Abstract
The class II transactivator (CIITA) is the key regulator of major histocompatibility complex (MHC) class II gene transcription. We demonstrate here that CIITA requires the ATPase subunit of an hSWI/SNF complex, brahma-related gene 1 (BRG-1), to activate transcription. When introduced into a cell line lacking BRG-1, CIITA was unable to activate cellular MHC class II genes. Reexpression of the wild-type but not an ATP-binding-deficient BRG-1 protein in this cell line restored the ability of CIITA to transactivate transcription of MHC class II genes. Interestingly, when the activity of CIITA was assayed in the BRG-1-deficient cell line by using a plasmid-based reporter assay, BRG-1 was not required for transcriptional activation, suggesting that the chromatin structure on the plasmid is such that BRG-1 is not necessary. Coimmunoprecipitation experiments were performed to determine if BRG-1 and CIITA proteins associate with each other in cells. We found that the two proteins coimmunoprecipitate and that amino acids 1 to 140 of CIITA are sufficient for binding. Taken together, these data suggest that BRG-1 and, very likely, an hSWI/SNF complex are required for transcription of MHC class II genes. The complex is likely recruited to MHC class II promoters, at least in part, by interaction with CIITA.
Expression of major histocompatibility complex (MHC) class II genes is regulated primarily at the level of transcription. The class II transactivator (CIITA) protein is the master switch for activating transcription of MHC class II genes, in that introduction of CIITA into most cell types results in transcription of these genes. To achieve this transcriptional effect, CIITA integrates the activities of a growing list of proteins recruited to promoters of MHC class II genes (19, 22). CIITA itself is brought to the promoters via interaction with sequence-specific DNA binding proteins. These include components of the heterotrimeric DNA binding complexes regulatory factor X (RFX) and nuclear factor Y (NF-Y) (13, 21, 35, 44, 58). RFX and NF-Y bind to the X1 and Y boxes, respectively, which are found in all MHC class II promoters (20). In addition, the cyclic AMP (cAMP) regulatory element binding protein CREB, which interacts with the X2 box of MHC class II promoters, has been implicated in recruiting CIITA (58).
Based upon numerous studies identifying additional CIITA binding proteins, it appears that CIITA regulates several aspects of transcription (19). CIITA has been found to functionally interact with several components of the general transcriptional machinery, including TATA-binding protein (TBP)-associated factors (18, 34). These interactions are likely to be important, ultimately, in the formation of preinitiation complexes at MHC class II promoters. Similarly, CIITA has been shown to interact with cyclin T1 and to require the activity of its associated kinase, cyclin-dependent kinase 9 (cdk9), to activate transcription (25). Presumably, CIITA promotes efficient transcriptional elongation through this interaction, a hypothesis supported by inhibition of MHC class II gene transcription by dominant-negative cdk9 (39).
There is mounting evidence that CIITA has the capacity to modulate chromatin structure on MHC class II genes. First, CIITA has been shown to interact with two proteins that have histone acetyltransferase (HAT) activity: CREB binding protein (CBP) (17, 29) and the p300/CBP-associated factor (p/CAF) (47). In fact, there is evidence that CIITA itself may have acetyltransferase activity (43). Consistent with these observations, acetylation of histones associated with the HLA-DRA promoter was found to be associated with the presence of CIITA at the promoter (4). However, a recent report suggested that the acetyltransferase activities of CBP and p/CAF were not necessary for them to serve as coactivators of CIITA (23). Nevertheless, MHC class II genes were activated in a number of tumor cell lines upon treatment with histone deacetylase inhibitors, in some cases in the absence of expression of the CIITA gene itself (33). Similarly, treatment of cells that lack the retinoblastoma protein with histone deacetylase inhibitors restored the ability of gamma interferon to activate MHC class II genes (42).
Aside from covalent modification of histones to alter chromatin structure, ATP-dependent chromatin remodeling complexes have garnered much attention for their role in transcriptional regulation (24). One subclass of these complexes is typified by the yeast SWI/SNF complex, which contains 11 protein subunits and has as its ATPase subunit the SWI2/SNF2 protein (7, 11). Also within this subgroup is the RSC complex, which has the Sth1/Nsp1 protein as its ATPase (8). The mammalian versions of these complexes are varied, and there are at least two and perhaps more versions (2, 30, 40, 52). However, they are defined by the two ATPase subunits that are present in various forms of the complex: brahma-related gene 1 (BRG-1) (26) and human brahma (hBRM) (36). A growing body of evidence supports the model in which gene-specific transcription factors recruit human SWI/SNF (hSWI/SNF) or similar complexes to promoters to facilitate transcription. For example, hSWI/SNF complexes have been found to associate with the nuclear hormone receptors (36, 38, 50, 51) β-catenin (3), MyoD (12), c-Myc (9), C/EBPβ (28), and EBNA2 (55). The association of hSWI/SNF with these factors is mediated through direct protein-protein interaction between the activator and one or more subunits of the hSWI/SNF complex. For instance, BRG-1 has been shown to bind directly to β-catenin (3), and hSNF5 was found to bind to c-Myc (9).
We were interested in whether hSWI/SNF complexes have a role in the transcription of MHC class II genes. We demonstrate here that CIITA is unable to activate the endogenous MHC class II genes in a cell line that lacks the hSWI/SNF ATPase BRG-1. Introduction of a BRG-1 cDNA into these cells restores the ability of CIITA to turn on MHC class II genes. We also found that CIITA associates with BRG-1, suggesting recruitment of hSWI/SNF to MHC class II promoters by CIITA.
MATERIALS AND METHODS
Cell culture, transfection, and luciferase assay.
