Abstract
We report for the first time an analysis of the ATPase activity of human DNA topoisomerase (topo) IIβ. We show that topo IIβ is a DNA-dependent ATPase that appears to fit Michaelis–Menten kinetics. The ATPase activity is stimulated 44-fold by DNA. The kcat for ATP hydrolysis by human DNA topo IIβ in the presence of DNA is 2.25 s–1. We have characterised a topo IIβ derivative which carries a mutation in the ATPase domain (S165R). S165R reduced the kcat for ATP hydrolysis by 7-fold, to 0.32 s–1, while not significantly altering the apparent Km. The specificity constant for the interaction between ATP and topo IIβ (kcat/Kmapp) showed a 90% reduction for βS165R. The DNA binding affinity and ATP-independent DNA cleavage activity of the enzyme are unaffected by this mutation. However, the strand passage activity is reduced by 80%, presumably due to reduced ATP hydrolysis. The mutant enzyme is unable to complement ts yeast topo II in vivo. We have used computer modelling to predict the arrangement of key residues at the ATPase active site of topo IIβ. Ser165 is predicted to lie very close to the bound nucleotide, and the S165R mutation could thus influence both ATP binding and ADP dissociation.
INTRODUCTION
Type II topoisomerases are essential enzymes which are required for the segregation of intertwined daughter chromosomes following replication (1–4). Their ability to control DNA topology is also important during transcription, replication and recombination (reviewed in 4). A number of anticancer drugs and antibiotics target the cleaved DNA intermediate of the topoisomerase (topo) II reaction cycle, leading to the formation of double-strand DNA breaks which can result in cell death (reviewed in 5,6).
The catalytic cycle of type II topoisomerases is ATP dependent. The ATPase domain is located at the N-terminus of the protein. DNA stimulates ATP hydrolysis by 4- to 20-fold, depending on the source of the enzyme and the reaction conditions (7–10).
Type II topoisomerases alter the topology of DNA by transiently cleaving one DNA helix, termed the gate helix, and transporting a second helix through the enzyme-bridged break (4,11,12). Extensive studies using pre-steady-state kinetic techniques on yeast DNA topo II have enabled Lindsley and co-workers to deduce how ATP binding and hydrolysis is coordinated with DNA cleavage and transport during the reaction cycle of eukaryotic topo II (13–18). The dimeric enzyme binds the gate segment and cleaves it, generating an intermediate where each monomer of topo II is covalently linked to one DNA strand via the active site tyrosine residue. The transported helix is thought to bind between the two N-terminal ATPase domains, and ATP binding causes these two domains to dimerise. One of the two bound ATP molecules is subsequently hydrolysed and then the transported segment is passed through the break in the gate segment. The ATPs are hydrolysed sequentially with the products of the first ATP hydrolysis reaction released prior to hydrolysis of the second ATP molecule. The transported helix is thought to exit via the C-terminal dimer interface, although it is not clear how this event correlates with hydrolysis of the second ATP molecule (13–18).
Human cells contain two isoforms of DNA topo II, α and β (reviewed in 19). The ATPase activity of human topo IIα has been reported (10). To date no data have been published on the ATPase activity of human DNA topo IIβ. In this paper we report the characterisation of the ATPase activity of human DNA topo IIβ. We also describe a mutated protein with reduced ATPase activity which results in loss of complementation activity.
MATERIALS AND METHODS
Wild-type human topo IIβ and the mutant protein βS165R were overproduced from plasmids YEphTOP2β-KLM (20) and YEphTOP2βS16SR (21), respectively, in strain JEL1 ΔtopI (α leu2 trp1 ura3-52 prb1-1122 pep4–3 Δhis3::PGAL 10–GAL4 Δtop1), kindly provided by R. Hanai (Department of Chemistry, Rikkyo University, Tokyo), then purified to >95% as described previously (21). Protein concentrations are expressed in terms of topo II dimers throughout this paper.
Assay for ATP hydrolysis activity
ATP hydrolysis was assayed by a coupled enzyme assay (9), which links ATP hydrolysis to NADH oxidation. Topo II was incubated with 1 mM ATP, supercoiled plasmid (pBR322- or pBluescript-derived), 2 mM phosphoenolpyruvate (PEP), 0.1 mM NADH, 5 U lactate dehydrogenase and 2.5 U pyruvate kinase in relaxation buffer at 30°C. The concentrations of topo II, ATP and DNA were varied as detailed in the figure legends. The rate of NADH oxidation was measured by the decrease in absorbance over 3 min at 340 nm, and is directly proportional to the rate of ATP hydrolysis. Absorbance was measured using a Shimadzu UV-1601 spectrophotometer. All components except topo II and ATP were in excess, as the doubling of the concentration of any component did not increase the rate of NADH oxidation (data not shown). No NADH oxidation was observed in the absence of ATP. Previous work with yeast topo II suggested that the ATP hydrolysis was not truly Michaelian (9). However our data appears to fit the Michaelis–Menten equation and, where appropriate, our data were fitted to the Michaelis–Menten equation using GraphPad Prism: rate = Vmax[S]/([S] + Kmapp), where Vmax is the maximum rate, [S] is substrate concentration and Kmapp is the apparent Michaelis constant. Errors in fitting the data to each equation are expressed as 95% confidence intervals. Yeast topo II does not co-purify with topo IIβ, so will not contribute to the ATPase activity assayed here.
