Abstract
Light-independent protochlorophyllide reduction leading to chlorophyll formation in the dark requires both chloroplast and nuclear gene expression in Chlamydomonas reinhardtii. Mutations in any one of the plastid (chlL, chlN, and chlB) or nuclear (y-1 to y-10) genes required for this process result in the phenotype of the yellow-in-the-dark or y mutants. Analysis of the chlL, chlN, and chlB transcript levels in both light- and dark-grown wild-type and y mutant cells showed that the y mutations have no effect on the transcription of these plastid genes. Protein gel blot analysis showed that the CHLN and CHLB proteins are present in similar amounts in light- and dark-grown wild-type cells, whereas CHLL is present only in wild-type cells grown in the dark or at light intensities ⩽15 μmol m−2 sec−1. Analysis of chlL transcript distribution on polysome profiles and rates of protein turnover in chloramphenicol-treated cells suggested that CHLL formation is most probably blocked at translation initiation or elongation. Furthermore, treatment of cells with metabolic inhibitors and uncouplers of photosynthetic electron transport showed that regulation of CHLL formation is linked to the physiologic status of the chloroplast. Similar to wild-type cells, y mutants contain nearly identical amounts of CHLN and CHLB when grown in either light or darkness. However, no CHLL is present in any of the y mutants except y-7, which contains an immunoreactive CHLL smaller than the expected size. Our findings indicate that CHLL translation is negatively photoregulated by the energy state or redox potential within the chloroplast in wild-type cells and that nuclear y genes are required for synthesis or accumulation of the CHLL protein.
INTRODUCTION
Two distinct mechanisms have become established for the reduction of protochlorophyllide (PChlide) to chlorophyllide (Chlide), a key step in the chlorophyll biosynthesis pathway. One mechanism, which is catalyzed by the enzyme NADPH:PChlide oxidoreductase (POR), depends completely on light for its activity (Reinbothe and Reinbothe, 1996; Timko, 1998). Light-dependent POR activity is present in cyanobacteria, green algae, and most nonvascular and vascular plants, and it is the only mechanism used for chlorophyll formation in angiosperms. The second mechanism, which is present in anoxygenic photosynthetic bacteria, cyanobacteria, nonvascular plants, ferns, and gymnosperms, can reduce PChlide to Chlide in a light-independent manner (Fujita, 1996; Armstrong, 1998). Organisms containing this PChlide reduction mechanism are all capable of chlorophyll formation in the dark. Although a large amount of information is now available on the regulation of POR biosynthesis and activity, little is known about the enzyme that mediates light-independent PChlide reduction (designated LIPOR), the factors that regulate its formation, and the enzyme's requirements for catalytic function.
Previous studies have shown that the products of three chloroplast genes (designated chlL, chlN, and chlB) and at least seven nuclear loci (designated y-1 to y-10) are required for light-independent PChlide reduction in the green alga Chlamydomonas reinhardtii (Sager, 1955; Timko, 1998). Cells with mutations in any of the three plastid genes or containing any one of the nuclear y mutations have identical phenotypes, referred to as yellow-in-the-dark, reflecting a loss of chlorophyll formation and the accumulation of the biosynthetic precursor PChlide. When grown in the light, however, these same cells synthesize chlorophyll and achieve a wild-type phenotype, a result of the presence of a functional light-dependent POR.
Although direct biochemical evidence is lacking, the products of the plastid chlL, chlN, and chlB genes are thought to constitute the subunits of an oligomeric complex that catalyzes light-independent PChlide reduction (Bauer et al., 1993; Fujita, 1996; Timko, 1998). The roles of the various y gene products in the process of light-independent PChlide reduction and their possible interactions with chl gene products are not known. The various y loci have been mapped for the Chlamydomonas genome. In complementation tests, heterozygous diploids produced from crosses of the y strains exhibited wild-type phenotypes, indicating that the mutations are recessive (Ford and Wang, 1980a, 1980b). No structural information is available for any y gene, and no specific function has been ascribed to any of their products. One possibility is that one or more of the various y genes encode additional subunits of the LIPOR complex or proteins involved in cofactor biosynthesis or provision of the reduction potential necessary in the LIPOR-catalyzed reaction. Alternatively, the y genes may encode proteins involved in regulating chl transcription or one or more post-transcriptional processes. Currently, an important body of evidence demonstrates the involvement of several distinct nuclear-encoded factors in the control of chloroplast gene transcription and post-transcriptional processes (Rochaix, 1996; Stern et al., 1997; Goldschmidt-Clermont, 1998).
In this study, we examine the expression of the plastid chlL, chlN, and chlB genes and the accumulation of their encoded proteins in light- and dark-grown wild-type and mutant strains of Chlamydomonas. We show that the expression of at least one chl gene product, CHLL, is negatively regulated by light in wild-type cells and that its accumulation in the dark is specifically blocked in the various y mutants by defects occurring post-transcriptionally.
RESULTS
Abundance of chlL, chlN, and chlB Transcripts in Wild-Type and y Cells
Wild-type Chlamydomonas cells contain little or no PChlide when grown in continuous light or darkness because both the light-dependent (POR) and light-independent (LIPOR) mechanisms of PChlide reduction leading to chlorophyll synthesis and accumulation are operational. In contrast, all known y mutants lack the ability to reduce PChlide in the dark but retain the capacity to synthesize and accumulate chlorophyll when exposed to light (Ford and Wang, 1980a, 1980b). When y mutants are transferred from the light into darkness, POR activity ceases and the cells rapidly accumulate PChlide (Cahoon and Timko, 1999). During the 96 hr immediately following transfer of light-grown cells to darkness, y-6-1 and y-7 showed the highest rates of PChlide accumulation, with the pigment reaching its maximum amounts at 24 hr after transfer. However, the total amount of PChlide accumulated per cell in these strains was low relative to that in the other y mutants. Strains y-1a, y-8, and y-10 accumulated the greatest amounts of Pchlide, and the high pigment levels were maintained for longer periods (Cahoon and Timko, 1999).