SW-13 cells (ATCC no. CCL-105), HeLa cells, and COS-1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum and antibiotics. RJ2.2.5 cells were maintained in RPMI-1640 with 10% fetal calf serum and antibiotics. Cells were transfected with Fugene-6 (Roche Molecular Biochemicals, Inc., Indianapolis, Ind.) essentially as described previously (37). A total of 1 μg of plasmid DNA was used in all transfections. When necessary, empty plasmid vector was used as a stuffer to maintain a total of 1 μg of DNA. For luciferase reporter experiments, all transfections received 10 ng of a plasmid (pRL-SV40; Promega, Inc., Madison, Wis.) that expresses the luciferase enzyme from Renilla reniformis, under the control of the simian virus 40 (SV40) promoter, as a transfection control. Forty-eight hours after transfection, cells were lysed, and firefly luciferase and Renilla luciferase activities were determined by using the DLR Dual Luciferase kit (Promega, Inc., Madison, Wis.) according to the manufacturer's instructions. All firefly luciferase data were normalized to the amount of Renilla luciferase expression. Mean values of two independent transfections are shown. The standard error of the means and P values were calculated with InStat software (GraphPad, Inc.).
Plasmids.
The plasmid pDRA-luc was constructed by subcloning the HLA-DRA promoter from pDRASCAT into plasmid pGL3Basic (Promega, Inc., Madison, Wis.), which contains the firefly luciferase gene, as described previously (37). The plasmid pCMV-CIITA was constructed by subcloning the CIITA cDNA from pSV-CIITA (17) into the plasmid pCDNA3 (InVitrogen, Inc., La Jolla, Calif.). The plasmid pCMV-C988 was created by subcloning the fragment of the CIITA cDNA up to the first HindIII site at nucleotide position 988 from pCMV-CIITA into HindIII-digested plasmid pCR3 (InVitrogen, Inc.). pCMV-C1644 was created by subcloning the fragment of the CIITA cDNA up to the SmaI site at nucleotide position 1688 into pCR3 digested with BamHI and SmaI. The plasmid pCMV-C2931 was created by subcloning the fragment of the CIITA cDNA up to the BamHI site at position 2931 from pCMV-CIITA into BamHI-digested plasmid pCR3. The plasmids pBJ5-BRG-1 and pBJ5-BRG-1(K798R) were gifts from Gerald Crabtree and were described previously (26).
RNA preparation and Northern analysis.
RNA was isolated with TRIzol reagent (InVitrogen, Inc.) essentially as described previously (10). RNA was resolved on 1% agarose-formaldehyde gels and transferred to nylon membranes (Immobilon-NY+; Millipore, Inc., Bedford, Mass.). HLA-DRA probes were made by PCR amplification of portions of the 4th and 5th exons of a human HLA-DRA cDNA (upstream primer, 5′-AGAGACTACAGAGAACGTGG; downstream primer, 5′-GACGAAAGTGACTCCAGTTCC) in the presence of [α-32P]ATP. The β-actin probes were made similarly (upstream primer, 5′-CAAGGTGTGATGGTGGGAATGG; downstream primer, 5′-CAGGATGGCGTGAGGGAGAGCA). Hybridization signals were detected with a PersonalFX phosphorimager (Bio-Rad, Inc., Hercules, Calif.) and quantified with QuantityOne software from the same manufacturer.
Coimmunoprecipitation.
COS-1 cells (106) were transfected with the indicated plasmids and lysed in 1.0 ml of lysis buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 1 mM EGTA, 0.2 mM NaVO3, 0.2 mM phenylmethylsulfonyl flouride, 0.5% NP-40) at 4°C for 3 h. Following sonication, the lysate was cleared by centrifugation at 14,000 × g for 15 min at 4°C. Anti-BRG-1 (H-88; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) or anti-hemagglutinin (anti-HA) (12CA5; Roche Molecular Biochemicals, Inc., Indianapolis, Ind.) antiserum was incubated for 1 h with 0.5 ml of the cleared lystate at 4°C. Twenty-five microliters of protein A/G PLUS-agarose (Santa Cruz Biotechnology, Inc.) was added, and the incubation continued for another 2 h. The beads were collected and washed four times in lysis buffer. The beads were boiled in Laemmli buffer, and the samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (7% polyacrylamide) followed by transfer to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore, Inc.). Twenty-five microliters of lysate was also run on the gels as a control to demonstrate the amount of expressed proteins present in the lysate. Blots were probed with the indicated primary antisera, followed by goat anti-mouse antiserum conjugated to horseradish peroxidase (Santa Cruz Biotechnology, Inc.). Detection was performed with Supersignal West Pico chemiluminescent substrate (Pierce, Inc., Rockford, Ill.) and BioMax Light-1 film (Kodak, Inc., Rochester, N.Y.).
GST fusion proteins and affinity purification of BRG-1.
Glutathione S-transferase (GST) and GST fusion proteins were generated as described (18, 27). Equivalent amounts of GST or GST fusion proteins were combined with 500 μg of nuclear extract from RJ2.2.5 cells, prepared by the method of Dignam et al. (14). The mixture was incubated overnight at 4°C with inversion and washed five times with wash buffer (50 mM Tris HCl [pH 8.0], 120 mM NaCl, 0.5% NP-40, 5 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride). The samples were boiled in Laemmli buffer and resolved by SDS-PAGE, followed by Western blot analysis as described above.
RESULTS
CIITA cannot activate MHC class II gene transcription in a cell line lacking the ATPase subunits of hSWI/SNF.
