Abstract
RNA editing in organelles of angiosperm plants results in alteration of Cs to Us in transcripts. In most editing sites analyzed in vitro or in vivo, sequences within approximately 30 nucleotides (nt) 5′ and 10 nt 3′ of the edited C have been found to be required for selection of the correct C editing target and for editing efficiency, but no consensus sequences have been identified. The effect of high-level expression of two different minigenes carrying either the rpoB-2 or the ndhF-2 editing site on editing was determined for all 31 known edited Cs in two transgenic tobacco lines. The editing efficiencies of both the corresponding rpoB and ndhF editing sites in the endogenous genes' transcripts and in several other genes' transcripts were reduced in the chloroplast transgenic plants. Conserved nucleotides could be identified in the sequences immediately 5′ of each overexpressed editing site and in the sites in the additional genes that experienced a competition effect, though the conserved nucleotides differ 5′ of rpoB-2 and 5′ of ndhF-2. Inspection of sequences surrounding chloroplast and mitochondrial editing sites reveals that they can be grouped into clusters which carry conserved nucleotides within 30 nt 5′ of the C target of editing. The data are consistent with a model in which the same trans factor recognizes several chloroplast or mitochondrial editing sites, depending on the particular sequence 5′ of the edited C.
Transcripts of vascular plant chloroplast and mitochondrial genomes are modified by C-to-U RNA editing (3, 10, 17, 26, 29). Angiosperm chloroplasts typically contain 25 to 31 known editing sites (28), while 441 sites have been detected in mitochondria of Arabidopsis, the only vascular plant with a mitochondrial genome that has been completely sequenced (11). RNA editing in angiosperm chloroplasts usually results in alterations of the second codon position in coding regions, changing predicted amino acids (28). Mitochondria exhibit predominantly coding-region editing, but editing in noncoding regions and silent third codon positions is also observed. In both chloroplasts and mitochondria, there is a preference for a 5′ pyrimidine and 3′ purine next to the C target of editing (2).
The requirements for chloroplast transcript editing have been probed in chloroplasts by introducing transgenes carrying sequences surrounding editing sites. Such experiments have established that 16 to 40 nucleotides immediately 5′ and 6 to 20 nucleotides 3′ are critical for editing. The amount of surrounding sequence required varies somewhat depending on the particular editing site. While 21 nucleotides surrounding the psbL editing site allowed efficient editing, 84 nucleotides surrounding two ndhB editing sites did not result in any transgene transcript editing (4). Though mitochondrial transformation is not yet possible, similar conclusions can be drawn by examining naturally occurring mitochondrial genomes which have undergone chance recombination events that have placed extra, truncated fragments of editing genes into coding regions. Analysis of several such accidental chimeric mitochondrial genes has revealed that a relatively small region will still undergo editing even after moving into completely different surrounding sequences (16, 26, 30). Furthermore, when the transferred sequences exhibit small changes relative to the intact, endogenous gene, the Cs in the chimeric transcripts are not edited (13, 18). The mitochondrial in vivo data are consistent with results from electroporation of wheat mitochondria with RNAs carrying various lengths of sequences surrounding the C259 coxII editing site. For example, editing of C259 in electroporated mitochondria required 16 nucleotides 5′ and 6 nucleotides 3′ of the editing target site (9).
Several lines of evidence have led to the concept that site-specific trans-acting factors exist for each individual editing site. Foremost among these was the discovery that high-level expression of a transgenic RNA containing a psbL editing site resulted in decrease in editing of the C in the endogenous psbL transcripts but not in decreased editing in four other sites that were assayed (7). Furthermore, inspection of the sequences immediately surrounding C targets of editing has not revealed any obvious consensus. Because of the large number of editing sites in mitochondria, and by analogy to trypanosome editing, the existence of guide RNAs for specific editing site recognition has been hypothesized. However, efforts to detect guide RNAs biochemically (unpublished data) and genetically (5) have not been successful. Furthermore, data from a chloroplast in vitro editing system implicate protein trans factors rather than RNA factors (15). Taken together, this information suggests that more than 400 proteins would be required for editing-site recognition in plant organelles, if each site is independent. Because editing occurs in an albino mutant lacking chloroplast ribosomes, the editing apparatus is thought to be nucleus encoded (33).
We decided to reinvestigate the question of independence of editing at different sites in chloroplasts. We have previously reported the construction of chloroplast-transgenic plants carrying sequences surrounding either an rpoB editing site and or an ndhF editing site (21, 22). Recently, we developed a sensitive and reliable poisoned primer extension (PPE) assay for the extent of plant organelle RNA editing (20). We examined the extent of editing in the 31 known tobacco chloroplast editing sites in two different tobacco lines that were homoplastomic for either an introduced rpoB or ndhF transgene. Contrary to the theory of independent editing, we discovered that high-level expression of each gene reduced editing at certain other sites. When we compared the sequences surrounding the introduced sites with sequences around the sites where editing was impaired, we could detect conserved elements immediately 5′ of the C target of editing. Our data are consistent with the sharing of editing trans factors in chloroplasts, and we propose that an analogous mechanism occurs in mitochondria, where similar putative cis elements can also be detected 5′ of edited Cs.
MATERIALS AND METHODS
Plant material.
Tobacco plants (Nicotiana tabacum cv. Petit Havana) were grown under sterile conditions on MS-agar medium (19) containing 30 g of sucrose per liter. The transplastomic plants used in this study were described previously (21, 22).
RNA extraction, cDNA synthesis, and PCR.
Total RNA was extracted with an RNeasy plant minikit (Qiagen) and treated with a DNA-free kit (Ambion). DNA-free RNA (1.5 μg) was reverse transcribed for 1 h at 37°C with an Omniscript kit (Qiagen) using random hexamers. cDNA samples were amplified by a standard protocol (5 min at 94°C followed by 40 cycles of 94°C for 30 s, 50 to 55°C for 30 s, and 72°C for 1 min) in a PTC-200 thermal cycler (MJ Research).
Cloning.
