Abstract
Alveolar rhabdomyosarcoma is a pediatric disease specified by the recurrent chromosome translocations t(2;13) and t(1;13). These translocations result in the formation of the PAX3-FKHR and PAX7-FKHR fusion genes, which are thought to play a causal role in the genesis of this disease. Although PAX3-FKHR exhibits transforming activity in immortalized fibroblast cell lines, a direct role of this fusion protein in tumorigenesis in vivo has not been shown. We determined whether expression of Pax3-FKHR in the mouse germ line would render these animals prone to the development of rhabdomyosarcomas. By targeting FKHR cDNA sequences into the Pax3 locus of embryonic stem cells, we used these cells to generate mice carrying a Pax3-FKHR knock-in allele. Despite low expression of the knock-in allele, heterozygous offspring of Pax3-FKHR chimeric mice showed developmental abnormalities. These included intraventricular septum defects, tricuspid valve insufficiency, and diaphragm defects, which caused congestive heart failure leading to perinatal death. In addition, Pax3-FKHR heterozygous offspring displayed malformations of some but not all hypaxial muscles. However, neither newborn heterozygous pups nor their chimeric parents showed any signs of malignancy. We conclude that the Pax3-FKHR allele causes lethal developmental defects in knock-in mice but might be insufficient to cause muscle tumors.
Recurrent chromosomal translocations occur in many hematopoietic malignancies and in some solid tumors and invariantly signify specific gene alterations that contribute to tumorigenesis. Translocation (2;13) and t(1;13) in alveolar rhabdomyosarcoma (ARMS) are two of the well-characterized translocations in solid tumors (47). ARMS and embryonal rhabdomyosarcoma (ERMS) together (RMS) constitute the third most common form of soft tissue sarcoma in children. RMS belongs to the family of small round blue cell tumors of childhood and exhibits features of skeletal muscle. Tumor cells are distinguished by their skeletal-muscle-like cross-striation and by their expression of muscle-specific proteins such as α-actin, MYOD, MYF5, myogenin, myosin heavy chain, desmin, myoglobin, and Z-band proteins (32). ERMS is prevalent in infants and young children, and the more aggressive ARMS occurs in children and adolescents (36). The latter has a poorer prognosis, and depending on the disease state at diagnosis, survival rates vary between 20 and 50% (1, 34).
ARMS is found predominantly in the extremities and trunk (39) and is typified by the above-mentioned karyotypic hallmarks, which create the PAX3-FKHR (4, 23, 44) and PAX7-FKHR (16) fusion genes, respectively. Both proteins encoded by these genes contain the paired and homeobox DNA binding domains of the PAX transcription factors, while their C-terminal transcription transactivation sequences have been replaced by the bisected DNA binding domain of FKHR and its strong transactivating sequences. Like PAX3 and PAX7, the fusion proteins can bind combined paired-homeodomain DNA binding sites, and in addition the fusion proteins bind homeodomain sites alone, a gain of function mediated by the FKHR transactivation domain (10). The fusion proteins are much stronger transcriptional transactivators than are PAX3 and PAX7 (5, 6).
Pax3 (26) is important in mammalian development, as exemplified by the phenotypic aberrations found in heterozygous and homozygous Splotch mice that carry inactivating mutations in Pax3 (7, 24, 48). This gene is expressed in the developing brain, the dorsal neural tube, and various neural crest-derived tissues, such as the developing outflow tract of the heart, dorsal root ganglia, Schwann cells, and melanocytes. Consistent with this expression pattern, heterozygous Splotch mice show pigment disturbances on the abdomen, feet, tail and head whereas homozygous fetuses display neural-tube closure defects, absence of dorsal root ganglia, truncus arteriosis, and absence of limb musculature. During development, the dorsal part of the developing early somite forms the dermomyotome, which then develops into two structures: the outer dermomyotome cell layer and the inner myotome layer. Whereas most body muscles develop from the myotomes (17, 18), the lateral dermomyotome, to which Pax3 expression becomes restricted, gives rise to the hypaxial muscles including those of the limbs, the diaphragm (7, 24, 48), and the tongue (14, 37). Consequently, and due to the inability of Pax3-null cells to migrate out of the dermomyotome (7, 15, 24, 48), Splotch homozygous mice fail to develop limb and diaphragm musculature (46), while the development of other hypaxial muscles is affected (7, 9, 24, 45). Despite considerable overlap in expression between Pax3 and Pax7 during somitic development (26, 31), Pax7 is essential for the specification of myogenic satellite cells (43), which give rise to the majority of nuclei within adult skeletal muscle (43). When fused to FKHR, both genes ensure the expression of the fusion protein in early myogenic cells, the likely precursors of ARMS.
Also, the Fkhr gene is highly expressed in developing and adult mouse muscle (22), but a specific role in muscle cell growth or differentiation has not been described.
PAX3-FKHR transforms both chicken and mouse fibroblasts in culture (27, 42) but its tumorigenic capacity in vivo is unknown. Expression of PAX3-FKHR in transgenic mice under the control of Pax3 regulatory sequences showed a dominant negative effect on normal Pax3 function, resulting in a phenotype partly resembling that of Splotch mice (3). However, the mice were not tumor prone, suggesting that expression of PAX3-FKHR alone is not sufficient to cause rhabdomyosarcoma. We used a knock-in approach to create a mouse model for alveolar rhabdomyosarcoma. We reasoned that expression of Pax3-FKHR under control of the endogenous Pax3 locus would mimic the situation in ARMS most faithfully. We report here that Pax3-FKHR chimeric mice and their heterozygous offspring do not develop muscle tumors. Pax3-FKHR heterozygous mice die around the time of birth as a result of respiratory and heart failure. These animals show numerous developmental abnormalities that could be explained by a dominant negative effect of Pax3-FKHR on Pax3 function.
MATERIALS AND METHODS
Creation of the knock-in construct.
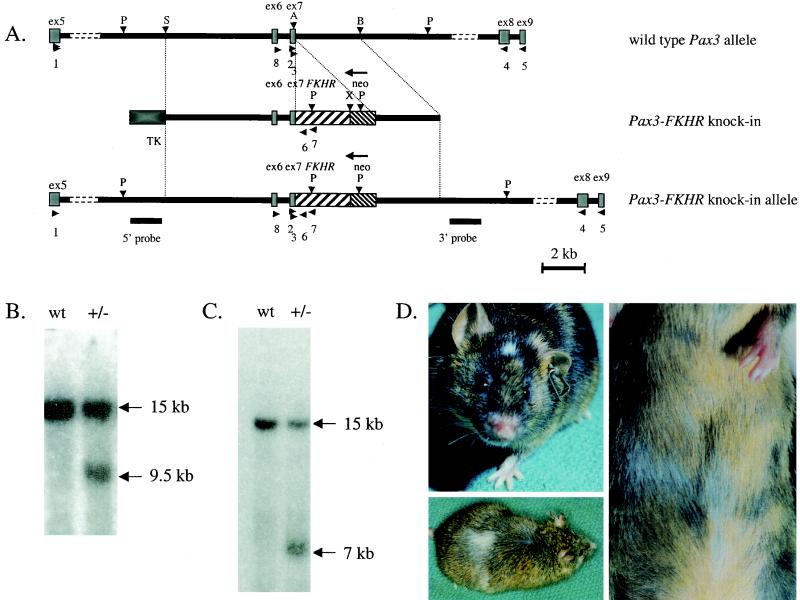
A mouse genomic P1 phage clone, which contained mouse Pax3 exons 6 and 7 and the surrounding sequences, was obtained from Incyte Genomics (Palo Alto, Calif.). A 6-kb fragment ranging from the SacI site within Pax3 intron 5 to the ApaI site at 50 bp upstream of the 3′ end of exon 7 (Fig. 1A) was fused in frame with a 1.4-kb ApaI-XhoI fragment from the PAX3-FKHR cDNA (33, 44), restoring the 3′ 50 bp of exon 7 of Pax3 and adding the 3′ half of human FKHR. For negative selection a herpes simplex virus (HSV) thymidine kinase (TK) gene under the control of the HSV TK promoter and polyomavirus enhancer was added upstream of the 5′ homology region of the targeting vector. A PGK-Neor selectable marker was cloned immediately 3′ of the ex7-FKHR fusion exon (Fig. 1A) in an orientation opposite to of that of the Pax3 gene. A 3.5-kb ApaI-BamHI genomic fragment was added 3′ of the Neo r marker, containing Pax3 genomic sequences downstream of the ApaI site in exon 7.
