Abstract
Peptide deformylases (PDFs) have been discovered recently in eukaryotic genomes, and it appears that N-terminal methionine excision (NME) is a conserved pathway in all compartments where protein synthesis occurs. This work aimed at uncovering the function(s) of NME in a whole proteome, using the chloroplast-encoded proteins of both Arabidopsis thaliana and Chlamydomonas reinhardtii as model systems. Dis ruption of PDF1B in A.thaliana led to an albino phenotype, and an extreme sensitivity to the PDF- specific inhibitor actinonin. In contrast, a knockout line for PDF1A exhibited no apparent phenotype. Photosystem II activity in C.reinhardtii cells was substantially reduced by the presence of actinonin. Pulse–chase experiments revealed that PDF inhibi tion leads to destabilization of a crucial subset of chloroplast-encoded photosystem II components in C.reinhardtii. The same proteins were destabilized in pdf1b. Site-directed substitutions altering NME of the most sensitive target, subunit D2, resulted in similar effects. Thus, plastid NME is a critical mechanism specifically influencing the life-span of photosystem II polypeptides. A general role of NME in modulating the half-life of key subsets of proteins is suggested.
Keywords: gene knockout/N-end rule/photosystem/protein degradation/stability
Introduction
N-terminal methionine excision (NME) is the major pathway causing diversity of N-terminal amino acids. As a result of NME, Gly, Ala, Pro, Cys, Ser, Thr or Val residues may be found at the N-terminus of proteins, in addition to Met (Meinnel et al., 1993). NME was originally described as a cytoplasmic co-translational pathway involving about two of every three proteins in any proteome. NME requires the sequential action of two enzymes: (i) peptide deformylase (PDF), the activity initially described as required for specifically removing the N-formyl group present on all nascent polypeptides synthesized in eubacteria (Giglione et al., 2000a), and (ii) methionine aminopeptidase (MAP), which removes methionine specifically in all organisms (Bradshaw et al., 1998). The removal of the N-formyl group is a prerequisite for the subsequent action of MAP (Solbiati et al., 1999).
In contrast with PDF, which acts on almost all polypeptides, MAP activity depends on the nature of the second residue in the target chain. If it is Gly, Ala, Pro, Cys, Ser, Thr or Val, the methionine is cleaved; other wise it is retained. Recently, nuclear-encoded organelle-targeted PDF and MAP have been identified in most genomes, including those of lower eukaryotes, mammals and insects (Giglione et al., 2000a; Meinnel, 2000; Bracchi-Ricard et al., 2001; Giglione and Meinnel, 2001b). At the same time, the basis of NME in higher and lower plants has been described and shown to involve three organellar MAPs, one mitochondria-targeted PDF (PDF1A) and one chloroplast- and mitochondria-targeted PDF (PDF1B) (Giglione et al., 2000b; Giglione and Meinnel, 2001a). These unexpected findings indicate that NME is a conserved pathway in all compartments where protein synthesis occurs.
There is a variety of evidence that NME is essential. First, NME is the target of several natural anti-cellular drugs such as actinonin, which is active in bacteria (Chen et al., 2000), and fumagillin, which is active in angiogenic cells, ameba and other human parasites (McCowen et al., 1951; Griffith et al., 1997; Sin et al., 1997; Liu et al., 1998; Zhang et al., 2002). Secondly, PDF and MAP are part of the ∼300-gene minimal genome requirement of eubacteria (Hutchison et al., 1999). Similarly, MAP is present in the extremely reduced genome of the eukaryotic parasite Encephalitozoon cuniculi (∼2000 proteins; Katinka et al., 2001). NME is such a highly conserved function that it is present even in organelles although organellar-encoded proteomes all include <100 proteins (Giglione et al., 2000b). Surprisingly, little is known about why NME is so extensively conserved, or what makes it important. Investigations of the function of cytoplasmic NME to date have been frustrating. Gene inactivation experiments in bacteria (Chang et al., 1989; Miller et al., 1989; Meinnel and Blanquet, 1994) and yeast (Li and Chang, 1995) led systematically to rapid cell death, making such studies difficult. Finally, the importance of NME in organelles is still entirely unclear, although recent pharmacological data suggested an essential function in plastid development (Giglione and Meinnel, 2001a; Serero et al., 2001b; Wiesner et al., 2001).
Here, we report a study of this process in plant plastids. Owing to the compaction of their genomes, there are only a few dozen protein targets of NME in plant organelles, and most of them are well characterized (Giglione and Meinnel, 2001a). The case of the chloroplast is of particular interest because, in contrast with mitochondria, protein synthesis, and consequently NME are not strictly necessary for higher plant growth when the growth medium is supplemented with a reduced carbon source (Maliga, 1984; Harris et al., 1994). Using two well-established model systems, Arabidopsis thaliana and Chlamydomonas reinhardtii, we elucidated the central function of the chloroplastic deformylation process in vivo. Here, we show that (i) PDF activity is essential for photosynthetic function, (ii) chloroplast PDF is the specific target of the antibiotic actinonin in vivo and (iii) NME is essential for biogenesis of photosystem II (PSII), primarily by stabilizing the D2 subunit. A general role of NME in modulating the half-life of key subsets of proteins is proposed.
Results
Isolation and characterization of pdf1b knockout mutants in A.thaliana
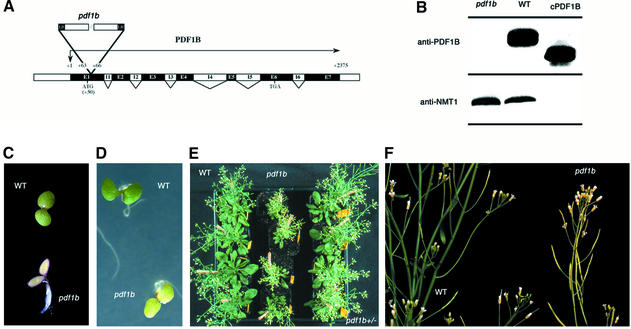
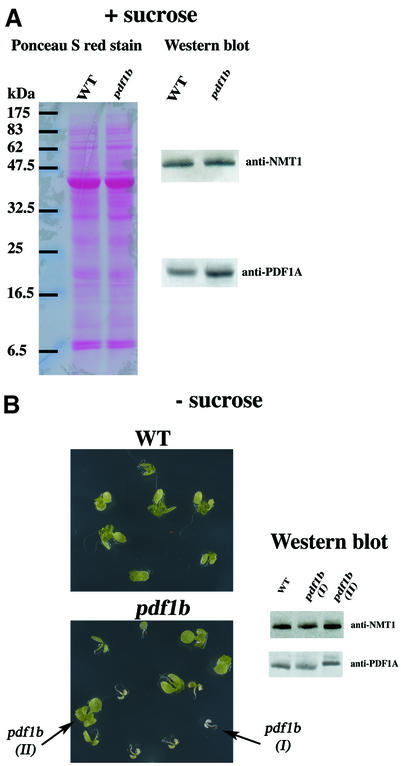
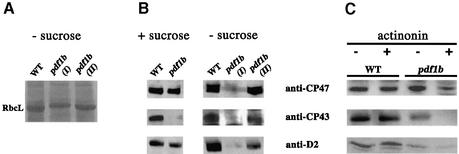
To determine the role of chloroplast deformylation in A.thaliana, we screened various T-DNA mutant collections for insertions in pdf1b. Only one mutant line was identified (Figure 1A). Homozygous pdf1b lines were obtained and western blotting experiments were performed to verify effective gene knockout (KO). pdf1b did not produce any PDF1B protein and therefore was a true pdf1b KO (Figure 1B). pdf1b displayed a pronounced albino phenotype at the early stage of development, i.e. 2 days after germination (Figure 1C). Later in development, the mutant recovered slightly, but still exhibited abnormally weak pigmentation with fewer and smaller leaves than the wild type and a dwarf phenotype (Figure 1D–F). The recovery varied between individual seeds from the same line. pdf1b mutants contained significantly more (>300%) PDF1A than the wild type (Figure 2A), suggesting that the overexpression of the second PDF gene compensated for the defect of the first. In sucrose-minus medium, in which the photosynthetic function of the plastid is required for plant growth, some pdf1b lines grew very slowly and displayed a pronounced albino phenotype without any rapid recovery, whereas others recuperated and greened (Figure 2B). Western blot analysis showed that individuals with a long-lasting albino phenotype contained an amount of PDF1A comparable with the wild type. In contrast, the fast-growing greener plantlets reproducibly had a higher content of PDF1A (Figure 2B). Thus the severe consequences associated with pdf1b disruption could be compensated epigenetically by overexpression of PDF1A.
