Abstract
Budding of transport vesicles in the Golgi apparatus requires the recruitment of coat proteins and is regulated by ADP ribosylation factor (ARF) 1. ARF1 activation is promoted by guanine nucleotide exchange factors (GEFs), which catalyze the transition to GTP-bound ARF1. We recently have identified a human protein, ARNO (ARF nucleotide-binding-site opener), as an ARF1-GEF that shares a conserved domain with the yeast Sec7 protein. We now describe a human Sec7 domain-containing GEF referred to as ARNO3. ARNO and ARNO3, as well as a third GEF called cytohesin-1, form a family of highly related proteins with identical structural organization that consists of a central Sec7 domain and a carboxy-terminal pleckstrin homology domain. We show that all three proteins act as ARF1 GEF in vitro, whereas they have no effect on ARF6, an ARF protein implicated in the early endocytic pathway. Substrate specificity of ARNO-like GEFs for ARF1 depends solely on the Sec7 domain. Overexpression of ARNO3 in mammalian cells results in (i) fragmentation of the Golgi apparatus, (ii) redistribution of Golgi resident proteins as well as the coat component β-COP, and (iii) inhibition of SEAP transport (secreted form of alkaline phosphatase). In contrast, the distribution of endocytic markers is not affected. This study indicates that Sec7 domain-containing GEFs control intracellular membrane compartment structure and function through the regulation of specific ARF proteins in mammalian cells.
The ADP ribosylation factor (ARF) proteins form a distinct group within the Ras superfamily of low molecular weight GTP-binding proteins. ARF1 initially was identified as a cofactor for the ADP ribosyltransferase activity of the A subunit of cholera toxin (CT-A) (1). The discovery that Saccharomyces cerevisiae arf1-defective strains are impaired in invertase secretion (2) provided evidence that ARF proteins are involved in secretion. Indeed, various studies subsequently have demonstrated that ARF1 is required for the recruitment of cytosolic proteins on the cytoplasmic face of Golgi stacks to form a coat (COP-I coat; refs. 3 and 4). The assembly of COP-I coats leads to sorting of cargo and budding of transport vesicles both in anterograde and retrograde directions (5). In addition, ARF1 deactivation by GTP-hydrolysis triggers coat disassembly, allowing uncoated vesicles to fuse with the acceptor membrane (6). ARF1 also has been shown to regulate AP1-adaptor recruitment onto trans-Golgi network membrane, facilitating clathrin binding to the adaptor complexes and budding of clathrin-coated vesicles (7, 8). These observations, together with the fact that ARF activation is coupled with phospholipase D stimulation and production of phosphatidic acid (9, 10), have led to the proposal that ARFs are central to the process of vesicle budding by coordinating coat recruitment, membrane vesiculation, subsequent coat disassembly, and vesicle fusion (11).
Like all G proteins, ARFs are only active when bound to GTP, and activation is catalyzed by guanine-nucleotide exchange factors (GEFs). Brefeldin A, which interferes with coat formation by preventing ARF1 binding on Golgi membranes (12), inhibits GDP/GTP exchange on ARF1 (13–15). Several GEF activities that stimulate GDP/GTP exchange on class I ARFs (ARF1 and ARF3) have been described (15, 16), but the significant breakthrough toward the identification of ARF1-GEF came recently from a genetic approach using S. cerevisae. This approach led to the cloning of two related ARF1-GEF encoding genes, referred to as GEA1 and GEA2 (17), and allowed the cloning of a human cDNA encoding ARNO (ARF nucleotide-binding-site opener), which was shown to promote guanine nucleotide exchange on human ARF1 (18). The proteins encoded by the GEA1/GEA2 genes and ARNO share a conserved region of roughly 200 aa with significant similarity to a portion of the S. cerevisiae SEC7 gene product (Sec7 domain). The Sec7 protein (Sec7p) is required for vesicular traffic through the Golgi apparatus in yeast (19), and overexpression of yeast ARF1 and ARF2 proteins can suppress sec7 mutant growth defect (20). Interestingly, the isolated Sec7 domain of ARNO is sufficient to promote ARF1 activation in vitro (18), indicating that this domain is responsible for the exchange activity (18). Two additional Sec7 domain-containing ARF-GEFs of molecular mass ≈200 kDa (21) and ≈47 kDa (22) have been isolated. The latter one, named B2–1/cytohesin-1 (Ch-1) shows extensive homologies with the predicted sequence of ARNO (84% identity) (23, 24). Finally, another close relative of ARNO, called GRP-1 (general receptor for phosphoinositides), recently has been identified on the basis of its high affinity for phosphoinositide-3,4,5-trisphosphate [PtdIns(3,4,5)P3] (25). Interestingly, in addition to a highly conserved Sec7 domain, ARNO, Ch-1, and GRP-1 share an almost identical pleckstrin homology (PH) domain. This domain is responsible for the high affinity of GRP-1 for PtdIns(3,4,5)P3 and mediates PtdIns(4,5)P2-dependent stimulation of ARNO GEF activity (18).
Thus, we are confronted with an expanding family encoding for Sec7 domain-containing proteins, some of them with demonstrated ARF-GEF activity. The targets of these GEFs are most likely members of the ARF subgroup, which consists of five closely related proteins in humans (26). Analysis of the distribution of the different ARF proteins has shown that in contrast to ARF1 and ARF3 (class I ARFs) and ARF4 and ARF5 (class II), which are associated with intracellular compartments, ARF6 (class III) is localized at the plasma membrane (27); two recent studies have independently come to the conclusion that ARF6 is associated with the plasma membrane and uncharacterized endosomal structures (28, 29). Therefore, by analogy with Rab proteins (30), different ARF proteins may be associated with distinct donor compartments where they may regulate specific budding events. Sec7 domain-containing GEFs may be responsible for this specificity by catalyzing guanine nucleotide exchange on specific ARF substrate(s) onto specific membrane compartments.
