Abstract
The Arabidopsis GNOM protein, a guanine nucleotide exchange factor (GEF) that acts on ADP ribosylation factor (ARF)–type G proteins, is required for coordination of cell polarity along the apical–basal embryo axis. Interallelic complementation of gnom mutants suggested that dimerization is involved in GNOM function. Here, direct interaction between GNOM molecules is demonstrated in vitro and by using a yeast two-hybrid system. Interaction was confined to an N-terminal domain conserved within a subgroup of large ARF GEFs. The same domain mediated in vitro binding to cyclophilin 5 (Cyp5), which was identified as a GNOM interactor in two-hybrid screening. Cyp5 displayed peptidylprolyl cis/trans–isomerase and protein refolding activities that were sensitive to cyclosporin A. Cyp5 protein accumulated in several plant organs and, like GNOM, was partitioned between cytosolic and membrane fractions. Cyp5 protein was also expressed in the developing embryo. Our results suggest that Cyp5 may regulate the ARF GEF function of the GNOM protein during embryogenesis.
INTRODUCTION
The GNOM gene was identified by multiple mutant alleles in a search for mutations affecting body organization of the Arabidopsis embryo (Mayer et al., 1991). All gnom alleles are defective in establishing the apical–basal axis of embryo polarity. The earliest defect observed in the gnom embryo, which is a perturbed division of the zygote, is followed by irregular cell division and elongation patterns at subsequent stages (Mayer et al., 1993). gnom seedlings lack a root, are defective in coordinated alignment of vascular cells, and display variably reduced apical structures (Mayer et al., 1993). These alterations have been attributed to defects in the establishment of coordinated cell polarity along the apical–basal axis in early embryos (Steinmann et al., 1999). Cloning of the GNOM gene (also called EMB30) revealed sequence similarity to the yeast vesicle trafficking protein Sec7p, including a central region called the Sec7 domain (Shevell et al., 1994; Busch et al., 1996). Proteins with Sec7 domains catalyze guanine nucleotide exchange on small GTP binding proteins of the ADP ribosylation factor (ARF) family required for vesicle coating in membrane trafficking (Chardin et al., 1996; Sata et al., 1998; Springer et al., 1999).
Guanine nucleotide exchange factors (GEFs) that act on ARFs (ARF GEFs) group into small and large family members, and large ARF GEFs include the yeast proteins Gea1p, Gea2p, and Sec7p (Chardin et al., 1996; Peyroche et al., 1996; Sata et al., 1998). GNOM is a membrane-associated, functional ARF GEF of the Gea type, suggesting its involvement in vesicular trafficking (Steinmann et al., 1999). Apart from the Sec7 domain, large ARF GEFs contain as yet functionally uncharacterized N- and C-terminal regions (Moss and Vaughan, 1998). In the case of GNOM, however, there is genetic evidence for subunit interaction, because gnom alleles with different mutations in the Sec7 domain exhibit full complementation (Busch et al., 1996). In this study, we demonstrate a physical interaction of GNOM molecules by using a yeast two-hybrid system and in vitro, thereby defining a novel N-terminal interaction domain that is conserved within a distinct subgroup of large ARF GEFs.
Large ARF GEFs, such as Gea1p, exist in high molecular weight complexes (Peyroche et al., 1996), but their interacting partners have not been characterized. We have screened for GNOM interactors by using the yeast two-hybrid system, and we have identified cyclophilin 5 (Cyp5) as an interacting protein. Cyp proteins catalyze cis/trans–isomerization of peptidylprolyl bonds, a rate-limiting step in protein folding, and their activity is inhibited by the immunosuppressive drug cyclosporin A (Fischer et al., 1989; Schönbrunner et al., 1991). Cyp proteins are involved in different signal transduction pathways, such as T cell activation and heat shock protein Hsp90–dependent signal transduction in glucocorticoid receptor regulation (Mattila et al., 1990; Bram and Crabtree, 1994; Duina et al., 1996).
Only a few studies have addressed the role of Cyp proteins during dimerization or oligomerization of target molecules. In the mouse glucocorticoid receptor complex, for example, Cyp40 interacts with the dimerization domain of Hsp90 (Carrello et al., 1999), and a yeast Cyp40 homolog, Cpr6, can reactivate the ATPase activity of Hsp90 in vitro (Prodromou et al., 1999). As an example of Cyp interaction with an oligomeric target in vivo, human CypA was shown to bind to the human immunodeficiency virus HIV-1 capsid protein Gag (Luban et al., 1993; Franke et al., 1994). Mutations in Gag abolish both CypA incorporation into virions and virion infectivity, mimicking the effects of cyclosporin A treatment of infected cells (Franke et al., 1994).
Our study identifies a conserved protein domain of a large ARF GEF that mediates both subunit interaction and Cyp binding. GNOM-interacting Cyp5 is a cyclosporin A–sensitive peptidylprolyl cis/trans–isomerase (PPIase) with protein refolding activity that is also expressed during embryogenesis. We propose that Cyp5 is a potential regulator of GNOM function in Arabidopsis embryogenesis.
RESULTS
Interaction of GNOM Subunits Mediated by an N-Terminal Domain
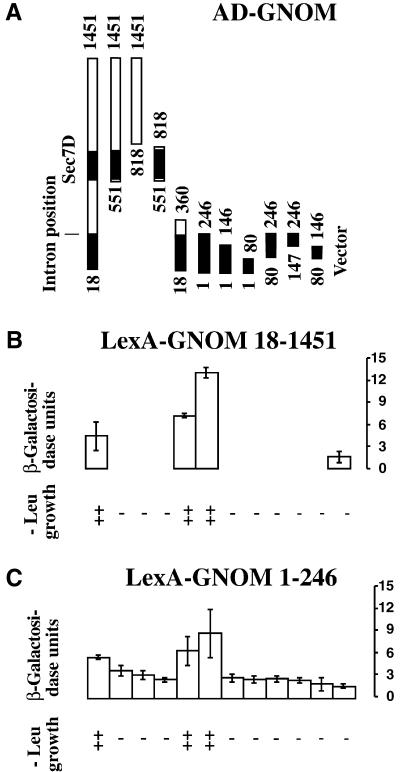
Genetic complementation between different gnom mutant alleles suggested that GNOM function involves physical interaction of GNOM subunits (Mayer et al., 1993; Busch et al., 1996). To examine this model, we generated a series of GNOM deletion constructs in interaction trap vectors of the yeast two-hybrid system (Gyuris et al., 1993). As shown in Figures 1A and 1B, interaction was observed between nearly full-length GNOM proteins lacking the first 17 amino acids. The region required for self-interaction was mapped to GNOM amino acids 1 to 246 (GNOM1–246) encoded by the first exon of the GNOM gene (Figure 1C; see also Figure 3A).
Figure 1.
Interaction between GNOM Subunits and Mapping of the Interaction Domain in Yeast Two-Hybrid Assays.
(A) GNOM fragments fused to an activation domain (AD–GNOM) tested for interaction with two LexA–GNOM fusions are represented by bars with amino acid positions indicated. Amino acids 1 to 246 encoded by the first exon and the Sec7 domain (Sec7D) are shaded. Vector, negative control.
(B) Interaction of AD–GNOM fragments with nearly full-length GNOM protein fused to a DNA binding domain (LexA–GNOM18–1451).
(C) Interaction of AD–GNOM fragments with N-terminal 246 amino acids of GNOM protein fused to a DNA binding domain (LexA–GNOM1–246).
Activation of leucine growth reporter (-Leu growth) is indicated by ++ or −; LacZ reporter activity is displayed as relative β-galactosidase units determined by liquid culture assay (Ausubel et al., 1995). Error bars represent standard deviations from three to five independent transformants.
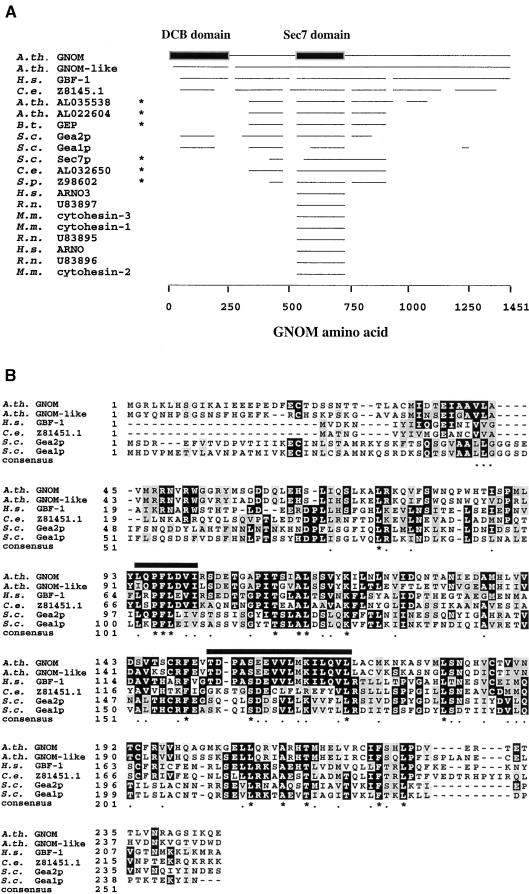
Figure 3.