FIG.1.
(A) Generation of the Pax3-FKHR knock-in allele by homologous recombination in ES cells. The top bar represents a physical map of the mouse Pax3 gene encompassing the area around exons 6 and 7 (gray boxes). The middle bar shows the targeting construct in which exon 7 of Pax3 is fused to exon 2 of FKHR (coarsely hatched box) and is followed by a Neor gene (finely hatched box) in the opposite transcriptional orientation. A TK selectable marker (black box) is present at the 5′ end. The bottom line represents the targeted Pax3-FKHR allele. Underneath the targeted allele, the positions of the external 5′ and 3′ probes for Southern blot hybridization are indicated. Horizontal arrowheads indicate the positions of primers, used for genotyping and RT-PCR (Table 1). P, PstI; S, SstI; A, ApaI; B, BamHI; X, XhoI. (B) Southern blot with the 5′ probe, showing wild-type (wt) and targeted Pax3-FKHR (+/−) alleles in a wild-type embryo and a Pax3-FKHR heterozygous littermate, respectively. (C) Southern blot with the 3′ probe showing wild-type (wt) and targeted Pax3-FKHR (+/−) alleles. (D) Middle (upper left) and highly chimeric (lower left and right) Pax3-FKHR chimeric mice, showing white spots on the head, belly, and back.
ES cells targeting and screening for homologously targeted clones.
The RW-4 mouse embryonic stem cells (ES) cells from Genome Systems were grown on irradiated mouse embryo fibroblast (MEF) feeders, transfected with the linearized Pax3-FKHR targeting construct (Fig. 1A), and selected with G418, as specified by the manufacturer. During the first 5 days, gancyclovir (working concentration, 2 μM) was added for negative selection. After 6 to 7 days of selection with G418, clones were picked and grown in 24-well plates. For Southern blot analysis, ES clones were grown for several days in 24-well gelatin-coated plates without MEFs. The cells were lysed in proteinase K buffer (20 mM Tris [pH 7.5], 20 mM EDTA 400 mM NaCl, 1% sodium dodecyl sulfate SDS, 400 μg of proteinase K per ml [Sigma]) at 55°C overnight, and the DNA was isolated using standard procedures.
Breeding chimeras.
Pax3-FKHR chimeric males were bred with C57BL/6 or NMRI females. Noon on the day of plug is E0.5.
Southern blot analyses.
Mouse genomic DNA was digested with PstI (Fig. 1A), fractionated on a 0.8% agarose gel, transferred to a Hybond nylon membrane (Amersham Life Science), and hybridized with 5′ and 3′ Pax3 genomic probes located outside the targeting construct (Fig. 1A).
PCR analyses.
To genotype offspring of the Pax3-FKHR chimeric mice, we used DNA from whole embryos, extraembryonal tissues, or tail snips. For direct PCR analysis, the tissues were lysed in 25 to 150 μl of proteinase K buffer, as previously described (47a). The PCR cocktail contained 2 μl of embryo lysate, 2 μl PCR buffer (Qiagen Inc.), 0.2 mM each deoxynucleoside triphosphate (Amersham Pharmacia Biotech Inc.), 0.5 μM each primers 2 and 7 (Table 1) and 0.025 U of HotStart Taq DNA polymerase (Qiagen Inc.) per ml in a total volume of 20 μl. Samples were incubated for 15 min at 95°C for activation of the Taq DNA polymerase and then amplified for 30 to 40 cycles (94°C for 30 s, 60°C for 30 s, and 72°C for 30 s). When necessary, nested PCR was performed under the same conditions. For the nested PCR, 2 μl of the undiluted PCR product or a 1:1,000 dilution (reverse transcriptase PCR [RT-PCR] of embryoid bodies) of the first PCR product was used with primers 3 and 6 (Table 1). PCR products were resolved by electrophoresis through 2% agarose gels. First and second nested rounds of PCR produced 677- and 563-bp Pax3-FKHR fragments, respectively.
TABLE 1.
Oligonucleotides used for PCR amplification of Pax3 and Pax3-FKHR sequences
| Name | Sequence | cDNA | Position (bp) |
|---|---|---|---|
| 1 | AGTCAGATGAAGGCTCCGATAT | Pax3 | 896-917 |
| 2 | GTGTCAGATCCCAGTAGCACCG | Pax3 | 1257-1279 |
| 3 | CAAAGCACTATTCCTTCGAACGC | Pax3 | 1317-1340 |
| 4 | CTGTAGCCTGCGGTGCTATAGG | Pax3 | 1673-1651 |
| 5 | CTTATGCAATATCTGGCTTGAGAT AATGAAAGG | Pax3 | 2245-2213 |
| 6 | CTCCAAGATCATCCTGTTCGGTC | PAX3-FKHR | 1609-1587 |
| 7 | AAAGTAGAGGCCATCTTTGCGGC | PAX3-FKHR | 1667-1645 |
| 8 | GCACTCTTACCAGCCCACGTCTATT | Pax3 | 1224-1248 |
| 9 | CGTTGACATCCGTAAAGACCTCTA | β-actin gene | 780-813 |
| 10 | TAAAACGCAGCTCAGTAACAGTCCG | β-actin gene | 1076-1052 |
RT-PCR analyses of RNA isolated from whole embryos, dissected embryonic structures, and differentiated embryoid bodies.
For RNA isolation, we used the RNeasy mini kit (Qiagen Inc.). RNA was isolated from single E10.5 embryos, E14.5 hearts, or one well of a 24-well plate of embryoid bodies (see below). We also dissected the hindbrain, heart, limb buds/somites, neural tubes, and pharyngeal arches from two Pax3-FKHR and two wild-type E10.5 embryos, combined the tissues of the same genotype, and isolated the RNA. RNA was purified on Qiagen columns and eluted in 50 μl (embryoid bodies) or 60 μl (embryos and dissected embryonic tissues) of water. Of these RNA solutions, we used 11 μl (whole embryos and heart), 5.5 μl (dissected embryonic tissues), or 5 μl (embryoid bodies) for reverse transcription (RT). RNA was mixed with 10 pmol of the specific primer (primer 5 for Pax3 RT, primer 7 for FKHR RT, and primer 10 for the β-actin [Table 1, Fig. 1A]) in a total volume of 12 μl, incubated at 65°C for 10 min, and chilled on ice for 5 min. Then 4 μl of 5× first-strand buffer (Life Technologies), 2 μl of 100 mM dithiothreitol (Promega), 1 μl each of 10 mM deoxynucleoside triphosphate (Fisher Scientific), 10 U of RNasin (Promega), and 200 U of Moloney murine leukemia virus RT (Life Technologies) were added. The reaction mixture was incubated at 37°C for 1 h, and the reaction was stopped by heat inactivation at 95°C for 5 min. A 2-μl aliquot of each reverse transcription reaction mixture was used for a subsequent PCR. For embryos and dissected embryonic tissues, a combination of primer 1 with primer 4 (for Pax3), 7 (first round for Pax3-FKHR), or 6 (seminested PCR for Pax3-FKHR) was used (Table 1; Fig. 1A). The first round of PCR for Pax3 produced a fragment of 777 bp, and the seminested second round PCR for Pax3-FKHR produced a fragment of 962 bp. For embryoid bodies, we used combinations of any of the following primer pairs: 8 and 4 (Pax3, 449-bp fragment), 8 and 7 (first round Pax3-FKHR, 692-bp fragment), 8 and 6 (seminested PCR for Pax3-FKHR, 634-bp fragment), and 9 and 10 (β-actin gene; 294-bp fragment) (Table 1; Fig. 1A).
Histology and immunohistochemistry.
Embryos and newborn mice were fixed in 4% paraformaldehyde in phosphate-buffered saline PBS for 12 to 48 h. Incisions on the back and abdomen of newborn mice were made prior to fixation. After fixation, the animals either were placed in 30% sucrose in PBS (for cryosections) for 24 to 48 h and then frozen in tissue-freezing medium (Triangle Biomedical Sciences) or were dehydrated and embedded in paraffin wax. Cryosections (10 μm) or paraffin sections (8 μm) of these specimens were placed on glass slides. After deparaffinizing sections in xylene and rehydrating them in PBS or removing the tissue-freezing medium from cryosections by washing in PBS, we stained them with hematoxylin-eosin or processed them for immunohistochemistry. Prior to immunohistochemical staining antigen retrieval was performed by heating the paraffin sections twice for 5 min each in 1.8 mM citric acid-8.2 mM sodium citrate (pH 6.0) in a microwave oven. Prior to incubations with antibodies, the cryosections were preblocked using an avidin-biotin blocking kit (Vector Laboratories, Inc). Whole-mount immunohistochemistry of mouse embryos was performed with a Pax3 antibody (see below), as described previously (19). Polyclonal anti-Pax3 antibodies were prepared as described previously (33) and diluted 1:500 prior to use. Specific antibody binding was detected using biotin-SP-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, Inc.). MF20 anti-myosin heavy-chain antibody was obtained from the Developmental Studies Hybridoma Bank, diluted 1:100 prior to use, and labeled with biotin using the InnoGenex labeling reagent. Biotin was detected with the Vestastain ABC-kit (Vector Laboratories, Inc). Slides were developed with Stable DAB (Research Genetics) or VIP substrate kits (Vector Laboratories, Inc).
