Abstract
The tomato Pti4 gene encodes a transcription factor that was identified on the basis of its specific interaction with the product of the Pto disease resistance gene in a yeast two-hybrid system. We show here that the Pti4 protein specifically binds the GCC-box cis element, which is present in the promoter region of many pathogenesis-related (PR) genes. Expression of the Pti4 gene in tomato leaves was rapidly induced by ethylene and by infection with Pseudomonas syringae pv tomato, and this induction preceded expression of GCC-box-containing PR genes. Although salicylic acid also induced Pti4 gene expression, it did not induce GCC-box PR genes. Rather, salicylic acid antagonized ethylene-mediated expression of GCC-box PR genes. We demonstrate that the Pti4 protein is specifically phosphorylated by the Pto kinase and that this phosphorylation enhances binding of Pti4 to the GCC box. In addition, induced overexpression of Pto and Pti4 in tomato leaves resulted in a concomitant increase in GCC-box PR genes. Our results support a model in which phosphorylation of the Pti4 protein by the Pto kinase enhances the ability of Pti4 to activate expression of GCC-box PR genes in tomato.
INTRODUCTION
A well-characterized plant defense response associated with pathogen attack is the expression of pathogenesis-related (PR) genes. PR genes are activated in both resistant and susceptible plants in response to pathogen attack. However, they often are expressed more rapidly and to a greater extent in incompatible interactions in which a resistant plant is challenged with an avirulent pathogen (Voisey and Slusarenko, 1989; van Kan et al., 1992; Jia and Martin, 1999). This implies that enhanced PR gene expression is mediated by a recognition event involving a disease resistance (R) gene and its cognate avirulence (avr) gene. The roles of PR genes in plant defense responses against pathogens have been widely investigated (Cutt and Klessig, 1992). The best-characterized plant defense genes include those that encode glucanases and chitinases. These enzymes appear to be antimicrobial, based on their ability to degrade cell wall components of pathogens (Mauch et al., 1988; Sela-Buurlage et al., 1993) and have been shown to enhance disease resistance when overexpressed in plants (Broglie et al., 1991; Zhu et al., 1994). Despite the established biological function in plant defense responses for some PR genes, the molecular mechanisms responsible for their expression in R–avr recognition remain largely unclear.
The expression of many defense-related genes is regulated by signaling molecules such as salicylic acid (SA) and ethylene. Increases in SA and its conjugates have been associated with the activation of defense genes in many plant species (reviewed in Dempsey et al., 1999). Compelling evidence for a key role of SA in defense gene regulation comes from analysis of tobacco and Arabidopsis NahG transformants in which SA is converted to an inactive form. The NahG plants fail to induce several PR genes such as PR-1, PR-2, and PR-5 and show increased susceptibility to pathogens (Delaney et al., 1994). Additional evidence for a role of SA in defense comes from certain Arabidopsis mutants in which the SA signaling pathway is blocked (Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997). These plants are unable to accumulate several PR gene transcripts when infected by pathogens and show increased susceptibility to disease. Most of the PR genes induced by SA encode acidic, extracellular forms of PR proteins (Friedrich et al., 1995). Another large class of PR genes encodes basic proteins, and many of these genes do not appear to be regulated by SA (Vidal et al., 1997).
Many PR genes that are induced by plant–pathogen interactions also are upregulated by exposure to ethylene (Deikman, 1997). Ethylene-regulated PR genes often encode basic proteins that are localized primarily in vacuoles (Eyal et al., 1993). Recent evidence indicates that expression of some of these PR genes is independent of SA but requires an ethylene-dependent signaling pathway. For example, the PDF1.2 gene, which encodes an antifungal peptide, is activated when Arabidopsis is infected by Alternaria brassicicola or exposed to ethylene; however, SA does not activate PDF1.2 (Penninckx et al., 1996). Activation of PDF1.2 was abolished in ethylene-insensitive mutants of Arabidopsis, whereas its induction by pathogens or ethylene was not affected in either NahG plants or a nonexpresser of PR genes (npr1) mutant (Penninckx et al., 1998).
An important approach to understanding PR gene regulation is to identify defense-responsive promoter regions and isolate their cognate DNA binding proteins. Several defense-related transcription factors have been identified by using this method, including the WRKY transcription factors (Rushton et al., 1996), a basic leucine zipper transcription factor G/HBF-1 (Dröge-Laser et al., 1998), and the ethylene-responsive element binding proteins (EREBPs; Ohme-Takagi and Shinshi, 1995). Isolation of these factors has provided new insights into PR gene regulation but also has raised the additional question of how these transcription factors themselves are regulated. In particular, because many PR genes are expressed rapidly during gene-for-gene incompatible interactions, it is important to understand the role of R genes in the regulation of defense-related transcription factors.
In tomato, the R gene Pto confers resistance to Pseudomonas syringae pv tomato strains expressing the avrPto gene. The molecular cloning of the Pto gene and the corresponding avirulence gene, avrPto, permits extensive investigation of the mechanism of R gene–mediated defense responses. Pto encodes an active serine/threonine protein kinase (Martin et al., 1993; Loh and Martin, 1995), and avrPto encodes a small hydrophilic protein that probably is delivered into the plant cell by way of a bacterial type III secretion system (Ronald et al., 1992; Alfano and Collmer, 1996). We and others have demonstrated that the Pto kinase interacts with AvrPto protein, and mutations in Pto or AvrPto that disrupt this interaction also abolish disease resistance (Scofield et al., 1996; Tang et al., 1996; Frederick et al., 1998). Therefore, a direct interaction between Pto and AvrPto determines gene-for-gene specificity in this plant–pathogen interaction. This recognition event appears to initiate multiple signal transduction pathways that lead to the hypersensitive response, oxidative burst, and enhanced expression of PR genes (Gu and Martin, 1998).
Several proteins with putative roles in Pto-mediated signaling pathways have been identified from a yeast two-hybrid screen by using the Pto kinase as bait (Zhou et al., 1995, 1997). Among them, Pti1 (for Pto-interacting 1), which encodes a serine/threonine kinase, appears to be a direct downstream component because Pto phosphorylates Pti1 but not vice versa. Overexpression of Pti1 in tobacco indicates that the protein is involved in the development of the hypersensitive response (Zhou et al., 1995). Three other genes encoding Pto-interacting proteins, Pti4, Pti5, and Pti6, encode related putative transcription factors; each has a DNA binding domain, a nuclear localization sequence, and a possible transactivation domain rich in acidic amino acids (Zhou et al., 1997). The proteins encoded by Pti4, Pti5, and Pti6 share extensive similarity with the amino acid sequences of EREBPs (Ohme-Takagi and Shinshi, 1995). Along with the EREBPs, Pti5 and Pti6 have been shown to bind the GCC-box cis element present in the promoter region of many ethylene-regulated PR genes (Ohme-Takagi and Shinshi, 1995; Zhou et al., 1995). Thus, the discovery of Pti4, Pti5, and Pti6 represents a definitive link between an R gene and the specific induction of genes involved in plant defense. In this article, we have further characterized the regulation of Pti4 gene expression and the relationship between the Pto kinase and the Pti4 protein.
RESULTS
Pti4 Protein Specifically Binds the GCC-Box cis Element
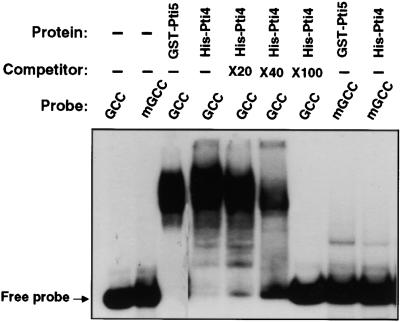
To examine whether Pti4 binds the same cis element as Pti5 and Pti6 (Zhou et al., 1997), we overexpressed Pti4 as a tagged protein with a region of six histidine residues in Escherichia coli and purified to near homogeneity by using a nickel–agarose affinity column (see Methods). The purified His-Pti4 protein was used in a mobility shift gel assay. As shown in Figure 1, in which glutathione S-transferase (GST)–Pti5 was used as a positive control, the His-Pti4 fusion protein was able to bind the GCC box. To test the specificity of this binding, we performed a competition experiment by adding the unlabeled GCC-box DNA to the mobility shift assay. The addition of unlabeled GCC-box to the binding assay decreased the binding of His-Pti4 to the labeled GCC-box, and 100-fold excess of unlabeled GCC-box DNA resulted in the complete loss of binding of the labeled GCC-box to the His-Pti4 protein. Furthermore, as was seen with GST-Pti5, the His-Pti4 protein did not bind a mutated GCC (mGCC) box containing two G-to-T substitutions in the GCCGCC core sequence (Ohme-Takagi and Shinshi, 1995; Zhou et al., 1997). From these results, we conclude that Pti4 specifically binds the GCC-box cis element.
Figure 1.
Pti4 Specifically Binds the GCC-Box Sequence.
A mobility shift assay was performed as described previously (Zhou et al., 1995), using 50 ng of purified His-Pti4 or GST-Pti5 fusion protein mixed with 5 fmol of 32P-labeled wild-type GCC-box or mGCC-box oligonucleotides (see Methods). The competitor consisted of the unlabeled wild-type GCC oligonucleotide. The notations ×20, ×40, and ×100 indicate that the amount of unlabeled wild-type GCC-box oligonucleotide added to these reactions was 20-, 40-, or 100-fold that of the labeled GCC-box oligonucleotide. The (−) signs indicate that no competitor, or no protein, was added to these reactions. The position of the unbound (Free) probe is shown.