Sequence Conservation of the GNOM DCB Domain.
(A) GNOM homologs identified by BLAST P search (BLAST plus BEAUTY, ungapped alignments, expect value 0.0001; Altschul et al., 1997). Horizontal lines aligned with GNOM amino acid positions (bottom) represent regions of sequence conservation >38%. Asterisks indicate sequences with higher overall homology to Sec7p. GenBank accession numbers are indicated, and other references are as follows: A.th. GNOM, Arabidopsis GNOM/EMB30 (Shevell et al., 1994; Busch et al., 1996); A.th.GNOM-like, Arabidopsis GNOM-like putative protein (Sato et al., 1998); H.s. GBF-1, human GBF1 (Mansour et al., 1998); B.t. GEP, bovine Sec7-like GEP (Morinaga et al., 1997); S.c. Gea1p and Gea2p, budding yeast Gea1p and Gea2p (Peyroche et al., 1996); H.s. ARNO and ARNO 3, human ARNO (Chardin et al., 1996) and ARNO3 (Franco et al., 1998); M.m. cytohesin-1, -2, and –3, mouse cytohesin-1, -2, and -3 (Klarlund et al., 1997; Kim et al., 1998); cytohesin-1/B2-1 (Liu and Pohajdak, 1992); R.n. U83895, U83896, and U83897, rat sec7 domain proteins (Telemenakis et al., 1997); C.e. Z8145.1 and AL032650, C. elegans protein; and S.p. Z98602, fission yeast protein.
(B) Clustal W (Thompson et al., 1994) alignment of GNOM DCB domain homology regions. Sequence identities (conservation) to the DCB domain are 70% (81%) for Arabidopsis GNOM-like putative protein amino acids 20 to 225, 38% (58%) for human GBF1 amino acids 34 to 227, 30% (53%) for Gea2p amino acids 88 to 230, 24% (53%) for Gea1p amino acids 87 to 226, and 26% (48%) for C.e. Z81451.1 amino acids 88 to 226 (Z81451.1 has a 33–amino acid insertion at position 138, which is not shown for simplicity of alignment). Black boxes indicate identity, and grey boxes indicate homology between at least three sequences. Consensus derived from identical and conserved amino acids of all six sequences is indicated as stars and dots. Black bars overline a conserved proline-phenylalanine-leucine (PFL) motif and a hydrophobic helix.
To analyze interaction in an independent test system, we performed in vitro protein binding assays, as displayed in Figure 2. A purified glutathione S-transferase (GST)–GNOM1–246 fusion protein pulled down the full-length GNOM protein from Arabidopsis protein extracts (Figure 2A). The same GST–GNOM fusion also bound to GNOM1–246 synthesized by in vitro translation (Figure 2B). As already observed during two-hybrid analysis (Figure 1), smaller subfragments of GNOM did not interact (J. Gadea and G. Jürgens, unpublished results), suggesting a structural requirement of the whole domain for binding. These results demonstrate a direct interaction between GNOM molecules mediated by a distinct N-terminal domain. For simplicity, we refer to this minimal domain as the dimerization domain.
Figure 2.
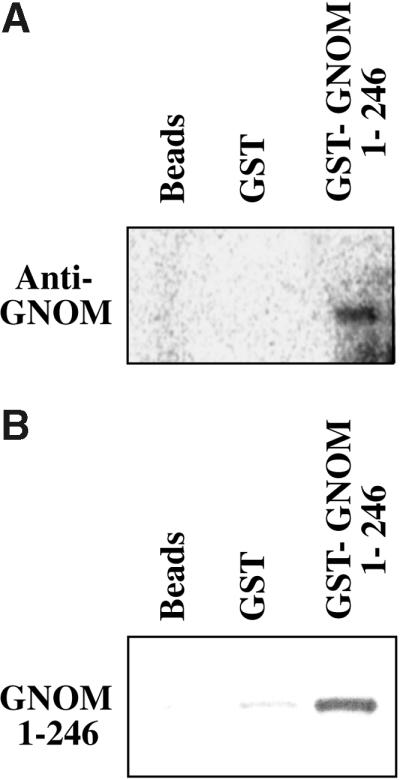
Interaction between GNOM Subunits in Vitro.
In vitro binding assays of the GST–GNOM1–246 fusion are shown.
(A) Arabidopsis protein extracts (protein gel blot; 165-kD band detected by anti-GNOM Sec7 antiserum).
(B) In vitro–translated 35S-methionine–labeled GNOM1–246 (autoradiograph of 15% SDS–polyacrylamide gel).
The controls were glutathione–Sepharose beads (beads) and GST-coupled beads (GST).
Sequence Conservation of the GNOM Dimerization Domain among Large ARF GEFs of the GNOM/Gea Type
In contrast to the well-characterized Sec7 domain of large ARF GEF proteins, little is known about the role of the N- and C-terminal regions of their proteins (Moss and Vaughan, 1998). By searching the databases, we identified five large ARF GEF sequences with significant homology to the GNOM dimerization domain, which are compared in Figure 3. In each case, the homologous region represents the N-terminal part of the protein, indicating a conserved position within the overall protein structure (Figure 3A). The GNOM dimerization domain is most similar in a GNOM-like putative protein identified by the Arabidopsis genome sequencing project, followed by human Golgi-specific brefeldin A–resistance factor 1 (GBF1), a Caenorhabditis elegans open reading frame, and yeast Gea1p and Gea2p proteins. By contrast, yeast Sec7p, protein sequences with higher overall homology to Sec7p, and mammalian small ARF GEFs did not display homology to the GNOM dimerization domain (Figure 3A). Coiled-coil domains, leucine zippers, or other known protein interaction motifs were not found in the GNOM dimerization domain. However, a predicted hydrophobic helix spanning amino acids 152 to 169, which is conserved among GNOM, the Arabidopsis GNOM-like putative protein, and human GBF1, contains leucine and valine in regularly spaced seven–amino acid intervals (Figure 3B). This hydrophobic region may act as a protein–protein interaction surface. In addition, all five GNOM/Gea–type sequences share a PFL motif flanked by conserved amino acids, giving the consensus sequence (L/I)XPFLX(V/I)(I/V). In summary, both the sequence and the N-terminal position of the GNOM dimerization domain are conserved within a distinct subgroup of large ARF GEFs, suggesting functional constraints.
Isolation of GNOM Interacting Proteins
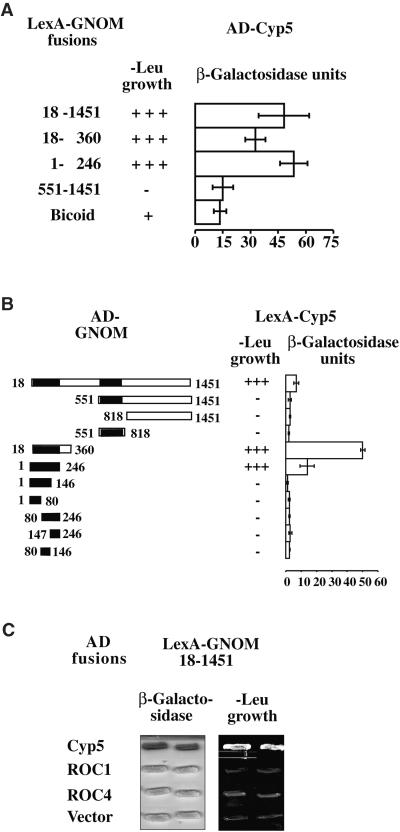
Genetic screening for Arabidopsis embryo pattern mutants identified GNOM as the earliest-acting zygotic gene currently known to be involved in apical–basal axis formation (Mayer et al., 1991, 1993). However, no genes encoding potential direct interactors have been identified by mutation, which prompted us to perform an interaction trap screening for presumptive regulators of GNOM function. The nearly full-length GNOM protein was used to screen two Arabidopsis cDNA libraries generated from suspension cells and young siliques (see Methods). In each case, 3.6 × 106 primary transformants were screened for galactose-dependent activation of leucine growth and lacZ reporters. Clones for strong interactors from the suspension cell library were grouped into 17 cDNA classes and subjected to further specificity tests. One of the GNOM interactors was a putative Cyp, Cyp5, which was represented by one clone from the cell suspension and 23 clones from the silique library. Among 17 different GNOM-interacting proteins tested against LexA–GNOM baits, only Cyp5 interacted with LexA–GNOM18–360, as shown in Figure 4A (also shown in M. Grebe and G. Jürgens, unpublished results). Interaction with Cyp5 was mediated by the GNOM dimerization domain, which was therefore designated the dimerization and Cyp binding (DCB) domain (Figure 4A; cf. Figure 3). Cyp5 was subjected to further domain mapping and chosen for more detailed characterization.