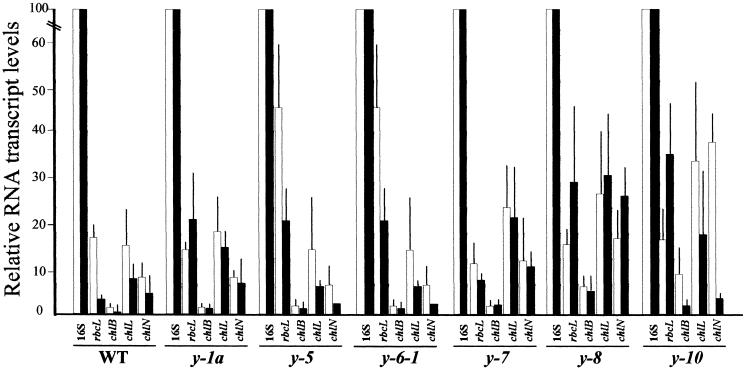
Because the various y mutants differ in their capacity to accumulate PChlide, we first examined whether any of the strains were altered in their ability to transcribe the plastid chlL, chlN, and chlB genes implicated in the light-independent Pchlide reduction process. As shown in Figure 1, chlL, chlN, and chlB transcripts are present in wild-type cells grown in the light and dark, although the steady state amounts of all three transcripts are approximately two- to fourfold greater in the light-grown cells. The increases in chlL and chlN transcripts observed here are consistent with our previous studies showing a small increase in chlB transcript abundance in light- versus dark-grown wild-type cells (Li et al., 1993).
Figure 1.
Steady State Amounts of chlL, chlN, and chlB Transcripts in Wild Type and y Mutant Strains of Chlamydomonas.
Shown are the results of semiquantitative RT-PCR analysis of chlL, chlN, and chlB transcripts in wild-type (WT) and y mutant strains of Chlamydomonas grown in continuous light (white bars) or darkness (black bars). In the experiment, rbcL transcript levels served as an internal control, and the relative abundance of each plastid transcript was normalized against the 16S rRNA. Given are the mean ±sd of at least three independent amplification reactions.
In general, the amounts of the chlL, chlN, and chlB transcripts observed in the various y mutants were similar to those found in wild-type cells. Only a few minor differences distinguished the various strains. Of particular note are the increases in the chlL, chlN, and chlB transcripts found in the light- and dark-grown y-7, y-8, and y-10 mutants relative to the wild type, and the markedly higher light-grown to dark-grown ratio of all three transcripts found in y-10 compared with the ratio found in the wild type and the other y mutants. No differences in the sizes of the chlL, chlN, or chlB transcripts present in the various mutant strains were apparent in comparison with those in wild-type cells, suggesting that no gross defects in RNA splicing or processing occur in the mutants. Consistent with previous reports that CHLN is encoded on a polycistronic message (Goldschmidt-Clermont, 1998), multiple-sized chlN transcripts were detected by RNA gel blot analysis (data not shown).
CHLL Preferentially Accumulates in the Dark and Is Missing in y Mutants
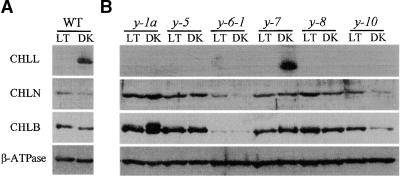
Because only minor differences in transcript levels were observed, we next examined the presence and relative abundance of the CHLL, CHLN, and CHLB proteins in the wild type and various y mutants. Extracts of total cellular proteins were prepared from wild-type cells and cells from the various y mutants grown under continuous light (50 to 100 μmol m−2 sec−1) or in darkness; after the proteins were fractionated by SDS-PAGE, the amounts of immunoreactive CHLL, CHLN, and CHLB protein present were determined by protein gel blot analysis. As shown in Figure 2, antisera specific for CHLL, CHLN, or CHLB detected the appropriate size of protein (i.e., 33, 67, or 80 kD, corresponding to CHLL, CHLN, or CHLB, respectively) in extracts prepared from wild-type cells grown in the dark. Whereas similar amounts of CHLN and CHLB protein were present in extracts prepared from wild-type cells grown in the light or dark, the CHLL protein was found only in extracts prepared from dark-grown wild-type cells. Only with prolonged exposure times was it possible to detect even small amounts of CHLL protein in extracts prepared from light-grown wild-type cells. Antiserum directed against the β subunit of coupling factor CF1-ATPase detected similar amounts of the protein in extracts from both dark- and light-grown cells, indicating that the loss of CHLL was not the result of nonspecific protein degradation. Interestingly, both CHLB and CHLL can be resolved into tightly spaced doublets under some electrophoresis conditions, suggesting that these proteins may undergo some form of post-translational modification or processing.
Figure 2.
Protein Gel Blot Analysis of CHLL, CHLN, and CHLB in Wild-Type and y Mutant Strains of Chlamydomonas.
Total cell extracts were prepared from light- (LT) and dark-grown (DK) (A) wild-type (WT) and (B) y mutant strains. The extracts were fractionated by SDS-PAGE, transferred to a nitrocellulose membrane, and reacted with antisera specific for one of the subunits of the light-independent PChlide reductase (CHLL, CHLN, or CHLB) or for the β subunit of the chloroplast coupling factor CF1-ATPase (β-ATPase), which served as a loading control.
In the six y mutants examined, the amounts of CHLN and CHLB were similar to those observed in wild-type cells grown under identical light or dark growing conditions. The only minor variation occurred with y-6-1 and y-1a. The amounts of CHLN and CHLB were lower in extracts prepared from both light- and dark-grown y-6-1 cells than in those from the wild-type cells. y-1a dark-grown cells contained a greater amount of the upper band of the CHLB doublet than did wild-type cells and the other y strains. In contrast, little to no CHLL was detected in the extracts of light- or dark-grown y mutants, suggesting that the nuclear mutations present in these strains may block CHLL formation or accumulation (or both). The only exception was y-7, which appeared to synthesize and accumulate an immunoreactive protein slightly smaller than the 33-kD CHLL observed in wild-type cells. The significance of this alteration is not known at present.