Ethylene Rapidly Increases Abundance of Pti4 but Not of Pti5 or Pti6 Transcripts
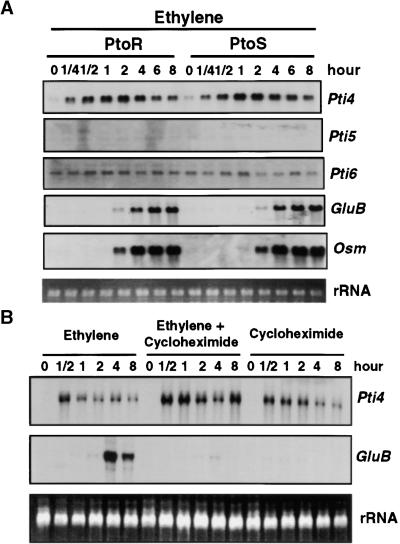
Pti4, Pti5, and Pti6 are similar to the tobacco EREBPs in both amino acid sequence and DNA binding properties (Ohme-Takagi and Shinshi, 1995; Zhou et al., 1997). We were interested, therefore, in whether ethylene is involved in the transcriptional regulation of the Pti4, Pti5, and Pti6 genes the way it is for some of the genes encoding EREBPs (Ohme-Takagi and Shinshi, 1995). For these experiments, we used two lines of the tomato cultivar Rio Grande that are isogenic except for the Pto locus: Rio Grande–PtoR (RG-PtoR) and Rio Grande–PtoS (RG-PtoS). RNA gel blot experiments (Figure 2A) revealed that in plants from both lines, Pti4 transcript abundance increased as early as 15 min after exposure to 10 ppm ethylene gas and continued to increase over the next 2 to 4 hr before gradually decreasing. Thus, Pto had no effect on the expression of the Pti4 gene in response to ethylene. In contrast to the rapid induction of Pti4 by ethylene, Pti5 and Pti6 were not responsive to ethylene treatment. The transcript levels of Pti5 and Pti6 remained low throughout the 8-hr ethylene treatment.
Figure 2.
Effect of Ethylene and Cycloheximide on Expression of Pti4, Pti5, Pti6, and GCC-Box-Containing PR Genes.
Fully expanded leaves from 4-week-old tomato plants were used for analysis of gene expression. Equal loading of lanes in all gels was verified by visualizing the rRNA in the RNA gels stained with ethidium bromide.
(A) RG-PtoR or RG-PtoS tomato plants were treated with 10 μL/L ethylene gas for the times indicated. Total RNAs were isolated from leaves and analyzed as duplicate RNA gel blots by hybridization with radiolabeled Pti4, Pti5, Pti6, GluB, or Osm probes.
(B) RG-PtoR plants were treated with ethylene (10 μL/L), cycloheximide (5 μg/mL), or a combination of both for the times indicated. Total RNAs were isolated from leaves and analyzed as duplicate RNA gel blots by hybridization with radiolabeled Pti4 or GluB probes.
A key role for the GCC box in mediating ethylene-responsive gene expression has been demonstrated in tobacco (Eyal et al., 1993; Ohme-Takagi and Shinshi, 1995). Recently, we identified a GCC box in the promoter regions of several tomato PR genes, including glucanase B (GluB), chitinase B (ChiB), and osmotin (Osm), and showed that expression of these genes was enhanced in an incompatible interaction involving Pto and avrPto (Jia and Martin, 1999). To determine whether these GCC-box-containing PR genes also were induced by ethylene, we hybridized the RNA gel blots with GluB and Osm probes (Figure 2A). The accumulation of GluB and Osm transcripts first was detected 2 hr after ethylene treatment and increased to a maximum by 4 to 6 hr. In a control experiment in which plants were similarly manipulated but not exposed to ethylene, neither Pti4 nor GluB transcripts were induced (data not shown). The first appearance of GluB and Osm transcripts thus was ∼1.5 hr after that of Pti4 transcripts. This result is consistent with Pti4 playing a role in the expression of these tomato GCC-box-containing PR genes. No difference in ethylene induction of GluB and Osm was observed between the RG-PtoR and RG-PtoS plants; therefore, the Pto gene does not appear to be involved in ethylene-regulated expression of these PR genes.
Effect of Cycloheximide on Ethylene Regulation of the Pti4 and GluB Genes
The rapid induction of Pti4 transcripts by ethylene suggests that de novo protein synthesis is not required for this process. This is typical for the induction of many immediate-early transcription factors, including those that appear to be involved in regulation of plant defense genes (Yang and Klessig, 1996). To examine whether de novo protein synthesis is involved in ethylene-induced Pti4 function, we blocked protein synthesis by pretreatment of tomato plants with 5 μg/mL cycloheximide for 30 min before applying ethylene. Cycloheximide commonly is observed to induce transcription factor genes (Suzuki et al., 1998; Yang and Klessig, 1996; Horvath et al., 1998), and as shown in Figure 2C, it increased Pti4 expression with timing and transcript levels similar to those induced by ethylene. Treatment with a combination of ethylene and cycloheximide further increased the abundance of Pti4 transcripts, possibly as a result of the additive effect of ethylene and cycloheximide. These findings suggest that transcriptional activation of Pti4 by ethylene occurs by way of preformed transactivator molecules in the plant cell. Alternatively, cycloheximide may inhibit the de novo protein synthesis of an unstable protein that negatively regulates the Pti4 transcripts, or cycloheximide might simply stabilize Pti4 transcripts (Li et al., 1994). Despite the increase in Pti4 transcripts in these experiments, GluB expression was not induced. In fact, cycloheximide treatment blocked the induction of GluB by ethylene, as observed after the combination treatment of cycloheximide and ethylene (Figure 2C). Therefore, induction of GluB by ethylene apparently requires de novo synthesis of the protein or proteins that activate its expression.
Effect of SA on Expression of Pti4, Pti5, Pti6, and GCC-Box PR Genes
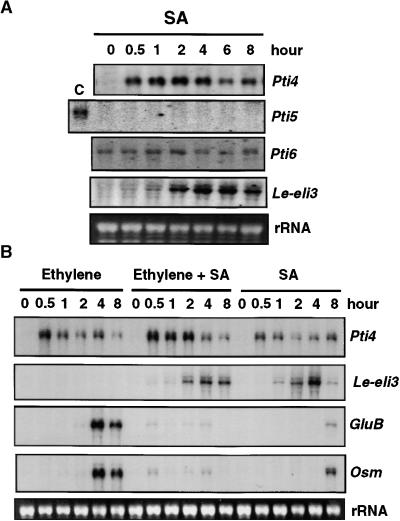
SA induces the expression of a wide array of plant genes that can be classified on the basis of how quickly they are induced (Dempsey et al., 1999). Several transcription factor genes, including tobacco EREBP-1, are immediate-early SA-responsive genes, being activated within 10 min of SA treatment (Horvath et al., 1998). We examined possible effects of SA on Pti4, Pti5, and Pti6 transcript abundance and found, as shown in Figure 3A, that Pti4 transcripts accumulated as early as 30 min after treatment, whereas Pti5 and Pti6 transcript levels were not responsive to SA. In a control experiment in which plants were treated with either water or 3-hydroxybenzoic acid (an inactive SA analog), induction of Pti4 transcripts was not detected (data not shown). To test whether SA-induced accumulation of Pti4 correlated with GCC-box PR gene accumulation, we hybridized an RNA gel blot with the GluB and Osm probes (Figure 3B). An increase in transcript abundance of these genes was not detected until 8 hr after treatment, and even then, the increase was negligible. To ensure that the SA-signaling pathway was activated by our SA treatment, we used a known SA-inducible gene, Le-eli3 (R.L. Thilmony and G.B. Martin, unpublished data), as a marker. Transcripts of Le-eli3 were detected 2 hr after treatment.
Figure 3.
Effect of SA and Ethylene on Expression of Pti4, Pti5, Pti6, Le-eli3, and the GCC-Box-Containing PR Genes GluB and Osm.
Fully expanded leaves from 4-week-old tomato plants were used for analysis of gene expression. Equal loading was verified by visualizing the rRNA in the RNA gels stained with ethidium bromide.
(A) RG-PtoR tomato plants were treated with 1 mM SA for the times indicated. Total RNAs were isolated from leaves and analyzed as duplicate RNA gel blots by hybridization with radiolabeled Pti4, Pti5, Pti6, or Le-eli3 probes. Note that lane C for Pti5 contains total RNA isolated from leaves of 8-week-old tomato plants in which the Pti5 transcript is detectable.
(B) RG-PtoR tomato plants were treated with ethylene (10 μL/L), SA (1 mM), or a combination of both for the times indicated. Total RNAs were isolated from leaves and analyzed as duplicate RNA gel blots by hybridization with radiolabeled Pti4, Le-eli3, GluB, or Osm probes.