PCR and RT-PCR products were cloned in the pCR2.1-TOPO vector (Invitrogen).
PPE.
PPE of RT-PCR products and determination of editing efficiency were conducted as previously described (20).
Primers.
Primers used for PCR and PPE are listed in Table 1.
TABLE 1.
Oligonucleotides used
| Name | Sequence (5′-3′) | Purposea |
|---|---|---|
| FAtpA | GAAAGGGGAGCGATGGAATACAC | PCR, atpA-1 and -2, 5′ |
| RAtpA | TTGAATAGGTCGGCGGATAAGAAG | PCR, atpA-1 and -2, 3′ |
| AtpA-1(T) | ATCGTGAACGACACACTTTAATCATTTATGATGAT | PPE, atpA-1 |
| AtpA-2(A) | GAGACATTTGGCGATAAGCTTGCGCTTG | PPE, atpA-2 |
| FATPF | GCCATCTGCCGGGAGTTTC | PCR, atpF-1, 5′ |
| RATPF | CGCCCTTTGCTGTTCAAACTG | PCR, atpF-1, 3′ |
| AtpF-1(T) | TTCGGGTTTAATACCGATATTTTAGCAACAAAT | PPE, atpF-1 |
| FndhA2 | AGCAGATGGGACAAAACTACTT | PCR, ndhA-2, 5′ |
| RndhA2 | CCAACGGGAGCAATACTT | PCR, ndhA-2, 3′ |
| NdhA-2(G) | GGCTAGAACAAGGTGATCACCAAAAGGAAT | PPE, ndhA-2 |
| FndhA5 | ATTTGGTTTATTTTACATTG | PCR, ndhA-5, 5′ |
| RndhA5 | TTACAGTGAAAGAAGTTGG | PCR, ndhA-5, 3′ |
| NdhA-5(T) | TATTTCTCTAGGTAATCTATTATTGACAACCTCGT | PPE, ndhA-5 |
| FndhB1 | TTTTTGGCCTAATTCTTCTTCTGA | PCR, ndhB-1 and -2, 5′ |
| RndhB1 | TTGCCCCACCCATGAGTAAATA | PCR, ndhB-1 and -2, 3′ |
| NdhB-1(G) | GAAATATAACCAAGGTATATCTTTTTGATCAGAGG | PPE, ndhB-1 |
| NdhB-2(A) | ATCCAGATAATAGGTAGGAGCATAAACTGAAACAT | PPE, ndhB-2 |
| FndhB2 | TACGGTCTAATGAGGCTACTA | PCR, ndhB-3, -4, -6, -7, -8, and -9, 5′ |
| RndhB2 | TCCCAATATCATGCTAAGAA | PCR, ndhB-3, -4, -6, -7, -8, and -9, 3′ |
| NdhB-3(G) | CCCCGGATGAACCATATAGCCAAGAGA | PPE, ndhB-3 |
| NdhB-4(G) | TTCTTGAAGCTCAATCTCTCCCCCGG | PPE, ndhB-4 |
| NdhB-6(T) | TCATTACCGTAGGAATTGGGTTCAAGCTTT | PPE, ndhB-6 |
| NdhB-7(C) | GGGTTCAAGCTTTCCCCAGCCCC | PPE, ndhB-7 |
| NdhB-8(C) | GTTGCTTTTCTTTCTGTTACTTCGAAAGTAGCTGC | PPE, ndhB-8 |
| NdhB-9(G) | TGAGAAATAAAAAGGAATATCGAAAATTCGAGTGG | PPE, ndhB-9 |
| FndhB3 | TCTTTAGCCCTATGTCTCTTATCC | PCR, ndhB-10, 5′ |
| RndhB3 | GGCTATCCTGAGCAATTGCA | PCR, ndhB-10, 3′ |
| NdhB-10(A) | CTGAGCAATTGCAATAATTGGGTTCATTGATA | PPE, ndhB-10 |
| FndhD | AATATTTTGAGCACGGGTTTTTA | PCR, ndhD-1 and -2, 5′ |
| RndhD | TGTGCTTCTCCATGGGTATCTG | PCR, ndhD-1 and -2, 3′ |
| NdhD-1(G) | TTGGAAAAACTACAATTATTGTTAACCAAGGAAAA | PPE, ndhD-1 |
| NdhD-2(G) | ATGATGAAAAAAAGTAAAAGGTCCCGAGACG | PPE, ndhD-2 |
| FndhF | ATGTTAATAGGAGCGGGACTTT | PCR, ndhF-2, 5′ |
| RndhF | TGGGCGGATTTAGCAACT | PCR, ndhF-2, 3′ |
| NdhF-2(T) | GATCGACCCACTTACTTCTATTATGT | PPE, ndhF-2 |
| FpetB | GTCGGCAAGTATGATGGTCCTA | PCR, petB-1, 5′ |
| RpetB | CTATAAAGGCCCAGAAATACCT | PCR, petB-1, 3′ |
| PetB-1(A) | CTATAAAGGCCCAGAAATACCTTGTTTACGTATCA | PPE, petB-1 |
| FpsbE | ATTCGATACTGGGTCATTCATAGC | PCR, psbE-1, 5′ |
| RpsbE | CTAAAACGATCTACTAAATTCATCGAG | PCR, psbE-1, 3′ |
| PsbE-1(G) | CTAAAACGATCTACTAAATTCATCGAGTTGTTCCAAAG | PPE, psbE-1 |
| FpsbL | TACCGTCTTTTTTTTGGGATC | PCR, psbL-1, 5′ |
| RpsbL | ATTTTGTTCGTTCGGGTTTGA | PCR, psbL-1, 3′ |
| PsbL-1(G) | AACATTTTGTTCGTTCGGGTTTGATTGTGT | PPE, psbL-1 |
| Frpl20 | AGAGATTTTCGTCGTTTGTG | PCR, rpl20-1, 5′ |
| Rrpl20 | ATTATTCTGGTGGATTCTTTC | PCR, rpl20-1, 3′ |
| Rpl20-1(G) | TATGATTTCGTTCGAAATCATATAAAGACAATTCC | PPE, rpl20-1 |
| FrpoA | GCTGTATTCATGCCTGTTCG | PCR, rpoA-1, 5′ |
| RrpoA | AATGCCCAATATCCGTTTTAC | PCR, rpoA-1, 3′ |
| RpoA-1(G) | GACATTTTGAGGCAATTATAGATCCTGGAAGG | PPE, rpoA-1 |
| FrpoB1 | GATAAAGGAAAGAGATGCTGTGTA | PCR, rpoB-1, -2, and -3, 5′ |
| RrpoB1 | TGTTCTGGGGTATATCAAGGTTC | PCR, rpoB-1, -2, and -3, 3′ |
| RpoB-1(G) | ATTTGATTGATCACAATTCTATATATTCCATTGAC | PPE, rpoB-1 |
