Abstract
Mammalian DNA polymerase μ (pol μ) is related to terminal deoxynucleotidyl transferase, but its biological role is not yet clear. We show here that after exposure of cells to ionizing radiation (IR), levels of pol μ protein increase. pol μ also forms discrete nuclear foci after IR, and these foci are largely coincident with IR-induced foci of γH2AX, a previously characterized marker of sites of DNA double-strand breaks. pol μ is thus part of the cellular response to DNA double-strand breaks. pol μ also associates in cell extracts with the nonhomologous end-joining repair factor Ku and requires both Ku and another end-joining factor, XRCC4-ligase IV, to form a stable complex on DNA in vitro. pol μ in turn facilitates both stable recruitment of XRCC4-ligase IV to Ku-bound DNA and ligase IV-dependent end joining. In contrast, the related mammalian DNA polymerase β does not form a complex with Ku and XRCC4-ligase IV and is less effective than pol μ in facilitating joining mediated by these factors. Our data thus support an important role for pol μ in the end-joining pathway for repair of double-strand breaks.
DNA polymerase μ (pol μ) is a recently described DNA polymerase that shows strong similarity in sequence (approximately 40% identity) and domain organization to terminal deoxynucleotidyl transferase (TdT) (1, 6). Together with the less closely related pol β, pol λ, and pol σ, these polymerases comprise the mammalian pol X family and thus possess similar carboxy-terminal domains with deoxynucleotidyl transferase activity. TdT, pol μ, and pol λ (but not pol β or pol σ) also possess an amino-terminal BRCA1 C terminus (BRCT) domain (1, and 6).
The biological role of pol μ is not yet clear (reviewed in references 27 and 31). A possible connection to somatic hypermutation of immunoglobulin genes has been suggested because pol μ is error prone under certain conditions and is expressed at high levels in the germinal centers of peripheral lymph nodes, where somatic hypermutation occurs (6). However, low levels of pol μ mRNA expression are observed in almost all cell types (1, 6), indicating that it may have a more general role in DNA metabolism.
In contrast to pol μ, TdT has a clear biological role. TdT expression is lymphoid restricted and contributes to antigen receptor diversity during V(D)J recombination (reviewed in reference 11). V(D)J recombination is a lymphoid-restricted genetic rearrangement required to assemble individual coding segments into mature antigen receptors (10). Recombination is initiated by introduction of DNA double-strand breaks (DSBs) adjacent to these coding segments. Resolution of these broken recombination intermediates requires factors implicated in the nonhomologous end-joining (referred to below as end-joining) pathway for general repair of DSBs, including the Ku heterodimer (Ku 70 and Ku 80), DNA-dependent protein kinase catalytic subunit (DNA-PKcs), and the XRCC4-ligase IV complex (X4-LIV). During V(D)J recombination, TdT randomly adds nucleotides to the ends of coding segments prior to their resolution (N-addition). N-addition in V(D)J recombination requires both Ku and DNA-PKcs (3, 26) as well as TdT. Association of TdT with Ku has also been observed in cells (15), suggesting that TdT is recruited to this pathway by Ku.
Similarity between TdT and pol μ in conjunction with the clear involvement of TdT in end-joining DSB repair during V(D)J recombination suggests that pol μ might also be involved in end joining. However, pol μ has a wider expression pattern than TdT and is a template-dependent polymerase. pol μ might therefore act more generally in end-joining DSB repair, much as has been observed for POL4, the only pol X family member in Saccharomyces cerevisiae (37). Mammalian end-joining assays both in cells and in cell extracts have demonstrated that ends with noncomplementary and partially complementary overhangs are most often repaired such that these overhangs are retained (5, 8, 13, 23, 30, 34, 36). DNA ends are aligned using whatever homology (if any) can be found within the overhangs, and resulting gaps are repaired by a DNA polymerase. This results in a reduced loss of flanking sequence at sites of DSB repair and requires polymerase activity in addition to core end-joining factors (Ku and XRCC4) in S. cerevisiae (37), mammalian cells (13), and mammalian cell extracts (5, 8).
Recent work indicates that pol μ may be uniquely well suited for this function. pol μ can realign primers with terminal mismatches by looping out any mismatched template nucleotide(s) (38). Therefore, pol μ may be able to direct synthesis at aligned partially complementary ends even when complementarity does not extend to the primer terminus, greatly increasing the spectrum of ends that can be joined by this pathway.
We report here evidence that pol μ is indeed involved in end-joining DSB repair. Cells respond to exogenous DSB-inducing agents both by increasing pol μ expression levels and by localization of pol μ to discrete nuclear foci. pol μ also forms a complex with the end-joining factors Ku and X4-LIV in a manner very similar to that of TdT, and this complex facilitates the ability of X4-LIV and Ku to join ends with partially complementary overhangs in vitro.
MATERIALS AND METHODS
Expression constructs.
The pol μ cDNA was amplified from a human thymus cDNA library (Clontech) using the N-terminal primer 5′ CCCCCATATGCTCCCCAAACGGCGGCGAG 3′ and the C-terminal primer 5′ CCCCTCGAGTCAGGCGTTTCTCTGCTCTGGAGGAAGG 3′. This product was introduced into pET28b (Novagen) to produce an N-terminal hexahistidine-tagged bacterial expression construct (pMH3) and was verified by sequence analysis to be identical in coding sequence to the previously reported pol μ sequence (6). A construct to express a pol μ-EGFP (enhanced green fluorescent protein) fusion protein in mammalian cells was then made by amplifying the coding sequence from pMH3 using primers 5′ CTTCGAATTCATGCACCACCACCACCACCACATGCTCCCCAAACGGCGGCGAGC 3′ and 5′ TTCGTCGACTGGGCGTTTCTCTGCTCTGGAGGAAGGT 3′ and inserting the PCR product into the mammalian expression vector pEGFPN1 (Clontech) to produce a pol μ coding sequence fused at its C terminus to the EGFP coding sequence.
A TdT cDNA (15) was amplified using the N-terminal primer 5′ ATGCTGCAGATGCACCACCACCACCACCACATGGATCCACCACGAGCG 3′ and the C-terminal primer 5′ ATGCTCGAGCTAGGCATTTCTTTCCCACGG 3′ to introduce a 5′ hexahistidine tag and was inserted into pFASTBAC1 (Invitrogen). This cDNA encodes a 1-amino-acid insertion (glutamine) at position 453 relative to the published human sequence. However, an analogous insertion is present in TdT sequences from almost all other species, arguing that the published form represents a rare polymorphism.
Cell culture.
Ramos cells (a human B lymphoma cell line) were maintained at 37°C in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. Human embryonic kidney (HEK) cells and Cos-7 cells were grown at 37°C in Dulbecco's modified Eagle medium supplemented with 10% FBS and penicillin-streptomycin.