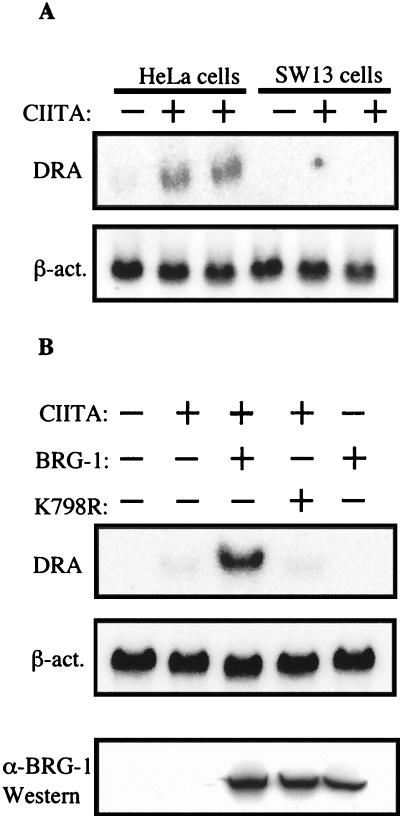
To address whether transcriptional activation of MHC class II genes requires SWI/SNF activity, we performed transient transfection experiments with the cell line SW-13. These cells do not express two of the ATPase subunits that have been found in preparations of hSWI/SNF, BRG-1, and hBRM (36). As a positive control for CIITA function, HeLa cells were transfected with a CIITA expression plasmid (pCMV-CIITA), followed by Northern analysis of HLA-DRA gene transcription. In HeLa cells transfected with vector alone, HLA-DRA transcript was not detected (Fig. 1A, lane 1). Transfection of HeLa cells with pCMV-CIITA resulted in transcriptional activation of the HLA-DRA gene (Fig. 1A, lanes 2 and 3). In contrast, transfection of pCMV-CIITA into the SW-13 cell line did not activate transcription of the HLA-DRA gene (Fig. 1A, lanes 3 to 6). The blot was reprobed with a β-actin probe to demonstrate the presence of RNA from SW-13 cells (Fig. 1A, lanes 10 to 12). Western analysis with anti-CIITA antiserum demonstrated that the CIITA protein was present at equivalent levels in HeLa and SW-13 cells transfected with pCMV-CIITA (data not shown). These data strongly suggested that CIITA requires either BRG-1 or hBRM protein to activate MHC class II genes.
FIG. 1.
CIITA requires the hSWI/SNF subunit BRG-1 to activate MHC class II genes. (A) CIITA can activate MHC class II genes in HeLa cells, but not SW-13 cells, which lack BRG-1 protein. HeLa or SW-13 cells were transfected with plasmid pCDNA3 (empty vector) or increasing amounts of plasmid pCMV-CIITA. Total RNA was isolated and analyzed by Northern blotting. Blots were probed sequentially with probes for HLA-DRA and β-actin. (B) The wild-type but not an ATPase-deficient version of BRG-1 can restore the ability of CIITA to activate MHC class II genes in SW-13 cells. SW-13 cells were transfected with pCMV-CIITA, pBJ5-BRG-1 (wild-type BRG-1) or pBJ5-BRG-1(K798R) (dominant-negative form of BRG-1) as indicated in the figure. Following transfection, total RNA was isolated and Northern analysis was performed as described for panel A. Western analysis of transfected cell protein extracts was performed with anti-BRG-1 antiserum to demonstrate the level of protein expressed from pBJ5-BRG-1(K798R) and pBJ5-BRG-1 (bottom panel).
Reexpression of BRG-1 in SW-13 cells restores the ability of CIITA to activate MHC class II genes.
To test the hypothesis that CIITA requires BRG-1 to activate transcription of MHC class II genes, we cotransfected the plasmid pBJ5-BRG-1, which expresses the BRG-1 protein (26), with pCMV-CIITA into SW-13 cells. We found that expression of BRG-1 restored the ability of CIITA to activate MHC class II genes. Transcriptional activation of both the HLA-DRA gene (Fig. 1B) and the HLA-DQB1 gene (data not shown) was restored. To confirm this need for BRG-1 for activation of MHC class II genes, we used an ATP-binding- and chromatin-remodeling-deficient form of BRG-1, in which a lysine residue at position 798 was mutated to arginine [BRG-1(K798R)] (26). When a plasmid expressing BRG-1(K798R) was cotransfected with pCMV-CIITA into SW-13 cells, transcription of MHC class II genes was not activated (Fig. 1B). The protein expression levels of wild-type and mutated BRG-1 were found to be essentially equal (Fig. 1B, bottom panel). These results clearly demonstrate that CIITA requires a functional BRG-1 protein to transactivate MHC class II genes.
Dominant-negative form of BRG-1 suppresses MHC class II transcription.
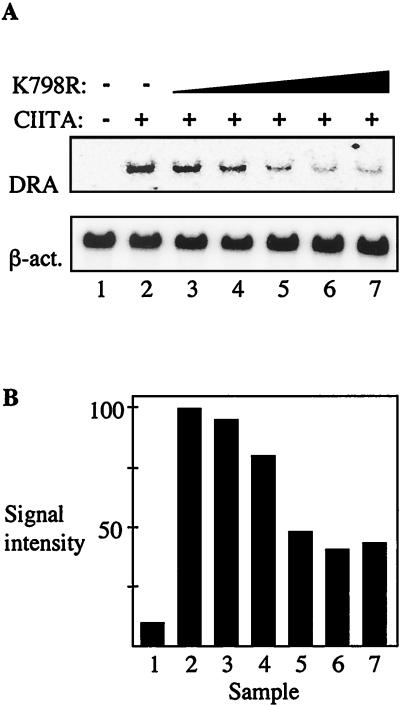
We wanted to confirm the role of BRG-1 in the activation of MHC class II. The BRG-1(K798R) mutant has previously been shown to have dominant-negative activity, suppressing the function of the wild-type protein (26). We employed this mutated version of BRG-1 in transfection experiments with HeLa cells. Cells were transfected with a constant amount of plasmid pCMV-CIITA and increasing amounts of pBJ5-BRG-1(K798R), followed by Northern analysis to detect MHC class II gene transcription. As the amount of dominant-negative BRG-1 increased, the activation of MHC class II gene transcription decreased (Fig. 2A). The band intensity was quantified with Quantity One software (Bio-Rad, Inc., Hercules, Calif.), and the signal for HLA-DRA was normalized to the signal for β-actin. At the highest ratio of BRG-1(K798R) to CIITA, the inhibition of MHC class II gene transcription was slightly greater than 50% (Fig. 2B). Cotransfection of wild-type BRG-1 with CIITA had no additional effect on the induction of MHC class II gene transcription, presumably due to abundant endogenous BRG-1 (data not presented). Western analysis indicated that BRG-1(K798R) had no effect on the levels of CIITA protein (data not presented). These data are consistent with a key role for the protein BRG-1 in the transcriptional activation of MHC class II genes.
FIG. 2.