Polysome Association of chlL Transcripts in Wild-Type and y Cells
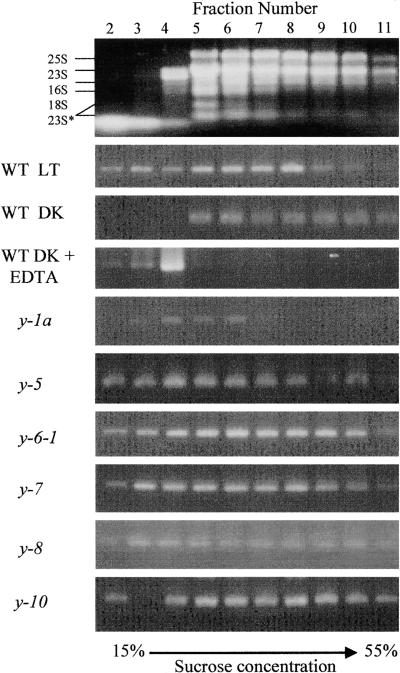
To determine whether the observed loss of CHLL in light-grown wild-type cells and dark-grown y mutants (except for y-7) resulted from a failure to synthesize the protein or from increased rates of protein turnover, we examined the distribution of transcripts encoding CHLL on polyribosomes within the cell. Polysome profiles were generated by sedimenting crude ribosomal extracts prepared from light- and dark-grown wild-type and y mutant cells through continuous sucrose gradients (15 to 55% [w/v]). After centrifugation, the gradients were divided into 12 equal fractions, the RNA was collected from each fraction, and the relative abundance of chlL transcripts in each fraction was analyzed by reverse transcription–polymerase chain reaction (RT-PCR).
Figure 3 shows the distribution of rRNAs from light-grown wild-type cells in the various gradient fractions after centrifugation. Fractions 5 to 11 correspond to the region of the gradient containing the majority of the polyribosomes, whereas fractions 2 to 6 contain predominately monosomes and unbound RNA transcripts. This pattern of rRNA distribution was typical for all of the extracts tested. Treatment of the crude extracts with EDTA before loading and centrifugation disrupted the ribosomes, such that the rRNAs were found only in fractions 2 to 6 (data not shown). After fractionation of extracts from light-grown wild-type cells, chlL transcripts were detected in fractions 2 to 10, with the highest concentrations in fractions 5 to 8 and in the upper portions of the gradient. In contrast, chlL transcripts in extracts from dark-grown wild-type cells were detected predominately in fractions 5 to 11, with no free chlL transcript found in the upper portion of the gradient. To confirm that the sedimentation pattern of chlL mRNA was the result of association with ribosomes, we treated extracts from dark-grown wild-type cells with 40 mM EDTA to dissociate polysomes before fractionation. As shown in Figure 3, after the EDTA treatment, chlL transcripts were restricted to fractions 2 to 4 near the top of the gradient.
Figure 3.
Association of chlL Transcripts with Polysomes in Light- and Dark-Grown Chlamydomonas Cells.
After sucrose gradient sedimentation of crude polysomal preparations isolated from light- (LT) and dark-grown (DK) wild-type (WT) and y mutant strains of Chlamydomonas, the gradients were divided into 12 fractions. An equal portion of the RNA purified from each fraction was analyzed by semiquantitative RT-PCR to determine the relative abundance of chlL transcripts. After PCR amplification, the reaction products were fractionated by agarose gel electrophoresis and stained with ethidium bromide. The first fraction, which represents the cell extract overlying the gradient, was typically void of RNA after centrifugation, and fraction 12 contained a debris pellet; neither fraction is presented here. At top is the distribution of rRNAs in the various fractions, with the sizes of the rRNAs labeled at left. The effects of EDTA treatment on transcript distribution are also shown. The 25S and 18S are cytoplasmic rRNAs, the 23S and 16S are plastid rRNAs, and the 23S* represents breakdown products of 23S (Rott et al., 1998).
Two different patterns of chlL transcript distribution were observed in extracts prepared from dark-grown y mutants. In y-1a, chlL transcripts were primarily restricted to the upper portion of the gradient (fractions 2 to 6). The presence of chlL transcripts primarily in the free RNA and monosomal portions of the gradients suggests that in this mutant, translation initiation or elongation may be retarded or blocked, leading to the failure to accumulate CHLL. In the other y mutants (e.g., y-5, y-6-1, y-7, y-8, and y-10), the distribution of chlL transcripts on the gradient is similar to that seen in light-grown wild-type cells, suggesting that a defect probably exists in either initiation efficiency or elongation of translation, or that the mutant has increased rates of CHLL turnover.
CHLL Turnover in Light- and Dark-Grown Cells
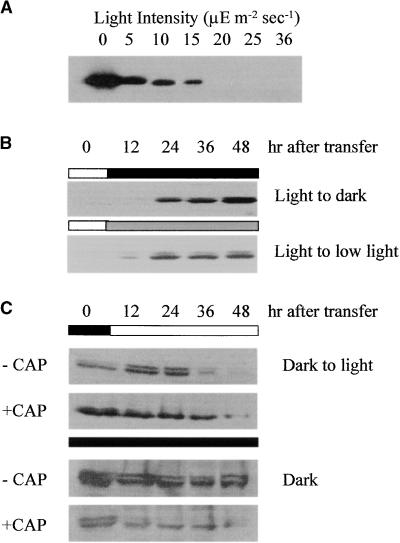
In light-grown wild-type cells, chlL transcripts appear to be at least partially polysome-associated; therefore, we next examined the relative rates of CHLL synthesis and turnover in light- versus dark-grown cells. Wild-type cells grown to early log phase under continuous high-intensity light conditions (50 to 100 μmol m−2 sec−1) did not accumulate CHLL. When these cells were transferred to growth conditions of low-intensity light, ranging from 5 to 36 μmol m−2 sec−1, only cells at the lower end of the intensity range (⩽15 μmol m−2 sec−1) accumulated CHLL (Figure 4A). Prolonged exposure of the autoradiographs revealed small amounts of CHLL in cells grown in light of ⩾20 μmol m−2 sec−1. As shown in Figure 4B, it takes ∼12 to 24 hr to detect a substantial accumulation of CHLL after transfer of the cells from high- to low-intensity light (5 μmol m−2 sec−1) or dark growth conditions. Synthesis and accumulation probably begin earlier, but the amounts of CHLL present were below the range detectable with the antiserum we used. The longer the cells remained under low light or dark growth conditions, the greater was the amount of CHLL detected in the cells.