| RpoB-2(C) | GGCACCATAATATCAGATTGGGGAGGAAG | PPE, rpoB-2 |
| RpoB-3(G) | CTCTAGAATTTCTCTTAGATTCAAACCCATAGCTG | PPE, rpoB-3 |
| FrpoB2 | ACTCCAGGTTCCTCGGGGTAAA | PCR, rpoB-6, 5′ |
| RrpoB2 | TTGCGGAGTAAATGGGCTTCTAA | PCR, rpoB-6, 3′ |
| RpoB-6(G) | ATCTTCATATACCAAACGCTCGCTAATAAGTACTG | PPE, rpoB-6 |
| FrpoC1 | CATCAACAGCTCCGAATTGGA | PCR, rpoC1-1, 5′ |
| RrpoC1 | TATGGGCCTAGCAAAAGAAAAATC | PCR, rpoC1-1, 3′ |
| RpoC1-1(G) | AGTGGCCCAAGCACTTATTTGTTGAGGAG | PPE, rpoC1-1 |
| FrpoC2 | AAAAATGGACCGCCCCTCAAA | PCR, rpoC2-2, 5′ |
| RrpoC2 | GCGCCCCATTCGTTCTGCT | PCR, rpoC2-2, 3′ |
| RpoC2-2(G) | GCTCGCAACAATCCAATAAGTTCTCCGGG | PPE, rpoC2-2 |
| Frps2 | AAGAGATGATGGAGGCAGGAGTTC | PCR, rps2-1, 5′ |
| Rrps2 | CTTTTCGGGAGACGGTTGAGT | PCR, rps2-1, 3′ |
| Rps2-1(G) | CGGGCCCTTATTGCAGCCCACT | PPE, rps2-1 |
| Frps14 | CAGAGGGAGAAGAAGAGGC | PCR, rps14-1 and -2, 5′ |
| Rrps14 | GCTCCTGGCAACAAACAT | PCR, rps14-1 and -2, 3′ |
| Rps14-1(C) | GGAACAGAAATATCATTCGATTCGTCGATCC | PPE, rps14-1 |
| Rps14-2(A) | CGATGAAGGCGTGTAGGTGCACTATTCC | PPE, rps14-2 |
| Fatp9 | GCGGGAGCTGCTATCGGTATT | PCR, atp9, 5′ |
| Ratp9 | ATCAAAAAGGCCATCATTAGG | PCR, atp9, 3′ |
| Atp9(C) | GCGGGAGCTGCTATCGGTATTGGAAAC | PPE, atp9 27-C3 |
| Fnad3 | CCAGCAAGGAGGGGAAGAACC | PCR, nad3, 5′ |
| Rnad3 | ACCGGAAGGATCGAAACCACATT | PCR, nad3, 3′ |
| Nad3(A) | AAGGAAGACCGAGTGGGAGCAAAGAAACTA | PPE, nad3 15-C2 |
| Frpl16 | TCAACCGAGGAACAAAACAAGAT | PCR, rpl16, 5′ |
| Rrpl16 | ACGATGGAAGTGCCCGATTAT | PCR, rpl16, 3′ |
| Rpl16(C) | CCTGCGGAAGTATCTACTCGTTACGGAATC | PPE, rpl16 13-C1 |
| PPrrn2 | AATACGAAGCGCTTGGATACAGTTGTAGGGA | PCR, transgene, 5′ |
| Trps16α3.1 | CTACCCCCCTTTTTGTATTTCCTTAATTTATTTCC | PCR, transgene, 3′ |
PCR, oligonucleotides used to amplify fragments containing editing sites; PPE, oligonucleotides used in PPE.
Sequences.
Information about the location of edited Cs is available in the works of Maier et al. (17) and Tsudzuki et al. (28).
RESULTS
Editing efficiency in wild-type tobacco leaf chloroplasts.
To date, 31 C-to-U editing sites have been found on the tobacco chloroplast genome. These sites are distributed on transcripts of 16 different genes. Table 2 lists all the editing sites using the nomenclature proposed by Tsudzuki et al. (28), which allows comparison between species.
TABLE 2.
RNA editing sites in tobacco chloroplasts
| Site | Positiona | Codonb | Amino acid change |
|---|---|---|---|
| atpA-1 | 791 | cCc | P to L |
| atpA-2 | 795 | ucC | None (S to S) |
| atpF-1 | 92 | cCa | P to L |
| ndhA-2 | 341 | uCa | S to L |
| ndhA-5 | 1073 | uCc | S to F |
| ndhB-1 | 149 | uCa | S to L |
| ndhB-2 | 467 | cCa | P to L |
| ndhB-3 | 586 | Cau | H to Y |
| ndhB-4 | 611 | uCa | S to L |
| ndhB-6 | 737 | cCa | P to L |
| ndhB-7 | 746 | uCu | S to F |
| ndhB-8 | 830 | uCa | S to L |
| ndhB-9 | 836 | uCa | S to L |
| ndhB-10 | 1481 | cCa | P to L |
| ndhD-1 | 2 | aCg | T to M |
| ndhD-2 | 383 | uCa | S to L |
| ndhF-2 | 290 | uCa | S to L |
| petB-1 | 611 | cCa | P to L |
| psbE-1 | 214 | Ccu | P to S |
| psbL-1 | 2 | aCg | T to M |
| rpl20-1 | 308 | uCa | S to L |
| rpoA-1 | 830 | uCa | S to L |
| rpoB-1 | 338 | uCu | S to F |
| rpoB-2 | 473 | uCa | S to L |
| rpoB-3 | 551 | uCa | S to L |
| rpoB-6 | 2000 | uCu | S to F |
| rpoC1-1 | 62 | uCa | S to L |
| rpoC2-2 | 3743 | uCa | S to L |
| rps2-1 | 248 | uCa | S to L |
| rps14-1 | 80 | uCa | S to L |
| rps14-2 | 149 | cCa | P to L |
Position (in nucleotides) is from the A of the initiator codon. Data are from the work of Tsudzuki et al. (28).
