Abstract
The yeast prion [PSI+] provides an epigenetic mechanism for the inheritance of new phenotypes through self-perpetuating changes in protein conformation. [PSI+] is a nonfunctional, ordered aggregate of the translation termination factor Sup35p that influences new Sup35 proteins to adopt the same state. The N-terminal region of Sup35p plays a central role in prion induction and propagation. The C-terminal region provides translation termination activity. The function of the highly charged, conformationally flexible middle region (M) is unknown. An M deletion mutant was capable of existing in either the prion or the nonprion state, but in either case it was mostly insoluble. Substituting a charged synthetic polypeptide for M restored solubility, but the prions formed by this variant were mitotically very unstable. Substituting charged flexible regions from two other proteins for M created variants that acquired prion states (defined as self-perpetuating changes in function transferred to them from wild-type [PSI+] elements), but had profoundly different properties. One was soluble in both the prion and the nonprion form, mitotically stable but meiotically unstable, and cured by guanidine HCl but not by alterations in heat shock protein 104 (Hsp104p). The other could only maintain the prion state in the presence of wild-type protein, producing Mendelian segregation patterns. The unique character of these M variants, all carrying the same N-terminal prion-determining region, demonstrate the importance of M for [PSI+] and suggest that a much wider range of epigenetic phenomena might be based on self-perpetuating, prion-like changes in protein conformation than suggested by our current methods for defining prion states.
Yeast prions represent a fundamentally different mechanism for the transmission of genetic information than DNA based inheritance. With prions, heritable changes in phenotype are produced by self-perpetuating changes in protein conformation rather than by any changes in nucleic acids (1–3). [PSI+] and other genetic elements of this type are called prions because of conceptual similarities between their modes of transmission and that postulated for the infectious agent of the mammalian prion diseases (1, 4). However, yeast prions play a different role in the biology of cells that harbor them. They are not generally pathogenic. Rather, they modify metabolism in an epigenetic manner that can be beneficial to the organism under certain circumstances (5, 6).
The protein determinant of [PSI+] is Sup35, a subunit of the translation termination factor (7). In [psi−] cells, which lack the prion, Sup35 protein (Sup35p) is soluble and functional. In [PSI+] cells, most Sup35p is found in self-perpetuating, ordered aggregates. In this state, the protein is nonfunctional. The reduced concentration of functional translation-terminator factor causes ribosomes to occasionally read-through stop codons (2, 3). Thus, the presence of [PSI+] is routinely monitored by suppression of nonsense-codon mutations in auxotrophic markers (8). The phenotype is heritable because Sup35p in the [PSI+] state influences newly synthesized Sup35p to adopt the same state, and because the protein is passed from mother cell to daughter during mitosis. When the daughter cell starts to make her own Sup35 proteins, they are influenced by preexisting [PSI+] complexes (inherited from the mother's cytoplasm) to undergo conformational conversion. Thus, the change in Sup35p function is inherited cytoplasmically.
Sup35p can be divided into three regions based on sequence analysis and functional investigations. The C-terminal region (C, amino acids 254–685) is responsible for the translation termination activity and is essential for viability (9–12). The N-terminal region (N, amino acids 1–123) is required for the induction and maintenance of [PSI+] (11–13). Deletion of N eliminates [PSI+], whereas even transient over expression of N induces [PSI+] (12). N is also responsible for the species barrier: in chimeric Sup35 proteins created from different species, the prion state is efficiently transferred only between proteins that share the same critical region of N (14–16). The role of the region between N and C (M, amino acids 124–253) remains unclear.
In inter-specific comparisons of Sup35p amino acid sequences, N and M are less conserved than C (7, 17). However, general features of these regions have been retained for long periods of evolution (14–16, 18). N regions from even distantly related Hemiascomycetes are rich in glutamine and asparagine residues (16, 19). M regions are highly charged, and their sequences are heavily biased toward a subset of charged amino acids (9, 16, 18, 19). In Saccharomyces cerevisiae, 42% of the residues in M are charged. All positively charged residues are lysines, and these cluster at the N-terminal end of M. The negatively charged residues, mostly glutamates, are concentrated at the C-terminal end.
Although [PSI+] is inherited in an orderly way, both mitotically and meiotically, it is metastable. [PSI+] cells occasionally give rise to [psi−] cells and vice versa as the [PSI+] conformation is lost or gained (20). The rate at which [PSI+] elements are lost greatly increases during growth on media containing guanidine hydrochloride (Gdn⋅HCl) (21, 22). The inheritance of [PSI+] is also modulated by several protein chaperones (23–26). This effect is most striking with heat shock protein 104 (Hsp104p), a protein remodeling factor. Both overexpression and deletion of the HSP104 gene cure cells of [PSI+] (23). Hsp104p must be present at an intermediate concentration for [PSI+] propagation.
The ability of Sup35p to exist in two stable and heritable conformations is fundamental to the conversion of cells from [psi−] to [PSI+] and vice versa. It is known that the crucial features of Sup35p required for this transition are confined to N and M: transferring N and M to a heterologous protein is sufficient to confer all of the prion behaviors of [PSI+] to that hybrid protein (27). Moreover, the transition from the nonprion to the prion state in vivo has been modeled in vitro by using the purified NM fragment of Sup35p. This polypeptide can exist for extended periods in the soluble state with a high degree of random coil and converts to a β-sheet-rich amyloid by seeded polymerization (28–30). To date, all of the critical elements for prion induction and propagation have been mapped to N. However, the M region (amino acids 124–253) is critical for solubility of NM in vitro. M alone remains soluble for months and cannot be seeded by preformed amyloids. On the other hand, N alone is soluble only in denaturing buffers (29, 31). This observation would suggest that M might play an important role in prion biology but its function has not been investigated in vivo. We created several alterations in the M region. They produced very different effects, demonstrating that M plays crucial roles in [PSI+] biology. In fact, the self-perpetuating prion states of these altered Sup35 proteins are so strikingly different from those of wild-type (WT) protein, they suggest that a much broader range of behaviors might involve prion-like changes in proteins than has previously been suspected.
Materials and Methods
Strains, Cultivation Procedures, and Genetic Analysis.
