F, lanes 9 and 10) as previously reported (32). In lsm1, lsm6, or lsm7-Δ strains the levels of most pre-tRNA species were not clearly different from the wild type (shown for lsm6-Δ and lsm1-Δ in Fig. 2A to F, lanes 11 to 16). An exception was pre-tRNAAla, which strongly accumulated in the lsm6-Δ and lsm7-Δ strains at 37°C (Fig. 2E, lanes 11 to 13, and data not shown). Surprisingly, similar accumulation was seen in a strain lacking Lsm1p (Fig. 2E, lanes 14 to 16), which is enriched in the cytoplasm (45).
RNA species larger than the predicted size of the wild-type PT (marked with asterisk; Fig. 2C-I, E, and F) were detected for tRNAPhe, tRNAPro, tRNAAla, and tRNAGly, in strains depleted of Lsm2p to Lsm5p or Lsm8p or after transfer of the lsm6-Δ, lsm7-Δ, or lsm1-Δ strain to 37°C (Fig. 2C to F and data not shown). Hybridization with probes specific for 3′-extended tRNAAla and tRNAGly showed that these correspond to aberrantly 3′-extended pre-tRNAs (data not shown). These may represent the products of read-through of the normal transcription termination site, which are rapidly degraded in wild-type cells but accumulate in the absence of Lsm proteins.
We conclude that Lsm proteins, particularly the essential proteins Lsm2p to Lsm5p and Lsm8p, are required for normal pre-tRNA maturation. Notably, the pattern of pre-tRNA accumulation was not identical for different tRNA species. This heterogeneity would be more consistent with the direct participation of Lsm proteins in pre-tRNA processing than an indirect role in reducing global processing efficiency.
Fusion proteins between Lsm2p and Lsm5p and the DNA binding domain of Gal4p, containing a strong nuclear localization signal, were previously constructed for use in two-hybrid analyses (13). These constructs confer partial temperature sensitivity for growth and cytoplasmic mRNA degradation when expressed in lsm2-Δ and lsm5-Δ backgrounds (19, 45). Growth of the lsm2ts or lsm5ts strains at 23°C resulted in strong pre-tRNA accumulation for tRNATyr, tRNAPro, and  (Fig. 2I to K, compare lanes 3 and 6 with lane 1), which was reduced by 3 h after transfer to 37°C (Fig. 2I to K, lanes 4 and 7). Accumulation of 3′-extended tRNAPro (marked with asterisk in Fig. 2J) was also observed. These observations support the role of Lsm2p and Lsm5p in pre-tRNA processing, although, as with cytoplasmic mRNA turnover, the basis of this defect in lsm2ts or lsm5ts strains is unclear.
(Fig. 2I to K, compare lanes 3 and 6 with lane 1), which was reduced by 3 h after transfer to 37°C (Fig. 2I to K, lanes 4 and 7). Accumulation of 3′-extended tRNAPro (marked with asterisk in Fig. 2J) was also observed. These observations support the role of Lsm2p and Lsm5p in pre-tRNA processing, although, as with cytoplasmic mRNA turnover, the basis of this defect in lsm2ts or lsm5ts strains is unclear.
Lsm complexes have been shown to be required for pre-mRNA splicing and for cytoplasmic mRNA degradation. However, no protein directly involved in these activities has previously been reported to be required for pre-tRNA processing. Moreover, no pre-tRNA processing defect was seen in strains inhibited for splicing due to genetic depletion of the splicing factor Prp8p or Syf3 under GAL control or carrying the ts-lethal prp2-1 allele (data not shown). The lsm2ts or lsm5ts strains showed only mild pre-mRNA splicing defects at any temperature (data not shown), but notably, splicing was more defective at 37 than at 23°C, in marked contrast to the pre-tRNA processing defect.
Similarly, no pre-tRNA processing defect was seen in strains defective in mRNA turnover due to the absence of the components of the exosome or the cytoplasmic exonuclease Xrn1p (data not shown). A double mutant strain, the prp2-1; xrn1-Δ strain, which is defective in both splicing and mRNA degradation, also failed to show any pre-tRNA processing defect (data not shown). Together these data indicate that pre-tRNA processing defects seen in the lsm mutant strains are probably not secondary consequences of defects in pre-mRNA splicing or mRNA degradation.
Lsm3p binds to pre-tRNAs.
