Abstract
The human genome is far smaller than originally estimated, and one explanation is that alternative splicing creates greater proteomic complexity than a simple count of open reading frames would suggest. The p53 homologue p63, for example, is a tetrameric transcription factor implicated in epithelial development and expressed as at least six isoforms with widely differing transactivation potential. In particular, p63α isoforms contain a 27-kDa C-terminal region that drastically reduces their activity and is of clear biological importance, since patients with deletions in this C terminus have phenotypes very similar to patients with mutations in the DNA-binding domain. We have identified a novel domain within this C terminus that is necessary and sufficient for transcriptional inhibition and which acts by binding to a region in the N-terminal transactivation domain of p63 homologous to the MDM2 binding site in p53. Based on this mechanism, we provide a model that explains the transactivation potential of homo- and heterotetramers composed of different p63 isoforms and their effect on p53.
The recent discovery of two homologues of the archetypal tumor suppressor protein p53, called p73 and p63 (p51/p40/p73L/ket) (1, 35, 39, 46, 49), has sparked speculations that surveillance of cellular integrity might be achieved through a network of these p53-like tumor suppressors (10). These speculations were further fueled by the observation that the p73 gene is localized to chromosome 1p36.3, a region that is frequently lost in neuroblastomas and other types of cancers (20, 43) while the p63 chromosomal location, 3q27-9, is deleted in some bladder cancers and amplified in some cervical, ovarian, lung, and squamous cell carcinomas (13, 49). Moreover, both p73 and p63 can bind to p53 DNA-binding sites in vitro and activate proteins under the control of a p53 promoter and induce apoptosis in vivo (18, 49).
Despite all this circumstantial evidence, however, only a very few mutations in p63 and p73 have been found in human tumors, and a direct link to carcinogenesis similar to that for p53 has so far not been established (17, 22, 27, 32, 43). Furthermore, knockout mouse studies of both proteins have revealed that they are involved in different biological processes. p73-knockout mice are characterized by chronic infections and developmental defects, including hippocampal dysgenesis, hydrocephalus, and abnormalities in the pheromone sensory pathway (51). p63-knockout mice show even more severe defects that include truncated limbs and a lack of all squamous epithelia (29, 50). Both phenotypes are very unlike that of the p53-knockout mice, which are without developmental defects but have an elevated susceptibility to tumorigenesis (9). These striking differences in biological function are surprising in light of the high homology among all three proteins that reaches 65% identity in the DNA-binding domain, with all amino acids that contact DNA being conserved. Most likely then, these different biological functions are connected to domains outside the DNA-binding domain unique to p73 and p63. Indeed, both proteins exist in multiple isoforms. In the case of p63, at least six different isotypes with widely differing transactivation potentials have been described elsewhere (49). Three of these isoforms are inactive due to the absence of the N-terminal transactivation (TA) domain (ΔN isoforms). The activity of the remaining isoforms (TA isoforms) is modulated by three C-terminal sequences, α, β, and γ, not found in p53. The smaller p63β and p63γ isoforms are transcriptionally active while the presence of the 27-kDa α C-terminal domain drastically reduces the activity of p63 (49). In addition, ΔNp63α, which is the major isoform found in cells, shows strong dominant-negative behavior toward transactivating p63 isoforms and moderate dominant-negative behavior toward p53. The overarching importance of the α C-terminal domain to the biological function of p63 is further emphasized by clinical observations. A human patient with a mutation that introduces a premature stop codon in the α C terminus shows skeletal reduction defects of the hands and feet, surprisingly similar to patients with a mutation in the DNA-binding domain (4). That this mutation in the α C terminus affects only the two α isoforms (TAp63α and ΔNp63α) and yet produces very similar abnormalities as do mutations that eliminate DNA binding of all six p63 isoforms suggests that α isoforms play a pivotal role in p63 processes.
We have identified an inhibitory domain within the α C terminus that is both necessary and sufficient for transcriptional inhibition. In this paper we demonstrate that this domain uses a new mode of inhibition by binding to a region in the N-terminal TA domain of p63 that is homologous to the MDM2 binding site in p53, masking residues that are important for transactivation. Based on this intramolecular inhibition, we provide a model that makes sense of many past experiments and explains how alternative splicing modulates the transactivation potential of homo- and heterotetramers composed of different p63 isoforms and how they regulate p53. Given the high homology between the α C termini of p63 and p73, the results presented here are also very likely relevant for p73.
MATERIALS AND METHODS
Cell culture and transactivation assays.
SAOS-2 cells were obtained from the American Type Culture Collection. Cells were maintained in Dulbecco modified Eagle medium with 10% fetal bovine serum at 37°C in a 5% CO2 atmosphere. Transactivation assays were carried out with the dual-luciferase reporter assay system (Promega) in SAOS-2 cells with 13 tandem repeats of an optimized p53-binding sequence (from Bert Vogelstein) subcloned into a firefly luciferase reporter vector (49). All experimental results were normalized for transfection efficiency and sample preparation variations with a pRL-CMV control Renilla luciferase plasmid (Promega). Experiments were performed in triplicate.
Expression vectors.
For the transactivation cell culture assays all p63 and p53 constructs and mutants were cloned into a pcDNA3 vector (Invitrogen) as XhoI-XbaI fragments and containing an N-terminal myc tag. Point mutants were made with a QuikChange (Stratagene) mutagenesis kit. Glutathione S-transferase (GST) fusion vectors were constructed by inserting BamHI-EcoRI fragments into a pGEX-6P-2 vector (Pharmacia Biotech). For the expression of the TIDΔ25 peptide, the construct was cloned into a pET-31b(+) vector (Novagen) as a single AlwNI fragment according to the kit instructions.
Immunoblotting.
Transfected cell lysates were resolved on a sodium dodecyl sulfate (SDS)-4 to 12% polyacrylamide gel electrophoresis (PAGE) gradient gel (NuPAGE Bis-Tris; Invitrogen) and transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore). p63 isoforms, mutants, and chimeras were detected with the 4A4 monoclonal p63 antibody and/or the 9E10 c-Myc antibody. Secondary anti-mouse immunoglobulin G antibodies, conjugated to horseradish peroxidase (Santa Cruz Biotechnology), were applied, followed by treatment with enhanced chemiluminescence (ECL) reagent (Amersham Biosciences) and exposure to Kodak Biomax ML film.
Immunofluorescence.
Transfection and immunofluorescence experiments were carried out with BHK cells attached to 18-mm-diameter round glass coverslips as described earlier (49). The 9E10 c-Myc-specific antibody was used as the primary antibody against the N-terminally myc-tagged TAp63α, followed by staining with a Cy3-conjugated goat anti-mouse secondary antibody. DNA was stained with the fluorochrome Hoechst 33278 (Sigma). myc-tagged TAp63α was stained with the c-Myc-specific antibody 9E10.
Peptide expression and purification.
The TIDΔ25 (residues 568 to 616) peptide was expressed as a fusion to ketosteroid isomerase and a His tag, separated from each by a methionine. The fusion was purified from inclusion bodies in a denaturing buffer (6 M guanidine-HCl, 0.5 M NaCl, 5 mM imidazole, and 20 mM Tris-HCl, pH 7.9) by Ni-affinity chromatography. The purified protein precipitated upon dialysis into water and was collected by centrifugation. The protein was then dissolved in 80% formic acid in a round-bottomed flask and treated with CNBr in the dark, in a fume hood, and under argon for 24 h. The solution was evaporated to dryness at 28°C and resuspended in 20 mM KPO4 (pH 7.4) and 100 mM NaCl overnight. The resulting supernatant was passed over a high-pressure liquid chromatograph under a CH3CN-H2O gradient, and the eluted fractions were assessed by mass spectrometry. The fraction containing the TIDΔ25, a 49-mer peptide, contained two species: one corresponded to the expected molecular mass of the peptide and the other was of the expected molecular mass plus six histidines. A subsequent Ni-affinity assay also indicated that the larger of the two peptide species still contained a C-terminal His tag.
GST pull-down assays.