Cells were either treated with 12 Gy of ionizing radiation (IR) delivered at approximately 1 Gy/min using a 60Co source, UV irradiated with 7 J/m2 at 0.86 J/m2/s, or grown in medium supplemented with 50 μM etoposide, 50 μg of mitomycin C/ml, or 50 μg of cisplatin/ml (Sigma-Aldrich). Cells were given a medium containing drugs for 1 h at 37°C, washed, returned to a standard medium, and harvested at the indicated times after treatment. The viability of the cells following each treatment was greater than 90%.
Immunological analysis and cellular localization.
A polyclonal antiserum to human pol μ was raised in rabbits by immunization with the peptide (CEEVERVRRSERYQT) and was affinity purified (Biogenes GmbH). This antiserum specifically recognized the 55-kDa human pol μ, as judged by Western blot analysis with both recombinant human antigen and a variety of human cell extracts, and did not cross-react with recombinant human TdT or pol β. Rabbit antisera raised against human XRCC4 (Serotec), human S15P p53 (Cell Signaling), and human γH2AX (Upstate Biotechnologies), as well as mouse monoclonal antibodies recognizing human Ku 80 (Ku15; Sigma), human Rad50 (2C6; Upstate Biotechnologies), human pol β (Neomarkers), and a pentahistidine epitope (Qiagen), were also used. A rhodamine-conjugated secondary antibody (Jackson Laboratories) was used to stain γH2AX foci.
For Western blot analysis, HEK cells were washed in phosphate buffered saline (PBS) and resuspended in extraction buffer (50 mM Tris [pH 7.5], 1 mM EDTA, 200 mM NaCl, 10% glycerol, 1% NP-40, and Complete protease inhibitor cocktail [Roche Biochemicals]). Cell extracts were sonicated for three 20-s pulses and then clarified by centrifugation at 15,000 × g for 15 min. One hundred micrograms of each clarified extract was electrophoresed on a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel and subjected to Western blot analysis using the primary antibodies described above, horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies, and an enhanced chemiluminescence (ECL) substrate (Amersham Pharmacia Biotech).
For cellular localization studies, Cos-7 cells grown on coverslips were transfected at 50% confluency as previously described (15). Cells were fixed with 4% paraformaldehyde in PBS for 10 min, washed with PBS, permeabilized with 0.2% Triton X-100, washed again with PBS, and incubated with 3% bovine serum albumin (BSA) and 10% FBS in PBS for 1 h. γH2AX was stained by incubation with a 1:400 dilution of an anti-γH2AX antibody for 45 min, followed by five 5-min PBS washes and subsequent incubation with a 1:400 dilution of a rhodamine-conjugated anti-rabbit antibody for 30 min, with a final 30-min PBS wash. Coverslips with fixed cells were mounted on slides in Vectashield mounting medium (Vector Laboratories), and red (γH2AX) or green (EGFP) fluorescence was detected using a Zeiss Axioskop microscope and charge-coupled device (CCD) camera with appropriate filters.
Immunoprecipitations shown in Fig. 3 were performed by using 200 μg of cell extract prepared as described above. Extracts were incubated overnight with each antibody at 4°C, followed by a 1-h incubation at 4°C with a 50% mixture of protein A- and protein G-Sepharose beads (Santa Cruz). The beads were washed four times with 1 ml of the extraction buffer. Recovered proteins were eluted by boiling in SDS-polyacrylamide gel electrophoresis sample buffer and were detected by Western blot analysis as described above.
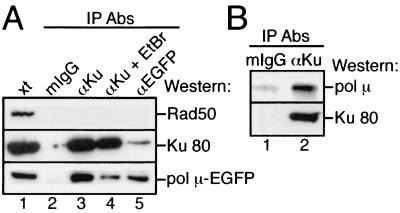
FIG. 3.
Association of pol μ with Ku. Protein complexes were immunoprecipitated (IP) from whole-cell extracts using mouse IgG (mIgG) or monoclonal antibodies (Abs) against Ku or EGFP. (A) Proteins immunoprecipitated from extracts (xt) from pol μ-EGFP-transfected HEK cells were characterized by Western blot analysis with anti-Rad50 (upper panel), anti-Ku 80 (middle panel) or anti-EGFP (lower panel) antibodies. When present, ethidium bromide (EtBr) was added to a concentration of 100 μg/ml. (B) Immunoprecipitated proteins from Ramos cell extracts were characterized by Western blot analysis with anti-pol μ antibodies (upper panel) or anti-Ku 80 antibodies (lower panel).
Immunoprecipitations shown in Fig. 6B were performed by mixing the indicated components, followed by incubation at 4°C for 30 min in a buffer containing 25 mM Tris (pH 7.5), 1 mM EDTA, 0.05% Triton X-100, 0.5 mg of BSA/ml, 100 mM NaCl, and 50 mM KCl. A polyclonal antiserum to XRCC4 (Serotec) was added as indicated, and reaction mixtures were incubated for 30 min at 4°C, followed by addition of 20 μl of a 50% protein A-Sepharose slurry and a final 1-h incubation at 4°C with constant mixing. Antibody-protein complexes were washed three times with a buffer containing 25 mM Tris (pH 7.5), 1 mM EDTA, 0.05% Triton X-100, and 100 mM NaCl; then they were washed one additional time in 10 mM Tris (pH 7.5)-1 mM EDTA. Recovered proteins were eluted by boiling in SDS-polyacrylamide gel electrophoresis sample buffer and detected by Western blot analysis as described above.
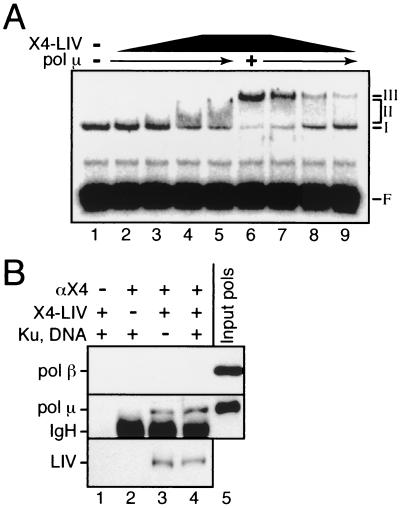
FIG. 6.
