Abstract
Intrinsic nucleosome dynamics termed “site exposure” provides spontaneous and cooperative access to buried regions of nucleosomal DNA in vitro. Two different mechanisms for site exposure have been proposed, one based on nucleosome translocation, the other on dynamic nucleosome conformational changes in which a stretch of the nucleosomal DNA is transiently released off the histone surface. Here we report on three experiments that distinguish between these mechanisms. One experiment investigates the effects on the accessibilities of restriction enzyme target sites inside nucleosomes when extra DNA (onto which the nucleosome may move at low energetic cost) is appended onto one end. The other two experiments test directly for nucleosome mobility under the conditions used to probe accessibility to restriction enzymes: one on a selected nonnatural nucleosome positioning sequence, the other on the well-studied 5S rRNA gene nucleosome positioning sequence. We find from all three assays that restriction enzymes gain access to sites throughout the entire length of the nucleosomal DNA without contribution from nucleosome translocation. We conclude that site exposure in nucleosomes in vitro occurs via a nucleosome conformational change that leads to transient release of a stretch of DNA from the histone surface, most likely involving progressive uncoiling from an end. Recapture at a distal site along DNA that has partially uncoiled would result in looped structures which are believed to contribute to RNA polymerase elongation and may contribute to spontaneous or ATP-driven nucleosome mobility. Transient open states may facilitate the initial entry of transcription factors and enzymes in vivo.
DNA target sites for gene regulatory proteins may be sterically occluded by packaging in nucleosomes in vivo, yet the cognate proteins are nevertheless still able to bind to them (11, 13, 36, 40, 50). How this is accomplished is not known. Entry of site-specific factors into chromatin may be facilitated in vivo by the action of nucleosome remodeling enzymes (14, 25, 26, 42, 45, 49), including histone acetylases and ATP-dependent machines that promote nucleosome translocation or conformational changes. A question inherent in this idea is how such factors would know which nucleosomes to remodel. Recent discoveries (6, 12, 26) suggest that in some cases remodeling factors are recruited to specific DNA target sites by other sequence-specific DNA binding proteins that bind first—raising the question of how those proteins gain access to their target sites.
We and others (8, 28) have suggested that spontaneous partial uncoiling or breathing motions of nucleosomal DNA could allow for the progressive invasion of a nucleosome starting from an end. Such conformational dynamics would allow for passive binding of proteins to target sites that, in the time average, are buried inside nucleosomes. This could also represent an initial step of entry into chromatin, with subsequent recruitment of remodeling factors acting to move or destabilize that histone octamer, thereby allowing a higher level of occupancy. Such nucleosome dynamics provide a physically plausible mechanism for the progression of processive enzymes such as polymerases and (potentially) ATP-dependent remodeling factors through nucleosomal DNA (16, 48). Rather than requiring such enzymes to actively pull DNA off the histone octamer surface, transient DNA uncoiling would allow for enzyme progression via a simple trapping of preexisting conformational fluctuations. This would constitute a Brownian-ratchet mechanism (4, 9).
In earlier studies (3, 28-31) it was shown that nucleosomes in vitro are in fact in such a dynamic conformational equilibrium: regions of the DNA that in the time average are bound on the histone octamer surface are nevertheless transiently released and freely accessible. We refer to this process as site exposure. Site exposure allows nucleosomal DNA to be accessible even to DNA binding proteins and enzymes (including most restriction enzymes) that, once bound, occlude the entire circumference of their DNA target sites. Isolated nucleosomes (28) and also test nucleosomes embedded in longer chains (22, 23) exhibit this behavior. The affinity of proteins for binding to nucleosomal DNA target sites are reduced by a factor equal to the position-dependent equilibrium constant for exposing the site, Keqconf (28, 30). In subsequent work (29) it was shown that there is a dramatic cooperativity inherent in nucleosomal site exposure. Arbitrary pairs of proteins strongly facilitate each other's binding to target sites that are buried within the same nucleosome. This cooperativity allows for far greater occupancy of nucleosomal target sites than would be achieved for a given limiting concentration of a single site specific DNA binding protein.
Two different mechanisms have been proposed to account for the observed site exposure equilibrium. Both mechanisms are nondissociative: they allow for transient DNA accessibility without full dissociation of DNA from the histone surface, as required by a large body of evidence on the behavior and stability of nucleosomes (46). One mechanism supposes that site exposure occurs via a transient partial uncoiling of DNA off the surface of the histone octamer, starting from one end of the nucleosomal DNA (8, 28, 30). In this mechanism, sites would be accessible during periods in which a transient uncoiling fluctuation exposes a sufficient length of the nucleosomal DNA. An alternative mechanism (31, 43) is based on the well-known phenomenon of nucleosome translocation (sliding) (21, 44). Histone octamers translocate nondissociatively along the DNA that wraps them; any particular site would be accessible during those periods that the octamer has moved a sufficient distance away so as to leave that site unoccupied.
Nucleosome translocation is often considered to be slow and in one study is shown to be visibly slowed by addition of 2 mM and even 0.2 mM Mg2+ (24). This argues against translocation as a possible mechanism for site exposure under physiological (Mg2+-containing) conditions. However, three lines of reasoning imply that nucleosome translocation is a real possibility that must be tested directly. One study (43) concluded that not only did nucleosome translocation occur under physiological conditions in vitro, but also nucleosome translocation was actually essential for site-specific factor binding. Moreover, the conditions used in those experiments included 7 mM Mg2+, in apparent contradiction to the results reported in reference 24. Second, while nucleosome translocation often is slow, the restriction enzyme digestion experiments are also slow (3, 28, 30). Experiments have not yet been devised to allow quantitative measurement of nucleosome translocation rates, which would be needed for a direct comparison with restriction enzyme digestion rates. Third, the data suggesting that nucleosome translocation is slow can be misleading. Studies carried out using two-dimensional gel analyses can be influenced by well-known gel caging effects (which, for example, suppress the dissociation of protein-DNA complexes, thereby allowing their resolution by gel electrophoresis), while studies carried out in free solution in general do not distinguish between two opposite limits—very slow translocation versus very rapid translocation in a dynamic positional equilibrium.
In summary, rather than excluding a nucleosome translocation mechanism for site exposure, existing studies either directly support that mechanism (43) or do not suffice to allow a direct test. New studies are needed.
MATERIALS AND METHODS
DNA constructs.
DNA construct 601.3 is derived from 601.2 (174 bp) (3); it incorporates the 145-bp nucleosomal region of 601.2 as mapped by exonuclease III plus 4 bp of 601.2 sequence on the left side and 3 bp on the right side (for the sequence as indicated in Fig. 1c —see below). Constructs 601.3, 601.3a, and 601.3b were prepared by PCR by using 601.2 cloned into the SmaI site of plasmid pGEM3Z as a template. Sequences 601.3a and 601.3b extend the sequences of 601.3 with an additional 11 bp of 601.2 DNA sequence on the right-hand end and 15 bp of 106.2 DNA sequence plus 40 or 90 bp of plasmid DNA sequence on the left. The primer pairs for PCR were as follows. Construct 601.3, left end, GCGGGCGCCCTGCAGAAGCTTG, and right end, GATGTATATATCTGACACGTGC; construct 601.3a, left end, TAATACGACTCACTATAGGGCG, and right end, TACATGCACAGGATGTATATATCT; construct 601.3b, left end, CGCCAGGGTTTTCCCAGTCA, and right end, TACATGCACAGGATGTATATATCT. For studies of nucleosome translocation on the 5S nucleosome positioning sequence (37, 38), we needed highly labeled DNA so that we would have adequate counts in nucleosomes after purification of disfavored (i.e., minor positioning) species. We found that a stretch at one end of the 5S sequence (which was well outside the region responsible for nucleosome positioning on this sequence [10]) inhibited the activity of T4 polynucleotide kinase. We therefore replaced this stretch with an equal length of sequence chosen by a random number generator. This was accomplished by PCR, using the original 256-bp EcoRI fragment (37) as a template, with the primers 5′-AATTCCAACGAATAA-CTTCCAGG-3′ and 5′-TTGAACGCAACCAGCCGGTAGATAAAGTCGGAGTGTAGTAGAGCGTTTGCCTACAACACCCGGTAT-3′ (the changed sequence is underlined). We refer to the resulting variant sequence as 5S′. All PCR products were purified by ion-exchange high-performance liquid chromatography as described earlier (3).