Dominant-negative BRG-1 can inhibit activation of MHC class II genes in HeLa cells. (A) HeLa cells were transfected with 20 ng of pCMV-CIITA and increasing amounts of pBJ5-BRG-1(K798R) (dominant-negative form of BRG-1) as indicated in the figure. The ratios of BRG-1 to CIITA are 1:1 (lane 3), 10:1 (lane 4), 20:1 (lane 5), 30:1 (lane 6), and 40:1 (lane 7). Total RNA was isolated, and Northern analysis was performed as described for Fig. 1A. (B) Band intensities of the blot shown in panel A were measured with QuantityOne software (Bio-Rad, Inc., Hercules, Calif.). The signal for HLA-DRA was normalized to the associated β-actin (β-act.) signal. The results obtained by transfection of pCMV-CIITA alone were set to 100.
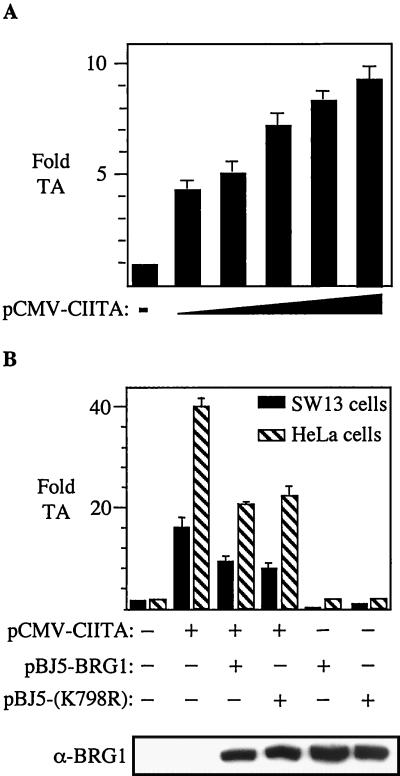
BRG-1 is not required for CIITA to activate an episomal HLA-DRA promoter.
We performed transfection experiments in SW-13 cells with the reporter plasmid pDRA-luc, which contains the HLA-DRA promoter linked to the luciferase gene. We have used this reporter plasmid previously in performing functional analyses of the CIITA protein (37). Interestingly, in the context of this assay, we found that CIITA could activate the HLA-DRA promoter moderately well in SW-13 cells (Fig. 3A). Cotransfection of the plasmid pBJ5-BRG-1 or pBRG-1(K798R) had a negative effect on the ability of CIITA to activate the HLA-DRA promoter in this assay in both SW-13 and HeLa cells (Fig. 3B). Since both the wild-type and dominant-negative BRG-1 produced this result, we are reluctant to claim that it is a functionally significant effect. The inclusion of BRG-1 or BRG-1(K798R) did not alter the levels of CIITA protein expressed from pCMV-CIITA, as determined by Western blotting (data not shown). Protein expression levels of wild-type and mutated BRG-1 were found to be essentially equal (Fig. 3B, bottom panel). It seems likely that since BRG-1 is involved in chromatin remodeling, the episomal HLA-DRA promoter may have a chromatin structure that does not require BRG-1 activity. The chromatin structure on the chromosomal HLA-DRA promoter may be significantly different from that on a transfected plasmid containing the promoter.
FIG. 3.
CIITA can activate an episomal HLA-DRA promoter in the absence of BRG-1. (A) SW-13 cells were cotransfected with the plasmid pDRA-luc, which contains the HLA-DRA promoter from −151 to +31, linked to the firefly luciferase gene along with either empty vector or increasing amounts of plasmid pCMV-CIITA. Fold transactivation (TA) was calculated by dividing the luciferase activity measured with pDRA-luc plus the indicated effector plasmid(s) by the activity obtained with the pDRA-luc plasmid alone. Error bars show the standard error of the mean for triplicate experiments. (B) Overexpression of BRG-1 or BRG-1(K798R) in HeLa or SW-13 cells has a similar, negative effect on HLA-DRA promoter activation by CIITA. The two cell lines were transfected with plasmids, as indicated below the histogram. Fold transactivation was calculated by dividing the luciferase activity measured with pDRA-luc plus the indicated effector plasmid(s) by the activity obtained with the pDRA-luc plasmid alone. Error bars represent the standard error of the mean for triplicate experiments. Solid bars, SW-13 cells; hatched bars, HeLa cells. Western analysis of transfected cell protein extracts was performed with anti-BRG-1 antiserum to demonstrate the level of protein expressed from pBJ5-BRG-1(K798R) and pBJ5-BRG-1 (bottom panel).
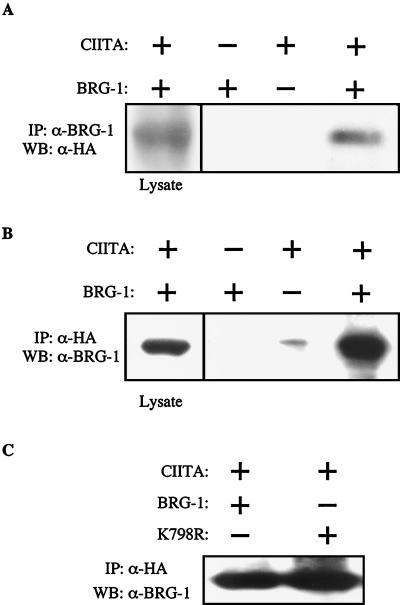
Association between the CIITA and BRG-1 proteins.