Several recent reports indicate that both SA-dependent and SA-independent pathways are responsible for activation of distinct sets of defense-related genes (Penninckx et al., 1996; Vidal et al., 1997; Thomma et al., 1998). In addition, certain components might be shared by both pathways. SA also has been reported to sensitize ethylene-inducible PR gene expression, and ethylene may enhance SA-induced expression of some PR genes (Lawton et al., 1994; Xu et al., 1994). To examine a possible interaction between SA and ethylene in the activation of Pti4, Pti5, and Pti6 and in the expression of PR genes containing GCC boxes, we treated tomato plants simultaneously with SA and ethylene. As shown in Figure 3B, this simultaneous treatment increased Pti4 transcripts by approximately twice that seen with either treatment alone and might reflect an additive effect. Ethylene did not activate Le-eli3 and did not affect Le-eli3 expression induced by SA in comparison with that induced by SA treatment alone. Interestingly, however, we found that SA substantially suppressed ethylene induction of GluB and Osm expression (Figure 3B). Together with our results shown in Figure 3A, we conclude that SA does not cause early induction of these PR genes and, in fact, has a negative effect on their activation by ethylene.
Induction of Pti4, Pti5, and Pti6 by the Wound Response Pathway
In tomato, ethylene is required for activation of proteinase inhibitor genes by the wound response pathway, although ethylene itself is not sufficient to induce wound-responsive gene expression (O'Donnell et al., 1996). Wounding of tomato leaves results in increased ethylene production (O'Donnell et al., 1996), and some PR genes are induced by mechanical wounding (Pastuglia et al., 1997). Furthermore, a GCC-box cis element is present in the promoter of at least some members of the 1-aminocyclopropane-1-carboxylate oxidase gene family, which is involved in ethylene biosynthesis (Jia and Martin, 1999). These observations suggest that Pti4, Pti5, and Pti6, singly or in combination, might be involved in the regulation of ethylene production in the plant and in turn play a role in wound-responsive gene expression. As shown in Figure 4A, wounding tomato leaves rapidly induced Pti4 transcripts but not Pti5 or Pti6 transcripts. Pti4 transcript abundance was very low, however, and quickly decreased to near background values just 6 hr after wounding. This corresponded to very low induction of GluB transcripts beginning 6 hr after wounding. By contrast, transcripts of proteinase inhibitor 2 gene (Pin2), a molecular marker for the wound response, increased markedly 4 hr after wounding and continued to accumulate to high amounts 8 hr after wounding. To further examine the possible involvement of Pti4 in the wound response pathway, we treated tomato plants with systemin, a well-characterized, potent signaling molecule involved in the wound response through the octodecanoid pathway (Schaller and Ryan, 1995). As shown in Figure 4B, systemin strongly increased Pin2 transcripts but it did not increase Pti4 transcripts. In related experiments, jasmonic acid also was shown not to induce Pti4 accumulation (Thara et al., 1999). Thus, the induction of Pti4 by wounding might not result from activation of the octadecanoid pathway. Rather, similar to tobacco EREBP genes, induction of Pti4 by wounding might result from a separate mechanism of wound-responsive transcriptional activation (Suzuki et al., 1998).
Figure 4.
Effect of Wounding or Systemin on Expression of Pti4, Pti5, Pti6, Pin2, and GluB.
Fully expanded leaves from 4-week-old tomato plants were used for analysis of gene expression. Equal loading was verified by visualizing the rRNA in the RNA gels stained with ethidium bromide. Pin II, Pin2.
(A) Tomato RG-PtoR plants were wounded by rubbing leaves with carborundum, after which total RNAs were isolated from leaves at the times indicated. Duplicate RNA gel blots were hybridized with radiolabeled Pti4, Pti5, Pti6, Pin2, or GluB probes. Note that lane C for Pti5 contains total RNA isolated from leaves of 8-week-old tomato plants in which Pti5 transcript is detectable.
(B) Tomato RG-PtoR plants were treated with 1 nM systemin, after which total RNAs were isolated from leaves at the times indicated. Duplicated RNA gel blots were hybridized with radiolabeled Pti4 or Pin2 probes.
Developmental and Organ-Specific Expression of Pti4, Pti5, and Pti6
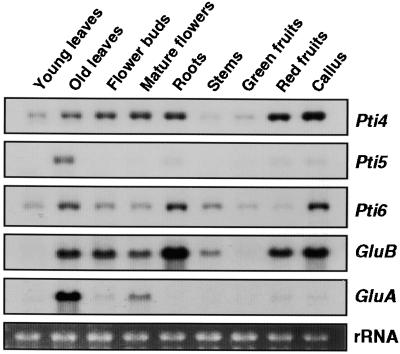
Expression of PR genes varies in different plant tissues and during different developmental stages (Cutt and Klessig, 1992). To investigate the possible roles of Pti4, Pti5, and Pti6 in regulation of PR gene expression in different tissues and organs at different developmental stages, we isolated tomato RNAs and analyzed them by RNA gel blot hybridization. As shown in Figure 5, Pti4 transcript abundance was less in young leaves than in old leaves. Low amounts of Pti4 transcripts also were found in stems and green fruits, whereas high amounts were detected in reproductive organs, roots, red fruits, and undifferentiated callus cells. In contrast, Pti5 transcripts were present only in low or undetectable amounts in all tissues, except for older leaves. The amounts of Pti6 transcripts were similar to those of Pti4 in all cases except red fruits, in which they were less abundant. Because Pti4 is induced by ethylene, the high amount of Pti4 transcripts in red fruits may have resulted from the high amounts of this hormone known to occur in ripening fruits. Consistent with this observation, Pti4 transcripts also were increased in senescent leaves and reproductive organs in which ethylene biosynthesis is increased (Abeles et al., 1992). However, because Pti5 and Pti6 are not induced by ethylene (Figure 2), the expression of these genes in some of the same tissues as Pti4 suggests that other developmental cues are involved in regulation of Pti5 and Pti6 expression.
Figure 5.
Expression of Pti4, Pti5, Pti6, GluB, and GluA in Various Tissues at Different Developmental Stages.
The third and fourth leaves of 3- to 4-week-old or 11- to 12-week-old tomato plants were harvested as young and old leaves, respectively. Unopened and fully opened flowers are referred to as flower buds and mature flowers, respectively. Root and stem tissues were harvested from 3- to 4-week-old plants. Green fruits were immature fruits, and red fruits were red ripe fruits. Total RNAs were isolated from these tissues, and duplicate RNA gel blots were hybridized with the Pti4, Pti5, Pti6, GluB, or GluA probes. Equal loading was verified by visualizing the rRNA in the RNA gel stained with ethidium bromide.
To examine the correlation of Pti4, Pti5, and Pti6 transcript levels with PR gene expression during development, we probed the same RNA gel blots with GluB and GluA. Expression of the GCC-box-containing gene GluB correlated well with Pti4 expression, whereas the GluA transcripts did not. The promoter region of GluA has not been characterized, but on the basis of these results, we speculate that it does not contain a GCC box.
Expression of Pti4 and Pti6 in Compatible or Pto-Mediated Incompatible Interactions
We observed previously that expression of GCC-box-containing PR genes is enhanced in an incompatible interaction involving Pto-containing tomato and the bacterial pathogen P. s. tomato expressing avrPto (Jia and Martin, 1999). Because the Pti4, Pti5, and Pti6 proteins all interact with Pto kinase in a yeast two-hybrid system and specifically bind the GCC box, we wanted to examine the possible mechanisms by which interaction of Pto kinase with these transcription factors regulates PR gene expression. We began by determining whether induction of Pti4, Pti5, or Pti6 transcripts correlated with an increase in PR gene expression during an incompatible interaction. Leaves of tomato cultivars RG-PtoR and RG-PtoS were vacuum-infiltrated with P. s. tomato DC3000 expressing the avrPto gene, and RNA was isolated over the time course shown in Figure 6A. Pti4 transcripts accumulated 30 min after inoculation with P. s. tomato, regardless of the expression of the Pto gene in the tomato leaves.
Figure 6.
Expression of Pti4, Pti6, GluB, and Osm in an Incompatible or Compatible Interaction Involving P. s. tomato.
(A) RG-PtoR or RG-PtoS tomato plants were vacuum-infiltrated with 2.2 × 107 colony-forming units per milliliter of P. s. tomato DC3000 expressing the avrPto gene in 10 mM MgCl2 and 0.005% Silwet.
(B) RG-PtoR tomato plants were vacuum-infiltrated with 10 mM MgCl2 and 0.005% Silwet as a control.
Leaf tissues were harvested at the indicated times after vacuum infiltration. Total RNAs were isolated, and duplicate RNA gel blots were hybridized with radiolabeled Pti4, Pti6, GluB, or Osm probes. Equal loading was verified by visualizing the rRNA in the RNA gels stained with ethidium bromide.
Experiments with virulent P. s. tomato T1 produced similar observations and further showed that this induction of Pti4 does not require SA or ethylene (Thara et al., 1999). In addition, Pti5 transcripts also are induced in both compatible and incompatible interactions with only slightly enhanced expression in the latter interaction (Thara et al., 1999). Pti6 transcripts were not induced by pathogen inoculation in either RG-PtoR or RG-PtoS plants (Figure 6A). As observed previously (Jia and Martin, 1999), expression of the GluB and Osm genes was enhanced substantially in the incompatible interaction (RG-PtoR) compared with their expression in the compatible interaction (RG-PtoS) (Figure 6A). These results indicate that enhanced expression of these GCC-box-containing PR genes during interactions with P. s. tomato is not directly correlated with the transcript amounts of Pti4, Pti5, and Pti6.