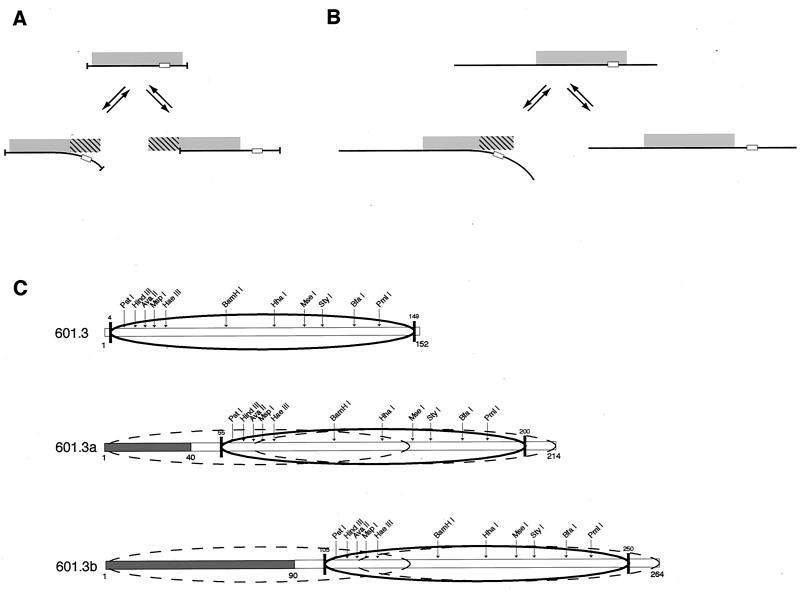
FIG. 1.
Uncoiling versus translocation mechanisms for DNA site exposure in a nucleosome. (A and B) Schematic illustration of the two mechanisms for nucleosome core particle length DNA (A) or long DNA (B). The left-hand path in each panel illustrates the partial uncoiling mechanism; the right-hand path illustrates the nucleosome translocation mechanism. The histone octamer is shown as a gray rectangle, and the DNA is shown as a line, with a particular target site shown within the nucleosomal DNA (small open rectangle). When the DNA is comparable in length to the 147-bp nucleosome core particle length, then both uncoiling and translocation of the histone octamer lead to net losses of histone-DNA contacts (hatched region of the histone octamer). In this case the two mechanisms are very similar, the major difference being only on which side of the (twofold rotationally symmetric) histone octamer the unsatisfied DNA-binding surface is to be found relative to the side on which a DNA target site is concomitantly made accessible. (C) DNA constructs designed to distinguish between the two mechanisms for site exposure. Sequence 601.3 derives from nucleosome positioning sequence 601.2 (3), after mapping of the nucleosome positioning on 601.2 using exonuclease III; sequences 601.3a and 601.3b extend 601.3 with additional nonnucleosomal 601.2 DNA (open bar) and plasmid DNA (shaded bar) appended. The predominant nucleosome position is illustrated by the heavy ellipse; on sequences 601.3a and 601.3b dashed ellipses indicate limiting alternative positions that could be achieved via nucleosome translocation while maintaining a full set of histone-DNA contacts.
Nucleosome reconstitution and analysis of equilibrium constants for site exposure.
The reconstitution, purification, and characterization of nucleosomes were all as described previously (1-3). We used the restriction enzyme kinetics method to measure Keqconf (1, 3, 27, 30). Absolute values of Keqconf for sequence 601.2 had been measured previously (3), obtained from ratios of the observed rate constants for restriction enzyme digestion on nucleosomes compared to naked DNA, scaled for the concentration of enzyme. As justified by the present results, we take the values of Keqconf for 601.3 to be identical to those for 601.2, since 601.3 differs only by lacking a few base pairs which are beyond the ends of the nucleosome. Values of Keqconf for 601.3a and 601.3b were measured relative to those for 601.3 as the ratios of the observed rate constants for digestion, with the same enzyme concentrations being used for all three samples. Some measurements were also made on an absolute scale, by comparison to the rates on naked DNA as described, to confirm the equivalence of Keqconf values for 601.3 and 601.2 (data not shown).
Mobility assays.
Studies of nucleosome mobility on sequence 601.3 and its derivatives were carried out as mock restriction digests by using the same procedures and conditions as for the restriction digests, except without enzyme. The strong positioning power of these sequences allows clean results to be obtained without a need to first purify a single positioning isomer. To distinguish between the possibilities of negligible total nucleosome mobility versus rapidly equilibrating nucleosome positions, the direct mobility assays are supplemented with the results of the restriction enzyme digests (see Discussion).
For studies on the 5S sequence, we devised a strategy that optimized the sensitivity for detection of any nucleosome mobility that would occur. Our strategy was to prepare an equilibrium population of nucleosome positions and then isolate nucleosomes at one of the least favored of these positions by native gel electrophoresis. Nucleosome mobility would then be seen as a redistribution of the purified disfavored isomer back toward the equilibrium set of positions, detectable both as loss of the starting disfavored position and repopulation of the strongly favored positions. Radiolabeled tracer amounts of the 5S′ DNA were reconstituted with chicken erythrocyte histone octamer and core particle DNA as the bulk DNA, at a molar ratio of DNA to histone octamer of 1 to 0.5 by gradual salt dialysis method as previously described (1-3). Nucleosomes at distinct positions on the DNA were separated from each other and from residual naked DNA by native gel electrophoresis in 5% polyacrylamide gels containing 1/3× Tris-borate EDTA (30 mM Tris-borate, 0.66 mM EDTA). Nucleosomes at major and minor positions were (separately) isolated from the gel by crushing and soaking individual gel bands and were then exchanged into 0.5× Tris-EDTA (TE) (TE is 10 mM Tris, 1 mM EDTA [pH 7.5]) and concentrated. Nucleosome mobility assays were carried out as mock restriction digests as described for the 601.3 sequences, using the four standard New England BioLabs (NEB) buffers (1, 2, 3, or 4). Samples (30 μl) contained tracer amounts of the gel-purified minor position 5S′ nucleosomes plus 100 nM unlabeled bulk chicken erythrocyte nucleosome core particles added as a carrier (gel purification removes the carrier nucleosomes that are present in our studies of restriction enzyme digestion) in the desired buffer. These were incubated in pairs at 0 or 37°C for 1 h (i.e., for significantly longer than the restriction enzyme digestion experiments). Additional samples were incubated in the same volume but in TE + 150 mM NaCl, with or without 10 mM MgCl2, again at 0 or 37°C for 1 h. Samples were loaded in 3% Ficoll and TE onto 5% native gels containing 1/3× Tris-borate EDTA, while the gels were running.
