Abstract
Box H/ACA small nucleolar ribonucleoprotein particles (H/ACA snoRNPs) play key roles in the synthesis of eukaryotic ribosomes. The ways in which these particles are assembled and correctly localized in the dense fibrillar component of the nucleolus remain largely unknown. Recently, the essential Saccharomyces cerevisiae Naf1p protein (encoded by the YNL124W open reading frame) was found to interact in a two-hybrid assay with two core protein components of mature H/ACA snoRNPs, Cbf5p and Nhp2p (T. Ito, T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki, Proc. Natl. Acad. Sci. USA 98:4569-4574, 2001). Here we show that several H/ACA snoRNP components are weakly but specifically immunoprecipitated with epitope-tagged Naf1p, suggesting that the latter protein is involved in H/ACA snoRNP biogenesis, trafficking, and/or function. Consistent with this, we find that depletion of Naf1p leads to a defect in 18S rRNA accumulation. Naf1p is unlikely to directly assist H/ACA snoRNPs during pre-rRNA processing in the dense fibrillar component of the nucleolus for two reasons. Firstly, Naf1p accumulates predominantly in the nucleoplasm. Secondly, Naf1p sediments in a sucrose gradient chiefly as a free protein or associated in a complex of the size of free snoRNPs, whereas extremely little Naf1p is found in fractions containing preribosomes. These results are more consistent with a role for Naf1p in H/ACA snoRNP biogenesis and/or intranuclear trafficking. Indeed, depletion of Naf1p leads to a specific and dramatic decrease in the steady-state accumulation of all box H/ACA snoRNAs tested and of Cbf5p, Gar1p, and Nop10p. Naf1p is unlikely to be directly required for the synthesis of H/ACA snoRNP components. Naf1p could participate in H/ACA snoRNP assembly and/or transport.
The nucleolus of eukaryotic cells contains a multitude of small nucleolar ribonucleoprotein particles (snoRNPs) that can be classified in three groups, based on the nature of their RNA component and associated proteins. The vast majority of these snoRNPs are either of the so-called box C/D or box H/ACA class (3, 21, 79). Only two related snoRNPs, MRP and RNase P, do not belong to those two groups. RNase P removes the 5′ leader segment of pre-tRNAs (19), while MRP performs the endonucleolytic cleavage of site A3 in the pre-rRNA (13, 49, 51, 57, 78, 84). Most box C/D and box H/ACA snoRNPs (thereafter termed C/D and H/ACA snoRNPs) respectively catalyze, within the pre-rRNA, the site-specific methylation of the 2′ oxygen of certain ribose moieties (2, 8, 10, 37, 39, 85, 92) and the conversion of specific uridines into pseudouridines (20, 62, 79, 87). In addition, some C/D snoRNPs are responsible for the site-specific methylation of spliceosomal U6 snRNA in the nucleolus (22, 93), while certain tissue-specific C/D snoRNPs may be involved in the modification of mRNAs (9). Moreover, few members of both snoRNP families are involved in certain pre-rRNA cleavage steps necessary for the production of mature rRNAs (31, 48, 58, 68, 76, 86, 91, 96). Quite remarkably, RNPs strongly related to H/ACA and/or C/D snoRNPs were recently found that are responsible for site-specific modifications of spliceosomal snRNAs transcribed by RNA polymerase II (14, 33). These particles accumulate within Cajal bodies and have hence been termed scaRNPs (for small-Cajal-body-specific RNPs) (14).
C/D snoRNPs all contain a small RNA component featuring the conserved C and D boxes (hence the name) and are associated with at least four proteins, Nop1p (in yeast) or fibrillarin (in higher eukaryotes) (3, 21, 63, 77, 88), Nop58p, Nop56p (23, 44, 52, 61, 103, 104), and Snu13p (yeast)/15.5-kDa or NHPX protein (human) (102). H/ACA snoRNPs derive their name from the nature of their RNA component (H/ACA snoRNA), characterized by the presence of two conserved sequence motifs, the H box and the ACA box, and a conserved secondary structure. This structure consists of two hairpins containing an irregular bulge (the “pseudouridylation pocket”) separated by a single-stranded hinge region containing the H box and followed by a single-stranded tail containing the 5′ACA3′ triplet situated 3 nucleotides from the 3′ end of the mature snoRNA (3, 21). The core of H/ACA snoRNPs contains four proteins; Cbf5p in yeast (5, 34, 43, 101, 105) (the orthologues of which are termed Nop60B in Drosophila [24, 71], Nap57 in rodents [54, 104], and dyskerin in humans [55]), Gar1p (3, 16, 21, 26), Nhp2p, and Nop10p (29, 40, 72, 101).
Except for the C/D snoRNA U3 in plants, which is transcribed by RNA polymerase III (38), C/D or H/ACA snoRNAs are transcribed by RNA polymerase II either as independent transcription units, polycistronic (i.e., comprising several small RNAs) transcripts, or as part of introns of pre-mRNAs (83). Intronic snoRNAs are released from pre-mRNAs by two alternative mechanisms. In most cases, intronic snoRNAs are produced from the debranched intron lariat, following splicing of the flanking exons (65, 70). Alternatively, some intronic C/D snoRNAs are released directly from the pre-mRNA in which they are embedded by endonucleolytic digestions performed by RNase III (6, 25, 99, 100). Release of mature snoRNAs from independent or polycistronic transcription units is initiated by an endonucleolytic, possibly cotranscriptional, digestion event in the 3′ portion of the primary transcript that requires the Nrd1p protein, the Sen1p helicase, and the cleavage factor IA activity of the RNA polyadenylation machinery (17, 56, 81). Sequences of different snoRNAs present on the same polycistronic transcript are separated from one another by RNase III-catalyzed endonucleolytic digestions (11, 12, 73). Precursor transcripts containing a single snoRNA may also be cleaved 5′ to the mature snoRNA sequence by RNase III (11). In all cases, final pre-snoRNA maturation steps always involve 3′-to-5′ exonucleolytic digestion performed by the exosome (1, 94) and, at least in the case of intronic or polycistronic snoRNAs, by 5′-to-3′ exonuclease digestion by Rat1p (70, 73) to the ends of the mature snoRNAs.
At what stage of the pre-snoRNA maturation process and where and how snoRNP proteins assemble with the sequences retained in the mature snoRNAs remain questions of debate and ongoing research. Snu13p may be the protein that initiates the assembly of C/D snoRNPs, since it interacts directly and specifically in vitro with the so-called “kink-turn” motif formed by the interaction between the C and D boxes (102). This hypothesis is strongly supported by the recent work of Omer et al., who have succeeded in reconstituting active archaeal C/D particles from purified components (64). They demonstrate that the obligatory order of in vitro RNP assembly is archaeal L7a (the archaeal orthologue of Snu13p; see also reference 41), archaeal NOP56, and archaeal fibrillarin (64). Information concerning H/ACA snoRNP assembly is far scarcer. Nhp2p is known to bind directly to RNA in vitro but is unlikely, on its own, to initiate the assembly of H/ACA snoRNPs (28). The nucleation steps of C/D and H/ACA snoRNP assembly are likely to occur during pre-snoRNA processing, possibly very early (75). Proteins bound to conserved boxes of the snoRNAs are believed to prevent exonucleases, generating the mature snoRNA termini, from degrading the body of snoRNAs. In addition, Nop1p is proposed in some cases to actively participate in pre-snoRNA processing by recruiting yeast RNase III (25). Gar1p has also been shown to interact directly with yeast RNase III, but the importance of this interaction for pre-snoRNA processing is unclear (90). Our understanding of how the particles reach the dense fibrillar component of the nucleolus, where they function, remains very limited. Most studies point to proteins bound to the conserved boxes of the snoRNAs as crucial to the nucleolar targeting of the particles (45-47, 59, 60, 75, 98). C/D snoRNPs transit through the Cajal bodies (in mammalian cells) (60, 83, 97) or the “nucleolar body” (97, 98) (in yeast) before reaching the dense fibrillar component of the nucleolus. Whether H/ACA snoRNPs follow the same route remains uncertain (83, 98).
In the present paper, we describe the characterization of Naf1p (encoded by the YNL124W open reading frame) as a predominantly nucleoplasmic protein specifically required for normal steady-state accumulation of H/ACA snoRNPs. Our results are compatible with a role for Naf1p in the assembly and/or the intranuclear trafficking of H/ACA snoRNPs.
MATERIALS AND METHODS
Strains, media, and plasmids.