The transactivation-inhibitory (TI) domain (residues 566 to 641 or 569 to 616) and the TA domain (residues 1 to 26, 1 to 34, or 1 to 141) of p63 were each expressed as a fusion protein with an N-terminal GST and a C-terminal His tag in Escherichia coli and purified by Ni-affinity and glutathione-affinity chromatography. The GST fusion protein bound to glutathione beads was incubated for an hour with different isoforms and mutants of p63 that were expressed with an in vitro transcription-translation system (Promega) and labeled with [35S]methionine or with a purified peptide (TIDΔ25). Unbound protein was removed by centrifugation through a 0.65-μm-pore-size filter unit (Amicon). The beads on the filter were washed four times with 200 μl of binding buffer (100 mM NaCl, 10% glycerol, 50 mM Tris-HCl, pH 8.0) by centrifugation. Finally, the proteins were eluted from the beads by incubation with hot SDS sample buffer and removed from the beads by centrifugation. After PAGE (4 to 12% NuPAGE Bis-Tris), the gels were vacuum dried at 80°C for 2 h and the amount of each protein bound to the beads was quantified with a PhosphorImager (Storm 840; Molecular Dynamics). Analysis of the band intensities was conducted with ImageQuant 1.2. Values reported are the fractions of protein bound compared to the amount of labeled protein loaded on the beads. As a negative control, the experiments were repeated with only GST bound to the beads. All experiments were performed in triplicate.
RESULTS
The last 71 residues of the α C terminus of p63 contain an inhibitory domain.
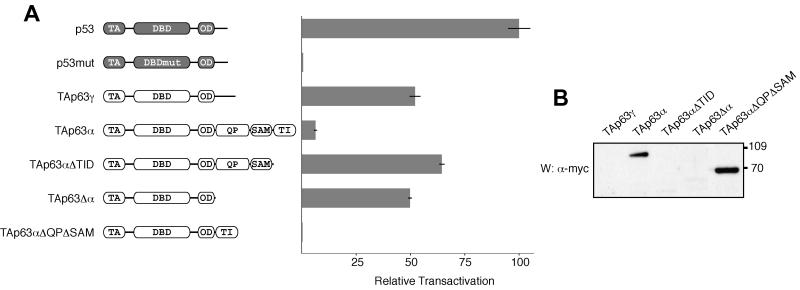
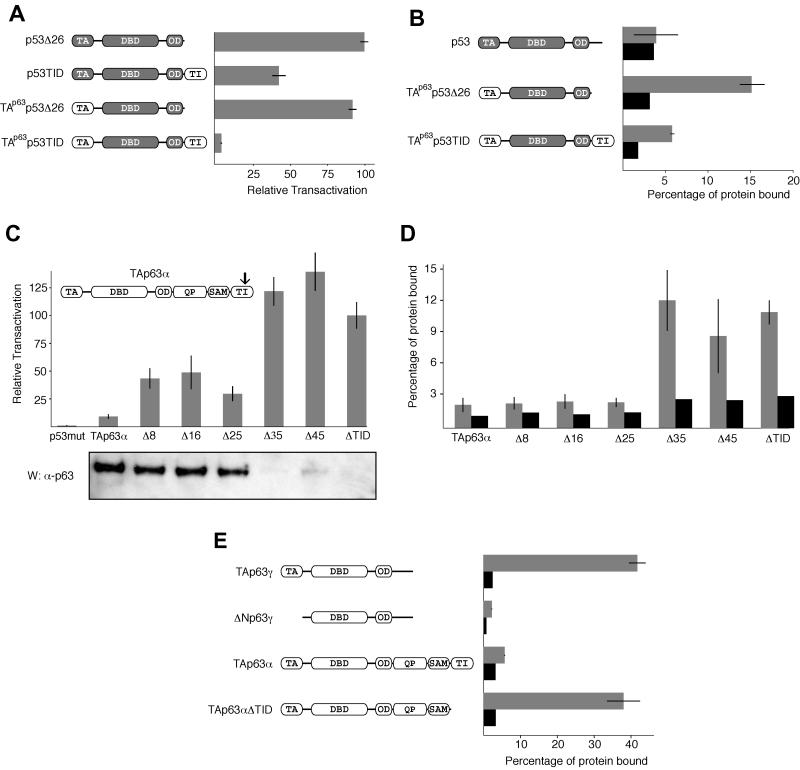
To identify the inhibitory domain in the large α C terminus and to investigate its mechanism of inhibition, we created C-terminal deletion mutants of TAp63α and monitored their transactivation with a luciferase assay in SAOS-2 cells. Wild-type TAp63α, the largest p63 isoform containing both the TA domain and the α C terminus, shows only very little transcriptional activity in these assays. Deleting the last 71 residues, however, restored the activity of the protein (TAp63αΔTID in Fig. 1A) to a level approaching that of a p53 control. This TI domain shows no homology to any known sequence, aside from the equivalent region of p73α, and is directly C-terminal to a SAM (sterile α motif) domain that was recently found in the α C terminus of both p63 and p73 (5, 28). Our finding agrees with an earlier study demonstrating transactivation activity for a mutant p63 identified in a human ectrodactyly-ectodermal dysplasia-clefting syndrome patient with a frameshift mutation toward the beginning of the SAM domain, creating an illegitimate 13-amino-acid peptide sequence followed by a premature stop codon after tyrosine-525 (4). Similar results have been obtained in a study with the p63-homologous protein p73 (36).
FIG. 1.
The last 71 residues of the α C terminus of p63 contain an inhibitory domain. (A) The relative transactivation levels of different p63 isoforms and deletion mutants were compared with those of the wild type and a DNA-binding-incompetent mutant of p53, as described in Materials and Methods. All values are scaled relative to the transactivation of wild-type p53, which was set to 100%. On the left the domain structure of the individual transcripts is indicated: TA (transactivation domain), DBD (DNA-binding domain), OD (oligomerization domain), QP (glutamine- and proline-rich domain), SAM (sterile alpha motif domain), TI (transactivation-inhibitory domain). TAp63αΔTID, residues 1 to 570; TAp63Δα, residues 1 to 397; TAp63αΔQPΔSAM, residues 1 to 397 and 568 to 641. (B) The lysates of p63-transfected cells used to measure relative transactivation (above) were resolved on an SDS-4 to 12% PAGE gradient gel, transferred to a PVDF membrane, and Western blotted with c-Myc-specific antibody 9E10. The bands that appear correspond to the expected molecular weights. The three species with high transactivation potential, TAp63γ, TAp63αΔTID, and TAp63Δα, are not visible, while the inactive species, TAp63α and TAp63αΔQPΔSAM, are abundant. Numbers at right are molecular weights in thousands.
To investigate if this TI domain is both necessary and sufficient for inhibition, we have deleted the entire α C terminus and then added the TI domain immediately following the oligomerization domain (TAp63αΔQPΔSAM). This transcript, which lacks the SAM domain as well as a glutamine- and proline-rich domain (QP domain, also known as TAD2), shows very low activity in our transactivation assays (Fig. 1A). In contrast, a deletion mutant lacking the α C terminus entirely (TAp63Δα) is as active as TAp63γ. To test the possibility that the inactivity of TAp63α and TAp63αΔQPΔSAM is due to low protein expression or rapid degradation, we examined the protein levels by Western blot analysis (Fig. 1B). Interestingly, the inactive forms, TAp63α and TAp63αΔQPΔSAM, are well expressed while the active isoforms, TAp63γ, TAp63αΔTID, and TAp63Δα, are barely detectable, indicating that the differences in activity cannot simply be explained by differences in protein levels. Our result is in accordance with findings that transactivating p63 isoforms have a much higher turnover rate than do inactive isoforms (34). Overall, the data presented in Fig. 1 suggest that the TI domain is both necessary and sufficient for inhibiting the activity of p63.
The TI domain does not inhibit nuclear localization.
