Abstract
One can imagine a variety of mechanisms that should result in self-perpetuating biological states. It is generally assumed that cytosine methylation is propagated in eukaryotes by enzymes that specifically methylate hemimethylated symmetrical sites (e.g., 5′CpG/GpC5′ or 5′CpNpG/GpNpC5′). Although there is wide support for this model, we and others have found examples of methylation that must be propagated by a different mechanism. Most methylated regions of the Neurospora genome that have been examined are products of repeat-induced point mutation, a premeiotic genome defense system that litters duplicated sequences with C⋅G to T⋅A mutations and typically leaves them methylated at remaining cytosines. In general, such relics of repeat-induced point mutation are capable of triggering methylation de novo. Nevertheless, some reflect a mechanism that can propagate heterogeneous methylation at nonsymmetrical sites. We propose that de novo and maintenance methylation are manifestations of a single mechanism in Neurospora, catalyzed by the DIM-2 DNA methyltransferase. The action of DIM-2 is controlled by the DIM-5 histone H3 Lys-9 methyltransferase, which in turn is influenced by other modifications of histone H3. DNA methylation indirectly recruits histone deacetylases, providing the framework of a self-reinforcing system that could result in propagation of DNA methylation and the associated silenced chromatin state.
Keywords: histone methylation‖chromatin‖RIP‖epigenetics‖ DNA methyltransferase
More than 25 years ago, Holliday and Pugh (1) and Riggs (2) pointed out that the symmetrical nature of methylated sites (5′CpG/GpC5′) observed in animals could allow for propagation of methylation patterns by a “maintenance methylase” specific for cytosines symmetrically opposed to 5-methylcytosines. In principle, a pattern of methylation, perhaps established early in development, could be faithfully propagated indefinitely. Results from transfection experiments in several animal systems are generally consistent with this model. Methylated transforming sequences tend to remain methylated through repeated cycles of DNA replication, whereas unmethylated transforming sequences typically remain unmethylated (3–6). Strong evidence for maintenance methylation also comes from observations in Arabidopsis (7). Findings that eukaryotic DNA methyltransferases (DMTs) such as Dnmt1 preferentially methylate hemimethylated CpG/GpC sites provided additional support for the model (8, 9). Nevertheless, some observations seem to conflict with the maintenance methylase model. The model predicts that in a clonal population of cells engaged only in maintenance methylation, (i) any given site should be either methylated or unmethylated in all cells, and (ii) methylation should be confined to symmetrical sequences. Violations of these predictions have been demonstrated in fungi, plants, and animals (10–23).
The fungus Ascobolus immersus offers a particularly clear example of propagation of methylation at nonsymmetrical sites and maintenance of heterogeneous methylation (24, 25). Sequences exposed to methylation induced premeiotically (MIP) in the sexual stage of the life cycle of Ascobolus retain their methylation through numerous vegetative cell divisions. As discussed below, DNA methylation that depends on preexisting methylation also has been found in the more thoroughly studied filamentous fungus, Neurospora crassa. Although mechanisms for maintenance methylation that do not rely on faithful copying at symmetrical sites can be imagined (e.g., see refs. 14, 26, 27), none have yet been demonstrated. Nevertheless, recent studies that have identified components of the methylation machine provide clues. In the picture that is emerging, DNA methylation patterns reflect a combination of superimposed mechanisms, at least some of which involve modifications of histones.