In situ hybridization.
Whole-mount E8.5 to E12.5 embryos were hybridized with digoxigenin-labeled Pax3 (bp 1224 to 1420) and FKHR (bp 2056 to 2656) cRNA probes. The hybridized probe was visualized using alkaline phosphatase-coupled anti-digoxigenin antibody and nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate substrate (Boehrenger Mannheim) as described previously (38).
BrdU incorporation.
Pregnant mice were injected with 200 μl of a 15-mg/ml solution of 5-bromo-2′-deoxyuridine (BrdU) (Roche) in 0.9% saline-0.007 N NaOH. The animals were sacrificed 1 h after injection and processed for cryosectioning as described above. Sections were stained with anti-BrdU antibody (Becton Dickinson) as specified by the manufacturer. BrdU antibody was detected with Vestastain ABC-kit (Vector laboratories, Inc.).
ISEL staining.
Apoptotic cells were detected on paraffin-embedded tissues or tissue cryosections using the Klenow-FragEL DNA fragmentation detection kit (Oncogene Research Products) as specified by the manufacturer.
Formation and myogenic differentiation of embryoid bodies.
Prior to the formation of embryoid bodies, ES cell cultures (2.5 × 106) were purged from mouse embryo fibroblasts feeders by preplating the cells for 1 h on gelatinized tissue culture plastic and then plating them onto 6-cm gelatinized petri dishes. After 3 days of culture in the presence of leukemia-inhibiting factor, the cells were transferred to 10-cm-diameter bacterial dishes and further incubated in 15 ml of ES cell medium without leukemia-inhibiting factor under gentle shaking. Every day, 1 ml of medium containing developing embryoid bodies was collected. After an additional 4 days of culture on non-tissue culture plastic, embryoid bodies were transferred to gelatin-coated 24-well plates (1 ml per well) and differentiated in the presence or absence of basic fibroblast growth factor (bFGF) (2.5 ng/ml) for 1 to 5 days.
Interventricular septum measurements.
The widths of interventricular septa were measured on serial sections of two wild-type pups, two live-born mutant pups, and two still-born mutant pups. The average width was calculated by adding the widths of each individual section of two animals and dividing by the number of sections. The standard deviation was calculated using Microsoft Excel.
RESULTS
Targeting of the Pax3 gene.
To obtain experimental evidence for a causal role of PAX3-FKHR in the occurrence of ARMS, we attempted to model this disease in the mouse by introducing the Pax3-FKHR gene into the germ line. At present, no animal model for ARMS exists. First we created several transgenic mouse lines carrying the PAX3-FKHR cDNA under the control of the MyoD promoter, the rat β-actin promoter, or the Pgk promoter, respectively (data not shown). Unlike the results recently reported for the fusion gene expressed under the control of the Pax3 promoter-enhancer (3), none of our transgenic lines expressed PAX3-FKHR mRNA in the limb muscles and none of these lines exhibited an abnormal phenotype. We reasoned that homologous recombination of Pax3-FKHR into the mouse Pax3 locus might overcome this problem and mimic the occurrence of the human t(2;13) chromosomal translocation in muscle precursors. The targeting construct (Fig. 1A) contained mouse Pax3 exons 6 and 7 and an additional 14 kb of surrounding sequences. Pax3 exon 7 was fused to exon 2 of human FKHR by replacing its 3′ part by human PAX3-FKHR sequences obtained from a full-length fusion cDNA (33). At the 3′ side of the fusion exon we placed a Neor selectable marker in the opposite transcriptional orientation. An HSV TK-negative selectable marker was added at the 5′ end of the targeting construct. After transfection and positive-negative selection of the ES cells, we obtained 11 independently targeted clones carrying the Pax3-FKHR fusion allele, as identified by Southern blot hybridization with both 5′ (Fig. 1B) and 3′ (Fig. 1C) external probes. All clones had a normal karyotype, and eight of these were injected into C57BL/6 blastocysts. Chimeric Pax3-FKHR offspring with medium to high contribution of agouti to the coat color showed white spots on the forehead, back, and belly (Fig. 1D), similar to those of Splotch heterozygous mice. This indicated that the neural crest-derived melanocyte precursors showed migration defects due to a failure of Pax3-FKHR to compensate for Pax3 hemizygosity or to a failure of the fusion gene to express.
Pax3-FKHR heterozygous mice die around the time of birth.
Of all the chimeric Pax3-FKHR mice (>50 animals), we generated, only those derived from ES cell clones 4 and 28 gave germ line transmission, as judged by the agouti coat color of the offspring. However, none of over 150 genotyped 14-day-old agouti offspring (P14) were positive for the knock-in allele, indicating that the presence of Pax3-FKHR was not compatible with postnatal life. Genotyping of embryos of timed pregnancies showed that pups heterozygous for the fusion gene (Fig. 1B and C) died perinatally.
Most of the Pax3-FKHR heterozygous embryos had a near-normal appearance. However, 10% of them (20 of 200) exhibited deformations of the central nervous system such as exencephaly, spina bifida, and overgrowth of some parts of the caudal neural tube, data that are reported elsewhere.
Analysis of Pax3-FKHR expression during early embryogenesis.
By using whole-mount immunostaining with our Pax3 antibody, we determined that the pattern of expression of Pax3 in E8.5-12.5 knock-in heterozygous embryos was the same as in wild-type littermates (Fig. 2A). Simultaneous whole- mount in situ hybridization of age-matched Pax3-FKHR embryos with a FKHR probe showed no coincidence between the FKHR and Pax3 signals in the neural tube and dermomyotome (data not shown), suggesting that the Pax3-FKHR allele was poorly expressed.
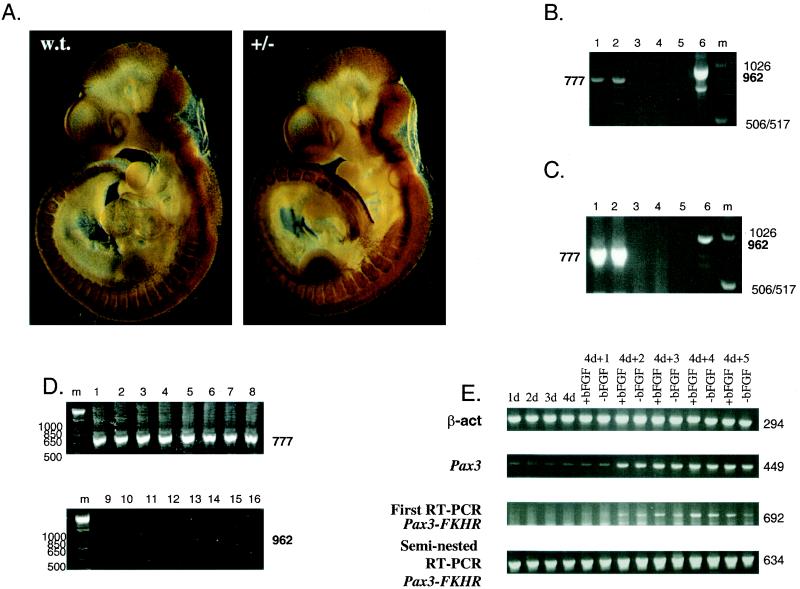
FIG.2.