Fig. 1. Characterization of the A.thaliana pdf1b line. (A) Schematic representation of pdf1b gene disruption in A.thaliana line pdf1b. The exon–intron structure is shown. Translation initiation and termination codons are indicated. The T-DNA inserts are represented with the 5′ border sequences of the insert (LB) labeled to indicate the orientation of the insertion. A three-base-pair deletion was observed at the site of the insertion as indicated. (B) Presence of PDF1B in wild-type and line pdf1b. Arabidopsis seeds were synchronized in the dark at 4°C for 2 days before sowing. Four hundred milligrams of 2-week-old shoots were homogenized and total proteins were extracted as described. Aliquots (250 µg) of protein were analyzed by SDS–PAGE; 250 ng of cPDF1B, the purified catalytic domain of PDF1B (Serero et al., 2001a), was run in parallel. The gels were blotted and analyzed by western blotting with anti-PDF1B and anti-NMT1 as a control. (C) Albino phenotype of 2-day-old pdf1b plantlet compared with wild type. (D) Intermediate phenotype of 3-day-old pdf1b plantlet compared with wild type. (E) Forty-five-day-old wild-type, pdf1b+/– and pdf1b plantlets, seen from the top. (F) Forty-five-day-old wild-type and pdf1b plantlets, seen from the side.
Fig. 2. KO pdf1b plants counteract the absence of PDF1B by increasing the level of PDF1A. (A) Expression of PDF1A in wild-type and pdf1b plants grown in 1% sucrose medium. One hundred and fifty micrograms of total protein was separated by SDS–PAGE, transferred to a nitrocellulose membrane and stained with Ponceau S red stain. The membrane was destained and probed using anti-PDF1A and anti-NMT1 antibodies. (B) Wild-type and pdf1b seeds were sown in a sucrose-minus growth medium, synchronized in the dark at 4°C for 2 days and then incubated in a growth cabinet. Seedlings were photographed 2 weeks later and each phenotype was collected separately (I, albino; II, greening). Aliquots (150 µg) of total protein extract were analyzed by 14% SDS–PAGE and western blotting as in (A).
Isolation and characterization of an A.thaliana pdf1a line: the PDF1A deficiency is fully compensated by PDF1B
The albino phenotype of line pdf1b and its reversibility indicate that the deformylation process is essential in the chloroplast by affecting the biogenesis of the photosynthetic apparatus. Moreover, this process seems to be so essential that the absence of PDF1B in KO pdf1b plants is counteracted by overproduction of PDF1A. This is consistent with PDF1A being routed to the chloroplast when overproduced, as observed in transient expression studies (Giglione and Meinnel, 2001a). Therefore, we investigated the effect of pdf1a inactivation.
Various T-DNA mutant collections were screened for insertions in pdf1a. Two independent mutant lines were identified (Figure 3A). Homozygous lines were obtained in each case and western blotting experiments, performed to check for effective gene knockout, showed that the line named pdf1a was a true PDF1A KO mutant (Figure 3B). Line CS15–187 still produced the PDF1A protein, probably because the T-DNA insertion was located in the promoter region too far (∼300 bases) from the transcribed sequence. Mutant pdf1a behaved similarly to the wild type and displayed no visible phenotype under standard growth conditions (data not shown). This was not unexpected, as both PDF1A and PDF1B are present in the mito chondria (Giglione et al., 2000b). These data suggest that PDF1B can fully compensate for the absence of PDF1A from mitochondria despite their biochemical differences (Serero et al., 2001a). Finally, PDF1B was not overproduced in line pdf1a (Figure 3B) unlike PDF1A in line pdf1b (Figure 2B).
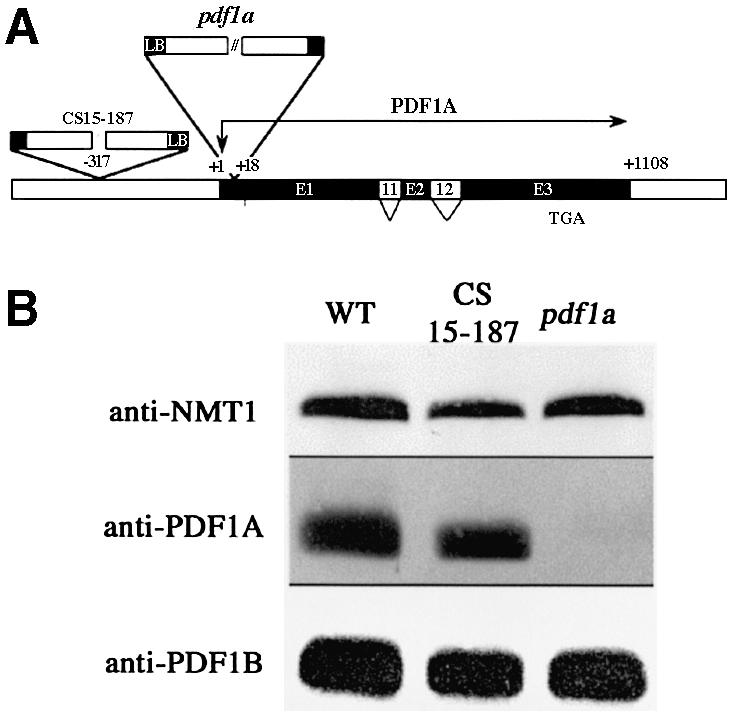
Fig. 3. Characterization of the A.thaliana pdf1a line. (A) Schematic representation of PDF1A gene disruption in A.thaliana lines CS1813– 43 and CS15–187. The exon–intron structure is shown as in Figure 1. Translation initiation and termination codons are indicated. The T-DNA inserts are represented with the 5′ border sequences of the insert (LB) labeled to indicate the orientation. (B) PDF1A protein in A.thaliana wild-type, pdf1a and CS15–187 lines. Aliquots (250 µg) of total protein extracts were analyzed by 14% SDS–PAGE. Western blots were performed with anti-PDF1A, anti-PDF1B and anti-NMT1 antibodies.
Deformylase activity is essential in the chloroplast: plastid PDF1B is the main target of actinonin in plants
Actinonin specifically inhibits bacterial PDF (Chen et al., 2000). In A.thaliana, it induces an albino phenotype at concentrations higher than 100 µM, indicating an alteration of plastid biogenesis (Giglione and Meinnel, 2001a; Serero et al., 2001b). Both plant PDFs are highly sensitive to actinonin in vitro (Serero et al., 2001a). Moreover, A.thaliana plantlets treated with actinonin only grow from germination to flowering on media containing a reduced carbon source; under these conditions no deleterious effects on development are observed. Thus it has been suggested that mitochondria were unaffected by the drug possibly because at submillimolar concentrations in the growth medium it could not penetrate these organelles (Serero et al., 2001a).
To determine whether the albino phenotype induced by actinonin is due to inhibition of plastid PDF in vivo and which PDF is the target of the drug, lines pdf1a and pdf1b were grown in the presence of various concentrations of actinonin. The sensitivity of the pdf1a line was the same as that of the wild type (Figure 4). Thus inhibition of PDF1A contributed little to the effect of actinonin in the wild type. In contrast, pdf1b was about three orders of magnitude more sensitive than the wild type to the drug (Figure 4). Pronounced bleaching was observed when pdf1b was challenged after the recovery stage with submicro molar concentrations of actinonin, whereas 100 µM was necessary to obtain the same effect in the wild type or in the pdf1a line. Thus actinonin treatment mimics PDF1B inactivation, indicating that plastid PDF1B is the major and primary target of actinonin in plants. The residual sensitivity of pdf1b to actinonin also indicated that the PDF1A protein overexpressed in line pdf1b was fully accessible and sensitive to the drug, consistent with a plastid localization.
Fig. 4. Line pdf1b is hypersensitive to actinonin. Plants were grown for 5 days in the presence of the indicated (top) concentration of actinonin (µM): (A) wild type; (B) pdf1a line; (C) pdf1b line.