RESULTS
Experimental design.
Existing studies by members of our group (3, 28-31) do not distinguish between translocation and uncoiling mechanisms for site exposure because those experiments utilized nucleosomes containing core particle length DNAs. For such short nucleosomal DNAs, the two distinct mechanisms for site exposure both lead to a similar net loss of histone-DNA contacts (Fig. 1a) and thus, presumably, to comparable free energy costs—which are the quantities measured by the restriction enzyme accessibility experiments.
In the present study, we carry out three distinct tests of the translocation mechanism. In the first of these we systematically vary the length of the nucleosomal DNA (Fig. 1b) so as to break the equivalence between the two mechanisms that existed in the earlier studies. By appending additional stretches of DNA for a nucleosome to translocate onto at low energetic cost, we eliminate most of the otherwise large free-energy penalty for nucleosome translocation, while leaving the free-energy penalty for uncoiling unchanged. We then test quantitatively for the effects of the added DNA on the equilibrium accessibilities of sites throughout the nucleosome. We make this experiment cleaner by using a selected nonnatural positioning sequence (derived from sequence 601 [17]) to where the nucleosomes prefer to be. As a consequence, there will still be a nonzero free-energy penalty for nucleosome translocation; but the remaining penalty will be far smaller than the cost of unsatisfied protein-DNA interaction surface measured earlier and will be measured directly in competitive nucleosome reconstitution assays (17, 18, 34, 35, 41).
This experiment takes advantage of the fact that site exposure takes place in a rapid preequilibrium regime (28, 30). The resulting digestion kinetics are first order in enzyme concentration, and are independent of the presence of additional enzymes for which recognition sites are lacking. An opposite limit, in which restriction enzymes trap slowly changing conformations (or locations) of nucleosomes, leads to kinetics that are zero-order in enzyme concentration and is ruled out by this and other observations (28, 30). We reconfirm below that this rapid preequilibrium condition is obeyed also with the longer DNA constructs used in the present study.
If nucleosome translocation is the dominant mechanism of site accessibility and thus is occurring in a rapid preequilibrium regime (that is, rapid in comparison to the restriction enzyme digestions), then a substantial decrease in the energetic cost of translocation necessarily will result in an exponential increase in the time-averaged accessibility of the initially buried DNA target site, with a correspondingly large (102- to 104-fold; see below) increase in the rate of restriction enzyme digestion. In contrast, if uncoiling is the dominant mechanism of site exposure, then the addition of extra DNA to one end of the nucleosome will leave the equilibrium accessibilities of buried nucleosomal DNA target sites unchanged.
The other two experimental tests of the translocation mechanism look directly for nucleosome translocation by two different methods. The first of these is simpler, but as will be seen below, requires the results of position-dependent accessibility studies (such as described above) in order to distinguish between two opposite limits, either (i) that an absence of apparent net mobility reflects an actual absence of any mobility or (ii) that an absence of apparent net mobility reflects an ongoing rapid dynamic positional equilibrium. We utilize this approach for the derivatives of the selected positioning sequence 601.
A second experiment allows direct observation of the rate and extent of nucleosome translocation. In this experiment we create a strongly disequilibrium distribution of initial nucleosome positions (i.e., nucleosomes located at a relatively disfavored site) and then watch nucleosome mobility as the nucleosomes try to reestablish equilibrium occupancy at the dominant position. The disequilibrium population required for this study is created by gel purification of a minor positioning species from an initial equilibrium distribution, which is prepared by our standard method of salt gradient dialysis. We utilize this approach for the 5S rRNA gene nucleosome positioning sequence.
As will be seen, our results from all three of these experiments contradict the predictions of the translocation mechanism. (i) The addition of linker DNA on sequence 601 leaves accessibility unchanged. This is a positive, quantitative measurement of no significant change, not a failure to detect changes that could exist. (ii) In accord with this finding, there is no significant mobility on sequence 601 on the timescale of the accessibility experiments, and this reflects an absence of any significant mobility, not just of net mobility. Finally, (iii) there is similarly no significant mobility on the 5S sequence on the timescale of the accessibility experiments, in contrast to an earlier report (43).
DNA templates.
These studies require a relatively homogeneous population of positioned nucleosomes. We took advantage of a family of DNA constructs derived from a high-affinity nonnatural nucleosome positioning sequence (clone 601) that had been isolated in an earlier SELEX experiment (17). Construct 601.2 (3) introduced a number of nucleotide substitutions into 601 so as to create sites for different restriction enzymes at locations along the entire nucleosomal length. The free energy of interaction of 601.2 with histone octamer in nucleosome reconstitution, the location of the single strongly preferred nucleosome position on this template, and the position-dependent equilibrium accessibility of sites along the nucleosome length have previously been reported (2, 3). The restriction enzyme sites located throughout 601.2 are used in studies described below to quantify the dynamic equilibrium accessibility of DNA target sites throughout the nucleosome.
Construct 601.3 (Fig. 1c) is identical to 601.2 except that it lacks a few base pairs from the short stretches of DNA extending beyond each nucleosome end of 601.2. The resulting sequence has a higher affinity than that of chemically random DNA or of the well-studied 5S rRNA gene nucleosome positioning sequence but has a comparable or lower affinity than other natural eukaryotic genomic sequences as well as other strong nonnatural nucleosome positioning sequences (Table 1). 601.3 was further modified by the addition of 51 or 101 bp of additional DNA on the left (plus 11 bp on the right) to create constructs 601.3a and 6012.3b, respectively.
TABLE 1.
Quantitative free-energy measurements
| DNA sequence | Mean ± SE of ΔΔG° (kcal mol−1)a | Reference |
|---|---|---|
| Lowest affinity known nonnatural sequence | +1.22 ± 0.10 | 41 |
| Bulk chicken genomic DNAb | +0.55 ± 0.03 | 18 |
| Chemically synthetic random DNA | +0.5 ± 0.13 | 17 |
| Sea urchin 5S RNA gene clone | ≡0.00 | |
| Mouse genome CAG clone | −0.78 ± 0.06 | 41 |
| 601.2 | −0.97 ± 0.05 | 2 |
| 601.3 | −0.95 ± 0.05 | 2 |
| Mouse genome TATA clone | −1.82 ± 0.12 | 41 |
| 601 | −2.9 ± 0.14 | 17 |
| Round 15 pool of selected sequencesc | −2.8 ± 0.11 | 17 |
+ΔΔG° values relative to the 5S reference sequence (37). ΔΔG°s reflect intrinsic properties of the molecules independent of the particular conditions used in the competition experiments (17, 18, 34, 35, 41).
Bulk chicken genomic DNA refers to 95% or more of the genome but omits the high-affinity tail (≤5%) of the genome (18).
Average affinity of the entire pool of surviving high-affinity sequences after 15 rounds of physical selection. The pool at this stage comprises ≈30 to 50 distinct sequence families (17).
Nucleosome reconstitution.