Edited nucleotides are in capitals.
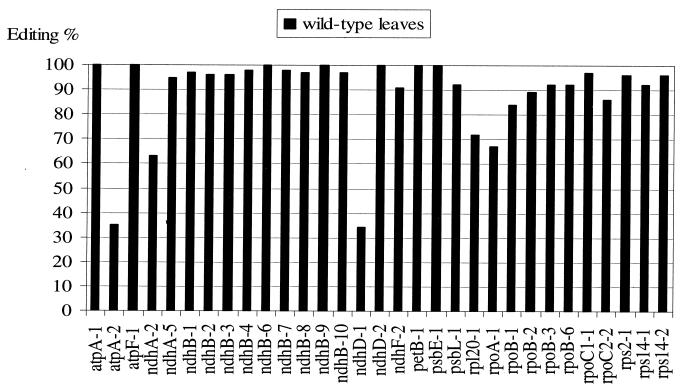
The editing efficiency of all editing sites was determined on transcripts isolated from leaves of 1-month-old tobacco plants (cv. Petit Havana). We used PPE to quantify the editing extent of each site. A minimum of two PPE assays were performed for each site, and a minimum of two different RNA preparations were assayed. No error bars are shown in Fig. 2 because the variations between samples and assays were very small, usually 1 to 2% and in no case greater than 5%.
FIG. 2.
Editing extent of 31 sites in young green leaf chloroplasts of wild-type tobacco. The editing percentage was determined for PPE products by quantifying the radioactivity associated with edited and unedited sites (ImageQuant software). The x axis represents the 31 editing sites listed in Table 2.
Like atpF-1 in Fig. 1, most sites are nearly fully edited in young tobacco leaves (Fig. 2). Some are edited at 80 to 90% (ndhF-2, psbL-1, rpoB-1, rpoB-2, and rpoC2-2), and five sites are clearly partially edited (atpA-2, ndhA-2, ndhD-1, rpl20-1, and rpoA-1). The lowest editing efficiency was found in atpA-2 and ndhD-1, which are edited at only 35 and 34%, respectively, in leaves of these plants. Editing at the atpA-2 site is silent, resulting in no change in the encoded amino acid. Low editing efficiencies of silent sites have also been observed in plant mitochondria (31). The low editing extent of ndhD transcripts means that the majority of ndhD transcripts are likely to be nontranslatable, as editing of the ndhD-1 site creates an AUG start codon.
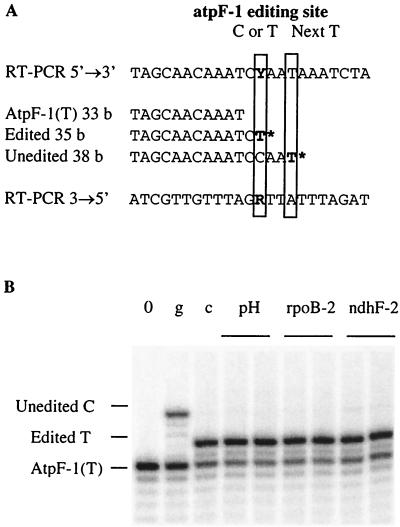
FIG. 1.
PPE assay of atpF-1. (A) PPE was performed on a site-specific RT-PCR product from a radiolabeled oligonucleotide. (A) Sequence surrounding atpF-1 and the 3′ part of atpF-1(T), the oligonucleotide used in this reaction. The primer extension is poisoned by dideoxynucleoside triphosphate incorporation, in this case a ddTTP (T*). When the site is edited, atpF-1(T) is extended only to the editing site, producing an extended oligonucleotide of 35 bases (b). When the site is not edited, the cDNA does not contain a T at the editing site, so atpF-1(T) is extended to the next T, producing a primer of 38 bases. (B) Separation on sequencing gel and phosphorimager exposure of the primer extension products. Lanes: pH, PPE from leaf extracts of wild-type tobacco; rpoB-2 and ndhF-2, PPE with transplastomic plants overexpressing rpoB-2 or ndhF-2; 0, PPE without template (this indicates the size of the radiolabeled oligonucleotide); g, PPE with a cloned unedited (genomic) PCR product; c, PPE with a cloned edited RT-PCR product.
Editing efficiency in transplastomic tobacco lines.
Transplastomic plants carrying sequences surrounding rpoB-2 or ndhF-2 as an additional copy in the plastid genome with an intact endogenous gene were previously generated (21, 22) and selected on antibiotic medium until homoplastomic. rpoB-2 tobacco carries a 92-nucleotide sequence from maize homologous to tobacco rpoB-2 (from −31 to +60); ndhF-2 tobacco carries a 35-nucleotide (from −27 to +7) sequence surrounding ndhF-2.