Strains GAL::naf1 and GAL::zz-naf1 were obtained as follows: two gene cassettes flanked on the 5′ side by a segment of the NAF1 promoter and on the 3′ side by a 5′ segment of the NAF1 open reading frame and containing either the HIS3 gene marker and the GAL10 promoter (cassette 1) or the same elements followed by the ZZ tag sequence (cassette 2) were PCR amplified by using plasmid pTL26 (42) and oligonucleotides YNL124-HIS3 (5′ AGCTTAAAGGTAAAGGAAAAAGATGGTAGGATGATAGGTAGGATGAAGCGGCTCTTGGCCTCCTCTAG 3′) and YNL124-pGAL (5′ CAAGTCCTGGTCGGGATTCTCCAAAGCCTTAGAAAACAAGTCATCGCTCATGATTACGAATTCCTTGAATTTTCAAA 3′) for cassette 1. For cassette 2, plasmid pTL27 (42) and oligonucleotides YNL124-HIS3 and YNL124-PROTA (5′ CAAGTCCTGGTCGGGATTCTCCAAAGCCTTAGAAAACAAGTCATCGCTCATATTCGCGTCTACTTTCGG 3′) were used. Cassettes 1 and 2 were integrated into strain YDL402 (42), creating strains GAL::naf1 and GAL::zz-naf1, respectively. To produce strain GAL::naf1/CBF5-TAP, the following procedure was followed: a gene cassette flanked on the 5′ side by 48 3′ terminal nucleotides from the CBF5 open reading frame and on the 3′ side by 47 nucleotides of the CBF5 terminator and containing the TAP tag sequence and the Kluyveromyces lactis TRP1 gene marker was amplified using plasmid pBS1479 (74) and oligonucleotides TAP-Cbf5/1 (5′ TCTGAAGACGGTGATTCTGAGGAAAAGAAATCTAAGAAATCTAAGAAATCCATGGAAAAGAGAAG 3′) and TAP-Cbf5/2 (5′ TCTAATCTAATAATAGAAAAAGTTTTTTGAAAAAAAGAAAGCTGTTATACGACTCACTATAGGG 3′). This cassette was integrated in strain GAL::naf1, creating strain GAL::naf1/CBF5-TAP. A strain expressing Naf1p-ZZ (CEN.PK; ura3-52; his3Δ1; leu2-3 112; trp1-289; YNL124w [44,1436]::kanMX4; pCD31) used for the immunoprecipitation experiments, the analysis of the sedimentation profile of Naf1p on a glycerol gradient, and the immunolocalization of Naf1p was produced as follows: the diploid strain CVLE083 (HE) (EUROSCARF accession number 30507D) (CEN.PK; MATa/α; ura3-52/ura3-52; his3Δ1/his3Δ1; leu2-3 112/leu2-3 112; trp1-289/trp1-289; YNL124w [44,1436]::kanMX4/YNL124W) was transformed with the centromeric plasmid pCD31, containing the NAF1-ZZ gene cassette expressed from the GAR1 promoter and terminator sequences. To produce pCD31, the NAF1 open reading frame flanked by BglII restriction sites was PCR amplified using yeast genomic DNA and oligonucleotides YNL124/1 (5′ CCCCCCAGATCTAATCATGAGCGATGACTTGTTTTCTAAGGCT 3′) and YNL124/2 (5′ CCCCCCAGATCTAGGGTTCTTGGATCTTGCTGATGCTGAT 3′). The resulting PCR fragment was digested with BglII and inserted into plasmid pHA113 (28) cut with BamHI, creating pCD31. Sporulation of the diploid strain (CEN.PK; MATa/α; ura3-52/ura 3/52; his3Δ1/his3Δ1; leu2-3 112/leu2-3 112; trp1-289/trp1-289; YNL124w [44,1436]::kanMX4/YNL124W; pCD31) was induced, tetrads were dissected, and a haploid strain (CEN.PK; ura3-52; his3Δ1; leu2-3 112; trp1-289; YNL124w [44,1436]::kanMX4; pCD31) was selected.
Saccharomyces cerevisiae strains were grown either in yeast-peptone medium (1% yeast extract, 1% peptone) supplemented with either 2% galactose, 2% raffinose, 2% sucrose, or 2% glucose as the carbon source or in YNB medium (0.17% yeast nitrogen base and 0.5% [NH4]2SO4) supplemented with 2% galactose, 2% raffinose, 2% sucrose, and the required amino acids. Escherichia coli DH5α strain (F', endA1, hsdr17 [rk− mk+], supE44, thi-1, recA1, gyrA [Nalr], relA1, Δ[lacIZYA-argF]U169, deoR, [φ80dlacΔ{lacZ}M15]) grown on Luria-Bertani (1% Bacto Tryptone, 0.5% Bacto-yeast extract, and 1% NaCl) liquid or solid media was used for all cloning procedures.
Production of anti-Nop10p antibodies.
An internal peptide from Nop10p (H2N CSAHPARFSPDDKY CONH2) was used to immunize a rabbit (performed by Eurogentec S.A.). The anti-Nop10p polyclonal serum obtained after 3 months reacted at 500-fold dilution with Nop10p from total yeast cellular extracts.
Production of total protein extracts.
Total protein extracts used for the Western blot analyses presented in Fig. 2A and 6B were produced as follows: a pellet corresponding to approximately 2 × 107 yeast cells was resuspended in 100 μl of ice-cold H2O. Twenty microliters of 100% trichloroacetic acid (TCA) and 100 μl of glass beads were then added. Yeast cells were broken by vigorous agitation at 4°C during 6 min. One milliliter of ice-cold 5% TCA was added, and the sample was centrifuged 15 min at 4°C and 16,000 × g in a microcentrifuge (Eppendorf 5415D). The supernatant was removed, and the pellet was resuspended in 80 μl of 100 mM Tris-HCl, pH 8.0, 4% sodium dodecyl sulfate (SDS), 20% glycerol, 0.04% bromophenol blue, and 200 mM dithiothreitol (DTT). Twenty microliters of 1 M Tris-HCl, pH 9.5, was added, and the sample was heated at 80°C for 5 min. The sample was then mixed by vigorous agitation for 5 min at 4°C and was heated again at 80°C for 5 min. Forty microliters of 100 mM Tris-HCl, pH 8.0, 4% SDS, 20% glycerol, 0.04% bromophenol blue, 200 mM DTT, and 10 μl of 1 M Tris-HCl, pH 9.5, was then added, and the sample was agitated vigorously for 5 min at 4°C. The sample was finally centrifuged 5 min at 4°C and 16,000 × g in a microcentrifuge (Eppendorf 5415D), and the supernatant was collected.
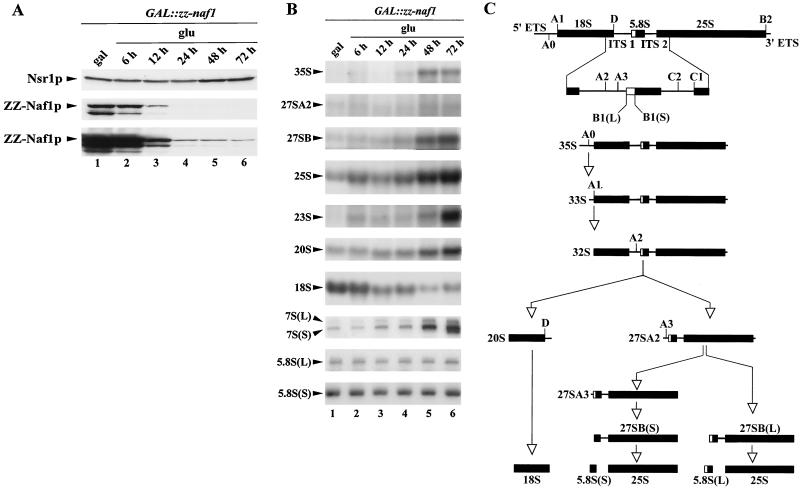
FIG. 2.
Naf1p depletion affects 18S rRNA accumulation. GAL::zz-naf1 cells were grown in a medium containing galactose (gal), raffinose, and sucrose, were washed, and were shifted to a medium containing glucose (glu). Cell aliquots were collected from the culture grown in galactose, raffinose, and sucrose (lanes 1) and after 6 (lanes 2), 12 (lanes 3), 24 (lanes 4), 48 (lanes 5) and 72 (lanes 6) h of growth in glucose-containing medium. Total proteins and RNAs were extracted from these samples for Western (A) and Northern (B) blot analysis. (A) Proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to a cellulose membrane. ZZ-Naf1p was detected by use of rabbit PAP and Nsr1p by using a monoclonal antibody. The third panel corresponds to an overexposed film to show the residual production of ZZ-Naf1p even after prolonged growth on glucose-containing medium. (B) High-molecular-weight RNAs were separated on a formaldehyde-agarose gel, and low-molecular-weight RNAs were separated on a denaturing 6% polyacrylamide gel. Gels were transferred to nylon membranes. Mature rRNAs and pre-rRNA processing intermediates were detected by use of specific oligonucleotide probes. (C) Cartoon of the pre-rRNA processing pathway. In wild-type cells, the 35S pre-rRNA is first cleaved at site A0 within the 5′ external transcribed spacer (5′ ETS), producing intermediate 33S, which is very rapidly cleaved at site A1, the 5′ end of 18S rRNA, to produce intermediate 32S. 32S is then cleaved at site A2 within the internal transcribed spacer 1 (ITS1), releasing 20S, the immediate precursor to 18S rRNA, and 27SA2. 27SA2 is then processed via two alternative pathways. It is either cut at site A3 to produce 27SA3, which is then trimmed by 5′-to-3′ exonucleases up to site B1(S), producing 27SB(S). Alternatively, it can be processed into 27SB(L) by an as-yet-unknown mechanism. 27SB(S) and 27SB(L) are then processed in the same manner to produce 25S and 5.8S(S) or 5.8S(L), respectively. In cells depleted of Naf1p, a fraction of 35S is directly cut at site A3, producing 23S that is degraded by the exosome. For a detailed review of the pre-rRNA processing pathway in yeast, see reference 96.
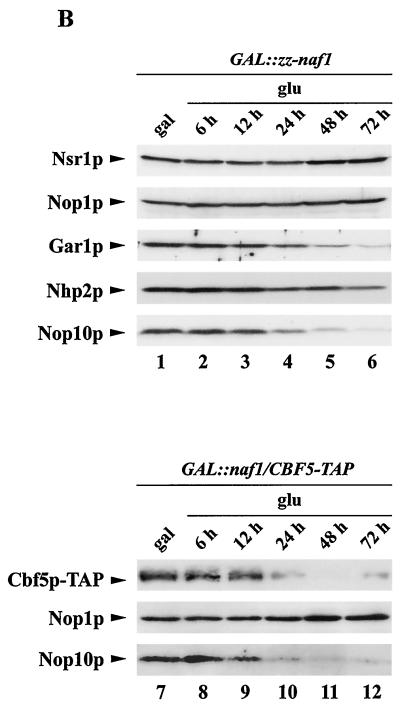
FIG. 6.