Association of pol μ with X4-LIV. (A) All reactions contained 90 nM 32P-labeled 60-bp DNA duplex and 5 nM Ku. F, free DNA probe; I, II, and III, species I, II, and III. pol μ (100 nM) was included as indicated (+). X4-LIV was added to a concentration of 25 nM (lanes 5 and 6), 10 nM (lanes 4 and 7), 5 nM (lanes 3 and 8), or 2.5 nM (lanes 2 and 9). (B) pol μ and pol β (each at 250 nM) were present in all immunoprecipitations. X4-LIV (50 nM), Ku (100 nM), and sheared calf thymus DNA (∼1 μM) were added as indicated (+). αX4, 1 μl of polyclonal anti-XRCC4 antisera. Input pols, a fraction of reaction input. Immunoprecipitated proteins were detected by Western blot analysis using an anti-pol β antibody (top panel) or an anti-histidine tag antibody (bottom two panels; hexahistidine tags are present on both ligase IV and pol μ). IgH, heavy chain of immunoprecipitating antibody (detected by a species cross-reaction of the anti-mouse Ig secondary antibody used for Western blot analysis).
Protein expression and purification.
pMH3 was introduced into BL21(DE3) pLysS (Novagen) cells, and a culture of these cells was grown to mid-log phase at 37oC. pol μ expression was induced by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the culture was grown for an additional 2 h at 30°C before harvesting.
A pellet from 1/2 liter of induced cells was then extracted in a buffer containing 50 mM NaPO4 (pH 8.0), 250 mM KCl, 10% glycerol, 0.2% Triton X-100, 10 mM imidazole, 7 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride (PMSF). The extract was clarified by centrifugation at 150,000 × g for 60 min, loaded onto a Ni-nitrilotriacetic acid (NTA) Superflow (Qiagen) column, and eluted with extraction buffer and 350 mM imidazole. pol μ-containing fractions from this column were pooled and loaded onto a gel filtration column equilibrated in 25 mM Tris (pH 8.0), 250 mM KCl, 10% glycerol, 0.05% Triton X-100, and 1 mM dithiothreitol (DTT). pol μ eluted as a single homogeneous species with a native molecular weight equivalent to a monomer (as determined by comparison to molecular weight standards) and was frozen in small aliquots and stored at −80°C.
A viral isolate for expression of the 5′ His-TdT pFASTBAC1 construct was prepared by the “Bac-to-Bac” method (Invitrogen). A pellet of approximately 2.5 × 108 Hi-5 cells (Invitrogen) infected with the 5′ His-TdT baculovirus was extracted in a buffer containing 50 mM NaPO4 (pH 8.0), 1 M KCl, 10% glycerol, 0.2% Triton X-100, 10 mM imidazole, 7 mM β-mercaptoethanol, 1 mM PMSF, 10 μM leupeptin, 1 μM pepstatin A, and 0.1 trypsin inhibitor unit (TIU) of aprotinin/ml. The extract was then clarified (20), loaded onto a Ni-NTA Superflow (Qiagen) column, washed with 4 column volumes of extraction buffer, and eluted with extraction buffer supplemented with 350 mM imidazole. TdT-containing fractions were first diluted 6.7-fold in a buffer containing 25 mM HEPES/KOH (pH 8.0), 10% glycerol, 0.2% Triton X-100, and 1 mM DTT and then loaded onto a Mono S cation exchange column (Amersham Pharmacia Biotech) equilibrated in 25 mM HEPES/KOH (pH 8.0), 150 mM KCl, 10% glycerol, 0.2% Triton X-100, and 1 mM DTT. The Mono S column was eluted with a linear gradient to 400 mM KCl over 20 column volumes, and TdT-containing fractions were pooled and frozen in small aliquots for storage at −80°C.
The cloning, expression, and purification of X4-LIV and Ku from insect cells has been described earlier (20). A TAP56 Escherichia coli strain to express human pol β was the gift of S. Wilson (National Institute of Environmental Health Sciences). pol β was purified from 1 liter of heat-induced cells as previously described (25).
Electrophoretic mobility shift assay (EMSA).
A radiolabeled 60-bp double-stranded (ds) DNA substrate was made by 5′ 32P end labeling DAR166 (5′-CAGCTGGGAATTCCATATGAGTACTGCAGATGCACTTGCTCGATAGATCTAACATGAGCC-3′) and annealing the labeled DNA to DAR167 (5′-GTAGGGCTCATGTTAGATCTATCGAGCAAGTGCATCTGCAGTACTCATATGGAATTCCCAGCTGAG-3′). The DNA was incubated with various protein combinations for 30 min at 4°C in a volume of 10 μl in standard reaction buffer (25 mM Tris [pH 8.0], 100 mM NaCl, 0.1 mM EDTA, 50 μg of BSA/ml, 0.05% Triton X-100, and 2 mM DTT). Samples were subjected to electrophoresis at 4°C for 40 min at 12 V/cm using a 3.5% polyacrylamide gel in a buffer containing 90 mM Tris-borate (pH 8.2) and 1 mM EDTA. The experiment for which results are shown in Fig. 5A was performed as described above, except that Ku and X4-LIV were incubated with the labeled DNA duplex for 10 min prior to the addition of polymerase, followed by an additional 30-min incubation on ice. The competition experiment for which results are shown in Fig. 5B was performed as described above, except that the polymerases were mixed prior to addition to the reaction mixture, which was incubated for 30 min at 4°C before addition of 0.2 μl of a mouse monoclonal antibody to TdT (clone 8-1 E4; Sigma), followed by a final 30-min incubation at 4°C.
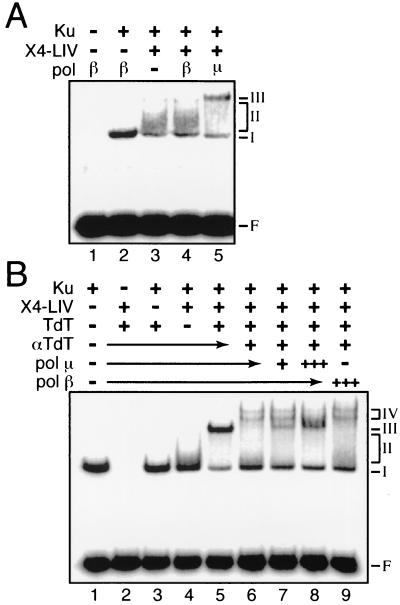
FIG. 5.
Participation of different pol X family members in complexes with end-joining factors. All reactions contained 90 nM 32P-labeled 60-bp DNA duplex. F, free DNA probe; I, II, III, and IV, species I, II, III, and IV. (A) pol β (100 nM), pol μ (100 nM), Ku (5 nM), and X4-LIV (25 nM) were included as indicated. (B) Ku (5 nM), X4-LIV (10 nM), TdT (25 nM), and α-TdT (0.2 μl) were added as indicated (+). pol μ or pol β was added at 25 nM (+) or 75 nM (+++).
Ligation assay.