Maintenance Methylation in Neurospora
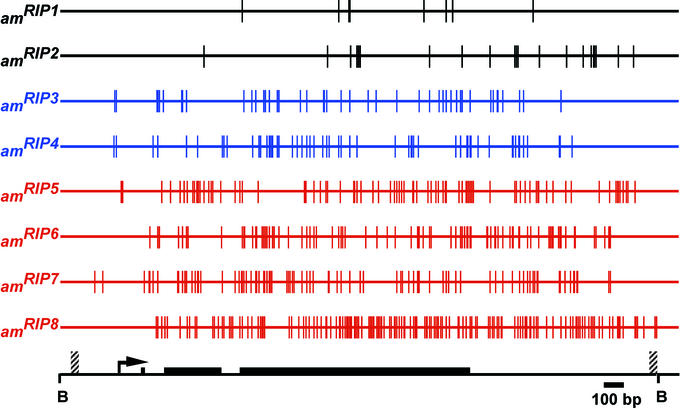
Ninety-eight percent of cytosines in the Neurospora genome are unmethylated, and most DNA methylation is found in relics of the genome defense system called repeat-induced point mutation (RIP; E.U.S., N. Tountas, S. Cross, B.S.M., J. Murphy, A. P. Bird, and M.F., unpublished work; refs. 25, 28, and 29). RIP detects duplicated sequences in the haploid genomes of specialized dikaryotic cells resulting from fertilization (30), and then riddles both copies of the duplicated sequence with C⋅G to T⋅A mutations (31). Frequently, but not invariably, sequences altered by RIP are left methylated, perhaps by the action of a putative DMT that is essential for RIP, RIP defective (RID) (32). This scenario is consistent with the possibility that the mutations occur by deamination of cytosines methylated in the sexual phase of the life cycle (29, 33). In principle, the methylation of relics of RIP that is observed in vegetative cells could either reflect creation of a signal for de novo methylation or could reflect propagation of methylation established earlier. To distinguish between these possibilities, eight alleles of the am gene that were generated by RIP were tested with two assays for their capacity to induce methylation de novo (34). First, alleles were demethylated by using the methylation inhibitor 5-azacytidine (5AC) and then scored for their ability to reestablish methylation. DNA from each allele also was cloned in bacteria, transformed into Neurospora to replace the am+ allele, and scored for de novo methylation. Equivalent results were obtained with both tests and are summarized in Fig. 1. The four alleles with the greatest number of mutations (58 to 158 in the 2.6-kb duplicated region) all triggered de novo methylation in vegetative cells, which is consistent with prior evidence that products of RIP can signal DNA methylation (35, 36). We were surprised to find, however, that two alleles (amRIP3 and amRIP4) that were initially methylated were incapable of inducing methylation de novo at their native chromosomal location (34). Apparently this methylation, which was not limited to symmetrical sites and was rather heterogeneous, depended on preexisting methylation established in the sexual phase by RIP.
Figure 1.
Mutations from RIP and methylation status of eight different am alleles. This figure was adapted from ref. 34 and readers should refer to that article for details of the experiments. Vertical lines indicate mutations. Alleles shown in black were not methylated. Alleles in blue were initially methylated but, after induced loss of methylation (with 5AC or by cloning and gene replacement), did not serve as de novo methylation signals. Alleles shown in red were methylated after the cross and served as de novo methylation signals. The horizontal line at the bottom represents the 2.7-kb region that was sequenced, including the 2.6-kb BamHI (B) fragment. Exons (thick bars) of the am gene, the extent of the duplication (striped bars), and the transcription start (arrow) are indicated.
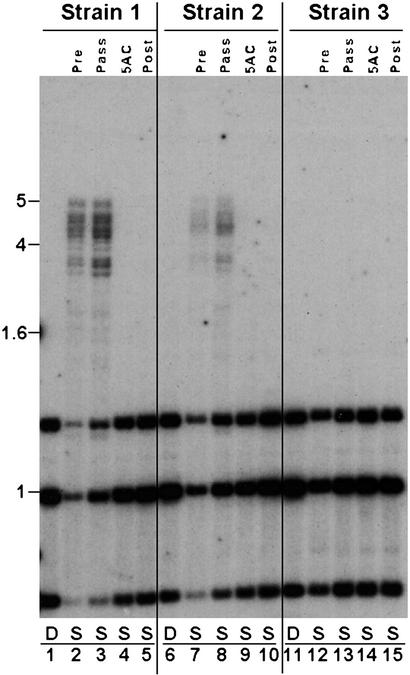
As a first step in exploring the mechanism of this maintenance methylation, we tested whether Neurospora would propagate methylation established in vitro. A 5.2-kb DNA fragment including the amRIP4 sequence was prepared by PCR using 2′-deoxyribosyl-5-methylcytosine triphosphate (dmCTP) in place of dCTP to ensure that every cytosine was methylated. Then, we introduced this, or an unmethylated control DNA, into an am+ strain by cotransformation with a fragment including the hygromycin resistance gene, hph, and enriched for transformants in which the native am gene had been replaced (37). Transformants bearing a single copy of the amRIP4 sequence in place of am+ were identified and analyzed by Southern hybridization. In contrast to our experience with a variety of other DNA fragments introduced at am, the methylation status of the in vitro-methylated sequences varied from transformant to transformant. Two of four transformants generated with methylated amRIP4 sequences showed stable incomplete methylation of these sequences (Fig. 2). Although heterogeneous, the observed methylation persisted through numerous cell cycles; only slight changes in the extent of methylation were observed after three serial passages of both strains (e.g., see Fig. 2, lanes 3 and 8). To verify that the methylation depended on preexisting methylation, i.e., that it was self-propagating, we treated each strain with 5AC for 48 h, verified that no detectable methylation remained (Fig. 2, lanes 4 and 9), and then grew the strains in the absence of the drug to ascertain whether the sequences would trigger methylation de novo. Methylation was not reestablished (Fig. 2, lanes 5 and 10), confirming the occurrence of a previously uncharacterized form of maintenance methylation that can propagate heterogeneous regional methylation. The loss of methylation that occurred in some of the transformants obtained with methylated DNA (Fig. 2, strain 3) suggests that establishment of stable methylation in our transformation system involves a stochastic step, perhaps associated with chromatin formation.