The aim of the present study was to define the substrate specificity of ARNO/Ch-1 GEFs and to determine their function in membrane trafficking. We report on the cloning of a third member of the human ARNO/Ch-1 family, which we refer to as ARNO3. We demonstrate that ARNO, Ch-1, and ARNO3 act on ARF1, but are inactive toward ARF6, in vitro. By overexpressing ARNO3 in mammalian cells we demonstrate that ARNO3 is involved in the control of Golgi structure and function in vivo.
EXPERIMENTAL PROCEDURES
Cloning of Human ARNO3 cDNA.
blast search (31) identified a partial sequence of a human expressed sequence tag (EST) from the Institute for Genomic Research (EST01394; ref. 32), which shared homology with ARNO and Ch-1. A 650-bp NcoI fragment from EST01394 was used to probe a human placenta cDNA library in λgt11 (CLONTECH). The largest clone (≈2.1-kb insert) was sequenced and found to encode a predicted protein of 399 aa referred to as ARNO3.
Expression and Purification of Recombinant Proteins.
Myristoylated ARF1 and ARF6 were produced and purified as described previously (33). As judged by SDS/PAGE, ARF proteins were >95% pure, and the myristoylated forms represented at least 80% of the purified products (data not shown). ARNO and Ch-1 cDNAs subcloned in pET11d (18) and ARNO3 cDNA inserted in pET3a were transformed into BL21 (DE3) Escherichia coli. Expression was induced by addition of 0.2 mM isopropyl β-d-thiogalactoside to 1-liter cultures (A600 ≈ 0.8 after ≈4 h at 37°C) for 2 h at 30°C. After lysis of the bacteria, lysates were cleared by ultracentrifugation (45 krpm for 90 min in Ti 60 Beckman rotor). The supernatants that contained about 10–20% of GEFs (as judged SDS/PAGE followed by Coomassie blue staining) were used directly as the source of GEFs in all guanine-nucleotide exchange experiments.
Guanosine 5′-[γ-thio]triphosphate ([35S]GTPγS) Binding Assay.
myrARF1 or myrARF6 (1 μM) were incubated at 37°C in 50 mM Hepes-NaOH (pH 7.5), 1 mM MgCl2, 1 mM DTT, 100 mM KCl, 10 μM [35S]GTPγS (≈1,000 cpm/pmol, NEN) supplemented with azolectin vesicles (1.5 g/liter, Sigma). Soluble bacterial protein lysates containing the different GEFs were added to a final concentration of 40 μg/ml of total proteins. The estimated GEF concentration in the assay was ≈100–200 nM. For experiments performed in the presence of the isolated ARNO Sec7 domain, the final concentration of the purified domain was 80 nM. At different time points, samples of 25 μl were removed, and radioactivity was measured as described (33).
Transient Overexpression of mycARNO3 and Immunofluorescence.
To construct pGEM-mycARNO3, an NdeI site spanning the initiation codon and an EcoRV site upstream of the stop codon of ARNO3 were introduced by PCR with the Pwo polymerase (Boehringer Mannheim) by using the primers 5′-gatcgatccatatgGATGAAGACGCGGGC and 5′-cgttcaaatgatatcgCTATTTTTTATTGGC, respectively (sites are underlined and ARNO3-derived sequences are in capital letters). The NdeI–EcoRV PCR fragment was inserted downstream the epitope for the anti-myc mAb 9E10 (34) in pGEM-1. For transient overexpression studies, BHK-21 or HeLa cells plated on 11-mm round coverslips were infected with T7 RNA polymerase recombinant vaccinia virus (VT7) (35) and then transfected with pGEM-mycARNO3 by using the Lipofectamine reagent (GIBCO/BRL) as previously described (36). Four hours after transfection cells were fixed in 3% paraformaldehyde and then permeabilized with 0.5% saponin as previously described (36). For β-COP and AP-2 labeling cells were fixed at −20°C for 5 min in methanol, followed by 30 sec in acetone. Confocal microscopy was performed with a Leica TCS 4D confocal microscope equipped with a mixed gas argon/krypton laser (Leica Laser Teknik). The following antibodies were used: mAb 9E10 against the myc-tag epitope (34), mAb against giantin (37), rabbit anti-p23 antibodies (38), rabbit anti-sec61β antibodies (provided by R. Hendriks, Zentrum für Molekulare Biologie der Universität, Heidelberg, Germany), mAb anti-β-COP (clone mAD; ref. 39), mAb anti-α-adaptin (clone AC1-M11 provided by M. S. Robinson, University of Cambridge, United Kingdom), and rabbit anti-ARNO antibodies raised against the purified recombinant protein expressed in E. coli (crossreacting with all three members of the ARNO family, data not shown). Texas red-conjugated phalloidin was purchased from Molecular Probes.
Transport of SEAP.
SEAP transport was studied in HeLa cells as described (40). Briefly, after infection with vaccinia VT7 virus, cells were cotransfected with pGEM-SEAP and pGEM-ARNO3 or pGEM-ARNO (18) or empty pGEM-1 vector for 4 h and 15 min. Cells were incubated in medium without methionine and cysteine for 15 min, pulsed for 10 min with 150 μCi/ml of 35S-Promix (Amersham) and chased for 2 h. Cells were solubilized in 20 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, and 0.2% BSA, and SEAP was immunoprecipitated by using antialkaline phosphatase antibody (Tebu, Le Perray-en-Yvelines, France). SEAP then was submitted to endoglycosidase H (endoH) digestion as previously described (40). Samples were analyzed by SDS/PAGE on 6–12% acrylamide gradient gels and autoradiographed.