The [PSI+] and [psi−] isogenic strain pair used were 74-D694a [MATa, ade1-14(UGA), trp1-289(UAG), his3Δ-200, ura3-52, leu2-3,112] and 74-D694α [MATα, ade1-14(UGA), trp1-289(UAG), his3Δ-200, ura3-52, leu2-3,112]. The HSP104 disruption strain used was 74-D694:hspΔ-1L [ade1-14(UGA), trp1-289(UAG), his3Δ-200, ura3-52, leu2-3,112, hsp104∷LEU2]. [ETA+] strain: SL1010-1A [ETA+] [psi−] [ade1-14 (UGA), his 5-2, met8-1, ura3-52, leu2-1(UAA) trp1 (1 or 289)] (The [ETA+] strain was a kind gift from S. W. Liebman, University of Illinois, Chicago).
Cells were grown at 30°C in rich media [yeast extract/peptone/dextrose (YPD)] until mid-logarithmic phase (32). [PSI+] was monitored by its ability to suppress the ade1-14 (UGA) mutation and allow cells to grow on synthetic medium (SC) lacking adenine (−ADE). The 74-D694 [psi−] cells are red and [PSI+] cells are white or pink on rich medium (13, 23). [PSI+] curing on medium containing Gdn⋅HCl, CuSO4 induction of [PSI+], and GFP fluorescence analysis of Sup35p aggregates were performed as described (33). Standard procedures for crossing, sporulation, and tetrad analysis were used (34).
Plasmid Construction.
The SUP35 M deletion (amino acids 124–253) was generated by site-directed mutagenesis using oligonucleotide primers and a QuikChange Site-Directed Mutagenesis kit (Stratagene). The primers used were 5′-CAC AGT CTC AAG GTA TGT TTG GTG GTA AAG-3′ and 5′-CTT TAC CAC CAA ACA TAC CTT GAG ACT GTG-3′. The template plasmid for mutagenesis was p316CUPSup35GFPSG. The middle region-deleted SUP35 gene fragment was then PCR amplified and inserted at the EcoRI/BamHI sites of the SUP35 integrative construct pJLI-sup35. PCR primers were as follows: primer A 5′-CG GAA TTC ATG TCG GAT TCA AAC CAA GG-3′ and primer B 5′-CGC GGA TCC TTA CTC GGC AAT TTT AAC-3′. Thus, in the resultant construct pJLI-sup35ΔM, amino acid 123 was fused directly to amino acid 254.
The KDG tripeptide repeats were inserted between SUP35 N and C by site-directed mutagenesis. Primers were as follows: 5′-CCA CAG TCT CAA GGT AAG GAT GGT AAG GAT GGT AAG GAT GGT AAG GAT GGT AAG GAT GGT AAG GAT GGT TTT GGT GGT AAA GAT-3′ and 5′-ATC TTT ACC ACC AAA ACC ATC CTT ACC ATC CTT ACC ATC CTT ACC ATC CTT ACC ATC CTT ACC ATC CTT ACC TTG AGA CTG TGG-3′. The SUP35 N-KDG6-C fragment was then PCR amplified and inserted into the SUP35 integrative construct pJLI-SUP35 at the EcoRI/BamHI sites.
The N9C substitution was created by amplifying the region of Hsp90p coding for amino acids 210–262 from genomic 74D-694 DNA by using the primers 5′-CAT CTA GAA AGG AAG TTG AAA AGG AAG-3′ and 5′-CTC CGC GGC TTG TTT AGT TCT TCT ATC TC-3′. The resulting fragment was inserted into pN164 by using the SacII and XbaI restriction sites. The NTC substitution was created by using the same strategy after amplification of amino acids 635–713 of human topoisomerase I from plasmid pKM10 (a kind gift from J. Champoux, University of Washington, Seattle). The primers used were 5′-GAT CTA GAG CAC CAC CAA AAA CTT TTG AG-3′ and 5′-CAC CGC GGC ACT GTG ATC CTA GGG TCC AG-3′. Fidelity of all constructs was confirmed by sequencing.
Yeast Transformation.
One milliliter of overnight culture was pelleted and resuspended in 0.1 ml LiAc-PEG-TE solution (40% wt/vol PEG 4000/100 mM LiAc/10 mM Tris⋅HCl, pH 7.5/1 mM EDTA). Cells were mixed with 10 μg/μl carrier DNA (salmon testis DNA, boiled for 10 min and rapidly chilled on ice) and 0.5 μg/μl transforming DNA and incubated overnight at room temperature. Cells were heat shocked at 42°C for 15 min, rapidly chilled on ice, spread directly onto synthetic defined medium selective for transformation, and incubated at 30°C.
Gene Replacement.
The two-step gene integration and replacement procedure was used to create M mutant strains as described (34, 35). Genotypes were confirmed by genomic PCR, and protein expression was confirmed by immunoblotting.
To disrupt HSP104 in [PSI+]ΔM and [PSI+]NKC strains, an hsp104∷URA3 disruption plasmid (pYSU2) was inserted in the genome. The plasmid was digested with EcoRI/HindIII to release the hsp104∷URA3 fragment, and the restriction digest was used to transform yeast cells. Transformants were selected on SC-URA medium, and the HSP104 disruption was confirmed by immunoblotting with anti-Hsp104p antibodies (mix of monoclonal 8-1 and 2-3; ref. 36). HSP104 was disrupted in [PSI+]NTC and SUP35N9C backgrounds by the short flanking homology method as described (37).
Sup35p Solubility Assay.
Cells grown to mid-logarithmic phase in liquid YPD medium were washed with 0.1 M EDTA (pH 8.0), resuspended in spheroplasting buffer (1.0 M sorbitol/0.1 M EDTA, pH 7.5/50 mM DTT/0.166 mg/ml zymolyase 100 T) and incubated at 30°C for 1 h. Cells were collected, washed with the spheroplasting buffer without zymolyase, and lysed by resuspension in lysis buffer (50 mM Tris⋅HCl, pH 7.2/10 mM KCl/100 mM EDTA/1 mM DTT/0.2% wt/vol SDS/1% vol/vol Triton X-100/1 mM benzamidine/2 mM phenylmethylsulfonyl fluoride/10 μg/ml leupeptin/2 μg/ml pepstatin A) with incubation on ice for 5 min. Lysates were subjected to centrifugation at 100,000 × g (48,000 rpm in a TLA-100 rotor, Beckman Coulter) for 20 min at 4°C. Supernatant and pellet fractions were analyzed by SDS/10% PAGE (NOVEX, San Diego) and transferred to a poly(vinylidene difluoride) membranes (Millipore). Membranes were incubated with primary antibody (antiSup35p- rabbit polyclonal antipeptide against amino acids 55–68) 1:1,000 in PBS buffer with 0.1% Tween 20 for 1 h, washed with PBS with 0.1% Tween 20, and incubated with goat anti-rabbit antibody conjugated to horseradish peroxidase (1:5,000), and immune complexes were visualized with enhanced chemiluminescence (ECL) reagent (Amersham Pharmacia).