These experiments showed that actinonin treatment at high concentrations in the wild-type line or at low con centrations in line pdf1b affected plastid development via complete inhibition of plastid PDF activity. This demonstrates (i) that NME is essential in plastids, and (ii) that actinonin is a suitable specific drug to block plastid NME.
Absence of plastid deformylation leads to a reduction of PSII efficiency in C.reinhardtii
We next investigated why deformylation is so important for plastid function, and whether blocking chloroplastic deformylation affects the behavior of all or only a subset of plastid proteins. The protein targets of PDF are restricted to the ∼80 plastid-encoded proteins, most of which under go N-deformylation (Giglione and Meinnel, 2001a). C.reinhardtii has dual PDF apparatus similar to that of land plants (Giglione and Meinnel, 2001a). This unicellular green alga features a single large plastid and has long been exploited for molecular studies of chloroplast function, as it allows powerful molecular genetics techniques to be used (Dent et al., 2001). These techniques include plastid transformation, which is not currently possible in Arabidopsis. Moreover, studies of photosynthesis in vivo, and particularly those of the light reactions, have benefited from a vast array of biophysical techniques used to examine specific components of the photosynthetic apparatus.
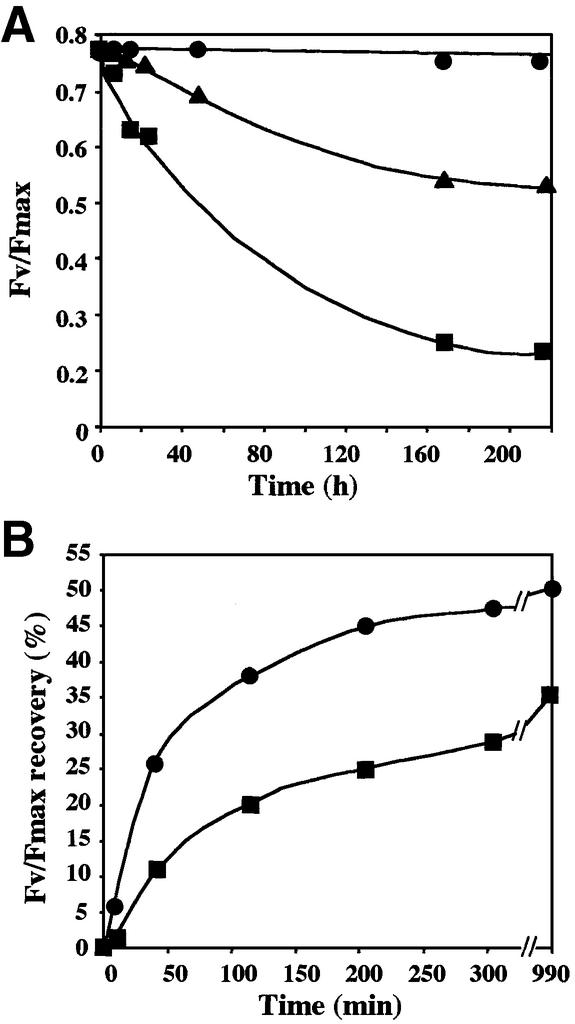
To test C.reinhardtii sensitivity to deformylation inhibition, cells were grown in the presence and the absence of actinonin, and the fluorescence induction kinetics was followed. There was a strong dose-dependent decrease of the variable fluorescence Fv/Fmax (Figure 5A), an indicator of PSII function (Lavergne and Briantais, 1995). When the culture reached stationary phase, PSII activity was reduced by >70%. In contrast, the other fluorescence parameters indicative of photosystem I or cytochrome b6f functions were unaffected, suggesting that the other components of the photosynthetic apparatus were insensitive to the block of deformylation.
Fig. 5. Actinonin leads to a reduction of PSII efficiency in C.reinhardtii. (A) Measurement of the variable fluorescence parameter Fv/Fmax upon actinonin addition (circles, 0 µM; triangles, 50 µM; squares, 500 µM) at time zero in early exponential-phase cultures. (B) Photo-inhibition experiments were carried out for 30 min when the variable fluorescence reached zero. The time-course of recovery in the absence (circles) or the presence (squares) of 0.5 mM actinonin was followed. To visualize only the fraction of recovery that depends on de novo protein synthesis (70%), we subtracted the Fv/Fmax measured at each time point in a duplicate sample incubated with 20 µg/ml chloramphenicol.
PSII activity is sensitive to high light treatment (photo-inhibition) (Aro et al., 1993). PSII recovery after photo-inhibition depends on two components, one involving post-translational repair of photodamaged subunits and the other relying on the synthesis of new proteins. To investigate the molecular effect of actinonin on PSII biogenesis, recovery experiments were followed after high light exposure in the presence of the drug. Each sample was analyzed in the presence and the absence of chloramphenicol, a specific inhibitor of organelle protein synthesis. PSII recovery was measured by monitoring the variable fluorescence Fv/Fmax (Figure 5B). Actinonin did not impair the protein-synthesis-independent (i.e. chloramphenicol-insensitive) recovery of PSII. In contrast, de novo synthesis of chloroplast-encoded PSII components appeared to be significantly reduced by the presence of the drug. We concluded that the inhibition of PSII in the presence of actinonin was due to a defect limiting the accumulation of one or several PSII components.
Inhibition of plastid deformylation in C.reinhardtii leads to rapid degradation of newly synthesized PSII core subunits
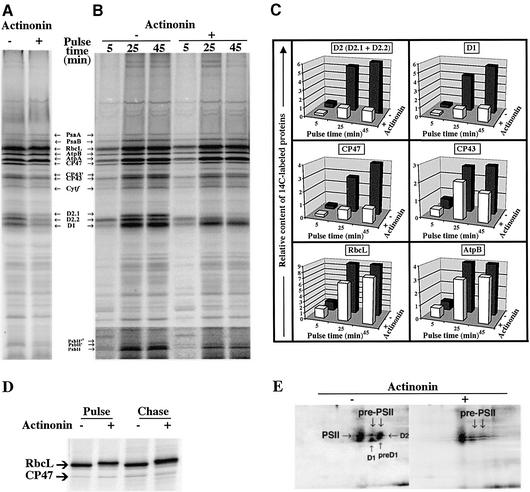
Pulse–chase experiments were carried out either immediately or 48 h after addition of actinonin. Labeling was performed in the presence of cycloheximide, so that only chloroplast-encoded proteins were labeled. In the sample treated for 48 h (Figure 6A), [14C]acetate incorporation into proteins was indistinguishable from that in untreated controls. This indicated that the plastid protein synthesis machinery, i.e. the components of the plastid RNA polymerases or those of the plastid ribosomes, was not affected by PDF inhibition. Therefore the inhibitory effect of actinonin was not due to a general block of protein synthesis. SDS–PAGE analysis of the various plastid-encoded (i.e. labeled) proteins showed that the large subunit of Rubisco (RbcL), the components of photo system I (PsaA and PsaB), ATPase (AtpA and AtpB) and cytochrome b6/f (Cytf) accumulated normally. These data were confirmed by western blotting analysis and chase experiments (data not shown). In contrast, the signals corresponding to several components of PSII, including D1, D2, CP43 and CP47 were significantly lower than in the control (Figure 6A).
Fig. 6. Inhibition of PDF induces a rapid degradation of plastid-encoded PSII subunits in C.reinhardtii. The experiments were performed in the presence (+) or the absence (–) of 0.5 mM actinonin. The location of several plastid proteins identified by western blot is indicated. (A) Samples were 45 min pulse-labeled with [14C]acetate 48 h after addition of actinonin. A phosphoimage of the urea-SDS–PAGE (12%–18% acrylamide) is shown. (B) Time-course pulse-labeling with [14C]acetate was performed 10 min after addition of actinonin. The contrast in the lower part of the gel was intensified to show PsbH and its derivatives, PsbH′ and PsbH”, more clearly. (C) Quantification of the data reported in (B) for CP43, CP47, D1, D2 (D2.1 + D2.2), RbcL and AtpB. (D) Pulse–chase labeling of whole cells with [35S]sulfate. A close-up of the phosphoimage of the SDS–PAGE (7.5%–15% acrylamide) in the region of the RbcL band is shown. AtpA and AtpB are not separated under these conditions and co-migrate just below RbcL. (E) Phosphoimage of [35S]sulfate-labeled membrane fractions analyzed by two-dimensional PAGE involving native (horizontal; top right) followed by denaturing conditions of separation (vertical). D1 and D2 were localized on the gels by western blotting analysis with specific antibodies (data not shown).