Analysis of steady-state levels of H/ACA snoRNP components and of the snR44 host gene mRNA in Naf1p-depleted cells. GAL::zz-naf1 (lanes 1 to 6) or GAL::naf1/CBF5-TAP (lanes 7 to 12) cells were grown in a medium containing galactose (gal), raffinose, and sucrose, were washed, and shifted to a medium containing glucose (glu). Cell aliquots were collected from the cultures grown in galactose, raffinose, and sucrose (lanes 1 and 7) and after 6 (lanes 2 and 8), 12 (lanes 3 and 9), 24 (lanes 4 and 10), 48 (lanes 5 and 11), and 72 (lanes 6 and 12) h of growth in glucose-containing medium. Total RNAs and proteins were extracted from these samples for Northern (A) and Western (B) blot analysis. A large set of RNAs was detected by use of complementary oligonucleotide probes. Phosphorimager scans of the Northern blots were used to quantify RNA levels. Values indicated are percentages of the figures obtained for RNAs originating from cells grown on galactose, raffinose, and sucrose. Cbf5p-TAP was detected using rabbit PAP. The remaining proteins were detected as described in previous figure legends.
Immunoprecipitations.
Total cellular extracts were produced from strains expressing either Naf1p-ZZ (see above), Cbf5p-ZZ (43), ZZ-Nop1p (kindly provided by T. Bergès; see reference 21), or the free ZZ tag (29). Cells frozen in liquid nitrogen were broken with dry ice in a kitchen blender (Osterizer). Aliquots of broken cell powder corresponding to 2 × 1010 cells were resuspended in 1 ml of 20 mM Tris-HCl, pH 8.0, 5 mM magnesium acetate [Mg(OAc)2], 0.2% Triton X-100, 150 mM potassium acetate (KOAc), 1 mM DTT, 1-U/μl RNasin (Promega), and protease inhibitors. Extracts were clarified by centrifuging 10 min at 16,000 × g in a microcentrifuge (Eppendorf 5415D). Aliquots of extracts corresponding to 10 mg of proteins were added to 70 μl of immunoglobulin G (IgG)-Sepharose beads (Amersham Pharmacia Biotech) in 1 ml of final volume of either the above buffer (20 mM Tris-HCl, pH 8.0, 5 mM Mg(OAc)2, 0.2% Triton X-100, 150 mM KOAc, 1 mM DTT, 1-U/μl RNasin [Promega], and protease inhibitors) or the same buffer, except that the KOAc concentration was adjusted to 500 mM. Immunoprecipitation was performed at 4°C for 1 h and 30 min on a shaking table. Beads were then washed five times with 1 ml of the buffer used for the immunoprecipitation (ice cold). The beads were divided in two aliquots for protein or RNA analysis. For protein analysis, the beads were washed further twice with 1 ml of ice-cold 150 or 500 mM sodium acetate, 20 mM Tris-HCl, pH 8.0, 5 mM Mg(OAc)2, 0.2% Triton X-100, 1 mM DTT, 1-U/μl RNasin (Promega), and protease inhibitors. After being washed, the beads were resuspended in 50 μl of 100 mM Tris-HCl, pH 8.0, 4% SDS, 20% glycerol, 0.04% bromophenol blue, and 200 mM DTT. For RNA analysis, the beads were washed additionally twice with the ice-cold buffer used for immunoprecipitation. One hundred sixty microliters of 4 M guanidinium isothiocyanate solution, 4 μl of glycogen, 80 μl of 100 mM sodium acetate (pH 5), 10 mM Tris-HCl (pH 8.0), 1 mM EDTA solution, 120 μl of phenol, and 120 μl of chloroform were added to the beads. The samples were thoroughly mixed, incubated 5 min at 65°C, and centrifuged 5 min at 4°C and at 16,000 × g in a microcentrifuge (Eppendorf 5415D). The aqueous phases were recovered and mixed with 120 μl of phenol and 120 μl of chloroform, and the samples were centrifuged 5 min at 4°C and 16,000 × g in a microcentrifuge (Eppendorf 5415D). RNAs from the aqueous phases were then precipitated with ethanol.
Fractionation of yeast extract on glycerol gradient.
A total cellular extract was prepared as described in the previous paragraph from the strain expressing Naf1p-ZZ (CEN.PK; ura3-52; his3Δ1; leu2-3 112; trp1-289; YNL124w [44,1436]::kanMX4; pCD31). Five hundred microliters of extract corresponding to 5 mg of proteins was loaded on a 10 to 30% glycerol gradient. Preparation of the gradient, loading of the extract, centrifugation, and collection of fractions were performed as described in references 4 and 95.
Western analysis.
Proteins from total extracts, obtained from gradient fractions after TCA precipitation or from immunoprecipitated pellets, were separated on SDS-12% polyacrylamide gels and transferred to Hybond-C extra membranes (Amersham Pharmacia Biotech). Gar1p, Nsr1p, and ribosomal protein L3 were detected as described in reference 4. Ribosomal protein S8 was detected by use of a rabbit polyclonal serum diluted 2,000-fold. Nhp2p was detected as described in reference 28. Nop10p was detected by use of the anti-Nop10 serum (see above) diluted 500-fold. Naf1p-ZZ and Cbf5p-TAP were detected using rabbit PAP (Dako) diluted 10,000-fold.
RNA extractions, Northern hybridizations, and 3′ end labeling of RNAs.
RNA extractions were performed as described in reference 89. RNA fractionations by polyacrylamide gel electrophoresis were performed as described in reference 29. Pre-rRNA precursors, mature rRNAs, RPS22B mRNA, and various small RNAs were detected using 32P-labeled oligodeoxynucleotide probes. Sequences of antisense oligonucleotides used to detect pre-rRNA precursors, mature rRNAs, snR10, snR11, snR31, snR33, snR36, snR42, snR46, U14, U24, U1, and MRP were reported in reference 29. The sequence of the oligonucleotide used to detect snR37 was reported in reference 28, the anti-TLC1 oligonucleotide was described in reference 15, and the anti-snR72 and anti-snR78 oligonucleotides were described in reference 73. The remaining oligonucleotides used were as follows: anti-RPS22B mRNA oligonucleotide 1, 5′ TGTACCACTACTAAAAACTTACTTAATAG 3′; anti-RPS22B mRNA oligonucleotide 2, 5′ AGCGAGTCATTTTTTACCTAATTACTA 3′; anti-U2, 5′ CCAACCCCACCCTACACCCCC 3′; anti-U3, 5′ ATGGGGCTCATCAACCAAGTTGG 3′; anti-U4, 5′ GACCATGAGGAGACGGTCTGG 3′; anti-U5, 5′ CAACACCCGGATGGTTCTGG 3′; anti-U6, 5′ CATCCTTATGCAGGGGAACTG 3′; anti-RNase P RNA, 5′ CGCCGTAGCGGGCGACAAGTC 3′; and anti-SRP RNA, 5′ CCCACCAGAAAGCCATTACAGCC 3′. Blots were hybridized with 5′ end-labeled oligonucleotide probes and were washed as described in reference 29. End labeling (3′) of RNAs with [5′-32P]pCp was performed as described in reference 21.
Pulse-chase analysis.
It was performed as indicated in reference 4, except that cells were grown in rich yeast-peptone medium and were shifted to minimal yeast nitrogen base medium only 4 h before the labeling.
Immunoelectron microscopy.
Detection of Naf1p-ZZ by immunoelectron microscopy was performed as described in reference 29.
RESULTS
Naf1p interacts with H/ACA snoRNP components.