Figure 4.
Specificity and Domain Mapping of Cyp5–GNOM Interaction in the Yeast Two-Hybrid System.
(A) Interaction of the activation domain (AD)–Cyp5 fusion with different LexA–GNOM fragments (amino acid positions indicated). Activation of -Leu growth is indicated by + or −; β-galactosidase activity is given as arbitrary units. Error bars represent standard deviations from five independent transformants. LexA-bicoid (Bicoid) is the negative control.
(B) LexA–Cyp5 interaction with AD–GNOM subfragments (amino acid positions indicated). -Leu growth and β-galactosidase assay are as given in (A).
(C) Specificity of GNOM–Cyp5 interaction compared with other Arabidopsis cyclophilins. LexA–GNOM fusion tested against AD fusions of full-length Cyp5, ROC1, ROC4, and AD vector pJG4.5 (Vector). β-Galactosidase activity and growth on -Leu medium of two transformants streaked on plates and grown for 3 days are shown.
Specific Interaction between GNOM and Cyp5 Is Mediated by the DCB Domain
To assess whether the GNOM–Cyp5 interaction was independent of the fusion context, we generated a LexA–Cyp5 fusion and analyzed its interaction with GNOM fragments fused to the transactivation domain. As shown in Figure 4B, the Cyp5–GNOM interaction was confirmed and mapped to GNOM1–246, whereas smaller subfragments of GNOM1–246 did not mediate interaction in the two-hybrid system. Specificity of the GNOM–Cyp5 interaction was addressed by analyzing possible interactions of GNOM with other members of the Cyp family, such as the related rotamase Cyp protein ROC1 and the more divergent ROC4 protein. In contrast to Cyp5, these two Cyp proteins did not interact with GNOM in the two-hybrid system (Figure 4C).
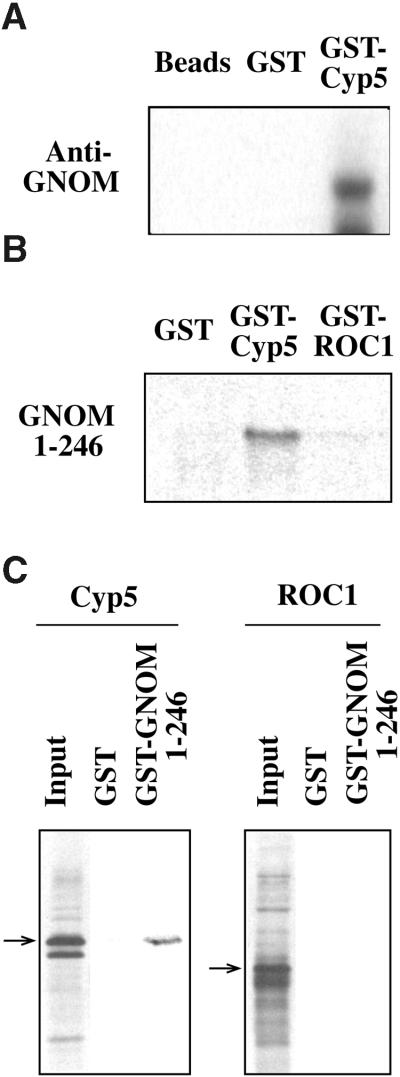
In the in vitro assay shown in Figure 5A, a purified GST–Cyp5 fusion protein pulled down full-length GNOM protein from Arabidopsis protein extracts. We also analyzed the binding of in vitro–translated ROC1 and Cyp5 to the GNOM DCB domain. The GST–GNOM1–246 fusion protein bound Cyp5 but not ROC1 (Figure 5C). Conversely, the in vitro–translated GNOM DCB domain interacted with a purified GST–Cyp5 fusion but not with the GST–ROC1 fusion protein (Figure 5B). Consistent with the two-hybrid results, in vitro–translated subfragments of the GNOM DCB domain did not interact with GST–Cyp5 (Figure 4B; J. Gadea and G. Jürgens, unpublished results). In summary, our results demonstrate that the GNOM–Cyp5 interaction is specific compared with other Arabidopsis Cyp proteins analyzed and suggest that Cyp5 binding and GNOM subunit interaction depend on similar structural features within the GNOM DCB domain.
Figure 5.
Specific GNOM and GNOM DCB Domain Binding to Cyp5 in Vitro.
(A) GST–Cyp5 binding of GNOM protein from Arabidopsis extracts. Protein gel blot analysis detected a 165-kD full-length GNOM band with anti-GNOM Sec7 antiserum (Anti-GNOM). Controls are beads or GST-coupled beads (GST) incubated with plant extract.
(B) and (C) Binding of 35S-methionine–labeled in vitro translation products to GST fusion proteins. Autoradiographs are of 15% SDS–polyacrylamide gels. In (B), specific binding of GNOM DCB domain (amino acids 1 to 246) to GST–Cyp5 is shown. Equal amounts of translation product were incubated with equal concentrations of GST, GST–ROC1, and GST–Cyp5. In (C), specific binding of Cyp5 translation product to GST–GNOM DCB domain (amino acids 1 to 246) is shown. Equal amounts of translation product (10% input) of ROC1 and Cyp5 (arrows) were incubated with equal amounts of GST and GST–GNOM amino acids 1 to 246.
Cyp5 Shares Features of Both Cytosolic and Secreted Cyp Proteins
Cyp proteins are cyclosporin A binding proteins (Handschumacher et al., 1984). Cyclosporin A inhibits both catalytic activities of Cyp proteins: peptidylprolyl cis/trans–isomerization (PPI) of oligopeptides and the refolding of protein substrates (Fischer et al., 1989). The GNOM-interacting Cyp5 is the product of the Arabidopsis AtCYP5 gene, which recently has been shown to encode a Cyp-like protein with an N-terminal endoplasmic reticulum–transport signal sequence (Saito et al., 1999). Searching for Cyp5-related proteins, we observed closest similarity with putative cytosolic Cyp proteins from nematodes, the plant Digitalis, and Arabidopsis rotamase Cyp protein ROC1, as presented in Figure 6. Specifically, Cyp5 and those homologs, as well as other plant cytosolic Cyp proteins, contain a seven–amino acid insertion, which seems to have originated early in eukaryotic evolution (Chou and Gasser, 1997).
Figure 6.
Alignment of Arabidopsis Cyp5 with Related Cyp Sequences.
Alignment of seven putative cytosolic eukaryotic Cyp proteins as identified and generated by BLAST P and BEAUTY search with multiple sequence alignment (Altschul et al., 1997). Dots indicate amino acid identity; dashes represent gaps. The Cyp5 sequence contains the predicted N-terminal endoplasmic reticulum (ER) transport signal (Saito et al., 1999) and PPIase consensus (PROSITE, underlined). Peptide antigen for rabbit immunization and two nonconserved amino acids (asterisks) involved in cyclosporin A binding of human CypA are underlined (Theriault et al., 1993). An ecotype polymorphism I30V between the Landsberg erecta and Columbia sequences is indicated (top line). The more divergent Arabidopsis ROC4 lacks a seven–amino acid insertion present in the other Cyp proteins.
The central region required for PPIase activity in functional Cyp proteins is more strongly conserved across species than between Cyp5 and Arabidopsis ROC1 (Figure 6). By overall sequence similarity, Cyp5 is more closely related to putative cytosolic Cyp proteins than to those involved in the secretory pathway (data not shown). Thus, Cyp5 appears to share some functional aspects with cytosolic Cyp proteins from other species. On the other hand, Cyp5 differs from its most similar Cyp proteins in two of nine conserved amino acids that are involved in cyclosporin A binding of human CypA (Theriault et al., 1993; Figure 6). It was thus not obvious whether Cyp5 has cyclosporin A–sensitive PPIase or protein refolding activities.