RESULTS
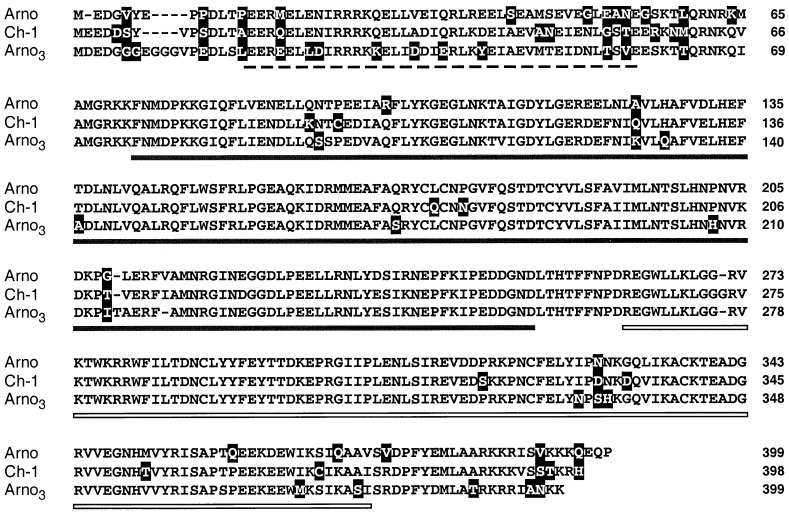
ARNO and Ch-1 are two closely related GEFs (84% identity) that act on ARF1 and ARF3 (18, 22). Both factors harbor a central domain of roughly 200 aa, showing extensive sequence homologies (42% identity) with a region of Sec7p (23, 41). By database search for new Sec7 domain-related sequences we have identified a human EST corresponding to a third member of the ARNO subfamily. The EST clone was used to probe a human placenta cDNA library, leading to the cloning of a 2,100-bp cDNA. This cDNA comprised an ORF coding for a deduced protein of 399 aa highly similar to ARNO and Ch-1, which we have referred to as ARNO3. Amino acid sequence comparison of ARNO, Ch-1, and ARNO3 showed that the three human proteins are highly conserved and consist of almost identical Sec7 and PH domains (Fig. 1). The most divergent portion among the three proteins is the N-terminal region, which contains a potential coiled coil-forming sequence (ref. 18 and Fig. 1). ARNO3 is probably the human homolog (99% identity) of the recently identified mouse GRP1 (25) and rat mSec7–3 (42) proteins. As reported for ARF-encoding genes that are expressed in all tissues (26), Northern blot analyses performed on total RNA from several human tissues revealed that ARNO and Ch-1 are ubiquitously expressed as a main transcript of 1.6 and 3.2 kb, respectively (data not shown). ARNO3 appeared to be expressed as a unique 4.5-kb transcript that was almost absent from liver, thymus, and peripheral blood lymphocytes (not shown). In conclusion, the ARNO family of SEC7-related genes comprises at least three members that are broadly expressed in various human tissues and encode for structurally very conserved proteins.
Figure 1.
Amino acid sequence alignment of the deduced sequences of ARNO, Ch-1, and ARNO3. Amino acid positions corresponding to nonconservative changes are highlighted. The coiled-coil (dashed line), Sec7 (thick line), and PH (double line) domains are indicated.
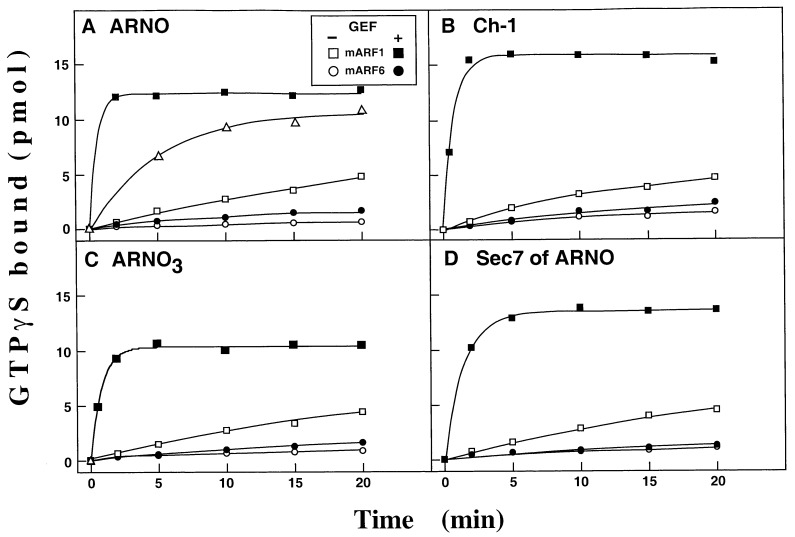
Because ARNO and Ch-1 are GEFs that act on class I ARFs (18, 22), we first examined whether ARNO3 showed similar activity. The data presented in Fig. 2 indicate that similar to ARNO (Fig. 2A) and Ch-1 (Fig. 2B), ARNO3 stimulated [35S]GTPγS binding on myrARF1 in the presence of azolectin vesicles (Fig. 2C). ARF6 is a member of the ARF subgroup but is structurally and functionally most distant from ARF1. Therefore it was important to examine the activity of ARNO-like GEFs toward ARF6. At physiological Mg2+ concentration (1 mM) and over a 20-min reaction period at 37°C, myrARF6 underwent a slow nucleotide exchange (Fig. 2 A–C, ○). Nucleotide exchange was strongly accelerated by lowering the free Mg2+ concentration to ≈1 μM, indicating that recombinant myrARF6 was functional (Fig. 2A, ▵). In physiological conditions (1 mM free Mg2+) none of the three ARNO/Ch-1 family members showed any guanine-nucleotide exchange activity toward myrARF6, demonstrating a specificity for ARF1 (Fig. 2 A–C, •). We next investigated whether the Sec7 domain could be responsible for this specificity. Indeed, the ARNO Sec7 domain is sufficient to promote guanine-nucleotide exchange on ARF1 (18) but its contribution to class I ARF specificity has not been assessed. Notably, ARNO-like proteins contain a PH domain that also could be involved in substrate specificity. This issue was addressed by assaying the activity of the ARNO Sec7 domain toward myrARF1 and myrARF6. As shown in Fig. 2D, the isolated ARNO Sec7 domain was active only on myrARF1, indicating that this domain is responsible for both the catalytic activity and the specificity for class I ARF proteins.
Figure 2.