Results
Sup35ΔMp Is Functional and Mostly Insoluble but Can Exist in Both Prion and Nonprion States.
Starting with [psi−] cells, the WT copy of SUP35 was replaced with a gene carrying a deletion of the middle region (SUP35ΔM). The strain contained a [PSI+]-suppressible nonsense mutation in the ADE1 gene. In this background, [PSI+] cells grow on synthetic media deficient in adenine (SC-ADE) and are white on rich media. [psi−] cells do not grow on SC-ADE and produce red colonies on rich media because of the buildup of a colored byproduct of adenine biosynthesis.
Recombinant strains containing the SUP35ΔM gene (n = 16) at the SUP35 locus were red on rich medium but showed a faint background of growth (dark red in color) on SC-ADE. Thus, at least some Sup35ΔM protein is functional in translation termination, but the protein is not as active as WT Sup35p (Fig. 1 A and C).
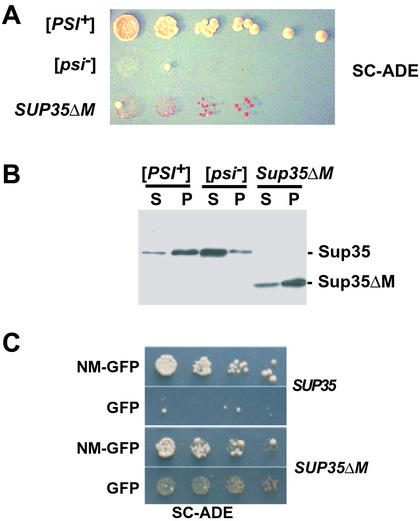
Figure 1.
Sup35ΔMp can convert to [PSI+]. (A) Five-fold serial dilution and growth of SUP35ΔM cells compared with WT [PSI+] and [psi−] cells on SC-ADE plates (30°C, 5 days). (B) Sup35p solubility assay of [PSI+], [psi−], and Sup35ΔMp. After cell lysis, high speed centrifugation and separation by SDS/10% PAGE Sup35ΔMp was detected by immunoblotting with anti-Sup35p antibody. S, supernatant fraction; P, pelleted fraction. (C) Induction of heritable [PSI+] factors in WT (SUP35) [psi−] and SUP35ΔM [psi−] strains. Five-fold serial dilutions of cells after 24-h induction of NM-GFP or GFP alone, plated on SC-ADE (30°C, 5 days).
Differential centrifugation of cell lysates revealed that a much smaller fraction of Sup35ΔMp was present in the supernatant after a 100,000 × g spin, compared with WT Sup35p in [psi−] strains (Fig. 1B). Coomassie blue staining demonstrated equal loading of total proteins in these fractions and revealed no detectable changes in the solubility of other proteins (data not shown). The partial insolubility of Sup35ΔMp explains the weak suppressor phenotype of SUP35ΔM cells because less Sup35p is available for translational termination than in WT cells. Clearly, the highly charged M region provides a solubilizing function for the Sup35 protein as a whole in vivo, as it does for the NM fragment in vitro.
Insolubility is a characteristic of Sup35p in the [PSI+] state (2) (Fig. 1B). The weak suppressor phenotype of SUP35ΔM could be due to a weak [PSI+] variant (see below). However the phenotype was unaffected by growth on Gdn⋅HCl, which efficiently cures [PSI+] (data not shown), suggesting that the insolubility of Sup35ΔMp might not be caused by prion formation. Also suggesting that the aggregates were not prion-like, the suppressor phenotype was recessive when SUP35ΔM cells were mated with WT [psi−] cells (data not shown). Moreover, differential centrifugation showed that aggregates of Sup35ΔMp in diploid cells did not cause WT Sup35p to fractionate into the pellet. That is, the Sup35ΔM protein in these cells could not recruit WT Sup35p to take on the [PSI+] state (data not shown). The aggregates had none of the characteristics of a prion and were more likely simply caused by loss of the solubilizing activity normally conferred by the highly charged M region.
Sup35ΔMp was, however, capable of acquiring a heritable [PSI+]-like state when SUP35ΔM cells were mated to WT [PSI+] cells. After mating, the strong suppressor phenotype of the [PSI+] parent was invariably dominant (data not shown), indicating that WT protein had converted Sup35ΔMp into a form that reduced its ability to function in translation. Moreover, the strong suppressor phenotype of the SUP35ΔM × [PSI+] diploid was cured by growth on medium containing Gdn⋅HCl (data not shown), indicating that Sup35ΔMp had acquired the [PSI+] state from WT protein, and could be cured of this state with Gdn⋅HCl, as is the WT protein.
Next, we asked whether Sup35ΔMp could acquire the prion state through another common mechanism, by transient over expression of NM. In previous studies, we used NM fused to GFP to monitor the formation and propagation of [PSI+] in living cells (2). Sup35p in the [PSI+] state has the capacity to capture NM-GFP and induce it to adopt the same state, forming GFP aggregates visible by fluorescence microscopy. Furthermore, overexpression of NM-GFP induces new [PSI+] element formation in [psi−] cells (2). This state is retained even when the NM-GFP plasmid is lost.