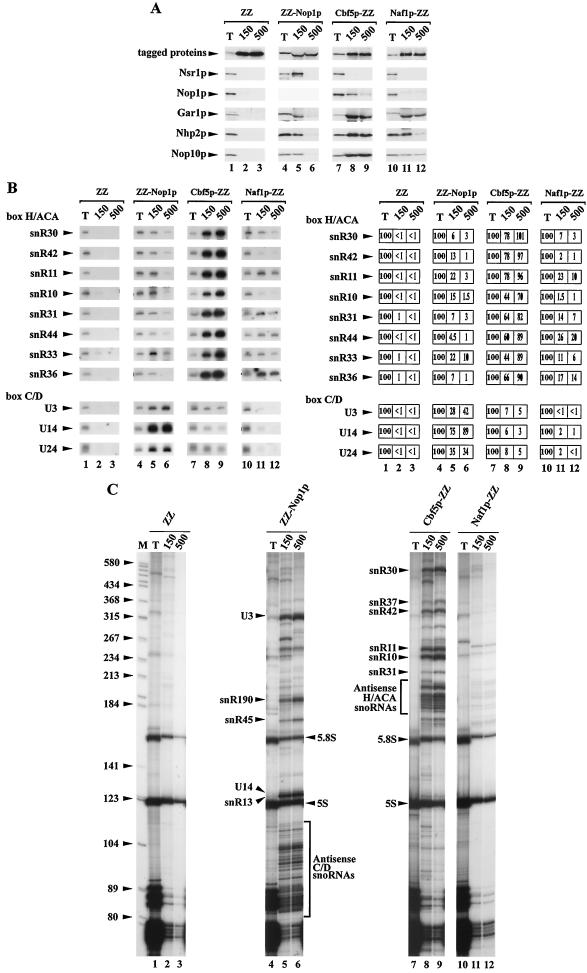
In the course of a comprehensive double-hybrid analysis of the S. cerevisiae proteome, Ito and colleagues found that the protein encoded by the YNL124W open reading frame (hereafter named Naf1p according to the nomenclature of the Saccharomyces Genome Database) interacts with both Cbf5p and Nhp2p, two core components of H/ACA snoRNPs (32). This result prompted us to assess by biochemical means the nature of the interaction between Naf1p and H/ACA snoRNP components. To that end, we constructed a yeast strain that expresses Naf1p-ZZ, i.e., a Naf1p protein tagged at its C terminus with two synthetic IgG-binding domains (called ZZ) derived from Staphylococcus aureus protein A. Strains expressing ZZ-tagged Nop1p (a core component of C/D snoRNPs), ZZ-tagged Cbf5p (a core component of H/ACA snoRNPs), or the ZZ tag alone were also used for control experiments. Immunoprecipitation experiments were carried out using total cellular extracts containing tagged proteins and IgG-Sepharose beads to which these avidly bind. Proteins and RNAs precipitated with the tagged proteins were analyzed by Western (Fig. 1A) and Northern (Fig. 1B) blot and [32P]pCp labeling (Fig. 1C) procedures, respectively. H/ACA snoRNP proteins Gar1p, Nhp2p, and Nop10p are precipitated with Naf1p-ZZ, albeit to a low extent (Fig. 1A, lanes 10 to 12). We estimate that approximately 5% of input Nhp2p or Nop10p and at least 10% of input Gar1p are precipitated with Naf1p-ZZ in 150 mM KOAc. Under these conditions, Nhp2p and Nop10p are precipitated with ZZ-Nop1p to the same extent (Fig. 1A, lanes 4 to 6). However, whereas the interaction between ZZ-Nop1p and Gar1p, Nhp2p, or Nop10p is strongly affected by an increase in KOAc concentration, the interaction between Naf1p-ZZ and Gar1p or Nop10p is more resistant. In addition, we note that the interaction between Naf1p-ZZ and Gar1p, Nhp2p, or Nop10p is specific. Under the conditions used, we detect no interaction between Naf1p-ZZ and Nop1p or Nsr1p, a nucleolar protein required for 18S rRNA production and efficient C/D snoRNA nucleolar targeting (Fig. 1A, lanes 10 to 12).
FIG. 1.
Analysis of the interactions between Naf1p-ZZ and H/ACA snoRNP components. Total extracts were produced in native conditions from yeast cells expressing either free ZZ tag (lanes 1 to 3), ZZ-Nop1p (lanes 4 to 6), Cbf5p-ZZ (lanes 7 to 9), or Naf1p-ZZ (lanes 10 to 12). Immunoprecipitation experiments were carried out using IgG-Sepharose beads in a buffer containing either 150 mM (lanes 2, 5, 8, and 11) or 500 mM (lanes 3, 6, 9, and 12) KOAc. After precipitation and being washed, beads were separated in two equal fractions. (A) Western blot analysis. Beads from one set of fractions were resuspended in protein-denaturing buffer. Aliquots of the resulting supernatants (lanes 2, 3, 5, 6, 8, 9, 11, and 12) and 1/20 of the corresponding amount of total proteins from the input extract (T and lanes 1, 4, 7, and 10) were submitted to SDS-polyacrylamide gel electrophoresis. Proteins were transferred to cellulose membranes. Tagged proteins were detected by use of rabbit PAP, Nsr1p by a monoclonal antibody, and Nop1p, Gar1p, Nhp2p, and Nop10p by polyclonal sera. Northern blot (B) and analysis (C) of [32P]pCp-labeled RNAs. RNAs retained on the beads of the second set of fractions were purified. Aliquots of precipitated RNAs (lanes 2, 3, 5, 6, 8, 9, 11, and 12) and 1/10 of the corresponding amount of total RNAs from the input extract (T and lanes 1, 4, 7, and 10) were submitted to denaturing 6% polyacrylamide gel electrophoresis. In panel B, separated RNAs were transferred to nylon membranes, and various H/ACA and C/D snoRNAs were detected by hybridization with specific oligonucleotide probes. Phosphorimager scans of the Northern blots were used to quantify snoRNA levels. The amounts of RNAs precipitated, expressed as percentages of input RNA levels, are indicated in the table on the right. In panel C, RNAs were 3′ end labeled with [32P]pCp and separated on a 6% sequencing gel. M, molecular weight markers (pBR322 digested with HaeIII and TaqI). Positions of RNAs inferred from their size are indicated.
As shown in Fig. 1B, lanes 10 to 12, several H/ACA snoRNAs tested (snR30, snR11, snR31, snR44, snR33, and snR36) are also precipitated with Naf1p-ZZ, with efficiencies ranging from 7 to 26%. Although significant, these efficiencies are much lower than those obtained in the case of Cbf5p-ZZ, with which H/ACA snoRNAs are precipitated almost quantitatively (Fig. 1B and C, lanes 7 to 9). Surprisingly, no significant immunoprecipitation of snR10 or snR42 with Naf1p-ZZ is obtained. Nevertheless, as revealed by the analysis of the pattern of precipitated [32P]pCp-labeled RNAs, the bulk of H/ACA snoRNAs in the size range of 180 to 250 nucleotides are precipitated with Naf1p-ZZ with efficiencies significantly above background (Fig. 1C, compare lanes 10 to 12 with lanes 1 to 3), whereas the bulk of C/D snoRNAs are not (Fig. 1B and C, lanes 10 to 12).
From these experiments, we conclude that Naf1p is not a core component of H/ACA snoRNPs. Nevertheless, these data suggest that Naf1p interacts specifically with several components of H/ACA snoRNPs, though in a weak and/or transient manner.
Depletion of Naf1p inhibits 18S rRNA accumulation.
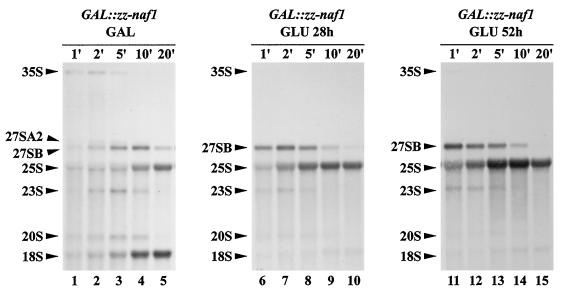
The weak interactions detected between Naf1p-ZZ and H/ACA snoRNP components suggest that Naf1p could be involved in one or several of the following processes: synthesis of H/ACA snoRNP components, assembly of H/ACA snoRNPs, intranuclear transport of these particles, regulation of their mode of action, and/or recycling of the particles. A defect in any one of these steps is likely to lead to an inhibition of the synthesis of pseudouridines within the pre-rRNA and of mature 18S rRNA. As a first step in assessing whether Naf1p is involved in one of the processes listed above, we analyzed the accumulation of rRNAs in cells in which production of Naf1p is inhibited. For that purpose, since Naf1p is essential for yeast viability (7), we constructed a yeast strain that conditionally expresses the Naf1p protein comprising a ZZ tag at the N terminus (ZZ-Naf1p protein), allowing its easy detection. The NAF1 open reading frame was tagged by the ZZ-encoding sequence and placed under the control of the regulated GAL1-10 promoter by homologous recombination, creating strain GAL::zz-naf1. This strain was propagated in a medium containing galactose, raffinose, and sucrose as carbon sources and was then shifted to a glucose-containing medium. As expected, the carbon source shift led to an increase in doubling time. After 24 h in glucose-containing medium, the doubling time of GAL::zz-naf1 cells had doubled, and after 48 h, it had tripled. After that time point, contrary to our expectations, the cells carried on dividing at roughly the same, reduced growth rate (doubling time of 9 h). Aliquots of GAL::zz-naf1 cells grown in galactose-containing medium or grown for 6, 12, 24, 48, or 72 h in glucose-containing medium were harvested. From these aliquots, total proteins and RNAs were extracted for Western (Fig. 2A) or Northern (Fig. 2B) blot analyses. The level of ZZ-Naf1p strongly diminishes between 6 and 24 h after transfer to glucose-containing medium (Fig. 2A, lanes 2 to 4). However, consistent with the fact that GAL::zz-naf1 cells continue growing even after 72 h in glucose-containing medium, complete depletion of ZZ-Naf1p was never achieved (Fig. 2A, panel showing overexposed ZZ-Naf1p). Nevertheless, the strong reduction in ZZ-Naf1p level obtained is correlated with a strong decrease in the steady-state level of mature 18S rRNA relative to the level of 25S rRNA (Fig. 2B, panels for 18S rRNA and 25S rRNA). According to phosphorimager quantification, the level of 18S rRNA, relative to that of 25S rRNA, is diminished by 95% after 48 h of growth in glucose-containing medium. The steady-state levels of various precursors to the mature rRNAs, relative to the 25S rRNA level, are less dramatically affected (for a cartoon of the pre-rRNA processing pathway, see Fig. 2C). We detect an accumulation of the 23S pre-rRNA that is produced when the 35S pre-rRNA is cleaved directly at site A3, without prior cleavage at sites A0, A1, and A2. Thus, this suggests that early cleavages of 35S pre-rRNA at sites A1 and A2, necessary for the production of 20S pre-rRNA (the immediate precursor to 18S rRNA), are somewhat impaired. The effects of a reduced ZZ-Naf1p level on pre-rRNA processing were also assessed by pulse-chase analysis using [methyl-3H]methionine. Such analysis (Fig. 3) clearly shows that synthesis of the 25S rRNA is not affected. In contrast, 18S rRNA synthesis is strongly inhibited, most probably as a direct result of reduced 20S pre-rRNA production detected by this approach.