Because Pti4 transcripts were moderately induced by wounding (Figure 4A), a mock inoculation was included as a control for comparing the difference of Pti4 transcripts induced by wounding and by vacuum infiltration of P. s. tomato. As shown in Figure 6B, Pti4 transcripts did increase during the early sampling points, but the transcript amounts declined markedly 4 hr after mock infiltration. This corresponded to a slight increase of GluB transcripts at the 4-, 6-, and 8-hr times. Plants inoculated with bacteria, in contrast, showed substantially greater and more prolonged expression of Pti4 transcripts (Figure 6A).
Pto Kinase Phosphorylates Pti4 Protein
In mammalian systems, the activity of transcription factors often is regulated by phosphorylation or dephosphorylation events (Hunter and Karin, 1992). Because the product of the Pto resistance gene is an active serine/threonine kinase (Loh and Martin 1995), it is possible that this kinase phosphorylates Pti4, Pti5, or Pti6 and thereby increases their activities. To examine this possibility, we conducted in vitro phosphorylation assays by using Pto kinase with Pti4, Pti5, and Pti6 as substrates. However, although expression and purification of GST-Pti5 and GST-Pti6 as described previously yielded proteins that were suitable for DNA binding assays (Zhou et al., 1997), they were neither sufficiently abundant nor pure enough for kinase assays (data not shown). Expression of Pti5 or Pti6 as a His-tagged fusion at the N terminus in E. coli also gave low yields. We finally succeeded in expressing and purifying Pti5 (but not Pti6) as a fusion protein with a His tag at its C terminus (see Methods). To test whether the Pti5-His fusion protein was functional, we performed a mobility shift assay. As shown in Figure 7B, the Pti5-His fusion protein specifically binds to a DNA fragment containing a wild-type GCC box but not to a mGCC box (Figure 7B).
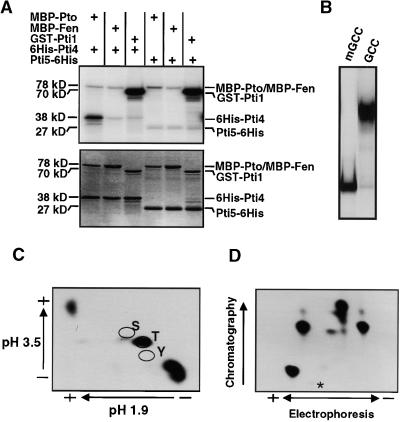
Figure 7.
Phosphorylation of the Pti4 Protein by Pto Kinase.
(A) His-Pti4, Pti5-His, MBP-Pto, MBP-Fen, and GST-Pti1 proteins were purified as described in Methods. Two micrograms of His-Pti4 or Pti5-His protein was incubated with 2 μg of MBP-Pto, MBP-Fen, or GST-Pti1 protein in an in vitro kinase assay. Total proteins from each reaction were separated by SDS-PAGE and analyzed by autoradiography. The autoradiograph (top) shows the phosphorylated proteins, and the Coomassie blue–stained gel (bottom) shows the protein profile. The (+) signs denote that the indicated protein was present in the reaction. Protein masses are shown in kilodaltons.
(B) A mobility shift assay was performed as described previously (Zhou et al., 1995), using 50 ng of purified Pti5-His fusion protein mixed with 5 fmol of 32P-labeled wild-type GCC-box or mGCC-box oligonucleotides (see Methods).
(C) Radiolabeled His-Pti4 protein that had been phosphorylated by Pto was transferred to a polyvinyl difluorine membrane, eluted, and hydrolyzed to its compositional amino acids as described in the Methods. The amino acids were separated by two-dimensional thin-layer chromatography. The (+) and (−) signs indicate the positions of the positive and negative electrodes. The locations of standards of serine (S), threonine (T), and tyrosine (Y) residues as determined by addition of ninhydrin are indicated. Radiolabeled, phosphorylated residues were detected by autoradiography.
(D) Radiolabeled His-Pti4 protein that had been phosphorylated by Pto was digested with trypsin, as described by Sessa et al. (1998). Peptides were separated horizontally by thin-layer electrophoresis at pH 4.7 and then vertically by ascending chromatography. The (+) and (−) signs indicate the positions of the positive and negative electrodes. Peptides were detected by autoradiography. The origin is indicated by an asterisk.
For kinase assays, Pto was overexpressed as a myelin basic protein (MBP)–Pto fusion in E. coli and purified as described previously (Loh and Martin, 1995). Figure 7A shows that MBP-Pto strongly phosphorylated the His-Pti4 fusion protein but did not phosphorylate the Pti5-His protein. To determine the specificity of Pto phosphorylation of Pti4, we included the serine/threonine kinases Fen and Pti1 as controls. Fen has 80% amino acid identity to Pto kinase; Pti1 is phosphorylated by Pto and plays a role in the hypersensitive response (Martin et al., 1994; Zhou et al., 1995). Substrates for Pti1 and Fen have not been identified. In our experiments, neither Fen nor Pti1 phosphorylated Pti4, indicating that phosphorylation of Pti4 by Pto is specific (Figure 7A). Moreover, the Pti5-His protein was not phosphorylated by Fen or Pti1. Coomassie blue staining indicated that similar amounts of each kinase and His-Pti4 or Pti5-His protein were used in each reaction (Figure 7A, bottom gel).
Pto Phosphorylates Pti4 on at Least Four Threonine Residues
Pto autophosphorylates serine/threonine residues by way of an intramolecular mechanism (Loh and Martin, 1995; Sessa et al., 1998). To determine which amino acids of Pti4 are phosphorylated by Pto, we subjected the Pto-phosphorylated Pti4 protein to phosphoamino acid analysis (Figure 7C). Pti4 was exclusively phosphorylated on threonine residues. To determine how many threonine residues on Pti4 were phosphorylated by Pto, we digested the phosphorylated Pti4 with trypsin and separated the resulting peptides by two-dimensional electrophoresis/chromatography. Figure 7D shows that four radioactive peptides were detected, indicating that at least four threonine residues in Pti4 are phosphorylated by Pto.
Phosphorylation of Pti4 by Pto Influences Its DNA Binding Activity in Vitro
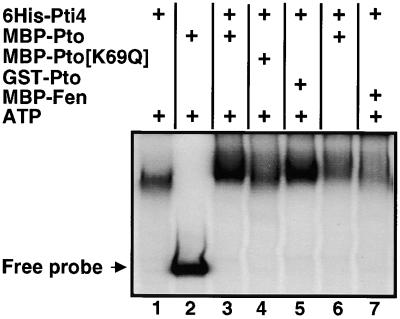
As shown in Figure 1, His-Pti4 specifically binds to the GCC box. To investigate whether phosphorylation of Pti4 affects its DNA binding activity in this assay, we first phosphorylated Pti4 by using the Pto kinase and then examined its binding to the GCC box by using a mobility shift assay. In preliminary experiments, we found that the presence of kinase buffer in the mobility shift assay reduced the binding of Pti4 to the GCC box; therefore, we increased the amount of this protein (to 0.25 μg) used in our experiments. As shown in Figure 8, phosphorylation of Pti4 with either of two different Pto fusion proteins, MBP-Pto or GST-Pto (Sessa et al., 1998), increased the binding of Pti4 to the GCC box. Control experiments conducted without the Pto kinase (lane1), with an inactive Pto protein (MBP-Pto [K69Q]; lane 4), or with the active kinase MBP-Fen, which does not phosphorylate Pti4 (lane 7), showed no effect on Pti4 binding of the GCC box. A kinase reaction of Pti4 with MBP-Pto in the absence of ATP also had no effect on Pti4 binding of the GCC box (lane 6). When the kinase buffer was used in the mobility shift assay, we observed unbound (free) probe only in the lane containing Pto (lane 2). We believe that the lack of free probe in the lanes containing Pti4 results from the excess amount of this protein used in the reactions; the protein may have bound loosely to the GCC box and dissociated during electrophoresis. This explanation is consistent with the slightly higher background observed in the lanes with Pti4 than in the lane with Pto alone (lane 2).
Figure 8.
Phosphorylation of Pti4 by Pto Affects Its DNA Binding Activity.
Components as indicated in each lane were preincubated at 25°C for 30 min in 10 μL of kinase buffer. For each kinase reaction, 0.25 μg of His-Pti4 and kinase was used. The final concentration of unlabeled ATP was 1 mM. After incubation, the volume in each tube was adjusted to 30 μL, with the mobility shift assay buffer containing the radiolabeled probe, and assayed as described in Methods. The (+) signs indicate that the corresponding protein, or ATP, was present in the reaction. The position of the unbound (Free) probe is shown.