SUP35ΔM cells were transformed with expression plasmids for GFP alone or NM-GFP. After 4 h of induction, intense coalescent foci were observed in many cells expressing NM-GFP, but never in cells expressing GFP alone (ref. 2 and data not shown). When plated to copper-free medium without selection for the plasmid, cells that had expressed NM-GFP produced colonies with a [PSI+] phenotype at a much higher frequency than those expressing GFP alone (Fig. 1C), suggesting that transient over expression of NM had converted the Sup35ΔMp to the prion state. This was confirmed by 4:0 segregation of the suppressor phenotype in crosses to WT [psi−] cells and curing by growth on medium containing Gdn⋅HCl (data not shown). Therefore, the Sup35ΔMp can exist in two different states (we designate them [PSI+]ΔM and [psi−]ΔM) that are genetically analogous to the [PSI+] and [psi−] states of the WT Sup35 protein. Unlike WT Sup35p, however, the protein is largely insoluble in both cases (Figs. 1B and 2B).
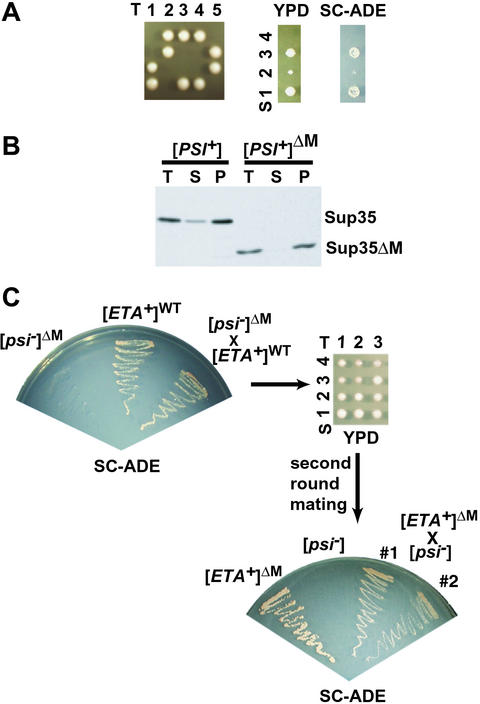
Figure 2.
Sup35ΔMp can maintain different [PSI+] variants. (A) Most SUP35ΔM spores from SUP35ΔM × WT [PSI+] crosses were not viable. (Left) Five representative tetrads from spores of a [PSI+] diploid from a SUP35ΔM [psi−] × SUP35 [PSI+] cross, dissected on YPD medium. (Right) Growth of one set of tetrads with one surviving SUP35ΔM spore on YPD and SC-ADE. T, tetrad; S, spore. (B) Sup35p solubility properties of the WT spore and the SUP35ΔM spore (methods as described in Fig. 1). T, total protein; S, supernatant; P, pellet. (C) Sup35ΔMp can form and propagate [ETA+]. (Upper Left) Growth of SUP35ΔM [psi−] haploid strain ([psi−]ΔM), WT [ETA+] haploid strain ([ETA+]WT), and the diploid from the cross of these two strains on SC-ADE medium. (Upper Right) Three representative tetrads dissected from the diploid SUP35ΔM[psi−] × SUP35[ETA+] on YPD medium. (Lower Right) Growth on SC-ADE of haploid parental strains and two diploid strains from a cross of a SUP35ΔM spore from SUP35ΔM [psi−] ([psi−]ΔM)/SUP35 [ETA+] ([ETA+]WT) diploid to WT [psi−].
Sup35ΔMp Can Maintain Different Prion Variants.
Although haploid [PSI+]ΔM strains were readily obtained by overexpressing NM-GFP, they were not readily obtained by other standard methods. For example, in one case SUP35ΔM was integrated at the site of WT SUP35 (in tandem with it) in a [PSI+] strain (i.e., both SUP35ΔM and SUP35 were present). These transformants retained the [PSI+] phenotype, as expected because WT Sup35p in the prion state converts Sup35ΔMp to that state. However, when one of the two genes was excised by selection against the URA3 marker that had been used for transformation, clones carrying only SUP35ΔM did not survive. SUP35ΔM derivatives were readily obtained when the initial insertion had been in a [psi−] background and other SUP35 [PSI+] variants are readily obtained by this method (35). Similarly, sporulating SUP35/SUP35ΔM [psi−] diploids yielded the expected number of viable SUP35ΔM spores, but sporulating SUP35/SUP35ΔM [PSI+] diploids yielded very few (Fig. 2A). Those that were recovered grew very slowly, even on rich media (Fig. 2A), and by differential centrifugation, virtually all of their Sup35ΔM protein was found in the pellet (Fig. 2B). Genetic crosses between these slow-growing strains and the WT [psi−] strain generated [PSI+] diploids (data not shown). Thus, Sup35ΔMp in the prion state can transmit that state to WT protein. But [PSI+] cells in which the only copy of Sup35p is Sup35ΔMp can have unexpected problems with viability.
One explanation is that Sup35ΔMp, like WT Sup35p, can form prion variants with different “strengths.” The phenomenon of different prion states (called prion “strains” or variants) that are strong, moderate, weak, and very weak is well characterized (13, 38–40). These variants are not caused by genetic differences, but are caused by epigenetic differences in the rates that prion variants capture and convert new Sup35p to the prion state. They have different levels of soluble Sup35p and different rates of translation termination (39–41). Because Sup35ΔMp is inherently less soluble than WT Sup35p, if it acquired a strong prion state there might be too little translation termination activity to keep cells viable. The haploid [PSI+]ΔM strains induced by transient overexpression of NM-GFP might represent weak variants, viable because a greater fraction of the Sup35ΔM protein remains soluble and active.
To test this possibility, we mated [psi−]ΔM cells to the weak [PSI+] variant [ETA+]. Conversion of Sup35ΔMp by this weak variant should leave a greater fraction of Sup35ΔMp in solution and produce more viable haploid SUP35ΔM [PSI+]ΔM spores. The diploid showed the same suppression of the ade1–14 marker as the [ETA+] haploid parent (Fig. 2C Left), suggesting that the Sup35ΔM protein had converted to a weak prion state. In contrast to the poor viability of SUP35ΔM spores after mating to strong [PSI+] strains (Fig. 2A), nearly all SUP35ΔM progeny from the [ETA+] were viable (Fig. 2C Upper Right). When these progeny were mated to WT [psi−] tester strains, the diploids exhibited the suppressor phenotype characteristic of [ETA+] strains (Fig. 2C Lower Right). Thus, Sup35ΔMp could acquire, maintain, and transmit the [ETA+] state to WT protein. The N region is sufficient to form prion variants of different strengths.