Cyp5 Catalyzes PPI and Protein Refolding in a Cyclosporin A–Sensitive Manner
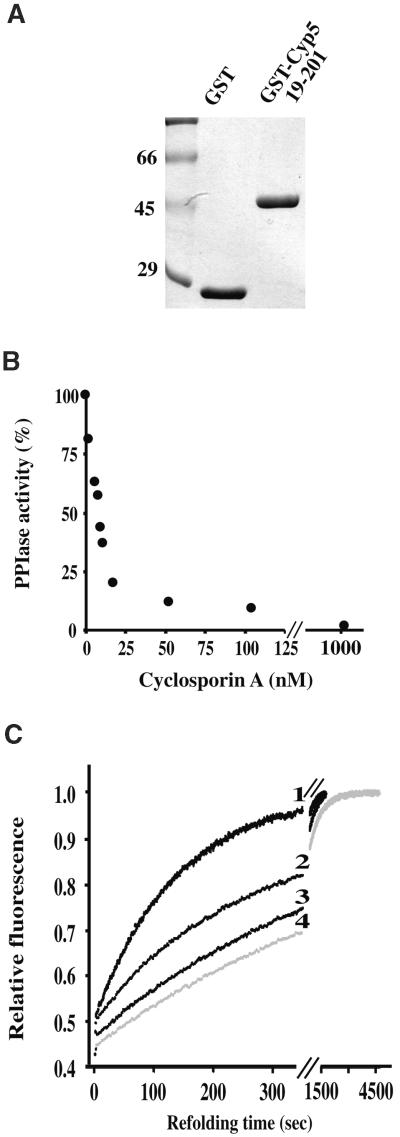
Cyp5 PPIase and protein refolding activities could be functionally relevant for Cyp5 interaction with GNOM or other proteins. Therefore, we analyzed both activities and their inhibition by cyclosporin A in vitro by using a purified GST–Cyp519–201 fusion protein lacking most of the hydrophobic endoplasmic reticulum transport signal, as shown in Figure 7A. We measured PPIase activity of GST–Cyp519–201 at a concentration of 3 nM, which was within the range at which first-order rate constants of the reaction increased linearly with enzyme concentration. The fusion protein displayed activity with a catalytic efficiency Kcat/Km of 5.7 × 106 M−1 sec−1 for the Suc-Ala-Ala-Pro-Phe-4-nitroanilide standard tetrapeptide substrate. Table 1 shows that the PPIase activity of Cyp5 was within the range of activities measured for other eukaryotic Cyp proteins. Catalytic activity was strongly inhibited by cyclosporin A with a calculated KI of 8 nM (Figure 7B). We analyzed protein refolding activity of the GST–Cyp519–201 fusion protein toward the artificial substrate RNase T1 (reduced carboxymethylated RNase T1, RCM-T1, variant S54G/P55N; Mücke and Schmid, 1994). The refolding of 0.7 μM RCM-T1 was accelerated approximately sixfold when 77 nM GST–Cyp519–201 fusion protein was added (Figure 7C). The GST–Cyp519–201 isomerase concentration was selected in a range at which first-order rate constants for catalysis showed linear dependence on enzyme concentration. The catalytic efficiency Kcat/Km was estimated to be 4.8 × 104 M−1 sec−1. The catalysis of protein refolding was inhibited to approximately half-maximal activity by 50 nM cyclosporin A (Figure 7C). In control experiments, the GST protein had no effect on PPIase or on protein refolding activity (Table 1; M. Grebe, J.-U. Rahfeld, and G. Jürgens, unpublished results). These results thus demonstrate that Cyp5 is a functional Cyp with cyclosporin A–sensitive PPIase and protein refolding activities, suggesting Cyp5 as a target for cyclosporin A action in Arabidopsis.
Figure 7.
Cyclosporin A Inhibition of Cyp5 PPIase and Protein Refolding Activities.
(A) A Coomassie blue–stained SDS–polyacrylamide gel with purified GST and GST–Cyp519–201 fusion used in PPIase and protein refolding assays. Numbers indicate molecular weight markers in kilodaltons.
(B) Inhibition of Cyp5 PPIase activity by different cyclosporin A concentrations. GST–Cyp519–201 (3 nM) was preincubated with varying concentrations of cyclosporin A, and PPIase activity was measured in 35 mM Hepes buffer, pH 7.8, at 10°C.
(C) Cyp5 catalysis of slow protein refolding of RNAse T1 (0.7 μM) and inhibition by cyclosporin A. The increase of fluorescence at 320 nm is shown as a function of the time of protein refolding. Curve 1 shows 77 nM Cyp5 without cyclosporin A; curve 2, 77 nM Cyp5 with 50 nM cyclosporin A; curve 3, 77 nM Cyp5 with 100 nM cyclosporin A; and curve 4, without Cyp5 and cyclosporin A. GST did not have an effect on refolding (not shown). Measurements were performed in 35 mM Hepes buffer, pH 7.8, at 10°C.
Table 1.
Comparison of Cyp5 and Other Cyp PPIase Activitiesa
| Source | Cyp Protein |
Kcat/Km (×10−6 M−1 sec−1) |
References |
|---|---|---|---|
| Arabidopsis | GST–Cyp5 | 5.7 | This study |
| GST | 0 | This study; Price et al. (1994) | |
| C. elegans | MBP–Cyp3 | 0.4 | Page et al. (1996) |
| C. elegans | MBP–Cyp6 | 8.4 | Page et al. (1996) |
| Human | Cyp A | 5.1 | Schönbrunner et al. (1991) |
| S. cerevisiae | Cytosolic | 3.3 | Schönbrunner et al. (1991) |
| Porcine kidney | (17 kD) | 5.9 | Schönbrunner et al. (1991) |
| Human | CypB | 6.3 | Price et al. (1994) |
| Human | GST–CypB | 4.9 | Price et al. (1994) |
| Maize | Microsomal | 25.0 | Sheldon and Venis (1996) |
| Maize | Cytosolic | 11.0 | Sheldon and Venis (1996) |
Shown are PPIase activities determined as catalysis of cis-trans interconversion of the Suc-Ala-Ala-Pro-Phe-4-nitroanilide substrate in protease-coupled assay. Except for budding yeast (S. cerevisiae) and maize Cyp proteins, which represent activities of native purified protein, all other activities were determined for recombinant protein or fusion protein. Kcat/Km (specific catalytic constant/Michaelis-Menten constant) with the unit (M−1 sec−1) gives the catalytic efficiency for the reaction.
Developmental and Subcellular Distribution of Cyp5 Protein
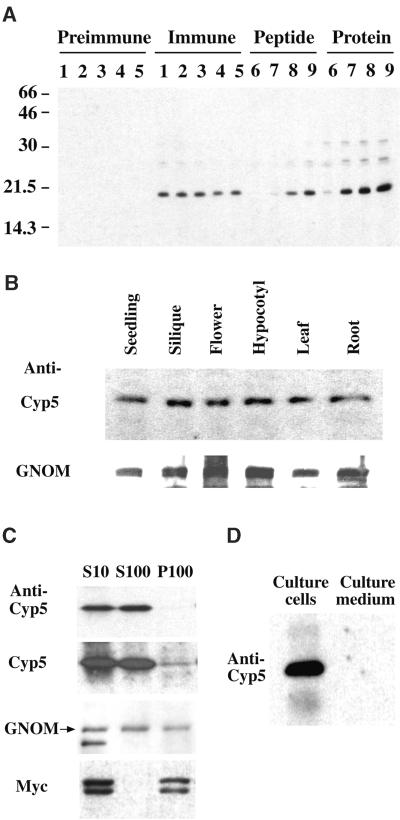
We raised an antiserum against a nonconserved Cyp5 C-ter-minal oligopeptide (see Figure 6) to determine the developmental expression and subcellular distribution of Cyp5. As shown in Figure 8A, the antiserum recognized a single band of 19 kD in siliques at high serum dilutions that was not detected by preimmune serum. The signal was diminished in a concentration-dependent manner by antiserum preincubation with the peptide antigen or the GST–Cyp519–201 fusion protein, indicating specific recognition of the Cyp5 protein (Figure 8A). The Cyp5-specific 19-kD band is consistent with the predicted molecular mass of 19.2 kD for the endoplasmic reticulum–processed Cyp5 protein, in contrast to the predicted 21.5 kD for the nonprocessed form. In vitro translation products of the nonprocessed and predicted processed form of Cyp5 showed the expected size difference (J. Gadea and G. Jürgens, unpublished results). Thus, Cyp5 appeared to be most abundant in its processed form.
Figure 8.
Expression and Subcellular Localization of Cyp5 Protein.
(A) Specificity of polyclonal anti-Cyp5 peptide antiserum. Shown is an immunoblot of total silique protein. Preimmune and immune serum dilutions are as follows: lane 1, 1:6000; lane 2, 1:9000; lane 3, 1:12,000; lane 4, 1:15,000; and lane 5, 1:18,000. Antiserum specificity determined by preincubation with peptide or purified GST–Cyp519–201 fusion protein is as follows: lane 6, 5 μg; lane 7, 500 ng; lane 8, 50 ng; and lane 9, 5 ng. Numbers at left indicate molecular weight markers in kilodaltons.
(B) Immunoblot of 30 μg of total protein from different Arabidopsis organs detected with 1:6000 dilutions of anti-Cyp5 peptide antiserum and anti-GNOM Sec7 antiserum (Steinmann et al., 1999). The 19-kD Cyp5 and 165-kD GNOM bands are shown.