Figure 2.
Maintenance of DNA methylation in Neurospora. A fully methylated 5.2-kb DNA fragment containing the amRIP4 allele was generated by PCR with dmCTP and targeted to the am locus, as described (37). Each transformant was analyzed by Southern hybridization to verify that transforming DNA had integrated properly and without extraneous copies (data not shown) and to assess methylation, as described (40). DNA from the original transformant (Pre), the transformant after three passages (Pass), the transformant after a 48-h treatment with 5-azacytidine (5AC; 24 μM added at 0 and 24 hours), and the transformant grown for 2 days (≈12–15 cell doublings) in the absence of 5AC after treatment with this drug (Post) was digested with the 5-mC-insensitive restriction endonuclease DpnII (D; shown for the original transformant only) or its 5-mC-sensitive isoschizomer Sau3AI (S), fractionated and probed for am. Results for three transformants are shown. Size markers (kb) are shown on the left.
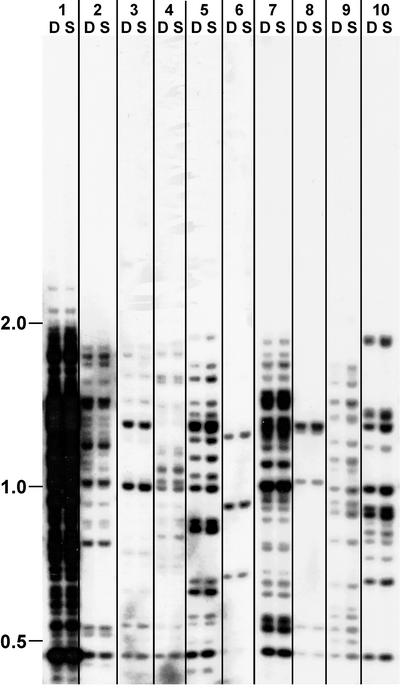
The observed fractional methylation at all restriction sites examined limits possible models to account for this propagation of methylation. Most surprisingly, the methylation did not noticeably spread; in every case examined, the methylation seemed to cover an ≈5-kb segment of DNA. We considered the possibility that the mutations from RIP, although insufficient to trigger methylation de novo, were nevertheless essential for propagation of methylation and, thereby, controlled the extent of methylation. To test this possibility, we performed a set of transformations with fully methylated am DNA lacking mutations by RIP. Because our gene-targeting scheme required selection for loss of am function, we used a nonfunctional am allele with an engineered ClaI site in place of the BglII site (plasmid pMS4). Transformants that integrated methylated DNA of this allele at the native am locus showed no propagation of the methylation (data not shown). Similarly, all cotransformants that integrated nonselected, fully methylated wild-type am DNA failed to retain noticeable methylation (Fig. 3). These findings support the hypothesis that the mutations by RIP play an essential role in the propagation of methylation on amRIP4 sequences. We considered the possibility that those alleles that could propagate methylation were those that could not be transcribed but found that even the heavily mutated alleles could support robust transcription when stripped of their methylation, which is consistent with the fact that their promoters were not mutated (38). We conclude that sequences differ in their ability to serve as substrates for maintenance methylation in Neurospora.
Figure 3.
Methylation of in vitro-methylated wild-type am alleles is not maintained. Transformants with ectopic copies of am were generated with fully methylated DNA by cotransformation with hph (data not shown). Ten arbitrary transformants bearing one or more copies of am are illustrated. Digestions with DpnII (D) and SauIIIAI (S) revealed no differences indicative of methylation, indicating that wild-type am sequences are unable to maintain methylation previously established in vitro and that a high copy number of gene fragments per se does not signal DNA methylation. Size markers (kb) are shown on the left.