The DSB substrate is a pair of dsDNA molecules: a 30-bp DNA duplex made by annealing 5′-32P-labeled SNM16 (5′-GATGTAATCCCTCGATGAGGTCTAGAACTGCAGTCTCA-3′) to 5′-phosphorylated DAR165 (5′-ACTGCAGTTCTAGACCTCATCGAGGGATTA-3′) and a 60-bp duplex made by annealing 5′-phosphorylated SNM15 (5′-CTGGGAATTCCATATGAGTACTGCAGATGCACTTGCTCGATAGATCTAACATGAGCC-3′) to DAR167. The gapped substrate (Gap) is made by annealing 5′-32P-labeled SNM16 and 5′-phosphorylated SNM15 to SNM17 (5′-GTAGGGCTCATGTTAGATCTATCGAGCAAGTGCATCTGCAGTACTCATATGGAATTCCCAGCTGAGACTGCAGTTCTAGACCTCATCGAGGGATTA-3′).
Ku and X4-LIV were preincubated with the DNA substrates in standard reaction buffer supplemented with 10% (wt/vol) polyethylene glycol (molecular mass, >8,000 kDa) and 25 μM each deoxynucleoside triphosphate for 15 min. Competitor (when present) and polymerase were then added, followed by an additional 15 min of preincubation. pol μ and pol β were added at the amounts indicated in the legend to Fig. 7. For T4 bacteriophage DNA polymerase or the Klenow fragment of E. coli polymerase I (both from New England Biolabs), we first determined that 1.5 × 10−3 U (1.5 mU) of these enzymes had activity equivalent to that of pol μ on the gap repair substrate; then we compared the effectiveness of this amount of these prokaryotic polymerases to that of pol μ in DSB repair reactions (S. A. Nick McElhinny and D. A. Ramsden, unpublished data). Reactions were initiated by addition of 5 mM Mg2+ and 0.1 mM ATP and were transferred to 37°C. Reactions were terminated and analyzed by denaturing gel electrophoresis as previously described (20). Radiolabeled DNA was detected and quantified with a PhosphorImager and ImageQuanNT software (Molecular Dynamics).
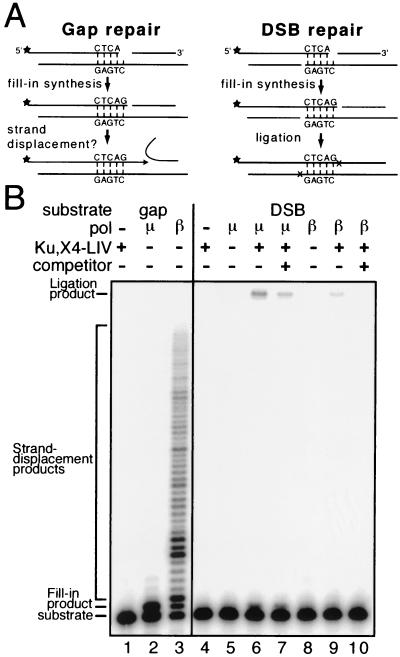
FIG. 7.
Relative activities of polymerases in gap and break repair. (A) Oligonucleotide sequences and substrate assembly as described in Materials and Methods. Stars indicate locations of 32P label. (B) 32P-labeled substrates at 5 nM (see panel A) were present in all reactions. pol μ (25 nM) or pol β (25 nM), Ku (5 nM), X4-LIV (50 nM), and salmon sperm DNA (1.5 μM) were added as noted. The reaction time was 1 min.
RESULTS
pol μ is part of the cellular response to DSBs.
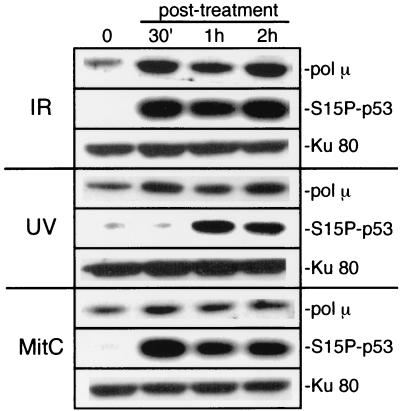
To determine whether pol μ is part of the cellular response to DNA damage, we first assessed pol μ protein expression in HEK cells following exposure to various DNA-damaging agents. We observed a three- to fivefold induction in levels of pol μ protein within 30 min of treatment with IR, a DNA DSB-inducing agent (Fig. 1, IR). pol μ was also induced upon exposure of a lymphoid cell line to IR (K. N. Mahajan and B. S. Mitchell, unpublished data). This finding is in contrast to previous work which argued that levels of properly spliced pol μ mRNA were reduced after exposure to this stimulus (1), and it raises the possibility that pol μ expression is regulated at the posttranscriptional level and that this regulation overcomes a reduction in properly spliced transcripts. Induction of pol μ expression was concurrent with induction of phosphorylation of p53 at serine 15, a previously described cellular response to DNA damage (32, 33) (Fig. 1, IR). The levels of another factor already implicated in repair of IR-induced breaks, Ku 80, remained essentially unchanged (Fig. 1, IR). Only low levels of induction were observed following treatment of HEK cells with UV radiation (Fig. 1, UV), and levels of pol μ protein remained relatively unchanged after treatment with DNA cross-linking agents such as mitomycin C (Fig. 1, MitC) or cisplatin (Mahajan and Mitchell, unpublished) at 50 μg/ml.
FIG. 1.
Effects of DNA-damaging agents on pol μ expression. HEK cells were exposed to either 12 Gy of IR, 7 J of UV irradiation/m2, or 50 μg of mitomycin C (MitC)/ml. Whole-cell extracts were prepared from cells that were left untreated (0) or treated for the indicated times. The blot was probed with anti-pol μ, anti-phosphoserine 15-p53 (S15P-p53), and anti-Ku 80 antibodies.