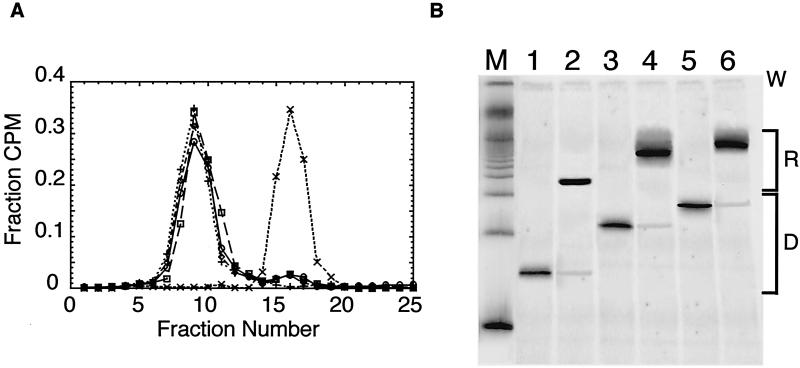
Constructs 601.3, 601.3a, and 601.3b were reconstituted into nucleosomes with purified chicken erythrocyte histones by salt gradient dialysis, and the resulting nucleosomes were purified by sucrose gradient ultracentrifugation (Fig. 2a). Reanalysis of the purified nucleosomes on sucrose gradients (Fig. 2a) and native gel electrophoresis (Fig. 2b) shows them to be largely free of contaminating naked DNA. The native gel results are consistent with predominant population of a single positioning isomer for all three constructs.
FIG. 2.
Purification and characterization of reconstituted nucleosomes. (A) Sucrose gradient purification and reanalysis by sucrose gradient. Nucleosomes are reconstituted by gradual salt dialysis and separated from naked DNA and any nonnucleosomal contaminants on 5 to 30% (wt/vol) sucrose gradients. ○, preparative run of reconstituted 601.3 nucleosomes; □, preparative run of reconstituted 601.3a nucleosomes; ⋄, preparative run of reconstituted 601.3b nucleosomes; +, reanalysis of gradient-purified 601.3 nucleosomes; ×, naked 601.3 DNA. (B) Native gel electrophoresis. W indicates the location of the loading wells, R indicates the mobilities of the reconstituted nucleosomes, and D indicates the mobilities of naked DNA. Lane M, 100 bp DNA marker; lanes 1, 3, and 5, naked DNA for constructs 601.3, 601.3a, and 601.3b, respectively; lanes 2, 4, and 6, the corresponding samples after reconstitution into nucleosomes and purification by sucrose gradient ultracentrifugation. Phosphorimager analysis of the gel reveals contamination of the nucleosomal samples by free DNA to be ≤2%. This small amount of naked DNA does not contribute to the measured Keqconf because it is digested to completion within the first time point, which is omitted from the subsequent analysis.
Effects of increased DNA length on equilibrium accessibilities of nucleosomal DNA target sites.
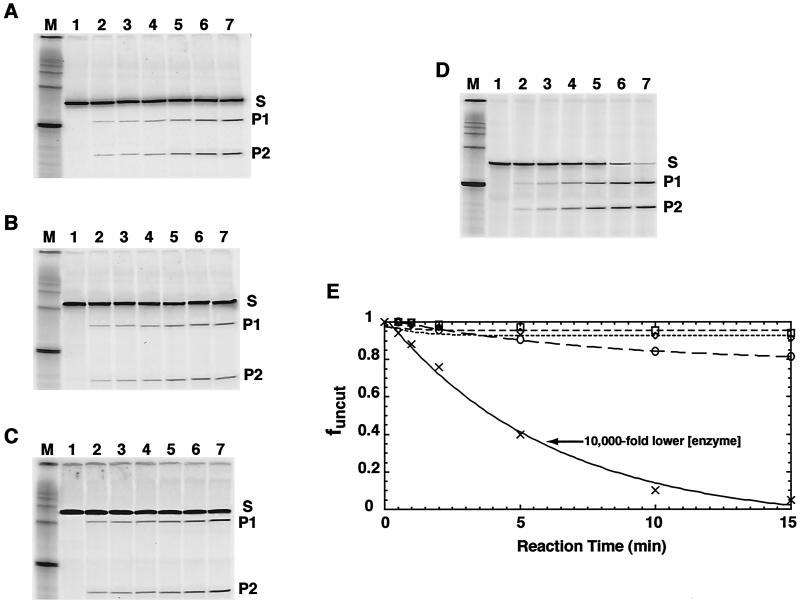
We used the restriction enzyme digestion kinetics method (27, 30) to determine the equilibrium constants for site exposure Keqconf at positions throughout the nucleosome for all three DNA constructs. Typical data are illustrated in Fig. 3.
FIG. 3.
Restriction enzyme digestion analyses of site exposure equilibria. Representative data are shown, probing site exposure at a site 100 to 105 bp from the 5′ end of the predominant core particle position. (A to C) Denaturing polyacrylamide gel analysis of the time course of digestion using the enzyme StyI at 1,000 units ml−1. (A) Reconstituted 601.3 nucleosomes; (B) 601.3a nucleosomes; (C) 601.3b nucleosomes. Lanes 1 through 7 are from samples removed at 0, 0.5, 1, 2, 5, 10, and 15 min, respectively, after reaction initiation. In each case, the substrate (S; 152 nucleotides [nt] for construct 601.3, 214 nt for 601.3a and 264 nt for 601.3b) is converted over time into two products (P1, 104 nt, and P2, 44 nt, for 601.3; P1, 154 nt, and P2, 56 nt, for 601.3a; P1, 204 nt, and P2, 56 nt, for 601.3b). The sizes of S, P1, and P2 expected from the DNA sequence are confirmed against the 100-bp DNA markers in lane M. (D) Naked 601.3 DNA digested with StyI at 0.1 units ml−1. (E) Quantitative analysis of the time course of digestion from the data in panels A through D, respectively. The fraction of DNA remaining uncut is plotted versus time. ○, 601.3 nucleosomes; ⋄, 601.3a nucleosomes; □, 601.3b nucleosomes; ×, 601.3 naked DNA. The superimposed lines represent the results of fits to a single exponential plus baseline (see Materials and Methods). Note that 10,000-fold-lower enzyme concentration is used in this naked DNA digestion and that values for Keqconf derive from observed rate constants for digestion scaled by the enzyme concentration. These data show that, were Keqconf to have increased 102- to 104-fold for 601.3a or 601.3b compared to Keqconf for 601.3 at certain sites, this would have been readily apparent.
The restriction enzyme buffers all include 10 mM Mg2+ for optimal enzyme activity, which in turn is required to allow reasonably quantitative results to be obtained, since the equilibrium accessibilities of the nucleosomal target sites are low. This [Mg2+] is greater than the 0.2 or 2 mM shown earlier to reduce (but not eliminate) nucleosome mobility at 37°C (24) but is comparable to the 7 mM utilized in the studies by Ura et al. (43), in which nucleosome mobility was reported both to occur and to be essential to allow site specific factor binding to buried nucleosomal target sites. We will consider in Discussion whether nucleosome mobility contributes to site accessibility at a lower [Mg2+].
Earlier studies (28, 30) showed that restriction enzyme digestion of nucleosomal DNA target sites takes place in a rapid preequilibrium limit. This is an important feature of the kinetic analysis. One key test is to determine the order of the reactions with respect to enzyme concentration. We carried out new studies to confirm that the preequilibrium limit applies also for these longer DNA templates. A 5.0-fold decrease in [BamHI] results in a 4.3-fold decrease in the rate of digestion of nucleosomes assembled on construct 601.3b; a 5.0-fold decrease in [StyI] leads to a 4.8-fold decrease in rate (data not shown). These results are consistent with first order kinetics. They rule out the possibility that the digestions simply trap slow nucleosome conformational changes including slow nucleosome translocation (see references 28 and 30 for a detailed discussion).