Pax3-FKHR expression in embryos during early gestation and midgestation and in differentiating embryoid bodies. (A) Whole-mount immunostaining of E10.5 wild-type and Pax3-FKHR littermate embryos, using an affinity-purified Pax3 antibody. (B) RT-PCR of heart RNA from E14.5 embryos; even lanes, wild-type RNA; odd lanes, mutant RNA; lanes 1 and 2, first-round RT-PCR for Pax3; lanes 3 and 4, RT-PCR for Pax3-FKHR; lanes 5 and 6, seminested PCR for Pax3-FKHR. (C) RT-PCR of whole RNA from E10.5 embryos. Lanes: 1 and 2, first-round RT-PCR for Pax3 with RNA of wild-type and Pax3-FKHR+/− embryos; 3 and 4, first-round RT-PCR for Pax3-FKHR with RNA of wild-type and Pax3-FKHR+/− embryos; 5 and 6, seminested RT-PCR for Pax3-FKHR with RNA of wild-type and Pax3-FKHR+/− embryos; m, 1-kb marker. (D) RT-PCR of RNA from dissected body parts of E10.5 embryos. Lanes: 1 to 8 first round of RT-PCR for Pax3 with RNA of wild-type (lanes 1 to 4) and Pax3-FKHR+/− (lanes 5 to 8) embryos; 9 to 16 seminested RT-PCR for Pax3-FKHR with RNA of wild-type (lanes 9 to 12) and Pax3-FKHR+/− (lanes 13 to 16) embryos; 1, 5, 9, and 13, neural tubes; 2, 6, 10, and 14 somites and limb buds; 3, 7, 11, and 15, pharyngeal arches; 4, 8, 12, 16, remaining parts of embryos; m, 1-kb marker. Primers: for Pax3, 1 and 4; for first-round Pax3-FKHR RT-PCR, 1 and 7; for seminested Pax3-FKHR PCR, 1 and 6 (Fig. 1A; Table 1). (E) RT-PCR of differentiating embryoid bodies. Lanes: 1d to 4d, RT-PCR of embryoid bodies, grown in suspension for 1 to 4 days; 4d+1 to 4d+5, RT-PCR of embryoid bodies differentiating for 1 to 5 days after growth in suspension, in the presence (+bFGF) or absence (−bFGF) of bFGF; top gel, β-actin gene; second gel from top, Pax3; third gel from top, Pax3-FKHR first-round PCR; bottom gel, Pax3-FKHR second-round PCR. Primers (Fig. 1; Table 1): β-actin gene, 9 and 10; Pax3, 4 and 8; first Pax3-FKHR PCR, 7 and 8; seminested Pax3-FKHR PCR, 6 and 8.
To determine whether the Pax3-FKHR transcript was present in knock-in heterozygotes, we performed RT-PCR with RNA isolated from whole E10.5 embryos. Expression of Pax3 (778 bp) but not Pax3-FKHR (963 bp) was easily detectable after one round of PCR in both wild-type and Pax3-FRKH embryos (Fig. 2C). After a second round of seminested PCR, the Pax3-FKHR product could be detected in the mutant E10.5 embryos (Fig. 2C). Simultaneous RT-PCR analysis of different dissected body structures of E10.5 embryos including hindbrain, limb buds and somites, neural tube, and pharyngeal arches (Fig. 2D) showed Pax3 expression in wild-type (Fig. 2D, lanes 1 to 4) and knock-in (lanes 5 to 8), heterozygous embryos, but expression of the Pax3-FKHR allele was below detectable levels in limb buds, pharyngeal arches, and the remainder of the dissected embryo (lanes 14 to 16) and was only marginally detectable in the neural tube (lane 13). The fact that the total amount of Pax3-FKHR RT-PCR product for all the dissected embryonic parts combined was smaller than that of the whole embryo can be explained by more overall loss of RNA due to a smaller initial amount of tissue. Overall, this result is in agreement with our initial observation using in situ hybridization, confirming that the Pax3-FKHR allele is poorly expressed.
Phenotype of Pax3-FKHR heterozygous embryos.
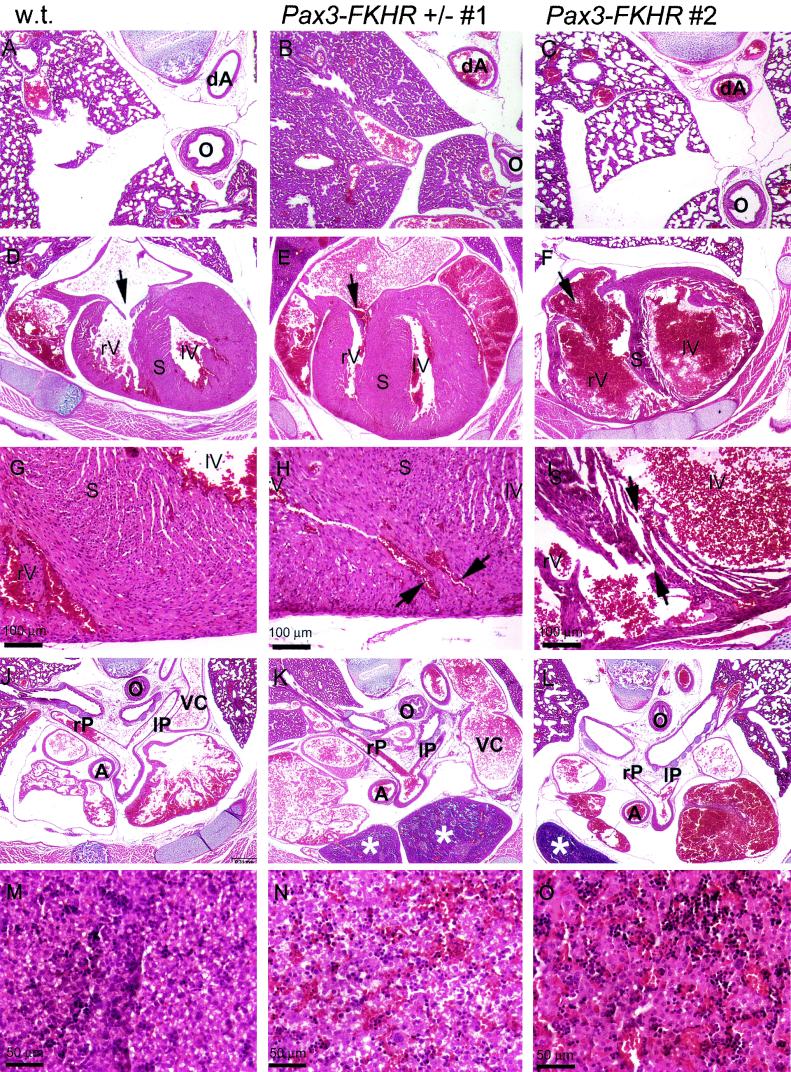
Crosses of Pax3-FKHR chimeras with C57BL/6 (clone 4) and NMRI (clone 28) females produced only 10 born Pax3-FKHR heterozygous pups (5 for ES clone 4 and 5 for ES clone 28), of which 7 (4 for ES clone 4 and 3 for ES clone 28) were dead at the time of observation, a few hours after birth. Histological analysis of three of these newborns (two for ES clone 4 and one for ES clone 28) showed that in contrast to wild-type littermates (Fig. 3A), their lung alveoli had a compact homogeneous appearance, indicating that the lungs had not inflated (Fig. 3B). Histological analysis of serial transverse sections of the thorax region of these heterozygous Pax3-FKHR embryos showed that the interventricular septum of the heart was abnormal, sometimes thicker (Fig. 3E, 666 ± 50 μm) but also sometimes thinner (see below) (Fig. 3F, 283 ± 54 μm) than that of wild-type littermates (Fig. 3D, 467 ± 79 μm). There were interventricular septum defects (VSDs) in the wall overlying the muscular part of the interventricular septum (Fig. 3H). In addition, the caval veins of the mutant hearts appeared overextended (compare Fig. 3J and K), indicating congestive heart failure.
FIG.3.
Hematoxylin-eosin staining of transverse sections of the thorax region of a P1 wild-type mouse (A, D, G, J, and M) and two Pax3-FKHR heterozygous mice (one dead at birth, [B, E, H, K, and N] and one alive [C, F, I, L, and O]). (A to C) Lungs. Lungs of a wild-type newborn (A), of a dead Pax3-FKHR newborn with noninflated lungs (B) and of a live Pax3-FKHR newborn with inflated lungs (C). (D to F) Heart. Interventricular septa and trabeculated ventricle walls of two mutant hearts appear aberrant (thicker [E] or thinner [F]) compared to those of a wild-type heart (D); the mutant heart (F) also has an abnormally large opening of the tricuspid valve (arrow). (G to I) Interventricular septum. VSD are evident in the hearts of the Pax3-FKHR knock-in mutants (arrows in panels H and I). (J to L) Rostral sections of the heart. The right and left pulmonary arteries are both open and attached to the correct lung; the thymus in mutants has an abnormal, more caudal location (asterisks in panels K and L); and the caval vein is overextended, indicating heart failure (compare VC in panels J and K). (M to O) Liver. The livers of Pax3-FKHR knock-in heterozygous embryos are engorged with blood (N and O), suggestive of heart failure. Abbreviations: rV, right ventricle; lV, left ventricle; S, septum; rP, right pulmonary trunk; lP, left pulmonary trunk; dA, decending aorta; O, esophagus.