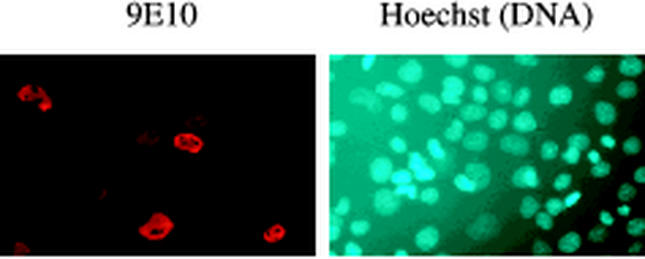
In agreement with results from other laboratories (5), our nuclear magnetic resonance investigation of the TI domain has shown that the TI domain lacks secondary structure, as characterized by a narrow range of proton chemical shifts. Unstructured sequences often contain signal peptides that target a protein to a certain cellular compartment. One possible mechanism by which the TI domain could inhibit the transcriptional activity of p63 is by sequestering it in the cytoplasm. To investigate if the TI domain prevents p63 from entering the nucleus, we have carried out immunofluorescence assays by expressing TAp63α in BHK cells. The results, shown in Fig. 2, demonstrate that TAp63α is nuclear, suggesting that the TI domain does not influence the subcellular localization of p63. We obtained similar results by fluorescence microscopy with cyan fluorescent protein-TAp63α fusion proteins in COS cells (data not shown).
FIG. 2.
The TI domain does not inhibit nuclear localization. Immunofluorescence was used to examine the cellular localization of TAp63α, as described in Materials and Methods. myc-tagged TAp63α was stained with the c-Myc-specific antibody 9E10 (left). The DNA was visualized with the fluorochrome Hoechst 33278 (right). The nuclear staining of TAp63α reveals that the TI domain does not inhibit nuclear localization of the p63 transcription factor.
The TI domain of p63 binds to the N-terminal TA domain.
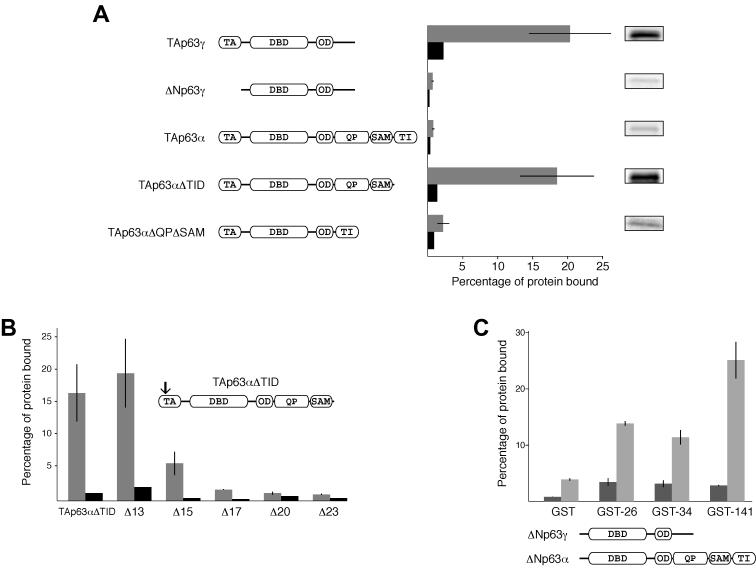
Several transcription factors contain domains that control their transactivation through an intramolecular mechanism (11, 12), mainly by inhibiting DNA binding activity. To investigate whether the TI domain inhibits the activity of p63 through an intramolecular mechanism, we bacterially expressed and purified a GST and TI domain fusion protein and conducted pull-down experiments on several different p63 isoforms 35S labeled in an in vitro transcription-translation system. While TAp63γ interacted with the GST-bound TI domain, ΔNp63γ did not (Fig. 3A), strongly suggesting that the TI domain in p63 binds to a region of the N-terminal TA domain. To further test this hypothesis, we conducted pull-down experiments with TAp63α, which contains both a TA and a TI domain. An intramolecular interaction between the two domains, as it might occur in TAp63α, should be more favorable than an intermolecular interaction with the external GST-fused TI domain. Indeed, in the pull-down experiment TAp63α bound very poorly. In accordance with this model, removal of the TI domain from TAp63α restored binding to a level comparable to that for TAp63γ, while a p63 form with the QP and the SAM domain removed (TAp63αΔQPΔSAM) also failed to bind (Fig. 3A). Comparison of the relative transactivation (Fig. 1A) with the results of these GST pull-down assays reveals a strong correlation that suggests that transactivation inhibition is linked to binding between the TI and TA domains.
FIG. 3.
The TI domain of p63 binds to the N-terminal TA domain. (A) GST-TID pull-down assay conducted on different p63 isoforms and deletion mutants. The gray bars show the percentages of protein bound to beads loaded with a GST-TID fusion protein, and the black bars show the amounts of protein bound to beads loaded with equal amounts of GST alone. On the right are representative bands corresponding to the levels of 35S-labeled protein bound to GST-TID, prior to quantification. Only p63 constructs with a free TA domain bind to the GST-TID. (B) GST-TID pull-downs of N-terminally truncated mutants of TAp63αΔTID. Removal of the first 15 amino acids results in a dramatic loss of binding, while removal of 17 or more amino acids destroys binding completely. The arrow indicates the approximate location of the truncations. (C) GST fusions to the first 26, 34, or 141 amino acids of TAp63 were used to assess binding to either ΔNp63γ (dark gray) or ΔNp63α (light gray). GST alone bound to neither well, while GST-26 and GST-34 did not bind to ΔNp63γ and bound with moderate affinity to ΔNp63α. GST-141 also failed to bind ΔNp63γ but bound with high affinity to ΔNp63α. Thus, the first 34 residues are necessary for binding, but the affinity is greater upon inclusion of additional amino acids in the TA domain.
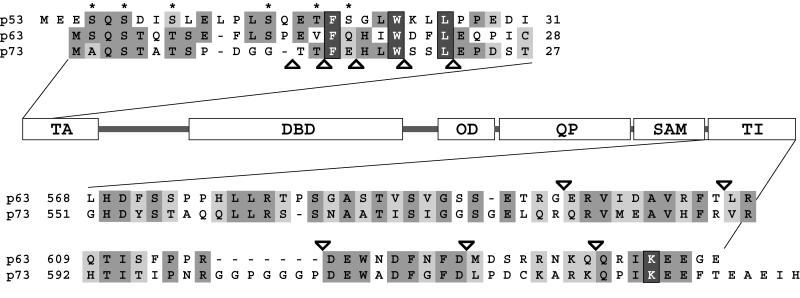
The TA region of p63, while poorly conserved overall with the TA of p53, contains a sequence that is homologous to the MDM2 binding site (23, 31). In particular, the three hydrophobic residues, F16, W20, and L23, which are deeply buried in a cleft in MDM2 in its complex with p53 (23), are conserved in p63 and p73 (Fig. 4). An interaction between MDM2 and the p73 protein has been reported in the literature, though, surprisingly, the interaction does not lead to degradation of p73 but inhibition of its transactivation potential (2, 8, 33, 48, 52). In the case of p63, several conflicting reports have been published. While some research groups do not find interaction between MDM2 and p63, others suggest that an interaction occurs that can either suppress transcriptional activity or enhance it (3, 19, 21, 25, 48). The sequence contacted by MDM2 has been shown elsewhere for p53 to be important for transactivation by binding to the TATA binding protein (14) as well as to several TATA binding protein-associated factors (TAFII40 and TAFII60) (26, 45). These observations, together with our finding that the TI domain binds the TA domain, suggest a mechanism by which the TI domain binds to the equivalent cofactor recruitment site in p63 and, by masking it, inhibits transactivation. As an initial test of this model, we have created N-terminal truncation mutants that deleted the first 13, 15, 17, 20, or 23 amino acids from the TA domain of TAp63αΔTID. In our TI domain interaction assay, the Δ13 mutant bound to the GST-TID fusion protein with an affinity comparable to that for p63 forms with the full-length TA domain (Fig. 3B). The Δ15 form bound with reduced affinity, and Δ17, which lacks F16, the first of the three putative key hydrophobic residues, does not bind. Further truncations, Δ20 and Δ23, also do not bind. These results suggest that the TI domain binds to the MDM2 binding sequence in the N-terminal acidic TA domain of p63 and inhibits activity by masking important residues that presumably contact the RNA transcriptional machinery.
FIG. 4.
The alignment of the amino termini of p53, TAp63, and TAp73 is shown above the domain structure model. The alignment of TI domains of p63α and p73α is shown below. The three conserved residues important for p53 binding to MDM2 and transcriptional cofactors are highlighted as white on dark gray. Asterisks indicate p53 residues shown to be phosphorylated under certain conditions. The SUMO-1-modified lysine is highlighted as white on dark gray. Triangles indicate truncation points at the N and C termini used in assays depicted in Fig. 3 and 6.