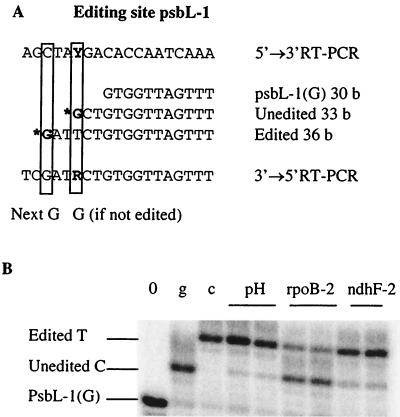
We previously reported that the minigenes in these lines are expressed at extremely high levels in comparison with the endogenous gene and that the editing efficiencies of the endogenous rpoB-2 and ndhF-2 sites were reduced (21, 22). When we analyzed 31 C-to-U editing sites, 23 showed no variation in editing extent in transplastomic lines compared to the wild-type tobacco, such as atpF-1 in Fig. 1. All the other sites exhibited an editing defect in one or the other of the two transplastomic lines, such as psbL-1 in Fig. 3.
FIG. 3.
Alteration of editing in transplastomic tobacco, determined by PPE assay of psbL-1. (A). Sequence surrounding psbL-1 and the 3′ part of psbL-1(G), the oligonucleotide used in this PPE. PPE is performed on a cDNA amplified with primers flanking psbL-1 from radiolabeled psbL-1(G). The primer extension is poisoned in this case by ddGTP (G*) incorporation. When the site is not edited, psbL-1(G) is extended only to the editing site, producing an extended oligonucleotide of 33 bases (b). When the site is edited, the complementary strand of the cDNA contains an A at the editing site, so psbL-1(G) is extended to the next G, producing a primer of 36 bases. (B). Separation on sequencing gel and phosphorimager exposure of the primer extension products. Lanes: pH, PPE with leaf extracts of wild-type tobacco; rpoB-2 and ndhF-2, PPE with transplastomic plants overexpressing rpoB-2 or ndhF-2; 0, PPE without template (this indicates the size of the radiolabeled oligonucleotide); g, PPE with a cloned unedited (genomic) PCR product; c, PPE with a cloned edited RT-PCR product.
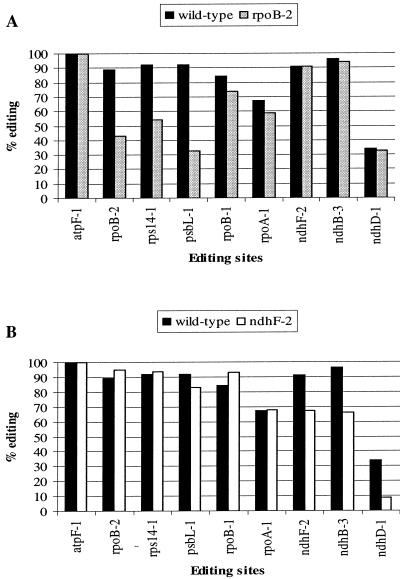
Five sites are less edited in rpoB-2 plants than in wild-type tobacco (Fig. 4A). In addition to a decrease in the editing level of the endogenous rpoB-2 (89 to 43%), a large reduction was also detected for psbL-1 (92 to 33%) and rps14-1 (92 to 54%). A weaker effect was observed for rpoA-1 (67 to 59%) and rpoB-1 (84 to 74%).
FIG. 4.
Competition effect. Overexpression of the rpoB-2 (A) or ndhF-2 (B) minigene induced a decrease in editing level in endogenous rpoB-2, rps14-1, psbL-1, rpoB-1, and rpoA-1 (A) and in endogenous ndhF-2, ndhB-3, and ndhD-1 (B).
Overexpression of ndhF-2 minigenes leads to a decrease in editing efficiency of three sites (Fig. 4B). The endogenous ndhF-2 editing level was reduced from 91 to 67%, editing of ndhB-3 decreased from 96 to 66%, and editing of ndhD-1 decreased from 34 to 9%. Thus, we observed a competition effect at sites other than the endogenous site that carried sequences identical to the transgene.
Identification of conserved sequences upstream of editing sites.
By analyzing the sequences surrounding the editing sites for which we observed a competition effect, we found conserved sequences that were absent in all other sites (Fig. 5). The −27/+7 sequence surrounding ndhF-2 present in the minigene is sufficient for the transgene transcripts to be edited at the usual C and for a competition effect to exist with ndhB-3 and ndhD-1. A block of eight nucleotides was found to be identical in the upstream sequence of ndhF-2 and ndhB-3, and six of these eight nucleotides were also present 5′ of ndhD-1. This conserved block, CUUxUxUU, could be a cis-acting element required for recognition by a factor operating at all three sites.
FIG. 5.
Putative cis-acting elements conserved in the upstream sequence of the editing sites showing a competition effect. Sites in bold are those introduced into the tobacco chloroplast genome. Gaps were added in the alignments to show similarity. Bold letters represent conserved nucleotides. Blocks of conserved nucleotides are shaded. Large capital Cs show C targets of editing. Nt, N. tabacum; Zm, Zea mays.
Likewise, we were able to detect sequence similarities between rpoB-2 and the sequences surrounding the four editing sites affected by overexpression of the −31/+60 rpoB-2 sequence (Fig. 5). Interestingly, one of the conserved nucleotides is an A at −20 that we previously reported to be important in editing of a minigene carrying −20/+6 rpoB-2 sequences. Transcripts of a −20/+6 rpoB-2 minigene are edited at 20% efficiency, but transcripts of a minigene in which the −20 A is replaced with T are not edited (23). As in the ndhF-2 cluster, the spacing of the conserved nucleotides varies between editing sites (Fig. 5). The five sequences all exhibit either an A or U immediately preceding the C target of editing (Fig. 5). Previously, Chaudhuri et al. (8) reported that altering the psbL-1 A to C resulted in loss of editing; perhaps either an A or U is required at this position (Fig. 5).
Similarities in the upstream sequences of other editing sites of tobacco chloroplast genome can also be detected. Clusters of two or three sequences emerge from alignments (Fig. 6A). For instance, a block of eight conserved residues, AAUUGGAU, is located in the nearest 5′ neighborhood of ndhD-2 and rpoC1-1; atpA-2 and ndhA-5 share a block of eight nucleotides out of nine in the 5′ vicinity of the C. To determine whether such editing-site sequence clusters might also exist in a monocot, we examined the sequences surrounding the 27 editing sites of the maize chloroplast genome. Clusters of two or three sites sharing some putative elements can be formed (Fig. 6B). A striking block of nine nucleotides, AAGUAGCUG, was found in the upstream sequence of atpA-3, ndhB-8, and rpl20-1.