GEF specificity of ARNO/Ch-1-like proteins for ARF1 versus ARF6. Measurement of GTPγS-binding to myrARF1 (□, ■) or myrARF6 (○, •) proteins (1 μM) in the absence (□, ○) or the presence (■, •) of bacterially expressed GEF (40 μg/ml of total soluble E. coli protein). (▵), spontaneous GTPγS binding on myrARF6 in low free Mg2+ concentration (1 μM). (A) ARNO, (B) Ch-1, (C) ARNO3, (D) purified isolated ARNO Sec7 domain (80 nM). Data shown are representative of three independent experiments.
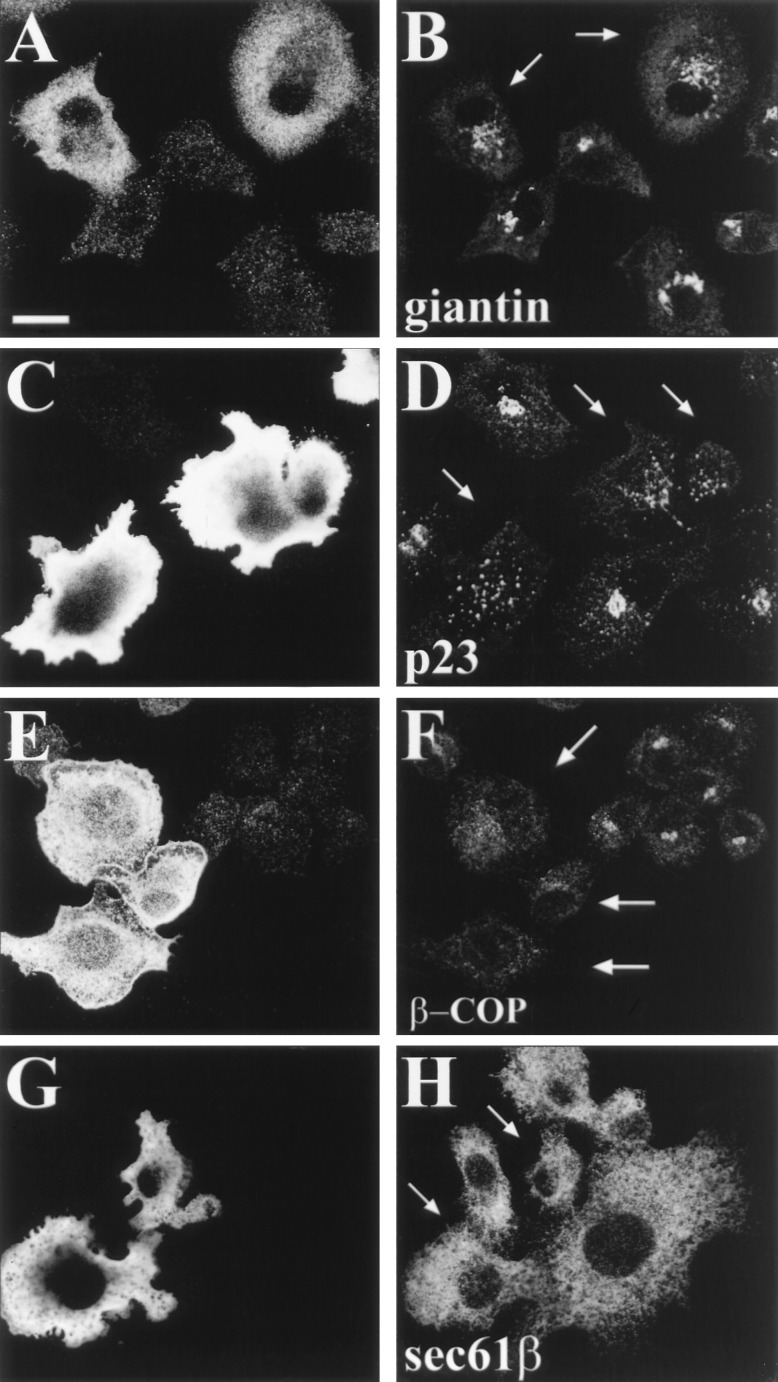
Overexpression of ARF1Q71L, a mutant that is preferentially in the GTP-bound form, results in morphological alterations of the Golgi apparatus in vivo (43–45). Therefore we have investigated the possibility that ARNO3 may regulate Golgi membrane structure by activating ARF1 in vivo. The consequences of ARNO3 expression on the morphology of the early secretory pathway were analyzed in BHK-21 cells transfected with N-terminally myc epitope-tagged ARNO3 (mycARNO3). After transfection, cells were fixed and double-labeled with anti-ARNO or anti-myc antibodies to visualize mycARNO3-expressing cells in combination with antibodies specific for various endoplasmic reticulum-Golgi resident proteins. Labeling with anti-ARNO (Fig. 3 A and E) or anti-myc antibodies (Fig. 3 C and G) gave a diffuse immunofluorescence staining throughout the cytoplasm, most likely representing a cytosolic protein. This assumption could be confirmed by permeabilizing the cells with saponin before fixation. In these conditions, excess of exogenous protein was washed out and mycARNO3 was detected only in vesicular structures that were spread in the entire cytoplasm (data not shown). We first looked at the intracellular distribution of giantin, a 400-kDa membrane protein that localizes to the Golgi complex (37). As shown in Fig. 3B, in nonexpressing cells, anti-giantin mAb decorated typical Golgi structures. In contrast, cells expressing mycARNO3 displayed a dispersed giantin staining that appeared in perinuclear aggregates that were heterogeneous in size (Fig. 3B, arrows). When cells were permeabilized before fixation, giantin-positive structures were negative for mycARNO3 staining (data not shown). In addition, overexpression of untagged ARNO3 induced a similar redistribution of giantin regardless of the presence of the myc tag (not shown). p23, a member of the p24 family of transmembrane proteins that associates with the cis-Golgi network/intermediate compartment of BHK cells (38), was similarly redistributed into perinuclear and vesicular structures in transfected cells (Fig. 3D, arrows). The observed change in Golgi morphology prompted an examination of the distribution of COP-1 coat proteins in ARNO3-overexpressing cells. As shown in Fig. 3F, β-COP became diffusely distributed in BHK-21 cells transfected with mycARNO3 (arrows), in contrast to its typical Golgi distribution in untransfected neighboring cells. By confocal microscopy, it was difficult to conclude whether this diffuse staining corresponded to β-COP redistributed to the cytosol or to small vesicular structures. Contrasting with this effect on Golgi structure, overexpression of mycARNO3 did not alter the distribution of sec61β, a resident endoplasmic reticulum marker (Fig. 3H).