To test whether the Lsm complex interacts directly with tRNA precursors, immunoprecipitation was performed with the C-terminally TAP-tagged Lsm3p construct (35) expressed under the endogenous promoter (Fig. 3). A similar construct was used previously to characterize interactions of Lsm3p (5). As previously reported, Lsm3-TAP coprecipitated U6 (Fig. 3C) and the RNase P RNA precursor (Fig. 3A) (1, 29, 40, 41), with weaker coprecipitation of scR1, the RNA component of the yeast signal recognition particle and very weak precipitation of mature P RNA (Fig. 3A and B). No clear coprecipitation was seen for mature tRNATrp or tRNATyr (Fig. 3E-II and F-II, lanes 4), which were recovered at the same level as in the control precipitation with a nontagged strain (Fig. 3, lanes 3). In contrast, precursors of 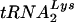 (Fig. 3D), tRNATyr (Fig. 3E-I), or tRNATrp (Fig. 3F-I), particularly the PT, were clearly coprecipitated with Lsm3-TAP. Notably, tRNA PT, which were most affected by Lsm depletion, were more efficiently precipitated. Other intron-containing pre-tRNA species were affected to a lesser extent by Lsm3p depletion and were also precipitated less efficiently (Fig. 3D to F). The efficiency of pre-tRNA precipitation was substantially lower than that for mature U6 but similar to the 1 to 3% recovery reported elsewhere for mRNA association of Lsm proteins (45). Approximately 30-fold-more cell equivalents were loaded for the precipitated (P) fractions than for the total (T) in Fig. 3. The low efficiency of coprecipitation of tRNA precursors with Lsm3p-TAP may reflect transient association in vivo. Alternatively, the Lsm-RNA association may not resist extended incubation (2.5 h at 4°C) and extensive washing during immunoprecipitation.
(Fig. 3D), tRNATyr (Fig. 3E-I), or tRNATrp (Fig. 3F-I), particularly the PT, were clearly coprecipitated with Lsm3-TAP. Notably, tRNA PT, which were most affected by Lsm depletion, were more efficiently precipitated. Other intron-containing pre-tRNA species were affected to a lesser extent by Lsm3p depletion and were also precipitated less efficiently (Fig. 3D to F). The efficiency of pre-tRNA precipitation was substantially lower than that for mature U6 but similar to the 1 to 3% recovery reported elsewhere for mRNA association of Lsm proteins (45). Approximately 30-fold-more cell equivalents were loaded for the precipitated (P) fractions than for the total (T) in Fig. 3. The low efficiency of coprecipitation of tRNA precursors with Lsm3p-TAP may reflect transient association in vivo. Alternatively, the Lsm-RNA association may not resist extended incubation (2.5 h at 4°C) and extensive washing during immunoprecipitation.
FIG. 3.
Lsm3p binds to pre-tRNAs. Immunoprecipitation of RNAs from a strain expressing TAP-tagged Lsm3p is shown. Lysates from the TAP-Lsm3 strain and the isogenic wild-type (WT) strain (YJV140) were immunoprecipitated with rabbit IgG agarose beads (Sigma). RNA was recovered from the lysate (T) and the immunoprecipitate (P) and analyzed by Northern hybridization. Probe names are indicated in parentheses. RNA species are shown on the right. Approximately 30-fold-more cell equivalents was loaded for the bound fractions than for the total and supernatant fractions.
Binding of Lhp1p to RNA substrates is reduced in strains depleted of Lsm3p or Lsm5p.
The Lsm complexes have been proposed previously to function as chaperones in RNP assembly (29), and we therefore determined whether they act to promote the association of Lhp1p with pre-tRNAs and other PolIII transcripts.
A C-terminal fusion between Lhp1p and two copies of the Z domain of Staphylococcus aureus protein A (see Materials and Methods) was expressed in GAL::HA-lsm3 and GAL::HA-lsm5 strains. Western blotting showed that HA-Lsm3p and HA-Lsm5p were below detectable levels in the 24-h samples while the level of Lhp1p-ProtA was unaffected (Fig. 4G). Northern hybridization showed that all fusion proteins were functional in RNA processing (data not shown, but see Fig. 4D). Lhp1p-ProtA immunoprecipitation was performed with extracts from strains grown in permissive RSG medium (0-h samples) and following transfer to glucose medium for 24 h to deplete Lsm3p or Lsm5p. Similar results were obtained for each double mutant strain, and data are shown only for Lhp1-ProtA/GAL::HA-lsm3. To ensure that inhibition of growth at late times after depletion of Lsm proteins is not responsible for the observed results, an analogous immunoprecipitation was performed with extract from Lhp1-ProtA/GAL::HA-lsm3 cells depleted of Lsm3p for 8.5 h, at which time there is very little growth defect (Fig. 1B). Data from this experiment are included in Fig. 4F. Control precipitations were performed on the Lhp1p-ProtA and GAL::HA-lsm3 single mutant strains. Approximately fourfold-more cell equivalents were loaded for the pellet fractions in Fig. 4A to E.