Results from many experiments such as those of Fig. 3 are summarized in Fig. 4 for sites throughout the nucleosome. All three constructs reveal an expected strong suppression of accessibility relative to naked DNA and strong dependence of Keqconf on the position within the nucleosome (3, 28), confirming that the assay is sensitive to relative accessibility over many orders of magnitude. Nevertheless, addition of 51 or 101 bp to the left end causes essentially no change in accessibility at most sites and no changes larger than twofold at any site. Averaging these data over all sites yields values of 1.1- ± 0.1-fold increases in Keqconf for both 601.3a and 601.3b.
FIG. 4.
Position-dependent equilibrium constants for site exposure (Keqconf) for the three DNA constructs, plotted on a log scale. 601.3, 601.3a, and 601.3b are represented by clear, shaded, and diagonally striped bars, respectively. Values for Keqconf range over 2 orders of magnitude at the sites probed yet are essentially invariant with addition of 55 or 105 bp of DNA beyond the end of the nucleosome.
These findings invalidate a key prediction of the translocation mechanism. Qualitatively, if translocation were occurring rapidly (in comparison to the timescale of the restriction enzyme reactions), then for the longest sequence used (601.3b), the possibility of translocation without net loss of any histone-DNA contacts (i.e., with little cost in free energy) would greatly increase the accessibility at all sites except PstI, HindIII, MspI, and HaeIII (Fig. 1c), which serve as negative controls. Sequence 601.3a would yield greatly enhanced accessibility at a smaller set of sites. In contrast to these predictions, we find that there are no significant changes in equilibrium accessibility at all sites for both longer constructs (Fig. 4).
For a proper quantitative analysis we must consider the free-energy costs of having unsatisfied DNA binding surface on the histone octamer and compare this to the free energy cost of nucleosome translocation. Because the restriction enzyme reactions occur in a rapid preequilibrium limit, the equilibrium accessibilities measured in these reactions relate to the free energy of the state in which the sites become accessible. The free energy cost of having unsatisfied DNA-binding surface is calculated from the free energy of site exposure, given by the formula ΔG° = −RT ln Keqconf, where R is the gas constant, T is the temperature, and the values Keqconf are obtained from our studies on nucleosome length DNA (3). The measured equilibrium constants Keqconf of 2 × 10−4 to 5 × 10−6 for sites near the ends and the middle of the nucleosomal DNA, respectively, yield free-energy costs of having unsatisfied DNA-binding surface of 5.1 to 7.3 kcal mol−1. The free-energy cost for transferring the histone octamer from the preferred position onto random sequence DNA is calculated as the difference in free energy of histone-DNA interactions for specific positioning versus random DNA, as measured in competitive nucleosome reconstitution assays (17, 18, 34, 35, 41). The needed data are summarized in Table 1. Transfer from the preferred location on sequence 601.3 (relative free energy = −0.95 kcal mol−1) to chemically synthetic random DNA (relative free energy = +0.5 kcal mol−1) or bulk chicken genomic DNA (relative free energy = +0.55 kcal mol−1) occurs with a modest change in free energy, 1.5 kcal mol−1 [ = (0.55 or 0.5) − (−0.95)].
Suppose we append to the left-hand end of a core particle length positioning sequence sufficient extra DNA so that leftward translocation of the histone octamer can now free up a site near the right-hand end of the positioning sequence while still maintaining contact with a full nucleosome length of DNA. The cost of this translocation will be at most this 1.5 kcal mol−1. (The cost will actually be reduced below this amount by an entropic gain for translocation to multiple sites; moreover, positioning DNA sequence will still comprise some of the nucleosomal DNA, potentially further decreasing the cost of this translocation. Including these effects would make for even larger differences between the two mechanisms but is not necessary to allow a decisive experimental test; hence, we shall not consider them further.) If this extra DNA were not appended, the cost of exposing a site just inside one end (which in this case would require a net loss of histone-DNA contacts) would have been the 5.1 kcal mol−1 measured in our earlier work.
Thus, if nucleosomes were rapidly translocating, to a degree sufficient to allow the observed site exposure preequilibrium, then providing a short additional stretch of DNA to one end must decrease the energetic cost of exposing sites near the other end of the nucleosome by 3.6 (= 5.1 − 1.5) kcal mol−1 or more, thereby increasing the equilibrium accessibility (Keqconf) at such sites at least ∼400-fold [= exponential (3.6/RT)]. Appending a longer stretch that allows access via translocation even to the middle of the original nucleosome position decreases the cost exposure via translocation by 5.8 (= 7.3 − 1.5) kcal mol−1 or more, which would increase the equilibrium accessibility at the middle of the nucleosome more than 104-fold.
Our finding that there are no significant changes (1.1- ± 0.1-fold) in equilibrium accessibility at all sites for both longer constructs (Fig. 4) is plainly inconsistent with these huge effects on accessibility inherent in the rapid nucleosome translocation mechanism. We conclude that rapid nucleosome translocation is not contributing to site accessibility in this system. The important question of why nucleosome translocation does not occur to a significant extent in this system is considered in Discussion.
Native gel electrophoretic mobility assay for net or equilibrating nucleosome translocation.
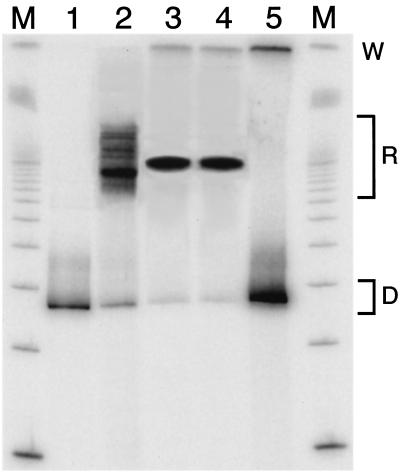
We consider next whether nucleosomes reconstituted on sequence 601.3b do in fact translocate to significant extents during mock restriction enzyme digestion reactions on this family of DNA constructs. These studies take advantage of the ability of native gel electrophoresis to resolve nucleosomes located at differing positions along a given DNA template (7, 20, 21). The (relatively weak) nucleosome positioning sequence from the 5S rRNA gene (37) gives rise to a set of bands having distinguishable mobilities (Fig. 5, lane 2), confirming the ability of this gel system to resolve such alternative positionings on DNA of this length. We detect particular alternative positions having occupancies of 0.5% or less (3, 28). Sequence 601.3b, which has a similar length (264 bp) positions nucleosomes more strongly (lane 3). The result of incubating this sample in a mock restriction enzyme digestion reaction (30 min, 37°C, in the buffer used for many of the digestion reactions) is shown in lane 4. The fraction of nucleosomes that have translocated to alternative positions is undetectably low, and in particular is much less than the 10 to 20% digested during the real reactions by this time point (Fig. 3).
FIG. 5.