Recruitment of hSWI/SNF to promoters by gene-specific transacting factors is often accomplished by the direct interaction between one or more subunits of hSWI/SNF and the factor. For example, BRG-1 has been shown to bind directly to β-catenin (3), and hSNF5 was found to bind to c-Myc (9). To determine if CIITA can associate with a complex containing BRG-1, we performed coimmunoprecipitation experiments with lysates from COS-1 cells transfected with pCMV-CIITA and/or pBJ5/BRG-1. Immunoprecipitation with anti-BRG-1 antiserum was followed by Western analysis with anti-HA antiserum to detect the HA-tagged CIITA protein expressed from pCMV-CIITA (17). If CIITA or BRG-1 alone (Fig. 4A) was expressed in the cells, then CIITA could not be detected by Western analysis. However, when both proteins were expressed, CIITA protein was found to be associated with BRG-1 (Fig. 4A). We performed the inverse experiment, in which anti-HA antiserum was used for immunoprecipitation and Western analysis was carried out with anti-BRG-1 antiserum. In this case, BRG-1 was detectable in the cells that received only CIITA (Fig. 4B). In this case, we detected the endogenous BRG-1 from the COS-1 cells. This explanation is supported by the fact that BRG-1 was not detected when CIITA was immunoprecipitated from SW-13 cells transfected with pCMV-CIITA alone (data not shown). The signal for BRG-1 was much stronger in cells that received both expression plasmids (Fig. 4B). Significantly, the association between CIITA and BRG-1 was not altered by the K798R mutation in dominant-negative BRG-1 (Fig. 4C).
FIG. 4.
BRG-1 and CIITA proteins coimmunoprecipitate. (A) COS-1 cells were transfected with pCMV-CIITA, pBJ5-BRG-1, or both plasmids. Anti-BRG-1 antiserum was used to immunoprecipitate (IP) BRG-1 and associated proteins from cellular lysates. Western blot (WB) analysis was performed with anti-HA antiserum, which recognizes the “flu tag” present on CIITA produced from pCMV-CIITA. The first lane contains cellular lysate from dually transfected cells. The band representing CIITA protein is indicated by an arrow. (B) Transfection was identical to that performed in panel A, but immunoprecipitation was performed with anti-HA antiserum and Western analysis was performed with anti-BRG-1 antiserum. The first lane contains cellular lysate from dually transfected cells. The band representing BRG-1 protein is shown. (C) The K798R mutation does not affect the interaction between BRG-1 and CIITA. The indicated expression plasmids were transfected into COS cells, and immunoprecipitation and Western blotting were performed with cellular lysates as in panel B.
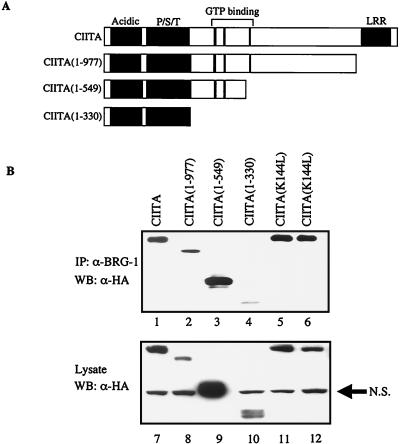
To identify the region of CIITA to which BRG-1 binds, plasmids expressing C-terminal deletions of CIITA were generated (Fig. 5A). These plasmids were cotransfected with pBJ5-BRG-1 into COS-1 cells, followed by coimmunoprecipitation with anti-BRG-1 antiserum and Western blotting with anti-HA antiserum to detect CIITA. Deletion of the C-terminal 582 amino acids of the CIITA protein (plasmid pCMV-C2931, containing amino acids 1 to 977) did not alter the binding of BRG-1 to CIITA (Fig. 5B, compare lane 1 to lane 2). Similarly, when the deletion was extended to amino acid 549 (plasmid pCMV-C1644, containing amino acids 1 to 549), the interaction was still sound (Fig. 5B, lane 3). Although it appears that more CIITA protein from pCMV-C1644 was coimmunoprecipitated than the other forms of CIITA, this seems to be due to higher levels of this protein being produced in the cell (Fig. 5B, lane 9).
FIG. 5.
Amino acids 1 to 548 of CIITA are required for the strongest interaction between CIITA and BRG-1. (A) C-terminal deletions of CIITA were generated by deletion of cDNA sequences (see Materials and Methods). Acidic, region rich in aspartic and glutamic acid residues (amino acids 26 to 140); P/S/T, region rich in proline/serine/threonine residues (amino acids160 to 319); LRR, leucine-rich region (amino acids 985 to 1086). (B) Western analysis with anti-HA antiserum of coimmunoprecipitation of CIITA with anti-BRG-1 antiserum (lanes 1 to 6) or of cellular lysates (lanes 7 to 12). pBJ5-BRG-1 was cotransfected with a plasmid expressing the indicated form of CIITA into COS-1 cells, followed by immunoprecipitation (IP) and Western blot (WB) analysis. CIITA(K141L) and CIITA(K144L) are full-length CIITA proteins that have point mutations that change the indicated lysine residues to leucine. N.S., nonspecific band detected in COS-1 cell extracts by anti-HA antiserum.
When an additional 218 amino acids were deleted from the C terminus of CIITA (plasmid pCMV-C988, containing amino acids 1 to 331), binding between CIITA and BRG-1 appeared diminished, although not completely extinguished (Fig. 5B, lane 4). Two lysine residues in CIITA, at positions 141 and 144, have been reported to be targets for acetylation (47). Since BRG-1 contains a bromodomain, a motif that has been reported to bind acetylated lysine (53), we tested whether these residues in CIITA were necessary for association with BRG-1. Mutation of either lysine residue within CIITA to leucine [CIITA(K141L) and CIITA(K144L)] did not reduce the association of the protein with BRG-1 (Fig. 5B, lanes 5 and 6).
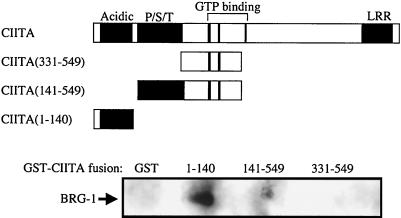
To confirm the association of CIITA and BRG-1 and further map the regions of CIITA required, we used a fusion protein between portions of CIITA and GST to purify BRG-1 from nuclear extracts. We expressed three different GST-CIITA fusion proteins in Escherichia coli representing amino acids 1 to 140, 141 to 549, and 331 to 549. The proteins were purified with glutathione-Sepharose and combined with nuclear extract from the CIITA−/− Burkitt's lymphoma cell line RJ2.2.5. In addition, the GST moiety alone was utilized in a similar binding reaction. Following extensive washing, proteins remaining bound to the GST or GST fusion proteins were analyzed by Western blotting with anti-BRG-1 antiserum. When GST alone or a GST-CIITA fusion containing amino acids 141 to 549 or 331 to 549 was employed in the binding reaction, BRG-1 protein was not detected by Western blotting (Fig. 6). However, when the GST-CIITA fusion containing amino acids 1 to 140 was used, the BRG-1 protein was detected (Fig. 6), indicating that the N terminus of CIITA is sufficient for association with the BRG-1 protein. We could not detect the hBRM protein in any of the lanes when we probed the blot with anti-hBRM antiserum (data not presented). We conclude from these data that CIITA can physically associate with BRG-1, fully consistent with the functional association demonstrated between the two proteins. The association is via the N-terminal 140 amino acids of CIITA, although the region of CIITA between amino acids 331 and 549 may contribute to efficient binding.