(C) Localization of Cyp5 protein by subcellular fractionation. Immunoblot of protein extracts from cell suspensions expressing the Golgi apparatus marker Myc-sialyl transferase (ST2-11) (Wee et al., 1998) subjected to differential centrifugation. S10, supernatant of 10,000g centrifugation; S100, cytosolic supernatant; P100, microsomal membrane pellet of 100,000g centrifugation. Protein gel blots were probed with an anti-Myc antibody (A14; Santa Cruz Biotechnologies, Santa Cruz, CA; 1:1000) for control of membrane integrity, as well as anti-GNOM Sec7 and anti-Cyp5 peptide antisera. The 19-kD Cyp5, 165-kD GNOM (arrow), and 46- and 48-kD ST2-11 (Myc) bands are shown with short and long exposure of Cyp5 detection.
(D) Localization of Cyp5 in cell suspension cultures. Cyp5 protein in extracts from suspension cells (Culture cells) and cell culture supernatant (Culture medium) on day 5 after passage was detected by immunoblotting with anti-Cyp5 peptide antiserum. The 19-kD Cyp5 band is shown. Culture supernatant was concentrated by ammonium sulphate precipitation (Saito et al., 1999).
The Cyp5 protein was detected in several tissues, including flowers and siliques (Figure 8B). The subcellular distribution of Cyp5 was analyzed by differential centrifugation of extracts from suspension cells expressing a Golgi apparatus marker, Myc-tagged sialyl transferase (ST2-11; Wee et al., 1998). Cyp5 localized mainly to the cytosolic fraction but was also present in the membrane fraction (Figure 8C). By contrast, the sialyl transferase marker was confined to the microsomal pellet, indicating that microsomal membrane integrity was preserved during extraction. To determine whether the processed Cyp5 protein was secreted from cells, we analyzed the medium from a suspension cell culture (Figure 8D). Upon 280-fold concentration of the culture supernatant, secreted Cyp5 protein could not be detected, whereas a strong signal was found in the cell extract when equal amounts of total protein were loaded. In summary, Cyp5 cofractionated with GNOM in cytosolic and membrane fractions, as would be required for their interaction in vivo.
Cyp5 Protein Localization in Embryogenesis
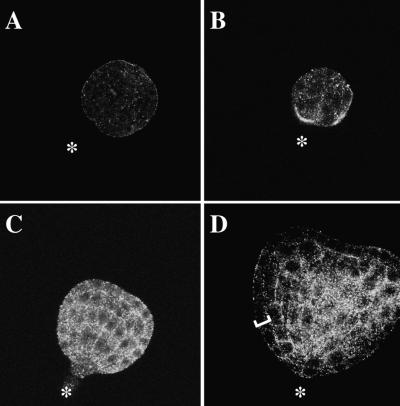
The Cyp5 protein was detected by indirect immunofluorescence in the developing embryo from the early globular stage, as shown in Figure 9. As a control for the specificity of the signal, embryos were stained with the preimmune serum and with the antiserum preincubated with native GST–Cyp519–201 protein. In both cases, no specific signal was observed (Figure 9A; data not shown). Early-globular-stage embryos gave a weak Cyp5 signal in all cells, displaying a punctate distribution in the cytoplasm (Figure 9B). At the late globular/transition stage, the signal became more intense in all cells (Figure 9C). From the early heart stage, epidermal cells displayed less Cyp5-specific staining than did the inner cells (Figure 9D), and this lessening of staining became more pronounced at the torpedo stage (data not shown). In summary, these results show that Cyp5 protein is expressed during Arabidopsis embryogenesis.
Figure 9.
Immunolocalization of Cyp5 during Embryogenesis.
Whole-mount preparations of embryos were stained with (A) preimmune serum (1:3000; control) or (B) to (D) anti-Cyp5 peptide antiserum (1:3000) followed by Cy3-conjugated secondary antibody (goat anti–rabbit; Dianova, Hamburg, Germany). Images represent internal optical sections generated by confocal microscopy. Stages of embryogenesis (Jürgens and Mayer, 1994) are shown in (A) to (D). Asterisks, basal end of embryo; bracket, epidermal cell layer.
(A) Mid-globular-stage embryo.
(B) Early-globular-stage embryo.
(C) Late globular/triangular–stage embryo.
(D) Early-heart-stage embryo. Note the low intensity of signal in the epidermal layer (bracket).
DISCUSSION
In this study, we demonstrated molecular interaction between identical subunits of GNOM, a large ARF GEF involved in apical–basal axis formation in the Arabidopsis embryo. The N-terminal domain required for interaction is conserved within a distinct subgroup of eukaryotic large ARF GEFs and is also essential for binding to the Cyp5 protein. Cyp5, a PPIase with protein refolding activity, is strongly expressed during Arabidopsis embryogenesis. These results suggest that Cyp5 may be a potential regulator of GNOM function.
Previous genetic studies revealed full complementation between gnom alleles with different mutations in the Sec7 domain (Busch et al., 1996), strongly suggesting that the molecular interaction of GNOM subunits reported here is functionally relevant during Arabidopsis embryogenesis. The GNOM DCB domain is conserved by sequence and N-terminal position among large ARF GEF proteins from several eukaryotes. These include the yeast proteins Gea1p and Gea2p, which can be functionally complemented by GNOM, and also a human GBF1 protein and a putative protein encoded by a C. elegans open reading frame (Mansour et al., 1998; Steinmann et al., 1999). Moreover, the intron following the coding sequence for the GNOM DCB domain is conserved in a homologous sequence identified in the Arabidopsis genome sequencing project. These findings suggest functional conservation of the DCB domain in the Gea/GNOM subgroup of eukaryotic large ARF GEFs. By contrast, the Sec7 subgroup represented by yeast Sec7p and Sec7p-related large ARF GEFs from Arabidopsis and other eukaryotes lack the DCB domain. Mammalian small ARF GEFs contain an N-terminal coiled-coil domain, which, in the case of human ARNO, mediates dimerization in vitro (Chardin et al., 1996). These findings add to the notion that dimerization may be a conserved feature of ARF GEF function and suggest that the coiled-coil domain of mammalian small ARF GEFs may play a role similar to that of the DCB domain of the GNOM/Gea–type large ARF GEFs.
After a search for GNOM interactors in the yeast two-hybrid system, we identified Cyp5 as a new protein that specifically bound to the GNOM DCB domain in yeast and in vitro. Cofractionation of GNOM and Cyp5 proteins in cytosol and membrane fractions from Arabidopsis extracts suggested multiple possible locations for in vivo interaction. To determine precisely the subcellular distribution of Cyp5 protein, one would have to perform immunolocalization at the electron microscopic level. Our attempts to do this were unsuccessful, because the anti-Cyp5 antiserum did not detect the epitope on ultrathin sections (T. Steinmann, H. Schwarz, and G. Jürgens, unpublished observations). However, cell fractionation experiments suggested that Cyp5 protein accumulates mainly in the cytosol but is also associated with membranes. We could not detect Cyp5 in the supernatant from cultured cell suspensions, which does not exclude the possibility that a small fraction of Cyp5 may be secreted from cells. Membrane localization of Cyp5 may be due to localization to endoplasmic reticulum subcompartments, as suggested by its functional N-terminal endoplasmic reticulum transport signal (Saito et al., 1999). The abundance of Cyp5 in the cytosol is in agreement with the cytoplasmic localization of mouse and rat secretory Cyp, CypB, which has been attributed to abortion of protein translocation after signal peptide cleavage (Arber et al., 1992; Schumacher et al., 1994).
The mechanism underlying the abortion of translocation has been described for the hepatitis B virus precore protein whose translocation is aborted after signal peptide cleavage, releasing the major part of the mature form into the cytosol (Garcia et al., 1988). Cyp5 may be localized to the cytosol in a similar way. Cyp5 interaction with cytosolic targets is also suggested by sequence similarity to cytoplasmic Cyp proteins from other eukaryotes. Together, our data suggest that the cytosol and possibly membrane fractions are cellular compartments relevant for the GNOM and Cyp5 interaction.
Saito et al. (1999) proposed that Cyp5 plays a role in postembryonic development rather than acting as a stress-responsive chaperone. Our study shows that Cyp5 protein is an enzyme with protein folding activities that is also expressed during embryogenesis, accumulating preferentially in inner cells at later stages. Early ubiquitous localization of Cyp5 coincides with irregular divisions affecting whole gnom embryos during early stages (Mayer et al., 1993). Moreover, epidermal cells are less strongly affected in gnom embryos (Mayer et al., 1993), which is consistent with ceasing Cyp5 localization in epidermal cells at the heart stage. Thus, expression of Cyp5 in cells affected in gnom mutant embryos is consistent with a presumed interaction of the GNOM and Cyp5 proteins, as suggested by their specific interactions in yeast and in vitro.