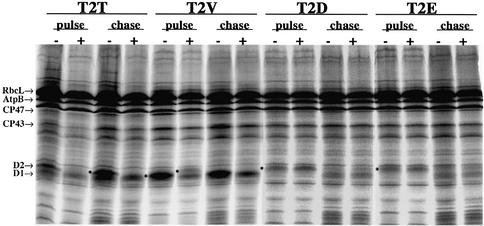
Time-course pulse-labeling experiments performed immediately after actinonin addition (Figure 6B and C) showed that the accumulation of the same set of PSII components was affected. At t = 5 min, the whole plastid protein set was synthesized normally in the presence of actinonin, except for a reduction of D2 labeling. After longer labeling times, the accumulation of each D1, D2, CP43 and CP47 was very much lower than in the wild type. This effect was dose dependent (data not shown). Despite the general reduction of D2 labeling in the actinonin-treated sample, the D2.2 band was converted to D2.1 by phosphorylation of the mature N-terminal threonine (Thr2) residue (Vener et al., 2001). This indicated that D2 accumulation, but not its capacity to be phosphorylated, was inhibited by actinonin. In another experiment, D2 synthesized in the presence of actinonin was completely degraded during 10 min chase experiments in the absence of the drug, in contrast with D1, CP43 and CP47, which were stable for >40 min (data not shown; see also Figure 8, T2T).
Fig. 8. Inhibiting D2 N-terminal methionine excision by changing its second residue leads to effects similar to those induced by actinonin. Pulse–chase experiments with the four C.reinhardtii psbD mutant strains were performed in the absence (–) or the presence (+) of 0.5 mM actinonin. Cells were pulse-labeled with [14C]acetate for 5 min; chase duration was 40 min. Chloroplast proteins were analyzed by urea-SDS–PAGE. Pulse experiments were carried out for 40 min. The positions of several plastid proteins are indicated. The relative position of D2 in various lanes of interest is labeled with an asterisk on the left-hand side of the corresponding band.
In addition to the PSII components D1, D2, CP43 and CP47, PsbH, a more peripheral component of the PSII complex, was also destabilized by actinonin. As described previously (de Vitry et al., 1991), the control experiment showed the appearance of two bands (PsbH′ and PsbH”) due to the phosphorylation of residues Thr2 and Thr4 (Gomez et al., 2002). In the presence of actinonin, the mobility of the unphosphorylated form was affected. The electrophoretic mobility of such small bacterial proteins is similarly shifted following actinonin treatment inhibiting PDF and thus inhibiting processing of the N-terminus (Apfel et al., 2001; Solbiati et al., 2002). Interestingly, only one additional band, of intermediate mobility, was detected (Figure 6B, bottom), suggesting an effect on one of the two phosphorylation events. In addition, the RbcL newly synthesized in the presence of actinonin had a mobility lower than normal, most clearly evidenced in a urea-free gel system (Figure 6D). This shift probably reflects an actinonin-induced block of the multiple pro cessing events that RbcL undergoes at its N-terminus. This processing, which is unique to RbcL in the plastid, is initiated by the PDF-catalyzed removal of N-formyl (see references in Giglione and Meinnel, 2001a). It is remarkable that the block of this processing is not accompanied by a reduction in protein accumulation.
Hence, of all the chloroplast-encoded proteins analyzed, only the five PSII proteins described above showed abnormally low accumulation. Since actinonin has no general effect on protein synthesis, this strongly suggests specific destabilization of these components. The stability of polypeptides engaged in multi-subunit protein complexes is largely a function of their ability to assemble with their partners (Choquet and Vallon, 2000). To analyze the effect of actinonin on PSII assembly more directly, we used two-dimensional native gel electrophoresis after [35S]sulfate pulse-labeling. A large proportion of the PSII-associated label was incorporated into a complex of ∼360 kDa (Figure 6E). This is consistent with the co-translational assembly of D1 into PSII centers. D2 is also incorporated directly into this complex, as expected from the comparable synthesis rates of the two polypeptides. In addition, lower molecular weight complexes containing various amounts of D1 and D2 (pre-PSII) were evidenced. That these latter are true assembly intermediates, originating from de novo assembled complexes, was supported by western blotting analysis and chase experiments (data not shown). The intensity of labeling associated with these assembly intermediates was reduced by actinonin treatment, probably due to enhanced sensitivity to degrada tion. We conclude that NME inhibition acts primarily by committing specific sensitive PSII subunits to the proteolytic pathway. In contrast, despite the alteration of its N-terminal processing, Rubisco assembly was found to proceed normally and all intermediary complexes were detected in normal amounts in the soluble fraction (data not shown).
ClpP is a major ATP-dependent protease involved in the N-end rule degradation mechanism of bacterial proteins (Tobias et al., 1991). We tested whether PSII degradation occurred via the plastid ClpP protease as in the case of cytochrome b6f (Majeran et al., 2000). ClpP is an essential gene in C.reinhardtii, and therefore we used a ClpP mutant with significantly reduced expression of the protein (Majeran et al., 2000). The rate of actinonin-dependent protein degradation in this Clp mutant was identical with that in the control (data not shown). Thus D2 and the other components of PSII were presumably degraded via a ClpP-independent proteolytic pathway.
Inhibition of chloroplast deformylation in A.thaliana leads to reduced accumulation of components of the PSII protein complex
To examine whether blocking the deformylation process in A.thaliana had similar effects to those in C.reinhardtii, the steady state protein content was analyzed in pdf1b Arabidopsis lines. No significant difference of content per net weight was observed for the whole protein set between the mutant and wild type (Figures 2A and 7A). However, the mobility of RbcL was shifted in pdf1b lines exhibiting the long-lasting albino phenotype (pdf1b I). This shift was not observed in the lines which managed to green (pdf1b II) (Figure 7A) and in which PDF1A expression was increased (see Figure 2). Western blotting analysis of the Arabidopsis KO line pdf1b using antibodies raised against D2, CP43 and CP47 was performed. As in C.reinharditii, accumulation of these protein components of PSII was lower in pdf1b than in wild-type seedlings. The effect was enhanced in line pdf1b I grown in the absence of sucrose (Figure 7B). The same defect of PSII accumulation was observed in pdf1b plants grown in the presence of 1 µM actinonin (Figure 7C). These consistent results validated the combined use of A.thaliana and C.reinhardtii as model systems to study the function of plastid NME.
Fig. 7. The accumulation of PSII components is affected in A.thaliana pdf1b lines as in actinonin-treated C.reinhardtii cells. Plants were grown for 15 days. Proteins were analyzed by SDS–PAGE. Western blot analysis was performed using anti-D2, anti-CP43 and anti-CP47 antibodies. (A) Wild-type and pdf1b lines were grown in the absence of sucrose. Plantlets were collected as described in Figure 2. Aliquots (150 µg) of total proteins were loaded on the gel, transferred onto a nitrocellulose membrane and stained with Ponceau S red stain. The position of RbcL is indicated. (B) The destained nitrocellulose membrane from (A) was analyzed by western blotting. (C) Wild-type and pdf1b lines were grown in the absence (–) or the presence (+) of 1 µM actinonin. One-hundred-milligram samples of membrane proteins were analyzed by western blotting.