Figure 4.
Protein Gel Blot Analysis of CHLL Accumulation and Turnover in Wild-Type Cells under Various Growth Conditions.
(A) The effect of light intensity on CHLL content in wild-type cells was examined by protein gel blot analysis. Total cellular extracts were prepared from the same number of cells grown in the dark or for 48 hr under the light intensity indicated. The extracts were fractionated by SDS-PAGE, transferred to a nitrocellulose membrane, and reacted with antiserum detecting CHLL.
(B) Accumulation of CHLL in wild-type cells at various times after transfer of the cultures from growth in the light (75 to 100 μmol m−2 sec−1) to darkness or low-light (5 μmol m−2 sec−1) growth conditions.
(C) Effects of chloramphenicol on CHLL formation and accumulation in wild-type cells. Dark-grown wild-type cells were treated with chloramphenicol (+CAP) (100 μg/mL), and the quantities of CHLL formed were determined by protein gel blot analysis at various times after continued growth in darkness or after transfer to high-intensity light (75 to 100 μmol m−2 sec−1) growth conditions. Control cells not treated with chloramphenicol (−CAP) were grown under the same conditions and sampled at the same times.
To determine the relative turnover rates of CHLL in wild-type cells, early log-phase cultures were grown in the dark for 48 hr to allow substantial accumulation of the CHLL protein. The cells were then transferred to high light conditions (50 to 100 μmol m−2 sec−1), which prevent CHLL accumulation. As shown in Figure 4C, the amounts of CHLL declined slowly after transfer of the cells from dark to light, with an apparent half-life of ∼24 hr. By 48 hr after transfer, little to no CHLL was detectable. Blocking translation on chloroplast ribosomes by treating the cells with chloramphenicol did not significantly affect the amounts of CHLL detected, indicating that the CHLL present was not the product of new protein synthesis. Cells maintained in the dark showed little change in CHLL content over the same time interval (Figure 4C). However, cells growing in the dark and treated with chloramphenicol showed a marked decrease in CHLL, with >50% gone by 6 to 12 hr after treatment. Similar results were found when cells were grown under low-intensity light (5 to 10 μmol m−2 sec−1). Little or no loss of CHLL was detected in untreated cells after 48 hr of growth under low light, whereas in the presence of chloramphenicol, CHLL was rapidly degraded (data not shown). Cumulatively, these data suggest that failure of wild-type cells to accumulate CHLL in the light probably results from decreased rates of protein synthesis rather than increased protein turnover. Furthermore, CHLL turnover in the dark or under very low light appears to be controlled by a nuclear-encoded, chloramphenicol-insensitive, dark-activated protease.
In contrast to what was observed for wild-type cells, y-7 cells did not accumulate CHLL under low light growth conditions. When grown under high-intensity light, or after treatment of dark-grown cells with chloramphenicol, y-7 lost CHLL in <3 hr (data not shown). These observations suggest that the improperly synthesized CHLL in y-7 degrades more quickly than the wild-type protein.
CHLL Accumulation Is Induced in the Light by Altering the Energy/Redox State of the Plastid
Given the observed preferential accumulation of CHLL in cells grown in darkness and the induction of its formation under low light intensities, we questioned whether CHLL expression was influenced by the metabolic (e.g., energy, redox) state of the cell or plastid. To address this question, we grew wild-type Chlamydomonas cells in the light in the presence or absence of various pharmacological compounds known to affect the energy or redox state of the chloroplast. By using these treatments, we hoped to create physiologic conditions in light-grown cells mimicking those found in dark-grown cells that were capable of CHLL accumulation.
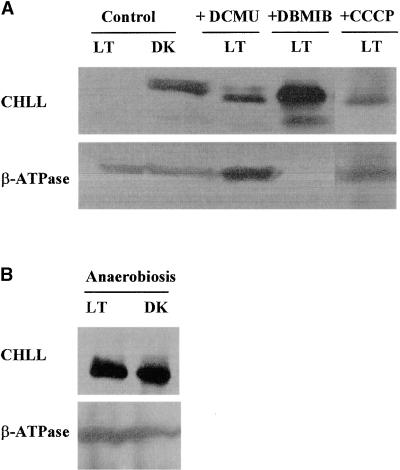
As shown in Figure 5A, wild-type cells grown in Tris-acetate-phosphate medium under light intensity >50 μmol m−2 sec−1 did not accumulate CHLL, whereas cells grown on the same medium in the dark readily accumulated the protein. The addition of carbonyl cyanide m-chlorophenylhydrazone (CCCP)—which is known to enhance the permeability of the thylakoid membrane and the uncoupling of ATP synthesis from photosynthetic electron transport (McCarty, 1980)—to the medium of light-grown cells resulted in the accumulation of CHLL in these cells by 48 hr after treatment. The amount of CHLL in these cells was approximately two- to threefold less than that observed in untreated cells in the dark.
Figure 5.
Effects of Various Cell Metabolic Inhibitors and Photosynthetic Electron Transport Uncouplers on CHLL Quantities in Wild-Type Chlamydomonas Cells.
Shown are the effects of various metabolic inhibitors and photosynthetic electron transport uncouplers on the synthesis or accumulation of CHLL. Wild-type Chlamydomonas cells were grown in Tris-acetate-phosphate medium in the light (LT; >50 μmol m−2 sec−1) or darkness (DK) in the presence (+) or absence (Control) of various pharmacological treatments. Whole-cell extracts were prepared, and CHLL quantities were measured by immunoblot analysis. As an internal control, the amounts of the β subunit of the chloroplast coupling factor CF1-ATPase (β-ATPase) were measured.
(A) CHLL content in untreated (Control) light- and dark-grown wild-type cells and light-grown wild-type cells 48 hr after treatment with 1 μM CCCP, 10 μM DCMU, or 10 μM DBMIB (+CCCP, +DCMU, and +DBMIB, respectively).