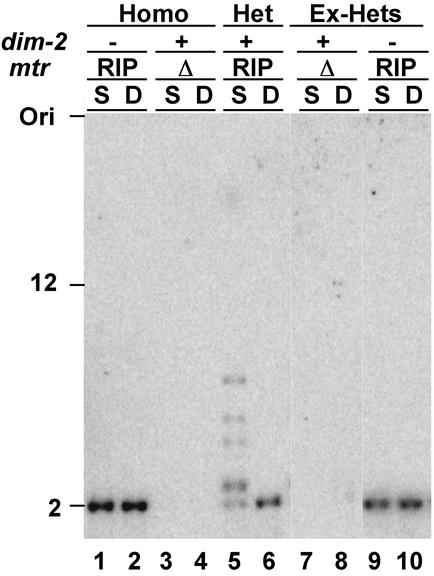
The findings described above raised the possibility that maintenance and de novo methylation are different manifestations of a common mechanism. Several observations support this idea. First, we found that amRIP4 DNA is frequently subjected to de novo methylation when it becomes integrated in multicopy arrays at ectopic sites (data not shown). This finding is in stark contrast to the case with am+ DNA, which remains free of methylation even when many copies were integrated into the genome (e.g., Fig. 3, transformant 1). Second, we found that amRIP4 DNA, but not am+ DNA, can induce methylation of a 200-bp segment of the ζ−η region that includes numerous mutations by RIP but is unable to trigger de novo methylation alone (H.T. and E.U.S., unpublished work). Third, we found that one DMT, DIM-2, is responsible for both de novo and maintenance methylation in vegetative cells of Neurospora (39). Mutants of dim-2 are devoid of DNA methylation in all tissues that can be assayed, including methylation in cells that are normally capable of rapid de novo methylation (40). To verify that DIM-2 is responsible for maintenance methylation, we first built a heterokaryon between a dim-2 deletion strain carrying an RIP-mutated allele of the mtr gene, which works as a de novo methylation signal, and a strain with a wild-type dim-2 allele carrying a deletion of the mtr gene, and confirmed de novo methylation (Fig. 4). Then, we isolated homokaryotic derivatives representing the two original constituents and showed that methylation was not maintained in the dim-2 isolate (Ex-Hets; Fig. 4, lanes 9 and 10).
Figure 4.
DIM-2 is the sole DMT required for DNA methylation in vegetative tissues. Heterokaryons (Het) were generated from two homokaryons (Homo): strain N1931, which has a deletion allele of dim-2 (dim-2−) and an RIP-mutated allele of the mtr gene (mtrRIP) and strain N534, which is dim-2+ and has mtr deleted (mtrΔ). DNA methylation was assayed by Southern blotting of genomic DNA digested with SauIIIAI (S) and DpnII (D). The RIP-mutated mtr allele is unmethylated in the dim-2 strain (lanes 1 and 2), but complementation allows for normal DNA methylation in the heterokaryon (lanes 5 and 6). The heterokaryon was broken, and resulting homokaryotic strains (Ex-Hets; lanes 7 to 10) were analyzed for DNA methylation. DNA methylation was not maintained in the absence of DIM-2 (lanes 9 and 10).
Clues to Involvement of Methyl/RIP-DNA Binding Proteins and Histone Deacetylation in DNA Methylation
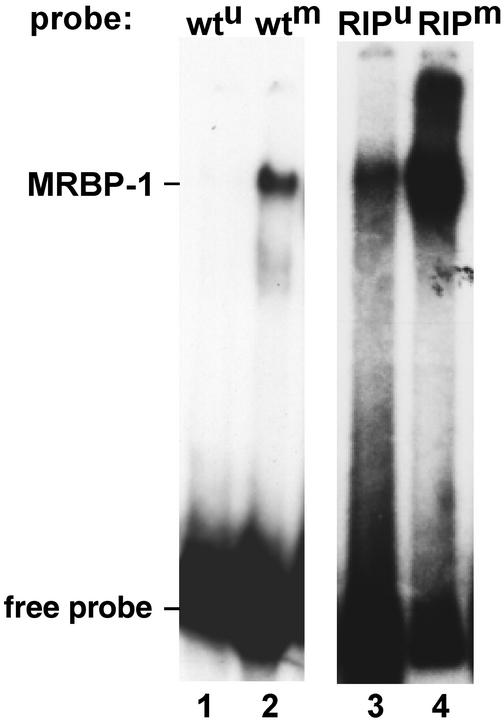
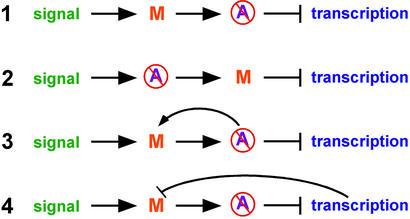
Two important, potentially related questions remain. How do mutations by RIP trigger DNA methylation? How does DNA methylation promote its own propagation? One possibility is that one or more proteins recognizes some feature(s) of sequences modified by RIP and induces DNA methylation. Clues to the existence of such proteins came from band-shift experiments with methylated DNA with mutations from RIP and cellular extracts of Neurospora. For example, an activity that we call methyl/RIP binding protein 1 (MRBP-1) binds strongly to methylated DNA with mutations by RIP (Fig. 5, lane 4) and less strongly to methylated native DNA (lane 2) or unmethylated mutated DNA (Fig. 5, lane 3; G.O.K., M.R.R., A. McCormack, L. David, and E.U.S., unpublished work). In principle, a factor such as MRBP-1 could directly or indirectly trigger DNA methylation. Recent observations favor the latter possibility. First, we found that treatment of Neurospora with the histone deacetylase (HDAC) inhibitor trichostatin A (TSA) can reduce methylation in some chromosomal regions (26). One model is that HDACs are recruited to regions