The M Region Promotes Mitotic Stability of the [PSI+] State.
On rich media, [PSI+]-mediated nonsense suppression is not required for growth, yet WT cells retain [PSI+] with high fidelity. In contrast [PSI+]ΔM strains lost the prion at a high rate (Fig. 3A). We asked whether we could restore stability simply by restoring solubility to the protein. To do this, a DNA fragment encoding a highly charged polypeptide rich in lysine and glutamic acid (6xKDG) was inserted in place of M creating the replacement SUP35NKC.
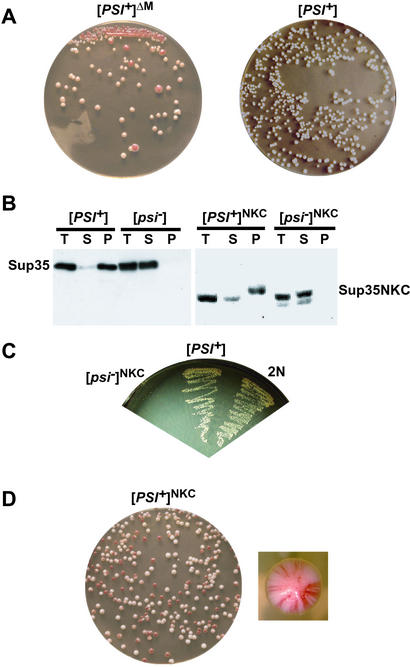
Figure 3.
The M region promotes mitotic stability of strong [PSI+]. (A) Appearance of red sectors ([psi−]) out of white colonies when [PSI+]ΔM cells were streaked onto YPD and incubated for several days (Left). Growth of [PSI+] cells on YPD shown for comparison (Right). (B) Sup35p solubility assay of the NKC mutant protein compared with that of WT Sup35p (methods as described in Fig. 1). (Left) WT [PSI+] and [psi−] strains. (Right) SUP35NKC [PSI+] and [psi−] ([PSI+]NKC and [psi−]NKC) haploid strains. (C) Mating SUP35NKC [psi−] ([psi−]NKC) to [PSI+] generated [PSI+] diploid. Growth of the two parental strains and a diploid (2N) progeny on SC-ADE is shown. (D) [PSI+]NKC cells were mitotically unstable. Cells were grown in liquid YPD for 16 h and plated onto YPD plates. Most colonies were red/white sectored (Left). A close-up image of one of the sectored colonies is shown (Right).
As expected, when the WT SUP35 gene was replaced by SUP35NKC in [psi−] cells, they retained a [psi−] phenotype. The solubility of the Sup35NKC protein in this state was comparable to that of WT Sup35p (Fig. 3B). To determine whether this protein could acquire the prion state, SUP135NKC mutants were mated to a typical strong prion strain (Fig. 3C) and the diploid strain showed the suppressor phenotype. Sporulation of this diploid (data not shown) produced haploid SUP35NKC [PSI+] cells ([PSI+]NKC). Most Sup35NKCp was soluble in [psi−]NKC strains, and most of the protein became insoluble when it adopted the [PSI+]NKC state (Fig. 3B). Unlike [PSI+]ΔM cells, [PSI+]NKC cells exhibited no growth defect when streaked on rich medium (data not shown). However, even though Sup35NKCp appeared to be as soluble as WT protein and produced no general growth disadvantage, the [PSI+]NKC phenotype was highly unstable (Fig. 3D). Thus, replacement of the M region with a charged polypeptide that increases its inherent solubility in vivo is not sufficient to restore stability to the prion state. M provides more than a simple solubilizing function to Sup35p. It also promotes the mitotic stability of [PSI+].
The M region is highly charged and, in the soluble state, circular dichromism spectroscopy shows it to have a highly flexible structure (≈60% random coil; A. Cashikar and T. Scheibel, personal communication). Our next alterations were to replace the M region with two naturally occurring polypeptides that, like WT M, are highly charged and are known to have conformational flexibility.
The Human Topoisomerase Linker Restores Mitotic Stability but Causes Meiotic Instability.
The human topoisomerase linker (T) has a percentage of charged residues similar to Sup35Mp. The linker has been characterized by x-ray crystallography and contains both structured and unstructured regions that link other functional domains of the protein (42). When an M to T replacement (SUP35NTC) was inserted in tandem with SUP35 in [psi−] cells, strains retaining only SUP35NTC were obtained at an equal frequency to WT. SUP35NTC strains were phenotypically identical to WT [psi−] cells with respect to growth on rich media and SC-ADE. Sup35NTCp could readily be converted to the prion state, [PSI+]NTC, by matings to WT [PSI+] strains. The haploid progeny of sporulation showed normal viability. [PSI+]NTC strains were also obtained after transient overexpression of Sup35NTCp from an inducible plasmid in the SUP35NTC background.
[PSI+]NTC was tested for other common prion properties including curability, mitotic stability, and non-Mendelian inheritance during meiosis (see Table 1). It was mitotically stable (Fig. 4A), capable of growth on SC-ADE (Fig. 4B) and was cured by growth on media containing Gdn⋅HCl, but was not cured by either the overexpression or the deletion of HSP104 (Table 1, Fig. 4C). To further characterize [PSI+]NTC and [psi−]NTC states, we analyzed the solubility of the Sup35NTC protein. In both [PSI+]NTC and [psi−]NTC cells, most Sup35NTCp was soluble after a 100,000 × g spin (Fig. 4D). To test the aggregation of Sup35p by using GFP, we expressed a plasmid with a fusion of the N and T regions to the GFP marker (NT-GFP) in [PSI+]NTC and [psi−]NTC cells. NT-GFP showed a diffuse fluorescence pattern in both strain types confirming that the protein does not form large aggregates (Fig. 4E). Therefore this protein can exist in states that are genetically analogous to the prion states of Sup35p, but in both states most of the protein remains soluble after centrifugation at 100,000 × g for 20 min.
Table 1.