(B) CHLL content in light- and dark-grown wild-type cells 48 hr after induction of anaerobic growth conditions by purging cultures with argon.
CHLL accumulation could also be induced in wild-type Chlamydomonas cells grown in the light by treatment with 3-(3,4-dichlorophenyl)-1,1-dimethyl-urea (DCMU) and dibromothymoquinone (DBMIB), compounds known to disrupt photosynthetic electron transport. DCMU blocks the transfer of electrons from quinone to plastoquinone (Trebst, 1980), thereby creating an abundance of oxidized plastoquinone and NADP+ and suppressing thioredoxin signaling (Reulland and Miginiac-Maslow, 1999). DBMIB blocks photosynthetic electron transport by interfering with the transfer of electrons from plastoquinone to cytochrome b6f (Trebst, 1980). In both cases, by 48 hr after treatment, wild-type cells accumulated CHLL in amounts roughly equivalent to those seen in dark-grown cells.
We also observed (Figure 5B) that placing light-grown wild-type cells under anaerobic stress conditions led to the induction of CHLL formation. To encourage anerobiosis, we grew cell cultures in sealed tubes purged with argon; the resulting amount of CHLL expression in the light was similar to that found in dark-grown cells. Cells grown anaerobically in the dark were not inhibited in their ability to synthesize CHLL.
DISCUSSION
The existence of the y mutants of Chlamydomonas has been known for several decades (Sager, 1955; Ford and Wang, 1980a, 1980b); however, the molecular basis for their phenotype has never been defined. Our studies detailed earlier in this article demonstrated conclusively that the mutations present in these various mutants affect genes encoding products that are involved in the proper post-transcriptional expression or accumulation of the CHLL subunit of the light-independent PChlide reductase. Furthermore, we showed that in at least one strain (y-1a), the product of the y gene may be involved in initiating chlL translation. Our studies also provided some new insights into how the process of light-independent PChlide reduction, leading to the formation of chlorophyll in the dark, is regulated and integrated with other biosynthetic activities of the cell.
Like most chloroplast genes (Gruissem and Tonkyn, 1993; Gillham et al., 1994; Mayfield et al., 1995; Rochaix, 1996; Stern et al., 1997; Goldschmidt-Clermont, 1998), chlL, chlN, and chlB are expressed constitutively, as shown by the fact that transcripts derived from these genes are present in both light- and dark-grown cells. The slightly greater amounts of chlL, chlN, and chlB transcripts in light-grown cells than in dark-grown cells could reflect subtle differences in the rates of transcription of the three plastid genes or in the stability of the messages under the two growth conditions. In this regard, rbcL transcripts (for the large subunit of ribulose bisphosphate carboxylase) are markedly more stable in the dark than in the light in both Chlamydomonas and tobacco (Salvador et al., 1993; Shiina et al., 1998). Thus, the pattern of transcript accumulation observed in the present studies for chlL, chlN, and chlB in dark-adapted Chlamydomonas cells is similar to that previously observed.
Post-transcriptional processes are well documented to play a key role in the control of chloroplast gene expression, and many chloroplast transcripts are preferentially translated in the light (Gruissem and Tonkyn, 1993; Gillham et al., 1994; Mayfield et al., 1995; Rochaix, 1996; Stern et al., 1997; Goldschmidt-Clermont, 1998). Our studies showed that in wild-type Chlamydomonas cells, CHLL expression is regulated post-transcriptionally by factors present or active only in dark-grown cells or in cells grown at very low light intensities. Negative photoregulation of translation within plastids has not been previously described.
The negative photoregulation of CHLL formation is consistent with the observations of Fujita and colleagues (Fujita, 1996; Fujita et al., 1998) regarding the coordination of the complementary activities of the light-dependent and light-independent mechanisms for PChlide reduction. Those researchers reported that in the cyanobacterium Plectonema boryanum, POR and LIPOR activities are inversely coordinately regulated by light intensity. Under high light intensity, the cyanobacterium uses the light-dependent POR exclusively in chlorophyll formation, whereas in the dark, the LIPOR mechanism is the sole mechanism in operation. At intermediate and low light intensities, both systems operate, the relative contribution of each being determined by the amount of light available. In wild-type Chlamydomonas cells, the abundance of the CHLL subunit increased with decreased light intensity, suggesting that availability of this subunit may play a role in controlling the relative activity of the LIPOR complex in the cell. Thus, as POR activity declines with decreasing light intensity, synthesis and accumulation of CHLL are upregulated. The exact mechanism controlling CHLL abundance in response to light availability is not known.
Control of plastid translation in response to light and developmental cues has been reported at both the initiation and the elongation steps (Gruissem and Tonkyn, 1993; Mayfield et al., 1995). This control is also known to require the participation of nuclear factors that interact directly with structural elements in the 5′ untranslated regions of specific chloroplast messages (e.g., psbC or psbA) to bring about their control activities (Danon and Mayfield, 1991, 1994a, 1994b; Hirose and Sugiura, 1996; Yohn et al., 1996, 1998; Kim and Mayfield, 1997; Alexander et al., 1998). In Chlamydomonas, at least four proteins have been identified that bind specifically to a stem–loop RNA structure in the 5′ untranslated region of the photosystem II A subunit (psbA) mRNA; the binding activity of these proteins coincides with the amount of psbA translation in dark- and light-grown cells (Danon and Mayfield, 1994b). Binding of these proteins to the psbA message is regulated by an ADP-dependent phosphorylation event, which is modulated by the redox potential within the plastid (Danon and Mayfield, 1994a; Yohn et al., 1996) or the availability of photosynthetically generated reduced thioredoxins (Hirose and Sugiura, 1996). The redox state of the plastoquinone pool has also been shown to affect the transcription rate of psbA (Pfannschmidt et al., 1999).