Complementation analysis
The viability of Saccharomyces cerevisiae strain JN394t2-4 (22) transformed with plasmids expressing wild-type or mutated topo IIβ was tested at 25 and 35°C on rich media containing glucose, raffinose or galactose as described previously (20).
Strand passage activity assays
Decatenation and relaxation assays were carried out in relaxation buffer (50 mM Tris–HCl pH 7.5, 0.5 mM EDTA, 1 mM DTT, 50 mM KCl, 30 µg/ml BSA) with 1 µg supercoiled plasmid DNA or 400 ng kinetoplast DNA, 1 mM ATP and 10 mM MgCl2 as described previously (21). Agarose gels were photographed with a Bio-Rad Geldoc 1000 imager and analysed using Tina v.2.08e software.
DNA binding and cleavage analysis
Gel retardation analysis was carried out as described previously (23). The DNA probes used were either a 40 bp linear substrate, or a four-way junction that includes sequences from the linear probe (23). Binding reactions were carried out for 45 min on ice in 20 µl of gel retardation (GR) assay buffer (50 mM Tris–HCl pH 7.5, 0.5 mM EDTA, 1 mM DTT, 50 mM KCl, 1% Triton X-100, 5% glycerol) containing 1 mg/ml BSA. Cleavage reactions contained 600 ng topo IIβ-derived protein and the 40 bp linear substrate which had been labelled at one end. Reactions were carried out in GR buffer with 10 mM MgCl2 or CaCl2 in the presence or absence of 10 µg/ml m-AMSA and/or 1 mM ATP, and processed as described previously (21).
Protein structure analysis
Protein structures were manipulated and displayed using QUANTA, release 4.1.1, version 95.0320 (Molecular Simulations Inc.) on a Silicon Graphics workstation. Residues 35–472 of wild-type topo IIβ were modeled onto residues 1–391 of gyrase B using the program Modeler (24).
RESULTS
Intrinsic and DNA-stimulated ATPase activity of human DNA topo IIβ
The ability of DNA topo IIβ to carry out ATP hydrolysis was assayed spectrophotometrically using a coupled enzyme assay (9). In the presence of 1 mM ATP, the rate of ATP hydrolysis by wild-type topo IIβ increased linearly with protein concentration, both in the presence and absence of DNA (data not shown). In the absence of DNA, the rate of ATP hydrolysis was 0.025 s–1 enzyme dimer–1, whereas the addition of excess supercoiled plasmid DNA increased the rate by 44-fold to 1.1 s–1 dimer–1, thus DNA stimulates the ATPase activity of DNA topo IIβ. This 44-fold stimulation by DNA is higher than the 10-fold stimulation seen with human topo IIα when analysed in a similar way (10).
Effect of DNA concentration on ATPase rate
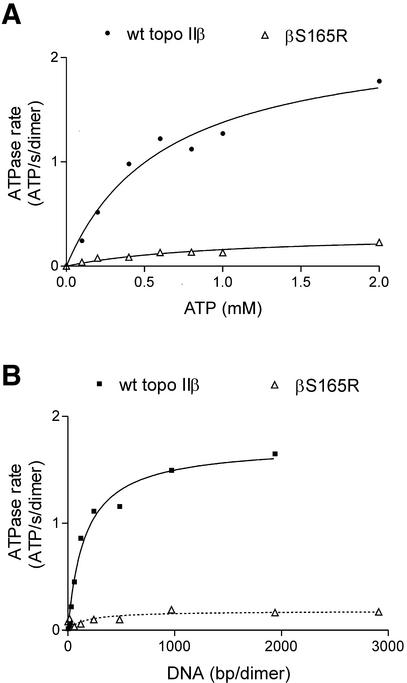
The effect on the wild-type topo IIβ ATPase activity of adding different concentrations of supercoiled plasmid is shown in Figure 1B (circles). Over 1000 bp of DNA per enzyme dimer were required for full stimulation of ATPase activity, after which the rate plateaus. The DNA concentration required to give half maximal stimulation of wild-type topo IIβ ATPase activity was 160 bp dimer–1 (16 µM). This DNA concentration most likely reflects the affinity of the enzyme for the plasmid DNA, rather than a requirement for a particular length of DNA. It is notable that unlike human topo IIα there is no peak of activity between 100 and 400 bp. This could be a function of different assay conditions or it could reflect a difference in the intrinsic properties of the β isoform, possibly related to their different biological roles.
Figure 1.
(A) Dependence of the topo IIβ and βS165R ATPase rates on ATP concentration. The rate of ATP hydrolysis is plotted against ATP concentration for 100 nM wt topo IIβ (circles) or βS165R (open triangles). DNA was present at 600 bp dimer–1 (60 µM). The curve drawn through the points is calculated from the Michaelis–Menten equation. Wild-type (wt) topo IIβ: kcat = 2.25 ± 0.5 s–1 dimer–1, Km = 0.63 ± 0.4 mM. S165R: kcat = 0.32 ± 0.14 s–1 dimer–1, Km = 1.0 ± 0.9 mM. (B) Dependence of topo IIβ and βS165R ATPase rates on DNA concentration. The rate of ATP hydrolysis is plotted against DNA concentration for 100 nM wt topo IIβ (circles) or βS165R (open triangles). ATP was present at 1 mM. A hyperbolic curve is fitted to each data set. wt topo IIβ: Vmax = 1.74 ± 0.2 s–1 dimer–1, Kd = 160 ± 65 bp dimer–1 (16 µM). For βS165R, Vmax = 0.18 ± 0.08 s–1 dimer–1, Kd = 163 bp dimer–1.