FIG. 3.
Pulse-chase analysis of pre-rRNA processing in ZZ-Naf1p-depleted cells. GAL::zz-naf1 cells were grown in a medium containing galactose (GAL), raffinose, and sucrose (lanes 1 to 5), were washed, and shifted to a medium containing glucose (GLU) for 28 h (lanes 6 to 10) or 52 h (lanes 11 to 15). Cells were labeled for 3 min with [methyl-3H]methionine, chased with an excess of cold methionine, and grown for a further 1 (lanes 1, 6, and 11), 2 (lanes 2, 7, and 12), 5 (lanes 3, 8, and 13), 10 (lanes 4, 9, and 14), and 20 (lanes 5, 10, and 15) min before collection of cells and freezing in liquid nitrogen. Extracted RNAs were separated on a formaldehyde-agarose gel and transferred to a nylon membrane. Labeled RNAs were detected by autoradiography. Pre-rRNA intermediates and mature rRNAs are indicated on the left.
Naf1p is located primarily in the nucleoplasm and displays little association with preribosomes in a glycerol gradient.
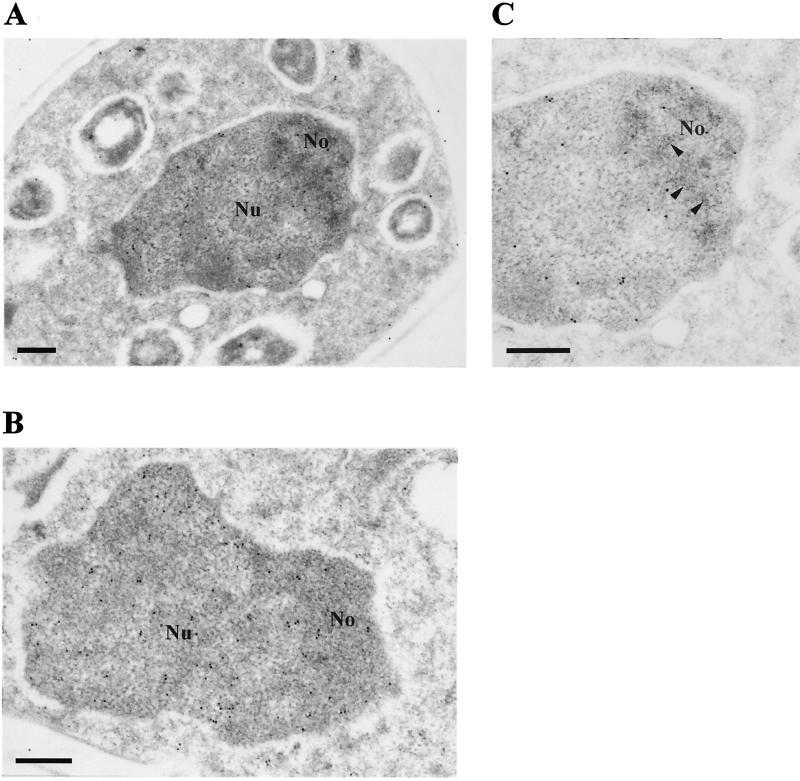
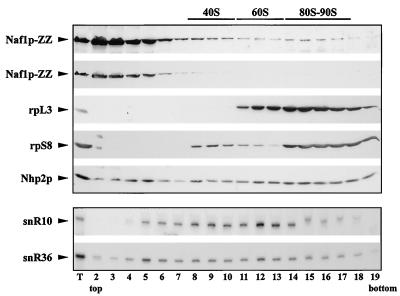
Naf1p could directly participate in 18S rRNA synthesis by interacting with H/ACA snoRNPs within the nucleolus, or it could be involved in 18S rRNA synthesis indirectly. To distinguish between these two possibilities, we determined the subcellular localization of Naf1p-ZZ by immunoelectron microscopy. Whereas the core H/ACA snoRNP proteins accumulate within the dense fibrillar component of the nucleolus (29), Naf1p-ZZ is found predominantly in the nucleoplasm (Fig. 4). A minor fraction of Naf1p-ZZ is found in the nucleolus but outside the dense fibrillar component. This result suggests that Naf1p is not directly involved in the early stages of pre-rRNA processing. To confirm this, we determined the distribution of Naf1p-ZZ in a glycerol gradient. The bulk of Naf1p-ZZ is found in the top five fractions of the gradient (Fig. 5, panel for Naf1p-ZZ). Thus, Naf1p-ZZ is found either free or associated with complexes of the size of snRNPs. Contrary to bona fide H/ACA snoRNP components (Fig. 5, panels for Nhp2p, snR10, and snR36) very little Naf1p-ZZ is found in fractions containing preribosomes.
FIG. 4.
Naf1p-ZZ accumulates mainly within the nucleoplasm. Shown are electron micrographs of a whole cell (×27,600 magnification) (A), a nucleus (×41,400 magnification) (B), and a nucleolus (×46,000 magnification) (C). Naf1p-ZZ was detected by electron microscopy after treatment of the grids with anti-protein A antibodies followed by incubation with colloidal gold-conjugated protein A. No, nucleolus; Nu, nucleoplasm. In panel C, a section of the nucleolar dense fibrillar component is indicated by arrow heads. Bar = 300 nm.
FIG. 5.
Sedimentation profile of Naf1p-ZZ in a glycerol gradient. A native extract from cells expressing Naf1p-ZZ was loaded on a 10 to 30% glycerol gradient and centrifuged for 10 h at 25,000 rpm in an SW41 Ti rotor. Nineteen fractions were collected and divided in two equal aliquots from which proteins or RNAs were extracted. Proteins were separated by SDS-polyacrylamide gel electrophoresis, while RNAs were separated by denaturing polyacrylamide gel electrophoresis. Proteins or RNAs were then transferred to cellulose or nylon membranes, respectively. Naf1p-ZZ and Nhp2p were detected by use of a polyclonal anti-Nhp2p serum, ribosomal protein L3 by a monoclonal anti-L3 antibody, and ribosomal protein S8 by a polyclonal anti-S8 serum. H/ACA snR10 and snR36 snoRNAs were detected using specific oligonucleotide probes. Lane numbers correspond to fraction numbers. Fraction 1, top of the gradient; fraction 19, bottom of the gradient; and T, protein or RNA aliquot from the nonfractionated input extract.
Depletion of Naf1p results in a specific decrease in H/ACA snoRNP levels.
The above data suggest that the possible roles of Naf1p are restricted to either synthesis of H/ACA snoRNP components, assembly, trafficking, and/or recycling of the particles. To narrow the spectrum of possible roles for Naf1p, we assessed the steady-state levels of H/ACA snoRNAs and H/ACA snoRNP proteins in cells depleted for Naf1p. RNA samples from GAL::zz-naf1 cells grown on galactose, raffinose, and sucrose as carbon sources or shifted to glucose containing medium for 6, 12, 24, 48, and 72 h (see above) were analyzed by Northern blot hybridizations performed with oligonucleotide probes complementary to a wide range of small RNAs (Fig. 6A). Relative amounts of RNAs assessed by quantification of phosphorimager scans of Northern blots are listed in the table in Fig. 6A. Note that the values have not been normalized to an internal standard, for lack of an obvious one. Strikingly, the steady-state levels of all H/ACA snoRNAs tested are strongly affected by ZZ-Naf1p depletion. All types of H/ACA snoRNAs, whether independently transcribed and possessing a methyl-2,2,7-guanosine cap (snR30, snR37, snR42, snR11, snR10, snR31, snR5, and snR33), independently transcribed and processed by Rnt1p (snR46 and snR36), or processed from an intron of a protein-encoding transcript (snR44), are roughly equally affected by ZZ-Naf1p depletion. The decrease in H/ACA snoRNA levels parallels the decrease in ZZ-Naf1p levels (Fig. 2A). Twelve and 24 h after the shift to glucose-containing medium, the levels of H/ACA snoRNAs are decreased on average by 50 and 85%, respectively (Fig. 6A, lanes 3 and 4). No such dramatic effect is observed for the other RNAs tested: C/D snoRNAs, spliceosomal snRNAs, RNase MRP and P RNAs, SRP RNA, and telomerase RNA (TLC1).
The effects of Naf1p depletion on the levels of Gar1p, Nhp2p, and Nop10p were tested by Western blot analysis of total protein extracts obtained from GAL::zz-naf1 cells, grown on galactose, raffinose, and sucrose or shifted to glucose-containing medium, using polyclonal sera. To monitor the effects of Naf1p depletion on Cbf5p levels, the same procedures were repeated using a strain (GAL::naf1/CBF5-TAP) that expresses native Naf1p conditionally and produces an epitope-tagged version of Cbf5p (Cbf5p-TAP). Steady-state levels of Gar1p, Nop10p (Fig. 6B, lanes 1 to 6), and Cbf5p-TAP (Fig. 6B, lanes 7 to 12) are lowered by Naf1p depletion. The level of Nhp2p is only very marginally reduced, and Nop1p accumulation is unaffected (Fig. 6B, lanes 1 to 6). We conclude that Naf1p depletion leads to a specific reduction in the levels of H/ACA snoRNP components.
The most straightforward interpretation of the above data is that Naf1p is involved either in the synthesis of H/ACA snoRNP components or in the assembly of the particles. Abortive assembly of H/ACA snoRNPs is likely to result in increased turnover of most of their constituents. Likewise, a defect in the synthesis of one H/ACA snoRNP component would lead to abortive assembly of the particles and hence would have the same consequences.