We observed that a greater proportion of the chlL message is loaded onto polysomes in dark-grown wild-type cells than in cells growing in the light, suggesting that one or more factors required for translation of this message may be limiting in light-grown cells. Furthermore, we showed that suppression of CHLL in the light depends on cellular energy levels or the availability of reduced cofactors generated by photosynthesis. CHLL production was induced in the light by treatment with CCCP and under anaerobic conditions. Prevention of electron flow by blocking oxidation/reduction at plastoquinone also resulted in CHLL formation in the light. These observations rule out the redox potential of the plastoquinone pool as the source of regulation—blocking either reduction or oxidation resulted in CHLL production—and suggest that a later step in photosynthetic electron flow, the plastidic ATP/ADP ratio, or thioredoxin signaling may be involved. Our findings indicate that the factors present in the Chlamydomonas genome that are involved in regulation of chlL translation, CHLL accumulation, or both are sensitive to metabolic energy levels within the cell. Their activity appears to be controlled by light or plastid redox potential (or possibly both) in a manner reciprocal to that observed for psbA.
By analogy to the previously reported mechanisms discussed earlier in this article, we believe that the products of the y genes function in a positive manner to promote chlL translation or CHLL accumulation in the dark and that these factors are either missing or inactive in the various y mutant strains. Defects in these proteins or in their ability to carry out their regulatory function in the various y mutants result in a loss of CHLL synthesis in the dark, leading to the inability to conduct light-independent PChlide reduction and chlorophyll formation. In the case of y-1a, the defect may be associated with translation initiation, whereas in the other strains, processes subsequent to translation initiation appear to be affected. Evidence for the latter type of regulation has been reported in Chlamydomonas. For example, both the nac1 and ac-115 mutants fail to accumulate the D2 protein of the photosystem II reaction center, despite the fact that high amounts of the psbD mRNA encoding this protein are ribosome bound (Kuchka et al., 1988; Wu and Kuchka, 1995; Goldschmidt-Clermont, 1998). The nuclear mutations in these strains have been suggested to affect translation elongation or to cause improper folding of the nascent D2 polypeptide, leading to its rapid degradation by a ClpAP-related protease (Goldschmidt-Clermont, 1998).
At this time, we cannot rule out the possibility that synthesis or accumulation of CHLL in some y strains may also be controlled by the availability of an essential cofactor, the formation or addition of which to the CHLL protein is controlled by the y gene product. Mutations in ccs1 encoding a putative heme ligase in Chlamydomonas, for example, lead to the degradation of the cytochrome b6f complex because of a lack of heme attachment (Xie and Merchant, 1996; Inoue et al., 1997). CHLL is a homolog of the nifH-encoded subunit of nitrogenase, the enzyme involved in fixation of atmospheric N2 (Suzuki and Bauer, 1992). Like NIFH, CHLL has been proposed to contain Rieske Fe-S centers and an ATP binding site (Suzuki and Bauer, 1992). The products of several different genes are required for the proper synthesis and insertion of the Rieske Fe-S center during the formation of a functional NIFH protein (Dean and Jacobson, 1992; Peters et al., 1995). Therefore, formation of a fully functional CHLL might require proper cofactor assembly. The loss of this assembly could lead to its failure to be synthesized or to rapid degradation after its release from the ribosome.
Unlike the other y mutants tested, y-7 accumulates an immunoreactive CHLL protein in the dark; however, the electrophoretic mobility of this protein differs from that of the wild type. The basis for this difference remains to be determined but could result from several factors, including a defect in protein processing or secondary modification.
Our studies described here are an initial step toward unraveling the complex mechanism controlling the synthesis and assembly of the functional LIPOR complex in wild-type cells and the factors regulating its activity in response to light. They also provide insight into the function of the y loci in LIPOR activity in Chlamydomonas.
METHODS
Algal Strains and Culture Conditions
Wild-type (cc134) cells and the various yellow-in-the-dark (y) mutant strains (i.e., y-1a, y-5, y-6-1, y-7, y-8, and y-10) of Chlamydomonas reinhardtii were obtained from the Chlamydomonas Genetic Center (Duke University, Durham, NC) and maintained on Tris-acetate-phosphate medium agar plates in the light (35 μmol m−2 sec−1; Harris, 1989). Unless otherwise noted, cells were grown in liquid Tris-acetate-phosphate medium at 22°C under continuous white light illumination (50 to 100 μmol m−2 sec−1) or in the dark. Dark-grown cells were handled under dim green light until frozen in liquid nitrogen.
Addition of Pharmacological Compounds
Carbonyl cyanide m-chlorophenylhydrazone (CCCP), 3-(3,4-dichlorophenyl)-1,1-dimethyl-urea (DCMU), and dibromothymoquinone (DBMIB; Sigma) were added to early log phase cc134 cultures to final concentrations of 1, 10, and 10 μM, respectively, and samples were taken 24 to 48 hr later. Light-grown cultures were maintained under continuous white light (>50 μmol m−2 sec−1) throughout the duration of the experiment. Dark-grown samples were grown under continuous light until early log phase, transferred to the dark, and acclimated for 48 hr, during which time the samples were treated with CCCP, DCMU, or DBMIB. Samples were taken 24 to 48 hr later.
Semiquantitative Reverse Transcription–Polymerase Chain Reaction Analysis of RNA
Total RNA was extracted with Tri-Reagent (Molecular Research Co., Cincinnati, OH) according to the manufacturer's protocol. Total RNA was treated with RNase-free DNase before use. Semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) was performed essentially as described by Beverly (1995). Total RNA (2 μg) was reverse transcribed with 200 units of Maloney murine leukemia virus reverse transcriptase (MMLV-RT; Boehringer Mannheim, Indianapolis, IN) in the presence of 25 ng of each of five primers (CHLLC, 5′-GCCGAAGTCATATTATTTTT-3′; CHLNC, 5′-CGGCATTGGTACATTCATTT-3′; CHLBC, 5′-GCCGACAAAGCTTCTTTT-3′; RBCLC, 5′-CACGTTCACCTTCTAGTTTA-3′; and 16SC, 5′-CCGCGGCTGCTGGCACAGAGTTA-3′) for 1 hr at 42°C. The first-strand cDNA was purified by phenol-chloroform extraction (Sambrook et al., 1989) and used to prime a Taq-polymerase catalyzed PCR reaction containing 50 μCi α-32P-dCTP, 25 ng of each of the four cDNA primers listed above, and the following gene-specific primers: CHLLA, 5′-CGGAAAAGGTGGTATTGGAAAAT-3′; CHLNA, 5′-GGCGGACCTTTTCAA-GGCAATT-3′; CHLBA, 5′-GCGTGTTAGCAGCTCTTTTAAAA-3′; RBCLA, 5′-GGTTCCACAAAACAGAAACTAAA-3′; and 16SA, 5′-GCAATTTGTGTAGTGGCGAA-3′. The reaction conditions were optimized by varying the amounts of the first-strand cDNA template, the annealing temperature, and the number of cycles of amplification to ensure that measurements were made in the linear range of the reaction (Beverly, 1995). The amplification products were quantified by using a PhosphorImager (model 445SI; Molecular Dynamics, Sunnyvale, CA), and the amounts of the transcripts for the three LIPOR subunits (chlL, chlN, and chlB) and the large subunit of ribulose bisphosphate carboxylase (rbcL) detected in each sample were normalized to a 16S rRNA standard.