Native gel mobility test for nucleosome translocation. Lanes 3 and 4, nucleosomes reconstituted with DNA template 601.3b (264 bp), prior to (lane 3) or after (lane 4) a mock restriction enzyme digestion (30 min, 37°C, HindIII buffer). Lane 2, a (relatively weak) natural nucleosome positioning sequence from the sea urchin 5S rRNA gene (37) (256 bp) yields reconstituted nucleosomes (designated R) at multiple positions on the DNA template, demonstrating that this gel system is capable of resolving such species when they exist. Quantitative studies show that fractions of 0.5% or less of the population having alternative mobilities are readily detected. The fraction of such species present after the mock digestion reaction (lane 4) is undetectably low, and much less than the 10 to 20% of the sample that is actually digested during such reactions when the enzyme is present. Lanes 1 and 5, naked 5S and 601.3b DNA, respectively. D, mobilities of the naked DNA; W, sample loading wells; lanes M, 100-bp ladder size standards.
These observations are consistent with two very different physical situations, but both lead to the same conclusion, namely, that nucleosome translocation on sequence 601 occurs slowly relative to the rate of the observed site exposure processes and thus, that translocation does not contribute significantly to site exposure equilibria on these DNAs. The two possible interpretations and their consequences are as follows. (i) Nucleosome translocation occurs negligibly slowly for this family of DNA constructs and under these conditions. The absence of significant net translocation reflects the absence of any significant extent of nucleosome translocation. If this interpretation is correct, it follows that nucleosome translocation does not contribute significantly to site exposure as probed by restriction enzyme accessibility. Alternatively, (ii) nucleosome translocation might be facile for this family of DNAs, with histone octamers equilibrating freely and rapidly between the set of alternative positions. In this scenario, there is no net translocation of nucleosomes during a mock digestion because the positioning is equilibrating freely both before and during the mock digestion reaction. But this hypothesis was ruled out by the results shown in Fig. 4 (see above).
In summary, this experiment (Fig. 5) and the measurement of the effects of added linker DNA on site exposure equilibria (Fig. 3 and 4) both point to the same conclusion: site exposure on this family of DNAs occurs by a mechanism distinct from nucleosome translocation.
Nucleosome translocation on the 5S positioning sequence.
The conclusion that site exposure on the 601 family of DNA sequences occurs without significant contribution from nucleosome mobility contradicts the conclusions of Ura et al. (43) (see the introduction), who studied accessibility and positioning on the 5S rRNA gene nucleosome positioning sequence.
These observations led us to directly investigate nucleosome mobility on the 5S sequence. Nucleosomes were reconstituted on a 256-bp derivative of the 5S gene positioning sequence (see Materials and Methods), and nucleosomes located at the dominant positions (fast mobility) and less-favored alternative positions (slow mobility) were separated from each other by native gel electrophoresis (Fig. 6a, lanes 2 and 3, respectively). This yields a high-disequilibrium population of nucleosomes that can be used for clear-cut analyses of nucleosome mobility in solution: nucleosome translocation allows the purified minor positions to redistribute back toward the equilibrium population—in particular, repopulating the major positions. Results from such studies are shown in Fig. 6b. We find that a negligible fraction of nucleosomes migrate away from their starting disfavored positions even after 1 h at 37°C in any of the four restriction enzyme buffers used, in clear contrast to the substantial fractions of nucleosomes that are cleaved during much shorter digestions under the same conditions, as can be seen from our data on sequence 601.3 (Fig. 3), or in earlier studies on the 5S sequence (27, 28, 30). The lack of significant nucleosome mobility during these experiments does not reflect an inability to detect mobility when it does occur, as can be seen from the significant redistribution that occurs after much longer (24 h) incubation under different conditions (Fig. 6b, lane 2).
FIG. 6.
Direct test for nucleosome mobility on the 5S nucleosome positioning DNA sequence. (A) Native gel purification of nucleosomes at the preferred and less-favored positions. Lane 1, starting equilibrium distribution of nucleosome positions prepared by salt gradient dialysis; lane 2, gel-purified major positioning isomers (for use as markers); lane 3, gel-purified minor positioning isomers. M, molecular weight markers; D, mobility of naked DNA; R, range of mobilities of reconstituted nucleosomes; W, loading well of the gel. (B) Tests for nucleosome mobility. Nucleosome mobility is manifested by net movement of nucleosomes from the initial disfavored positions (slower mobility) to the dominant (most-favored) major positions (faster mobility). Lanes 1 and 3, purified major isomers, serving as a marker for the electrophoretic mobility of nucleosomes that have moved to the dominant positions; lane 2, positive control showing nucleosome mobility occurring over long times (24 h, TE + 150 mM NaCl, 0°C); lanes 5, 7, 9, and 11, samples after 1 h of incubation at 37°C in NEB restriction enzyme digestion buffers no. 1, 2, 3, and 4, respectively; lanes 4, 6, 8, and 10, same as lanes 5, 7, 9, and 11, except incubated at 0°C. A negligible fraction of nucleosomes move away from the minor positions in 1 h at 37°C in any of the four NEB restriction enzyme buffers, far fewer than are cleaved by restriction enzymes over much shorter times.
We conclude from these results that even for the 5S positioning sequence, nucleosome mobility occurs to a negligible extent on the 1-h timescale under the quasiphysiological conditions under which the restriction enzyme digestion experiments are carried out.
DISCUSSION
Spontaneous site exposure in vitro occurs via a mechanism that is distinct from nucleosome translocation.
All three experiments in the present study—the analysis of the effects of added linker DNA on site exposure equilibria and the two direct tests for nucleosome mobility on sequence 601.3 and on the 5S sequence—point to the same conclusion: nucleosome mobility does not contribute significantly to spontaneous nucleosomal site exposure as probed by restriction enzyme digestion in vitro. These results are obtained when using both nonnatural and natural nucleosome positioning sequences; and the nonnatural sequences used have affinities falling within the range of natural sequences (Table 1) and do not affect the viability of yeast (Saccharomyces cerevisiae) when incorporated into the yeast genomic DNA (A. Thåström and J. Widom, unpublished data). Consequently, we conclude that our results are representative of the behavior of natural DNA.
Alternative mechanisms based on nucleosome conformational changes are consistent with these and other data. Given a fixed nucleosome position, appending increasing lengths of DNA at one end of the nucleosome would simply be of no consequence to site exposure occurring via uncoiling, since the nucleosomes are not free to explore that extra DNA.
Other considerations place additional constraints on the nature of the site exposure process. Site exposure occurs spontaneously; it is evidently driven by thermal fluctuations. Since even buried target sites become accessible, conformational changes leading to site exposure must involve the displacement of a stretch of DNA off the histone surface. Such a displacement of DNA off the histone surface must be only partial, as it is not accompanied by significant nucleosome dissociation or DNA exchange (46). Coupled enzymatic assays show that access to sites anywhere within a nucleosome can occur on a timescale of seconds or faster, likely much faster (31). The relatively low measured equilibrium constants for site exposure imply that rates for the reverse (recapture) processes are fast compared to site exposure itself. The progressive decrease in Keqconf with distance inward from an end (3, 28) and the linkage between binding sites as manifested by the observed cooperativity (29) are both consistent with the uncoiling (breathing) model, in which uncoiling initiates at either end and proceeds inward toward the middle of the nucleosome. Such uncoiling would probably occur in stepwise fashion corresponding to loss of successive contacts with the DNA backbone at each DNA helical turn (19).
Contribution of nucleosome mobility to site accessibility at lower [Mg2+].
As mentioned in Results, the restriction enzyme buffers used in the present study all include 10 mM Mg2+ for optimal enzyme activity, which in turn is required to allow reasonably quantitative results to be obtained. Since Mg2+ has been reported in an earlier study (24) (although not in another [43]) to reduce nucleosome mobility, the question arises whether nucleosome mobility might contribute to site accessibility at a lower [Mg2+] which may be more physiological.