FIG. 4.
Lhp1p binds less efficiently to RNA substrates in the absence of Lsm proteins. (A to E) The strains GAL::HA-lsm3 (lanes 1 to 3), Lhp1p-ProtA (lanes 4 to 6), and Lhp1p-ProtA/GAL::HA-lsm3 (lanes 7 to 12) were grown at 30°C in RSG medium (lanes 1 to 9) or transferred to glucose medium for 24 h (lanes 10 to 12). Lysates were immunoprecipitated with IgG agarose, and RNA was recovered from the lysate (T), the immune supernatant (S), and the immunoprecipitate (P) and analyzed by Northern hybridization. Probe names are indicated in parentheses. RNA species are shown on the left. Due to the fact that depletion of Lsm proteins results in altered levels of some of the analyzed RNA species, different exposure times were used for different panels to adequately visualize RNAs. Approximately fourfold-more cell equivalents are loaded for the bound material. The arrowhead indicates the 5′ processed, 3′ unprocessed pre-tRNATyr. (F) Graphic representation of efficiency of immunoprecipitation by Lhp1p of RNAs from panels A to E in the Lhp1-ProtA/GAL::HA-lsm3 strain before (0 h) and after (8.5 and 24 h) depletion of Lsm3p. Values for each RNA species after depletion are expressed relative to the value before depletion, which is arbitrarily set at 1. Immunoprecipitation efficiency was calculated based on PhosphorImager quantification of Northern hybridization data from panels A to E. (G) Levels of Lsm3p, Lsm5p, and Lhp1p during Lsm depletion. Western blots of total protein were decorated with anti-HA to detect both HA-Lsm3p and HA-Lsm5p and with anti-protein A to detect Lhp1p-ProtA, following growth in galactose medium (0-h samples) or 24 h after transfer to glucose medium.
Data from Northern analyses (Fig. 4A to E) were quantified with a PhosphorImager and are presented graphically (Fig. 4F) and numerically (Table 2). Lhp1p-ProtA efficiently coprecipitated pre-tRNAs, pre-5S rRNA, pre-P RNAs, U6 snRNA, and scR1 RNA (Fig. 4, lanes 4 to 6). Precipitation was also efficient in the Lhp1p-ProtA/GAL::lsm strains in permissive medium (Fig. 4, lanes 7 to 9), although a mild reduction in precipitation was visible compared to the single mutant, possibly related to overexpression of Lsm3p and Lsm5p from the GAL promoter. After depletion of Lsm3p or Lsm5p for 24 h, the immunoprecipitation efficiency dropped for all RNAs except U6. Similar but less pronounced effects were observed for samples depleted for 8.5 h. The largest effects were on the signal recognition particle RNA, scR1, and pre-P RNA (76- and 18-fold, respectively), with threefold reductions for pre-5S (Fig. 4E and F) and the tRNATyr and  PT (shown for tRNATyr in Fig. 4D and F; similar results were observed for
PT (shown for tRNATyr in Fig. 4D and F; similar results were observed for 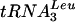 [Table 2]). Binding of Lhp1p to 5′ processed, 3′ unprocessed pre-tRNATyr and
[Table 2]). Binding of Lhp1p to 5′ processed, 3′ unprocessed pre-tRNATyr and 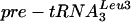 was also reduced three- to fourfold upon depletion of Lsm3p or Lsm5p (indicated with an arrowhead in Fig. 4D). Accurate quantitation of these precursors in the supernatant samples was not feasible, however, due to their proximity to the abundant unspliced but 5′ and 3′ processed form, which is not precipitated. All of these are RNA PolIII transcripts with terminal poly(U) tracts that would have been predicted to bind Lhp1p in the absence of cofactors. Mature P RNA and tRNA were not detectably coprecipitated with Lhp1p-ProtA-tagged strains, but some coprecipitation of mature 5S rRNA was observed, which was reduced approximately twofold on depletion of Lsm3p or Lsm5p. In contrast, immunoprecipitation of U6 was elevated on depletion of Lsm3p or Lsm5p, consistent with competition for the 3′ poly(U) tract as previously proposed (1, 34). We conclude that binding of Lhp1p to many but not all RNA substrates is less efficient in cells lacking Lsm proteins.