Figure 3.
Overexpression of mycARNO3 induces fragmentation of the Golgi apparatus and β-COP redistribution. BHK-21 cells were transfected with the mycARNO3 construct and processed for immunofluorescence. Cells were double-labeled with anti-ARNO antibodies (A and E) or anti-myc tag mAb (C and G) in combination with anti-giantin (B), anti-p23 (D), anti-β-COP (F), and anti-sec61β (H). The arrows indicate mycARNO3-expressing cells. (Bar = 15 μm.)
Altogether, these results indicate that ARNO3 overexpression induces major morphological alterations of the Golgi apparatus. We next investigated whether Golgi function also was altered. SEAP, used as a marker of the exocytic pathway, was coexpressed with mycARNO3 or ARNO, and the sensitivity of intracellular SEAP to endoH was monitored by pulse–chase experiments. In control cells, two forms of SEAP were resolved (Fig. 4). The upper band was endoH resistant and represents mature SEAP molecules that have passed cis/medial Golgi compartments (40). The lower band was fully sensitive to endoH and corresponds to high mannose oligosaccharide-containing molecules still present in compartments before the cis/medial Golgi. In contrast, in cells expressing mycARNO3 or ARNO, no endoH-resistant form could be detected, indicating that SEAP did not reach cis/medial Golgi compartments. These observations indicate that overexpression of ARNO or ARNO3 leads to similar inhibition of transport and maturation of secreted proteins.
Figure 4.
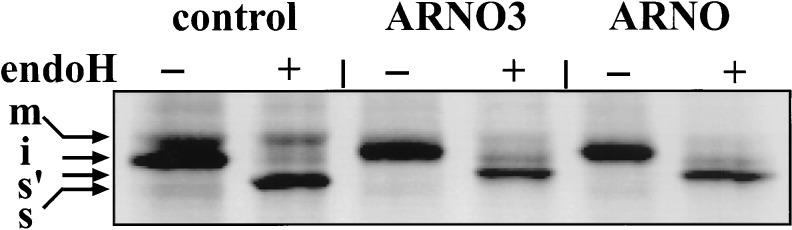
mycARNO3 and ARNO overexpression inhibit intracellular transport of SEAP. HeLa cells were cotransfected with pGEM-SEAP and pGEM-mycARNO3 or pGEM-ARNO or pGEM-1 plasmids. Cells were pulsed for 10 min, chased for 2 h, and solubilized. SEAP was immunoprecipitated and treated or not with endoH. m, the mature form of SEAP (endoH-resistant); i, the immature form still associated with the endoplasmic reticulum; s, the endoH-digested immature form, and s′, the endoH-digested form detected only in ARNO3- and ARNO-expressing cells, which probably is O-glycosylated.
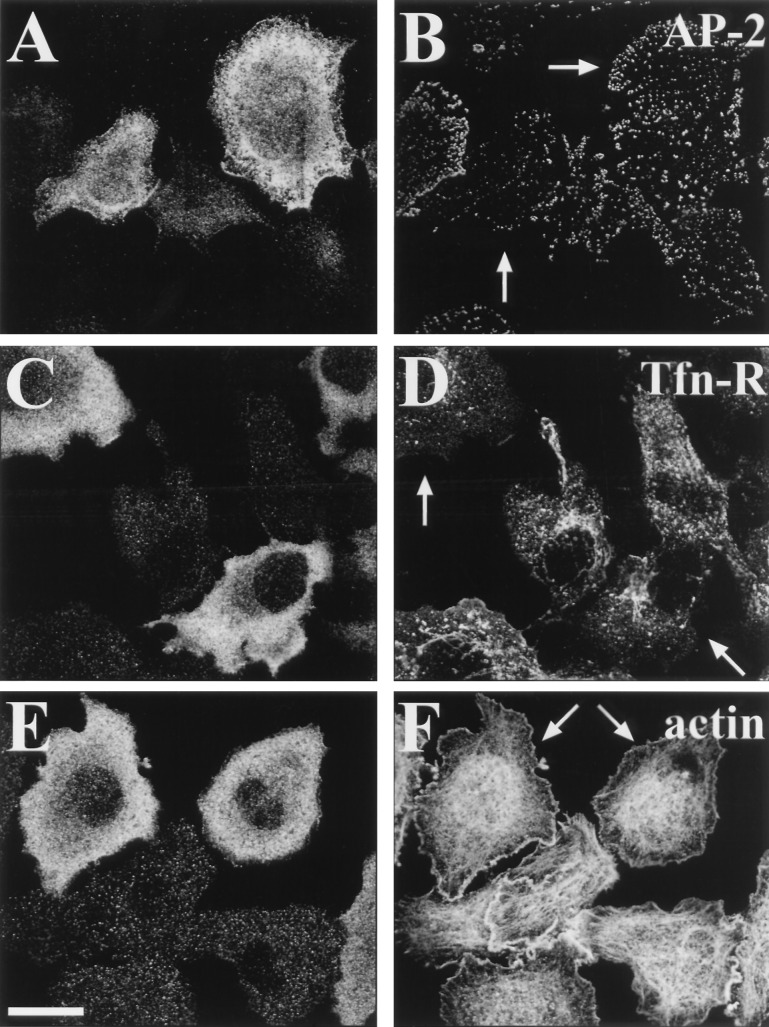
To investigate whether ARNO3 overexpression affected specifically secretory compartments, we analyzed the intracellular distribution of different markers of the endocytic pathway. As shown in Fig. 5, the distribution of plasma membrane AP-2 adaptor complexes and of the transferrin receptor (Tfn-R) appeared unaffected by overexpression of mycARNO3 (Fig. 5 B and D). In addition, cell shape and actin cytoskeleton organization were unchanged in mycARNO3-expressing cells (Fig. 5F). Altogether, these findings indicate that overexpression of ARNO3 affects Golgi morphology without affecting other intracellular compartments.