Immunoblot Analysis of Protein Expression
SDS-PAGE and protein gel blot analysis were performed as previously described (Li and Timko, 1996) using polyclonal antisera raised against the purified CHLL, CHLN, and CHLB proteins and the enhanced chemiluminescence detection kit (Amersham UK). Polyclonal antiserum recognizing the Chlamydomonas CHLL protein was produced in rabbits by a commercial vendor (Alpha Diagnostic International, San Antonio, TX); recombinant CHLL protein produced by overexpression in Escherichia coli was used as antigen. The recombinant, full-length CHLL protein was produced by expression in pET19b (Novagen, Madison, WI), an 890-bp DNA fragment encompassing the chlL coding region. The overexpressed protein was purified by SDS-PAGE before its use in immunizations. Antisera directed against CHLN and CHLB were provided by J.-D. Rochaix (University of Geneva, Switzerland) and Y. Fujita (Osaka University, Japan), respectively; R. McCarty (Johns Hopkins University, Baltimore, MD) provided antiserum recognizing the β subunit of coupling factor CF1-ATPase.
Examination of RNA Distribution in Polysomal Profiles after Continuous Sucrose Gradient Centrifugation
Polysome profiles were generated by sucrose gradient centrifugation as described by Cohen et al. (1998), with the following modification. Samples were treated with 40 mM EDTA for 10 min on ice. To determine the relative abundance of the various transcripts in the polysomal fractions, we performed RT-PCR as described above, except that no α-32P-dCTP was added to the reaction. Reaction products were separated in 1% agarose gels containing 0.5 × Tris-acetate-EDTA buffer, stained with ethidium bromide, and visualized by UV illumination.
Acknowledgments
We thank Reginald Garrett for his comments on the manuscript, Jianmin Wang for construction of the pET-chlL expression plasmid, and Jean-David Rochaix and Yuichi Fujita for providing antisera against the CHLN and CHLB proteins, respectively. This work was supported by a grant from the National Science Foundation (Grant No. MCB-9818037) awarded to M.P.T. A.B.C. was supported in part by a National Institutes of Health Training Grant (No. 5T32 GM08136).
References
- Alexander, C., Faber, N., and Klaff, P. (1998). Characterization of protein-binding to the spinach chloroplast psbA mRNA 5′ un-translated region. Nucleic Acids Res. 26, 2265–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, G.A. (1998). Greening in the dark: Light-independent chlorophyll biosynthesis from anoxygenic photosynthetic bacteria to gymnosperms. J. Photochem. Photobiol. B 43, 87–100. [Google Scholar]
- Bauer, C.E., Bollivar, D.W., and Suzuki, J.Y. (1993). Genetic analysis of photopigment biosynthesis in eubacteria: A guiding light for algae and plants. J. Bacteriol. 175, 3919–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverly, S.M. (1995). Enzymatic amplification of RNA by PCR. In Short Protocols in Molecular Biology, 3rd ed, F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, eds (New York: John Wiley), pp. 15.13–15.15.
- Cahoon, A.B., and Timko M.P. (1999). Effects of nuclear y mutations on expression of plastid genes required for light-independent chlorophyll formation in Chlamydomonas. In The Chloroplast: From Molecular Biology to Biotechnology, J.H. Argyroudi-Akoyunoglou and H. Senger, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 195–200.
- Cohen, A., Yohn, C.B., Bruick, R.K., and Mayfield, S.P. (1998). Translational regulation of chloroplast gene expression in Chlamy-domonas reinhardtii. Methods Enzymol. 297, 192–208. [Google Scholar]
- Danon, A., and Mayfield, S.P. (1991). Light regulated translational activators: Identification of chloroplast gene–specific mRNA binding proteins. EMBO J. 10, 3993–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon, A., and Mayfield, S.P. (1994. a). Light regulated translation of chloroplast messenger RNAs through redox potential. Science 266, 1717–1719. [DOI] [PubMed] [Google Scholar]
- Danon, A., and Mayfield, S.P. (1994. b). ADP-dependent phosphorylation regulates RNA binding in vitro: implications in light-modulated translation. EMBO J. 13, 2227–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, D.R., and Jacobson, M.R. (1992). Biochemical genetics of nitrogenase. In Biological Nitrogen Fixation, G. Stacey, R.H. Burris, and H.J. Evans, eds (New York: Chapman and Hall), pp. 763–834.