was also reduced three- to fourfold upon depletion of Lsm3p or Lsm5p (indicated with an arrowhead in Fig. 4D). Accurate quantitation of these precursors in the supernatant samples was not feasible, however, due to their proximity to the abundant unspliced but 5′ and 3′ processed form, which is not precipitated. All of these are RNA PolIII transcripts with terminal poly(U) tracts that would have been predicted to bind Lhp1p in the absence of cofactors. Mature P RNA and tRNA were not detectably coprecipitated with Lhp1p-ProtA-tagged strains, but some coprecipitation of mature 5S rRNA was observed, which was reduced approximately twofold on depletion of Lsm3p or Lsm5p. In contrast, immunoprecipitation of U6 was elevated on depletion of Lsm3p or Lsm5p, consistent with competition for the 3′ poly(U) tract as previously proposed (1, 34). We conclude that binding of Lhp1p to many but not all RNA substrates is less efficient in cells lacking Lsm proteins.
TABLE 2.
Immunoprecipitation efficiency of Lhp1pa
| RNA | Value for strain (time in glucose):
|
|||
|---|---|---|---|---|
| GAL::lsm3 (0 h) | Lhp1p- ProtA | Lhp1p-ProtA/ GAL::lsm3 (0 h) | Lhp1p-ProtA/ GAL::lsm3 (24 h) | |
| scR1 | 0.2 | 44.2 | 30.3 | 0.4 |
| Pre-RNase P RNA | 0.2 | 15.6 | 6.4 | 0.35 |
| RNase P RNA | 0.2 | 1.4 | 0.85 | 0.6 |
| U6 | 0.6 | 16.6 | 7.7 | 22.1 |
| tRNATyr-PT | 0.7 | 77.4 | 68.4 | 24.1 |
| tRNA3Leu-PT | 0.9 | 64.2 | 48.4 | 13.1 |
| tRNA3Leu | 0.1 | 0.4 | 0.3 | 0.1 |
| Pre-5S | 0.4 | 77.3 | 63 | 21 |
| 5S | 0.5 | 3.9 | 2.4 | 1.1 |
The significance of the observed coprecipitation of mature 5S with Lhp1p-ProtA is not clear, but we note that Lhp1p was reported previously to coprecipitate with two proteins (Bud2p and Cdc95p) identified in pre-60S ribosomal complexes predicted to contain 5S (15). The efficiency of coprecipitation of scR1 RNA with Lhp1p-ProtA was quite high, around 44%, suggesting an extended period of interaction. This is supported by the copurification of Lhp1p with Srp54p (15).
To further test functional interactions between Lhp1p and Lsm proteins, pre-tRNA processing was analyzed in strains carrying lhp1-Δ together with either GAL::lsm3 or GAL::lsm5. Data are shown for the intron-containing pre-tRNATyr (Fig. 5A) and pre-tRNAPro (Fig. 5B); similar results were observed for other intron-containing tRNAs (tRNAPhe and  ), dicistronic
), dicistronic  -tRNAAsp, and monocistronic tRNAGly. In RSG medium, the double mutant strains resembled the lhp1-Δ strain (Fig. 5, lanes 5, 10, and 15), in which the 5′ processed, 3′ unprocessed pre-tRNA is absent and the level of other pre-tRNA species is reduced. Depletion of either Lsm3p or Lsm5p from the strain lacking Lhp1p led to strong accumulation of PT, which became progressively shortened and more heterogeneous in length at later time points. These species were previously observed in strains lacking only Lhp1p (Fig. 4B, lane 16) and attributed to exonucleolytic trimming in its absence (51). The 3′-truncated PT were strongly accumulated on depletion of the Lsm proteins in the presence or absence of Lhp1p (Fig. 5A and B, lanes 2 to 4). Moreover, the level of these 3′-truncated pre-tRNAs in the Lsm-depleted strains was substantially higher than in the lhp1-Δ strain (Fig. 5, compare lanes 3 and 15), showing that the effects are not solely due to reduced Lhp1p binding. This strong accumulation of exonuclease digestion intermediates indicates that both the endonuclease that normally carries out 3′ processing and the 3′ exonuclease(s) that functions by default process the pre-tRNAs less efficiently in the absence of an Lsm complex.