Retention of the N-terminal methionine of D2 by changing the second amino acid is enough to induce D2 instability and mimics the effects of actinonin
D1, D2, CP47 and PsbH have been shown to under go N-formyl-methionine removal in plants and algae (http://www.isv.cnrs-gif.fr/tm/maturation/images/chloro.html; Giglione and Meinnel, 2001a and references cited therein). Inhibition of PDF activity by actinonin treatment is expected to result in N-formyl-methionine retention. To determine whether destabilization of D2, the most sensitive target, was due to the presence of the formyl group or of the N-terminal methionine, various C.reinhardtii mutants of D2, predicted to retain or lose their first methionine, were constructed by modifying the nature of the second codon of the psbD gene. The T2V D2 mutant was expected to undergo normal cleavage of its N-formyl-methionine, whereas mutants T2D and T2E should lose only the N-formyl group, but not the methionine (for rules of methionine cleavage in plants, see Giglione and Meinnel, 2001a, and references cited therein). We found by fluorescence induction analysis and western blotting (data not shown) that the T2E substitution led to loss of PSII activity and phototrophy, whereas the T2V mutant had normal PSII activity and proteins, as described previously (Fleischmann and Rochaix, 1999). In the T2D mutant, PSII accumulation was severely reduced, but the cells were able to grow phototrophically.
Pulse–chase experiments were performed with each mutant line and with a ‘T2T’ control strain carrying only the silent base substitution at –1 that was present in all the constructs. During the pulse, the T2T and T2V strains produced normal amounts of the D2 protein (labeled with an asterisk), which remained stable during the chase (Figure 8). In this gel system, the D2 protein in the T2V mutant migrated faster than the corresponding protein in the T2T mutant or in the wild type, and co-migrated with D1. When the pulse was performed in the presence of actinonin, the D2 protein in the T2V mutant became as unstable as in the T2T control, as evidenced by a reduced 14C signal. The T2V D2 protein showed a clear upward mobility shift in the presence of actinonin, probably resulting from the retention of the formyl-methionine caused by the actinonin-induced block of PDF.
In contrast, both T2D and T2E mutants showed similarly lower 14C signals corresponding to D2, independent of the presence of actinonin (Figure 8). The D2 protein in these strains was slightly shifted upwards, and this shift was also independent of the presence of actinonin. This suggests that the effect is due to the presence of the initial methionine and not of the formyl group. Both T2D and T2E D2 proteins disappeared completely during the chase, indicating that they were highly unstable. During the chase, these strains displayed slightly reduced rates of synthesis of D1 and CP47 but not of CP43, characteristic of the translational regulation of D1 and CP47 in D2-less strains (de Vitry et al., 1989). In contrast, the reduced D2 signal was clearly attributable to early post-translational destabilization, since this protein was synthesized normally in all PSII-less mutants. Overall, our data indicated that instability of D2 can be induced by the retention of the N-terminal methionine alone, caused either by substituting the second residue or by blocking deformylation.
Discussion
Little is currently known about why NME is a conserved pathway, essential for cell survival. In the cytoplasm of eukaryotes, a subset of key proteins starting with Gly undergo a post-translational N-myristoylation (Johnson et al., 1994). N-myristoylation needs to be initiated by MAP action to unmask glycine as the first residue. Disruption of the N-myristoyl transferase gene is lethal in yeast, demonstrating the essential character of N-myristoylation and thus explaining the crucial role of NME in the cytoplasm of eukaryotes (Duronio et al., 1989). However, N-myristoylation does not occur in bacteria or in either of the two organelles of bacterial origin, the mitochondrion and the chloroplast. Other post-translational modifications, such as N-acetylation, N-methylation or O-phosphorylation of N-terminal residues, which have been described in the organelles or the cytoplasm of eukaryotes, could also explain why NME is essential. Such post-translational modifications are rare in bacteria and, when occurring, are not essential for cell growth. This suggests that these N-terminal modifications, including N-myristoylation, appeared later than NME during evolution and were grafted onto a subset of proteins already undergoing NME. Hence, the essential nature of NME is probably not a consequence of these processes.
A decade ago, Varshavsky (1996) elaborated the N-end rule, i.e. the mechanism that links protein half-life to the nature of its N-terminal amino acid. This rule was suggested by a series of experiments exploring the metabolic fate of fusion proteins between ubiquitin and a reporter protein such as Escherichia coli β-galactosidase. The stabilizing amino acids were all those residues unmasked by NME, including methionine. Although the N-end rule pathway seems to be conserved, different versions operate in different organisms. There are also examples of proteins escaping the rule as a result of small changes, involving post-translational modifications, at the N-terminus itself. For instance, some proteins with an N-terminal cysteine, like the GTPase-activating protein RGS4, may undergo oxidation of their first side-chain and be degraded by the N-end rule pathway after N-terminal arginylation (Davydov and Varshavsky, 2000; Kwon et al., 2002). The half-lives of the proto-oncogene c-mos and the ribosomal protein P1 depend on the phosphorylation state of Ser2 (Nishizawa et al., 1992; Nusspaumer et al., 2000). The D1 protein of PSII is phosphorylated in higher plants as part of its repair cycle during photo-inhibition (Aro et al., 1993; Vener et al., 2001). In C.reinhardtii, the N-terminal Thr2 of subunit D2 is phosphorylated but its phosphorylation is unnecessary for PSII biogenesis (Andronis et al., 1998; Fleischmann and Rochaix, 1999; this work). Similarly, the half-lives of homoserine trans-succinylase and hypoxanthine phosphoribosyltransferase depend on their N-terminal residues but not on their phosphorylation state (Johnson et al., 1988; Biran et al., 2000). Thus N-terminal features other than the destabilizing residues defined by the N-end rule can determine the half-life of proteins. In this context, an attractive hypothesis to explain the importance of NME is its putative involvement in protein half-life.
Using A.thaliana and C.reinhardtii as model systems and actinonin, a drug which specifically inhibits chloroplast PDF (Serero et al., 2001a; this work), we revealed why NME is such an important and conserved process in a whole proteome, the plastid-encoded proteins. Inhibition of N-formyl-methionine removal caused progressive loss of photosynthesis activity. This was due to a particular subset of proteins, the core components of PSII, becoming highly unstable. The D2 subunit appears as the primary most sensitive target whose destabilization impairs PSII biogenesis, as demonstrated by substitutions altering its N-terminus (Figure 8). However, PSII is the only protein complex of the chloroplast for which biogenesis is impaired. All the other complexes, including ATPase, cytochrome b6f, photosystem I, ribosome, RNA polymerase and Rubisco appear to assemble and function normally. Other plastid-encoded PSII components that undergo NME but were not probed in this study (the products of psbE, psbF, psbK and psbL) could also be destabilized by the same mechanism. In Arabidopsis, a cascade mechanism originating from the same destabilization of PSII most likely leads to the progressive loss of the other chlorophyll proteins and results in the albino phenotype. This effect is similar to the case of cytochrome b6f disruption in tobacco (Monde et al., 2000). It may involve an enhanced sensitivity of D1 to NME in higher plants, as suggested by its higher turnover and the crucial role of its N-terminus as compared with C.reinhardtii.
What is the mechanism by which D2 is destabilized? NME inhibition may act indirectly by preventing D2 O-phosphorylation. However, our data indicate that this is not the case. In addition, substituting Thr2 in D2 with a non-phosphorylatable residue (e.g. T2V) does not impair PSII function (Fleischmann and Rochaix, 1999; this work). D2 destabilization is observed when a substitution in its amino acid sequence (T2D or T2E) prevents MAP action. Thus retention of the N-terminal methionine, independent of its N-formylation, is enough to cause early D2 degradation. It could be also argued that D2 degradation associated with methionine retention is primarily caused by its inability to incorporate into PSII complexes. Our two-dimensional electrophoresis data do not support this interpretation; unassembled D2 was not observed, although it would have been expected if assembly was prevented. Four other targets, namely D1, CP43, CP47 and PsbH, were degraded in addition to D2. The degradation of these proteins may have been due to destabilization by the same mechanism as affected D2. Nevertheless, the individual subunits of PSII are only fully stabilized when a minimum core involving D1, D2, CP43, CP47 and cytochrome b-559 is formed (de Vitry et al., 1989; van Wijk et al., 1997; Wollman et al., 1999). Chase experiments show that, in contrast with the D2 subunit, D1, CP43 and CP47 synthesized in the presence of actinonin are stable. Therefore destabilization of the other core PSII components is most likely to be a consequence of the absence of D2, their major assembly partner. Furthermore, the partial recovery of PSII activity after photo-inhibition (Figure 5B) suggests that at least a fraction of the D1 produced in the presence of actinonin can be stabilized.