Overexpression of Pto Overcomes SA Inhibition of Expression of PR Genes Having a GCC Box
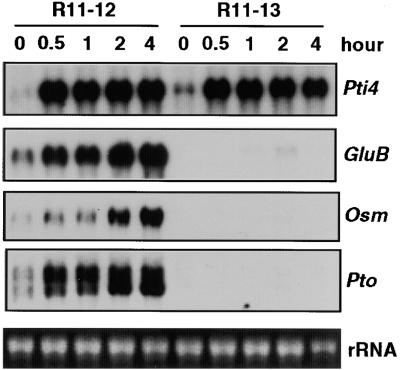
The phosphorylation of Pti4 by Pto raises the possibility that this post-translational modification affects the activity of Pti4 in the induction of PR genes with GCC boxes. One way to test this hypothesis is to increase the expression of Pto and Pti4 concomitantly and to determine whether there is a direct correlation between this increase and the expression of PR genes having a GCC box. To perform these experiments, we used a pair of tomato lines, R11-12 and R11-13, derived from a tomato line carrying a 35S::Pto transgene. R11-12 is homozygous for the transgene, and R11-13 is a sibling line that by segregation has lost the transgene (Tang et al., 1999). We took advantage of three observations in these experiments: (1) Pti4 is inducible by SA (Figure 3A); (2) SA does not induce expression of PR genes having the GCC box and, in fact, suppresses ethylene induction of GluB and Osm expression (Figure 3B); and finally, (3) SA is known to increase expression from the cauliflower mosaic virus 35S promoter that controls expression of Pto in R11-12 (Qin et al., 1994).
Tomato lines R11-12 and R11-13 plants were treated with SA, and their RNA was isolated over a 4-hr period. Figure 9 shows that transcript abundance of Pti4 increased within 30 min and reached similar amounts in both lines. As expected, SA also induced expression of the 35S::Pto transgene in line R11-12, and Pto transcripts reached their maximum at 2 hr after SA treatment. Importantly, increases in the transcripts for GluB and Osm occurred only in the Pto-overexpressing line, and the timing of this increase correlated with the appearance of the Pti4 and Pto transcripts. As described previously (Tang et al., 1999), some PR genes are constitutively expressed in R11-12 and other Pto-overexpressing lines when the plants are older (>6 weeks) or grown under high light conditions. We purposely used younger plants (4 weeks old) grown under lower light conditions to avoid this constitutive expression (see Methods). Thus, although Pto and PR gene transcripts were low at time 0, treatment with SA greatly increased the amounts of their transcripts (Figure 9). In summary, the ability of Pto overexpression to overcome the SA inhibition of PR gene expression (Figure 3) and the phosphorylation of Pti4 by Pto support a role for Pti4 in the activation of PR genes having a GCC box.
Figure 9.
Expression of Pti4, the GCC-Box-Containing PR Genes, and Pto Induced by SA in Tomato Plants with or without a 35::Pto Transgene.
A tomato line that is homozygous for a 35S::Pto transgene (R11-12) and a sibling line lacking the transgene (R11-13) were treated with 1 mM SA, and leaves were harvested at the times indicated. Total RNAs were isolated, and duplicate RNA gel blots were hybridized with radiolabeled Pti4, GluB, Osm, or Pto probes. Equal loading was verified by visualizing the rRNA in the RNA gel stained with ethidium bromide.
DISCUSSION
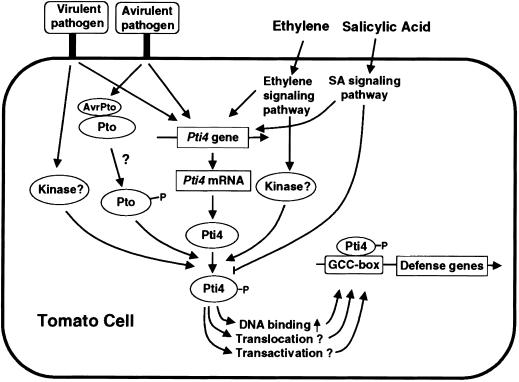
To identify components involved in Pto-mediated signal transduction pathways leading to disease resistance, we previously had used the product of the Pto disease resistance as a bait in the yeast two-hybrid system to search for Pti proteins. Among them, we found that Pti4, Pti5, and Pti6 encode putative transcription factors strikingly similar to those of the EREBPs both in structure and GCC-box binding properties (Figure 1; Zhou et al., 1997). These observations prompted us to investigate the responsiveness of one of these genes, Pti4, to ethylene and other possible inducers and to investigate the relationship between the Pto kinase and the Pti4 protein. As depicted in Figure 10, our results indicate that Pti4 is a component of an ethylene signaling pathway leading to expression of several ethylene-regulated genes. Although SA rapidly induced transcription of Pti4, it antagonized ethylene-regulated expression of GCC-box-containing PR genes. Overexpression of Pto and concomitant induction of Pti4 after SA application correlated with increased expression of GCC-box-containing PR genes. Moreover, the Pto kinase specifically phosphorylated Pti4 on a few of the threonine residues, phosphorylation that enhanced Pti4 binding of the GCC box. Given these observations, we postulate that phosphorylation of Pti4 by Pto modulates Pti4 activity, which then increases expression of PR genes during resistance to bacterial speck disease (i.e., more than that observed in a compatible interaction; Figure 10).
Figure 10.
Proposed Model for Activation of GCC-Box-Containing PR Genes by Transcription Factor Pti4 in Pto-Mediated and Other Defense Response Signaling Pathways.
?, mechanistic steps that are still conjectural; P, phosphorylation.
Pti4 Is Associated with Ethylene Regulation of GCC-Box-Containing PR Genes
Pti4 transcripts rapidly accumulated in response to ethylene, an increase that preceded expression of the GCC-box-containing GluB and Osm genes. Furthermore, transcriptional activation of GluB by ethylene required de novo synthesis of protein factors, which might be required for interacting with GCC-box cis elements. However, no synthesis of new proteins was required for the rapid accumulation of Pti4 transcripts induced by ethylene. This suggests that Pti4 is an immediate-early response gene, the product of which is required for GCC-box cis element binding to activate PR genes in the ethylene signaling pathway (Figure 10).
The correlation of the expression of Pti4 and PR genes having a GCC box in plant tissues known to produce increased amounts of ethylene provides further support for a role of Pti4 in ethylene-regulated PR gene expression. In addition, in preliminary experiments with transgenic plants overexpressing the Pti4 transgene, we have found that several ethylene-regulated PR genes are constitutively expressed (Y.-Q. Gu and G.B. Martin, unpublished data). In tobacco, induction of EREBP genes by wounding is insufficient for activation of GCC-box-containing PR genes (Suzuki et al., 1998), and an Arabidopsis EREBP, AtEBP, has been shown to cross-couple with a basic leucine zipper transcription factor (Büttner and Singh, 1997). It is possible therefore that the activity of Pti4 is modulated by interaction with other transcription factors in the activation of GCC-box promoters.
Pti4, Pti5, and Pti6 proteins belong to a large family of APETALA2/EREBP–type transcription factors in plants. This family of transcription factors has been proposed to play various roles in plant growth, development, and responses to different environmental stimuli (Okamuro et al., 1997). The differential expression of Pti4, Pti5, and Pti6 in various plant tissues implies that they respond to different endogenous signals and have distinct roles in plants. Thus, it is not surprising that Pti4, but not Pti5 or Pti6, responds to ethylene treatment. Recently, several genes were isolated from an Arabidopsis cDNA library by using tobacco EREBP-1 as a probe; only one, ethylene-responsive factor 1 (ERF1), was induced rapidly by ethylene, and its role in regulating ethylene-inducible genes was demonstrated (Solano et al., 1998). Perhaps Pti4 is a tomato functional homolog of ERF1 in mediating ethylene-regulated expression of PR genes containing a GCC box.
SA Induces Pti4 but Antagonizes Ethylene-Induced PR Gene Expression
SA plays an important signaling role in activation of PR genes in plant defense responses during pathogen attack. Genes activated by SA can be categorized into immediate-early and late genes, based on their response time to SA treatment (Dempsey et al., 1999). Some immediate-early SA-responsive genes encode transcription factors that when activated, may be responsible for the expression of late SA-responsive genes. For example, transcripts of the tobacco myb1 transcription factor gene rapidly accumulate when plants are treated with SA; then Myb1 proteins specifically bind the H-box cis element in the promoter of PR-1a, a late SA-responsive gene (Yang and Klessig, 1996). SA also might regulate PR gene expression by modulating the activity of transcription factors. For example, SA treatment has been shown to increase the ASF-1 element binding activity present in tobacco nuclear extracts (Stange et al., 1997). This enhanced binding activity is likely mediated by protein phosphorylation events because treatment of SA-induced tobacco nuclear extracts with alkaline phosphatase decreases ASF-1 binding activity. Currently, the mechanism by which SA modulates the activity of these transcription factors remains unclear.
In this study, we found that although SA rapidly increased Pti4 transcripts, it did not induce expression of GCC-box-containing PR genes. This suggests that SA might have a negative effect on the expression of GCC-box-containing PR genes. In support of this hypothesis, we found that SA treatment suppressed the ethylene induction of these PR genes. One possible mechanism for this suppression is a blockage of ethylene biosynthesis, given that SA antagonizes the activity of 1-aminocyclopropane-1-carboxylate oxidase, an enzyme that catalyzes the production of ethylene (Fan et al., 1995). That mechanism, however, is unlikely to be operative here because ethylene was provided constantly during the simultaneous treatment with SA. Alternatively, SA might negatively modulate components involved in the ethylene signaling pathway, thereby suppressing the expression of GCC-box-containing PR genes. Given the observation that simultaneous treatment with ethylene and SA further increased the amount of Pti4 RNA, perhaps SA exerts a negative effect on the activity of the Pti4 protein (Figure 10). The mechanism responsible for such negative regulation, if one exists, remains to be determined.