Three mutant pups (one for ES clone 4 and two for ES clone 28) survived for at least 2 h after birth but suffered from respiratory insufficiency, exhibiting cyanosis and labored, irregular breathing. Histological analyses of sections of the thorax region of two of the live-born mutants (one from ES clone 4 and one from ES clone 28) revealed phenotypic abnormalities similar to those described above. These pups showed several small VSDs within the muscular part of the interventricular septa, some of which were present at the position where the septum attaches to the interventricular groove (Fig. 3I). The live-born animals also had an abnormally large opening of the right tricuspid valve (Fig. 3F). The differences were that the lungs had inflated (Fig. 3C) and the interventricular septum and ventricle walls were thinner (compare Fig. 3D and F).
In contrast to Splotch homozygous mice, which have persistent truncus arteriosus (20), the outflow tract of the heart appeared to be normal in all heterozygous knock-in Pax3-FKHR mutants examined. The right and left pulmonary arteries were both open and attached to the correct lung (Fig. 3J to L, rP and lP). Nonetheless, their livers were engorged with blood (compare Fig. 3N and O with M), which is suggestive of heart failure. The Pax3-FKHR mutants also had an abnormal, more caudal location of the left and right lobes of the thymus (Fig. 3K and L).
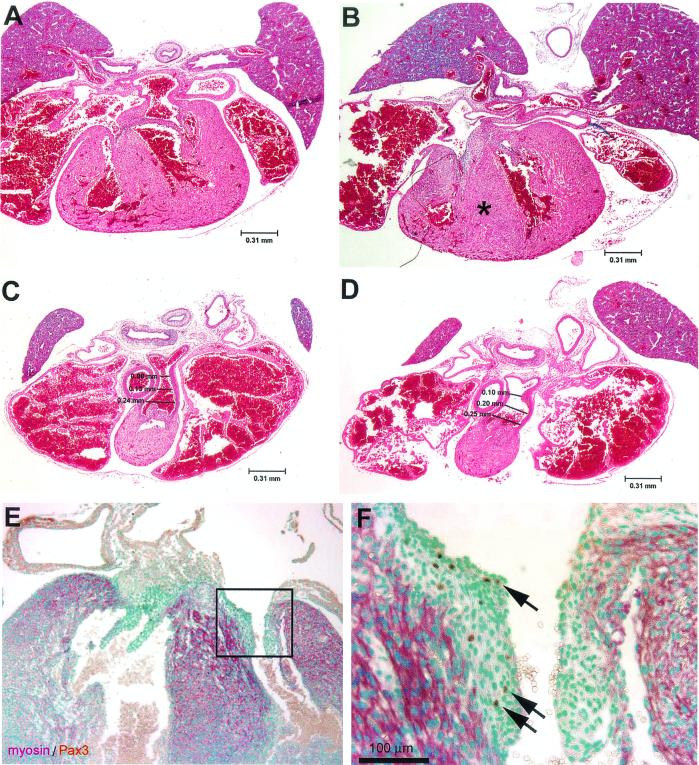
Examination of the embryos at earlier stages of development showed that the heart phenotype in Pax3-FKHR heterozygous embryos first became apparent at E16.5. Serial transverse sections through the heart of E16.5 Pax3-FKHR and wild-type embryos revealed that the muscular component of the interventricular septum was already abnormally thickened (Fig. 4A and B), while the trabeculated walls of the left and right ventricular chambers of the heart appeared normal (Fig. 4A and B). Another difference from wild-type littermates was that the lumen of the pulmonary trunk portion of the outflow tract was already dilated (compare numbers in Fig. 4C and D).
FIG. 4.
Anatomy of interventricular septum and outflow tract of the hearts of E16.5 wild-type and Pax3-FKHR heterozygous embryos and Pax3 expression. (A to D) Hematoxylin and eosin staining of transverse sections of wild-type (A and C) and mutant (B and D) hearts. The muscular component of the interventricular septum of the heterozygous mutant is already abnormally thickened (asterisk in panel B); the outflow tract of the heart of the Pax3-FKHR mouse is appropriately divided, but the lumen of the pulmonary trunk portion of the outflow tract is dilated (compare panels C and D). (E) Pax3 plus Pax3-FKHR expression in the heart of an E16.5 Pax3-FKHR embryo. (F) Higher magnification of the boxed region of panel E. Green, nuclear staining with methylene green; purple, staining with α-myosin heavy chain; brown, staining with anti-Pax3.
In addition to the heart phenotype, sagittal sections of the trunk region showed that the muscles of the diaphragm of the Pax3-FKHR heterozygotes were underdeveloped (Fig. 5A to D). At higher magnification (Fig. 5B and D), the muscle fibers of the mutant diaphragm appeared both shorter and thinner. Transverse sections revealed that some of the diaphragms also contained gaps in their musculature (Fig. 5E). Serial sagittal sections of E13.5 mutant and wild-type diaphragms showed that their size and cellularity were similar at this developmental stage (Fig. 5F and G), making it unlikely that the difference observed later during gestation was the result of a migration defect of early muscle precursors into the developing diaphragm.
FIG. 5.
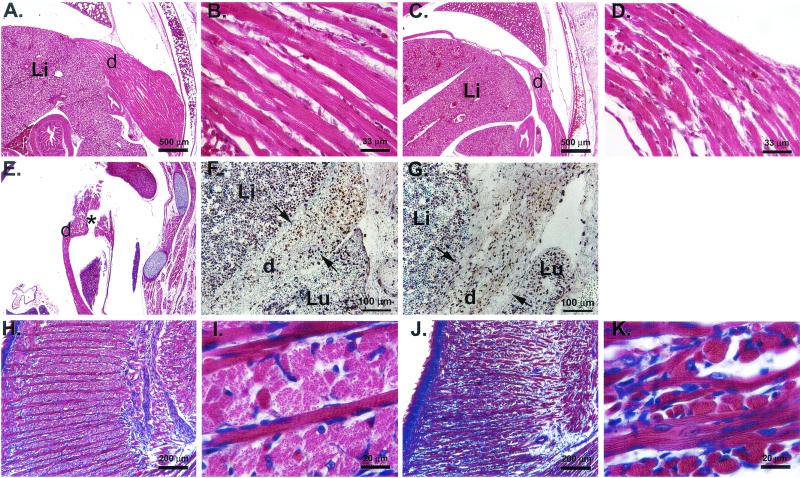
Muscles of the diaphragm and tongue of Pax3-FKHR heterozygous pups are abnormal around the time of birth but not at E13.5. (A to D) Hematoxylin and eosin staining of similar transverse sections of P1 wild-type (A and B) and Pax3-FKHR knock-in (C and D) diaphragms showing a much reduced musculature of the mutant diaphragm. (E) Sagittal section of a P1 Pax3-FKHR diaphragm showing gaps (asterisk). (F and G) BrdU incorporation and Pax3 and Pax3-FKHR expression in diaphragms of E13.5 wild-type (F) and Pax3-FKHR knock-in (G) pups. Green, nuclear staining with methylene green; purple, staining with anti-BrdU; brown, staining with anti-Pax3. (H to K) Hematoxylin and eosin staining of similar sagittal sections of the tongue of P1 wild-type (H and I) and Pax3-FKHR mutant (J and K) pups. Abbreviations: d, diaphragm; Li, liver; Lu, lung.
We also investigated whether there was a difference in the number of Pax3-expressing (antibody staining), dividing (BrdU incorporation), and apoptotic (in situ end-labeling (ISEL) staining) cells by staining sections of wild-type and mutant diaphragms at E13.5. No differences were detected (Fig. 5F and G and data not shown). Because the muscles of the limbs, tongue, and diaphragm are all derived from the lateral dermomyotome (7, 11, 45), we also analyzed histological sections of the limb and tongue muscles of these animals. The limb muscles of the mutants were indistinguishable from those of their wild-type littermates (data not shown), but the muscles of the tongue were underdeveloped and disorganized in structure (Fig. 5H and J). This was especially clear from the appearance of the lateral muscle component, whose diameter appeared to be much smaller than that in the wild-type littermates (Fig. 5I and K). We also compared serial sagittal sections of the tongue of E13.5 wild-type and mutant embryos, and similarly to E13.5 diaphragms, we noticed no significant differences in size and cellularity at this stage, nor did we see a difference in the number of Pax3-expressing, dividing (BrdU incorporation), and apoptotic (ISEL staining) cells (data not shown). Thus, the presence of the Pax3-FKHR allele impaired the normal development of some but not all hypaxial muscles, but the differences are probably not caused by a migration defect.