M substitution mutants and their properties
| Region length; % charge; no. of positive amino acid residues that are lysine | [PSI+] inducibility
|
Segregation pattern [PSI+]:[psi−] | Stable in mitosis | Gdn⋅HCl curable | Protein solubility
|
Hsp104 curability
|
|||
|---|---|---|---|---|---|---|---|---|---|
| Sup35 N terminus over express | Mate to [PSI+] | [PSI+] | [psi−] | ΔHSP104 | Hsp104 over express | ||||
| WT; 130 aa; 42%; 24 of 24 | + | + | 4:0 | + | + | − | + | + | + |
| ΔM | + | + | 4:0 | − | + | − | − | + | + |
| KDG6(NKC); 18 aa; 67%; 6 of 6 | + | + | 4:0 | − | + | − | + | N.T. | N.T. |
| HuTop I (NTC); 79 aa; 44%; 17 of 21 | + | + | 4:0 17% 3:1 56% 2:2 22% 1:3 4% | + | + | + | + | − | − |
| Hsp90 (N9C); 53 aa; 77%; 17 of 17 | − | + | 2:2* | +* | +* | −* | + | +* | − |
N.T., not tested.
Tested in heterozygous [PSI+] diploid (WT/N9C).
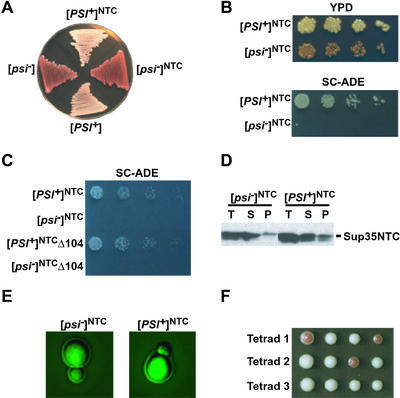
Figure 4.
Substitution of human topoisomerase I linker for the Sup35p M region causes meiotic instability. (A) [psi−]NTC and [PSI+]NTC strains containing the linker from human topoisomerase I in place of the M linker of Sup35p were plated onto YPD; [PSI+] and [psi−] were plated for comparison. (B) Growth phenotype of SUP35NTC cells on YPD and SC-ADE. (C) Deletion of HSP104 did not affect the suppressor state of [PSI+]NTC cells. (D) Sup35p solubility assay of [PSI+]NTC and [psi−]NTC (methods as described in Fig. 1). (E) Expression of NT-GFP in [psi−]NTC and [PSI+]NTC did not induce aggregation. (F) Tetrad dissection of a [PSI+]NTC diploid shows varying numbers of [PSI+]NTC and [psi−]NTC spores.
The [PSI+]NTC state was dominant in crosses to [psi−]NTC cells, indicating that it readily converted soluble Sup35NTCp to the prion state. When [PSI+]NTC homozygous diploids were sporulated, the frequency of meiotic transmission of the suppressor phenotype to offspring was not always 4:0, the ratio typical for WT [PSI+] diploids, but it was clearly non-Mendelian (Fig. 4F and Table 1). This segregation pattern was similar to that of another yeast prion, [URE3] (43). These findings suggest that the highly charged M region also influences the accurate propagation of [PSI+] elements through meiosis. The different effects of M substitutions on mitotic and meiotic stability suggest that the mechanisms for maintaining meiotic and mitotic stability are, at least in part, distinct.
Substitution of the Hsp90p Linker for M Causes Another Distinct Genetic Behavior.
The other M substitution we tested was derived from S. cerevisiae Hsp90 protein. This highly charged region (amino acids 210–262 of the polypeptide sequence) connects the two stably folded domains of Hsp90p and is degraded by even very short treatments with proteases, suggesting it is not inherently a tightly folded polypeptide (44). As with SUP35NTC, when M was replaced by this portion of the HSP90 coding sequence (SUP35N9C), [psi−] cells retained a nonsuppressor state (data not shown).
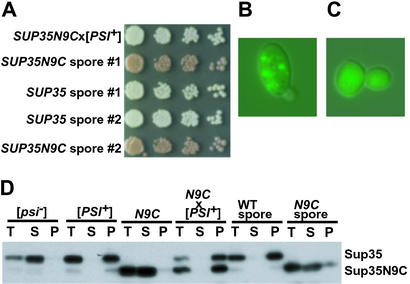
In contrast to SUP35NTC, SUP35ΔM, and SUP35NKC cells, a suppressor state could not be induced in haploid SUP35N9C cells by overexpression of polypeptides containing the N region (data not shown). The protein could, however, acquire a [PSI+]-like state when SUP35N9C cells were mated to WT [PSI+] cells (Fig. 5A). The diploid strain had many other characteristics of [PSI+] strains, including a suppressor phenotype that was eliminated by plating to media containing Gdn⋅HCl (see Table 1). It also showed strong mitotic stability. But surprisingly, sporulation of this diploid always produced two [PSI+] colonies with a SUP35 genotype and two SUP35N9C with the [psi−] phenotype (Fig. 5A).
Figure 5.
Substitution of the Hsp90p linker for the Sup35p M region causes distinct genetic behavior. (A) Diploid [PSI+] cells with the SUP35/N9C genotype grew white on YPD (top row). On sporulation of this diploid, two red colonies and two white colonies were always obtained (following rows). (B) Expression of N9-GFP in the SUP35/N9C diploid [PSI+] strain causes aggregation. (C) Expression of N9-GFP in haploid cells expressing only N9C did not cause aggregation. (D) Sup35p solubility assay of N9C indicates that it pelleted only in the presence of Sup35p in the [PSI+] state. The protein returned to the soluble state after sporulation (methods as described in Fig. 1).
These observations suggested that Sup35N9Cp could enter a [PSI+]-like state, but could only acquire that state from preformed [PSI+] elements and could not thereafter retain it on its own (when separated by sporulation from the WT protein). To more fully characterize these transitions we examined the solubility of the Sup35N9C protein in the haploid SUP35N9C strain, the diploid SUP35/SUP35N9C [PSI+] strain and the haploid progeny of sporulation. The Sup35N9C protein was almost entirely soluble in the SUP35N9C parent, but was insoluble (as was WT Sup35p) in the heterozygous [PSI+] diploid (Fig. 5D). After sporulation, Sup35N9Cp became soluble once again in the SUP35N9C haploid progeny, whereas the insoluble prion state was maintained in SUP35 progeny (Fig. 5D, right lanes). This result was confirmed by the presence of small foci in the [PSI+] diploid after 2 h of expression of an N9-GFP fusion protein (Fig. 5B). In contrast, N9-GFP fluorescence in the nonsuppressed haploid SUP35N9C remained diffuse (Fig. 5C). Thus, Sup35N9Cp can readily enter a [PSI+]-like state under the influence of WT protein in that state, but it cannot maintain that state on its own.