Proteins involved in signal transduction, such as receptor kinases, are known to be activated by dimerization (Weiss and Schlessinger, 1998). By analogy, GNOM dimerization may be required for ARF GEF activity because GNOM function is restored by genetic interaction of certain gnom mutant alleles producing full-length protein that is inactive on its own (Busch et al., 1996; Steinmann et al., 1999). The DCB domain required for GNOM subunit interaction also binds to Cyp5. So what role might Cyp5 play in the presumed regulation of the ARF GEF activity of GNOM? The Cyp5 protein displayed PPIase and protein refolding in vitro activities, which were effectively inhibited by cyclosporin A, thus identifying Cyp5 as a new potential cyclosporin A target in Arabidopsis. These observations suggest a role for Cyp5-mediated protein folding in regulating GNOM activity, although the details of this interaction remain to be determined.
Interactions between cyclophilins and respective dimerizing or oligomerizing target molecules have been analyzed in a few cases. A competitive activation mechanism has been implicated in the interaction of yeast Hsp90 with the cyclophilin-40 homolog Cpr6 (Prodromou et al., 1999). Cpr6 displaces the inhibitory cochaperone STI1 in a competitive manner, restoring Hsp90 ATPase activity in vitro (Prodromou et al., 1999). The mouse STI1 homolog, Hop, and Cyp40 have been shown to bind to the Hsp90 dimerization domain (Carrello et al., 1999). However, this Cyp40 interaction involves tetratricopeptide repeat domains that are not found in Cyp5. Another example is the cyclosporin A–sensitive interaction of oligomeric HIV-1 capsid protein Gag with human CypA, which is required for virion infectivity (Luban et al., 1993; Franke et al., 1994). Although the PPIase domain of CypA is essential for this interaction, the exact underlying mechanism is not known. However, CypA has been suggested to be involved in Gag protein complex disassembly (Luban, 1996). These examples may illustrate how Cyp5 might regulate GNOM activity. Further in vivo analysis of GNOM DCB domain–Cyp5 interaction will give insight into the developmental function of Cyp-mediated protein folding and regulation of large ARF GEF activity in Arabidopsis embryogenesis. Mechanisms underlying GNOM dimerization may also reveal how large ARF GEF activity is regulated in other eukaryotic systems.
METHODS
Cloning Constructs for Interaction Assays
The analysis and mapping of GNOM–GNOM and GNOM–Cyp5 interactions required modifications of interaction trap vectors pJG4-5 and pEG202 (Gyuris et al., 1993). The NotI sites were eliminated from both vectors by NotI restriction digest, fill in, and ligation. The derived vectors were subjected to EcoRI restriction digest, followed by fill in and insertion of the NotI linker d(TTGCGGCCGCAA) (New England Biolabs, Beverly, MA). The vectors were designated pMG5 (pJG4-5 derivative) and pMG8 (pEG202 derivative). A NotI linker was inserted into the Bsu36I site preceding codon 18 of GNOM cDNA clone c96 (Busch et al., 1996). The cDNA fragments NotI-ScaI and NotI-BstXI, from which protruding 3′ ends had been removed, were inserted into pMG5 and pMG8, both of which had been digested with XhoI, filled in, and digested with NotI. The resulting constructs expressed the DNA binding fusion proteins LexA–GNOM18–1451 and LexA–GNOM18–360 and the activation domain (AD) fusion proteins AD–GNOM18–1451 and AD–GNOM18–360.
Plasmids expressing LexA–GNOM551–1451 and AD–GNOM551–1451 fusion proteins were generated by inserting a BstXI-ScaI cDNA fragment, from which protruding 3′ ends had been removed, into the filled-in EcoRI sites of pEG202 and pJG4-5. The vector pJG4-5 was modified by inserting the ClaI linker d(CATCGATG) (New England Biolabs) into the filled-in XhoI site and was designated pMG2. After removal of protruding 3′ termini, a BstXI-ClaI cDNA fragment was inserted into the filled-in EcoRI and the ClaI site of pMG2 generating the AD–GNOM551–818 fusion expression construct. pJG4-5 was modified by digestion with EcoRI, filling in, and insertion of a ClaI linker, resulting in vector pMG1. A ClaI-ScaI cDNA fragment was inserted into the ClaI and filled-in XhoI site of pMG1, generating a construct expressing the AD–GNOM818–1451 fusion. Coding regions for GNOM1–246 and subfragments were amplified with Pwo-DNA polymerase (peQlab, Erlangen, Germany) by using polymerase chain reaction primers with EcoRI and XhoI restriction sites; they were cloned into pJG4-5 and pEG202 for two-hybrid experiments, pBluescript KS+ (Stratagene, La Jolla, CA) for in vitro translation, or pGEX4T-1 (Pharmacia, Braunschweig, Germany) for expression of glutathione S-transferase (GST) fusions. Expression from pJG4-5 derivatives was tested by immunoblotting using 12CA5 anti-HA (Boehringer Mannheim, Mannheim, Germany) or anti-GNOM Sec7 sera (Steinmann et al., 1999). DNA binding of fusion proteins from pEG202 derivatives was tested by repression assay (Ausubel et al., 1995) and expression tested with anti-GNOM Sec7 antibody (Steinmann et al., 1999). All steps involved standard molecular cloning procedures (Sambrook et al., 1989).
Construction of Interaction Trap cDNA Libraries
cDNA libraries from an Arabidopsis thaliana (ecotype Columbia) cell suspension culture and young siliques (ecotype Landsberg erecta) were generated in pJG4-5. RNA isolation and poly(A)+ mRNA generation involved standard procedures (Sambrook et al., 1989). Oligo(dT)-primed cDNA synthesis and directional cloning were performed using a Stratagene cDNA synthesis kit according to the manufacturer's instructions. Reverse transcriptase was replaced by Superscript reverse transcriptase (GIBCO BRL Life Technologies, Eggenstein, Germany). Two million primary transformants were recovered for the silique library, and 3 × 107 were recovered for the cell suspension library. Plasmid DNA was prepared from both libraries by standard procedures (Sambrook et al., 1989). Insert sizes averaged 1.2 kb, and inserts >3 kb were present in both libraries.
Search for GNOM Interacting Proteins
pMG8 expressing a LexA–GNOM18–1451 fusion protein served as bait to screen the cDNA libraries. Materials and procedures for testing the baits, GNOM–GNOM interactions, quantative β-galactosidase assays, interaction trap screening, isolation of plasmids, and specificity tests were as described previously (Gyuris et al., 1993; Ausubel et al., 1995). Specifically, using the GNOM protein as a bait, yeast strain EGY48, and the lacZ reporter pSH18-34, 3.5 × 106 primary clones from each library transformation were replated to 2 × 107 colonies on Leu− medium. One thousand five hundred eighty-five clones growing on Leu− medium up to day 4 were restreaked for further testing. Two hundred and fourteen clones of the silique library and 170 clones of the suspension library screen displayed activation of both reporters. Grouping of galactose-dependent colonies from the suspension library screen into 17 classes of cDNAs was achieved by cross-hybridization of amplified plasmid inserts. The longest clone from each group was sequenced using a Sequenase version 2.0 kit (Amersham, Braunschweig, Germany) and rehybridized to amplified plasmid inserts from clones of the silique screen. The plasmid with Cyp5 cDNA, isolated as a full-length clone, was retransformed and tested against GNOM constructs and different arbitrary proteins including Lexa-bicoid (pRFHM1; Ausubel et al., 1995; data not shown). Cyp5 cDNA sequence was cloned into pEG202. Coding regions of ROC1 and ROC4 (Lippuner et al., 1994) were amplified from cDNA by using Pwo-DNA polymerase (peQlab) and polymerase chain reaction primers with restriction sites and subcloned into pJG4-5.
Expression and Purification of GST Fusion Proteins
The cDNA sequences encoding the Cyp5 and rotamase Cyp1 (ROC1) proteins were cloned into pGEX4T-1 (Pharmacia) for expression of GST fusions and into pBluescript KS+ for in vitro translation. The amplified sequence encoding Cyp519–201 was cloned into pGEX4T-1. Expression of GST fusions in Escherichia coli DH5α was induced with 0.1 mM isopropyl thiogalactoside for 3 hr at 37°C. Bacterial lysis, coupling of fusion proteins to glutathione–Sepharose (Pharmacia), washes, and elution were performed according to the manufacturer's instructions and standard protocols (Ausubel et al., 1995). Briefly, for large-scale purification of GST fusion proteins, glutathione–Sepharose columns (2-mL bed volume) were prepared, and extracts were cleared by ultracentrifugation, passed through columns, and washed three times with PBS and 1% Triton X-100 and twice with PBS. GST fusions were eluted with 10 mM reduced glutathione. Eppendorf (Hamburg, Germany) centrifugal filter tubes were used for removing glutathione by PBS washes and for protein concentration.