Determination of kcat and Km values for the ATPase reaction of DNA topo IIβ
The ATPase activity was assayed as a function of increasing ATP concentration and the data were fitted to the Michaelis–Menten equation, as illustrated by the best fit curve in Figure 1A (solid curve). Lindsley and co-workers have demonstrated that ATP hydrolysis by yeast topoisomerase II is not truly Michaelian, but follows a complex scheme of sequential hydrolysis of the two ATP molecules (9,13–18). At concentrations of ATP >350 µM, however, the Michaelis–Menten equation is a good approximation for the steady-state reaction (9,13). The kcat for topo IIβ at 30°C was 2.25 ± 0.5 s–1. The apparent Michaelis constant (Kmapp), was 0.63 ± 0.4 mM for wild-type DNA topo IIβ, which is comparable to the 0.56–0.78 mM reported for DNA topo IIα (10). The kcat is similar to the maximum rate of 2.17 s–1 dimer–1 observed for topo IIα (10). DNA gyrase and calf thymus topo II also have comparable turnover rates of 1 s–1 dimer–1 and 1.1 s–1 dimer–1, respectively. These are all lower than the kcat of the yeast enzyme, with rates of >7 s–1 dimer–1 (7–10).
Production of DNA topo IIβS165R, with a mutation in the ATPase domain
During production of human DNA topo IIβ expression constructs an XbaI cloning site was introduced, causing a serine to arginine change at S165R (21). This enzyme was unable to complement yeast strains bearing a temperature-sensitive mutation at the non-permissive temperature, whether grown on rich or minimal media, or in the presence or absence of glucose, galactose or raffinose. Figure 2 shows a qualitative assay with strain JN394top2-4 bearing plasmids encoding topo II from yeast, human topo IIα, human topo IIβ or human topo IIβS165R grown on rich medium containing glucose. Quantitative assays using equivalent numbers of cells indicate that topo IIβ complements as well as topo IIα and yeast topo II in this strain, whereas βS165R does not complement at all (data not shown). These results indicate that βS165R had not retained sufficient activity to fulfil its roles in vivo, unlike wild-type topo IIβ (20). We wished to determine the reason for this lack of complementation. Circular dichroism analysis showed that the mutation did not induce any gross structural changes in the topo IIβ (data not shown).
Figure 2.
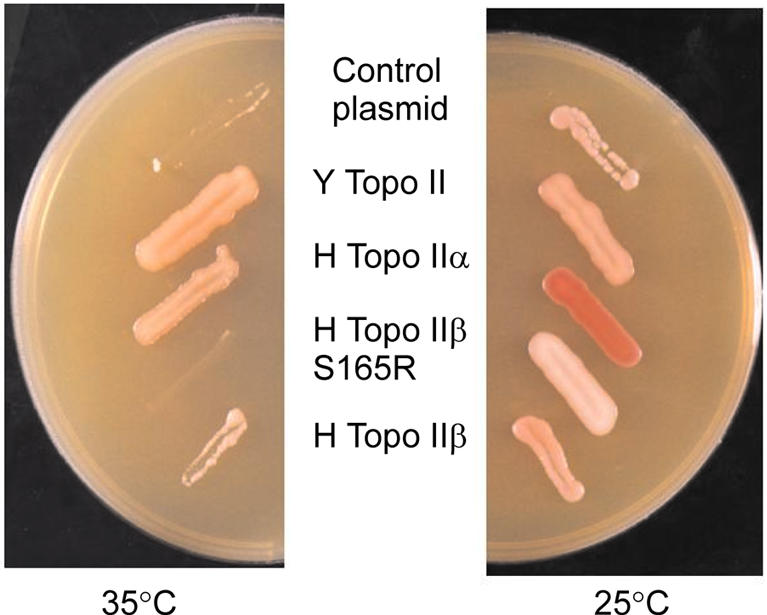
βS165R does not complement temperature-sensitive yeast topo II. In vivo complementation of yeast strain JN394top2-4. Yeast transformants expressing plasmid-borne type II topoisomerases were streaked onto YPDA plates and incubated at 25 or 35°C as indicated. A number of proteins were tested (plasmid names in parentheses); vector control (YCP50), yeast topo II (GIT2), human topo IIα (YEpWOB6), human topo IIβS165R (YEphTOP2βS165R) or human topo IIβ wt (YEphTOP2βKLM). Human topo IIβS165R was unable to complement JN394top2-4 at 35°C.