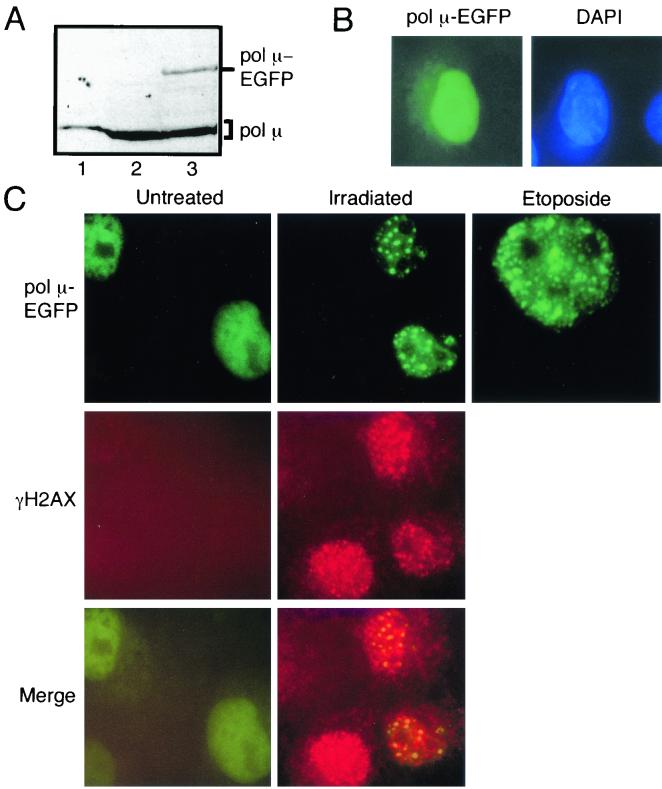
Factors required for DSB repair frequently form nuclear foci at sites of DNA damage (12, 16, 19). A construct expressing a pol μ-EGFP fusion protein was therefore used to determine if pol μ formed similar foci in response to IR. The transfected pol μ-EGFP fusion protein was observed in 50 to 80% of all cells (determined by GFP fluorescence), and Western blot analysis of bulk-transfected cells (Fig. 2A) determined that after correction for transfection efficiency, cellular levels of this protein are comparable to those of endogenous pol μ. The pol μ-EGFP fusion protein was also effectively localized to the nucleus (Fig. 2B). Treatment of pol μ-EGFP-expressing cells with two DSB-inducing agents, IR and etoposide, induced formation of discrete nuclear foci within 30 min (Mahajan and Mitchell, unpublished), and these foci increased in number until 2 h after treatment (Fig. 2C, pol μ-EGFP). Foci were not observed in untreated pol μ-EGFP-expressing cells (Fig. 2C) or in treated cells expressing only the EFGP portion of the fusion protein (Mahajan and Mitchell, unpublished).
FIG. 2.
Cellular localization of pol μ. (A) Levels of the pol μ-EGFP fusion protein and those of endogenous pol μ were compared by Western blot analysis, using the pol μ-specific antibody, of 100 μg of extract from HEK cells transfected either with the vector alone (lane 2) or with pol μ-EGFP (lane 3). Fifty nanograms of recombinant pol μ is shown as a marker (lane 1); note that recombinant pol μ migrates slightly more slowly than endogenous pol μ due to the hexahistidine tag. (B) The cellular distribution of the pol μ-EGFP fusion protein was determined by comparing GFP fluorescence to 4′,6′-diamidino-2-phenylindole (DAPI) staining (used to delineate nuclei) in cells transfected with pol μ-EGFP. (C) Cells transfected with pol μ-EGFP were assessed for GFP fluorescence (pol μ-EGFP) without treatment (Untreated), 2 h after treatment with IR (Irradiated), or 8 h after treatment with etoposide (Etoposide). Also shown are untreated and irradiated cells stained in parallel with antibodies to γH2AX (stained red by a rhodamine-conjugated secondary antibody), as well as merged images of pol μ-EGFP and γH2AX fluorescence (Merge).
The histone variant H2AX is phosphorylated at serine 139 in response to IR (29). Foci of this form of H2AX, termed γH2AX, have previously been shown to be specific for sites of DSBs in irradiated cells (28). Parallel immunofluorescence for γH2AX focus formation in pol μ-EGFP-transfected cells indicates that IR-dependent γH2AX foci are also formed under these conditions (Fig. 2C, γH2AX). Most importantly, the majority (>60%) of pol μ-EGFP foci are coincident with γH2AX foci (Fig. 2C, Merge). Cells therefore respond to DSB-inducing agents both with increased levels of pol μ protein and with frequent localization of pol μ to foci that also contain a marker of DSB sites.
pol μ associates with factors required for end-joining DSB repair.
This evidence argues that pol μ is part of the cellular response to DSB-inducing agents and suggests a role in DSB repair. Because the related polymerase TdT associates with Ku (15), we asked whether pol μ might similarly associate with Ku, thus implicating this factor specifically in the Ku-dependent end-joining pathway for DSB repair. Ku and pol μ-EGFP were coimmunoprecipitated from pol μ-EGFP-transfected cells with antibodies to either Ku or EGFP but not with irrelevant mouse immunoglobulin G (IgG) control antibodies (Fig. 3A; compare lanes 3 and 5 to lane 2). To determine if this association is DNA dependent, we treated extracts with 100 μg of ethidium bromide/ml, which disrupts the ability of Ku to interact with DNA (14). Ethidium bromide treatment reduced, but did not eliminate, recovery of pol μ-EGFP after immunoprecipitation of Ku (Fig. 3A; compare lane 3 to lane 4). Similar effects of ethidium bromide treatment are observed in coimmunoprecipitations of Ku with the end-joining factors DNA-PKcs (35) and ligase IV (20). Association of pol μ-EGFP and other end-joining factors with Ku is therefore stimulated in the presence of DNA, but these associations are not wholly dependent on DNA. Another DNA-binding protein, Rad50, was not coimmunoprecipitated with either Ku or pol μ-EGFP under these conditions (Fig. 3A), nor was Ku coimmunoprecipitated with antibodies to EGFP in cells expressing only the EGFP portion of the fusion protein (Mahajan and Mitchell, unpublished), further demonstrating the specificity of this interaction. Endogenous pol μ was also coimmunoprecipitated with Ku in untransfected cells (Fig. 3B; compare lane 1 to lane 2), indicating that association of pol μ with Ku is not dependent on the presence of the EGFP tag.
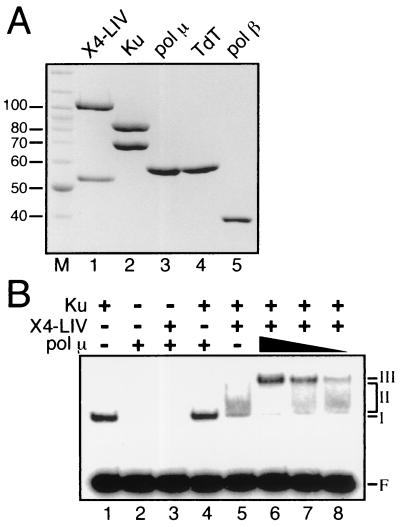
To further characterize the interaction between pol μ and end-joining factors, an EMSA was performed using purified recombinant proteins (Fig. 4A) and a 32P-labeled 60-bp DNA duplex probe. A DNA-protein complex with retarded mobility was detected upon addition of Ku alone (Fig. 4B, lane 1, species I) but not with pol μ alone (Fig. 4B, lane 2), X4-LIV alone (20), or pol μ and X4-LIV (Fig. 4B, lane 3). Similarly, addition of pol μ does not alter the mobility of Ku-bound DNA (Fig. 4B, lane 4). Association of pol μ with Ku alone is thus apparently insufficient for stable recruitment of pol μ to DNA in EMSAs, which is likely indicative of a greater stringency of these assays relative to coimmunoprecipitations (Fig. 3) under the conditions used here. A previously characterized complex formed between X4-LIV and Ku-bound DNA is detectable but is also unstable under the conditions and with the X4-LIV concentration (25 nM) used in this assay (Fig. 4B, lane 5, species II) (20). However, addition of pol μ (100 nM) to reactions containing both Ku and X4-LIV resulted in formation of a novel, more stable species with reduced mobility (Fig. 4B, lane 6, species III) compared to that of the species containing only Ku and X4-LIV. This end-joining complex, containing Ku, X4-LIV, and pol μ, is present even at low concentrations of pol μ (25 nM [Fig. 4B, lane 8]). pol μ is thus stably recruited to DNA only in the presence of both Ku and X4-LIV.