Naf1p is unlikely to be involved in the production of H/ACA snoRNP proteins. We have checked that the steady-state levels of GAR1 and NOP10 mRNAs in GAL::zz-naf1 cells grown 48 h in glucose-containing medium are not reduced compared to those of cells grown on galactose/raffinose/sucrose (data not shown). Yet at that time point, Gar1p and Nop10p levels have already substantially declined (Fig. 6B, lanes 5 and 11). A role for Naf1p in protein synthesis is not very plausible, given that it accumulates within the nucleus. To investigate whether Naf1p could be involved in the transcription as such of H/ACA snoRNA sequences, we assessed the accumulation, in the course of Naf1p depletion, of the mature mRNA of the snR44 host gene, RPS22B, which encodes ribosomal protein rpS22B. There is in fact another gene, RPS22A, which encodes the ribosomal protein rpS22A, which is identical to rpS22B but for 1 amino acid. The oligonucleotides used to detect RPS22B mRNA cannot hybridize to RPS22A mRNA. As shown in Fig. 6A, there is no significant reduction of the RPS22B mRNA level after 48 h of growth in glucose-containing medium. Since production of mature snR44 has been shown to be mostly dependent upon splicing of its host pre-mRNA (65), we conclude that defective transcription of the SNR44 sequence is most probably not the explanation for the drastic reduction in snR44 steady-state levels observed during Naf1p depletion.
DISCUSSION
The processes and trans-acting factors involved in the regulation of H/ACA snoRNP assembly, intranuclear trafficking, and activity remain largely unknown. We are particularly interested in these issues. Our attention was therefore drawn to the Naf1p factor when it was reported that it interacts in a double-hybrid test with two core components of H/ACA snoRNPs, Nhp2p and Cbf5p (32). In accordance with those results, we find by immunoprecipitation experiments that Naf1p interacts in cell extracts, directly or indirectly, with several components of H/ACA snoRNPs. We believe that the interaction is specific because we fail to detect any significant immunoprecipitation with epitope-tagged Naf1p of components of C/D snoRNPs or of the control Nsr1p protein. Our conclusion that Naf1p physically associates with H/ACA snoRNP components is reinforced by the recent finding that Naf1p coprecipitates with Cbf5p tagged with the Flag epitope (30). The efficiencies of precipitation of H/ACA snoRNP components with Naf1p are clearly low, which is probably indicative of a transient and/or labile association.
Naf1p is found mainly in the nucleoplasm, cannot be detected in the dense fibrillar component of the nucleolus, and sediments in a glycerol gradient above all as a free protein or as part of a complex of the size of free snRNPs. This suggests strongly that (i) Naf1p is not a core component of active mature H/ACA snoRNPs, consistent with a previous proposal that most mature H/ACA snoRNPs contain only the Cbf5p, Gar1p, Nhp2p, and Nop10p proteins (29) and that (ii) Naf1p is not directly involved in pre-rRNA processing and modification. We are fully aware that the Naf1p presence in the dense fibrillar component of the nucleolus may have gone undetected, and we observe that traces of Naf1p sediment with preribosomes. Nevertheless, the likeliest interpretation of the 18S rRNA accumulation defect detected in Naf1p-depleted cells remains that it is the consequence of the reduction in snR10 and snR30 H/ACA snoRNP levels. Indeed, the most striking phenotype resulting from Naf1p depletion is a dramatic and specific decrease in the accumulation of all H/ACA snoRNP components except Nhp2p. Remarkably, none of the other small RNAs tested, in particular none of the C/D snoRNAs, show a similar decrease in steady-state levels. The very specific reduction in H/ACA snoRNA levels following Naf1p depletion was also demonstrated by the groups of G. Chanfreau and D. Tollervey (P. K. Yang, G. Rotondo, T. Porras, P. Legrain, and G. Chanfreau, submitted for publication; and A. Fatica, M. Dlaki, and D. Tollervey, submitted for publication). This accumulation defect is probably not due to defective transcription, since independently transcribed H/ACA snoRNAs and the snR44 intronic RNA are equally affected. In the case of snR44, our results strongly suggest that no defective transcription (either initiation, elongation, or termination) is occurring: snR44 production has been shown to be dependent upon splicing of its host pre-mRNA (65), and we failed to detect a significant decrease in the steady-state accumulation of the corresponding mRNA. Defective pre-snoRNA processing in Naf1p-depleted cells is not very probable either. The activity of the enzymatic machinery (i.e., splicing machinery, Rnt1p endonuclease, Rat1p exonuclease, and exosome) as such is not impaired, since it is involved in processing precursors to both H/ACA and C/D snoRNAs. Yet no defective processing of C/D snoRNAs is detected. For example, we notice that the pattern of bands detected with the U24 or snR78 probes reflecting heterogeneous end formation by exonucleases is not altered by Naf1p depletion. We cannot of course exclude the possibility that the processes that target the processing machinery to H/ACA snoRNA precursors are impaired. We note, however, that when probes complementary to mature H/ACA snoRNA sequences are used, no obvious accumulation of H/ACA snoRNA precursors was observed. Finally, although the possibility is formally open, impaired production of H/ACA snoRNP proteins is unlikely, for the following reasons: we have established that the steady-state levels of GAR1 and NOP10 mRNAs are not decreased by the time the steady-state accumulation of the proteins that they encode is already significantly diminished. Because Naf1p accumulates within the nucleus, the possibility that Naf1p intervenes in cytoplasmic translation is very slight. A role for Naf1p in the nuclear import of Cbf5p, Gar1p, and Nop10p can still be envisaged.
We favor the idea that Naf1p is involved in H/ACA snoRNP assembly and/or in H/ACA snoRNP transport from the nucleoplasm to the nucleolus. A role for Naf1p in intranuclear trafficking is consistent with its localization in the nucleoplasm and within the nucleolus and with its weak association with mature H/ACA snoRNAs. Intriguingly, the localization of Naf1p in the nucleolus in close vicinity to the dense fibrillar component is reminiscent of that of the nucleolar body, the yeast equivalent of the Cajal body, through which C/D snoRNPs are proposed to transit before reaching the dense fibrillar component (97, 98). No evidence has been provided so far that H/ACA snoRNPs transit through the nucleolar body. If they do, their routing process to and from the nucleolar body is likely to require trans-acting factors different from those for C/D snoRNPs (98). Naf1p is thus a possible candidate.
The presence of some Naf1p within the nucleolus may reflect, rather than a transport function, an involvement of this protein in late H/ACA snoRNP assembly steps. Indeed, in the case of the U3 snoRNP, for example, recent work strongly suggests that final U3 snoRNP assembly occurs in the nucleolar body (in yeast) or the Cajal body (in mammals) (97). Because Naf1p is predominantly found in the nucleoplasm, a role for this protein in putative early snoRNP assembly steps taking place at or in the vicinity of pre-snoRNA transcription sites is also a very attractive hypothesis. A role for Naf1p in particle assembly is consistent with the observation that depletion of Cbf5p, Nhp2p, or Nop10p, which obviously prevents assembly of complete particles, has the same consequences as does Naf1p depletion on accumulation of H/ACA snoRNAs and Gar1p (29, 43). In the absence of particle assembly, the body of H/ACA snoRNAs normally protected by bound proteins may be degraded by the exonucleases that remove the flanking sequences and/or may be turned over by a discard pathway distinct from the normal processing pathway. Cbf5p, Gar1p, and Nop10p form a very stable complex (A. Henras and M. Caizergues-Ferrer, unpublished observation) that may be targeted for degradation when stable association with H/ACA snoRNAs is prevented. How could Naf1p promote particle assembly? The most striking motifs present in Naf1p are, in the N-terminal part, a serine-rich domain and, at the C terminus, a large proline- and glutamine-rich domain somewhat reminiscent of a domain found in the RNA-binding protein Nrd1p (80). Thus, it is conceivable that the C-terminal part of Naf1p could be involved in binding to H/ACA snoRNAs at an early stage. The serine-rich domain of Naf1p, which also contains a high proportion of negatively charged residues, is predicted to be highly phosphorylated by casein kinase II. It could recruit one or several H/ACA snoRNP proteins by providing a binding platform for exposed basic domains of these proteins, such as the C-terminal KKE repeat domain of Cbf5p, the putative amphipathic alpha-helix at the N terminus of Nhp2p, and/or the overall basic small Nop10p protein.