with numerous mutations by RIP, perhaps by MRBP-1 or a similar factor, and that the resulting hypoacetylated state leads directly or indirectly to methylation of DNA. Fig. 6 illustrates several possible relationships among DNA methylation, histone acetylation, and transcription. Model 1, which suggests that hypoacetylation is a simple consequence of DNA methylation accounts for indications from animal systems that methyl-DNA binding proteins and DMTs recruit HDACs (see ref. 41) and a preliminary observation that mutation of dim-2 leads to hyperacetylation of histone H4 (data not shown) but does not account for the observed hypomethylation induced by TSA. Conversely, model 2 does not account for the effects of DNA methylation on histone acetylation. The selectivity of demethylation by TSA could reflect an absence of histone acetyltransferases in regions that do not lose methylation in response to TSA (e.g., in model 3) or may reflect an indirect effect, e.g., through transcription (model 4).
Figure 5.
Gel mobility shift assay using partially purified methyl-RIP binding protein-1 (MRBP-1). MRBP-1 was purified by chromatography on CM Macroprep, SP Fast Flow Sepharose, and heparin Sepharose (data not shown) and incubated 10 min on ice [in 10 mM Hepes, pH 7.9/50 mM KCl/1 mM EDTA/5 mM DTT/10% (vol/vol) glycerol] with 1–2 ng of a 250-bp probe (third exon of am) prepared in methylated (m) or unmethylated (u) form by PCR from template with either a wild-type (wt) or RIP-mutated (RIP) allele. Each reaction contained 1 μg of poly dI⋅dC as competitor and was analyzed by electrophoresis in 4% polyacrylamide gels containing 0.5× TBE buffer (52).
Figure 6.
Four potential relationships of methylation signal, DNA methylation (M), histone hypoacetylation (slashed A), and transcription. Arrows and blocked lines indicate stimulation and inhibition, respectively. See text for details.
Preliminary information on the relationship between histone acetylation and DNA methylation came from chromatin immunoprecipitation with antibodies prepared against tetra-acetylated histone H4 to assess acetylation of histone H4 molecules associated with particular genomic regions. A region whose DNA methylation is reduced by TSA (amRIP) showed corresponding hyperacetylation, as expected, whereas regions whose DNA methylation was unaffected by TSA (e.g., Ψ63) showed no change in histone acetylation (data not shown). This finding is consistent with the idea that not all DNA regions have the capability to become hyperacetylated, possibly because they lack targets for histone acetyltransferases. Interestingly, we also identified an unmethylated DNA region that is hypoacetylated, suggesting that hypoacetylation of histone H4 per se does not directly lead to DNA methylation, which conflicts with models 2 and 3 (Fig. 6).
The DIM-5 Histone Methyltransferase Integrates Histone Code Information and Controls DNA Methylation
Results from investigating a Neurospora mutant, dim-5, which is completely defective in DNA methylation, provided the strongest evidence that histones are intimately involved in DNA methylation (42). Isolation and characterization of the dim-5 gene showed that it encodes a histone H3 methyltransferase specific for Lys-9 (ref. 42 and data not shown). To confirm that histone H3 is the biologically significant target of DIM-5, we constructed mutant histone H3 genes that replaced Lys-9 with Leu or Arg and introduced the mutant genes into a strain bearing an allele of the bacterial hph gene that was silenced by DNA methylation (43). The transformants showed reactivation of hph and dramatic loss of methylation of all chromosomal regions examined (42). Thus methylation of histone H3 controls DNA methylation in Neurospora.