Discussion
We have demonstrated that the M region of Sup35p makes important and diverse contributions to genetic and biochemical properties of [PSI+]. Sup35p mutants with a deletion of the M region or with substitutions in place of M can form prions, but these states are strikingly distinct from WT [PSI+] and from each other. A wide variety of prion states and behaviors can be conferred on the same C-terminal functional domain and N-terminal prion domain by intervening “auxiliary” sequences.
Prion proteins such as PrP and Sup35p aggregate when adopting the prion conformation (2, 37, 45, 46). However, large-scale aggregation is neither necessary nor sufficient for entry into the prion state. (The former has also been suggested by the analysis of certain URE3 prion variants, ref. 47.) We have shown that M helps maintain Sup35p in the soluble state and, as a result, Sup35ΔMp is found in the pellet after centrifugation of cell lysates, regardless of its prion state. This finding confirms the special nature of prion state protein. Differences between the Sup35ΔM protein in [PSI+]ΔM cells and in [psi−]ΔM cells are not simply a difference between aggregated and nonaggregated states. This point is also demonstrated by our experiments with Sup35NTCp. No aggregated state was detectable in [PSI+]NTC cells. The prion state of Sup35NTCp may well involve higher-order complexes, but if so, they are clearly different in character from the large complexes of WT Sup35p in the [PSI+] state.
The M region is also important for the stabilization of [PSI+] during cell division. Cells with either the SUP35ΔM or the SUP35NKC replacement genotype could enter a prion state, but this state was not well maintained during mitotic division. Cells with the SUP35NTC genotype could also enter the prion state, and [PSI+]NTC was mitotically stable. However, [PSI+]NTC was not propagated after meiosis with the same fidelity as [PSI+]. Thus, the propagation of prion elements is quite sensitive to changes in the M region. Requirements for the maintenance of the prion during mitotic and meiotic cell division are distinct and M contributes to them both.
Altering the M region also had important consequences for prion curing. Because Hsp104p function is sensitive to Gdn⋅HCl, it has been suggested that Gdn⋅HCl treatment cures cells through the inactivation of Hsp104p (48–51), but this hypothesis is controversial (48, 50, 52). We have identified a prion state, [PSI+]NTC, which can be cured by growth on Gdn⋅HCl but cannot be cured by HSP104 deletion (Table 1). This finding indicates that curing by Gdn⋅HCl is not solely caused by Hsp104p inactivation. The results also suggest that some feature of M strongly influences interactions with Hsp104p. In this respect, it is intriguing that 24 of 24 of the positively charged amino acids in M are lysines (Table 1). Polylysine binds Hsp104p with high affinity and triggers a series of changes in ATP hydrolysis and Hsp104p conformation (36). The deletion/insertion mutations we used also exhibit some conformational flexibility and lysine richness. However, the effect of Hsp104p on the prion-state conversions of these proteins differed. There must be something more than the mere presence of lysines and conformational flexibility that influences the interactions of the M region with Hsp104p. For example, length, specific sequence elements, residue spacings, and conformational predisposition could all influence these interactions.
The Sup35N9C protein showed an intriguing genetic property that was entirely unexpected. Unlike other M alterations, Sup35N9Cp was incapable of entering a [PSI+]-like state until cells expressing it were mated to cells already containing WT Sup35p in that state. Further, Sup35N9Cp could not support the prion state on its own. Once the two proteins were separated by sporulation, SUP35N9C cells reverted to a [psi−] phenotype and the Sup35N9C protein returned to the soluble state. Thus, Sup35N9Cp can participate in a heritable phenotypic change caused by a protein-only mechanism that exhibits Mendelian segregation, a striking departure from ordinary prion behavior.
The M region is not required for entry into the prion state. Yet in the context of proteins with the same N-terminal prion-determining region and C-terminal functional region, substitution of M with artificial linker regions confers a rich variety of genetic and biochemical characteristics to the prion state. Proteins with very different physical properties can undergo self-perpetuating conformational changes in state, but produce similar phenotypes (Sup35ΔMp, Sup35NTCp); proteins with similar physical characteristics can display very different genetic properties (WT Sup35p, Sup35N9Cp). Indeed, were it not for the fact that (i) the prion-determining nature of the N region of Sup35p has been extensively characterized by genetic, biochemical and cell biological methods, and (ii) the self-perpetuating changes in function obtained with each of our different mutants were acquired from WT Sup35p that was in its well-characterized prion state, it might be hard to argue that the changes in function we observed were caused by self-perpetuating prion conformations.
We have created prion variants with unusual properties by deliberate, artificial manipulations. We have no direct evidence that proteins with such properties exist in nature. However, given the divergence of prion sequences, which is particularly great in the M region, it seems reasonable to suppose that proteins that have such properties might well have appeared. Thus, we suggest that there may be many prion-like, self-perpetuating states not recognizable as such by the prion-defining criteria used to date. It may be that more epigenetic changes in biology are caused by prion-like processes than previously realized.
Acknowledgments
We thank J. Shorter for comments on the manuscript. This work was supported by the National Institutes of Health and the Howard Hughes Medical Institute. N.S. was supported by National Institutes of Health Grants 5 T32 GM07183-24 and 5 T32 GM07281.
Abbreviations
- Gdn⋅HCl
guanidine hydrochloride
- YPD
yeast extract/peptone/dextrose
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Self-Perpetuating Structural States in Biology, Disease, and Genetics,” held March 22–24, 2002, at the National Academy of Sciences in Washington, DC.