To determine whether this conserved TA region is sufficient for association with the TI domain, we created three GST fusion proteins with either the first 26, first 34, or first 141 amino acids of the TA region of p63. All three fusions contained the conserved F, W, and L and were used to pull down either ΔNp63γ or ΔNp63α (Fig. 3C). In accordance with our model of a TA-TI interaction, none of the GST fusions bound to the γ form of ΔNp63 and all bound the α form. However, GST fused to the first 141 residues of TAp63 bound to ΔNp63α better than did either of the two shorter fusions, suggesting that the MDM2 interaction site identified through homology with p53 is necessary for association but that binding improves when sequences outside the MDM2 interaction site are included.
The conserved hydrophobic residues within the TA domain are important both for binding the TI domain and for transactivation.
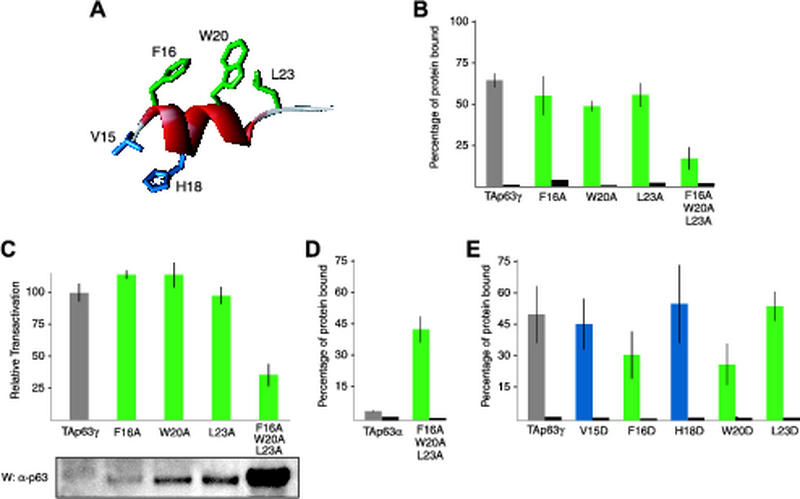
To address the role of the three putative key hydrophobic side chains, F16, W20, and L23 (Fig. 5A), in binding the TI domain, we replaced each individually with alanine in TAp63γ and performed pull-down assays with the GST-TI domain fusion protein. These experiments demonstrated only a slight reduction in binding affinity for all three single mutants relative to that for wild-type TAp63γ (Fig. 5B). We also tested the transcriptional activity of all three mutants to examine whether binding and activity are correlated as in the previous domain deletion experiments (Fig. 1A and 3A), and we found that the activity of the single mutants, like the binding affinity, was basically unchanged (Fig. 5C). Mutating all three residues simultaneously to alanine, however, had a significant effect on binding as well as on transactivation, reducing both by about 70%. Western blots show a slight accumulation of the single mutants and a very large buildup of the triple mutant, whereas the wild-type protein is barely detectable (Fig. 5C). Based on these results, a much weaker interaction between the triply mutated TA domain and the TI domain in TAp63α is expected. A weaker intramolecular interaction within TAp63α should result in stronger binding to an external, unmutated TA domain. Indeed, in a GST pull-down assay with the first 141 amino acids of the TA domain (GST-141), the triply mutated TAp63α showed strong binding while wild-type TAp63α did not bind (Fig. 5D). This triply mutated TAp63α is also completely inactive in the transactivation assay (data not shown). Thus, the same hydrophobic side chains involved in promoting the transactivation of p63 in the absence of a TI domain are also involved in the stability of the protein and in binding the TI domain.
FIG. 5.
Conserved residues F16 and W20 are important both for binding to the TI domain and for transactivation. (A) A model of the putative MDM2 binding helix within the TA domain of p63. Hydrophobic residues on one face of the helix thought to be involved in binding to transcriptional cofactors are colored green. A pair of residues on the opposite face are colored blue. The structure is adapted from the crystal structure of MDM2 bound to a peptide from the TA domain of p53 (23). (B) GST pull-down assays on TAp63γ containing TA domain mutations to alanine. The gray bar shows the percentage of wild-type TAp63γ bound to beads loaded with a GST-TID fusion protein, the green bars show the percentages of bound mutant TAp63γ, and the black bars show the amounts of TAp63γ, wild type or mutant, bound to beads loaded with GST only. (C) The relative transactivation levels of wild-type TAp63γ and TA domain mutants were compared. All values are scaled relative to the transactivation of wild-type TAp63γ, which was set to 100% and appears gray. The transactivation levels of the mutants are shown in green. The lysates of transfected cells used to measure relative transactivation were resolved and Western blotted with the anti-p63 4A4 antibody. The bands are of the same size and correspond to the expected molecular weight. (D) GST pull-down assays on TAp63α containing a triple mutation in the TA domain. The gray bar shows the percentage of wild-type TAp63α bound to beads loaded with GST fused to the first 141 amino acids of the TA domain, while the green bar indicates the percentage of TAp63α with F16A, W20A, and L23A mutations bound. The black bars show the amounts of protein bound to beads loaded with GST only. (E) GST pull-down assays on TAp63γ containing TA domain mutations to aspartate. The gray bar shows the percentage of wild-type TAp63γ bound to beads loaded with a GST-TID fusion protein, the green and blue bars show the percentages of mutant TAp63γ bound, and the black bars show the amounts of protein bound to beads loaded with GST only.
Mutating hydrophobic residues to aspartic acid instead of alanine can have more dramatic effects since burying a charged side chain in a hydrophobic environment is energetically highly unfavorable. Figure 5E shows that mutating F16 and W20 to aspartic acid reduces the affinity by almost 50% while mutating L23 again does not affect the binding. As expected, mutating V15 and H18, two residues that are located on the opposite face of the putative binding helix (Fig. 5A), does not affect the binding affinity either. All mutation data combined suggest that F16 and W20 are important amino acids that contribute to binding of the TI domain, while L23 as well as amino acids on the opposite face of the helix (V15 and H18) is not involved. The weak effect of mutating either F16 or W20 alone to alanine, however, also suggests that other regions within the first 141 amino acids of TAp63 may be involved in binding, in agreement with our previous findings (Fig. 3C).
The TI domain is a specific inhibitor of the p63 TA domain.
Given the conservation of these three hydrophobic residues and their importance in the association with the TI domain, we created chimeric proteins to test whether the TI domain might also act on p53. We began by adding the TI domain to the C terminus of the oligomerization domain of p53 (after removing p53's own 26-amino-acid C-terminal regulatory domain). In transactivation assays this chimeric protein (p53TID) showed a >50% reduction of activity relative to that of p53Δ26 (Fig. 6A). We tested if this partial inhibition is due to binding of the TI domain to the p53 MDM2 binding domain by GST pull-down assays. These experiments showed no binding of p53 above that of the GST control (Fig. 6B), suggesting that the interaction between the p63 TI domain and the p53 TA domain is weak at best. In contrast, a chimeric protein in which the TA domain of p53 has been replaced with p63's TA domain (TAp63p53Δ26) is readily pulled down by the GST-TID (Fig. 6B) and is highly active in the dual-luciferase transactivation assays (Fig. 6A). The double chimera of p53 with the TA domain of p63 and the TI domain of p63 (TAp63p53TID), however, binds to GST-TID only weakly (Fig. 6B) and is severely repressed in transactivation assays (Fig. 6A), similar to TAp63α. When Western blot analyses are performed, all the chimeras are undetectable with the exception of TAp63p53TID, which is barely visible after long exposures (data not shown). These activity and binding results suggest that the TI domain of p63 preferentially interacts with the TA domain of p63, even if both are presented in the context of a different protein. The TA domain of p53, however, does not appear to interact with the inhibitory domain.
FIG. 6.