Investigators using kinase and phosphatase inhibitors have demonstrated that protein phosphorylation/dephosphorylation plays a vital role in the SA-signaling pathways that lead to the activation of PR genes (Conrath et al., 1997). Recently, a tobacco 48-kD mitogen-activated protein kinase, the activity of which is induced by SA, was found to be involved in plant defense response pathways (Zhang et al., 1998). These results suggest that a phosphorylation/dephosphorylation cascade involved in SA signaling might change the activity of transcription factors in mediating PR gene expression. In addition, the negative regulatory role of SA on DNA binding proteins has been reported (Buchel et al., 1996). The binding activity of GT-1–like protein, which binds several fragments of tobacco PR-1a promoter, was severely decreased in nuclear extracts from SA-treated leaves (Buchel et al., 1996). Therefore, Pti4 could be a target for SA-activated protein kinases/phosphatases, and modulation of Pti4 by those enzymes may downregulate its activity.
Possible Role of Pti4 Phosphorylation by Pto in Regulation of PR Genes
R gene–dependent transcript accumulation of defense-related genes has been observed in several plant–pathogen interaction systems (e.g., Kiedrowski et al., 1992; Reuber and Ausubel, 1996). We found previously that expression of GCC-box-containing PR genes was enhanced by Pto–avrPto recognition (Figure 6A; Jia and Martin, 1999). Because Pti4, Pti5, and Pti6 bind specifically to the GCC-box element, Pto–avrPto recognition possibly could cause rapid transcriptional activation of these genes, the products of which then might mediate expression of GCC-box-containing PR genes. However, our results indicate that although Pti4 transcripts rapidly accumulate after pathogen attack, this induction of Pti4 is not specific for Pto–avrPto recognition but occurs in both compatible and incompatible interactions. In addition, although the abundance of Pti5 transcripts is slightly enhanced in an incompatible interaction (Thara et al., 1999), constitutive expression of Pti5 in transgenic tomato plants does not result in increased expression of GCC-box PR genes (Y.-Q. Gu and G.B. Martin, unpublished data). Moreover, Pti6 was not transcriptionally activated by pathogen attack. Thus, the enhanced expression of GCC-box-containing PR genes in the avrPto–Pto incompatible interaction is not correlated simply with increased Pti4, Pti5, and Pti6 transcripts. An alternative possibility is that recognition of avrPto by Pto plays a role in modulating the activity of Pti4, Pti5, and Pti6 proteins. This finding is consistent with the observation that the Pto kinase physically interacts with Pti4, Pti5, and Pti6 proteins (Zhou et al., 1997) and with our present results demonstrating that Pto phosphorylates Pti4.
Phosphorylation of transcription factors is a common mechanism for regulation of gene expression in eukaryotes (Hunter and Karin, 1992). However, regulation of plant transcription factors by phosphorylation has been reported in only a few cases (Yu et al., 1993; Després et al., 1995; Subramaniam et al., 1997; Dröge-Laser et al., 1998). In all of these cases, the identity of the kinases involved is not known. Several lines of evidence suggest that phosphorylation of Pti4 by the Pto kinase plays an important role in Pto-mediated regulation of expression of GCC-box-containing PR genes during plant resistance to the bacterial speck pathogen. First, a kinase-deficient Pto that does not confer disease resistance fails to interact with Pti4 in a yeast two-hybrid system (Jia et al., 1997; Zhou et al., 1997). Second, the phosphorylation of Pti4 by Pto is highly specific; two other kinases, although closely related, do not phosphorylate the Pti4 protein. Third, Pti4 transcripts accumulate in both compatible and incompatible Pseudomonas spp–tomato interactions, but enhanced expression of GCC-box-containing PR genes is seen only in the incompatible interaction involving Pto and AvrPto. Fourth, phosphorylation of Pti4 by Pto enhances its binding to the GCC box. Finally, the negative effect of SA on the expression of GCC-box-containing PR genes is overcome by overexpression of Pto and concomitant expression of Pti4. Our experiments did not exclude a role for Pti5 and Pti6 (or other transcription factors) in Pto-mediated regulation of PR genes. However, because Pti5 is not phosphorylated by Pto in vitro, and because we were not successful in isolating sufficient Pti6 for phosphorylation analysis, we were unable to investigate further any possible mechanisms whereby Pto might regulate the activity of these proteins.
The increased binding of phosphorylated Pti4 to the GCC box provides one explanation for the enhanced expression of PR genes during disease resistance. How AvrPto might be involved in this event, however, remains unclear. Pto–avrPto recognition occurs by a physical interaction of the two proteins (Scofield et al., 1996; Tang et al., 1996), an interaction that might stimulate Pto kinase activity and in turn allow phosphorylation of Pti4. Alternatively, a small amount of the Pto kinase might exist in a complex with a negative regulator such that interaction with AvrPto would release active Pto for Pti4 phosphorylation. Our results with Pto overexpression (in the absence of AvrPto) favor the latter explanation (Figure 9). Further support for this model can be derived from the observation that Pto kinase autophosphorylates by means of an intramolecular mechanism and phosphorylates the Pti1 and Pti4 substrates in the absence of AvrPto (Figure 7; Zhou et al., 1995; Sessa et al., 1998).
Pto phosphorylates at least four threonine residues in Pti4 (and no serine residues). The Pti4 protein contains 10 threonine residues (Zhou et al., 1997); two of these are located in the putative DNA binding domain, two in a possible transactivation domain, and one near a nuclear localization sequence. Phosphorylation is known to affect cellular localization, binding activity to cognate cis elements, transactivation potential, and turnover of transcription factors (Hunter and Karin, 1992; Papavassiliou et al., 1992). An alteration in any of these processes potentially could affect Pti4 regulation of PR gene expression. We have shown that Pto phosphorylation of Pti4 enhances the binding of the transcription factor to the GCC box. However, we cannot exclude the possibility that other mechanisms (e.g., localization or transactivation) also play an important role in modulating Pti4 activity. By mapping the Pti4 phosphorylation sites and examining their functional relevance, we should gain further insights into the role of Pto kinase in activating defense genes during the incompatible interaction.
METHODS
Plant Materials, Growth Conditions, and Treatment
Tomato (Lycopersicon esculentum) cultivars Rio Grande–PtoR (RG-PtoR) and Rio Grande–PtoS (RG-PtoS), which are near-isogenic tomato lines for the Pto locus, were grown in the greenhouse with 16 hr of light and temperatures of 26 to 28°C. Three- to 4-week-old tomato plants of similar sizes were used for the experiments. For the experiments with Pto-transgenic line R11-12 and its sibling line R11-13, plants were grown in the greenhouse with low light intensity, and seedlings of 3-week-old plants were used for treatment. Under this growth condition, the R11-12 plants did not show the spontaneous cell death caused by a combination of overexpression of Pto and high light intensity (Tang et al., 1999).
The bacterial pathogen expressing the avrPto gene, Pseudomonas syringae pv tomato DC3000, was grown at 30°C with shaking in King's medium B (Martin et al., 1993) containing 100 μg/mL rifampicin. Bacteria were pelleted by centrifugation, washed once with 10 mM MgCl2, and resuspended at a concentration of 2.2 × 107 colony-forming units per milliliter in 10 mM MgCl2 and 0.005% Silwet (Union Carbide, Southbury, CT). Leaves of RG-PtoR plants that were infiltrated at this inoculum level displayed a hypersensitive response ∼12 hr after inoculation. RG-PtoS leaves infiltrated with this same inoculum showed disease symptoms beginning at 12 hr after inoculation. The infiltration of plants was performed according to Jia et al. (1997). Plants were submerged in the bacterial suspension in a bell jar, and a vacuum was applied for 2 min and then released rapidly to ensure uniform infiltration of leaves.
Ethylene treatment of plants was performed in a plexiglass chamber. A volume of ethylene was injected into the chamber to give a final concentration of 10 μL/L. For chemical compound treatments, solutions were prepared as follows: salicylic acid (SA) and cycloheximide were directly dissolved in water at concentrations of 1 mM and 5 μg/mL, respectively; a systemin stock solution was prepared by dissolving synthetic mature systemin peptide (a gift from C.A. Ryan, Washington State University, Pullman) in water at 1 mg/mL; a working solution was made by diluting the stock solution 5000-fold to give a final concentration of 200 ng/mL. The compounds were applied to tomato leaves by stem feeding with the method of O'Donnell et al. (1996). Wounding was performed by rubbing tomato leaves with carborundum according to the method described by Zhang and Klessig (1998). For all the experiments, leaf tissues were harvested at the specified times, frozen in the liquid nitrogen, and stored at −80°C before isolation of RNA.