FIG. 4.
Protein-DNA complexes involving pol μ, Ku, and X4-LIV. (A) Coomassie blue-stained SDS-PAGE gel of purified recombinant proteins. Lane M, marker; lane 1, 4 μg of X4-LIV; lane 2, 4 μg of Ku heterodimer; lane 3, 2 μg of pol μ; lane 4, 2 μg of TdT; lane 5, 2 μg of pol β. (B) All reactions contained 90 nM 32P-labeled 60-bp DNA duplex. F, free DNA probe; I, II, and III, species I, II, and III. Ku (5 nM) and X4-LIV (25 nM) were included as indicated (+). pol μ was added at a concentration of 100 nM (lanes 2, 3, 4, and 6), 50 nM (lane 7), or 25 nM (lane 8).
The specificity for pol μ in the formation of this complex was then assessed by comparison to parallel experiments using purified TdT and pol β, two other pol X family members. Under the conditions used in this assay, pol β is unable to bind to DNA on its own (Fig. 5A, lane 1), in a complex with Ku (Fig. 5A, lane 2), or, in contrast to pol μ, even in a complex with Ku and X4-LIV (Fig. 5A; compare lane 3 to lanes 4 and 5). However, TdT behaves identically to pol μ in our gel shift assay, as it also forms a stable complex on DNA only in the presence of both Ku and X4-LIV (Fig. 5B, lane 5, species III). This species could be further retarded by antibodies to TdT (Fig. 5B, lane 6, species IV), Ku, or X4-LIV (Nick McElhinny and Ramsden, unpublished), confirming the presence of all three of these components in species III.
We performed a competition assay to more quantitatively compare the abilities of different pol X family members to participate in complexes with Ku and X4-LIV. An antibody to TdT supershifts the TdT-Ku-X4-LIV complex (Fig. 5B, species IV), allowing us to distinguish complexes containing TdT from complexes containing pol μ (note the appearance of species III in addition to species IV in Fig. 5B, lane 7). Although the two complexes are apparent when TdT and pol μ are present in equimolar amounts (Fig. 5B, lane 7), when a threefold excess of pol μ over TdT is present, species IV (α-TdT-TdT-Ku-X4-LIV-DNA) is largely converted to species III (pol μ-Ku-X4-LIV-DNA) (Fig. 5B; compare lane 6 to lane 8). This is consistent with recruitment of pol μ in place of TdT to Ku- and X4-LIV-bound DNA. In contrast, pol β is unable to compete with TdT for Ku- and X4-LIV-bound DNA (Fig. 5B; compare lane 6 to lane 9). The ability of pol μ to readily compete with TdT suggests that these two polymerases have similar affinities for Ku- and X4-LIV-bound DNA. If pol μ is also expressed with TdT in lymphocyte precursors, pol μ should therefore compete with TdT for ends in these cells (barring a contribution from additional interactions mediated by other cellular factors not tested here).
pol μ and TdT both appear to stabilize the interaction between X4-LIV and Ku-bound DNA (Fig. 4B, compare lane 5 to lane 6; Fig. 5B, compare lane 4 to lane 5). This was explored further by comparing, in the presence and absence of pol μ, the abilities of X4-LIV to form a complex with Ku at different X4-LIV concentrations. Efficient recruitment of X4-LIV to Ku-bound DNA in the absence of pol μ requires more than 25 nM X4-LIV (Fig. 6A, lane 5) (20), but in the presence of pol μ, recruitment of X4-LIV is readily observed with as little as 2.5 nM X4-LIV (Fig. 6A, lane 9). Similar results are observed when TdT is used in place of pol μ (Fig. 5B, lane 5) (Nick McElhinny and Ramsden, unpublished). pol μ and TdT thus improve the affinity of X4-LIV for Ku-bound DNA more than 10-fold, and we demonstrated above that X4-LIV is required for stable recruitment of pol μ or TdT to Ku-bound DNA (Fig. 4B and 5B). This suggested that TdT and pol μ might associate directly with X4-LIV. We confirmed this supposition by demonstrating that purified pol μ, but not pol β, could be coimmunoprecipitated with X4-LIV irrespective of the presence of Ku and DNA (Fig. 6B; compare lane 3 to lane 4). Similar results were obtained when TdT was substituted for pol μ (Nick McElhinny and Ramsden, unpublished), arguing that the ability to interact with X4-LIV is a specific property of the closely related pol X family members TdT and pol μ.
pol μ facilitates end joining in vitro.
To determine if pol μ could provide some functional advantage to DSB repair, we compared the activity of pol μ to that of pol β in activity assays. The intrinsic polymerase activity of pol β was greater than that of pol μ on all substrates tested, including a model gap repair substrate (Fig. 7A; Fig. 7B, compare lane 2 to lane 3) and other substrates containing larger gaps or recessed ends (Nick McElhinny and Ramsden, unpublished). Interestingly, pol μ was also generally less capable than pol β of extending synthesis beyond a gap, reflecting a greatly reduced ability to displace the downstream strand (Fig. 7B; compare lane 2 to lane 3).