The TI domain is a specific inhibitor of the p63 TA domain. (A) Relative transactivation of different chimeras composed of swapped domains of p63 and p53. p53Δ26, residues 1 to 361 of p53; p53TID, residues 1 to 361 of p53 and 568 to 641 of p63; TAp63p53Δ26, residues 1 to 127 of p63 and 92 to 361 of p53; TAp63p53TID, residues 1 to 127 of p63, 92 to 361 of p53, and 568 to 641 of p63. Shaded segments represent p53 domains, and nonshaded segments represent p63 domains. (B) GST pull-down assay on different p53/p63 chimeras. The gray bars show the percentages of protein bound to beads loaded with a GST-TID fusion protein, and the black bars show the amounts of protein bound to beads loaded with GST only. (C) The relative transactivation levels of TAp63α with various C-terminal truncations were compared with that for a DNA-binding-incompetent mutant of p53. All values are scaled relative to the transactivation of TAp63αΔTID, which was set to 100%. TAp63α mutants lacking the last 8 (Δ8), 16 (Δ16), or 25 (Δ25) amino acids show a moderate increase in activity relative to full-length TAp63α. Δ35, Δ45, and ΔTID (missing the last 71 residues) have high activity. The arrow indicates the approximate location of the truncations. The lysates of transfected cells used to measure relative transactivation were resolved on an SDS-4 to 12% PAGE gradient gel, transferred to a PVDF membrane, and Western blotted with the anti-p63 4A4 antibody. The bands correspond to the expected molecular weights, gradually decreasing from left to right. (D) Results of GST-TID pull-down assays with C-terminally truncated forms of TAp63α. GST-TID fails to bind TAp63α presumably because the intramolecular association is more favorable than the same association in trans. Removal of the last 35 or more residues leads to a sudden and dramatic improvement in binding most likely caused by a disruption of the intramolecular TI-TA domain interaction. (E) GST pull-down assay with a truncated TI domain. The affinities of different p63 isoforms for a truncated form of the TI domain lacking the last 25 amino acids (GST-TIDΔ25) were measured. The results are very similar to the results obtained with GST fused to the full-length TI domain.
The TI domain contains two subdomains that inhibit transcription through different mechanisms.
The modest reduction of transactivation of the p53TID chimera, in spite of the absence of a detectable interaction between the TI domain and p53, prompted us to look into alternative mechanisms of transactivation repression. Since it remains possible that a weak association between p53 and the TI domain is sufficient to account for the loss of activity, we looked for a correlation between disruption of the intramolecular TA-TI domain interaction and relief of inhibition through a series of C-terminal truncations of TAp63α. Removal of 8, 16, or 25 amino acids all led to a small increase in transcriptional activity relative to TAp63α. Further removal of amino acids (Δ35 and Δ45) restored the transactivation to levels comparable to those for TAp63αΔTID. Western blot analysis of these transactivation assays showed high protein levels for TAp63α and the Δ8, Δ16, and Δ25 deletion mutants, while Δ35, Δ45, and ΔTID are hardly detectable (Fig. 6C), reconfirming the inverse correlation of protein level with transcriptional activity observed earlier (Fig. 1).
To investigate the molecular mechanism by which the relief of inhibition occurs in these C-terminal truncations, we performed pull-down assays (Fig. 6D). The Δ8, Δ16, and Δ25 deletion mutants did not show any increase in binding affinity to the GST-TID fusion protein relative to TAp63α. Such an increase in binding would be expected if the C-terminal truncation disrupted the native intramolecular TA-TI domain interaction, allowing the GST-TID to bind in trans. Removal of 35 or more amino acids from the C terminus, however, restored binding affinity to levels comparable to those for TAp63αΔTID. These results clearly demonstrate that the last 25 amino acids in the TI domain of p63 are unnecessary for binding to the TA domain. We confirmed these results by performing pull-down assays with the Δ25 truncation mutant of the TI domain fused to GST (GST-TIDΔ25). This construct was able to bind TAp63γ and TAp63αΔTID, but not ΔNp63γ or TAp63α, with relative affinities that are indistinguishable from those for GST fused to the full-length TI domain (Fig. 6E). Thus, short C-terminal deletions (Δ8, Δ16, and Δ25) partially relieve inhibition, though the intramolecular TA-TI domain interaction remains intact, implying that the TI domain may contain two subdomains with different inhibitory mechanisms. This interpretation is further supported by our Western blot analysis of the protein levels in the transactivation assays. In agreement with our earlier results (Fig. 1), all p63 forms that are inactive due to binding of the TI domain to the TA domain accumulate in the cell and can be detected as strong bands in Western blots. In contrast, all active p63 forms in which the TA domain is not masked by the TI domain are barely detectable, most likely due to rapid turnover of the protein. Further support for our model of two different subdomains within the TI domain is based on sequence comparison between p63 and p73. The sequence of p73 that shows high homology to the TI domain of p63 has a polyglycine insertion 25 amino acids from the C terminus (Fig. 4). This insertion coincides with the position beyond which further removal of amino acids eliminates the TA-TI domain interaction and may mark a boundary between these inhibitory subdomains.
The last 25 amino acids, on the other hand, seem to inhibit the activity of p63 by an additional mechanism that is different from the binding of the TI domain to the TA domain. Interestingly, the last eight amino acids in p63 contain a conserved lysine that becomes covalently modified by the ubiquitin homologue SUMO-1 in p73 (30) (Fig. 4). Covalent attachment of ubiquitin or SUMO-1 has been shown elsewhere to affect protein stability and can affect their turnover rate inside cells (6). Sumoylation might, therefore, explain the inhibitory effect of the TI domain on p53 as well as the partial relief of inhibition by the Δ8, Δ16, and Δ25 mutants. Since the activities of the Δ8, Δ16, and Δ25 truncation mutants are relatively similar, this additional inhibitory effect is most likely localized in the last eight amino acids. If this model is correct, removing the last eight amino acids from the C terminus of the TI domain in the p53TID chimera should relieve the partial inhibition with this chimera that we observed as shown in Fig. 6A. Indeed, in transactivation assays the p53TIDΔ8 construct showed a transcriptional activity that is indistinguishable from that of wild-type p53.
Thus, the combination of the results of the pull-down assays, transactivation assays, Western blot analysis, and sequence comparison strongly suggests that the first ∼50 residues of the TI domain inhibit transactivation by binding to the TA domain, while the last 25 residues may be involved in associations with other proteins that modulate stability and activity of p63. Interestingly, a human patient with split-hand-split-foot malformation caused by a nonsense mutation at Q634 has been identified elsewhere (47). This mutation creates a form of p63α that lacks the last eight amino acids (Δ8 form) and demonstrates, in accordance with our results, the importance of even just the last eight amino acids for p63's biological function.
To investigate the exact mechanism by which binding of the TI domain to the TA domain inhibits transcriptional activity, we investigated the interaction of the first 141 amino acids of the TA domain with several cofactors of the transcriptional machinery (TAFII32, TAFII40, TAFII60, TAFII70, TAFII250, CBP, and p300) both by pull-down assays with GST-141 and by immunoprecipitation. None of these factors bound significantly above background levels, suggesting either that different cofactors are involved in p63 transcriptional activity or that posttranslational modifications, in particular phosphorylation, might be necessary to establish an interaction.
The TI domain binds directly to the TA domain.
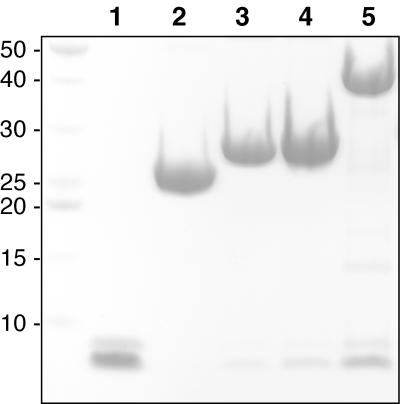
The pull-down experiments described so far were performed in rabbit reticulocyte lysates as part of the in vitro transcription-translation kit used to label the p63 isoforms with 35S. To explore the possibility that there might be a rabbit protein in the lysates that mediates the TA-TI domain association, we conducted pull-downs exclusively with purified proteins made recombinantly in E. coli. The GST-TA fusions were used to assess binding to the first 49 residues of the TI domain (TIDΔ25), and the results were assessed by Coomassie blue staining (Fig. 7). In agreement with our results shown in Fig. 3C, the GST alone did not bind the TI peptide, but fusions of the GST to TA peptides of increasing length did. Once again, the GST fused to the first 141 residues bound better than did GST fused to the first 26 or 34 residues. This experiment demonstrates that the TA-TI domain interaction is indeed direct and also suggests that no posttranslational modification is required for either domain to drive the association.