In this work, we demonstrate that NME is directly involved in determining the half-life of various key proteins in a whole proteome. Methionine at the N-ter minus of some proteins, for instance in D2, may act as a destabilizing residue. Therefore, NME, not only in the plastid, but also in the cytoplasm and the mitochondria, may be a general mechanism for modulating protein life-span, probably in concert with features in the immediate vicinity of the N-terminal residue. The machinery responsible for this degradation remains unknown. Nevertheless, this process does not seem to involve either ubiquitination, a mechanism that does not occur in the plastid, or the Clp protease. The plastid contains a number of proteases in addition to Clp, including FtsH and DegP (Adam and Clarke, 2002). Recently, several have been implicated in D1 protein turnover but their function remains largely uncharacterized (Lindahl et al., 2000; Haussuhl et al., 2001). They might be part of the proteolytic process that we observed as a result of NME inactivation. Deformylation and N-methionine removal (Meinnel et al., 1993), and part of PSII assembly (Zhang et al., 1999), are co-translational processes. However, degradation of the PSII components due to N-terminal methionine retention appears to be largely post-translational, as evidenced by the reduced D2 accumulation as labeling time is increased (Figure 6B and C) and in chase experiments (Figure 8).
The involvement of NME as a checkpoint in the modulation of the half-life of crucial components of organelles suggests that it could have a similar function in mitochondria. However, the function and importance of mitochondrial NME, especially in animal cells, remain to be identified. Unfortunately, functional analysis of NME is complicated by the fact that mitochondrial function is essential for cell survival.
Materials and methods
Growth conditions
Arabidopsis thaliana (WS-2 ecotype) seeds were sterilized and sown on 0.5× Murashige and Skoog (ICI) medium supplemented with 0.8% agarose (Difco, Detroit, MI) and 1% sucrose (ICI) unless otherwise stated. Petri dishes were incubated in a growth chamber (22°C, 16 h of daylight; light intensity, 100 µE/m2/s1) for up to 5 weeks. Arabidopsis thaliana lines were propagated under greenhouse conditions as described. Wild-type C.reinhardtii (WTS34 mt+, derived from strain 137c) and psbD mutant strains were grown on Tris-acetate phosphate (TAP) medium (Harris, 1989) at 25°C under 30 µE/m2/s1 continuous illumination or in minimum medium at 100 µE/m2/s1. Actinonin was purchased from Sigma, dissolved in methanol and stored at –20°C in aliquots at 260 mM.
Isolation and characterization of T-DNA insertion mutants
PCR-based identification of T-DNA insertions in PDF1A and PDF1B was performed at the Arabidopsis Knockout Facility (Madison, WI). The screening protocol is available at http://www.biotech.wisc.edu/NewServicesAndResearch/Arabidopsis/. DNA pools from the A.thaliana libraries of T-DNA insertion mutants were obtained from the Arabi dopsis Biological Resource Center, Ohio State University, Columbus, OH. Line CS15–187 carried a single insertion consisting of one single T-DNA. Line pdf1a carried a single insertion consisting of one left-border to right-border plus part of the T-DNA plasmid. Line pdf1b carried a single insertion consisting of two concatemeric left-border to left-border T-DNA molecules.
Site-directed mutagenesis in C.reinhardtii chloroplast
The wild-type strain was transformed by tungsten particle bombardment (Boynton et al., 1988) with a home-built pressurized-helium device. Transformants were grown on TAP plates containing spectinomycin and streptomycin as described previously (Kuras and Wollman, 1994) until homoplasmy was achieved, as assessed by polymerase chain reaction (PCR) analysis. The plasmids pSK-141, pSK-142, pSK-145 and pSK-146 (kind gifts from J.D. Rochaix, Geneva, Switzerland) were used to transform WTS34, yielding transformants 141(T2V), 142(T2D), 145(T2E) and 146(T2T), respectively.
In vivo 14C and 35S pulse–chase labeling experiments in C.reinhardtii
The protocol was derived from Kuras and Wollman (1994). Cells grown in TAP medium were harvested, washed and resuspended in high-salt minimal medium to a chlorophyll concentration of 22 µg/ml. The cell suspension was incubated in minimum medium at 25°C with agitation for 15 min at an incident light intensity of 20 µE/m2/s. The cells were further incubated for 10 min with agitation in the absence or the presence of actinonin (5–1000 µM). Five minutes prior to the addition of the radioactive label, the cytoplasmic protein-synthesis inhibitor cyclo heximide (12 µg/ml final concentration) was added. Na[1–14C]acetate (74 MBq; 2.1 GBqCi/mmol, 0.37 GBq/ml; Amersham) was then added, and labeling was carried out for 5–45 min as indicated. Aliquots of 2 ml were diluted into 20 ml of pre-chilled TAP and centrifuged at 6000 r.p.m. (rotor JA-20, Beckman) for 5 min. Pellets were washed with 1 ml of 10 mM HEPES containing anti-protease inhibitors, resuspended in 30 µl of a buffer containing 50 mM DTT and 0.1 M Na2CO3, frozen in liquid nitrogen and conserved at –80°C. For the chase, the labeled cells were diluted into 20 ml of TAP medium, rinsed and incubated for an additional 10–40 min in the presence of 20 µg/ml chloramphenicol before harvesting.
For two-dimensional PAGE analysis, labeling was carried out in a similar manner except that cells were suspended overnight in sulfur-free TAP medium and [14C]acetate was replaced by Na235SO4 (46 MBq; 37 TBq/mmol, 1.33 GBq/ml; Amersham). Cells were disrupted with a French Press, and the cell extract was ultracentrifuged at 80 000 r.p.m. for 40 min at 4°C (rotor TLA100–2; Beckman). The supernatant was used for soluble protein analysis. The pellet was resuspended in 3 ml of buffer A [5 mM HEPES–KOH pH 7.5, 10 mM EDTA, 0.2 mM phenylmethyl sulfonyl fluoride (PMSF), 1 mM benzidine, 5 mM ε-caproic acid, 1.8 M sucrose], poured into SW45.1 centrifuge tubes (Beckman) and layered with 3 ml of buffer B (same as A but with 1.3 M sucrose), and 5 ml of buffer C (same as A but with 0.5 M sucrose). Centrifugation (30 000 r.p.m.) was carried out for 1 h at 4°C. Both membrane interfaces were recovered, pooled and frozen.
Protein analysis
Protein concentrations were measured with the BC assay protein quantification kit (Uptima). Bovine serum albumin was used as the protein standard. Chlorophyll concentration was measured according to Porra et al. (1989). The ImageQuant program and the data obtained with a phosphoimager (Storm 840; Amersham) were used for all other quantification procedures.
Arabidopsis thaliana samples were frozen in liquid nitrogen and disrupted in an MM 300 mixer mill (Qiagen). To extract total protein, the samples were resuspended in a pre-chilled 50 mM HEPES buffer (pH 7.2) containing 150 mM NaCl, 10% glycerol, 1.5 mM MgCl2, 1% Triton, 1 mM EGTA, 2 mM PMSF and an anti-protease mixture (Roche), and incubated under agitation at 4°C for 45 min to allow solubilization. The supernatant was separated from the insoluble fraction by centrifugation at 13 000 r.p.m. at 4°C for 1 h. The procedure described by Schagger and von Jagow (1991) was used to extract membrane proteins. Protein extracts were analyzed by SDS–PAGE.
Chlamydomonas reinhardtii labeled protein samples were resuspended in 1 volume of Laemmli loading buffer, and analyzed on 12%–18% or 7.5%–15% acrylamide gels containing SDS and urea when indicated (Piccioni et al., 1981). The gels were stained with Coomassie Blue, dried and used for autoradiography. Soluble and thylakoid membrane fractions (solubilized with 1.6% N-dodecyl-maltoside, Sigma) were analyzed by two-dimensional PAGE, using colorless native or blue native PAGE respectively (Schagger and von Jagow, 1991; Schagger et al., 1994) in the first dimension and urea-SDS–PAGE in the second. Gels were transferred onto nitrocellulose and exposed in a phosphoimager before revelation with antibodies.