References
- 1.Wickner R B, Masison D C, Edskes H K. Yeast. 1995;11:1671–1685. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- 2.Patino M M, Liu J J, Glover J R, Lindquist S. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 3.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 4.Lindquist S. Cell. 1997;89:495–498. doi: 10.1016/s0092-8674(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 5.Eaglestone S S, Cox B S, Tuite M F. EMBO J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.True H L, Lindquist S L. Nature (London) 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 7.Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox B S, Tuite M F, McLaughlin C S. Yeast. 1988;4:159–178. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- 9.Kushnirov V V, Ter-Avanesyan M D, Telckov M V, Surguchov A P, Smirnov V N, Inge-Vechtomov S G. Gene. 1988;66:45–54. doi: 10.1016/0378-1119(88)90223-5. [DOI] [PubMed] [Google Scholar]
- 10.Kushnirov V V, Ter-Avanesian M D, Dagkesamanskaia A R, Chernov Iu O, Inge-Vechtomov S G, Smirnov V N. Mol Biol. 1990;24:1037–1041. [PubMed] [Google Scholar]
- 11.Ter-Avanesyan M D, Kushnirov V V, Dagkesamanskaya A R, Didichenko S A, Chernoff Y O, Inge-Vechtomov S G, Smirnov V N. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 12.Ter-Avanesyan M D, Dagkesamanskaya A R, Kushnirov V V, Smirnov V N. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derkatch I L, Chernoff Y O, Kushnirov V V, Inge-Vechtomov S G, Liebman S W. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chernoff Y O, Galkin A P, Lewitin E, Chernova T A, Newnam G P, Belenkiy S M. Mol Microbiol. 2000;35:865–876. doi: 10.1046/j.1365-2958.2000.01761.x. [DOI] [PubMed] [Google Scholar]
- 15.Kushnirov V V, Kochneva-Pervukhova N V, Chechenova M B, Frolova N S, Ter-Avanesyan M D. EMBO J. 2000;19:324–331. doi: 10.1093/emboj/19.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santoso A, Chien P, Osherovich L Z, Weissman J S. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- 17.Stansfield I, Jones K M, Kushnirov V V, Dagkesamanskaya A R, Poznyakovski A I, Paushkin S V, Nierras C R, Cox B S, Ter-Avanesyan M D, Tuite M F. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kushnirov V V, Ter-Avanesyan M D, Didichenko S A, Smirnov V N, Chernoff Y O, Derkach I L, Novikova O N, Inge-Vechtomov S G, Neistat M A, Tolstorukov I I. Yeast. 1990;6:461–472. doi: 10.1002/yea.320060603. [DOI] [PubMed] [Google Scholar]
- 19.Nakayashiki T, Ebihara K, Bannai H, Nakamura Y. Mol Cell. 2001;7:1121–1130. doi: 10.1016/s1097-2765(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 20.Cox B S. Heredity. 1965;20:505–521. [Google Scholar]
- 21.Singh A, Helms C, Sherman F. Proc Natl Acad Sci USA. 1979;76:1952–1956. doi: 10.1073/pnas.76.4.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuite M F, Mundy C R, Cox B S. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chernoff Y O, Lindquist S L, Ono B, Inge-Vechtomov S G, Liebman S W. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 24.Jung G, Jones G, Wegrzyn R D, Masison D C. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kushnirov V V, Kryndushkin D S, Boguta M, Smirnov V N, Ter-Avanesyan M D. Curr Biol. 2000;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- 26.Newnam G P, Wegrzyn R D, Lindquist S L, Chernoff Y O. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Lindquist S. Science. 2000;287:661–664. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- 28.DePace A H, Santoso A, Hillner P, Weissman J S. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 29.Glover J R, Kowal A S, Schirmer E C, Patino M M, Liu J J, Lindquist S. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 30.Serio T R, Cashikar A G, Kowal A S, Sawicki G J, Moslehi J J, Serpell L, Arnsdorf M F, Lindquist S L. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 31.King C Y, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K. Proc Natl Acad Sci USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 33.Chernoff Y O, Uptain S M, Lindquist S L. Methods Enzymol. 2002;351:499–538. doi: 10.1016/s0076-6879(02)51867-x. [DOI] [PubMed] [Google Scholar]
- 34.Guthrie C, Fink G R, editors. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 35.Liu J J, Lindquist S. Nature (London) 1999;400:573–576. doi: 10.1038/23048. [DOI] [PubMed] [Google Scholar]
- 36.Cashikar A G, Schirmer E C, Hattendorf D A, Glover J R, Ramakrishnan M S, Ware D M, Lindquist S L. Mol Cell. 2002;9:751–760. doi: 10.1016/s1097-2765(02)00499-9. [DOI] [PubMed] [Google Scholar]
- 37.Sondheimer N, Lindquist S. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 38.Liebman S W, All-Robyn J A. Curr Genet. 1984;8:567–573. doi: 10.1007/BF00395701. [DOI] [PubMed] [Google Scholar]
- 39.Zhou P, Derkatch I L, Uptain S M, Patino M M, Lindquist S, Liebman S W. EMBO J. 1999;18:1182–1191. doi: 10.1093/emboj/18.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uptain S M, Sawicki G J, Caughey B, Lindquist S. EMBO J. 2001;20:6236–6245. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kochneva-Pervukhova N V, Chechenova M B, Valouev I A, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Yeast. 2001;18:489–497. doi: 10.1002/yea.700. [DOI] [PubMed] [Google Scholar]
- 42.Stewart L, Redinbo M R, Qiu X, Hol W G, Champoux J J. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 43.Wickner R B. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 44.Scheibel T, Siegmund H I, Jaenicke R, Ganz P, Lilie H, Buchner J. Proc Natl Acad Sci USA. 1999;96:1297–1302. doi: 10.1073/pnas.96.4.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Mol Cell Biol. 1997;17:2798–2805. doi: 10.1128/mcb.17.5.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prusiner S B, Scott M R. Annu Rev Genet. 1997;31:139–175. doi: 10.1146/annurev.genet.31.1.139. [DOI] [PubMed] [Google Scholar]
- 47.Fernandez-Bellot E, Guillemet E, Ness F, Baudin-Baillieu A, Ripaud L, Tuite M, Cullin C. EMBO Rep. 2002;3:76–81. doi: 10.1093/embo-reports/kvf011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreira P C, Ness F, Edwards S R, Cox B S, Tuite M F. Mol Microbiol. 2001;40:1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- 49.Glover J R, Lindquist S. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 50.Jung G, Masison D C. Curr Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- 51.Jung G, Jones G, Masison D. Proc Natl Acad Sci USA. 2002;99:9936–9941. doi: 10.1073/pnas.152333299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wegrzyn R D, Bapat K, Newnam G P, Zink A D, Chernoff Y O. Mol Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]