-tRNAAsp, and monocistronic tRNAGly. In RSG medium, the double mutant strains resembled the lhp1-Δ strain (Fig. 5, lanes 5, 10, and 15), in which the 5′ processed, 3′ unprocessed pre-tRNA is absent and the level of other pre-tRNA species is reduced. Depletion of either Lsm3p or Lsm5p from the strain lacking Lhp1p led to strong accumulation of PT, which became progressively shortened and more heterogeneous in length at later time points. These species were previously observed in strains lacking only Lhp1p (Fig. 4B, lane 16) and attributed to exonucleolytic trimming in its absence (51). The 3′-truncated PT were strongly accumulated on depletion of the Lsm proteins in the presence or absence of Lhp1p (Fig. 5A and B, lanes 2 to 4). Moreover, the level of these 3′-truncated pre-tRNAs in the Lsm-depleted strains was substantially higher than in the lhp1-Δ strain (Fig. 5, compare lanes 3 and 15), showing that the effects are not solely due to reduced Lhp1p binding. This strong accumulation of exonuclease digestion intermediates indicates that both the endonuclease that normally carries out 3′ processing and the 3′ exonuclease(s) that functions by default process the pre-tRNAs less efficiently in the absence of an Lsm complex.
FIG. 5.
Processing of tRNA precursors in the absence of Lhp1p and Lsm proteins. Shown are results of Northern analysis of tRNA processing in GAL::lsm3/lhp1-Δ and GAL::lsm5/lhp1-Δ double mutants. RNA was extracted from GAL-regulated constructs following transfer from permissive RSG medium to repressive glucose medium for the times indicated. The lhp1-Δ strain was grown on glucose medium at 30°C. Probe names are indicated in parentheses. Names of tRNAs are shown on the left, and schematic representations of tRNA species are shown on the right. tRNAPro precursors designated with a and b represent two forms of PT (32), whereas the asterisk shows the position of the shortened PT in the lhp1-Δ strain. A darker exposure of pre-tRNAPro in the lhp1-Δ strain is shown in a separate panel (lane 16). WT, wild type.
Some accumulation of pre-tRNAs in double mutants was detected 2 h after transfer to glucose medium, and accumulation was substantial by 8 h (Fig. 5, lanes 6 and 11).
tRNA degradation products accumulate in Lsm-depleted cells.
Depletion of Lsm proteins was accompanied by the appearance of slightly faster migrating forms of all tRNA species examined, suggesting that the mature tRNAs were undergoing partial truncation (Fig. 2). Shorter, truncated tRNAs, which we assume to be degradation intermediates, were readily observed for all species at late times of depletion of Lsm2p to Lsm5p or Lsm8p (shown for mature tRNAPro and  in Fig. 6C and D for GAL::lsm2, GAL::lsm3, lsm6-Δ, and lsm1-Δ). Some evidence for degradation of pre-tRNATyr and pre-tRNAPro was obtained (Fig. 6A and B), but this was much less clear than the accumulation of mature tRNA fragments. Degradation products accumulated most strongly upon depletion of the essential Lsm proteins but were also detectable at lower levels in the lsm6-Δ, lsm7-Δ, and lsm1-Δ strains. This supports the role of both nuclear and cytoplasmic Lsm proteins in degradation of stable RNA species, in addition to their recently reported roles in mRNA degradation (4, 5, 19, 45).
in Fig. 6C and D for GAL::lsm2, GAL::lsm3, lsm6-Δ, and lsm1-Δ). Some evidence for degradation of pre-tRNATyr and pre-tRNAPro was obtained (Fig. 6A and B), but this was much less clear than the accumulation of mature tRNA fragments. Degradation products accumulated most strongly upon depletion of the essential Lsm proteins but were also detectable at lower levels in the lsm6-Δ, lsm7-Δ, and lsm1-Δ strains. This supports the role of both nuclear and cytoplasmic Lsm proteins in degradation of stable RNA species, in addition to their recently reported roles in mRNA degradation (4, 5, 19, 45).
FIG. 6.
Depletion of Lsm proteins leads to accumulation of tRNA degradation intermediates. Strains were grown and RNA was prepared as described for Fig. 2. Probe names are indicated in parentheses. WT, wild type.
DISCUSSION
Pre-tRNA processing was strongly affected by depletion of the essential Lsm proteins, Lsm2p to Lsm5p and Lsm8p, whereas the absence of Lsm1p, Lsm6p, or Lsm7p resulted in much weaker phenotypes. This is different from the stability of U6, which requires all seven proteins, Lsm2p to Lsm8p (29), but similar observations have been made for pre-rRNA processing (J. Kufel, J. Beggs, and D. Tollervey, unpublished observations). It may be that the lack of any essential protein prevents Lsm complex formation, while nonessential proteins can partially replace each other and form complexes that retain substantial activity. During U6 synthesis and in mRNA turnover and translation, Lsm complexes have been proposed previously to act as chaperones that modify the structure of RNP complexes, and a bacterial Sm-like protein functions to promote correct RNA-RNA interactions (29, 30, 45, 47, 52). The absence of Lsm proteins does not inhibit accumulation of mature tRNAs but alters the pattern of processing intermediates. The absence of the yeast La protein, Lhp1, also perturbed pre-tRNA processing without reducing tRNA levels (51). Accumulation of pre-tRNAs in the lsm strains closely resembles the processing pattern of mutant pre-tRNACGASer (sup61-10 allele), in which a mutation in the anticodon stem disturbs tRNA structure and renders processing strictly dependent on Lhp1p (26, 51). Roles in promoting correct formation of RNP complexes would be consistent with the consequences of Lsm protein depletion for pre-tRNA processing.