FIG. 6.
Affinity isolation of BRG-1 by the N-terminal portion of CIITA. GST, or GST-CIITA fusion proteins were expressed in E. coli and purified with glutathione-Sepharose. The recombinant proteins were combined with nuclear extract from RJ2.2.5 cells and allowed to interact overnight. Following extensive washing, the bound proteins were analyzed by Western blot analysis with anti-BRG-1 antiserum. The top of the figure is a diagram of the regions of CIITA used in the GST fusion proteins. Acidic, region rich in aspartic and glutamic acid residues (amino acids 26 to 140); P/S/T, region rich in proline, serine, and threonine residues (amino acids160 to 319); LRR, leucine-rich region (amino acids 985 to 1086).
DISCUSSION
Once CIITA has associated with MHC class II promoters via interactions with DNA binding factors (13, 21, 35, 44, 58), it activates transcription of MHC class II genes by recruiting and coordinating at least three different activities: (i) components of the general transcriptional apparatus, such as TFIID; (ii) cyclin T1 and its associated kinase to promote processivity of RNA polymerase II; and (iii) factors and complexes that have a role in chromatin remodeling. This final set not only involves proteins that have HAT activity, but, together with the results presented here, also includes an ATP-dependent chromatin-remodeling complex. CIITA requires at least one of the ATPase subunits of hSWI/SNF, BRG-1, to activate transcription of MHC class II genes. It seems likely that the hSWI/SNF complex is recruited to MHC class II genes by a direct interaction between CIITA and one of the subunits of the BRG-1-containing hSWI/SNF complex. Presumably, the presence of hSWI/SNF would result in an alteration in the chromatin structure of MHC class II genes such that transcription is facilitated.
The identification of the role of hSWI/SNF in CIITA function leads to an interest in the potential complementary activity of the associated and endogenous acetyltransferase activities of CIITA (17, 23, 29, 46, 47). Chromatin-remodeling complexes and HATs have been found to functionally complement each other in a number of other systems. For example, it was recently reported that BRG-1 and CBP synergistically activate MHC class I genes through the enhancer A (6). Similarly, SWI/SNF and HAT p300 activities were found to act sequentially during activation of transcription by retinoic acid receptor (15). Likewise, recruitment of BRG-1 and SWI/SNF to the beta interferon enhanceosome requires nucleosome acetylation and the presence of CBP. The acetylation of a nucleosome positioned over the transcriptional start site did not alter chromatin structure; however, it was required for efficient recruitment of BRG-1 (1). The relationship between HAT, which occurs concurrently with the arrival of CIITA at MHC class II promoters (4), and the function of BRG-1 in MHC class II transcription remains to be determined.
Two lysine residues in CIITA (lysines 141 and 144) have been shown to be acetylated, and mutation of these residues reduces the ability of CIITA to activate transcription (47). Recent evidence has suggested that the cooperation between HATs and chromatin-remodeling complexes may center on the interaction between bromodomains present in subunits of SWI/SNF and acetylated lysine residues. Acetylated lysines either on histones or on transcriptional activator proteins would presumably serve as “docking” sites for chromatin-remodeling complexes (56). However, this appears not to be the case for the association between CIITA and BRG-1, since mutation of lysine 141 or lysine 144 of CIITA did not disrupt the coimmunoprecipitation between CIITA and BRG-1. In addition, we found that the amino-terminal 140 amino acids of CIITA are sufficient to mediate association between the two proteins. However, the most robust association between CIITA and BRG-1 also required the region of CIITA between amino acids 331 and 541.
Interestingly, by using a reporter plasmid containing the HLA-DRA promoter, we found that BRG-1 did not have an effect on transcription of the HLA-DRA promoter in transient transfection assays. Expression of wild-type or ATPase-defective versions of BRG-1 had a minor inhibitory effect on the activation of the HLA-DRA promoter by CIITA. The apparent irrelevance of BRG-1 in this experimental assay is possibly due to chromatin structure on the episomal promoter not being assembled into a specific array compared to that on the chromosomal genes. This observation is particularly important in light of a recent report of the HAT activity of CBP and P/CAF not being required for these proteins to act as cofactors for CIITA (23). Since these experiments were performed exclusively with a plasmid-based reporter system, it would be interesting to determine if the results would have been similar for the transcription of endogenous genes.
An additional connection between hSWI/SNF and MHC class II transcription centers on the retinoblastoma (Rb) protein. A number of reports have demonstrated that in tumor cell lines lacking Rb protein, induction of MHC class II genes by gamma interferon is defective, even though CIITA itself is induced in most cases (31, 32, 49). Additionally, in Rb−/− murine embryonic fibroblasts, the MHC class II Abeta gene was uninducible by CIITA (57). It has also been demonstrated the Rb facilitates occupancy of conserved elements in the HLA-DRA promoter (41). Therefore, since Rb has been found to interact with BRG-1 (16), it may play a role in the functional interaction between CIITA and hSWI/SNF. Rb has been found to potentiate transcriptional activation by the glucocorticoid receptor (45), which also is coactivated by SWI/SNF (36, 51). Further investigation is needed to determine the exact role Rb may have in CIITA function.