Taken together, the likely cause of perinatal death of the knock-in pups was congestive heart failure resulting from a combination of the VSD, tricuspid valve insufficiency, and diaphragm defects.
Expression of Pax3 and Pax3-FKHR in the heart.
To correlate Pax3 and Pax3-FKHR expression with the heart phenotype of mutant embryos, serial transverse sections of E16.5 (Fig. 4E and F) and E13.5 (data not shown) hearts were stained with Pax3 antibodies. This detected a small number of Pax3-positive cells in the mitral and tricuspid valve region of the membranous component of the interventricular septum in both wild-type and Pax3-FKHR heterozygous hearts. This, together with the described heart phenotype, led us to examine the expression of the Pax3-FKHR allele in the heart at midgestation. RT-PCR analysis of RNA from hearts of E14.5 wild-type and Pax3-FKHR heterozygous mice using Pax3 and FKHR primers of similar efficiency (data not shown) detected expression of Pax3 (Fig. 2B, lanes 1 and 2) but not of Pax3-FKHR (lanes 3 and 4) after the first round of PCR amplification. A subsequent round of seminested PCR revealed the presence of the Pax3-FKHR product in the heart of the knock-in but not of the wild-type mice (Fig. 2B, lanes 5 and 6), as confirmed by DNA sequence analysis of the bands. Thus, the Pax3-FKHR knock-in allele was expressed in the heart, albeit at a lower level than the normal Pax3 allele.
In vitro differentiation of Pax3-FKHR knock-in ES cells.
To determine whether the expression of the targeted Pax3-FKHR allele was intrinsically repressed, we generated aggregation embryoid bodies with our Pax3-FKHR ES cell lines (41). After being plated onto gelatin-coated plastic in the presence or absence of bFGF, the embryoid bodies differentiated spontaneously and generated different cell lineages (including myogenic cells) (41), in which we detected the expression of both Pax3 and Pax3-FKHR (Fig. 2E). Again, based on the observation that the Pax3 and FKHR primers were equally efficient in PCR (results not shown), we estimated that at the beginning of differentiation, the amount of Pax3-FKHR PCR product was two- to threefold smaller than that of Pax3 product but at day 4 the amounts were almost equal. This suggested that expression of the Pax3-FKHR allele is overall lower but not intrinsically compromised by the FKHR and/or Neor sequences.
Interestingly, the amount of Pax3-FKHR RT-PCR product diminished on day 5 of differentiation, indicating a possible delayed loss of Pax3-FKHR-expressing cells.
Absence of tumors in Pax3-FKHR chimeric mice and heterozygous pups.
None of the Pax3-FKHR chimeric mice (>50 mice) developed muscle tumors during the 1.5 years that they were maintained. Also, the heterozygous pups that were born showed no signs of tumorigenic growth. In view of the fact that the Pax3-FKHR allele is poorly expressed, this is not a surprising result. Therefore, a conditional knock-in allele would be a better approach to assess the tumorigenicity of Pax3-FKHR in vivo.
DISCUSSION
Expression of the Pax3-FKHR fusion during embryogenesis.
The majority of ARMSs are characterized by the t(2;13) chromosomal translocation (47), which results in the expression of the chimeric transcription factor PAX3-FKHR. The expression of the fusion protein is under the control of PAX3 regulatory sequences. The type of myogenic precursor in which the t(2;13) occurs is still unknown, but we hypothesize that it must be a primitive precursor that is likely to express PAX3, because gene transcription is thought to precede recombination so that the recombination machinery can gain access to its targets (8). Primitive muscle precursors during embryonic development and in the developing muscles after birth express PAX3 and are therefore possible targets for this translocation. In planning our experiments, we hypothesized that homologous recombination of Pax3-FKHR into the mouse Pax3 locus would mimic the t(2;13) translocation in humans and that this would result in expression of the chimeric transcription factor in all Pax3-positive tissues, including embryonic and postnatal muscle precursors. This in turn might have resulted in the occurrence of muscle tumors in heterozygous knock-in mice, provided that expression of Pax3-FKHR alone would be sufficient for tumorigenic transformation. However, none of our chimeric mice or their heterozygous progeny developed tumors. In situ hybridization and RT-PCR of whole-embryo RNA revealed that the Pax3-FKHR allele was expressed at a considerably lower level than the wild-type Pax3 allele during early embryogenesis (Fig. 2B). Expression of Pax3-FKHR in dissected E10.5 embryonic structures (limb buds and somites, neural tube, and pharyngeal arches) was just at or below the levels detectable by the assay system (Fig. 2D). It should be stressed that despite the low level of expression of the Pax3-FKHR allele, the phenotypic aberrations in our heterozygous knock-in mutants were identical in mice derived from two independently targeted ES cell clones. Most importantly, the defects were quite different from those of mice heterozygous for any of the known Splotch alleles, including those that deleted the entire Pax3 gene (25). Splotch heterozygotes show only pigmentation defects and occasionally spina bifida (21). Therefore, we are confident that the low expression of Pax3-FKHR is sufficient to create the observed phenotypic aberrations.
One of the reasons for the low expression of the fusion gene could be the presence of the Pgk-Neor cassette, which has been shown to sometimes attenuate the expression of the targeted gene (28). Our in vitro differentiation experiments with the Pax3-FKHR knock-in ES cells showed that expression of the allele was two- to threefold lower than that of the wild-type allele at the beginning of differentiation and almost equal on day 4 of differentiation. This result suggested that in the in utero developing embryo, selection might take place against Pax3-FKHR expression. This could occur through apoptosis of cells that start expressing Pax3-FKHR or, alternatively, by silencing of the fusion allele, through hypermethylation, implying that its regulation in the developing embryo is different from that in differentiating embryoid bodies in vitro. Differences in methylation at identical loci between different ES cell clones and tetraploid embryos derived from these clones have been reported (29). This shows that epigenetic modifications such as the methylation status of a gene are unstable in ES cells and could therefore explain the observed discrepancy in the level of Pax3-FKHR expression in our heterozygous embryos and ES cells. Although we did not study the methylation status of the fusion locus in embryos and ES cells, we prefer this explanation over that of massive apoptosis of Pax3-FKHR-expressing cells in the early embryo. This is based on the observation that ISEL staining of transverse serial sections of entire E9 wild-type and Pax3-FKHR embryos (data not shown) failed to show elevated levels of apoptotic cells in the dermomyotomes and dorsal neural tube of the mutants (both expressing high levels of Pax3). Obviously the suppression of Pax3-FKHR is not complete and varies among tissues, with the highest expression being found in the developing heart, somewhat lower expression being found in the neural tube, and low expression being found in the dermomyotome, limb buds, and branchial arches.
Forced expression of Pax3-FKHR in primary myoblasts cultured in vitro leads to massive apoptosis on induction of differentiation of these cells (I. Lagutina and A. Hollenbach, unpublished results). Increased apoptosis was also detected in PAX3-FKHR-expressing rhabdomyosarcomas (12). Our analysis of the diaphragm and tongue from E13.5 Pax3-FKHR mice showed that the size and cellularity of these structures were similar to those of wild-type littermates, and we could not find evidence for increased apoptosis of Pax3-FKHR cells at this developmental stage. Our in vitro culture experiments of primary myoblasts showed that cells expressing Pax3-FKHR grow faster than normal myoblasts (A. Hollenbach and I. Lagutina, unpublished results), a Pax3-FKHR effect also reported for rhabdomyosarcomas and cell lines (2, 12). It is therefore unlikely that the diminished musculature of the tongue and diaphragm of newborn Pax3-FKHR mice is caused by reduced growth of the muscle precursors. We propose that gradual apoptosis during later stages of muscle differentiation might cause the poor development of the diaphragm and tongue musculature, although we have no direct experimental support for this explanation.