Figure 5.
mycARNO3 overexpression has no effect on the endocytic pathway. BHK or HeLa (C and D) cells were transfected with mycARNO3. Cells were double-labeled with anti-ARNO3 antibodies to visualize the transfected cells (A, C, and E) in combination with antibodies against the α-adaptin component of plasma membrane AP-2 adaptor complexes (B), to anti-transferrin receptor (Tfn-R)(D), or with phalloidin to visualize F-actin distribution (F). The arrows indicate mycARNO3-expressing cells. (Bar = 15 μm.)
DISCUSSION
In this study we report on the identification of ARNO3, a member of a family of human proteins that includes the closely related ARNO and Ch-1 (18, 22). These three proteins are highly related (77% of identical positions within the three primary sequences) and show a similar overall organization consisting of (i) an amino-terminal domain of ≈40 aa, which may adopt a coiled-coil structure involved in homodimerization (18) and possibly heterodimerization; (ii) a 200-aa central domain homologous to a portion of Sec7p, a protein involved in secretion that is genetically linked to ARFs in S. cerevisiae (20, 41); (iii) finally, a carboxy-terminal PH domain that mediates PtdIns(4,5)P2-dependent stimulation of ARNO exchange activity (18) and shows high affinity for PtdIns(3,4,5)P3 in the case of GRP-1, the mouse homolog of ARNO3 (25). By using bacterially expressed proteins and an in vitro GTP-binding assay, we observed that all three members of the ARNO family activate ARF1 but are ineffective on ARF6 or on the more distantly related RhoA, Rac1, and CDC42Hs Rho family GTP-binding proteins (data not shown). In addition, we demonstrated that substrate specificity of ARNO-like GEFs resides solely in the Sec7 domain, which is also the catalytic domain that stimulates guanine-nucleotide exchange (18). These findings, together with the observation that Ch-1 is a GEF active on ARF3 but not on ARF5 (class II ARF) (22), indicate that ARNO-like GEFs are specific for class I ARF proteins.
ARF1 is associated with Golgi membranes (2, 46) as well as Golgi-derived COP-coated vesicles (3), and its role in coat formation on early secretory compartments is firmly established (for review see ref. 47). In addition, expression of ARF1Q71L, a mutant form defective for GTP hydrolysis, leads to Golgi vesiculation (44) by preventing coat dissociation and vesicle fusion (6). Therefore, we investigated the possibility that ARNO3 overexpression could result in some alterations of the Golgi complex by activating ARF1 in vivo. Indeed, we observed that Golgi resident proteins—giantin, p23, and coat components (β-COP)—are redistributed into large vesicular structures in ARNO3-ovexpressing cells. Concomitant to its effect on Golgi morphology, ARNO3 overexpression was accompanied by an inhibition of intracellular transport of SEAP. Therefore, the Golgi remnants that are formed in ARNO3-expressing cells and that do not contain exogenous mycARNO3 (data not shown) are unable to support maturation and transport of secreted proteins.
Altogether these findings indicate that the effects of ARNO3 on both morphology and function of the Golgi are mediated by activation of ARF1 on Golgi membranes. Interestingly, it has been shown that overexpression of wild-type ARF1 has no effect on Golgi morphology, arguing that this abundant cellular protein is not a limiting component of the budding machinery (44). A reasonable hypothesis is that ARF1 activation by GDP/GTP exchange is a limiting step in the budding process. Overexpression of ARNO3 should result in ARF1-GTP accumulation onto Golgi membranes. Incubation of Golgi membranes in the presence of ARF1Q71I and GTP enhances membrane association of β-COP in vitro (45). Similarly, ARNO3-mediated ARF1 activation may induce COP-1 coatomer binding and favor the budding of coated vesicles, leading to the observed fragmentation of the Golgi compartment that is typical of both ARF1Q71L-transfected (44) and ARNO3-transfected cells. Therefore, ARNO3 overexpression could modify the balance between budding and fusion events that is required for maintaining the integrity of the Golgi complex. The novel equilibrium that is reached in ARNO3-overexpressing cells and results in Golgi fragmentation may be fixed by other limiting steps of the system such as GTP hydrolysis that involves a Golgi-associated ARF1-GAP (48). A complete interpretation of the effects of ARNO3 overexpression is complicated by the fact that COP-I-coated vesicles operate in transport both in anterograde direction across Golgi stack (ref. 49 and references herein) and in retrograde retrieval of endoplasmic reticulum residents (49, 50). In addition, we cannot exclude the possibility that ARF3, which is a potential substrate for ARNO3, also may participate in some effects of ARNO3 overexpression in vivo.
Surprisingly, Frank et al. (51) recently have reported that ARNO promotes nucleotide exchange on ARF6 in vitro. These findings contradict our own observations as well as recent data by Klarklung et al. (52) showing that ARNO-like GEFs activate ARF1 but not ARF6. In addition, Frank et al. described an association of ARNO with the cell periphery that was interpreted to mean that ARNO is involved in regulating ARF6 activity in vivo (51). However, we think that this latter observation has to be taken with caution because analysis of the distribution of ARNO was performed on cells that were fixed before permeabilization, in conditions where the major pool of ARNO is cytosolic, making it difficult to analyze the membrane association of ARNO. Overexpression of ARF6 or ARF6Q67L induces a redistribution of the transferrin receptor (Tfn-R) to the cell surface (28) and triggers extensive plasma membrane invaginations and actin cytoskeletal rearrangements (29, 53, 54). On the contrary, ARF6 overexpression has no effect on β-COP distribution (29). These observations are consistent with a role for ARF6 in regulating endocytosis and actin cytoskeleton at the cell periphery. We have examined the distribution of endocytic markers (AP-2, Tfn-R) and actin cytoskeleton organization in ARNO3-transfected cells and did not observe any significant modification. In conclusion, our morphological, functional, and biochemical data indicate that ARNO3 does not activate ARF6 at the cell periphery but regulates ARF1 function in the secretory pathway.