One particularly intriguing observation is that BRG-1 is mutated in a variety of tumor cell lines (54). This clearly has a primary effect on deregulating the cell cycle of these cells (48). However, an additional benefit to the survival of these cells may be the loss of efficient CIITA function in cells lacking BRG-1. Although far from certain, if expression of MHC class II proteins by tumors does have a role in antitumor immunity (5), then the loss of induction of these genes due to an absence of BRG-1 protein would provide a significant growth advantage to the tumor.
We present data here that clearly demonstrate the importance of BRG-1 in MHC class II gene transcription. This functional role of BRG-1 also translates into a physical association with CIITA. Currently, we are uncertain whether the two proteins bind directly to each other, or whether the interaction is between CIITA and one of the other subunits in hSWI/SNF. This functional relationship provides additional insight into the mechanism that activates transcription of MHC class II genes.
Acknowledgments
We thank Gerald Crabtree for the BRG-1 expression plasmids. We are also grateful to Tomasz Kordula for careful reading of the manuscript and helpful suggestions.
This work was supported by institutional funds from Cleveland State University and the Lerner Research Institute of the Cleveland Clinic Foundation.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. [DOI] [PubMed] [Google Scholar]
- 3.Barker, N., A. Hurlstone, H. Musisi, A. Miles, M. Bienz, and H. Clevers. 2001. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 20:4935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beresford, G. W., and J. M. Boss. 2001. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat. Immunol. 2:652-657. [DOI] [PubMed] [Google Scholar]
- 5.Blanck, G. 1999. HLA class II expression in human tumor lines. Microbes Infect. 1:913-918. [DOI] [PubMed] [Google Scholar]
- 6.Brockmann, D., O. Lehmkuhler, U. Schmucker, and H. Esche. 2001. The histone acetyltransferase activity of PCAF cooperates with the brahma/SWI2-related protein BRG-1 in the activation of the enhancer A of the MHC class I promoter. Gene 277:111-120. [DOI] [PubMed] [Google Scholar]
- 7.Cairns, B. R., N. L. Henry, and R. D. Kornberg. 1996. TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol. Cell. Biol. 16:3308-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, S. W., K. P. Davies, E. Yung, R. J. Beltran, J. Yu, and G. V. Kalpana. 1999. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat. Genet. 22:102-105. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski, P. 1993. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques 15:532-534, 536-537. [PubMed] [Google Scholar]
- 11.Cote, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53-60. [DOI] [PubMed] [Google Scholar]
- 12.de la Serna, I. L., K. A. Carlson, and A. N. Imbalzano. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27:187-190. [DOI] [PubMed] [Google Scholar]
- 13.DeSandro, A. M., U. M. Nagarajan, and J. M. Boss. 2000. Associations and interactions between bare lymphocyte syndrome factors. Mol. Cell. Biol. 20:6587-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dilworth, F. J., and P. Chambon. 2001. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene 20:3047-3054. [DOI] [PubMed] [Google Scholar]
- 16.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 17.Fontes, J. D., S. Kanazawa, D. Jean, and B. M. Peterlin. 1999. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol. Cell. Biol. 19:941-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fontes, J. D., B. Jiang, and B. M. Peterlin. 1997. The class II trans-activator CIITA interacts with the TBP-associated factor TAFII32. Nucleic Acids Res. 25:2522-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fontes, J. D., S. Kanazawa, N. Nekrep, and B. M. Peterlin. 1999. The class II transactivator CIITA is a transcriptional integrator. Microbes Infect. 1:863-869. [DOI] [PubMed] [Google Scholar]
- 20.Glimcher, L. H., and C. J. Kara. 1992. Sequences and factors: a guide to MHC class-II transcription. Annu. Rev. Immunol. 10:13-49. [DOI] [PubMed] [Google Scholar]
- 21.Hake, S. B., K. Masternak, C. Kammerbauer, C. Janzen, W. Reith, and V. Steimle. 2000. CIITA leucine-rich repeats control nuclear localization, in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome, and MHC class II gene transactivation. Mol. Cell. Biol. 20:7716-7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harton, J. A., and J. P.-Y. Ting. 2000. Class II transactivator: mastering the art of major histocompatibility complex expression. Mol. Cell. Biol. 20:6185-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harton, J. A., E. Zika, and J. P. Ting. 2001. The histone acetyltransferase domains of CBP and pCAF are not necessary for cooperativity with CIITA. J. Biol. Chem. 276:38715-38720. [DOI] [PubMed] [Google Scholar]
- 24.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6:D1054-D1064. [DOI] [PubMed] [Google Scholar]
- 25.Kanazawa, S., T. Okamoto, and B. M. Peterlin. 2000. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity 12:61-70. [DOI] [PubMed] [Google Scholar]
- 26.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 27.Klemm, R. D., J. A. Goodrich, S. Zhou, and R. Tjian. 1995. Molecular cloning and expression of the 32-kDa subunit of human TFIID reveals interactions with VP16 and TFIIB that mediate transcriptional activation. Proc. Natl. Acad. Sci. USA 92:5788-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowenz-Leutz, E., and A. Leutz. 1999. A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4:735-743. [DOI] [PubMed] [Google Scholar]
- 29.Kretsovali, A., T. Agalioti, C. Spilianakis, E. Tzortzakaki, M. Merika, and J. Papamatheakis. 1998. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol. Cell. Biol. 18:6777-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon, H., A. N. Imbalzano, P. A. Khavari, R. E. Kingston, and M. R. Green. 1994. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370:477-481. [DOI] [PubMed] [Google Scholar]
- 31.Lu, Y., M. E. Tschickardt, B. J. Schmidt, and G. Blanck. 1997. IFN-gamma inducibility of class II transactivator is specifically lacking in human tumour lines: relevance to retinoblastoma protein rescue of IFN-gamma inducibility of the HLA class II genes. Immunol. Cell Biol. 75:325-332. [DOI] [PubMed] [Google Scholar]