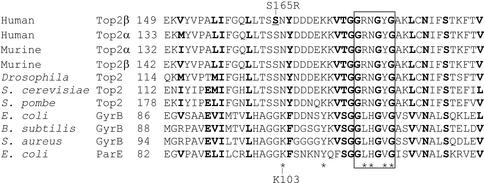
Ser165 lies in the ATPase domain, which is conserved from prokaryotes to eukaryotes. The N-terminal ATPase domain of human DNA topoisomerase IIβ (21) is 21% identical to the ATPase domain of Escherichia coli DNA gyrase B (25). Figure 3 shows an alignment of the region surrounding Ser165 (conserved residues in bold). Many of the key residues at the active site of gyrase B are conserved in eukaryotic topo II, including the catalytic residue Glu42 (26), residue Asn46 and the glycine-rich loop (Gly114, Gly117 and Gly119) in subdomain 1. This glycine-rich loop (GXXGXG) corresponds closely to the phosphate-binding loop (P-loop or Walker A box) present in many adenine and guanine nucleotide-binding proteins (reviewed in 27). It plays an important role in nucleotide binding, as shown by X-ray crystallography (28), and site-directed mutagenesis (9). Several other residues are also involved in nucleotide binding by gyrase B (29,30); these residues are marked with an asterisk in Figure 3, for example, Lys103, which forms a salt bridge with the β-phosphate of bound AMPPNP (28). However, as shown in the alignment in Figure 3, the nucleotide-binding residue Lys103 is not conserved between prokaryotic and eukaryotic enzymes, and neither are the residues immediately preceding it. Multiple sequence alignments indicate that in eukaryotic DNA topo II the amino acid residue that corresponds to Lys103 in gyrase B is an asparagine residue, Asn166 in human DNA topo IIβ. Many of the residues preceding Asn166 are conserved between eukaryotic type II topoisomerases, but not with the prokaryotic enzymes. Two conserved residues from subdomain 2 are also within hydrogen bonding distance of the bound ATP: Gln335 and Lys337. Site-directed mutagenesis has revealed that only Lys337 is critical for ATPase activity (31,32). In the present study we have investigated whether the inability of βS165R to complement yeast topo II is due to a deficiency in its ATPase activity.
Figure 3.
Alignment of the amino acid sequences surrounding Ser165 of topo IIβ. Conserved residues are indicated in bold. Asterisks indicate residues in E.coli gyr B which make hydrogen bonds to AMPPNP: Lys103, Tyr109, Leu115, His116, Val118 and Gly119. The box includes residues of the highly conserved glycine loop, Gly114, Gly117 and Gly119.
ATPase activity of human DNA topo IIβ S165R
In the presence of 1 mM ATP the rate of DNA-stimulated ATP hydrolysis by βS165R was found to be directly dependent on the protein concentration (data not shown). The rate of ATP hydrolysis was 0.072 s–1 dimer–1, 15-fold lower than the 1.1 s–1 of the wild-type enzyme. ATP hydrolysis by βS165R in the absence of DNA was too low to measure reproducibly.
Effect of DNA concentration on ATPase rate
ATP hydrolysis was assayed over a range of DNA concentrations to determine whether the βS165R mutant had an altered response to DNA. These results are shown in Figure 1B (open triangles). Over 1000 bp of DNA per enzyme dimer were required for full stimulation of ATPase activity of βS165R, the same as seen for wild-type topo IIβ. The DNA concentration to achieve half maximal stimulation was not significantly different for wild-type topo IIβ (160 bp dimer–1) and βS165R (163 bp dimer–1). This suggests that the reduced ATPase activity of βS165R does not arise from a lower affinity for DNA than the wild-type enzyme.
The S165R mutation reduces the kcat for ATP hydrolysis but does not significantly affect the Kmapp
To test if the reduced rate of ATP hydrolysis by βS165R arose from an altered affinity for ATP, DNA-stimulated ATP hydrolysis was assayed over a range of ATP concentrations. The data can be fitted to the Michaelis–Menten equation, as illustrated by the best fit curve in Figure 1A (open triangles). The kcat for βS165R was 0.32 ± 0.14 s–1, 7-fold lower than the kcat for wild-type topo IIβ (2.25 ± 0.5 s–1). These data indicate that the S165R mutation has reduced the rate of ATP hydrolysis to 14% of that of the wild-type topo IIβ. The two proteins had similar values for the apparent Michaelis constant (Kmapp), at 0.63 ± 0.4 and 1.0 ± 0.9 mM for wild-type topo IIβ and βS165R, respectively. The specificity constant for the interaction between ATP and topo IIβ (kcat/Kmapp) is thought to be dependent solely on rate constants involved in ATP binding and has no dependence on the rate constants for ATP hydrolysis and product release (13). kcat/Kmapp was 3.57 × 103 M–1 s–1 for wild-type topo IIβ and 0.32 × 103 M–1 s–1 for βS165R, a reduction of 90%. This suggests that some aspect of ATP binding has been altered in the βS165R protein.