Defects in pre-tRNA processing were detected prior to growth inhibition, and coprecipitation of pre-tRNAs was seen with TAP-tagged Lsm3p, consistent with direct effects. Lsm proteins affect pre-mRNA splicing and degradation; however, the tRNA processing phenotypes reported here appear to be specific for lsm mutants, since they were not observed in strains deficient in either mRNA splicing or mRNA degradation. The function of Lsm proteins in pre-tRNA processing is further supported by two-hybrid interactions (13, 48) that were reported between Lsm8p and the putative RNA helicase Sen1p, which acts as a positive effector of the tRNA splicing endonuclease (10, 36, 49), and between Lsm2p and Tpt1p, the 2′-phosphotransferase that functions in tRNA splicing (9). These interactions would be consistent with the Lsm proteins promoting pre-tRNA splicing by aiding the recruitment of splicing cofactors.
We also observed aberrant 3′-extended pre-tRNAs in Lsm-depleted cells, which were substantially longer than the normal PT. These are likely to represent products of transcriptional read-through that would normally have been rapidly degraded in wild-type cells. A role for the essential Lsm proteins in tRNA degradation is supported by the observation of truncated tRNA fragments in depleted strains. It is currently unclear whether these fragments arise from tRNAs that would normally have been degraded in wild-type cells, but without clear intermediates, in which case the detection of degradation intermediates may reflect a loss of nuclease processivity. Alternatively, the absence of a functional Lsm complex may provoke tRNA degradation, perhaps due to problems in RNA folding that would otherwise have been corrected by an Lsm-associated chaperone activity. In other experiments, degradation of the mature rRNAs was also observed (J. Kufel, J. Beggs, and D. Tollervey, unpublished observations), indicating that this phenomenon is not restricted to tRNAs. In the case of mRNAs, the Lsm1p-Lsm7p complex is reported to confer protection against 3′ degradation by the cytoplasmic exosome, a complex of 3′→5′ exonucleases (19), while promoting 5′ degradation by the 5′→3′ exonuclease Xrn1p (45). Two-hybrid interactions have been reported between Lsm2p and Lsm8p and the exosome component Mtr3p and between Lsm2p, Lsm4p, Lsm8p, and Xrn1p (13, 48), whereas proteomic analysis revealed association of Lsm8p with another component of the exosome, Rrp42p (22). These are consistent with direct association of the Lsm2p-Lsm8p complex with the RNA degradation machinery. Moreover, defects in 5.8S rRNA synthesis seen in strains depleted of any of the essential Lsm proteins are consistent with reduced processivity of both the exosome and the 5′→3′ exonucleases Rat1p and Xrn1p (J. Kufel, J. Beggs, and D. Tollervey, unpublished observations).
An alternative explanation for the pre-tRNA accumulation seen in the Lsm protein-depleted strains might be that the accumulated species are misfolded or otherwise defective pre-tRNAs that would otherwise have been targeted for rapid degradation. We think that this is less likely, since it would imply a substantial level of pre-tRNA degradation in wild-type cells. Moreover, a similar phenotype was not seen in strains lacking components of the exosome complex (unpublished observations).
Pre-tRNAs also undergo extensive covalent nucleotide modification, and a large number of different modified nucleotides have been identified (39) which play important roles in tRNA function (reviewed in several chapters of reference 18). At least in some cases there is evidence for interactions among the tRNA modification, splicing, and export pathways. The requirements for Lsm proteins in pre-tRNA localization, tRNA modification, and export have not yet been addressed.
Interactions between Lsm complexes and Lhp1p.
All RNA PolIII PT are believed to associate with La/Lhp1p, via their 3′ poly(U) tracts. La (and presumably Lhp1p) binds to 3′-terminal poly(U) tracts in vitro (43), but efficient binding to complex RNP substrates in vivo may be more dependent on cofactors. The binding of Lhp1p to large RNA PolIII transcripts, scR1 and the pre-P RNA, was drastically reduced (76- and 18-fold, respectively) by depletion of Lsm3p or Lsm5p for 24 h, with 11- and 5.5-fold reduction after just 8.5 h. The association of Lhp1p with smaller PolIII transcribed RNAs, pre-5S and tRNA PT, was reduced to a lesser extent, with a threefold reduction following Lsm depletion for 24 h. The synthetic lethality of mutations in LSM5 to LSM8 in combination with lhp1-Δ (33) supports their functional interaction.