We next assessed the relative activities of these polymerases in an end-joining assay, performed in the presence of Ku and X4-LIV. This substrate (Fig. 7A, DSB repair) possesses partially overlapping ends that, when aligned, produce a 1-nucleotide gap similar to that of the gap repair substrate, but in a context that tests the ability to couple gap repair mediated by the polymerase to end joining mediated by Ku and X4-LIV. Although our data suggest that pol μ is intrinsically a less active polymerase than pol β, pol μ was more than 5-fold (average from five replicate experiments, 7.2-fold ± 1.3-fold) more active than pol β in facilitating repair of the DSB repair substrate (Fig. 7B; compare lane 6 to lane 9). Strikingly, addition of competitor DNA abolished the activity of pol β on this substrate (Fig. 7B; compare lane 9 to lane 10), while the activity of pol μ was reduced only twofold (Fig. 7B; compare lane 6 to lane 7). No end-joining activity was detected in experiments using prokaryotic T4 bacteriophage polymerase or the Klenow fragment of E. coli DNA polymerase I, indicating that these polymerases were even less effective than pol β (Nick McElhinny and Ramsden, unpublished). Additional factors (e.g., DNA-PKcs and Artemis [18]) will further facilitate end joining in cells and may even help direct polymerase activity (5, 17). However, the complex described above (including only Ku, X4-LIV, and pol μ or TdT) already provides a clear functional advantage for pol μ over other DNA polymerases in coupling gap repair synthesis to end-joining activity, arguing that pol μ plays an important role in facilitating repair of partially overlapping ends in cells.
DISCUSSION
We have shown here that total cellular pol μ levels increase after exposure of cells to IR (Fig. 1). We have also shown that EGFP-tagged pol μ localizes to discrete nuclear foci upon treatment with either of two DSB-inducing agents, IR and etoposide (Fig. 2C). Moreover, after IR treatment, pol μ-EGFP foci are largely coincident with γH2AX foci. Formation of γH2AX foci has been correlated with sites of DSBs in IR-treated cells (28), is a prerequisite for focus formation by other repair factors (22), and facilitates DSB repair by the end-joining pathway in S. cerevisiae (7). Using both cell extracts and purified proteins, we further demonstrated that pol μ associates with factors required for repair of DSBs by the end-joining pathway (Fig. 3 and 6B), requires end-joining factors for stable recruitment to DNA (Fig. 4, 5, and 6), and facilitates the activity of end-joining factors in vitro (Fig. 7). Taken together, this evidence argues that pol μ is employed specifically in the end-joining pathway for repair of DSBs introduced by exogenous DNA-damaging agents. However, in light of the fact that focus formation was also observed in response to UV irradiation (Mahajan and Mitchell, unpublished), other DNA damage repair pathways may also recruit pol μ.
pol μ and TdT form stable complexes on DNA with Ku and X4-LIV that appear indistinguishable, with respect to both factor requirements and affinity (Fig. 5B). These complexes require X4-LIV, and the presence of either polymerase enhances the affinity of X4-LIV for Ku-bound DNA (Fig. 6A). These two effects are presumably due to the abilities of pol μ and TdT to interact with X4-LIV in the absence of Ku and DNA (Fig. 6B). Physical association of pol μ and TdT with X4-LIV will couple the catalytic activities of these factors; this may guard against unproductive polymerization during end joining by allowing ligation to occur immediately upon completion of the minimum synthesis required to form a ligation substrate. This association should increase both the speed and the accuracy of cellular end joining and is likely to be particularly important in controlling the template-independent activity of TdT. The advantages inherent in such an arrangement suggest that the activities of other processing factors during end joining may also be coupled to ligation activity in the same way.
In contrast to TdT and pol μ's apparently equivalent abilities to form complexes with end-joining factors, these polymerases clearly have distinct catalytic activities and cell type distributions. TdT is a template-independent polymerase found only in precursor lymphocytes and has been definitively linked to diversification during V(D)J recombination (11). On the other hand, pol μ has a much wider tissue distribution (possibly ubiquitous) and is template dependent (6), arguing for a more general role in end joining. Assays for end joining in end-joining-proficient cells and cell extracts have determined that a template-dependent polymerase is frequently employed to fill in gaps when noncomplementary or partially complementary ends are joined, enabling joining of such ends with minimal loss of sequence (5, 8, 13, 23, 30, 34, 36). The results of our in vitro end-joining assay (Fig. 7) are consistent with pol μ being well suited to fulfill this role. However, there may be some level of redundancy with other DNA polymerases (e.g., pol α [24] and pol λ [9]) in mammals.
In addition to assisting in repair of DSBs induced by exogenous DNA-damaging agents, pol μ likely also participates in repair of DSBs that are intermediates in the normal cellular processes of V(D)J recombination and immunoglobulin class switching. Our data indicate that pol μ is an efficient competitor for TdT at DNA ends in vitro and, if coexpressed in developing lymphocytes, might help regulate the frequency or extent of TdT-mediated N-additions during V(D)J recombination.
pol μ has also been argued to play a role in somatic hypermutation (6, 31). Somatic hypermutation of immunoglobulin genes might also progress through a DSB intermediate (4, 21); thus, our data implicating pol μ in DSB repair might be construed as consistent with a role for pol μ in somatic hypermutation. However, DSBs in somatically hypermutating cells are not observed in the G1 phase of the cell cycle (21), when most end joining occurs, and mice with mutations in the end-joining factor DNA-PKcs have normal levels of somatic hypermutation (2). Further investigation is necessary to determine if pol μ contributes to somatic hypermutation and, if so, whether pol μ is linked to this process through the end-joining pathway.
Acknowledgments
K. N. Mahajan and S. A. Nick McElhinny contributed equally to this work.
We thank Melissa Hayden for help in cloning human pol μ, S. Wilson (NIEHS) for providing the pol β-expressing strain, R. Prasad (NIEHS) for advice in expression and purification of pol β, and B. Bagnell for image analysis. We also thank T. Kunkel (NIEHS), N. P. Mahajan (UNC), and members of the Ramsden and Mitchell laboratories for helpful discussions, as well as E. Perkins for critical reading of the manuscript.
This work was supported by Public Health Service grants RO1 CA 84442-01 to D.A.R. and RO1 CA34085 to B.S.M. K.N.M is supported by a Rothrock-Thomas Fund grant, and S.A.N.M. is supported by a National Science Foundation Graduate Research Fellowship.