The SMN complex, containing the SMN protein, has also been proposed to be involved in H/ACA snoRNP assembly in some eukaryotes, since the SMN protein can interact with the GAR domains of Gar1p (69, 82). In addition, the SMN protein has also been shown to interact with the GAR domain of fibrillarin, suggesting that the SMN complex could also be involved in C/D snoRNP assembly (35, 69, 82). Moreover, a role for the SMN complex, so far not detected in S. cerevisiae, in the assembly of spliceosomal snRNPs has been well documented in higher eukaryotes (18, 50, 82). Thus, this complex probably exerts a role that is broader than and different from that of Naf1p. Indeed, the hypothesis that the SMN complex could play in higher eukaryotes the role of Naf1p in lower eukaryotes is contradicted by the fact that Schizosaccharomyces pombe contains both an SMN complex (27, 66, 67) and a protein, encoded by the SPBC30D10.15 open reading frame, significantly related to S. cerevisiae Naf1p (29% identity, 48% similarity over 438 amino acids), which very likely constitutes its S. pombe orthologue. In addition to SMN, two non-snoRNP proteins, Rvb2p (the yeast orthologue of the mammalian helicase p50) and Srp40p (the yeast orthologue of Nopp140p), have also been proposed to be required, directly or indirectly, for H/ACA snoRNP assembly and localization (36, 53, 104). Like Naf1p depletion, depletion of Srp40p (in a yeast strain lacking the LES2 gene) inhibits H/ACA snoRNA accumulation, while that of C/D snoRNAs remains unaffected (104). Unlike Naf1p, however, Srp40p and Rvb2p seem also to be linked to C/D snoRNPs (36, 61). The mammalian orthologue of Srp40p, Nopp140p, interacts with C/D snoRNP components, and Rvb2p is required for normal accumulation of both C/D and H/ACA snoRNAs. In addition, Rvb2p has been implicated in various processes, including chromatin remodeling (36, 61). Finally, no interaction has been described between Rvb2p or Srp40p and Naf1p. For all these reasons, Srp40p and Rvb2p are unlikely to directly cooperate with Naf1p. In contrast, a good candidate for a Naf1p partner is the essential Shq1p protein. By a double-hybrid test, Shq1q and Naf1p have been shown to interact (32). Moreover, Shq1p associates with Flag-Cbf5p (30) and we have established that Shq1p remains associated (directly or indirectly) with TAP-tagged Gar1p after tandem affinity purification (C. Dez, C. Froment, and V. Dossat, unpublished observation). The functional importance of the interactions detected between Shq1p, Naf1p and H/ACA snoRNP components has in fact been demonstrated by the group of G. Chanfreau, who have shown that Shq1p, like Naf1p, is required for H/ACA snoRNA accumulation (Yang et al., submitted).
Acknowledgments
We are very grateful to P. Yang and G. Chanfreau (University of California at Los Angeles) and to A. Fatica and D. Tollervey (University of Edinburgh) for communicating results before publication. We thank E. Vanrobays for pointing out to us the results of Ito et al. (32) and for numerous discussions and helpful advice, and we thank P.-E. Gleizes and N. Gas, in whose lab detection of Naf1p-ZZ by electron microscopy was performed, for help with the interpretation of the electron microscopy photographs. The gifts of plasmids pBS1479 from B. Séraphin (CNRS, Gif-sur-Yvette, France) and pTL26 from D. Lafontaine (Université Libre de Bruxelles), of a strain expressing ZZ-Nop1p from T. Bergès (Université de Poitiers), of strains YDL402 and YDL524 expressing Cbf5p-ZZ from D. Lafontaine, of monoclonal anti-Nsr1p antibody from J. Woolford (Carnegie-Mellon University), and of monoclonal anti-L3 antibody and polyclonal anti-S8 serum from J. Warner (Albert Einstein College of Medicine) are gratefully acknowledged. We are thankful to members of the Ferrer lab for help and numerous discussions. We thank Y. de Préval for synthesis of oligonucleotides, D. Villa for artwork, and A. Rivals for expert technical assistance.
C.D. is supported by a grant from the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche. This work was financed by la Ligue Nationale contre le Cancer, the CNRS, and Université Paul Sabatier, Toulouse, France.
Footnotes
We dedicate this work to the memory of Albert Henry (1910-2002), Université Libre de Bruxelles et Académie Royale de Belgique.
REFERENCES
- 1.Allmang, C., J. Kufel, G. Chanfreau, P. Mitchell, E. Petfalski, and D. Tollervey. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18:5399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachellerie, J.-P., and J. Cavaillé. 1997. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 22:257-261. [DOI] [PubMed] [Google Scholar]
- 3.Balakin, A. G., L. Smith, and M. J. Fournier. 1996. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 86:823-834. [DOI] [PubMed] [Google Scholar]
- 4.Bousquet-Antonelli, C., E. Vanrobays, J.-P. Gélugne, M. Caizergues-Ferrer, and Y. Henry. 2000. Rrp8p is a yeast nucleolar protein functionally linked to Gar1p and involved in pre-rRNA cleavage at site A2. RNA 6:826-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadwell, C., H. J. Yoon, Y. Zebarjadian, and J. Carbon. 1997. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol. Cell. Biol. 17:6175-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caffarelli, E., M. Arese, B. Santoro, P. Fragapane, and I. Bozzoni. 1994. In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis. Mol. Cell. Biol. 14:2966-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capozzo, C., F. Sartorello, F. Dal Pero, M. D'Angelo, A. Vezzi, S. Campanaro, and G. Valle. 2000. Gene disruption and basic phenotypic analysis of nine novel yeast genes from chromosome XIV. Yeast 16:1089-1097. [DOI] [PubMed] [Google Scholar]
- 8.Cavaillé, J., and J.-P. Bachellerie. 1998. SnoRNA-guided ribose methylation of rRNA: structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic Acids Res. 26:1576-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavaillé, J., K. Buiting, M. Kiefmann, M. Lalande, C. I. Brannan, B. Horsthemke, J.-P. Bachellerie, J. Brosius, and A. Huttenhofer. 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 97:14311-14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavaillé, J., M. Nicoloso, and J.-P. Bachellerie. 1996. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 383:732-735. [DOI] [PubMed] [Google Scholar]
- 11.Chanfreau, G., P. Legrain, and A. Jacquier. 1998. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284:975-988. [DOI] [PubMed] [Google Scholar]
- 12.Chanfreau, G., G. Rotondo, P. Legrain, and A. Jacquier. 1998. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17:3726-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu, S., R. H. Archer, J. M. Zengel, and L. Lindahl. 1994. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc. Natl. Acad. Sci. USA 91:659-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darzacq, X., B. E. Jady, C. Verheggen, A. M. Kiss, E. Bertrand, and T. Kiss. 2002. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 21:2746-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dez, C., A. Henras, B. Faucon, D. Lafontaine, M. Caizergues-Ferrer, and Y. Henry. 2001. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 29:598-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dragon, F., V. Pogacic, and W. Filipowicz. 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20:3037-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatica, A., M. Morlando, and I. Bozzoni. 2000. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 19:6218-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer, U., Q. Liu, and G. Dreyfuss. 1997. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 90:1023-1029. [DOI] [PubMed] [Google Scholar]
- 19.Frank, D. N., and N. R. Pace. 1998. Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem. 67:153-180. [DOI] [PubMed] [Google Scholar]
- 20.Ganot, P., M.-L. Bortolin, and T. Kiss. 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89:799-809. [DOI] [PubMed] [Google Scholar]
- 21.Ganot, P., M. Caizergues-Ferrer, and T. Kiss. 1997. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 11:941-956. [DOI] [PubMed] [Google Scholar]
- 22.Ganot, P., B. E. Jády, M.-L. Bortolin, X. Darzacq, and T. Kiss. 1999. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol. 19:6906-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautier, T., T. Bergès, D. Tollervey, and E. Hurt. 1997. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 17:7088-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giordano, E., I. Peluso, S. Senger, and M. Furia. 1999. minifly, a Drosophila gene required for ribosome biogenesis. J. Cell Biol. 144:1123-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giorgi, C., A. Fatica, R. Nagel, and I. Bozzoni. 2001. Release of U18 snoRNA from its host intron requires interaction of Nop1p with the Rnt1p endonuclease. EMBO J. 20:6856-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girard, J.-P., H. Lehtonen, M. Caizergues-Ferrer, F. Amalric, D. Tollervey, and B. Lapeyre. 1992. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 11:673-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hannus, S., D. Buhler, M. Romano, B. Séraphin, and U. Fischer. 2000. The Schizosaccharomyces pombe protein Yab8p and a novel factor, Yip1p, share structural and functional similarity with the spinal muscular atrophy-associated proteins SMN and SIP1. Hum. Mol. Genet. 9:663-674. [DOI] [PubMed] [Google Scholar]
- 28.Henras, A., C. Dez, J. Noaillac-Depeyre, Y. Henry, and M. Caizergues-Ferrer. 2001. Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic Acids Res. 29:2733-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henras, A., Y. Henry, C. Bousquet-Antonelli, J. Noaillac-Depeyre, J.-P. Gélugne, and M. Caizergues-Ferrer. 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 17:7078-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 31.Hughes, J. M., and M. Ares, Jr. 1991. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10:4231-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jady, B. E., and T. Kiss. 2001. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 20:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang, W., K. Middleton, H. J. Yoon, C. Fouquet, and J. Carbon. 1993. An essential yeast protein, CBF5p, binds in vitro to centromeres and microtubules. Mol. Cell. Biol. 13:4884-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones, K. W., K. Gorzynski, C. M. Hales, U. Fischer, F. Badbanchi, R. M. Terns, and M. P. Terns. 2001. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 276:38645-38651. [DOI] [PubMed] [Google Scholar]
- 36.King, T. H., W. A. Decatur, E. Bertrand, E. S. Maxwell, and M. J. Fournier. 2001. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell. Biol. 21:7731-7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20:3617-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiss, T., C. Marshallsay, and W. Filipowicz. 1991. Alteration of the RNA polymerase specificity of U3 snRNA genes during evolution and in vitro. Cell 65:517-526. [DOI] [PubMed] [Google Scholar]
- 39.Kiss-Laszlo, Z., Y. Henry, J.-P. Bachellerie, M. Caizergues-Ferrer, and T. Kiss. 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85:1077-1088. [DOI] [PubMed] [Google Scholar]
- 40.Kolodrubetz, D., and A. Burgum. 1991. Sequence and genetic analysis of NHP2: a moderately abundant high mobility group-like nuclear protein with an essential function in Saccharomyces cerevisiae. Yeast 7:79-90. [DOI] [PubMed] [Google Scholar]