The less efficient association between Lhp1p and RNAs observed in the absence of Lsm3p and Lsm5p was not due to depletion of Lhp1p, the level of which was unaltered, or to nonspecific inhibition of RNA binding, since the precipitation of the U6 snRNA was substantially increased following Lsm depletion. Lhp1p has been proposed previously to “hand on” U6 to the Lsm2p-Lsm8p complex (21), and in the absence of Lsm proteins, Lhp1p may remain associated with U6 for a longer period.
A correlation was seen between the pre-tRNAs that were coprecipitated with Lsm3p-TAP and the pre-tRNAs that accumulated in the absence of Lsm proteins, consistent with a direct role for the Lsm complex in their processing. However, the low coprecipitation efficiency suggests that the association of Lsm complexes with tRNA precursors is transient. This, and the reduction of Lhp1p binding to its substrates in the absence of Lsm proteins, is consistent with an Lsm complex functioning as a chaperone in the assembly of pre-tRNA/protein complexes. Potential roles would include facilitating the interaction of pre-tRNAs with processing enzymes and cofactors, including Lhp1p, and/or increasing their specific activities.
Acknowledgments
We thank Phil Mitchell for critical reading of the manuscript.
This work was supported by the Wellcome Trust.
REFERENCES
- 1.Achsel, T., H. Brahms, B. Kastner, A. Bachi, M. Wilm, and R. Lührmann. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18:5789-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achsel, T., H. Stark, and R. Lührmann. 2001. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl. Acad. Sci. USA 98:3685-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltrame, M., and D. Tollervey. 1992. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 11:1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeck, R., B. Lapeyre, C. E. Brown, and A. B. Sachs. 1998. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol. 18:5062-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouveret, E., G. Rigaut, A. Shevchenko, M. Wilm, and B. Séraphin. 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19:1661-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers, J. C., M. G. Kurilla, and J. D. Keene. 1983. Association between the 7S RNA and the lupus La protein varies among cell types. J. Biol. Chem. 258:11438-11441. [PubMed] [Google Scholar]
- 7.Collins, B. M., S. J. Harrop, G. D. Kornfeld, I. W. Dawes, P. M. Curmi, and B. C. Mabbutt. 2001. Crystal structure of a heptameric Sm-like protein complex from Archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol. 309:915-923. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, M., L. H. Johnston, and J. Beggs. 1995. Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 14:2066-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Culver, G. M., S. M. McCraith, S. A. Consaul, D. R. Stanford, and E. M. Phizicky. 1997. A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J. Biol. Chem. 272:13203-13210. [DOI] [PubMed] [Google Scholar]
- 10.DeMarini, D. J., M. Winey, D. Ursic, F. Webb, and M. R. Culbertson. 1992. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2154-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelke, D. R., P. Gegenheimer, and J. Abelson. 1985. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J. Biol. Chem. 260:1271-1279. [PubMed] [Google Scholar]
- 12.Frendewey, D., T. Dingermann, L. Cooley, and D. Soll. 1985. Processing of precursor tRNAs in Drosophila. Processing of the 3′ end involves an endonucleolytic cleavage and occurs after 5′ end maturation. J. Biol. Chem. 260:449-454. [PubMed] [Google Scholar]
- 13.Fromont-Racine, M., A. E. Mayes, A. Brunet-Simon, J. C. Rain, A. Colley, I. Dix, L. Decourty, N. Joly, F. Ricard, J. D. Beggs, and P. Legrain. 2000. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast 17:95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furter, R., M. Snaith, D. E. Gillespie, and B. D. Hall. 1992. Endonucleolytic cleavage of a long 3′-trailer sequence in a nuclear yeast suppressor tRNA. Biochemistry 31:10817-10824. [DOI] [PubMed] [Google Scholar]
- 15.Gavin, A.-C., M. R. Bösche, O. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A.-M. Michon, C.-M. Cruciat, M. Remor, C. Höfert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M.-A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Séraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 16.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficient transformation of intact yeast cells. Nucleic Acids Res. 20:1425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottschalk, A., G. Neubauer, J. Banroques, M. Mann, R. Lührmann, and P. Fabrizio. 1999. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 18:4535-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grosjean, H., and R. Benne. 1998. Modification and editing of RNA. ASM Press, Washington, D.C.