FIG. 7.
The TI domain binds directly to the TA domain. The GST-TA fusions used in Fig. 3C were used to assess binding to a recombinantly expressed and purified TIDΔ25 peptide (residues 568 to 616). Lane 1 contains the purified TIDΔ25 peptide. The doublet is due to the presence (higher molecular weight) or absence (lower molecular weight) of a C-terminal six-histidine tag as indicated by both mass spectrometry and nickel affinity (see Materials and Methods). The histidine tag does not appear to affect the affinity of the TIDΔ25 peptide for the TA domain. The peptide was incubated with either GST, GST-26, GST-34, or GST-141 fusion proteins bound to glutathione-Sepharose, washed, eluted with hot SDS sample buffer, and run in lanes 2, 3, 4, and 5, respectively. The very high molecular weight bands correspond to the GST fusions and occur at roughly equivalent concentrations. The low-molecular-weight doublet indicates the fraction of TIDΔ25 still bound to the fusions following the washes. GST alone does not bind a detectable amount of peptide, while GST-26 and GST-34 bind with moderate affinity and GST-141 binds with high affinity. Numbers at left are molecular weights in thousands.
DISCUSSION
Our experiments demonstrate that p63 and p53 employ different mechanisms to regulate transcriptional activity. p53 also contains a regulatory domain at its extreme C terminus which is, in contrast to the TI domain of p63, only 26 amino acids long. More importantly, the mechanism of inhibition seems to be different, as the inhibitory domain of p53 is involved in regulating the sequence-specific binding affinity of the DNA-binding domain (15, 16, 24) while the TI domain of p63 binds to the N-terminal TA domain. The closely related protein p73 shows a very similar domain structure, including a TI domain and a SAM domain in its α C terminus. This high sequence homology between the two proteins (Fig. 4) predicts that the results shown here are relevant not only for p63 but also for p73.
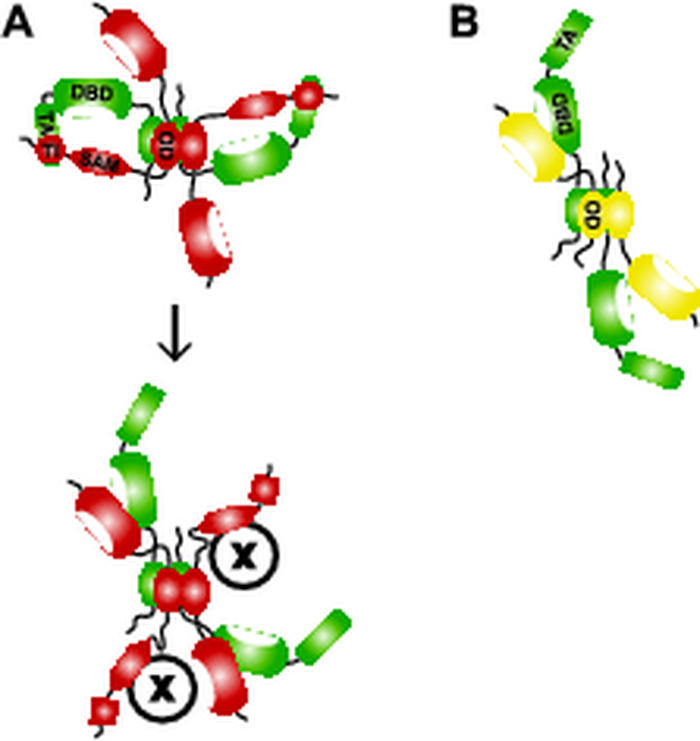
ΔNp63γ and ΔNp63α display a similar and moderate dominant-negative effect toward p53 but radically different effects toward transactivating TAp63γ (49). TAp63γ is, if at all, only weakly repressed by ΔNp63γ but very strongly repressed by ΔNp63α, the major p63 isoform in cells. In light of our results, these different effects of p63 isoforms on p53 and TAp63γ can now be interpreted. The TI domain of ΔNp63α cannot interact intramolecularly since it lacks its own TA domain. However, p63 tetramerizes (7), and formation of hetero-oligomers between TAp63γ and ΔNp63α could allow the TI domain to interact with the TA domain of another protein within the tetramer. A 2:2 complex of ΔNp63α and TAp63γ would, therefore, reduce the number of active TA domains in the tetramer to zero (Fig. 8A), powerfully inhibiting the activity of TAp63γ. ΔNp63γ, however, lacks the TI domain, and a 2:2 complex with TAp63γ will leave two active TA domains (Fig. 8B). Thus, by controlling the ratios of the different isoforms through alternative splicing, the activity of p63 may be fine-tuned.
FIG. 8.
Models of p63 heterotetramers. (A) In a tetramer consisting of two TAp63γ (green) and two ΔNp63α (red) monomers, the TI domains of ΔNp63α can interact with and inhibit both TA domains of the TAp63γ monomers. Activation of this tetramer could occur by recruiting another protein, e.g., a kinase that disrupts the TI-TA domain interaction. Binding of kinase or the other factor, illustrated as “X,” could be facilitated by the SAM domain. (B) Tetramerization of two TAp63γ monomers (green) with two ΔNp63γ monomers (yellow) leaves both TA domains active.
With respect to p53, ΔNp63γ and ΔNp63α show very similar levels of inhibition. Earlier results have suggested that p53 and p63 cannot form hetero-oligomers (7), and the inhibitory effects of ΔNp63 isoforms on p53 were attributed to competition for DNA-binding sites (49). Other research laboratories have reported a direct interaction between p53 and p63 in their DNA-binding domains (38). In addition, Pozniak et al. have shown that ΔN isoforms of p73 play an antiapoptotic role and are direct antagonists of p53 in sympathetic neurons (37). They attribute this antagonistic role of p73 to direct binding to p53. While our results suggest that the TI domain of p63 cannot directly bind to the TA domain of p53 and inhibit its transactivation potential, they are consistent with a model based on competition for DNA-binding sites and do not contradict a possible association through the DNA-binding domains. The combined results of all research groups are also consistent with a model in which p63 and p73 are involved in regulatory networks with p53 and control p53's apoptotic function in neurons and epithelial stem cells. Most likely, however, the TI domain is used only for the direct regulation of the transcriptional control of p63 and maybe p73, but not in interactions with p53.
Our results explain why TAp63α shows a low level of transactivation relative to that of TAp63γ and explain the strong dominant-negative effect of ΔNp63α on transactivating p63 isoforms. Based on our data, it is now possible to integrate a wealth of other studies about p63 regulation into one consistent model. In this model, a high concentration of ΔNp63α immediately inactivates any small concentration of active p63 isoforms and keeps them in an inhibited complex. Complex formation of active p63 isoforms with ΔNp63α increases the lifetime of the TA domain-containing forms while keeping them in an inactive state. To activate the transcriptional potential of p63, only the inhibitory interaction between the TI and the TA domains has to be relieved, thereby avoiding time-consuming protein expression. In this way, differential splicing of the p63 gene can create a transcriptional regulatory complex that allows the cell to control the activity of p63 in a fast, switch-like manner.
Several lines of evidence support such a model. First, ΔNp63α is the most abundant isoform found in epithelial cells, and it has been demonstrated that even a mutant ΔNp63α incapable of binding DNA retains the strong dominant-negative effect toward active p63 isoforms, while losing its dominant-negative effect on p53. This suggests that the inhibition of active p63 isoforms by ΔNp63α is not due to competition for DNA-binding sites but occurs through direct interaction between the isoforms. Second, several reports have demonstrated that p63 isoforms with an active TA domain are barely detectable by immunochemistry (40, 49), and the TA domain has been implicated in the responsibility for fast degradation (34). In contrast, ΔNp63 isoforms, which lack the TA domain, seem to be more stable. Further supporting these observations, mutations made in the putative MDM2 binding helix of p63 dramatically increase stability of the protein (Fig. 5C), and notably these same residues are involved in an association with the TI domain (Fig. 3B and C, 5B, and 7). Thus, when sites within the TA domain responsible for fast degradation are masked by the TI domain, protein stability and protein levels should increase, and indeed, our results indicate that TA isoforms containing the TI domain accumulate to significantly higher levels than those without (Fig. 1B and 6C). According to our model, this masking and stabilization could also occur in trans in hetero-oligomeric complexes containing ΔNp63α and TAp63γ/β. Finally, the central, biological role that the α C terminus plays in p63 is emphasized by the analysis of p63 mutations in human patients. Surprisingly, patients with mutations in the α C terminus that partially abolish the inhibitory function of the TI domain and create transcriptionally active p63α forms show very similar phenotypes as do patients with mutations in the DNA-binding domain that abolish DNA binding of all p63 isoforms (4). While the mechanism of how loss of inhibition in the α isoforms and loss of DNA binding in all isoforms lead to similar phenotypes is currently unknown, it shows that the inhibitory function in the TI domain is of paramount importance.