FIG. 6.
Chloroplast editing sites in tobacco (Nt) (A) and in maize (Zm) (B) sharing short sequences in their upstream regions. (C) Similarities and differences in sequences upstream of edited Cs in tobacco and the corresponding C or T in Arabidopsis thaliana (At). Gaps were introduced in the alignments to show similar sequences. Bold letters represent conserved nucleotides, and blocks of conserved nucleotides are shaded. Large capital Cs show C targets of editing.
If the conserved nucleotides upstream of C targets of editing are important for editing-site recognition in one species, then their presence should be nonessential in a species that has a genomic T rather than a C at the site. We show two comparisons of sequences surrounding edited Cs in tobacco that are not edited in Arabidopsis due to the presence of a genomic T. In both rpoB-2 and psbL-1, a conserved nucleotide present in a tobacco editing-site cluster is not conserved in the unedited Arabidopsis transcript (Fig. 6C). However, when editing occurs in both Arabidopsis and tobacco, the conserved nucleotides are present in both species. The conserved nucleotide sequences upstream of ndhF-2, ndhB-3, and ndhD-1 exist in both tobacco (Fig. 5) and Arabidopsis (Fig. 6C), indicating that this is a conserved editing-site cluster in these two dicots.
Comparison of 5′ sequences between chloroplasts and mitochondria.
We wondered whether an analogous situation might exist in mitochondria, in which multiple sites share cis elements and therefore possibly recognition factors. We considered whether mitochondrial sites might exhibit some of the same putative 5′ elements and therefore possibly be recognized by a nucleus-encoded factor targeted to both organelles. We compared sequence data available in the tobacco and closely related petunia genera and discovered that elements similar to those found in the rpoB-2 site could be detected in several mitochondrial genes (Fig. 7).
FIG. 7.
Mitochondrial editing sites with upstream regions exhibiting similarity to either rpoB-2 or ndhF-2. These editing sites are known to be edited in tobacco (N. tabacum [Nt]) or in a related species (Nicotiana sylvestris [Ns]) or genus (Petunia hybrida [Ph]). Mitochondrial editing sites are named x-C1, where x represents the codon position and 1 is the location of the C within the codon. Underlined sites are those tested for a competition effect. The asterisk in coxII-2 shows the first editing site, 148-C2, of exon 2, which must be edited to U in order to form the conserved nucleotide. Site names in bold are those introduced in tobacco chloroplast genome (Zm, Zea mays). Gaps were introduced in the alignments to illustrate similarities. Bold letters represent conserved nucleotides, and blocks of conserved nucleotides are shaded. Large capital Cs show C targets of editing.
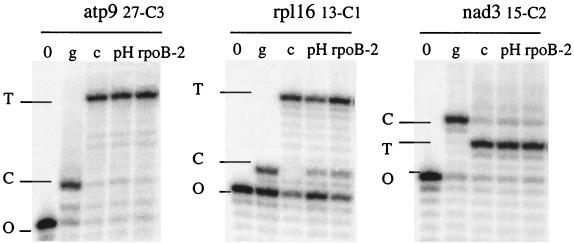
If the same factor is dually targeted to both organelles, then overexpression in chloroplasts would not be expected to affect editing in mitochondria, unless some feedback mechanism causes enhanced distribution of the factor to the transgenic chloroplasts or alters synthesis of the factor. To investigate this possibility, we assayed editing of three sites in mitochondrial transcripts of atp9, rpl16, and nad3 in the chloroplast transgenic plants carrying the rpoB-2 minigene. These three sites carry elements similar to those present in the rpoB-2 competition cluster (Fig. 7). Nevertheless, the extent of editing was similar in the mitochondria from the wild type and the rpoB-2 transformants (Fig. 8).
FIG. 8.
Editing in mitochondrial sites is not altered in tobacco overexpressing rpoB-2 despite sequence similarities in their upstream sequences. A PPE assay was performed on atp9 27-C3, rpl16 13-C1, and nad3 15-C2. Lanes: pH PPE with leaf extracts of wild-type tobacco; rpoB-2, PPE with transplastomic plants overexpressing rpoB-2; 0, PPE without template showing the radiolabeled oligonucleotide (O); g, PPE with a cloned unedited (C) PCR product; c, PPE with a cloned edited (T) RT-PCR product.
DISCUSSION
When editing of a large number of edited Cs was discovered in plant mitochondria, there was speculation as to whether editing factors specific to each individual editing site exist or whether several editing sites that exhibit limited sequence similarity might be recognized by a single factor, such as a guide RNA (12). Unfortunately, this possibility has not been testable in plant mitochondria because of the absence of mitochondrial transformation methods and the lack of a reliable extract capable of supporting editing in vitro. In chloroplasts, because overexpression of a single editing site in transgenic plants was observed to affect only the editing extent of the homologous gene's transcripts (7), it has become generally accepted that there is a single factor for each chloroplast editing site. However, previous studies were incomplete in that they examined editing of only a few endogenous genes' transcripts in addition to the transcripts of the chloroplast gene homologous to the overexpressed transgene. Our finding that high-level expression of one editing site decreases editing at several other sites favors the hypothesis of families of editing sites in plant organelles. Based on cross-competition, it is likely that one editing factor is responsible for recognition of several sites, though our data do not exclude the possibility that particular editing factors exist that operate only on a single site. Binding of chloroplast proteins to 5′ untranslated regions also exhibited cross-competition when assayed in gel shift experiments (24). Thus, the action of RNA-binding proteins on multiple sequences may not be unusual in plant organelle gene regulation.