- 19.He, W., and R. Parker. 2001. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics 158:1445-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermann, H., P. Fabrizio, V. A. Raker, K. Foulaki, H. Horning, H. Brahms, and R. Lührmann. 1995. SnRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interaction. EMBO J. 14:2076-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herschlag, D. 1995. RNA chaperones and the RNA folding problem. J. Biol. Chem. 270:20871-20874. [DOI] [PubMed] [Google Scholar]
- 22.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S.-L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schaddorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickowa, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sørensen, J. Matthiesen, R. S. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. V. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 23.Kambach, C., S. Walke, R. Young, J. M. Avis, E. de la Fortelle, V. A. Raker, R. Lührmann, J. Li, and K. Nagai. 1999. Crystal structure of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 5:375-387. [DOI] [PubMed] [Google Scholar]
- 24.Kufel, J., B. Dichtl, and D. Tollervey. 1999. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lafontaine, D., and D. Tollervey. 1996. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 24:3469-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long, K. S., T. Cedervall, C. Walch-Solimena, D. A. Noe, M. J. Huddleston, R. S. Annan, and S. L. Wolin. 2001. Phosphorylation of the Saccharomyces cerevisiae La protein does not appear to be required for its functions in tRNA maturation and nascent RNA stabilization. RNA 7:1589-1602. [PMC free article] [PubMed] [Google Scholar]
- 27.Longtine, M. S., A. R. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 28.Lygerou, Z., P. Mitchell, E. Petfalski, B. Séraphin, and D. Tollervey. 1994. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 8:1423-1433. [DOI] [PubMed] [Google Scholar]
- 29.Mayes, A. E., L. Verdone, P. Legrain, and J. D. Beggs. 1999. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 18:4321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moller, T., T. Franch, P. Hojrup, D. R. Keene, H. P. Bachinger, R. G. Brennan, and P. Valentin-Hansen. 2002. Hfq. A bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9:23-30. [DOI] [PubMed] [Google Scholar]
- 31.Mura, C., D. Cascio, M. R. Sawaya, and D. S. Eisenberg. 2001. The crystal structure of a heptameric archaeal Sm protein: implications for the eukaryotic snRNP core. Proc. Natl. Acad. Sci. USA 98:5531-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Connor, J. P., and C. L. Peebles. 1991. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:425-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pannone, B. K., S. Do Kim, D. A. Noe, and S. L. Wolin. 2001. Multiple functional interactions between components of the Lsm2-Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics 158:187-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pannone, B. K., D. Xue, and S. L. Wolin. 1998. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 17:7442-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 36.Rasmussen, T. P., and M. R. Culbertson. 1998. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6885-6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinke, J., and J. A. Steitz. 1985. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res. 13:2617-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rinke, J., and J. A. Steitz. 1982. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell 29:149-159. [DOI] [PubMed] [Google Scholar]
- 39.Rozenski, J., P. F. Crain, and J. A. McCloskey. 1999. The RNA Modification Database: 1999 update. Nucleic Acids Res. 27:196-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salgado-Garrido, J., E. Bragado-Nilsson, S. Kandels-Lewis, and B. Séraphin. 1999. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 18:3451-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Séraphin, B. 1995. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 14:2089-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Séraphin, B., and M. Rosbash. 1989. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59:349-358. [DOI] [PubMed] [Google Scholar]
- 43.Stefano, J. E. 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36:145-154. [DOI] [PubMed] [Google Scholar]
- 44.Stevens, S. W., and J. Abelson. 1999. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl. Acad. Sci. USA 96:7226-7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tharun, S., W. He, A. E. Mayes, P. Lennertz, J. D. Beggs, and R. Parker. 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404:515-518. [DOI] [PubMed] [Google Scholar]
- 46.Tollervey, D. 1987. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 6:4169-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Törö, I., S. Thore, C. Mayer, J. Basquin, B. Séraphin, and D. Suck. 2001. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 20:2293-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockson, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 49.Ursic, D., K. L. Himmel, K. A. Gurley, F. Webb, and M. R. Culbertson. 1997. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 25:4778-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venema, J., and D. Tollervey. 1996. RRP5 is required for the formation of both 18S and 5.8S rRNA in yeast. EMBO J. 15:5701-5714. [PMC free article] [PubMed] [Google Scholar]
- 51.Yoo, C. J., and S. L. Wolin. 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89:393-402. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9:11-22. [DOI] [PubMed] [Google Scholar]