- 41.Kuhn, J. F., E. J. Tran, and E. S. Maxwell. 2002. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 30:931-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lafontaine, D., and D. Tollervey. 1996. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 24:3469-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lafontaine, D. L., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12:527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lafontaine, D. L., and D. Tollervey. 1999. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA 5:455-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lange, T. S., A. Borovjagin, E. S. Maxwell, and S. A. Gerbi. 1998. Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 17:3176-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lange, T. S., M. Ezrokhi, F. Amaldi, and S. A. Gerbi. 1999. Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA. Mol. Biol. Cell 10:3877-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lange, T. S., M. Ezrokhi, A. V. Borovjagin, R. Rivera-Leon, M. T. North, and S. A. Gerbi. 1998. Nucleolar localization elements of Xenopus laevis U3 small nucleolar RNA. Mol. Biol. Cell 9:2973-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li, H. V., J. Zagorski, and M. J. Fournier. 1990. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindahl, L., and J. M. Zengel. 1995. RNase MRP and rRNA processing. Mol. Biol. Rep. 22:69-73. [DOI] [PubMed] [Google Scholar]
- 50.Liu, Q., U. Fischer, F. Wang, and G. Dreyfuss. 1997. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 90:1013-1021. [DOI] [PubMed] [Google Scholar]
- 51.Lygerou, Z., C. Allmang, D. Tollervey, and B. Séraphin. 1996. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science 272:268-270. [DOI] [PubMed] [Google Scholar]
- 52.Lyman, S. K., L. Gerace, and S. J. Baserga. 1999. Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA 5:1597-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meier, U. T. 1996. Comparison of the rat nucleolar protein nopp140 with its yeast homolog SRP40. Differential phosphorylation in vertebrates and yeast. J. Biol. Chem. 271:19376-19384. [PubMed] [Google Scholar]
- 54.Meier, U. T., and G. Blobel. 1994. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J. Cell Biol. 127:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitchell, J. R., E. Wood, and K. Collins. 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402:551-555. [DOI] [PubMed] [Google Scholar]
- 56.Morlando, M., P. Greco, B. Dichtl, A. Fatica, W. Keller, and I. Bozzoni. 2002. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol. Cell. Biol. 22:1379-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morrissey, J. P., and D. Tollervey. 1995. Birth of the snoRNPs: the evolution of RNase MRP and the eukaryotic pre-rRNA-processing system. Trends Biochem. Sci. 20:78-82. [DOI] [PubMed] [Google Scholar]
- 58.Morrissey, J. P., and D. Tollervey. 1993. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell. Biol. 13:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Narayanan, A., A. Lukowiak, B. E. Jady, F. Dragon, T. Kiss, R. M. Terns, and M. P. Terns. 1999. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J. 18:5120-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narayanan, A., W. Speckmann, R. Terns, and M. P. Terns. 1999. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell 10:2131-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Newman, D. R., J. F. Kuhn, G. M. Shanab, and E. S. Maxwell. 2000. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA 6:861-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ni, J., A. L. Tien, and M. J. Fournier. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89:565-573. [DOI] [PubMed] [Google Scholar]
- 63.Ochs, R. L., M. A. Lischwe, W. H. Spohn, and H. Busch. 1985. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol. Cell 54:123-133. [DOI] [PubMed] [Google Scholar]
- 64.Omer, A. D., S. Ziesche, H. Ebhardt, and P. P. Dennis. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. USA 99:5289-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ooi, S. L., D. A. Samarsky, M. J. Fournier, and J. D. Boeke. 1998. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA 4:1096-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Owen, N., C. L. Doe, J. Mellor, and K. E. Davies. 2000. Characterization of the Schizosaccharomyces pombe orthologue of the human survival motor neuron (SMN) protein. Hum. Mol. Genet. 9:675-684. [DOI] [PubMed] [Google Scholar]
- 67.Paushkin, S., B. Charroux, L. Abel, R. A. Perkinson, L. Pellizzoni, and G. Dreyfuss. 2000. The survival motor neuron protein of Schizosacharomyces pombe. Conservation of survival motor neuron interaction domains in divergent organisms. J. Biol. Chem. 275:23841-23846. [DOI] [PubMed] [Google Scholar]
- 68.Peculis, B. A., and J. A. Steitz. 1993. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell 73:1233-1245. [DOI] [PubMed] [Google Scholar]
- 69.Pellizzoni, L., J. Baccon, B. Charroux, and G. Dreyfuss. 2001. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 11:1079-1088. [DOI] [PubMed] [Google Scholar]
- 70.Petfalski, E., T. Dandekar, Y. Henry, and D. Tollervey. 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Phillips, B., A. N. Billin, C. Cadwell, R. Buchholz, C. Erickson, J. R. Merriam, J. Carbon, and S. J. Poole. 1998. The Nop60B gene of Drosophila encodes an essential nucleolar protein that functions in yeast. Mol. Gen. Genet. 260:20-29. [DOI] [PubMed] [Google Scholar]
- 72.Pogacic, V., F. Dragon, and W. Filipowicz. 2000. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 20:9028-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qu, L.-H., A. Henras, Y.-J. Lu, H. Zhou, W.-X. Zhou, Y.-Q. Zhu, J. Zhao, Y. Henry, M. Caizergues-Ferrer, and J.-P. Bachellerie. 1999. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol. 19:1144-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Séraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 75.Samarsky, D. A., M. J. Fournier, R. H. Singer, and E. Bertrand. 1998. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 17:3747-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Savino, R., and S. A. Gerbi. 1990. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 9:2299-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schimmang, T., D. Tollervey, H. Kern, R. Frank, and E. C. Hurt. 1989. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 8:4015-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmitt, M. E., and D. A. Clayton. 1993. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:7935-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith, C. M., and J. A. Steitz. 1997. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell 89:669-672. [DOI] [PubMed] [Google Scholar]
- 80.Steinmetz, E. J., and D. A. Brow. 1998. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl. Acad. Sci. USA 95:6699-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Steinmetz, E. J., N. K. Conrad, D. A. Brow, and J. L. Corden. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413:327-331. [DOI] [PubMed] [Google Scholar]
- 82.Terns, M. P., and R. M. Terns. 2001. Macromolecular complexes: SMN—the master assembler. Curr. Biol. 11:R862-R864. [DOI] [PubMed] [Google Scholar]
- 83.Terns, M. P., and R. M. Terns. 2002. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 10:17-39. [PMC free article] [PubMed] [Google Scholar]
- 84.Tollervey, D. 1995. Genetic and biochemical analyses of yeast RNase MRP. Mol. Biol. Rep. 22:75-79. [DOI] [PubMed] [Google Scholar]
- 85.Tollervey, D. 1996. Small nucleolar RNAs guide ribosomal RNA methylation. Science 273:1056-1057. [DOI] [PubMed] [Google Scholar]
- 86.Tollervey, D. 1987. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 6:4169-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tollervey, D., and T. Kiss. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9:337-342. [DOI] [PubMed] [Google Scholar]
- 88.Tollervey, D., H. Lehtonen, M. Carmo-Fonseca, and E. C. Hurt. 1991. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 10:573-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tollervey, D., and I. W. Mattaj. 1987. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 6:469-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tremblay, A., B. Lamontagne, M. Catala, Y. Yam, S. Larose, L. Good, and S. A. Elela. 2002. A physical interaction between Gar1p and Rnt1p is required for the nuclear import of H/ACA small nucleolar RNA-associated proteins. Mol. Cell. Biol. 22:4792-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tycowski, K. T., M. D. Shu, and J. A. Steitz. 1994. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science 266:1558-1561. [DOI] [PubMed] [Google Scholar]
- 92.Tycowski, K. T., C. M. Smith, M. D. Shu, and J. A. Steitz. 1996. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl. Acad. Sci. USA 93:14480-14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tycowski, K. T., Z. H. You, P. J. Graham, and J. A. Steitz. 1998. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell 2:629-638. [DOI] [PubMed] [Google Scholar]
- 94.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vanrobays, E., P.-E. Gleizes, C. Bousquet-Antonelli, J. Noaillac-Depeyre, M. Caizergues-Ferrer, and J.-P. Gélugne. 2001. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 20:4204-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 97.Verheggen, C., D. L. Lafontaine, D. Samarsky, J. Mouaikel, J.-M. Blanchard, R. Bordonné, and E. Bertrand. 2002. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 21:2736-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Verheggen, C., J. Mouaikel, M. Thiry, J.-M. Blanchard, D. Tollervey, R. Bordonné, D. L. Lafontaine, and E. Bertrand. 2001. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 20:5480-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Villa, T., F. Ceradini, and I. Bozzoni. 2000. Identification of a novel element required for processing of intron-encoded box C/D small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:1311-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Villa, T., F. Ceradini, C. Presutti, and I. Bozzoni. 1998. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol. Cell. Biol. 18:3376-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Watkins, N. J., A. Gottschalk, G. Neubauer, B. Kastner, P. Fabrizio, M. Mann, and R. Luhrmann. 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4:1549-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Watkins, N. J., V. Ségault, B. Charpentier, S. Nottrott, P. Fabrizio, A. Bachi, M. Wilm, M. Rosbash, C. Branlant, and R. Luhrmann. 2000. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103:457-466. [DOI] [PubMed] [Google Scholar]
- 103.Wu, P., J. S. Brockenbrough, A. C. Metcalfe, S. Chen, and J. P. Aris. 1998. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J. Biol. Chem. 273:16453-16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang, Y., C. Isaac, C. Wang, F. Dragon, V. Pogacic, and U. T. Meier. 2000. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol. Biol. Cell 11:567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zebarjadian, Y., T. King, M. J. Fournier, L. Clarke, and J. Carbon. 1999. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 19:7461-7472. [DOI] [PMC free article] [PubMed] [Google Scholar]