It seems likely that DIM-5 serves to integrate information relevant to whether DNA in a particular chromosome region should be methylated. In vitro assays of DIM-5 activity on various substrates showed that the enzyme is strongly inhibited by methylation of Lys-4, phosphorylation of Ser-10 or acetylation of Lys-14 of histone H3 (E. Schmidt, H.T., and E.U.S., unpublished work), similar to observations with other K9 histone H3 methyltransferases (44–46). This provides a possible mechanistic explanation for our observation that an inhibitor of an HDAC can inhibit DNA methylation (26). Interestingly, and quite unexpectedly, DIM-5 showed strong activity on a peptide dimethylated at Lys-9 (data not shown). This contrasts with available information on previously described H3 histone methyltransferase (44, 45) and raises the possibility that trimethyl-K9 is the specific mark for cytosine methylation in Neurospora.
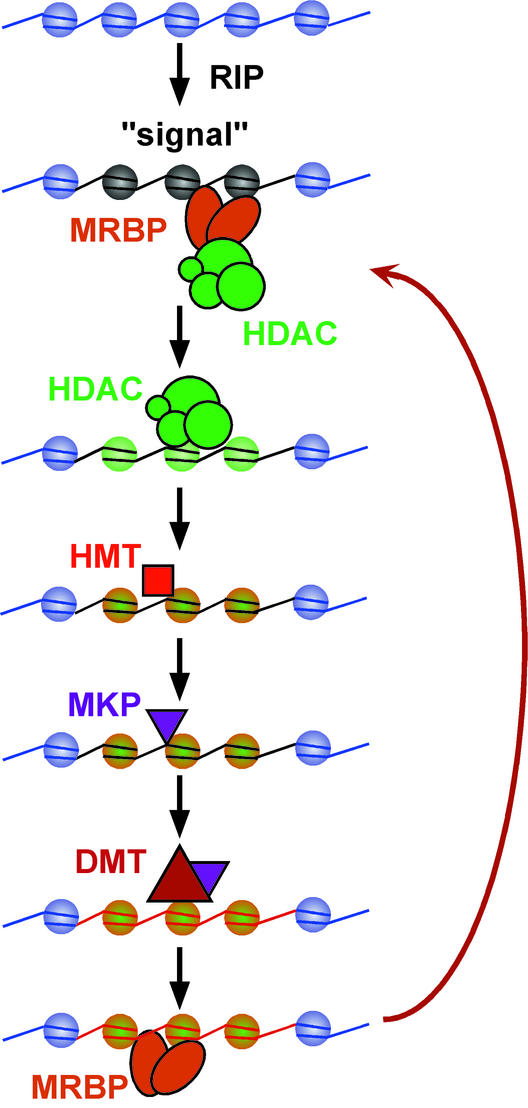
Our working model (Fig. 7) summarizes the likely elements of the DNA methylation machinery of Neurospora. The aberrant base composition resulting from RIP may be recognized by MRBP-1 or a similar factor that recruits an HDAC complex. Based on findings in other eukaryotes that have revealed connections between such complexes and chromatin remodeling factors (CRFs; ref. 47) and on indications from plants (48) and animals (49, 50) that putative CRFs are involved in DNA methylation, it seems likely that one or more such factors are involved in this step. At present, we do not know which amino acid residues of histones must be hypoacetylated to allow for the action of DIM-5, but they probably include K9 and K14 of histone H3 (44) and perhaps one or more of K5, K8, K12, and K16 of histone H4. In addition, we have preliminary evidence that DIM-5, or a DIM-5 complex, is sensitive to other modifications, including the methylation, phosphorylation, and acetylation state of K4, S10, and K14 of histone H3, respectively (data not shown), consistent with observations on histone methyltransferases from other organisms (44, 45, 46). The product of DIM-5 may be recognized by a chromo-domain protein, such as a homologue of HP1 (47). There is at least one candidate for such a protein in the Neurospora proteome (www-genome.wi.mit.edu/annotation/fungi/Neurospora/). The hypothetical Neurospora methyl-lysine recognition protein (MKP) may directly or indirectly recruit the DMT, DIM-2. Conceivably, DIM-2 could directly recruit chromatin modification factors, as in animals (51). In any case, once methylated, the DNA should attract methyl-binding proteins, such as MRBP-1, which would again recruit chromatin modification enzymes, strengthening the epigenetic loop and resulting in the observed propagation of DNA methylation.
Figure 7.