For western blotting, proteins were electro-transferred onto nitrocellulose membranes and stained with Ponceau S red stain (Carlo Erba) to check for effective homogeneous transfer. Destained blots were developed with the ECL detection kit (Amersham). Rabbit antisera against A.thaliana (AtPDF1A and AtPDF1B), both purified as described previously (Serero et al., 2001a), and anti-A.thaliana N-myristoyl tranferase 1 (NMT1) (B.Boisson and T.Meinnel, unpublished results) were raised at Eurogentech (Herstal, Belgium) and further purified before use. Antisera against tobacco CP43, CP47 and D2 proteins were a kind gift from R.Barbato (Novara, Italy). The antisera against the plastid proteins from C.reinhardtii have been described by Hamel et al. (2000).
Fluorescence transients
Fluorescence induction was measured at room temperature on cells grown in standard conditions or photo-inhibited at 800 µE/m2/s (0.5 µg/ml chlorophyll in TAP medium, 30 min) and allowed to recover at 6 µE/m2/s (0–990 min). Cells were dark-adapted for 3 min and transferred to 1 ml cuvettes and placed in a home-built fluorimeter (Joliot et al., 1998); fluorescence induction was recorded over 1.4 s. Initial fluorescence (F0) was recorded at the onset of illumination and maximum fluorescence (Fmax) after addition of 3-(3,4-dichlorophenyl)-1,1-dimethylurea which blocks electron donation beyond the primary acceptor QA. The nor malized variable fluorescence
Fv/Fmax = (Fmax – F0)/Fmax
is a relative measure of the light-reducible QA, and hence of the number of active PSII centers.
Acknowledgments
Acknowledgements
We thank A.R.Grossman and F.-A.Wollman for a critical review of the manuscript, and R.Barbato and J.D.Rochaix for gifts of materials. C.Phan is acknowledged for help in generating the C.reinhardtii psbD mutant strains. This article is dedicated to Marco Meinnel. This work was supported by CNRS (UPR2355, UPR1261 and ATIPE, MCT and PCV grants to T.M.), the Association pour la Recherche sur le Cancer (ARC, Villejuif, France) and the Fondation pour la Recherche Médicale.
References
- Adam Z. and Clarke,A.K. (2002) Cutting edge of chloroplast proteolysis. Trends Plant Sci., 7, 451–456. [DOI] [PubMed] [Google Scholar]
- Andronis C., Kruse,O., Deak,Z., Vass,I., Diner,B.A. and Nixon,P.J. (1998) Mutation of residue threonine-2 of the D2 polypeptide and its effect on photosystem II function in Chlamydomonas reinhardtii. Plant Physiol., 117, 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfel C.M., Locher,H., Evers,S., Takacs,B., Hubschwerlen,C., Pirson,W., Page,M.G. and Keck,W. (2001) Peptide deformylase as an antibacterial drug target: target validation and resistance development. Antimicrob. Agents Chemother., 45, 1058–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro E.M., Virgin,I. and Andersson,B. (1993) Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta, 1143, 113–134. [DOI] [PubMed] [Google Scholar]
- Biran D., Gur,E., Gollan,L. and Ron,E.Z. (2000) Control of methionine biosynthesis in Escherichia coli by proteolysis. Mol. Microbiol., 37, 1436–1443. [DOI] [PubMed] [Google Scholar]
- Boynton J.E. et al. (1988) Chloroplast transformation in Chlamydo monas with high velocity microprojectiles. Science, 240, 1534–1538. [DOI] [PubMed] [Google Scholar]
- Bracchi-Ricard V., Nguyen,K.T., Zhou,Y., Rajagopalan,P.T., Chakrabarti,D. and Pei,D. (2001) Characterization of an eukaryotic peptide deformylase from Plasmodium falciparum. Arch. Biochem. Biophys., 396, 162–170. [DOI] [PubMed] [Google Scholar]
- Bradshaw R.A., Brickey,W.W. and Walker,K.W. (1998) N-terminal processing: the methionine aminopeptidase and N α-acetyl transferase families. Trends Biochem. Sci., 23, 263–267. [DOI] [PubMed] [Google Scholar]
- Chang S.Y., McGary,E.C. and Chang,S. (1989) Methionine amino peptidase gene of Escherichia coli is essential for cell growth. J. Bacteriol., 171, 4071–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.Z. et al. (2000) Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry, 39, 1256–1262. [DOI] [PubMed] [Google Scholar]
- Choquet Y. and Vallon,O. (2000) Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie, 82, 615–634. [DOI] [PubMed] [Google Scholar]
- Davydov I.V. and Varshavsky,A. (2000) RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J. Biol. Chem., 275, 22931–22941. [DOI] [PubMed] [Google Scholar]
- Dent R.M., Han,M. and Niyogi,K.K. (2001) Functional genomics of plant photosynthesis in the fast lane using Chlamydomonas reinhardtii. Trends Plant Sci., 6, 364–371. [DOI] [PubMed] [Google Scholar]
- de Vitry C., Olive,J., Drapier,D., Recouvreur,M. and Wollman,F.A. (1989) Posttranslational events leading to the assembly of photosystem II protein complex: a study using photosynthesis mutants from Chlamydomonas reinhardtii. J. Cell Biol., 109, 991–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vitry C., Diner,B.A. and Popot,J.L. (1991) Photosystem II particles from Chlamydomonas reinhardtii. J. Biol. Chem., 266, 16614–16621. [PubMed] [Google Scholar]
- Duronio R.J., Towler,D.A., Heuckeroth,R.O. and Gordon,J.I. (1989) Disruption of the yeast N-myristoyl transferase gene causes recessive lethality. Science, 243, 796–800. [DOI] [PubMed] [Google Scholar]
- Fleischmann M.M. and Rochaix,J.D. (1999) Characterization of mutants with alterations of the phosphorylation site in the D2 photosystem II polypeptide of Chlamydomonas reinhardtii. Plant Physiol., 119, 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglione C. and Meinnel,T. (2001a) Organellar peptide deformylases: universality of the N-terminal methionine cleavage mechanism. Trends Plant Sci., 6, 566–572. [DOI] [PubMed] [Google Scholar]
- Giglione C. and Meinnel,T. (2001b) Peptide deformylase as an emerging target for antiparasitic agents. Emerg. Therap. Targets, 5, 41–57. [DOI] [PubMed] [Google Scholar]
- Giglione C., Pierre,M. and Meinnel,T. (2000a) Peptide deformylase as a target for new generation, broad spectrum antimicrobial agents. Mol. Microbiol., 36, 1197–1205. [DOI] [PubMed] [Google Scholar]
- Giglione C., Serero,A., Pierre,M., Boisson,B. and Meinnel,T. (2000b) Identification of eukaryotic peptide deformylases reveals universality of N-terminal protein processing mechanisms. EMBO J., 19, 5916–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez S.M., Nishio,J.N., Faull,K.F. and Whitelegge,J.P. (2002) The chloroplast grana proteome defined by intact mass measurements from liquid chromatography mass spectrometry. Mol. Cell Proteomics, 1, 46–59. [DOI] [PubMed] [Google Scholar]
- Griffith E.C., Su,Z., Turk,B.E., Chen,S., Chang,Y.-H., Wu,Z., Biemann,K. and Liu,J. (1997) Methionine aminopeptidase (type 2) is the common target for angiogenesis AGM-1470 and ovalicin. Chem. Biol., 4, 461–471. [DOI] [PubMed] [Google Scholar]
- Hamel P., Olive,J., Pierre,Y., Wollman,F.A. and de Vitry,C. (2000) A new subunit of cytochrome b6f complex undergoes reversible phosphorylation upon state transition. J. Biol. Chem., 275, 17072–17079. [DOI] [PubMed] [Google Scholar]
- Harris E.H. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego, CA. [DOI] [PubMed]
- Harris E.H., Boynton,J.E. and Gillham,N.W. (1994) Chloroplast ribosomes and protein synthesis. Microbiol. Rev., 58, 700–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussuhl K. Andersson,B. and Adamska,I. (2001) A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J., 20, 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C.A., Peterson,S.N., Gill,S.R., Cline,R.T., White,O., Fraser,C.M., Smith,H.O. and Venter,J.C. (1999) Global transposon mutagenesis and a minimal Mycoplasma genome. Science, 286, 2165–2169. [DOI] [PubMed] [Google Scholar]
- Johnson D.R., Bhatnagar,R.S., Knoll,L.J. and Gordon,J.I. (1994) Genetic and biochemical studies of protein N-myristoylation. Annu. Rev. Biochem., 63, 869–914. [DOI] [PubMed] [Google Scholar]
- Johnson G.G., Kronert,W.A., Bernstein,S.I., Chapman,V.M. and Smith,K.D. (1988) Altered turnover of allelic variants of hypoxanthine phosphoribosyltransferase is associated with N-terminal amino acid sequence variation. J. Biol. Chem., 263, 9079–9082. [PubMed] [Google Scholar]
- Joliot P., Beal,D. and Delosme,R. (1998) In vivo measurements of photosynthetic activity: methods. In Rochaix,J.-D., Goldschmidt-Clermont,M. and Merchant,S. (eds), The Molecular Biology of Chloroplast and Mitochondria in Chlamydomonas. Kluwer, Dordrecht, The Netherlands, pp. 433–449.