- Ford, C., and Wang, W.-Y. (1980. a). Three new yellow loci in Chlamydomonas reinhardtii. Mol. Gen. Genet. 179, 259–263. [DOI] [PubMed] [Google Scholar]
- Ford, C., and Wang, W.-Y. (1980. b). Temperature sensitive yellow mutants of Chlamydomonas reinhardtii. Mol. Gen. Genet. 180, 5–10. [Google Scholar]
- Fujita, Y. (1996). Protochlorophyllide reduction: A key step in the greening of plants. Plant Cell Physiol. 37, 411–421. [DOI] [PubMed] [Google Scholar]
- Fujita, Y., Takagi, H., and Hase, T. (1998). Cloning of the gene encoding arotochlorophyllide reductase: The physiological significance of the co-existence of light-dependent and -independent protochlorophyllide reduction systems in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 39, 177–185. [DOI] [PubMed] [Google Scholar]
- Gillham, N.W., Boynton, J.E., and Hauser, C.R. (1994). Translational regulation of gene expression in chloroplasts and mitochondria. Annu. Rev. Genet. 28, 71–93. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M. (1998). Coordination of nuclear and chloroplast gene expression in plant cells. Int. Rev. Cytol. 177, 115–180. [DOI] [PubMed] [Google Scholar]
- Gruissem, W., and Tonkyn, J.C. (1993). Control mechanisms of plastid gene expression. Crit. Rev. Plant Sci. 12, 19–55. [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press). [DOI] [PubMed]
- Hirose, T., and Sugiura, M. (1996). cis-Acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: Development of an in vitro translation system from tobacco chloroplasts. EMBO J. 15, 1687–1695. [PMC free article] [PubMed] [Google Scholar]
- Inoue, K., Dreyfuss, B.W., Kindle, K.L., Stern, D.B., Merchant, S., and Sodeinde, O.A. (1997). Ccs1, a nuclear gene required for the post-translational assembly of chloroplast c-type cytochromes. J. Biol. Chem. 272, 31747–31754. [DOI] [PubMed] [Google Scholar]
- Kim, J.M., and Mayfield, S.P. (1997). Protein disulfide isomerase as a regulator of chloroplast translational activation. Science 27, 1954–1957. [DOI] [PubMed] [Google Scholar]
- Kuchka, M.R., Mayfield, S.P., and Rochaix, J.-D. (1988). Nuclear mutations specifically affect the synthesis and/or degradation of the chloroplast-encoded D2 polypeptide of photosystem II in Chlamydomonas reinhardtii. EMBO J. 7, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and Timko, M.P. (1996). The pc-1 phenotype of Chlamy-domonas reinhardtii results from a deletion of mutation in the nuclear gene for NADPH:protochlorophyllide oxidoreductase. Plant Mol. Biol. 30, 15–37. [DOI] [PubMed] [Google Scholar]
- Li, J., Goldschmidt-Clermont, M., and Timko, M.P. (1993). Chloroplast encoded chlB is required for light-independent protochlorophyllide reductase activity in Chlamydomonas reinhardtii. Plant Cell 5, 1817–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield, S.P., Yohn, C.B., Cohen, A., and Danon, A. (1995). Regulation of chloroplast gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 147–166. [Google Scholar]
- McCarty, R.E. (1980). Delineation of the mechanism of ATP synthesis in chloroplasts: Use of uncouplers, energy transfer inhibitors, and modifiers of coupling factor 1. Methods Enzymol. 69, 719–728. [Google Scholar]
- Peters, J.W., Fisher, K., and Dean, D.R. (1995). Nitrogenase structure and function: A biochemical–genetic perspective. Annu. Rev. Microbiol. 49, 335–366. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt, T., Nilsson, A., and Allen, J.F. (1999). Photosynthetic control of chloroplast gene expression. Nature 397, 626–628. [Google Scholar]
- Reinbothe, S., and Reinbothe, C. (1996). Regulation of chlorophyll biosynthesis in angiosperms. Plant Physiol. 111, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reulland, E., and Miginiac-Maslow, M. (1999). Regulation of chloroplast enzyme activities by thioredoxins: Activation or relief from inhibition? Trends Plant Sci. 4, 136–141. [DOI] [PubMed] [Google Scholar]
- Rochaix, J.D. (1996). Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol. Biol. 32, 327–341. [DOI] [PubMed] [Google Scholar]
- Rott, R., Levy, H., Drager, R.G., Stern, D.B., and Schuster, G. (1998). 3′-Processed mRNA is preferentially translated in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 18, 4605–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager, R. (1955). Inheritance in the green alga Chlamydomonas reinhardtii. Genetics 40, 476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador, M.L., Klein, U., and Bogorad, L. (1993). Light-regulated and endogenous fluctuations of chloroplast transcript levels in Chlamydomonas. Regulation by transcription and RNA degradation. Plant J. 3, 213–219. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Shiina, T., Allison, L., and Maliga, P. (1998). rbcL transcript levels in tobacco plastids are independent of light: Reduced dark transcription rate is compensated by increased mRNA stability. Plant Cell 10, 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, D.B., Higgs, D.C., and Yang, J.J. (1997). Transcription and translation in chloroplasts. Trends Plant Sci. 2, 308–315. [Google Scholar]
- Suzuki, J.Y., and Bauer, C.E. (1992). Light-independent chlorophyll biosynthesis: Involvement of the chloroplast gene chlL (frxC). Plant Cell 4, 929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timko, M.P. (1998). Pigment biosynthesis: Chlorophylls, heme, and carotenoids. In Molecular Biology of Chlamydomonas: Chloroplasts and Mitochondria, J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 403–420.
- Trebst, A. (1980). Inhibitors in electron flow: Tools for the functional and structural localization of carriers and energy conservation sites. Methods Enzymol. 69, 674–715. [Google Scholar]
- Wu, H.Y., and Kuchka, M.R. (1995). A nuclear suppressor overcomes defects in synthesis of the chloroplast psbD gene product caused by mutation in two distinct nuclear genes of Chlamy-domonas. Curr. Genet. 27, 263–269. [DOI] [PubMed] [Google Scholar]
- Xie, Z., and Merchant, S. (1996). The plastid encoded ccsA gene is required for heme attachment to chloroplast c-type cytochromes. J. Biol. Chem. 271, 4632–4639. [DOI] [PubMed] [Google Scholar]
- Yohn, C.B., Cohen, A., Danon, A., and Mayfield, S.P. (1996). Altered mRNA binding activity and decreased translation initiation in a nuclear mutant lacking translation of the chloroplast psbA mRNA. Mol. Cell. Biol. 16, 3560–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn, C.B., Cohen, A., Danon, A., and Mayfield, S.P. (1998). A poly(A) binding protein functions in the chloroplast as a message-specific translation factor. Proc. Natl. Acad. Sci. USA 95, 2228–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]