Expression of the Pax3-FKHR allele during the first 4 days of differentiation of knock-in ES cells was comparable to that of the wild-type Pax3 allele (Fig. 2E), and that of Pax3-FKHR but not Pax3 mRNA appeared to diminish on day 5 of differentiation. This indicated a possible loss of Pax3-FKHR-expressing cells. This result would be in agreement with our explanation above, in which differentiating Pax3-FKHR myogenic cells in embryos undergo apoptosis.
It is noteworthy that the musculature of the limbs was not similarly affected. An explanation might be that in our mice the differentiating myoblasts in the developing limb are less sensitive to the effect of Pax3-FKHR than are those in the tongue and diaphragm. Alternatively, the muscles of the tongue and diaphragm might express more Pax3-FKHR during development than to those of the limbs.
Our data obtained with primary myoblasts do not support a direct transcriptional regulation of Pax3 by Pax3-FKHR (unpublished results). Therefore it is likely that inhibition of Pax3 function is mediated by altered regulation of downstream targets as suggested by cDNA array analysis of NIH 3T3 cells (32). These genes may include Six1, Eya2, and Mox1 (40), which are involved in the early steps of skeletal muscle differentiation.
Cardiac failure of Pax3-FKHR heterozygous mice.
The presence of a single copy of the Pax3-FKHR fusion gene caused perinatal death. In all cases, mutant animals exhibited signs of cardiac failure exemplified by a blood-engorged liver (compare Fig. 3N and O with Fig. 3M) and/or overextension of the caval veins (Fig. 3J and K), and all mutants exhibited VSDs. However, differences in the time of death between mutants correlated with the severity of diaphragm defects: mutants dying just before or during birth showed gaps in the diaphragm musculature (Fig. 5C), whereas those that survived for several hours did not. We infer that these gaps further impair the function of the diaphragm, thereby disabling the creation of negative pressure within the pleural cavity and preventing inflation of the lungs (Fig. 3B).
Small VSDs are unlikely to be the direct cause of death: human patients with small VSDs have an excellent prognosis, and in many cases these defects close spontaneously during the first 2 years of life (35). An abnormally large opening of the tricuspid valve in these mutants (Fig. 3F) indicated tricuspid valve insufficiency, which alone usually does not result in perinatal death. However, the combination of these defects (Fig. 3A and B) and the underdeveloped diaphragm may well cause respiratory and cardiac failure and subsequent death.
Origin of Pax3-expressing cells in the embryonal heart.
Pax3 is expressed during midgestation in the dorsal part of the neural tube, which gives rise to migrating neural crest cells. A specific neural crest cell population, termed the cardiac neural crest, is responsible for morphogenesis of the outflow region of the developing heart, and absence of Pax3 results in severe outflow tract defects (13). Experiments using labeled cells derived from cardiac neural crest revealed that these cells are absent in the atrioventricular canal and in the mature mitral and tricuspid valves (30). In our study we observed a small number of Pax3- and Pax3-FKHR-expressing cells in the valve region of the membranous component of the interventricular septa of E13.5 and E16.5 hearts and in the mitral and tricuspid valves of E16.5 embryos (Fig. 4). Although we do not know whether these cells have a neural crest origin, we speculate that Pax3-FKHR expression in these cells negatively affects valve function and might directly contribute to the abnormally large opening of the tricuspid valve in these mutants.
Differences and similarities between knock-in and transgenic Pax3-FKHR mice.
In the transgenic-mouse study by Anderson et al. (3), amplified Pax3 enhancer/promoter driven Pax3-FKHR transgenes did escape silencing. This may occur because not all Pax3 regulatory sequences were present in the transgenic construct, because of the absence of a Neor selectable marker, or because of integration of the transgenes into ectopic chromosomal sites. A different regulation of the Pax3-FKHR transgenes is further suggested by the observation that the transgenic lines were viable and therefore did not suffer from the respiratory failure described here. This suggests that Pax3-expressing cells contributing to the heart and diaphragm were not affected or were much less affected than in our knock-in animals.
Due to the low level of expression of the PAX3-FKHR knock-in allele, we cannot conclude whether expression of PAX3-FKHR alone is sufficient to cause rhabdomyosarcoma. However, given the fact that the transgenic model of Anderson et al. (3) also fails to develop rhabdomyosarcomas, it suggests that PAX3-FKHR expression alone might be insufficient to cause tumors.
Acknowledgments
We thank Steven Skapek for critical reading of the manuscript; Linda Mann, Oleg Lagutin, and Jeff Wiggle for help and technical advice; and Craig McPherson for the initial genotyping of offspring of Pax3-FKHR chimeras. We thank Christie Nagy for blastocyst injections.
This work was supported by Public Health Service grants HL60714 and HL60104 to S.J.C. and CA71907 to G.C.G, Cancer Center (CORE) support grant CA-21765, and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital.
REFERENCES
- 1.Anderson, J., T. Gordon, A. McManus, T. Mapp, S. Gould, A. Kelsey, H. McDowell, R. Pinkerton, J. Shipley, and K. Pritchard-Jones. 2001. Detection of the PAX3-FKHR fusion gene in paediatric rhabdomyosarcoma: a reproducible predictor of outcome? Br. J. Cancer 85:831-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J., A. Ramsay, S. Gould, and K. Pritchard-Jones. 2001. PAX3-FKHR induces morphological change and enhances cellular proliferation and invasion in rhabdomyosarcoma. Am. J. Pathol. 159:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, M. J., G. D. Shelton, W. K. Cavenee, and K. C. Arden. 2001. Embryonic expression of the tumor-associated PAX3-FKHR fusion protein interferes with the developmental functions of Pax3. Proc. Natl. Acad. Sci. USA 98:1589-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, F. G., N. Galili, J. Holick, J. A. Biegel, G. Rovera, and B. S. Emanuel. 1993. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 3:113-117. [DOI] [PubMed] [Google Scholar]
- 5.Bennicelli, J. L., S. Advani, B. W. Schafer, and F. G. Barr. 1999. PAX3 and PAX7 exhibit conserved cis-acting transcription repression domains and utilize a common gain of function mechanism in alveolar rhabdomyosarcoma. Oncogene 18:4348-4356. [DOI] [PubMed] [Google Scholar]
- 6.Bennicelli, J. L., R. H. Edwards, and F. G. Barr. 1996. Mechanism for transcriptional gain of function resulting from chromosomal translocation in alveolar rhabdomyosarcoma. Proc. Natl. Acad. Sci. USA 93:5455-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bober, E., T. Franz, H. H. Arnold, P. Gruss, and P. Tremblay. 1994. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development 120:603-612. [DOI] [PubMed] [Google Scholar]
- 8.Boehm, T., L. Mengle-Gaw, U. R. Kees, N. Spurr, I. Lavenir, A. Forster, and T. H. Rabbitts. 1989. Alternating purine-pyrimidine tracts may promote chromosomal translocations seen in a variety of human lymphoid tumours. EMBO J. 8:2621-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckingham, M. 2001. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 11:440-448. [DOI] [PubMed] [Google Scholar]
- 10.Cao, Y., and C. Wang. 2000. The COOH-terminal transactivation domain plays a key role in regulating the in vitro and in vivo function of Pax3 homeodomain. J. Biol. Chem. 275:9854-9862. [DOI] [PubMed] [Google Scholar]
- 11.Christ, B., and C. P. Ordahl. 1995. Early stages of chick somite development. Anat. Embryol. (Berlin) 191:381-396. [DOI] [PubMed] [Google Scholar]
- 12.Collins, M. H., H. Zhao, R. B. Womer, and F. G. Barr. 2001. Proliferative and apoptotic differences between alveolar rhabdomyosarcoma subtypes: a comparative study of tumors containing PAX3-FKHR or PAX7-FKHR gene fusions. Med. Pediatr. Oncol. 37:83-89. [DOI] [PubMed] [Google Scholar]