The S165R mutation does not affect DNA binding or ATP-independent cleavage
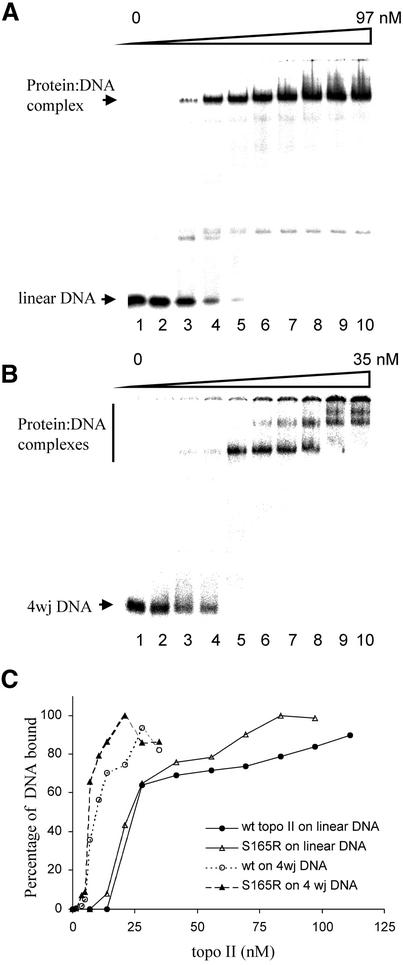
The data in Figure 1B suggested that the affinity for DNA was not altered by the S165R mutation. To confirm this, gel retardation assays (23) were used to investigate whether the S165R mutation had altered the binding of topo IIβ to DNA. Titration curves showed that βS165R was as effective as wild-type topo IIβ in binding to both a linear 40 bp duplex containing a single topo II cleavage site (Fig. 4A and C) and to a synthetic four-way junction of related sequence (Fig. 4B and C) (23). Furthermore, the complexes of wild-type topo IIβ and βS165R with DNA showed very similar stabilities in response to increasing salt concentration (data not shown). These results indicate that the ability of topo IIβ to bind either linear or four-way junction DNA was not altered by the S165R mutation.
Figure 4.
Wild-type topo IIβ and βS165R have similar DNA binding affinities. A gel retardation assay was employed to study DNA binding. (A) Linear DNA probe was incubated with 0, 6.9, 13.9, 20.8, 27.8, 41.7, 55.6, 69.4, 83.3 and 97.2 nM βS165R in lanes 1–10, respectively. (B) Four-way junction substrate was incubated with 0, 1.4, 3.5, 4.9, 6.9, 10.4, 13.9, 20.8, 27.8 and 34.7 nM βS165R in lanes 1–10, respectively. (C) Quantification of DNA binding. Filled circles, wt topo IIβ on linear DNA; open triangles, βS165R on linear DNA; open circles, wt topo IIβ on four-way junction DNA; filled triangles, βS165R on four-way junction DNA. The protein–DNA complexes were quantified and are plotted as a percentage of the input DNA against protein concentration.
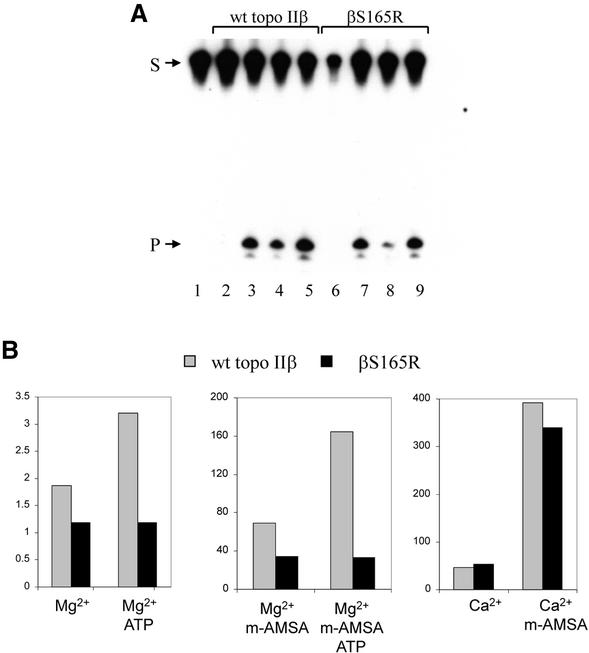
The linear substrate used in the gel retardation assay contains a single topo IIβ cleavage site and cleavage at this site is stimulated by the topo II poison m-AMSA. Cleavage by wild-type topo II and βS165R was assayed in the absence of ATP, and in the presence of Mg2+ (Fig. 5A, lanes 2 and 6), Mg2+ with m-AMSA (Fig. 5A, lanes 3 and 7), Ca2+ (Fig. 5A, lanes 4 and 8) or Ca2+ with m-AMSA (Fig. 5A, lanes 5 and 9; see also Fig. 5B). Magnesium ions support both DNA cleavage and ATP hydrolysis by topo II, whereas calcium ions can only support DNA cleavage. Under all four conditions βS165R (Fig. 5A, lanes 6–9) had similar DNA cleavage activity to wild-type topo IIβ (Fig. 5A, lanes 2–5). The ability of βS165R to carry out ATP-independent site-specific cleavage both in the presence and absence of m-AMSA was therefore unchanged with respect to the wild-type enzyme. The addition of ATP increased Mg2+-dependent cleavage by wild-type topo IIβ by 2-fold, both with and without m-AMSA. However, cleavage by βS165R was not affected by ATP (Fig. 5B).
Figure 5.