Many questions remain to be answered. First, the effects of ARNO3 overexpression are restricted to Golgi compartments, suggesting that ARF1 activation by ARNO3 occurs specifically on Golgi membranes. What are the determinant(s) of this specificity? ARNO-like GEFs share a conserved PH domain that interacts with membrane phosphoinositides (18, 25) and may be involved in transient association of the GEFs with intracellular membrane targets. Protein–protein interactions also may contribute to this specificity. In this respect, it is worth mentioning that whereas brefeldin A inhibits ARF1 activation on Golgi membranes (13, 14) it does not inhibit the exchange activity of recombinant ARNO-like GEFs (18). This finding suggests that brefeldin A-sensitive component(s) associate with ARNO-like GEFs in vivo to allow specific association of the GEFs with Golgi membranes. Second, expression analyses reveal that the three ARNO-like genes are widely expressed in different human tissues, suggesting that there may be cell type(s) expressing more than one ARNO-like GEF. Biochemical (GEF activity) and functional (SEAP transport) analyses indicate that ARNO-like proteins play overlapping roles. The existence of several ARF1-GEFs may be related to the fact that ARF1 controls the recruitment of different coats (AP1-adaptors, COP-I coat) to distinct compartments of the early exocytic pathway. Finally, the biological function of ARF3, which is also a substrate of Ch-1 (22), and its involvement in vesicular transport have not yet been addressed. Each ARNO-like GEF family member may be involved in the activation of ARF1/ARF3 at distinct intracellular locations to control the recruitment of specific coat proteins. A detailed and comparative analysis of the intracellular distribution of ARNO, Ch-1, and ARNO3 should help to clarify these important issues.
Acknowledgments
We are indebted to Drs. G. Banting, J. Gruenberg, H. P. Hauri, R. Hendriks, T. Kreis, A. LeBivic, and M. S. Robinson for generously providing antibodies for this study. J. Gruenberg, W. Hempel, and Dr. R. G. Parton are thanked for helpful discussions and comments on this manuscript. This work was supported by Institut National de la Santé et de la Recherche Médicale and Centre National de la Recherche Scientifique institutional funding and specific grants from La Ligue Nationale contre le Cancer and Le Groupement des Entreprises Françaises dans la Lutte contre le Cancer. M.F. was supported by a postdoctoral fellowship from the Association pour la Recherche sur le Cancer.
ABBREVIATIONS
- ARF
ADP ribosylation factor
- ARNO
ARF nucleotide-binding-site opener
- Ch-1
cytohesin-1
- endoH
endoglycosidase H
- EST
expressed sequence tag
- GEF
guanine exchange factor
- [35S]GTPγS
guanosine 5′-[γ-thio]triphosphate
- PH
pleckstrin homology
- SEAP
secreted form of alkaline phosphatase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ223957).
References
- 1.Kahn R A, Gilman A G. J Biol Chem. 1984;259:6228–6234. [PubMed] [Google Scholar]
- 2.Stearns T, Willingham M C, Botstein D, Kahn R A. Proc Natl Acad Sci USA. 1990;87:1238–1242. doi: 10.1073/pnas.87.3.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serafini T, Orci L, Amherdt M, Brunner M, Kahn R A, Rothman J E. Cell. 1991;67:239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- 4.Orci L, Palmer D J, Amherdt M, Rothman J E. Nature (London) 1993;364:732–734. doi: 10.1038/364732a0. [DOI] [PubMed] [Google Scholar]
- 5.Rothman J E, Wieland F T. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 6.Tanigawa G, Orci L, Amherdt M, Ravazzola M, Helms J B, Rothman J E. J Cell Biol. 1993;123:1365–1371. doi: 10.1083/jcb.123.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stamnes M A, Rothman J E. Cell. 1993;73:999–1005. doi: 10.1016/0092-8674(93)90277-w. [DOI] [PubMed] [Google Scholar]
- 8.Traub L M, Ostrom J A, Kornfeld S. J Cell Biol. 1993;123:561–573. doi: 10.1083/jcb.123.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown H A, Gutowski S, Moomaw C R, Slaughter C, Sternweis P C. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- 10.Cockcroft S, Thomas G M, Fensome A, Geny B, Cunningham E, Gout I, Hiles I, Totty N F, Truong O, Hsuan J J. Science. 1994;263:523–526. doi: 10.1126/science.8290961. [DOI] [PubMed] [Google Scholar]
- 11.Bednarek S Y, Orci L, Schekman R. Trends Cell Biol. 1996;6:468–473. doi: 10.1016/0962-8924(96)84943-9. [DOI] [PubMed] [Google Scholar]
- 12.Orci L, Tagaya M, Amherdt M, Perrelet A, Donaldson J G, Lippincott-Schwartz J, Klausner R D, Rothman J E. Cell. 1991;64:1183–1195. doi: 10.1016/0092-8674(91)90273-2. [DOI] [PubMed] [Google Scholar]
- 13.Donaldson J G, Finazzi D, Klausner R D. Nature (London) 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- 14.Helms J B, Rothman J E. Nature (London) 1992;360:352–354. doi: 10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- 15.Franco M, Chardin P, Chabre M, Paris S. J Biol Chem. 1996;271:1573–1578. doi: 10.1074/jbc.271.3.1573. [DOI] [PubMed] [Google Scholar]