Stylized working model for the induction and maintenance of DNA methylation in Neurospora crassa. The precise nature of the signal for DNA methylation resulting from heavy mutagenesis by RIP (indicated by black DNA on gray nucleosomes) is unknown but seems to consist, at least in part, of structural features of A⋅T-rich DNA (H.T. and E.U.S., unpublished work; ref. 53). The signal may be recognized by a factor such as MRBP-1, which recognizes RIP-mutated and/or methylated DNA (Fig. 5). Such a factor may directly or indirectly (e.g., through chromatin remodeling factors) recruit one or more HDACs. Histone hypoacetylation (green nucleosomes) of particular residues is postulated to be one prerequisite for DIM-5 histone methyltransferase (HMT) activity. Methylated K9 (orange nucleosomes), which results from the action of DIM-5 on histone H3 (Fig. 7), is presumably recognized by a chromo-domain protein (47) or some other methyl-lysine recognition protein (MKP). The MKP then would directly or indirectly cause the DMT, DIM-2 (39), to methylate (red) the associated DNA. Once methylated, the DNA may bind more strongly to MBRP and/or other methyl-DNA binding proteins, and these, in turn, could again recruit HDAC complexes, resulting in a maintenance loop (red arrow). Whether the various factors that are (for simplicity) shown transiently associated with chromatin actually remain associated with the chromatin is not yet established.
Acknowledgments
Funding was provided by grants from the Human Frontiers Science Program (RG304/95M) and the National Institutes of Health (GM35690) (to E.U.S.).
Abbreviations
- DMT
DNA methyltransferase
- RIP
repeat-induced point mutation
- MRBP-1
methyl/RIP binding protein 1
- 5AC
5-azacytidine
- HDAC
histone deacetylase
- TSA
trichostatin A
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Self-Perpetuating Structural States in Biology, Disease, and Genetics,” held March 22–24, 2002, at the National Academy of Sciences in Washington, DC.
References
- 1.Holliday R, Pugh J E. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 2.Riggs A D. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 3.Harland R. Proc Natl Acad Sci USA. 1982;79:2323–2327. doi: 10.1073/pnas.79.7.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein R, Gruenbaum Y, Pollack Y, Razin A, Cedar H. Proc Natl Acad Sci USA. 1982;79:61–65. doi: 10.1073/pnas.79.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wigler M, Levy D, Perucho M. Cell. 1981;24:33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- 6.Lorincz M C, Schubeler D, Groudine M. Mol Cell Biol. 2001;21:7913–7922. doi: 10.1128/MCB.21.23.7913-7922.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeddeloh J A, Bender J, Richards E J. Genes Dev. 1998;12:1714–1725. doi: 10.1101/gad.12.11.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bestor T H, Ingram V M. Proc Natl Acad Sci USA. 1985;82:2674–2678. doi: 10.1073/pnas.82.9.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolden A H, Nalin C M, Ward C A, Poonian M S, Weissbach A. Mol Cell Biol. 1986;6:1135–1140. doi: 10.1128/mcb.6.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selker E U, Stevens J N. Proc Natl Acad Sci USA. 1985;82:8114–8118. doi: 10.1073/pnas.82.23.8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selker E U, Fritz D Y, Singer M J. Science. 1993;262:1724–1728. doi: 10.1126/science.8259516. [DOI] [PubMed] [Google Scholar]
- 12.Toth M, Müller U, Doerfler W. J Mol Biol. 1990;214:673–683. doi: 10.1016/0022-2836(90)90285-T. [DOI] [PubMed] [Google Scholar]
- 13.Woodcock D M, Crowther P J, Diver W P. Biochem Biophys Res Commun. 1987;145:888–894. doi: 10.1016/0006-291x(87)91048-5. [DOI] [PubMed] [Google Scholar]
- 14.Selker E U. Trends Biochem Sci. 1990;15:103–107. doi: 10.1016/0968-0004(90)90193-f. [DOI] [PubMed] [Google Scholar]
- 15.Yisraeli J, Szyf M. In: DNA Methylation: Biochemistry and Biological Significance. Razin A, Cedar H, Riggs A D, editors. New York: Springer; 1984. pp. 353–378. [Google Scholar]
- 16.Turker M S, Swisshelm K, Smith A C, Martin G M. J Biol Chem. 1989;264:11632–11636. [PubMed] [Google Scholar]
- 17.Finnegan E J, Brettell R I S, Dennis E S. In: DNA Methylation: Molecular Biology and Biological Significance. Jost J P, Saluz H P, editors. Basel: Birkäuser; 1993. pp. 218–261. [Google Scholar]
- 18.Kishimoto N, Sakai H, Jackson J, Jacobsen S E, Meyerowitz E M, Dennis E S, Finnegan E J. Plant Mol Biol. 2001;46:171–183. doi: 10.1023/a:1010636222327. [DOI] [PubMed] [Google Scholar]