Overexpression and Purification of Recombinant His-Pti4 and Pti5-His Proteins
For expression and purification of Pti4 protein, the coding sequence was amplified by polymerase chain reaction (PCR), introducing a BamHI site in front of the ATG codon and immediately after the stop codon. The PCR product was gel purified, digested with BamHI, and ligated in frame into the His-tagged pEQ9 expression vector (Qiagen, Chatsworth, CA). The resulting plasmid, pEQ9Pti4, was sequenced to ensure that no PCR-induced errors were introduced into the Pti4 sequence. DH12S Escherichia coli cells were transformed with pEQ9Pti4 and grown to an OD600 of 0.6. Bacteria were induced with 1 mM isopropyl β-d-thiogalactopyranoside for 5 hr at 37°C. Most of the His-Pti4 fusion protein was insoluble, being localized in inclusion bodies (data not shown). Therefore, the bacterial pellet was resuspended in buffer A (50 mM Tris-HCl, pH 7.5, and 200 mM NaCl) containing 8 M urea. The supernatant was loaded onto a nickel–agarose column and washed with 20 column volumes of 8 M urea in buffer A. Protein (His-Pti4) was renatured on the column by further washing with a urea gradient from 8 to 0 M in buffer A. Protein (His-Pti4) was eluted from the column with 300 mM imidazole in buffer A and then dialyzed against buffer A overnight at 4°C.
For overexpression and purification of Pti5 protein, the coding sequence was amplified by PCR, introducing an NcoI site in front of the ATG and immediately before the stop codon. This PCR product was cloned in-frame into the NcoI site of pET-15b expression vector (Novagen, Madison, WI). The resulting plasmid, pETPti5, from which Pti5 was expressed with 6 histidines at its C terminus, was transformed into E. coli BL21. The procedure for induction and purification of Pti5-His fusion protein was the same as that for His-Pti4. Purification of GST-Pti5 and the gel mobility shift assays were performed as described previously (Zhou et al., 1997). The sequence of the GCC oligonucleotide was 5′-CATAAGAGCCGCCACTAAAATAAGACCGATCAAATAAGAGCCGCCAT-3′ and that of the mutated mGCC oligonucleotide was 5′-CATAAGATCCTCCACTAAAATAAGACCGAT-CAAATAAGATCCTCCAT-3′ (the boldface type indicates the nucleotides that differ between the two oligonucleotides).
RNA Isolation and RNA Gel Blot Hybridization
RNA isolation was performed according to Perry and Francki (1992) with some modifications. Approximately 1 g of frozen tissue was ground in liquid nitrogen with a mortar and pestle to a fine powder. The leaf powder was extracted by vigorous vortex mixing for 30 sec in a mixture of 2 mL of water-saturated phenol and an equal volume of extraction buffer (100 mM Tris-HCl, pH 8.0, 100 mM LiCl, 10 mM EDTA, and 1% SDS) preheated at 75°C. Two milliliters of 24:1 (v/v) chloroform–isoamyl alcohol was added to the sample and vortex mixed again for 30 sec. The mixture was centrifuged at 12,000g for 10 min at 4°C, and 2 mL of supernatant was transferred to a new 15-mL tube. RNA was precipitated by adding an equal volume of 4 M LiCl and incubating in a −70°C freezer for 30 min. RNA was pelleted by centrifugation at 12,000g for 20 min at 4°C, washed once with 70% ethanol, and resuspended in nuclease-free water. Ten micrograms of total RNA from each sample was separated by electrophoresis on a 1.2% formaldehyde–agarose gel, transferred to Hybond C membrane, and immobilized by UV light cross-linking.
RNA gel blot hybridizations were performed according to the manufacturer's instructions (Amersham). Briefly, the Hybond C membrane was prehybridized for at least 4 hr in hybridization solution containing 50% formamide, 2 × SSPE (1 × SSPE is 0.15 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA, pH 7.4), and 5 × Denhardt's solution (1 × Denhardt's solution is 0.02% Ficoll, 0.02% PVP, and 0.02% BSA) (Ausubel et al., 1987). Hybridization was initiated by adding denatured DNA probes labeled with 32P-dATP by using a random hexamer labeling kit (Ambion, Austin, TX). After a minimum 16-hr incubation, the membrane was washed three times (20 min each time) at 55°C in 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) (Ausubel et al., 1987) plus 0.1% SDS. Radioactivity was detected by autoradiography.
In Vitro Kinase Assay, Phosphoamino Acid Analysis, and Peptide Mapping
For the phosphorylation assays, His-Pti4 and Pti5-His were purified as described above, and the myelin basic protein (MBP)–Pto, glutathione S-transferase (GST)–Pti1, MBP-Fen, and GST-Pto kinases were expressed and purified from E. coli as previously described (Loh and Martin, 1995; Zhou et al., 1995; Sessa et al., 1998). The phosphorylation assay was performed in solution at room temperature for 20 min. Each reaction contained 2 μg of kinase and 4 μg of His-Pti4 in a 50-μL volume with final concentrations of 50 mM Tris-HCl, pH 7.0, 1 mM DTT, 10 mM MnCl2, 20 μM unlabeled ATP, and 4 μCi of γ-32P-ATP (6000 Ci/mmol; Amersham Pharmacia Biotech). The reaction was stopped by adding 15 μL of 4 × SDS sample buffer (1 × SDS sample buffer is 62.5 mM Tris-HCl, 5% glycerol, and 2.5% β-mercaptoethanol) and boiling for 10 min before loading onto an SDS–polyacrylamide gel. After electrophoresis, the gel was stained with Coomassie Brilliant Blue G 250, dried, and subjected to autoradiography. Phosphoamino acid analysis and peptide mapping of phosphorylated Pti4 by Pto were performed by the procedure described by Sessa et al. (1998)(2000).
Acknowledgments
We thank Young Jin Kim and Ramesh Kantety for help with the ethylene treatment experiments and Guido Sessa and Mark D'Ascenzo for help with phosphorylation assays. We thank Bud Ryan for generously providing systemin and the pin2 gene. We thank Ying-Tsu Loh, Roger Thilmony, and Dan Kliebenstein for helpful discussions, and Adam Bogdanove, Xiaohua He, Yuko Nakajima, and Bob Tuori for critical reading of the manuscript. This research was supported by a grant from the Monsanto Corporation, by National Science Foundation Grant No. MCB-96-30635 and No. 98-96308, and by the David and Lucile Packard Foundation.
References
- Abeles, F.B., Morgan, P.W., and Saltveit, M.E., Jr. (1992). Ethylene in Plant Biology. (San Diego, CA: Academic Press).
- Alfano, J.R., and Collmer, A. (1996). Bacterial pathogens in plants: Life up against the wall. Plant Cell 8, 1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Smith, J.A., Seidman, J.G., and Struhl, K., eds (1987). Protocols in Molecular Biology. (New York: John Wiley).
- Broglie, K.E., Chet, I., Holliday, M., Cressman, R., Biddle, P., Knowlton, S., Mauvais, C.J., and Broglie, R. (1991). Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254, 1194–1197. [DOI] [PubMed] [Google Scholar]
- Buchel, A.S., Molemkamp, R., Bol, J.F., and Linthorst, H.J.M. (1996). The PR-1a promoter contains a number of elements that bind GT-1–like nuclear factors with different affinity. Plant Mol. Biol. 30, 493–504. [DOI] [PubMed] [Google Scholar]
- Büttner, M., and Singh, K. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC-box DNA-binding protein, interacts with an Ocs element binding protein. Proc. Natl. Acad. Sci. USA 94, 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath, U., Silva, H., and Klessig, D.F. (1997). Protein dephosphorylation mediates salicylic acid–induced expression of PR-1 genes in tobacco. Plant J. 11, 747–757. [Google Scholar]
- Cutt, J.R., and Klessig, D.F. (1992). Pathogenesis-related proteins. In Genes Involved in Plant Defense, T. Boller and F. Meins, eds (New York: Springer-Verlag), pp. 209–243.