REFERENCES
- 1.Aoufouchi, S., E. Flatter, A. Dahan, A. Faili, B. Bertocci, S. Storck, F. Delbos, L. Cocea, N. Gupta, J. C. Weill, and C. A. Reynaud. 2000. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. 28:3684-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bemark, M., J. E. Sale, H. J. Kim, C. Berek, R. A. Cosgrove, and M. S. Neuberger. 2000. Somatic hypermutation in the absence of DNA-dependent protein kinase catalytic subunit (DNA-PK(cs)) or recombination-activating gene (RAG)1 activity. J. Exp. Med. 192:1509-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogue, M. A., C. Wang, C. Zhu, and D. B. Roth. 1997. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity 7:37-47. [DOI] [PubMed] [Google Scholar]
- 4.Bross, L., Y. Fukita, F. McBlane, C. Demolliere, K. Rajewsky, and H. Jacobs. 2000. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity 13:589-597. [DOI] [PubMed] [Google Scholar]
- 5.Chen, S., K. V. Inamdar, P. Pfeiffer, E. Feldmann, M. F. Hannah, Y. Yu, J. W. Lee, T. Zhou, S. P. Lees-Miller, and L. F. Povirk. 2001. Accurate in vitro end joining of a DNA double strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: effect of Ku on repair fidelity. J. Biol. Chem. 276:24323-24330. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez, O., J. F. Ruiz, T. Lain de Lera, M. Garcia-Diaz, M. A. Gonzalez, T. Kirchhoff, A. C. Martinez, A. Bernad, and L. Blanco. 2000. DNA polymerase μ (Pol μ), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 19:1731-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Downs, J. A., N. F. Lowndes, and S. P. Jackson. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408:1001-1004. [DOI] [PubMed] [Google Scholar]
- 8.Feldmann, E., V. Schmiemann, W. Goedecke, S. Reichenberger, and P. Pfeiffer. 2000. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 28:2585-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Diaz, M., K. Bebenek, R. Sabariegos, O. Dominguez, J. Rodriguez, T. Kirchhoff, E. Garcia-Palomero, A. J. Picher, R. Juarez, J. F. Ruiz, T. A. Kunkel, and L. Blanco. 2002. DNA polymerase λ, a novel DNA repair enzyme in human cells. J. Biol. Chem. 277:13184-13191. [DOI] [PubMed] [Google Scholar]
- 10.Gellert, M. 1997. Recent advances in understanding V(D)J recombination. Adv. Immunol. 64:39-64. [DOI] [PubMed] [Google Scholar]
- 11.Gilfillan, S., C. Benoist, and D. Mathis. 1995. Mice lacking terminal deoxynucleotidyl transferase: adult mice with a fetal antigen receptor repertoire. Immunol. Rev. 148:201-219. [DOI] [PubMed] [Google Scholar]
- 12.Haaf, T., E. I. Golub, G. Reddy, C. M. Radding, and D. C. Ward. 1995. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. USA 92:2298-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabotyanski, E. B., L. Gomelsky, J. O. Han, T. D. Stamato, and D. B. Roth. 1998. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 26:5333-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai, J. S., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA 89:6958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahajan, K. N., L. Gangi-Peterson, D. H. Sorscher, J. Wang, K. N. Gathy, N. P. Mahajan, W. H. Reeves, and B. S. Mitchell. 1999. Association of terminal deoxynucleotidyl transferase with Ku. Proc. Natl. Acad. Sci. USA 96:13926-13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maser, R. S., K. J. Monsen, B. E. Nelms, and J. H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17:6087-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mickelsen, S., C. Snyder, K. Trujillo, M. Bogue, D. B. Roth, and K. Meek. 1999. Modulation of terminal deoxynucleotidyltransferase activity by the DNA-dependent protein kinase. J. Immunol. 163:834-843. [PubMed] [Google Scholar]
- 18.Moshous, D., I. Callebaut, R. de Chasseval, B. Corneo, M. Cavazzana-Calvo, F. Le Deist, I. Tezcan, O. Sanal, Y. Bertrand, N. Philippe, A. Fischer, and J. P. de Villartay. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105:177-186. [DOI] [PubMed] [Google Scholar]
- 19.Nelms, B. E., R. S. Maser, J. F. MacKay, M. G. Lagally, and J. H. Petrini. 1998. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280:590-592. [DOI] [PubMed] [Google Scholar]
- 20.Nick McElhinny, S. A., C. M. Snowden, J. McCarville, and D. A. Ramsden. 2000. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 20:2996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papavasiliou, F. N., and D. G. Schatz. 2000. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature 408:216-221. [DOI] [PubMed] [Google Scholar]
- 22.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 23.Pfeiffer, P., S. Thode, J. Hancke, and W. Vielmetter. 1994. Mechanisms of overlap formation in nonhomologous DNA end joining. Mol. Cell. Biol. 14:888-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pospiech, H., A. K. Rytkonen, and J. E. Syvaoja. 2001. The role of DNA polymerase activity in human non-homologous end joining. Nucleic Acids Res. 29:3277-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasad, R., A. Kumar, S. G. Widen, J. R. Casas-Finet, and S. H. Wilson. 1993. Identification of residues in the single-stranded DNA-binding site of the 8-kDa domain of rat DNA polymerase β by UV cross-linking. J. Biol. Chem. 268:22746-22755. [PubMed] [Google Scholar]
- 26.Purugganan, M. M., S. Shah, J. F. Kearney, and D. B. Roth. 2001. Ku80 is required for addition of N nucleotides to V(D)J recombination junctions by terminal deoxynucleotidyl transferase. Nucleic Acids Res. 29:1638-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynaud, C. A., S. Frey, S. Aoufouchi, A. Faili, B. Bertocci, A. Dahan, E. Flatter, F. Delbos, S. Storck, C. Zober, and J. C. Weill. 2001. Transcription, beta-like DNA polymerases and hypermutation. Philos. Trans. R. Soc. Lond. B 356:91-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 30.Roth, D. B., and J. H. Wilson. 1986. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell. Biol. 6:4295-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz, J. F., O. Dominguez, T. Lain de Lera, M. Garcia-Diaz, A. Bernad, and L. Blanco. 2001. DNA polymerase μ, a candidate hypermutase? Philos. Trans. R. Soc. Lond. B 356:99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shieh, S. Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 33.Siliciano, J. D., C. E. Canman, Y. Taya, K. Sakaguchi, E. Appella, and M. B. Kastan. 1997. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 11:3471-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, J., C. Baldeyron, I. De Oliveira, M. Sala-Trepat, and D. Papadopoulo. 2001. The influence of DNA double-strand break structure on end-joining in human cells. Nucleic Acids Res. 29:4783-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suwa, A., M. Hirakata, Y. Takeda, S. A. Jesch, T. Mimori, and J. A. Hardin. 1994. DNA-dependent protein kinase (Ku protein-p350 complex) assembles on double-stranded DNA. Proc. Natl. Acad. Sci. USA 91:6904-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thode, S., A. Schafer, P. Pfeiffer, and W. Vielmetter. 1990. A novel pathway of DNA end-to-end joining. Cell 60:921-928. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, T. E., and M. R. Lieber. 1999. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase β (Pol4)-dependent pathway. J. Biol. Chem. 274:23599-23609. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, Y., X. Wu, F. Yuan, Z. Xie, and Z. Wang. 2001. Highly frequent frameshift DNA synthesis by human DNA polymerase μ. Mol. Cell. Biol. 21:7995-8006. [DOI] [PMC free article] [PubMed] [Google Scholar]