- 19.Pelissier T, Thalmeir S, Kempe D, Sanger H L, Wassenegger M. Nucleic Acids Res. 1999;27:1625–1634. doi: 10.1093/nar/27.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goubely C, Arnaud P, Tatout C, Heslop-Harrison J S, Deragon J M. Plant Mol Biol. 1999;39:243–255. doi: 10.1023/a:1006108325504. [DOI] [PubMed] [Google Scholar]
- 21.Lindroth A M, Cao X, Jackson J P, Zilberman D, McCallum C M, Henikoff S, Jacobsen S E. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 22.Ramsahoye B H, Biniszkiewicz D, Lyko F, Clark V, Bird A P, Jaenisch R. Proc Natl Acad Sci USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vu T H, Li T, Nguyen D, Nguyen B T, Yao X M, Hu J F, Hoffman A R. Genomics. 2000;64:132–143. doi: 10.1006/geno.1999.6094. [DOI] [PubMed] [Google Scholar]
- 24.Rossignol J-L, Faugeron G. In: Gene Silencing in Higher Plants and Related Phenomena in Other Eukaryotes. Meyer P, editor. Vol. 197. Heidelberg: Springer; 1994. p. 26. [Google Scholar]
- 25.Selker E U. Adv Genet. 2002;46:439–450. doi: 10.1016/s0065-2660(02)46016-6. [DOI] [PubMed] [Google Scholar]
- 26.Selker E U. Proc Natl Acad Sci USA. 1998;95:9430–9435. doi: 10.1073/pnas.95.16.9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rountree M R, Bachman K E, Herman J G, Baylin S B. Oncogene. 2001;20:3156–3165. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- 28.Selker E U, Garrett P W. Proc Natl Acad Sci USA. 1988;85:6870–6874. doi: 10.1073/pnas.85.18.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selker E U. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- 30.Selker E U, Cambareri E B, Jensen B C, Haack K R. Cell. 1987;51:741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- 31.Cambareri E B, Jensen B C, Schabtach E, Selker E U. Science. 1989;244:1571–1575. doi: 10.1126/science.2544994. [DOI] [PubMed] [Google Scholar]
- 32.Freitag M, Williams R, Kothe G O, Selker E U. Proc Natl Acad Sci USA. 2002;99:8802–8807. doi: 10.1073/pnas.132212899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watters M K, Randall T A, Margolin B S, Selker E U, Stadler D R. Genetics. 1999;153:705–714. doi: 10.1093/genetics/153.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer M J, Marcotte B A, Selker E U. Mol Cell Biol. 1995;15:5586–5597. doi: 10.1128/mcb.15.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selker E U, Richardson G A, Garrett-Engele P W, Singer M J, Miao V. Cold Spring Harbor Symp Quant Biol. 1993;58:323–329. doi: 10.1101/sqb.1993.058.01.038. [DOI] [PubMed] [Google Scholar]
- 36.Cambareri E B, Singer M J, Selker E U. Genetics. 1991;127:699–710. doi: 10.1093/genetics/127.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao V P W, Singer M J, Rountree M R, Selker E U. Mol Cell Biol. 1994;14:7059–7067. doi: 10.1128/mcb.14.11.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rountree M R, Selker E U. Genes Dev. 1997;11:2383–2395. doi: 10.1101/gad.11.18.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouzminova E A, Selker E U. EMBO J. 2001;20:4309–4323. doi: 10.1093/emboj/20.15.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foss H M, Roberts C J, Claeys K M, Selker E U. Science. 1993;262:1737–1741. doi: 10.1126/science.7505062. [DOI] [PubMed] [Google Scholar]
- 41.Dobosy J R, Selker E U. Cell Mol Life Sci. 2001;58:721–727. doi: 10.1007/PL00000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamaru H, Selker E U. Nature (London) 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 43.Irelan J T, Selker E U. Genetics. 1997;146:509–523. doi: 10.1093/genetics/146.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakayama J, Rice J C, Strahl B D, Allis C D, Grewal S I. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 45.Rea S, Eisenhaber F, O'Carroll D, Strahl B D, Sun Z W, Schmid M, Opravil S, Mechtler K, Ponting C P, Allis C D, Jenuwein T. Nature (London) 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 46.Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis C D, Tempst P, Reinberg D. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richards E J, Elgin S C. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 48.Jeddeloh J A, Stokes T L, Richards E J. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 49.Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibbons R J, McDowell T L, Raman S, O'Rourke D M, Garrick D, Ayyub H, Higgs D R. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 51.Rountree M R, Bachman K E, Baylin S B. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 53.Miao V P, Freitag M, Selker E U. J Mol Biol. 2000;300:249–273. doi: 10.1006/jmbi.2000.3864. [DOI] [PubMed] [Google Scholar]