- Deikman, J. (1997). Molecular mechanism of ethylene regulation of gene transcription. Physiol. Plant. 100, 561–566. [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friendrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey, D.A., Shah, J., and Klessig, D.F. (1999). Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci. 18, 547–575. [Google Scholar]
- Després, C., Subramaniam, R., Matton, D.P., and Brisson, N. (1995). The activation of the potato PR-10a gene requires the phosphorylation of the nuclear factor PBF-1. Plant Cell 7, 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge-Laser, W., Kaiser, A., Lindsay, W.L., Kalkier, B.A., Loake, G.J., Doerner, P., Dixon, R.A., and Lamb, C. (1998). Rapid stimulation of a soybean protein-serine kinase that phosphorylates a novel bZIP DNA-binding protein, G/HBF-1, during the induction of early transcription-dependent defenses. EMBO J. 16, 726–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal, Y., Meller, Y., Lev-Yadum, S., and Fluhr, R. (1993). A basic-type PR-1 promoter directs ethylene responsiveness, vascular and abscission zone-specific expression. Plant J. 4, 225–234. [DOI] [PubMed] [Google Scholar]
- Fan, X., Mattheis, J.P., and Fellman, J.K. (1995). Inhibition of apple ACC oxidase activity and respiration by acetylsalicylic acid. Plant Physiol. 108 (suppl.), 69.. [Google Scholar]
- Frederick, R.D., Thilmony, R.L., Sessa, G., and Martin, G.B. (1998). Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol. Cell 2, 241–245. [DOI] [PubMed] [Google Scholar]
- Friedrich, L., Vernooij, B., Gaffney, T., Morse, A., and Ryals, J. (1995). Characterization of tobacco plants expressing a bacterial salicylate hydroxylase gene. Plant Mol. Biol. 19, 959–968. [DOI] [PubMed] [Google Scholar]
- Horvath, D.M., Huang, D.J., and Chua, N.-H. (1998). Four classes of salicylate-induced tobacco genes. Mol. Plant-Microbe Interact. 11, 895–905. [DOI] [PubMed] [Google Scholar]
- Hunter, T., and Karin, M. (1992). The regulation of transcription by phosphorylation. Cell 70, 375–387. [DOI] [PubMed] [Google Scholar]
- Jia, Y., and Martin, G.B. (1999). Rapid transcript accumulation of pathogenesis-related genes during an incompatible interaction in bacterial speck disease-resistant tomato plants. Plant Mol. Biol. 40, 455–465. [DOI] [PubMed] [Google Scholar]
- Jia, Y., Loh, Y.-T., Zhou, J., and Martin, G.B. (1997). Allele of Pto and Fen occur in bacterial speck-suspectible and fenthion-insensitive tomato and encode active protein kinases. Plant Cell 9, 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski, S., Kawalleck, P., Hahlbrock, K., Somssich, I.E., and Dangl, J. (1992). Rapid activation of a novel plant defense gene is strictly dependent on the Arabidopsis RPM1 disease resistance. EMBO J. 11, 4677–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton, K.A., Potter, S.L., Uknes, S., and Ryals J. (1994). Acquired resistance signal transduction in Arabidopsis is ethylene independent. Plant Cell 6, 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Strabala, T.J., Hagen, G., and Guilfoyle, T. (1994). The soybean SAUR open reading frame contains a cis element responsible for cycloheximide-induced mRNA accumulation. Plant Mol. Biol. 24, 715–723. [DOI] [PubMed] [Google Scholar]
- Loh, Y.-T., and Martin, G.B. (1995). The Pto bacterial resistance gene and the Fen insecticide sensitivity gene encode functional protein kinases with serine/threonine specificity. Plant Physiol. 108, 1735–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G.B., Brommonschenkel, S., Chunwongse, J., Frary, A., Ganal, M.W., Spivey, R., Wu, T., Earle, E.D., and Tanksley, S.D. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262, 1432–1436. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Frary, A., Wu, T., Brommonschenkel, S., Chunwongse, J., Earle, E.D., and Tanksley, S.D. (1994). A member of the Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell 6, 1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch, F., Mauch-Mani, B., and Boller, T. (1988). Antifungal hydrolase in pea tissue. II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol. 88, 936–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, P.J., Calvert, C., Atzorn, R., Wasternack, C., Leyser, H.M.O., and Bowles, D.J. (1996). Ethylene as a signal mediating the wound response of tomato plants. Science 274, 1914–1917. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro, J.K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavassiliou, A.G., Treier, M., Chavrier, C., and Bohmann, D. (1992). Targeted degradation of c-Fos, but not v-Fos by a phosphorylation-dependent signal on c-Jun. Science 258, 1941–1944. [DOI] [PubMed] [Google Scholar]
- Pastuglia, I., Roby, D., Dumas, C., and Cock, J.M. (1997). Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase gene in Brassica oleracea. Plant Cell 9, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Eggermont, K., Terras, F.R., Thomma, B.P.H.J., De Samblanx, G.W., Buchala, A., Métraux, L.-P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8, 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Thomma, B.P.H.J., Buchala, A., Metraux, J.-P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, L.K., and Francki, R.I.B. (1992). Insect-mediated transmission of mixed and reassorted cucumovirus genomic RNAs. J. Gen. Virol. 73, 2105–2114. [DOI] [PubMed] [Google Scholar]
- Qin, X.-F., Holuigue, L., Horvath, D.M., and Chu, N.-H. (1994). Immediate-early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell 6, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber, T.L., and Ausubel, F.M. (1996). Isolation of Arabidopsis genes that differentiate between resistance response mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell 8, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald, P.C., Salmeron, J.M., Oldroyd, G.E.D., and Staskawicz, B.J. (1992). The cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J. Bacteriol. 174, 1604–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J., Torres, J.T., Parniske, M., Wernert, P., Hahlbrock, K., and Somssich, I.E. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR-1 genes. EMBO J. 15, 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Schaller, A., and Ryan, C.A.. (1995). Systemin—A polypeptide defense signal in plants. Bioessays 18, 27–33. [DOI] [PubMed] [Google Scholar]
- Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.W., and Staskawicz, B.J. (1996). Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274, 2063–2065. [DOI] [PubMed] [Google Scholar]
- Sela-Buurlage, M., Ponstein, A.S., Bres-Vloemans, S.A., Melchers, L.S., van den Elzen, P.J.M., and Cornelissen, B.J.C. (1993). Only specific tobacco (Nicotiana tabacum) chitinase and β-1,3-glucanases exhibit antifungal activity. Plant Physiol. 101, 857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa, G., D'Ascenzo, M., Loh, Y.T., and Martin, G.B. (1998). Biochemical properties of two protein kinases involved in disease resistance signaling in tomato. J. Biol. Chem. 273, 15860–15865. [DOI] [PubMed] [Google Scholar]
- Sessa, G., D'Ascenzo, M., and Martin, G.B. (2000). Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto-Pto–mediated hypersensitive response. EMBO J. 19, in press. [DOI] [PMC free article] [PubMed]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid–insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSIVE-FACTOR1. Genes Dev. 12, 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stange, C., Ramirez, I., Gomez, I., Jordana, X., and Holuigue, L. (1997). Phosphorylation of nuclear proteins directs binding to salicylic acid–responsive elements. Plant J. 11, 1315–1324. [DOI] [PubMed] [Google Scholar]
- Subramaniam, R., Despres, C., and Brisson, N. (1997). A functional homolog of mammalian protein kinase C participates in the elicitor-induced defense response in potato. Plant Cell 9, 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Suzuki, N., Ohme-Takagi, M., and Shinshi, H. (1998). Immediate early induction of mRNAs for ethylene-responsive transcription factors in tobacco leaf strips after cutting. Plant J. 15, 657–665. [DOI] [PubMed] [Google Scholar]
- Tang, X., Frederick, R.D., Zhou, J., Halterman, D.A., Jia, Y., and Martin, G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and the Pto kinase. Science 274, 2060–2063. [DOI] [PubMed] [Google Scholar]
- Tang, X., Xie, M., Kim, Y.J., Zhou, J., Klessig, D.F., and Martin, G.B. (1999). Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell 11, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thara, V.K., Tang, X., Gu, Y., Martin, G., and Zhou, J.-M. (1999). Pseudomonas syringae pv. tomato induces the expression of tomato EREBP-like genes Pti4 and Pti5 independent of ethylene, salicylate and jasmonate. Plant J. 20, 475–484. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Eggermont, K., Penninckx, I.A.M.A., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kan, J.A.L., Joosten, M.H.A.J., Wagemakers, C.A.M., van den Berg-Velthuis, G.C.M., and de Wit, P.J.G.M. (1992). Differential accumulation of mRNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium fulvum. Plant Mol. Biol. 20, 513–527. [DOI] [PubMed] [Google Scholar]
- Vidal, S., Ponce de Leon, I., Denecke, J., and Palva, E.T. (1997). Salicylic acid and the plant pathogen Erwinia carotovora induce defense genes via antagonistic pathways. Plant J. 11, 115–123. [Google Scholar]
- Voisey, C.R., and Slusarenko, A.J. (1989). Chitinase mRNA and enzyme activity in Phaseolus vulgaris (L.) increase more rapidly in response to avirulent than to virulent cells of Pseudomonas syringae pv. phaseolicola. Physiol. Mol. Plant Pathol. 35, 403–412. [Google Scholar]
- Xu, Y., Chang, P.-F.L., Liu, D., Narasimhan, M., Raghothama, K.G., Hasegawa, P.M., and Bressan, R.A. (1994). Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6, 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., and Klessig, D.F. (1996). Isolation and characterization of a tobacco mosaic virus–inducible myb oncogene homolog from tobacco. Proc. Natl. Acad. Sci. USA 93, 14972–14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L.M., Lamb, C.L., and Dixon, R.A. (1993). Purification and biochemical characterization of proteins which bind to the H-box cis element implicated in transcriptional activation of plant defense genes. Plant J. 3, 805–816. [DOI] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (1998). The tobacco wound–activated mitogen-activated protein kinase is encoded by SIPK. Proc. Natl. Acad. Sci. USA 95, 7225–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Du, H., and Klessig, D.F.. (1998). Activation of the tobacco kinase by both a cell wall–derived carbohydrate elicitor and purified proteinaceous elicitins from Phytophthora spp. Plant Cell 10, 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Loh, Y.-T., Bressan, R.A., and Martin, G.B. (1995). The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83, 925–935. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Tang, X., and Martin, G.B. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis element of pathogenesis-related genes. EMBO J. 16, 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q., Maher, E.A., Masoud, S., Dixon, R.A., and Lamb, C.J. (1994). Enhanced protection against fungal attack by constitutive co-expression of chitinase and glucanase in transgenic tobacco. BioTechnology 12, 807–812. [Google Scholar]