The question remains how p63 becomes active. Our results indicate that disruption of the inhibitory interaction between the TI and the TA domains would trigger p63's transactivation potential. Such a trigger could be phosphorylation of either of the two domains, and several known sites of phosphorylation in the TA domain of p53 are conserved in p63 (Fig. 4). As discussed above, potential regulatory sites for the TA-TI interaction might be located in the last 25 amino acids of the TI domain. Alternatively, since SAM domains are protein-protein interaction modules that are found in a large variety of different protein classes, the SAM domain that is located directly N-terminal to the TI domain could recruit, for example, kinases (Fig. 8). A recent analysis of mutations found in the SAM domain of p63 in human patients (28) has suggested a binding site for a potential interaction partner that is different from the SAM-SAM domain dimerization interface found in several SAM domain structures so far (41, 42, 44). This opens the possibility that the p63 SAM domain might interact with a binding partner that is not another SAM domain. Studies to address these questions are being carried out.
Acknowledgments
We thank Nilesh Shah for assistance with the cyanogen bromide cleavage reactions, Alessander Guimaraes for generating the luciferase reporter, and Holly Ingraham for helpful discussions.
Financial support was provided by an institutional research grant from the American Cancer Society (AC-02-8), the UCSF Innovations in Basic Sciences Award, and the Sandler Family supporting foundation. Z.S. was supported by the Burroughs Wellcome Fund. H.C.L., H.D.O., and A.E.K. were supported by an NIH training grant (GM08284).
REFERENCES
- 1.Augustin, M., C. Bamberger, D. Paul, and H. Schmale. 1998. Cloning and chromosomal mapping of the human p53-related KET gene to chromosome 3q27 and its murine homologue Ket to mouse chromosome 16. Mamm. Genome 9:899-902. [DOI] [PubMed] [Google Scholar]
- 2.Balint, E., S. Bates, and K. H. Vousden. 1999. Mdm2 binds p73 alpha without targeting degradation. Oncogene 18:3923-3929. [DOI] [PubMed] [Google Scholar]
- 3.Calabro, V., G. Mansueto, T. Parisi, M. Vivo, R. A. Calogero, and G. L. Mantia. 2002. The human MDM2 oncoprotein increases the transcriptional activity and the protein level of the p53 homolog p63. J. Biol. Chem. 277:2674-2681. [DOI] [PubMed] [Google Scholar]
- 4.Celli, J., P. Duijf, B. C. Hamel, M. Bamshad, B. Kramer, A. P. Smits, R. Newbury-Ecob, R. C. Hennekam, G. van Buggenhout, A. van Haeringen, C. G. Woods, A. J. van Essen, R. de Waal, G. Vriend, D. A. Haber, A. Yang, F. McKeon, H. G. Brunner, and H. van Bokhoven. 1999. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 99:143-153. [DOI] [PubMed] [Google Scholar]
- 5.Chi, S. W., A. Ayed, and C. H. Arrowsmith. 1999. Solution structure of a conserved carboxy-terminal domain of p73 with structural homology to the SAM domain. EMBO J. 18:4438-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciechanover, A. 1998. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17:7151-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davison, T. S., C. Vagner, M. Kaghad, A. Ayed, D. Caput, and C. H. Arrowsmith. 1999. p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J. Biol. Chem. 274:18709-18714. [DOI] [PubMed] [Google Scholar]
- 8.Dobbelstein, M., S. Wienzek, C. Koenig, and J. Roth. 1999. Inactivation of the p53-homologue p73 by the mdm2-oncoprotein. Oncogene 18:2101-2106. [DOI] [PubMed] [Google Scholar]
- 9.Donehower, L. A., M. Harvey, B. L. Slagle, M. J. McArthur, C. A. Montgomery, J. S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215-221. [DOI] [PubMed] [Google Scholar]
- 10.Flores, E. R., K. Y. Tsai, D. Crowley, S. Sengupta, A. Yang, F. McKeon, and T. Jacks. 2002. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416:560-564. [DOI] [PubMed] [Google Scholar]
- 11.Giovane, A., A. Pintzas, S. M. Maira, P. Sobieszczuk, and B. Wasylyk. 1994. Net, a new ets transcription factor that is activated by Ras. Genes Dev. 8:1502-1513. [DOI] [PubMed] [Google Scholar]
- 12.Hagman, J., and R. Grosschedl. 1992. An inhibitory carboxy-terminal domain in Ets-1 and Ets-2 mediates differential binding of Ets family factors to promoter sequences in the mb-1 gene. Proc. Natl. Acad. Sci. USA 89:8889-8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hibi, K., B. Trink, M. Patturajan, W. H. Westra, O. L. Caballero, D. E. Hill, E. A. Ratovitski, J. Jen, and D. Sidransky. 2000. AIS is an oncogene amplified in squamous cell carcinoma. Proc. Natl. Acad. Sci. USA 97:5462-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horikoshi, N., A. Usheva, J. Chen, A. J. Levine, R. Weinmann, and T. Shenk. 1995. Two domains of p53 interact with the TATA-binding protein, and the adenovirus 13S E1A protein disrupts the association, relieving p53-mediated transcriptional repression. Mol. Cell. Biol. 15:227-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hupp, T. R., and D. P. Lane. 1994. Allosteric activation of latent p53 tetramers. Curr. Biol. 4:865-875. [DOI] [PubMed] [Google Scholar]
- 16.Hupp, T. R., A. Sparks, and D. P. Lane. 1995. Small peptides activate the latent sequence-specific DNA binding function of p53. Cell 83:237-245. [DOI] [PubMed] [Google Scholar]
- 17.Ichimiya, S., Y. Nimura, H. Kageyama, N. Takada, M. Sunahara, T. Shishikura, Y. Nakamura, S. Sakiyama, N. Seki, M. Ohira, Y. Kaneko, F. McKeon, D. Caput, and A. Nakagawara. 2001. Genetic analysis of p73 localized at chromosome 1p36.3 in primary neuroblastomas. Med. Pediatr. Oncol. 36:42-44. [DOI] [PubMed] [Google Scholar]
- 18.Jost, C. A., M. C. Marin, and W. G. Kaelin. 1997. p73 is a simian p53-related protein that can induce apoptosis. Nature 389:191-194. [DOI] [PubMed] [Google Scholar]
- 19.Kadakia, M., C. Slader, and S. J. Berberich. 2001. Regulation of p63 function by MDM2 and MDMX. DNA Cell Biol. 20:321-330. [DOI] [PubMed] [Google Scholar]
- 20.Kaghad, M., H. Bonnet, A. Yang, L. Creancier, J.-C. Biscan, A. Valent, A. Minty, P. Chalon, J.-M. Lelias, X. Dumont, P. Ferrara, F. McKeon, and D. Caput. 1997. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90:809-819. [DOI] [PubMed] [Google Scholar]
- 21.Kojima, T., Y. Ikawa, and I. Katoh. 2001. Analysis of molecular interactions of the p53-family p63 gene products in a yeast-two-hybrid system: homotypic and heterotypic interactions and association with p53-regulatory factors. Biochem. Biophys. Res. Commun. 281:1170-1175. [DOI] [PubMed] [Google Scholar]
- 22.Kovalev, S., N. Marchenko, S. Swendeman, M. LaQuaglia, and U. M. Moll. 1998. Expression level, allelic origin, and mutation analysis of the p73 gene in neuroblastoma tumors and cell lines. Cell Growth Differ. 9:897-903. [PubMed] [Google Scholar]