Three lines of evidence suggest that the elevated [Mg2+] used here does not change the nature of our results. First, although only qualitative data are available, the results reported in reference 24 revealed similar reductions in nucleosome mobility in both 0.2 and 2 mM Mg2+ at 37°C; hence, there is no indication that the higher [Mg2+] used by Ura et al. (43) or us should have further effects, nor is there any indication that nucleosome mobility is facile even in subphysiological (0.2 mM) concentrations of Mg2+. Second, our own direct assays of nucleosome mobility on the 5S′ sequence, comparable to those shown in Fig. 6 except carried out in 0 mM Mg2+, 37°C, revealed negligible nucleosome mobility on the timescale of several hours (data not shown). Third, other direct studies on nucleosome mobility, monitoring reequilibration of nucleosome positions on sequence 601 in 1.5 mM Mg2+, 30°C, following the action of the ISWI protein (which drives nucleosomes to the ends of DNA fragments [15]), again show negligible nucleosome mobility on the hour timescale, although substantial repositioning occurs over a 24-h incubation (Chin and Widom, unpublished). In summary, nucleosome mobility on the 5S sequence and on selected high-affinity sequences such as 601 is slow relative to rates of site accessibility measured here regardless of whether the [Mg2+] is lower or higher than physiological.
A kinetic barrier for nucleosome translocation.
We consider next the important question of why nucleosome translocation appears not to occur to a significant extent, even though, were it to occur, it would greatly lower the free-energy cost involved in another process (namely, site exposure) that does occur.
Such behavior represents the essence of the difference between equilibria and kinetics. It can indeed be the case that nucleosome translocation, were it able to occur at a sufficient rate, would provide a lower energy path for access to DNA target sites inside nucleosomes, yet the system may be kinetically blocked from ever being able to attain such states. Alternative mechanisms, such as a partial DNA uncoiling or other conformational change, which might involve greater net free-energy cost but happen to have lower free-energy barriers, would then dominate the actual behavior of the system.
Mechanisms for nucleosome translocation and for DNA looping during nucleosome transcription.
The fact that site exposure occurs rapidly in comparison to nucleosome translocation suggests that, rather than nucleosome translocation being the mechanism of site exposure, the opposite situation may obtain: partial DNA uncoiling might be a mandatory initial step in the detailed mechanism for nucleosome translocation. Partially uncoiled DNA may sometimes be recaptured (rewrapped) starting from a point displaced along the DNA, so as to form a bulged loop; diffusion of such loops off the other end of the nucleosome would lead to a net translocation. Thus, we suggest that partial uncoiling may represent a first step in mechanisms for both spontaneous (28, 33) and ATP-powered catalyzed (32, 33) nucleosome translocation. Such looped structures are also believed to play a role in transcriptional elongation (5, 39), allowing the histone octamer to step around an elongating polymerase without dissociating.
Site exposure as a potential initial step in nucleosome invasion in vivo.
We and others suggest that site exposure occurring via partial DNA unwrapping starting from an end is a possible explanation for how gene regulatory proteins may gain initial entry to DNA target sites that are sterically occluded in nucleosomes (8, 28). Cooperative effects that arise from the binding of multiple DNA binding proteins to DNA inside the same nucleosome allow for synergistic nucleosome invasion (29, 47). Subsequent action of histone acetylases or deacetylases or of ATP-dependent chromatin remodeling enzymes that are recruited by site-specifically bound regulatory proteins (6, 26) can lead to quantitative changes in Keqconf (1, 27) or possibly displace the histone octamer altogether, in either case thereby changing the time-averaged occupancy achieved by regulatory proteins at that same and other nearby target sites.
Relation to other studies.
We consider finally the possible reasons for the discrepancy between our results and those of Ura et al. (43) concerning the mechanisms of site accessibility on the 5S sequence. One possible reason may be differences in the detailed solution conditions used. We consider this explanation to be unlikely, as many different conditions are examined in the present study with consistent results obtained for all of them; and the conditions studied by Ura et al. fall generally within the range studied here. A second possible explanation is that the details of the DNA sequence used might be of great importance. Our 5S rRNA sequence comes from an organism (sea urchin) different from that used by Ura et al. (Xenopus); moreover, we replaced a small stretch of it (outside the region reported to be responsible for its nucleosome positioning properties [10]) with a random nucleotide sequence. We consider this explanation too to be unlikely. The 601 and 5S sequences studied here are completely unrelated to each other, yet their behavior is equivalent. Moreover, as described above, their affinity for histones and corresponding nucleosome positioning power falls within the normal range of natural DNAs, and even higher affinity sequences can be incorporated into the genomes of living yeast cells without evident adverse consequence.
Rather, we consider that the most likely explanation for the discrepancy between these reports is that the Ura et al. study confuses the presence of multiple nucleosome positions with nucleosome mobility. The data presented in that study demonstrate that increasing the length of the template DNA increased the mispositioning of nucleosomes (i.e., the occupancy of alternative positions) on it. The study then equates increased mispositioning with mobility. But these are two entirely different properties, so such an equivalence cannot properly be made. Multiple positioning is an equilibrium property, whereas nucleosome mobility is a kinetic property. There are innumerable examples of processes that are favored at equilibrium but nevertheless do not occur. Our results on the 5S sequence highlight the difference. Figure 6a, lane 1, illustrates that many distinct nucleosome positions are significantly occupied at equilibrium; but in Fig. 6b, lanes 3 to 11 demonstrate that the rate of migration from less-favored to more-favored positions is negligibly small in a diversity of quasiphysiological conditions. No data supporting the interpretation of increased mobility (as distinct from mispositioning) are provided in the study by Ura et al. Thus, the actual results of that study are not in conflict with ours; only the interpretation of that earlier work is in conflict with our results.
In summary, the three distinct experiments reported here are all in agreement, and moreover are not contradicted by other results in the literature. Nucleosome mobility (translocation) contributes negligibly to spontaneous (uncatalyzed) nucleosomal site exposure in vitro. Site exposure must occur by a nucleosome conformational change that displaces a stretch of DNA off the nucleosome surface while leaving the register of the histone octamer along the DNA unchanged.
Acknowledgments
This work was supported by a grant from the NIH (to J.W.) and by an NIH Cell and Molecular Basis of Disease Traineeship (to J.D.A.).
We acknowledge with gratitude the use of instruments in the Keck Biophysics Facility, which was established with a grant from the W. M. Keck Foundation. We thank members of our laboratory for valuable discussion.
REFERENCES
- 1.Anderson, J. D., P. T. Lowary, and J. Widom. 2001. Effects of histone acetylation on the dynamic equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 307:977-985. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. D., and J. Widom. 2001. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol. Cell. Biol. 21:3830-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, J. D., and J. Widom. 2000. Sequence- and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 296:979-987. [DOI] [PubMed] [Google Scholar]
- 4.Astumian, R. D., and M. Bier. 1994. Fluctuation driven ratchets: molecular motors. Phys. Rev. Lett. 72:1766-1769. [DOI] [PubMed] [Google Scholar]
- 5.Bednar, J., V. M. Studitsky, S. A. Grigoryev, G. Felsenfeld, and C. L. Woodcock. 1999. The nature of the nucleosomal barrier to transcription: direct observation of paused intermediates by electron cryomicroscopy. Mol. Cell 4:377-386. [DOI] [PubMed] [Google Scholar]
- 6.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 7.Dong, F., J. C. Hansen, and K. E. van Holde. 1990. DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc. Natl. Acad. Sci. USA 87:5724-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenfeld, G. 1996. Chromatin unfolds. Cell 86:13-19. [DOI] [PubMed] [Google Scholar]
- 9.Feynman, R. P., R. B. Leighton, and M. Sands. 1963. The Feynman lectures on Physics, vol. I. Addison-Wesley, Reading, Mass.