Sharing of trans factors may help explain how new editing sites evolve. How an editing apparatus could arise that would permit editing of many hundreds of plant mitochondrial editing sites has particularly been puzzling. If one factor is responsible for multiple chloroplast and/or mitochondrial editing sites, models for evolution of editing are more readily formulated. If each of the nine putative editing-site clusters shown in Fig. 5 and 6 is recognized by one factor per cluster, then we need to invoke the existence of only nine chloroplast editing factors rather than the 20 factors predicted by the one-factor-per-site independent-editing model. The number of required mitochondrial editing recognition factors could also be greatly reduced by action of the same factor on multiple sites. In addition to the gene cluster shown in Fig. 7, a number of other groups of mitochondrial sequences with some conserved 5′ elements can be detected by sequence inspection (data not shown).
The similarities between chloroplast and mitochondrial sites suggests one scenario for the evolution of new editing sites. If an editing site, recognized by a nucleus-encoded factor, has arisen in the mitochondrion, a low level of mistargeting could allow some of the protein to enter the chloroplast. If the editing factor by chance recognizes a chloroplast sequence, a T-to-C mutation could then be tolerated as a result of a low level of editing by the dually targeted factor. Selection pressure to optimize editing efficiency could then result in further alteration of the cis-acting chloroplast elements or in the nucleus-encoded factor. Similarly, a chloroplast editing factor might “leak” into the mitochondrion and result in recognition of new editing sites. Likewise, within the same organelle, once an editing factor recognizes one site, either DNA mutations at other sites near potential C targets of editing could allow recognition of new sites, or chance similarities could result in editing at multiple sites.
Though we observed that excess rpoB-2 causes a significant reduction in psbL-1 editing, when Chaudhuri et al. (7) overexpressed psbL-1 in transgenic tobacco, using the same minigene regulatory sequences, they did not detect any effect on editing of rpoB-2. There are several possible reasons for this discrepancy. First, Chaudhuri et al. did not provide data assessing the degree of homoplasmicity of the transgenic plants they examined. If their plants contained a mixture of transformed and untransformed chloroplasts, reduction in rpoB-2 editing may not have been detectable because the wild-type chloroplasts present would have normal, high levels of rpoB-2 editing. Second, the method Chaudhuri et al. used to examine rpoB-2 editing was bulk sequencing of amplified cDNAs, a technique less accurate than the PPE method we employed. Third, it is possible that a trans factor that recognizes both psbL-1 and rpoB-2 has higher affinity for the rpoB-2 site, so that rpoB-2 is preferentially edited with respect to psbL-1 despite the presence of excess psbL-1 editing sites in transgenic chloroplasts.
The competition effect that we observed at sites other than rpoB-2 and ndhF-2 allowed us to detect putative cis-acting editing motifs. Without the functional information that now allows us to group different editing sites into units, it was not previously possible to detect conserved nucleotides with any confidence. Detecting the common elements we describe (Fig. 5) also would have been difficult because of the lack of conservation of spacing from the editing site to the conserved nucleotides. As a result, experiments to test the importance of particular nucleotides have required guesswork, with no sound basis for selecting particular nucleotides to be altered for in vivo or in vitro assays. The conserved sequences in clustered sites we report here are obvious targets for future mutational analysis. Nevertheless, the conserved 5′ nucleotides are not the only ones essential to editing. At some sites, nucleotides 3′ of the edited C have been found to be necessary. For example, in tobacco psbL, a minigene with sequence from −63 to +10 was 70% edited but a −62/+1 minigene transcript was not edited at all in vivo (7).
A high percentage of AU was observed among the conserved nucleotides found upstream of target Cs. In tobacco mRNAs, A and U represent 73% of the conserved residues in the region 5′ of edited Cs. The AT content of the tobacco genome is 62%, so the presence of 73% AU is significantly different from the average. Motifs in mRNA recognized by a variety of RNA binding proteins are often AU rich (6, 32). The mammalian C-to-U editing enzyme Apobec-1 has been shown to bind with high affinity to an AU-rich consensus sequence (1). Furthermore, AU-rich sequences are known to be important in chloroplast RNA cleavage and stability (25) and in chloroplast translation (14).
We were able to detect sequences 5′ of several mitochondrial genes that are highly similar to the conserved sequences in the rpoB-2 competition cluster (Fig. 5 and 7). Though we detected no reduction in mitochondrial editing due to overexpression of an editable transcript in the chloroplast, the question of whether the same factor might sometimes recognize both chloroplast and mitochondrial sites remains. A number of nucleus-encoded proteins are known to be targeted to both chloroplasts and mitochondria. Arguing against sharing of factors is an experiment in which mitochondrial sequences were introduced into transgenic chloroplasts (27). None of the seven sites in exon 2 of coxII were edited when introduced into chloroplasts, even though one of the sites (Fig. 7) has some of the conserved sequence elements detected in the rpoB-2 competition cluster. However, in the coxII 154-C2 site, which is similar to the rpoB-2 sequence (Fig. 7), the conserved U in the −19 AU sequence must be created by editing. If the C at −18 is not edited in chloroplasts, then the downstream C may not be recognizable even if an rpoB-2-like factor is present. Nevertheless, because mitochondrial transcripts undergo editing at many more sites than those of chloroplasts, not all recognition factors are likely to be shared between plastids and mitochondria. Determining whether a subset of editing factors are dually targeted and function in both organellar compartments will require further experimentation.
Acknowledgments
We thank Martha Reed and Sangbom Lyi for the transplastomic tobacco plants and Nemo Peeters for instruction in PPE.
This work was supported by NIH grant R01GM50723 to M.R.H.
REFERENCES
- 1.Anant, S., and N. O. Davidson. 2000. An AU-rich sequence element (UUUN[A/U]U) downstream of the edited C in apolipoprotein B mRNA is a high-affinity binding site for Apobec-1: binding of Apobec-1 to this motif in the 3′ untranslated region of c-myc increases mRNA stability. Mol. Cell. Biol. 20:1982-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bock, R. 2002. RNA editing in plant mitochondria and chloroplasts, p. 38-60. In B. L. Bass (ed.), RNA editing. Oxford University Press, Oxford, England.