- 23.Kussie, P. H., S. Gorina, V. Marechal, B. Elenbaas, J. Moreau, A. J. Levine, and N. P. Pavletich. 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274:948-953. [DOI] [PubMed] [Google Scholar]
- 24.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 25.Little, N. A., and A. G. Jochemsen. 2001. HDMx and MDM2 can repress transcription activation by p53 but not by p63. Oncogene 20:4576-4580. [DOI] [PubMed] [Google Scholar]
- 26.Lu, H., and A. J. Levine. 1995. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc. Natl. Acad. Sci. USA 92:5154-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mai, M., H. Huang, C. Reed, C. Qian, J. S. Smith, and B. Alderete. 1998. Genomic organization and mutation analysis of p73 in oligodendrogliomas with chromosome 1 p-arm deletions. Genomics 51:359-363. [DOI] [PubMed] [Google Scholar]
- 28.McGrath, J. A., P. H. Duijf, V. Dötsch, A. D. Irvine, R. de Waal, K. R. Vanmolkot, V. Wessagowit, A. Kelly, D. J. Atherton, W. A. Griffiths, S. J. Orlow, A. van Haeringen, M. G. Ausems, A. Yang, F. McKeon, M. A. Bamshad, H. G. Brunner, B. C. J. Hamel, and H. van Bokhoven. 2001. Hay-Wells syndrome is caused by missense mutations in the SAM domain of p63. Hum. Mol. Genet. 10:221-229. [DOI] [PubMed] [Google Scholar]
- 29.Mills, A. A., B. Zheng, X.-J. Wang, H. Vogel, D. R. Roop, and A. Bradley. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398:708-713. [DOI] [PubMed] [Google Scholar]
- 30.Minty, A., X. Dumont, M. Kaghad, and D. Caput. 2001. Covalent modifications of p73α by SUMO-1. J. Biol. Chem. 275:36316-36323. [DOI] [PubMed] [Google Scholar]
- 31.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 32.Nomoto, S., N. Haruki, M. Kondo, H. Konishi, T. Takahashi, and T. Takahashi. 1998. Search for mutations and examination of allelic expression imbalance of the p73 gene at 1p36.33 in human lung cancers. Cancer Res. 58:1380-1383. [PubMed] [Google Scholar]
- 33.Ongkeko, W. M., X. Q. Wang, W. Y. Siu, A. W. Lau, K. Yamashita, A. L. Harris, L. S. Cox, and R. Y. Poon. 1999. MDM2 and MDMX bind and stabilize the p53-related protein p73. Curr. Biol. 9:829-832. [DOI] [PubMed] [Google Scholar]
- 34.Osada, M., R. Inaba, H. Shinohara, M. Hagiwara, M. Nakamura, and Y. Ikawa. 2001. Regulatory domain of protein stability of human p51/TAp63, a p53 homologue. Biochem. Biophys. Res. Commun. 283:1135-1141. [DOI] [PubMed] [Google Scholar]
- 35.Osada, M., M. Ohba, C. Kawahara, C. Ishioka, R. Kanamaru, I. Katoh, Y. Ikawa, Y. Nimura, A. Nakagawara, M. Obinata, and S. Ikawa. 1998. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat. Med. 4:839-843. [DOI] [PubMed] [Google Scholar]
- 36.Ozaki, T., M. Naka, N. Takada, M. Tada, S. Sakiyama, and A. Nakagawara. 1999. Deletion of the COOH-terminal region of p73α enhances both its transactivation function and DNA-binding activity but inhibits induction of apoptosis in mammalian cells. Cancer Res. 59:5902-5907. [PubMed] [Google Scholar]
- 37.Pozniak, C. D., S. Radinovic, A. Yang, F. McKeon, D. R. Kaplan, and F. D. Miller. 2000. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science 289:304-306. [DOI] [PubMed] [Google Scholar]
- 38.Ratovitski, E. A., M. Patturajan, K. Hibi, B. Trink, K. Yamaguchi, and D. Sidransky. 2001. p53 associates with and targets ΔNp63 into a protein degradation pathway. Proc. Natl. Acad. Sci. USA 98:1817-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Senoo, M., N. Seki, M. Ohira, S. Sugano, M. Watanabe, S. Inuzuka, T. Okamoto, M. Tachibana, T. Tanaka, Y. Shinkai, and H. Kato. 1998. A second p53-related protein, p73L, with high homology to p73. Biochem. Biophys. Res. Commun. 248:603-607. [DOI] [PubMed] [Google Scholar]
- 40.Signoretti, S., D. Waltregny, J. Dilks, B. Isaac, D. Lin, L. Garraway, A. Yang, R. Montironi, F. McKeon, and M. Loda. 2000. p63 is a prostate basal cell marker and is required for prostate development. Am. J. Pathol. 157:1769-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smalla, M., P. Schmieder, M. Kelly, A. T. Laak, G. Krause, L. Ball, M. Wahl, P. Bork, and H. Oschkinat. 1999. Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites. Protein Sci. 8:1954-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stapleton, D., I. Balan, T. Pawson, and F. Sicheri. 1999. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol. 6:44-49. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, H., S. Ichimiya, Y. Nimura, M. Watanabe, M. Furusato, and S. Wakui. 1998. Mutation, allelotyping, and transcription analyses of the p73 gene in prostatic carcinoma. Cancer Res. 58:2076-2077. [PubMed] [Google Scholar]
- 44.Thanos, C. D., K. E. Goodwill, and J. U. Bowie. 1999. Oligomeric structure of the human EphB2 receptor SAM domain. Science 283:833-836. [DOI] [PubMed] [Google Scholar]
- 45.Thut, C., J. L. Chen, R. Klemm, and R. Tjian. 1995. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science 267:100-104. [DOI] [PubMed] [Google Scholar]
- 46.Trink, B., K. Okami, L. Wu, V. Sriuranpong, J. Jen, and D. Sidransky. 1998. A new human p53 homologue. Nat. Med. 4:747-748. [DOI] [PubMed] [Google Scholar]
- 47.van Bokhoven, H., B. C. Hamel, M. Bamshad, E. Sangiorgi, F. Gurrieri, P. H. Duijf, K. R. Vanmolkot, E. van Beusekom, S. E. van Beersum, J. Celli, G. F. Merkx, R. Tenconi, J. P. Fryns, A. Verloes, R. A. Newbury-Ecob, A. Raas-Rotschild, F. Majewski, F. A. Beemer, A. Janecke, D. Chitayat, G. Crisponi, H. Kayserili, J. R. Yates, G. Neri, and H. G. Brunner. 2001. p63 gene mutations in EEC syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am. J. Hum. Genet. 69:481-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, X., T. Arooz, W. Y. Siu, C. H. Chiu, A. Lau, K. Yamashita, and R. Y. Poon. 2001. MDM2 and MDMX can interact differently with ARF and members of the p53 family. FEBS Lett. 490:202-208. [DOI] [PubMed] [Google Scholar]
- 49.Yang, A., M. Kaghad, Y. Wang, E. Gillett, M. D. Fleming, V. Dötsch, N. C. Andrews, D. Caput, and F. McKeon. 1998. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2:305-316. [DOI] [PubMed] [Google Scholar]
- 50.Yang, A., R. Schweitzer, D. Sun, M. Kaghad, N. Walker, R. T. Bronson, C. Tabin, A. Sharpe, D. Caput, C. Crum, and F. McKeon. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398:714-718. [DOI] [PubMed] [Google Scholar]
- 51.Yang, A., N. Walker, R. Bronson, M. Kaghad, M. Oosterwegel, J. Bonnin, C. Vagner, H. Bonnet, P. Dikkes, A. Sharpe, F. McKeon, and D. Caput. 2000. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404:99-103. [DOI] [PubMed] [Google Scholar]
- 52.Zeng, X., L. Chen, C. A. Jost, T. Maya, D. Keller, X. Wang, W. G. Kaelin, M. Oren, J. Chen, and H. Lu. 1999. MDM2 suppresses p73 function without promoting p73 degradation. Mol. Cell. Biol. 19:3257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]