DNA cleavage by wt topo IIβ and βS165R. (A) Cleavage reactions contained linear DNA probe and 56 nM each protein: lanes 2–5, wt topo IIβ; lanes 6–9, βS165R. Cleavage was assayed in the presence of: 10 mM Mg2+ (lanes 1, 2 and 6), 10 mM Mg2+ and m-AMSA (lanes 3 and 7), 10 mM Ca2+ (lanes 4 and 8) or 10 mM Ca2+ and m-AMSA (lanes 5 and 9). The uncleaved 40 bp substrate is indicated by S, and the 21 bp labelled cleavage product is indicated by P. Labelling of both DNA strands revealed that wt topo IIβ and βS165R cleaved the two strands equally (data not shown). (B) Histograms comparing cleavage by wt topo IIβ (grey) and βS165R (black) in the presence of 10 mM Mg2+ with and without ATP (left), 10 mM Mg2+ and m-AMSA with and without ATP (middle) and 10 mM Ca2+ or 10 mM Ca2+ and m-AMSA (right). Results are expressed relative to background and are derived from a different experiment to that shown in (A).
The strand passage activity of the βS165R mutant is reduced by >80%
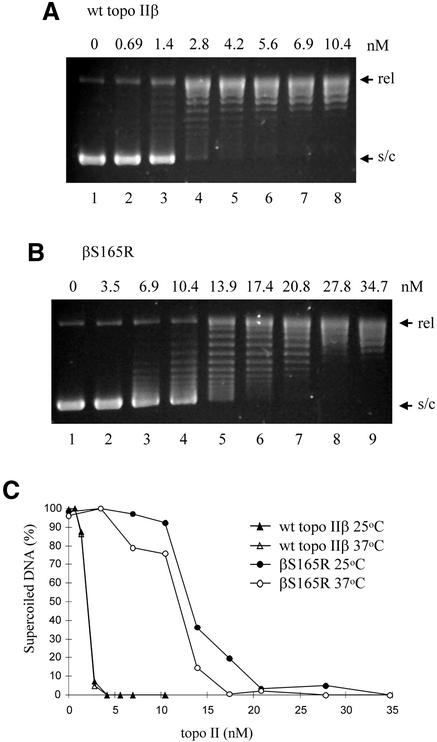
The strand passage activities of wild-type topo IIβ and βS165R were assayed in order to determine whether the 7-fold reduction in the ATP hydrolysis activity of βS165R had affected the overall rate of catalysis. Figure 6 shows relaxation assays performed with increasing concentrations of wild-type topo IIβ (Fig. 6A) or βS165R (Fig. 6B). Wild-type topoIIβ is more processive than βS165R in relaxation assays. The protein concentration required to relax 50% of the supercoiled plasmid DNA was calculated to be 1.9 nM for wild-type topo IIβ and 11.7 nM for βS165R, and thus the mutant protein has 16% of the wild-type relaxation activity. Both wild-type topo IIβ and βS165R had the same or greater activity at 37 compared with 25°C, indicating that the activity of βS165R is not temperature sensitive (Fig. 6C).
Figure 6.
βS165R has reduced relaxation activity compared to wt topo IIβ. Increasing concentrations of wt topo IIβ (A) or βS165R (B) were incubated with 1 µg supercoiled plasmid DNA (s/c) in the presence of ATP for 30 min at 37°C. Relaxed plasmid is indicated (rel). (A) Lanes 1–8 contained 0, 0.69, 1.4, 2.8, 4.2, 5.6, 6.9 and 10.4 nM protein, respectively. (B) Lanes 1–9 contained 0, 3.5, 6.9, 10.4, 13.9, 17.4, 20.8, 27.8 and 34.7 nM protein, respectively. (C) The relaxation activity of wt topo IIβ (triangles) and βS165R (circles) was assayed at both 25 (filled symbols) and 37°C (open symbols). The percentage of supercoiled DNA remaining is plotted against protein concentration.
Decatenation assays with increasing concentrations of protein also showed that βS165R had only 20% of wild-type strand passage activity (Fig. 7A and B). Fifty per cent of the kDNA was decatenated by 0.97 nM topo IIβ whereas 4.7 nM βS165R was required for the same level of activity. The dependence of decatenation on Mg2+ concentration was also examined, and both wild-type topo IIβ and βS165R had near maximal activity between 5 and 30 mM Mg2+ (33; data not shown).
Figure 7.