- 3.Bock, R. 2000. Sense from nonsense: how the genetic information of chloroplasts is altered by RNA editing. Biochimie 82:549-557. [DOI] [PubMed] [Google Scholar]
- 4.Bock, R., M. Hermann, and H. Kossel. 1996. In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J. 15:5052-5059. [PMC free article] [PubMed] [Google Scholar]
- 5.Bock, R., and P. Maliga. 1995. In vivo testing of a tobacco plastid DNA segment for guide RNA function in psbL editing. Mol. Gen. Genet. 247:439-443. [DOI] [PubMed] [Google Scholar]
- 6.Burd, C. G., and G. Dreyfuss. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265:615-621. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri, S., H. Carrer, and P. Maliga. 1995. Site-specific factor involved in the editing of the psbL mRNA in tobacco plastids. EMBO J. 14:2951-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhuri, S., and P. Maliga. 1996. Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J. 15:5958-5964. [PMC free article] [PubMed] [Google Scholar]
- 9.Farre, J. C., G. Leon, X. Jordana, and A. Araya. 2001. cis recognition elements in plant mitochondrion RNA editing. Mol. Cell. Biol. 21:6731-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giege, P., and A. Brennicke. 2001. From gene to protein in higher plant mitochondria. C. R. Acad. Sci. Ser. III 324:209-217. [DOI] [PubMed] [Google Scholar]
- 11.Giege, P., and A. Brennicke. 1999. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA 96:15324-15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gualberto, J. M., J. H. Weil, and J. M. Grienenberger. 1990. Editing of the wheat coxIII transcript: evidence for twelve C to U and one U to C conversions and for sequence similarities around editing sites. Nucleic Acids Res. 18:3771-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanson, M. R., C. A. Sutton, and B. Lu. 1996. Plant organelle gene expression: altered by RNA editing. Trends Plant Sci. 1:57-64. [Google Scholar]
- 14.Hirose, T., and M. Sugiura. 1996. Cis-acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: development of an in vitro translation system from tobacco chloroplasts. EMBO J. 15:1687-1695. [PMC free article] [PubMed] [Google Scholar]
- 15.Hirose, T., and M. Sugiura. 2001. Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system. EMBO J. 20:1144-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubo, N., and K. Kadowaki. 1997. Involvement of 5′ flanking sequence for specifying RNA editing sites in plant mitochondria. FEBS Lett. 413:40-44. [DOI] [PubMed] [Google Scholar]
- 17.Maier, R. M., P. Zeltz, H. Kossel, G. Bonnard, J. M. Gualberto, and J. M. Grienenberger. 1996. RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 32:343-365. [DOI] [PubMed] [Google Scholar]
- 18.Mulligan, R. M., M. A. Williams, and M. T. Shanahan. 1999. RNA editing site recognition in higher plant mitochondria. J. Hered. 90:338-344. [DOI] [PubMed] [Google Scholar]
- 19.Murashige, T., and F. Skoog. 1962. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant 15:473-479. [Google Scholar]
- 20.Peeters, N. M., and M. R. Hanson. 2002. Transcript abundance supercedes editing efficiency as a factor in developmental variation of chloroplast gene expression. RNA 8:497-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed, M. L., and M. R. Hanson. 1997. A heterologous maize rpoB editing site is recognized by transgenic tobacco chloroplasts. Mol. Cell. Biol. 17:6948-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed, M. L., S. M. Lyi, and M. R. Hanson. 2001. Edited transcripts compete with unedited mRNAs for trans-acting editing factors in higher plant chloroplasts. Gene 272:165-171. [DOI] [PubMed] [Google Scholar]
- 23.Reed, M. L., N. M. Peeters, and M. R. Hanson. 2001. A single alteration 20 nt 5′ to an editing target inhibits chloroplast RNA editing in vivo. Nucleic Acids Res. 29:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robida, M. D., P. M. Merhige, and M. J. Hollingsworth. 2002. Proteins are shared among RNA-protein complexes that form in the 5′ untranslated regions of spinach chloroplast mRNAs. Curr. Genet. 41:53-62. [DOI] [PubMed] [Google Scholar]
- 25.Rott, R., V. Liveanu, R. G. Drager, D. Higgs, D. B. Stern, and G. Schuster. 1999. Altering the 3 UTR endonucleolytic cleavage site of a Chlamydomonas chloroplast mRNA affects 3-end maturation in vitro but not in vivo. Plant Mol. Biol. 40:679-686. [DOI] [PubMed] [Google Scholar]
- 26.Smith, H. C., J. M. Gott, and M. R. Hanson. 1997. A guide to RNA editing. RNA 3:1105-1123. [PMC free article] [PubMed] [Google Scholar]
- 27.Sutton, C. A., O. V. Zoubenko, M. R. Hanson, and P. Maliga. 1995. A plant mitochondrial sequence transcribed in transgenic tobacco chloroplasts is not edited. Mol. Cell. Biol. 15:1377-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsudzuki, T., T. Wakasugi, and M. Sugiura. 2001. Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol. 53:327-332. [DOI] [PubMed] [Google Scholar]
- 29.Wakasugi, T., T. Tsudzuki, and M. Sugiura. 2001. The genomics of land plant chloroplasts: gene content and alteration of genomic information by RNA editing. Photosynth. Res. 70:107-118. [DOI] [PubMed] [Google Scholar]
- 30.Williams, M. A., B. M. Kutcher, and R. M. Mulligan. 1998. Editing site recognition in plant mitochondria: the importance of 5′-flanking sequences. Plant Mol. Biol. 36:229-237. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, R. K., and M. R. Hanson. 1996. Preferential RNA editing at specific sites within transcripts of two plant mitochondrial genes does not depend on transcriptional context or nuclear genotype. Curr. Genet. 30:502-508. [DOI] [PubMed] [Google Scholar]
- 32.Xu, N., C. Y. Chen, and A. B. Shyu. 1997. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell. Biol. 17:4611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeltz, P., W. R. Hess, K. Neckermann, T. Borner, and H. Kossel. 1993. Editing of the chloroplast rpoB transcript is independent of chloroplast translation and shows different patterns in barley and maize. EMBO J. 12:4291-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]