- Katinka M.D. et al. (2001) Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature, 414, 450–453. [DOI] [PubMed] [Google Scholar]
- Kuras R. and Wollman,F.A. (1994) The assembly of cytochrome b6/f complexes: an approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J., 13, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y.T., Kashina,A.S., Davydov,I.V., Hu,R.G., An,J.Y., Seo,J.W., Du,F. and Varshavsky,A. (2002) An essential role of N-terminal arginylation in cardiovascular development. Science, 297, 96–99. [DOI] [PubMed] [Google Scholar]
- Lavergne J. and Briantais,J.-M. (1995) Photosystem II heterogeneity. In Ort,D.R. and Yocum,C.F. (eds), Advances in Photosynthesis: Oxygenic Photosynthesis: The Light Reactions. Kluwer, Dordrecht, The Netherlands, pp. 265–287.
- Li X. and Chang,Y.H. (1995) Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine aminopeptidases. Proc. Natl Acad. Sci. USA, 92, 12357–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M., Spetea,C., Hundal,T., Oppenheim,A.B., Adam,Z. and Andersson,B. (2000) The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell, 12, 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Widom,J., Kemp,C.W. and Clardy,J. (1998) Structure of human methionine aminopeptidase-2 complexed with fumagillin. Science, 282, 1324–1327. [DOI] [PubMed] [Google Scholar]
- McCowen M.C., Callender,M.E. and Lawlis,J.,Jr (1951) Fumagillin (H-3), a new antibiotic with amebicidal properties. Science, 113, 202–203. [DOI] [PubMed] [Google Scholar]
- Majeran W., Wollman,F.A. and Vallon,O. (2000) Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b6f complex. Plant Cell, 12, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga P. (1984) Isolation and characterization of mutants in plant cell culture. Annu. Rev. Plant Physiol., 35, 519–542. [Google Scholar]
- Meinnel T. (2000) Peptide deformylase of eukaryotic protists: a target for new antiparasitic agents? Parasitol. Today, 16, 165–168. [DOI] [PubMed] [Google Scholar]
- Meinnel T. and Blanquet,S. (1994) Characterization of the Thermus thermophilus locus encoding peptide deformylase and methionyl-tRNA(fMet) formyltransferase. J. Bacteriol., 176, 7387–7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Mechulam,Y. and Blanquet,S. (1993) Methionine as translation start signal: a review of the enzymes of the pathway in Escherichia coli. Biochimie, 75, 1061–1075. [DOI] [PubMed] [Google Scholar]
- Miller C.G., Kukral,A.M., Miller,J.L. and Movva,N.R. (1989) pepM is an essential gene in Salmonella typhimurium. J. Bacteriol., 171, 5215–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monde R.A., Zito,F., Olive,J., Wollman,F.A. and Stern,D.B. (2000) Post-transcriptional defects in tobacco chloroplast mutants lacking the cytochrome b6/f complex. Plant J., 21, 61–72. [DOI] [PubMed] [Google Scholar]
- Nishizawa M., Okazaki,K., Furuno,N., Watanabe,N. and Sagata,N. (1992) The ‘second-codon rule’ and autophosphorylation govern the stability and activity of Mos during the meiotic cell cycle in Xenopus oocytes. EMBO J., 11, 2433–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusspaumer G., Remacha,M. and Ballesta,J.P. (2000) Phosphorylation and N-terminal region of yeast ribosomal protein P1 mediate its degradation, which is prevented by protein P2. EMBO J., 19, 6075–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccioni R.G., Bennoun,P. and Chua,N.H. (1981) A nuclear mutant of Chlamydomonas reinhardtii defective in photosynthetic photo phosphorylation. Characterization of the algal coupling factor ATPase. Eur. J. Biochem., 117, 93–102. [DOI] [PubMed] [Google Scholar]
- Porra R.J., Thompson,W.A. and Kriedemann,P.E. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta, 975, 384–394. [Google Scholar]
- Schagger H. and von Jagow,G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem., 199, 223–231. [DOI] [PubMed] [Google Scholar]
- Schagger H., Cramer,W.A. and von Jagow,G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem., 217, 220–230. [DOI] [PubMed] [Google Scholar]
- Serero A., Giglione,C. and Meinnel,T. (2001a) Distinctive features of the two classes of eukaryotic peptide deformylases. J. Mol. Biol., 314, 695–708. [DOI] [PubMed] [Google Scholar]
- Serero A., Giglione,C. and Meinnel,T. (2001b) Seeking new targets for antiparasitic agents. Trends Parasitol., 17, 7–8. [DOI] [PubMed] [Google Scholar]
- Sin N., Meng,L., Wang,M.Q., Wen,J.J., Bornmann,W.G. and Crews,C.M. (1997) The anti-angiogenic agent fumagillin covalently binds and inhibits the methionine aminopeptidase, MetAP-2. Proc. Natl Acad. Sci. USA, 94, 6099–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solbiati J., Chapman-Smith,A., Miller,J.L., Miller,C.G. and Cronan,J.E.,Jr (1999) Processing of the N termini of nascent polypeptide chains requires deformylation prior to methionine removal. J. Mol. Biol., 290, 607–614. [DOI] [PubMed] [Google Scholar]
- Solbiati J., Chapman-Smith,A. and Cronan,J.E.,Jr (2002) Stabilization of the biotinoyl domain of Escherichia coli acetyl-CoA carboxylase by interactions between the attached biotin and the protruding ′thumb′structure. J. Biol. Chem., 277, 21604–21609. [DOI] [PubMed] [Google Scholar]
- Tobias J.W., Shrader,T.E., Rocap,G. and Varshavsky,A. (1991) The N-end-rule in bacteria. Science, 254, 1374–1377. [DOI] [PubMed] [Google Scholar]
- van Wijk K.J., Roobol-Boza,M., Kettunen,R. Andersson,B. and Aro,E.M. (1997) Synthesis and assembly of the D1 protein into photosystem II: processing of the C-terminus and identification of the initial assembly partners and complexes during photosystem II repair. Biochemistry, 36, 6178–6186. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. (1996) The N-end rule: functions, mysteries, uses. Proc. Natl Acad. Sci. USA, 93, 12142–12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vener A.V., Harms,A., Sussman,M.R. and Vierstra,R.D. (2001) Mass spectrometric resolution of reversible protein phosphorylation in photosynthetic membranes of Arabidopsis thaliana. J. Biol. Chem., 276, 6959–6966. [DOI] [PubMed] [Google Scholar]
- Wiesner J., Sanderbrand,S., Beck,E. and Jomaa,H. (2001) Seeking new targets for antiparasitic agents. Trends Parasitol., 17, 7–8. [DOI] [PubMed] [Google Scholar]
- Wollman F.A., Minai,L. and Nechushtai,R. (1999) The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim. Biophys. Acta, 1411, 21–85. [DOI] [PubMed] [Google Scholar]
- Zhang L., Paakkarinen,V., van Wijk,K.J. and Aro,E.M. (1999) Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem., 274, 16062–16067. [DOI] [PubMed] [Google Scholar]
- Zhang P., Nicholson,D.E., Bujnicki,J.M., Su,X., Brendle,J.J., Ferdig,M., Kyle,D.E., Milhous,W.K. and Chiang,P.K. (2002) Angiogenesis inhibitors specific for methionine aminopeptidase 2 as drugs for malaria and leishmaniasis. J. Biomed. Sci., 9, 34–40. [DOI] [PubMed] [Google Scholar]