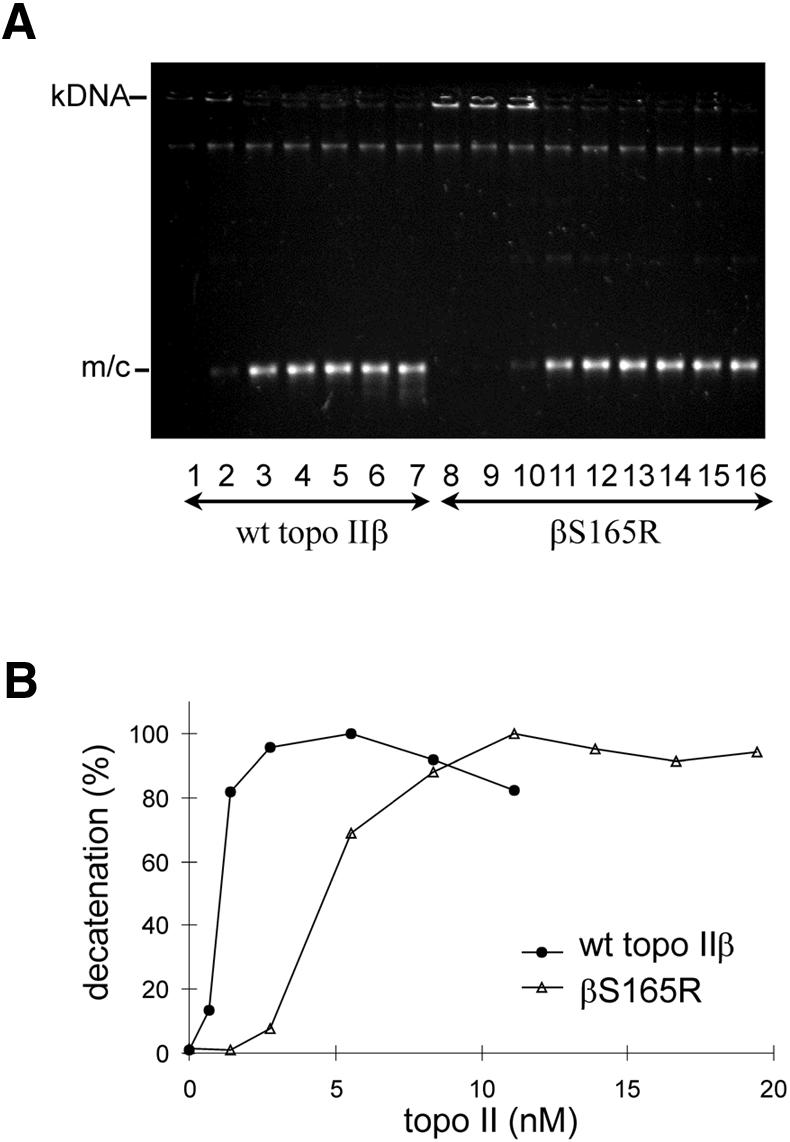
βS165R has reduced decatenation ability compared to wt topo IIβ. Topo II was incubated with 400 ng kinetoplast DNA (kDNA) in the presence of ATP at 37°C for 30 min. (A) Minicircles released by topo II activity are indicated (m/c). Lanes 1–7: 0, 0.7, 1.4, 2.8, 5.6, 8.3 and 11.1 nM wt topo IIβ; lanes 8–16: 0, 1.4, 2.8, 5.6, 8.3, 11.1, 13.9, 16.7 and 19.4 nM βS165R. These data are plotted as percentage decatenation against concentration of wt topo IIβ (filled circles) and βS165R (open triangles) in (B).
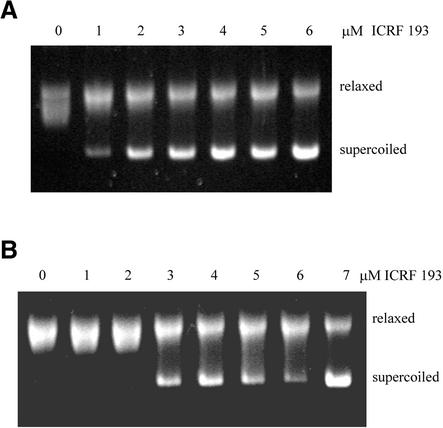
Relaxation assays were also carried out in the presence of the topo II inhibitor ICRF-193, which stabilises the closed clamp form of the enzyme (34). Increasing concentrations of ICRF-193 were added to relaxation assays containing wild-type topo IIβ (Fig. 8A) or βS165R (Fig. 8B). βS165R was less sensitive to inhibition by ICRF-193. The concentration of ICRF-193 required for 50% inhibition of activity (IC50) was 1.9 µM for wild-type topo IIβ and 3.5 µM for βS165R.
Figure 8.
Inhibition of relaxation activity by ICRF-193. Relaxation reactions were performed with (A) wt topo IIβ in the presence of 0, 1, 2, 3, 4, 5 or 6 µM ICRF-193 as indicated or (B) βS165R in the presence of 0, 1, 2, 3, 4, 5, 6 or 7 µM ICRF-193 as indicated.
Modelling of the ATPase domains of wild-type topo IIβ and βS165R
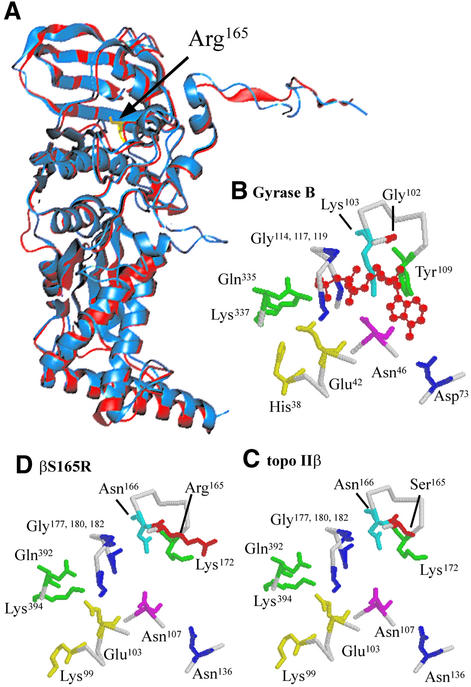
The ATPase domain of DNA topo IIβ is only 21% identical to that of E.coli DNA gyrase B, and although the X-ray crystal structure of the gyrase B ATPase domain has been determined, there is no structural information for this domain of the eukaryotic type II enzymes. To investigate whether the ATPase domains of eukaryotic and prokaryotic type II topoisomerases could be structurally similar, the ATPase domain of wild-type topo IIβ was modelled onto the ATPase domain of DNA gyrase B (28). Superimposition of the modelled topo IIβ onto gyrase B (Fig. 9A) reveals that the structures are very similar, although small differences in the shapes of many of the intervening loops can be observed.