- 16.Tsai S C, Adamik R, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1994;91:3063–3066. doi: 10.1073/pnas.91.8.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peyroche A, Paris S, Jackson C L. Nature (London) 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- 18.Chardin P, Paris S, Antonny B, Robineau S, Béraud-Dufour S, Jackson C L, Chabre M. Nature (London) 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- 19.Franzusoff A, Schekman R. EMBO J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deitz S B, Wu C, Silve S, Howell K E, Melancon P, Kahn R A, Franzusoff A. Mol Cell Biol. 1996;16:3275–3284. doi: 10.1128/mcb.16.7.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morinaga N, Tsai S C, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1996;93:12856–12860. doi: 10.1073/pnas.93.23.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meacci E, Tsai S C, Adamik R, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1997;94:1745–1748. doi: 10.1073/pnas.94.5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, Pohajdak B. Biochim Biophys Acta. 1992;1132:75–78. doi: 10.1016/0167-4781(92)90055-5. [DOI] [PubMed] [Google Scholar]
- 24.Kolanus W, Nagel W, Schiller B, Zeitlmann L, Godar S, Stockinger H, Seed B. Cell. 1996;86:233–242. doi: 10.1016/s0092-8674(00)80095-1. [DOI] [PubMed] [Google Scholar]
- 25.Klarlund J K, Guilherme A, Holik J J, Virbasius J V, Chawla A, Czech M P. Science. 1997;275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- 26.Tsuchiya M, Price S R, Tsai S C, Moss J, Vaughan M. J Biol Chem. 1991;266:2772–2777. [PubMed] [Google Scholar]
- 27.Cavenagh M M, Whitney J A, Carroll K, Zhang C J, Boman A L, Rosenwald A G, Mellman I, Kahn R A. J Biol Chem. 1996;271:21767–21774. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- 28.D’Souza-Schorey C, Li G, Colombo M I, Stahl P D. Science. 1995;267:1175–1178. doi: 10.1126/science.7855600. [DOI] [PubMed] [Google Scholar]
- 29.Peters P J, Hsu V W, Ooi C E, Finazzi D, Teal S B, Oorschot V, Donaldson J G, Klausner R D. J Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zerial M, Stenmark H. Curr Opin Cell Biol. 1993;5:613–620. doi: 10.1016/0955-0674(93)90130-i. [DOI] [PubMed] [Google Scholar]
- 31.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Adams M D, Dubnick M, Kerlavage A R, Moreno R, Kelley J M, Utterback T R, Nagle J W, Fields C, Venter J C. Nature (London) 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- 33.Franco M, Chardin P, Chabre M, Paris S. J Biol Chem. 1995;270:1337–1341. doi: 10.1074/jbc.270.3.1337. [DOI] [PubMed] [Google Scholar]
- 34.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuerst T R, Niles E G, Studier E G, Moss B. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guillemot J C, Montcourrier P, Vivier E, Davoust J, Chavrier P. J Cell Sci. 1997;110:2215–2225. doi: 10.1242/jcs.110.18.2215. [DOI] [PubMed] [Google Scholar]
- 37.Linstedt A D, Hauri H P. Mol Biol Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rojo M, Pepperkok R, Emery G, Kellner R, Stang E, Parton R G, Gruenberg J. J Cell Biol. 1997;139:1119–1135. doi: 10.1083/jcb.139.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pepperkok R, Scheel J, Horstmann H, Hauri H P, Griffiths G, Kreis T E. Cell. 1993;74:71–82. doi: 10.1016/0092-8674(93)90295-2. [DOI] [PubMed] [Google Scholar]
- 40.Martinez O, Schmidt A, Salamero J, Hoflack B, Roa M, Goud B. J Cell Biol. 1994;127:1575–1588. doi: 10.1083/jcb.127.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Achstetter T, Franzusoff A, Field C, Schekman R. J Biol Chem. 1988;263:11711–11717. [PubMed] [Google Scholar]
- 42.Telemenakis I, Benseler F, Stenius K, Sudhof T C, Brose N. Eur J Cell Biol. 1997;74:143–149. [PubMed] [Google Scholar]
- 43.Dascher C, Balch W E. J Biol Chem. 1994;269:1437–1448. [PubMed] [Google Scholar]
- 44.Zhang C J, Rosenwald A G, Willingham M C, Skuntz S, Clark J, Kahn R A. J Cell Biol. 1994;124:289–300. doi: 10.1083/jcb.124.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teal S B, Hsu V W, Peters P J, Klausner R D, Donaldson J G. J Biol Chem. 1994;269:3135–3138. [PubMed] [Google Scholar]
- 46.Donaldson J G, Kahn R A, Lippincott-Schwartz J, Klausner R D. Science. 1991;254:1197–1199. doi: 10.1126/science.1957170. [DOI] [PubMed] [Google Scholar]
- 47.Schekman R, Orci L. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 48.Cukierman E, Huber I, Rotman M, Cassel D. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- 49.Orci L, Stamnes M, Ravazzola M, Amherdt M, Perrelet A, Sollner T H, Rothman J E. Cell. 1997;90:335–349. doi: 10.1016/s0092-8674(00)80341-4. [DOI] [PubMed] [Google Scholar]
- 50.Letourneur F, Gaynor E C, Hennecke S, Demolliere C, Duden R, Emr S D, Riezman H, Cosson P. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 51.Frank S, Upender S, Hansen S H, Casanova J E. J Biol Chem. 1998;273:23–27. doi: 10.1074/jbc.273.1.23. [DOI] [PubMed] [Google Scholar]
- 52.Klarlund J K, Rameh L E, Cantley L C, Buxton J M, Holik J J, Sakelis C, Patki V, Corvera S, Czech M P. J Biol Chem. 1998;273:1859–1862. doi: 10.1074/jbc.273.4.1859. [DOI] [PubMed] [Google Scholar]
- 53.Radhakrishna H, Klausner R D, Donaldson J G. J Cell Biol. 1996;134:935–947. doi: 10.1083/jcb.134.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D’Souza-Schorey C, Boshans R L, McDonough M, Stahl P D, Van Aelst L. EMBO J. 1997;16:5445–5454. doi: 10.1093/emboj/16.17.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]