- 32.Lu, Y., G. D. Ussery, M. M. Muncaster, B. L. Gallie, and G. Blanck. 1994. Evidence for retinoblastoma protein (RB) dependent and independent IFN-gamma responses: RB coordinately rescues IFN-gamma induction of MHC class II gene transcription in noninducible breast carcinoma cells. Oncogene 9:1015-1019. [PubMed] [Google Scholar]
- 33.Magner, W. J., A. L. Kazim, C. Stewart, M. A. Romano, G. Catalano, C. Grande, N. Keiser, F. Santaniello, and T. B. Tomasi. 2000. Activation of MHC class I, II, and CD40 gene expression by histone deacetylase inhibitors. J. Immunol. 165:7017-7024. [DOI] [PubMed] [Google Scholar]
- 34.Mahanta, S. K., T. Scholl, F. C. Yang, and J. L. Strominger. 1997. Transactivation by CIITA, the type II bare lymphocyte syndrome- associated factor, requires participation of multiple regions of the TATA box binding protein. Proc. Natl. Acad. Sci. USA 94:6324-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masternak, K., A. Muhlethaler-Mottet, J. Villard, M. Zufferey, V. Steimle, and W. Reith. 2000. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 14:1156-1166. [PMC free article] [PubMed] [Google Scholar]
- 36.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mudhasani, R., and J. D. Fontes. 2002. Inhibition of class II trans-activator function by HIV-1 tat in mouse cells is independent of competition for binding to cyclin T1. Mol. Immunol. 38:539-546. [DOI] [PubMed] [Google Scholar]
- 38.Nie, Z., Y. Xue, D. Yang, S. Zhou, B. J. Deroo, T. K. Archer, and W. Wang. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20:8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamoto, H., K. Asamitsu, H. Nishimura, N. Kamatani, and T. Okamoto. 2000. Reciprocal modulation of transcriptional activities between HIV-1 tat and MHC class II transactivator CIITA. Biochem. Biophys. Res. Commun. 279:494-499. [DOI] [PubMed] [Google Scholar]
- 40.O'Neill, D., J. Yang, H. Erdjument-Bromage, K. Bornschlegel, P. Tempst, and A. Bank. 1999. Tissue-specific and developmental stage-specific DNA binding by a mammalian SWI/SNF complex associated with human fetal-to-adult globin gene switching. Proc. Natl. Acad. Sci. USA 96:349-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osborne, A., M. Tschickardt, and G. Blanck. 1997. Retinoblastoma protein expression facilitates chromatin remodeling at the HLA-DRA promoter. Nucleic Acids Res. 25:5095-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osborne, A., H. Zhang, W.-M. Yang, E. Seto, and G. Blanck. 2001. Histone deacetylase activity represses gamma interferon-inducible HLA-DR gene expression following the establishment of a DNase I-hypersensitive chromatin conformation. Mol. Cell. Biol. 21:6495-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raval, A., T. K. Howcroft, J. D. Weissman, S. Kirshner, X. S. Zhu, K. Yokoyama, J. Ting, and D. S. Singer. 2001. Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Mol. Cell 7:105-115. [DOI] [PubMed] [Google Scholar]
- 44.Scholl, T., S. K. Mahanta, and J. L. Strominger. 1997. Specific complex formation between the type II bare lymphocyte syndrome-associated transactivators CIITA and RFX5. Proc. Natl. Acad. Sci. USA 94:6330-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh, P., J. Coe, and W. Hong. 1995. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature 374:562-565. [DOI] [PubMed] [Google Scholar]
- 46.Sisk, T. J., T. Gourley, S. Roys, and C. H. Chang. 2000. MHC class II transactivator inhibits IL-4 gene transcription by competing with NF-AT to bind the coactivator CREB binding protein (CBP)/p300. J. Immunol. 165:2511-2517. [DOI] [PubMed] [Google Scholar]
- 47.Spilianakis, C., J. Papamatheakis, and A. Kretsovali. 2000. Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Mol. Cell. Biol. 20:8489-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strobeck, M. W., K. E. Knudsen, A. F. Fribourg, M. F. DeCristofaro, B. E. Weissman, A. N. Imbalzano, and E. S. Knudsen. 2000. BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl. Acad. Sci. USA 97:7748-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tschickardt, M. E., Y. Lu, M. Jacim, G. D. Ussery, V. Steimle, B. Mach, and G. Blanck. 1995. RB and a novel E2F-1 binding protein in MHC class II deficient B-cell lines and normal IFN-gamma induction of the class IL transactivator CIITA in class II non-inducible RB-defective tumor lines. Int. J. Cancer 62:461-465. [DOI] [PubMed] [Google Scholar]
- 50.Wallberg, A. E., E. M. Flinn, J. A. Gustafsson, and A. P. Wright. 2000. Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain. Biochem. Soc. Trans. 28:410-414. [PubMed] [Google Scholar]
- 51.Wallberg, A. E., K. E. Neely, A. H. Hassan, J.-Å. Gustafsson, J. L. Workman, and A. P. H. Wright. 2000. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor τ1 activation domain. Mol. Cell. Biol. 20:2004-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 53.Winston, F., and C. D. Allis. 1999. The bromodomain: a chromatin-targeting module? Nat. Struct. Biol. 6:601-604. [DOI] [PubMed] [Google Scholar]
- 54.Wong, A. K., F. Shanahan, Y. Chen, L. Lian, P. Ha, K. Hendricks, S. Ghaffari, D. Iliev, B. Penn, A. M. Woodland, R. Smith, G. Salada, A. Carillo, K. Laity, J. Gupte, B. Swedlund, S. V. Tavtigian, D. H. Teng, and E. Lees. 2000. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 60:6171-6177. [PubMed] [Google Scholar]
- 55.Wu, D. Y., G. V. Kalpana, S. P. Goff, and W. H. Schubach. 1996. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 70:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, W., S. Kadam, B. M. Emerson, and J. J. Bieker. 2001. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Krüppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol. 21:2413-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, X., S. Pattenden, and R. Bremner. 1999. pRB is required for interferon-gamma-induction of the MHC class II abeta gene. Oncogene 18:4940-4947. [DOI] [PubMed] [Google Scholar]
- 58.Zhu, X.-S., M. W. Linhoff, G. Li, K.-C. Chin, S. N. Maity, and J. P.-Y. Ting. 2000. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol. Cell. Biol. 20:6051-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]