In Vitro Binding Assays
In vitro binding assays (Ausubel et al., 1995) were modified for requirements of precipitation from Arabidopsis extracts. Glutathione–Sepharose beads (25-μL bed volume) with 1 to 3 μg of coupled GST fusion protein were blocked with 1% milk powder in PBS and 1% Triton X-100 for 30 min, washed with PBS and 1% Triton X-100, blocked with 2% BSA in PBS and 1% Triton X-100 for 30 min, and washed once with PBS. Native protein extracts were prepared from Arabidopsis suspension culture cells (Steinmann et al., 1999). Protein extraction and binding assays were performed in the cold (6°C). Suspensions filtered through Miracloth (Calbiochem, Bad Soden, Germany) were frozen in liquid nitrogen and homogenized. Three volumes of bead binding (BB) buffer (50 mM potassium phosphate, pH 7.5, 150 mM KCl, and 1 mM MgCl2) containing proteinase inhibitor mix (Sigma, Deisenhofen, Germany), 1 mM phenylmethylsulfonyl fluoride (Sigma), and 1 μg/mL pepstatin A (Sigma) were added, followed by further homogenization and sonication, as applied for bacterial protein extracts (Ausubel et al., 1995). After centrifugation at 12,000g for 20 min, protein concentration of supernatant was determined (Bradford, 1976). Total Arabidopsis protein (150 to 200 μg) in 150 μL of BB buffer was supplemented to give a reaction mixture of 300 μL containing 2 mM DTT, 5% glycerol, 1% Triton X-100, and proteinase inhibitors (see above). This was added to glutathione–Sepharose beads (25-μL bed volume) carrying 1 to 3 μg of fusion protein. Reactions were incubated for 1 hr at 6°C under gentle rotation. After three washes with BB buffer, 5% glycerol, and 1% Triton X-100, beads were resuspended in 25 μL of Laemmli buffer (Laemmli, 1970), and proteins were eluted at 95°C for 5 min. Beads were pelleted, and supernatants (24 μL) were split for duplicate SDS-PAGE, allowing Coomassie Brilliant Blue R 250 staining and immunodetection with anti-GNOM Sec7 antiserum at 1:6000 dilution (Steinmann et al., 1999). For binding assays with in vitro translation products, transcription and translation (TNT) reactions, including 35S-methionine (ICN, Eschwege, Germany), were performed using a TNT T7-coupled wheat germ extract system (Promega, Mannheim, Germany) according to the manufacturer's instructions. A 20-μL in vitro translation reaction was added to 1 to 3 μg of a coupled GST fusion protein supplemented with 300 μL BB buffer, 1% Triton X-100, and 5% glycerol. Binding reactions were performed on ice under agitation for 1 hr. Samples were washed, eluted, and subjected to SDS-PAGE. Gels were analyzed by using Coomassie Brilliant Blue R 250 for staining; then they were dried and autoradiographed.
Measurement of Peptidylproplyl cis/trans–Isomerase Activity
Peptidylproplyl cis/trans–isomerase (PPIase) activity of fusion proteins was determined by protease-coupled assay by using tetrapeptide substrate Suc-Ala-Ala-Pro-Phe-4-nitroanilide (Fischer et al., 1989). Reactions were performed in 35 mM Hepes (Sigma), pH 7.8, at 10°C, and monitored at 390 nm. Equipment and methods for calculation of enzyme activity were as described previously (Hani et al., 1999). Reactions containing 2 × 10−5 M substrate and desired concentrations of fusion protein (0 to 30 nM) were started by addition of chymotrypsin to a final concentration of 13 μM. Inhibition of PPIase activity was measured by incubating cyclosporin A (Calbiochem) with the enzyme in the reaction mixture for 5 min before starting the reaction by addition of the substrate and chymotrypsin.
Protein Refolding Experiments
For protein refolding experiments, ribonuclease T1 variant RCM-T1 (Ser-54-Gly/Pro-55-Asn) was prepared as described by Mücke and Schmid (1994). RCM-T1 was unfolded in 0.1 M Tris-HCl, pH 8.0, at 15°C. The acceleration of the refolding rate of RCM-T1 was determined (Mücke and Schmid, 1994). Briefly, refolding was initiated at 15°C by a 67-fold dilution of unfolded protein to final conditions of 0.1 M Tris-HCl, pH 8.0, 2 M NaCl, 0.7 μM RCM-T1, and the desired concentrations of GST–Cyp519–201 or GST diluted in the same buffer. Refolding reactions were monitored by the change in protein fluorescence at 320 nm (10-nm band width) after excitation at 268 nm (1.5-nm band width) by using a Hitachi F-3010 fluorescence spectrophotometer (Hitachi, Tokyo, Japan). Small contributions of GST–Cyp5 fusion protein, GST, or cyclosporin A to fluorescence were determined and subtracted from the values in individual experiments.
Generation of Antibodies to Cyp5, Immunoblotting, and Immunolocalization
A Cyp5-specific nonconserved peptide YKIEAEGKQSGTPKS (amino acids 175 to 189) was selected for antibody generation. This peptide was synthesized with an additional N-terminal cysteine, purified, and coupled to KLH at Eurogentec (Seraing, Belgium). Freund's adjuvant containing 3 mg of the KLH peptide was injected into rabbit followed by a booster shot with the same amount of incomplete Freund's adjuvant 4 weeks later. Serum was collected and tested as described by Lauber et al. (1997). For specificity control, antipeptide antiserum was preincubated with peptide or the purified native GST–Cyp519–201 fusion in 2% BSA and PBS for the time used for blot incubation (1 hr) before immunodetection. Competition assays for immunocytochemistry included preincubation of 1 mL of 1:3000 antipeptide serum in 2% BSA with 20 μg of GST–Cyp519–201 coupled to 100 μL of glutathione–Sepharose beads. Anti-Cyp5 antibodies were removed by pelleting beads, and supernatant was used as a control in immunostaining. Materials and procedures for cell fractionation, immunoblot analysis, immunolocalization, and image processing were described previously (Lauber et al., 1997; Steinmann et al., 1999).
Plant Growth and Cell Culture
Arabidopsis wild-type (Landsberg erecta) and gnom alleles, conditions for plant growth, and generation and maintainance of Arabidopsis suspension cell cultures were described previously (Mayer et al., 1991, 1993; Busch et al., 1996; Steinmann et al., 1999).
NOTE ADDED IN PROOF
Since this manuscript was accepted, K. Jackson and D. Soell ([1999]. Mutations in a new Arabidopsis cyclophilin disrupt interaction with protein phosphatase 2A. Mol. Gen. Genet. 262, 830–838) have reported the cloning of a new Arabidopsis cyclophilin, ROC7, most closely related to Cyp5, as an interaction partner for the protein phosphatase 2A, RCN1.
Acknowledgments
We thank Roger Brent for providing the yeast strains and vectors for the interaction trap version of the two-hybrid system, Jörg Grosshans and Stefan Sigrist for advice on analysis of protein interactions, Paul Dupree for the Myc-sialyl transferase line, Franz X. Schmid for a gift of the RNase T1 variant, Jörg Fanghänel for help on RCM-T1 refolding assays, Agnes Hepp and Roger Grau for technical assistance, Heinz Schwarz for advice on antibody generation and for immunizing rabbits, and Niko Geldner, Maren Heese, Michael Lenhard, Ulrike Mayer, Birgit Schwab, and Georg Strompen for critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft through Grant No. Ju 179/3-4 and a Leibniz award to G.J.
References
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber, S., Krause, K.H., and Caroni, P. (1992). S-cyclophilin is retained intracellularly via a unique COOH-terminal sequence and colocalizes with the calcium storage protein calreticulin. J. Cell Biol. 116 113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K., eds (1995). Current Protocols in Molecular Biology. (New York: John Wiley).