- 10.FitzGerald, P. C., and R. T. Simpson. 1985. Effects of sequence alterations in a DNA segment containing the 5S RNA gene from Lytechinus variegatus on positioning of a nucleosome core particle in vitro. J. Biol. Chem. 260:15318-15324. [PubMed] [Google Scholar]
- 11.Fragoso, G., S. John, M. S. Roberts, and G. L. Hager. 1995. Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev. 9:1933-1947. [DOI] [PubMed] [Google Scholar]
- 12.Goldmark, J. P., T. G. Fazzio, P. W. Estep, G. M. Church, and T. Tsukiyama. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423-433. [DOI] [PubMed] [Google Scholar]
- 13.Iyer, V., and K. Struhl. 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 14:2570-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 15.Langst, G., E. J. Bonte, D. F. V. Corona, and P. B. Becker. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843-852. [DOI] [PubMed] [Google Scholar]
- 16.Lorch, Y., J. W. LaPointe, and R. D. Kornberg. 1987. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49:203-210. [DOI] [PubMed] [Google Scholar]
- 17.Lowary, P. T., and J. Widom. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19-42. [DOI] [PubMed] [Google Scholar]
- 18.Lowary, P. T., and J. Widom. 1997. Nucleosome packaging and nucleosome positioning of genomic DNA. Proc. Natl. Acad. Sci. USA 94:1183-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 20.Luger, K., T. J. Rechsteiner, A. J. Flaus, M. M. Y. Waye, and T. J. Richmond. 1997. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272:301-311. [DOI] [PubMed] [Google Scholar]
- 21.Meersseman, G., S. Pennings, and E. M. Bradbury. 1992. Mobile nucleosomes—a general behavior. EMBO J. 11:2951-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owen-Hughes, T., R. T. Utley, J. Cote, C. L. Peterson, and J. L. Workman. 1996. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science 273:513-516. [DOI] [PubMed] [Google Scholar]
- 23.Owen-Hughes, T., and J. L. Workman. 1996. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J 15:4702-4712. [PMC free article] [PubMed] [Google Scholar]
- 24.Pennings, S., G. Meersseman, and M. E. Bradbury. 1991. Mobility of positioned nucleosomes on 5S rDNA. J. Mol. Biol. 220:101-110. [DOI] [PubMed] [Google Scholar]
- 25.Peterson, C. L., and C. Logie. 2000. Recruitment of chromatin remodeling machines. J. Cell. Biochem. 78:179-185. [DOI] [PubMed] [Google Scholar]
- 26.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 27.Polach, K. J., P. T. Lowary, and J. Widom. 2000. Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J. Mol. Biol. 298:211-223. [DOI] [PubMed] [Google Scholar]
- 28.Polach, K. J., and J. Widom. 1995. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 254:130-149. [DOI] [PubMed] [Google Scholar]
- 29.Polach, K. J., and J. Widom. 1996. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J. Mol. Biol. 258:800-812. [DOI] [PubMed] [Google Scholar]
- 30.Polach, K. J., and J. Widom. 1999. Restriction enzymes as probes of nucleosome stability. Methods Enzymol. 304:278-298. [DOI] [PubMed] [Google Scholar]
- 31.Protacio, R. U., K. J. Polach, and J. Widom. 1997. Coupled enzymatic assays for the rate and mechanism of DNA site-exposure in a nucleosome. J. Mol. Biol. 274:708-721. [DOI] [PubMed] [Google Scholar]
- 32.Richmond, T. J., and J. Widom. 2000. Nucleosome and chromatin structure. In S. Elgin and J. L. Workman (ed.), Chromatin structure and gene expression: frontiers in molecular biology 2/e. Oxford University Press, Oxford, United Kingdom.
- 33.Schiessel, H., J. Widom, R. F. Bruinsma, and W. M. Gelbart. 2001. Polymer reptation and nucleosome repositioning. Phys. Rev. Lett. 86:4414-4417. [DOI] [PubMed] [Google Scholar]
- 34.Shrader, T. E., and D. M. Crothers. 1989. Artificial nucleosome positioning sequences. Proc. Natl. Acad. Sci. USA 86:7418-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shrader, T. E., and D. M. Crothers. 1990. Effects of DNA sequence and histone-histone interactions on nucleosome placement. J. Mol. Biol. 216:69-84. [DOI] [PubMed] [Google Scholar]
- 36.Simpson, R. T., S. Y. Roth, R. H. Morse, H.-G. Patterton, J. P. Cooper, M. Murphy, M. P. Kladde, and M. Shimizu. 1993. Nucleosome positioning and transcription. Cold Spring Harbor Symp. Quant. Biol. 58:237-245. [DOI] [PubMed] [Google Scholar]
- 37.Simpson, R. T., and D. W. Stafford. 1983. Structural features of a phased nucleosome core particle. Proc. Natl. Acad. Sci. USA 80:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson, R. T., F. Thoma, and J. M. Brubaker. 1985. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell 42:799-808. [DOI] [PubMed] [Google Scholar]
- 39.Studitsky, V. M., G. A. Kassavetis, E. P. Geiduschek, and G. Felsenfeld. 1997. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science 278:1960-1963. [DOI] [PubMed] [Google Scholar]
- 40.Svaren, J., and W. Horz. 1997. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci. 22:93-97. [DOI] [PubMed] [Google Scholar]
- 41.Thåström, A., P. T. Lowary, H. R. Widlund, H. Cao, M. Kubista, and J. Widom. 1999. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 288:213-229. [DOI] [PubMed] [Google Scholar]
- 42.Travers, A. 1999. An engine for nucleosome remodeling. Cell 96:311-314. [DOI] [PubMed] [Google Scholar]
- 43.Ura, K., J. J. Hayes, and A. P. Wolffe. 1995. A positive role for nucleosome mobility in the transcriptional activity of chromatin templates: restriction by linker histones. EMBO J. 14:3752-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Holde, K. E. 1989. Chromatin. Springer-Verlag, New York, N.Y.
- 45.Varga-Weisz, P. D., and P. B. Becker. 1998. Chromatin-remodeling factors: machines that regulate? Curr. Biol. 10:346-353. [DOI] [PubMed] [Google Scholar]
- 46.Widom, J. 1999. Equilibrium and dynamic nucleosome stability. Methods Mol. Biol. 119:61-77. [DOI] [PubMed] [Google Scholar]
- 47.Widom, J. 1998. Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 27:285-327. [DOI] [PubMed] [Google Scholar]
- 48.Widom, J. 1997. Transcription through nucleosomes. Science 278:1899-1901. [DOI] [PubMed] [Google Scholar]
- 49.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 50.Zhu, Z., and D. J. Thiele. 1996. A specialized nucleosome modulates transcription factor access to a C. glabrata metal responsive promoter. Cell 87:459-470. [DOI] [PubMed] [Google Scholar]