- 13.Conway, S. J., R. E. Godt, C. J. Hatcher, L. Leatherbury, V. V. Zolotouchnikov, M. A. Brotto, A. J. Copp, M. L. Kirby, and T. L. Creazzo. 1997. Neural crest is involved in development of abnormal myocardial function. J. Mol. Cell. Cardiol. 29:2675-2685. [DOI] [PubMed] [Google Scholar]
- 14.Couly, G. F., P. M. Coltey, and N. M. Le Douarin. 1993. The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development 117:409-429. [DOI] [PubMed] [Google Scholar]
- 15.Daston, G., E. Lamar, M. Olivier, and M. Goulding. 1996. Pax-3 is necessary for migration but not differentiation of limb muscle precursors in the mouse. Development 122:1017-1027. [DOI] [PubMed] [Google Scholar]
- 16.Davis, R. J., C. M. D'Cruz, M. A. Lovell, J. A. Biegel, and F. G. Barr. 1994. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 54:2869-2872. [PubMed] [Google Scholar]
- 17.Denetclaw, W. F., Jr., B. Christ, and C. P. Ordahl. 1997. Location and growth of epaxial myotome precursor cells. Development 124:1601-1610. [DOI] [PubMed] [Google Scholar]
- 18.Denetclaw, W. F., and C. P. Ordahl. 2000. The growth of the dermomyotome and formation of early myotome lineages in thoracolumbar somites of chicken embryos. Development 127:893-905. [DOI] [PubMed] [Google Scholar]
- 19.Dent, J. A., A. G. Polson, and M. W. Klymkowsky. 1989. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development 105:61-74. [DOI] [PubMed] [Google Scholar]
- 20.Franz, T. 1989. Persistent truncus arteriosus in the Splotch mutant mouse. Anat. Embryol. (Berlin) 180:457-464. [DOI] [PubMed] [Google Scholar]
- 21.Franz, T., R. Kothary, M. A. Surani, Z. Halata, and M. Grim. 1993. The Splotch mutation interferes with muscle development in the limbs. Anat. Embryol. (Berlin) 187:153-160. [DOI] [PubMed] [Google Scholar]
- 22.Furuyama, T., T. Nakazawa, I. Nakano, and N. Mori. 2000. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 349:629-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galili, N., R. J. Davis, W. J. Fredericks, S. Mukhopadhyay, F. J. Rauscher, 3rd, B. S. Emanuel, G. Rovera, and F. G. Barr. 1993. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 5:230-235. [DOI] [PubMed] [Google Scholar]
- 24.Goulding, M., A. Lumsden, and A. J. Paquette. 1994. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development 120:957-971. [DOI] [PubMed] [Google Scholar]
- 25.Goulding, M., S. Sterrer, J. Fleming, R. Balling, J. Nadeau, K. J. Moore, S. D. Brown, K. P. Steel, and P. Gruss. 1993. Analysis of the Pax-3 gene in the mouse mutant splotch. Genomics 17:355-363. [DOI] [PubMed] [Google Scholar]
- 26.Goulding, M. D., G. Chalepakis, U. Deutsch, J. R. Erselius, and P. Gruss. 1991. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 10:1135-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollenbach, A. D., J. E. Sublett, C. J. McPherson, and G. Grosveld. 1999. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO J. 18:3702-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holzenberger, M., P. Leneuve, G. Hamard, B. Ducos, L. Perin, M. Binoux, and Y. Le Bouc. 2000. A targeted partial invalidation of the insulin-like growth factor I receptor gene in mice causes a postnatal growth deficit. Endocrinology 141:2557-2566. [DOI] [PubMed] [Google Scholar]
- 29.Humpherys, D., K. Eggan, H. Akutsu, K. Hochedlinger, W. M. Rideout III, D. Biniszkiewicz, R. Yanagimachi, and R. Jaenisch. 2001. Epigenetic instability in ES cells and cloned mice. Science 293:95-97. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, X., D. H. Rowitch, P. Soriano, A. P. McMahon, and H. M. Sucov. 2000. Fate of the mammalian cardiac neural crest. Development 127:1607-1616. [DOI] [PubMed] [Google Scholar]
- 31.Jostes, B., C. Walther, and P. Gruss. 1990. The murine paired box gene, Pax7, is expressed specifically during the development of the nervous and muscular system. Mech. Dev. 33:27-37. [DOI] [PubMed] [Google Scholar]
- 32.Khan, J., R. Simon, M. Bittner, Y. Chen, S. B. Leighton, T. Pohida, P. D. Smith, Y. Jiang, G. C. Gooden, J. M. Trent, and P. S. Meltzer. 1998. Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res. 58:5009-5013. [PubMed] [Google Scholar]
- 33.Lam, P. Y., J. E. Sublett, A. D. Hollenbach, and M. F. Roussel. 1999. The oncogenic potential of the Pax3-FKHR fusion protein requires the Pax3 homeodomain recognition helix but not the Pax3 paired-box DNA binding domain. Mol. Cell. Biol. 19:594-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarville, M. B., S. L. Spunt, and A. S. Pappo. 2001. Rhabdomyosarcoma in pediatric patients: the good, the bad, and the unusual. Am. J. Roentgenol. 176:1563-1569. [DOI] [PubMed] [Google Scholar]
- 35.Moss, A. 1989. Heart disease in infants, children, and adolescents, 4th ed. The Williams & Wilkins Co., Baltimore.
- 36.Newton, W. A., Jr., E. A. Gehan, B. L. Webber, H. B. Marsden, A. J. van Unnik, A. B. Hamoudi, M. G. Tsokos, H. Shimada, D. Harms, D. Schmidt, et al. 1995. Classification of rhabdomyosarcomas and related sarcomas. Pathologic aspects and proposal for a new classification—an Intergroup Rhabdomyosarcoma Study. Cancer 76:1073-1085. [DOI] [PubMed] [Google Scholar]
- 37.Noden, D. M. 1983. The embryonic origins of avian cephalic and cervical muscles and associated connective tissues. Am. J. Anat. 168:257-276. [DOI] [PubMed] [Google Scholar]
- 38.Oliver, G., A. Mailhos, R. Wehr, N. G. Copeland, N. A. Jenkins, and P. Gruss. 1995. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development 121:4045-4055. [DOI] [PubMed] [Google Scholar]
- 39.Pappo, A. S., D. N. Shapiro, W. M. Crist, and H. M. Maurer. 1995. Biology and therapy of pediatric rhabdomyosarcoma. J. Clin. Oncol. 13:2123-2139. [DOI] [PubMed] [Google Scholar]
- 40.Ridgeway, A. G., and I. S. Skerjanc. 2001. Pax3 is essential for skeletal myogenesis and the expression of Six1 and Eya2. J. Biol. Chem. 276:19033-19039. [DOI] [PubMed] [Google Scholar]
- 41.Rohwedel, J., V. Maltsev, E. Bober, H. H. Arnold, J. Hescheler, and A. M. Wobus. 1994. Muscle cell differentiation of embryonic stem cells reflects myogenesis in vivo: developmentally regulated expression of myogenic determination genes and functional expression of ionic currents. Dev. Biol. 164:87-101. [DOI] [PubMed] [Google Scholar]
- 42.Scheidler, S., W. J. Fredericks, F. J. Rauscher III, F. G. Barr, and P. K. Vogt. 1996. The hybrid PAX3-FKHR fusion protein of alveolar rhabdomyosarcoma transforms fibroblasts in culture. Proc. Natl. Acad. Sci. USA 93:9805-9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seale, P., L. A. Sabourin, A. Girgis-Gabardo, A. Mansouri, P. Gruss, and M. A. Rudnicki. 2000. Pax7 is required for the specification of myogenic satellite cells. Cell 102:777-786. [DOI] [PubMed] [Google Scholar]
- 44.Shapiro, D. N., J. E. Sublett, B. Li, J. R. Downing, and C. W. Naeve. 1993. Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res. 53:5108-5112. [PubMed] [Google Scholar]
- 45.Tajbakhsh, S., and M. Buckingham. 2000. The birth of muscle progenitor cells in the mouse: spatiotemporal considerations. Curr. Top. Dev. Biol. 48:225-268. [DOI] [PubMed] [Google Scholar]
- 46.Tremblay, P., S. Dietrich, M. Mericskay, F. R. Schubert, Z. Li, and D. Paulin. 1998. A crucial role for Pax3 in the development of the hypaxial musculature and the long-range migration of muscle precursors. Dev. Biol. 203:49-61. [DOI] [PubMed] [Google Scholar]
- 47.Trent, J., J. Casper, P. Meltzer, F. Thompson, and J. Fogh. 1985. Nonrandom chromosome alterations in rhabdomyosarcoma. Cancer Genet. Cytogenet. 16:189-197. [DOI] [PubMed] [Google Scholar]
- 47a.Van Deursen, J., J. Boer, L. Kasper, and G. Grosueld. 1996. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup 214. EMBO J. 15:5574-5583. [PMC free article] [PubMed] [Google Scholar]
- 48.Williams, B. A., and C. P. Ordahl. 1994. Pax-3 expression in segmental mesoderm marks early stages in myogenic cell specification. Development 120:785-796. [DOI] [PubMed] [Google Scholar]