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Bram, R.J., and Crabtree, G.R. (1994). Calcium signaling in T cells stimulated by a cyclophilin B-binding protein. Nature 371 355–358. [DOI] [PubMed] [Google Scholar]
- Busch, M., Mayer, U., and Jürgens, G. (1996). Molecular analysis of the Arabidopsis pattern formation gene GNOM: Gene structure and intragenic complementation. Mol. Gen. Genet. 250 681–691. [DOI] [PubMed] [Google Scholar]
- Carrello, A., Ingley, E., Minchin, R.F., Tsai, S., and Ratajczak, T. (1999). The common tetratricopeptide repeat acceptor site for steroid receptor-associated immunophilins and hop is located in the dimerization domain of Hsp90. J. Biol. Chem. 274 2682–2689. [DOI] [PubMed] [Google Scholar]
- Chardin, P., Paris, S., Antonny, B., Robineau, S., Beraud-Dufour, S., Jackson, C.L., and Chabre, M. (1996). A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature 384 481–484. [DOI] [PubMed] [Google Scholar]
- Chou, I.T., and Gasser, C.S. (1997). Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol. Biol. 35 873–892. [DOI] [PubMed] [Google Scholar]
- Duina, A.A., Chang, H.C., Marsh, J.A., Lindquist, S., and Gaber, R.F. (1996). A cyclophilin function in Hsp90-dependent signal transduction. Science 274 713–715. [DOI] [PubMed] [Google Scholar]
- Fischer, G., Wittmann-Liebold, B., Lang, K., Kiefhaber, T., and Schmid, F.X. (1989). Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature 337 476–478. [DOI] [PubMed] [Google Scholar]
- Franco, M., Boretto, J., Robineau, S., Monier, S., Goud, B., Chardin, P., and Chavrier, P. (1998). ARNO 3, a Sec7-domain guanine nucleotide exchange factor for ADP ribosylation factor 1, is involved in the control of Golgi structure and function. Proc. Natl. Acad. Sci. USA 95 9926–9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, E.K., Yuan, H.E., and Luban, J. (1994). Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372 359–362. [DOI] [PubMed] [Google Scholar]
- Garcia, P.D., Ou, J.H., Rutter, W.J., and Walter, P. (1988). Targeting of the hepatitis B virus precore protein to the endoplasmic reticulum membrane: After signal peptide cleavage translocation can be aborted and the product released into the cytoplasm. J. Cell Biol. 106 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris, J., Golemis, E., Chertkov, H., and Brent, R. (1993). Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75 791–803. [DOI] [PubMed] [Google Scholar]
- Handschumacher, R.E., Harding, M.W., Rice, J., Drugge, R.J., and Speicher, D.W. (1984). Cyclophilin: A specific cytosolic binding protein for cyclosporin A. Science 226 544–547. [DOI] [PubMed] [Google Scholar]
- Hani, J., Schelbert, B., Bernhardt, A., Domdey, H., Fischer, G., Wiebauer, K., and Rahfeld, J.-U. (1999). Mutations in a peptidylprolyl-cis/trans-isomerase gene lead to a defect in 3′-end formation of a pre-mRNA in Saccharomyces cerevisiae. J. Biol. Chem. 274 108–116. [DOI] [PubMed] [Google Scholar]
- Jürgens, G., and Mayer, U. (1994). Arabidopsis. In Embryos: Color Atlas of Development, J.B.L. Bard, ed (London: Wolfe Publishing), pp. 7–21.
- Kim, H.S., Chen, Y., and Lonai, P. (1998). Complex regulation of multiple cytohesin-like genes in murine tissues and cells. FEBS Lett. 433 312–316. [DOI] [PubMed] [Google Scholar]
- Klarlund, J.K., Guilherme, A., Holik, J.J., Virbasius, J.V., Chawla, A., and Czech, M.P. (1997). Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science 275 1927–1930. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lauber, M.H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W., and Jürgens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner, V., Chou, I.T., Scott, S.V., Ettinger, W.F., Theg, S.M., and Gasser, C.S. (1994). Cloning and characterization of chloroplast and cytosolic forms of cyclophilin from Arabidopsis thaliana. J. Biol. Chem. 269 7863–7868. [PubMed] [Google Scholar]
- Liu, L., and Pohajdak, B. (1992). Cloning and sequencing of a human cDNA from cytolytic NK/T cells with homology to yeast SEC7. Biochim. Biophys. Acta 1132 75–78. [DOI] [PubMed] [Google Scholar]
- Luban, J. (1996). Absconding with the chaperone: Essential cyclophilin–Gag interaction in HIV-1 virions. Cell 87 1157–1159. [DOI] [PubMed] [Google Scholar]
- Luban, J., Bossolt, K.L., Franke, E.K., Kalpana, G.V., and Goff, S.P. (1993). Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73 1067–1078. [DOI] [PubMed] [Google Scholar]
- Mansour, S.J., Herbrick, J.A., Scherer, S.W., and Melancon, P. (1998). Human GBF1 is a ubiquitously expressed gene of the Sec7 domain family mapping to 10q24. Genomics 54 323–327. [DOI] [PubMed] [Google Scholar]
- Mattila, P.S., Ullman, K.S., Fiering, S., Emmel, E.A., McCutcheon, M., Crabtree, G.R., and Herzenberg, L.A. (1990). The actions of cyclosporin A and FK506 suggest a novel step in the activation of T lymphocytes. EMBO J. 9 4425–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, U., Torrez-Ruiz, R.A., Berleth, T., Misera, S., and Jürgens, G. (1991). Mutations affecting body organization in the Arabidopsis embryo. Nature 353 402–407. [Google Scholar]
- Mayer, U., Büttner, G., and Jürgens, G. (1993). Apical–basal pattern formation in the Arabidopsis embryo: Studies on the role of the gnom gene. Development 117 149–162. [Google Scholar]
- Morinaga, N., Moss, J., and Vaughan, M. (1997). Cloning and expression of a cDNA encoding a bovine brain brefeldin A–sensitive guanine nucleotide-exchange protein for ADP-ribosylation factor. Proc. Natl. Acad. Sci. USA 94 12926–12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, J., and Vaughan, M. (1998). Molecules in the ARF orbit. J. Biol. Chem. 273 21431–21434. [DOI] [PubMed] [Google Scholar]
- Mücke, M., and Schmid, F.X. (1994). Folding mechanism of ribonuclease T1 in the absence of the disulfide bonds. Biochemistry 33 14608–14619. [DOI] [PubMed] [Google Scholar]
- Page, A.P., MacNiven, K., and Hengartner, M.O. (1996). Cloning and biochemical characterization of the cyclophilin homologues from the free-living nematode Caenorhabditis elegans. Biochem. J. 317 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche, A., Paris, S., and Jackson, C.L. (1996). Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature 384 479–481. [DOI] [PubMed] [Google Scholar]
- Price, E.R., Jin, M., Lim, D., Pati, S., Walsh, C.T., and McKeon, F.D. (1994). Cyclophilin B trafficking through the secretory pathway is altered by binding of cyclosporin A. Proc. Natl. Acad. Sci. USA 91 3931–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou, C., Siligardi, G., O'Brien, R., Woolfson, D.N., Regan, L., Panaretou, B., Ladbury, J.E., Piper, P.W., and Pearl, L.H. (1999). Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18 754–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, T., Niwa, Y., Ashida, H., Tanaka, K., Kawamukai, M., Matsuda, H., and Nakagawa, T. (1999). Expression of a gene for cyclophilin which contains an amino-terminal endoplasmic reticulum–targeting signal. Plant Cell Physiol. 40 77–87. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sata, M., Donaldson, J.G., Moss, J., and Vaughan, M. (1998). Brefeldin A–inhibited guanine nucleotide-exchange activity of Sec7 domain from yeast Sec7 with yeast and mammalian ADP ribosylation factors. Proc. Natl. Acad. Sci. USA 95 4204–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S., Kaneko, T., Kotani, H., Nakamura, Y., Asamizu, E., Miyajima, N., and Tabata, S. (1998). Structural analysis of Arabidopsis thaliana chromosome 5. IV. Sequence features of the regions of 1,456,315 bp covered by nineteen physically assigned P1 and TAC clones. DNA Res. 5 41–54. [DOI] [PubMed] [Google Scholar]
- Schönbrunner, E.R., Mayer, S., Tropschug, M., Fischer, G., Takahashi, N., and Schmid, F.X. (1991). Catalysis of protein folding by cyclophilins from different species. J. Biol. Chem. 266 3630–3635. [PubMed] [Google Scholar]
- Schumacher, A., Westermann, B., Osborn, M., and Nordheim, A. (1994). The N-terminal signal peptide of the murine cyclophilin mCyP-S1 is required in vivo for ER localization. Eur. J. Cell Biol. 63 182–191. [PubMed] [Google Scholar]
- Sheldon, P.S., and Venis, M.A. (1996). Purification and characterization of cytosolic and microsomal cyclophilins from maize (Zea mays). Biochem. J. 315 965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevell, D.E., Leu, W.M., Gillmor, C.S., Xia, G., Feldmann, K.A., and Chua, N.H. (1994). EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77 1051–1062. [DOI] [PubMed] [Google Scholar]
- Springer, S., Spang, A., and Schekman, R. (1999). A primer on vesicle budding. Cell 97 145–148. [DOI] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Gälweiler, L., Palme, K., and Jürgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318. [DOI] [PubMed] [Google Scholar]
- Telemenakis, I., Benseler, F., Stenius, K., Südhof, T.C., and Brose, N. (1997). Rat homologues of yeast Sec7p. Eur. J. Cell Biol. 74 143–149. [PubMed] [Google Scholar]
- Theriault, Y., Logan, T.M., Meadows, R., Yu, L., Olejniczak, E.T., Holzman, T.F., Simmer, R.L., and Fesik, S.W. (1993). Solution structure of the cyclosporin A/cyclophilin complex by NMR. Nature 361 88–91. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee, E.G.-T., Sharrier, D.J., Prime, T.A., and Dupree, P. (1998). Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell 10 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, A., and Schlessinger, J. (1998). Switching signals on or off by receptor dimerization. Cell 94 277–280. [DOI] [PubMed] [Google Scholar]