Figure 9.
Modelling of the ATPase domain of DNA topo IIβ onto the X-ray crystal structure of E.coli DNA gyrase B ATPase domain. (A) Superimposition of the modelled ATPase domain of topo IIβS165R (blue) (amino acids 35–472) onto that of gyrase B (red) (amino acids 1-391). (B–D) Arrangement of the key residues round the active sites of (B) gyrase B, (C) the modelled structure of wt topo IIβ and (D) the modelled structure of βS165R. The bound AMPPNP is shown in red in (B).
The arrangement of key residues around the active site of gyrase B is maintained in the modelled structure of wild-type topo IIβ, as shown in Figure 9B and C. The nucleotide-binding residues Asn46 (Asn107 in topo IIβ), Asp73 (Asn136 in topo IIβ) and the glycine loop are conserved between gyrase B and topo IIβ, and the positions of these residues within the active sites are almost identical. Gln335 (Gln392) and Lys337 (Lys394) from subdomain 2 also have very similar positions in gyrase B and topo IIβ. This is consistent with previous observations that the residues equivalent to Gly117 of the glycine-rich loop and Gln335 and Lys337 of subdomain 2 play very similar roles in both topo II and DNA gyrase (9,32).
The nucleotide-binding residues Lys103 and Tyr109 correspond to Asn166 and Lys172 in topo IIβ, and the locations of these residues are very similar, although the exact positions of the different side chains are a little different. The catalytic residue Glu42 (Glu103 in topo IIβ) has the same position in gyrase B and topo IIβ. The residue corresponding to His38, which is important for aligning and polarising Glu42, is Lys99 in topo IIβ. However, the side chain of Lys99 is not directed towards Glu103 in the topo IIβ model, and is thus not suitable for aligning the active glutamate residue. Lys99 lies just after a loop of 21 residues which has no counterpart in gyrase B. Modelling the shape and orientation of this loop is less accurate than modelling regions where topo IIβ and gyrase B align well, and it is possible that assigning an incorrect structure to the loop could result in a misalignment of the residues adjacent to it, including Lys99. Lys99 is conserved in all eukaryotic type II topoisomerases, and there do not appear to be any other nearby residues which could perform this function instead.
The modelling was repeated using the βS165R sequence to investigate whether the mutation could have introduced significant structural alterations within the ATPase domain. The resulting structure was virtually identical to that of wild-type topo IIβ. This suggests that the reduction in ATPase activity observed for this mutant is unlikely to arise from gross structural changes. The residues at the active site of βS165R are predicted to have identical positioning to those in topo IIβ (compare Fig. 9C and B). The substituted arginine residue does not affect the positioning of any other residues and does not appear to protrude into the predicted AMPPNP binding site, although it would be in close proximity to the bound nucleotide.
DISCUSSION
This paper reports for the first time the kinetics of ATP hydrolysis by human topo IIβ. The kcat for topo IIβ at 30°C was 2.25 s–1, which is comparable to the 2.17 s–1 dimer–1 previously reported for human topo IIα at optimal DNA concentrations at 37°C (10). DNA gyrase and calf thymus topo II have comparable rates of 1 s–1 and 1.1 s–1, respectively (7,8), and yeast and Drosophila topo II have the highest rates of ATP hydrolysis, at 7 s–1 and 5–13 s–1 (9,35–37).
The rate of ATP hydrolysis was stimulated 44-fold by supercoiled plasmid DNA, which is greater than the 10-fold stimulation seen for topo IIα. Type II topoisomerases from other species also show DNA stimulation of between 4- and 20-fold. For example, yeast topo II and DNA gyrase both show 20-fold stimulation of ATP hydrolysis by DNA, whilst calf thymus topo II shows only a 4-fold stimulation by DNA. However, the calf thymus enzyme studied was not full length, but a mixture of 140 and 125 kDa proteolytic fragments which may not have exactly the same characteristics as the full-length enzyme (7–10).
It is notable that as the DNA concentration is increased, topo IIβ ATPase activity does not spike at 100–400 bp dimer–1, unlike topo IIα (10,38). This hyperstimulation phenomenon has been interpreted by Campbell and Maxwell (38) as arising from protein–protein interactions between topo II dimers that are bound adjacent to each other on one piece of DNA. The difference in the DNA hyperstimulation between human DNA topo IIα and DNA topo IIβ could result from a difference in their abilities to interact with adjacent dimers that reflect isoform-specific cellular functions. For example, topo IIα has been shown to associate with sites of DNA replication, whereas topo IIβ does not (39,40). Alternatively, the presence of a spike in ATPase activity at particular DNA concentrations may have some dependence on reaction conditions. For example, with topo IIα Hammonds and Maxwell (10) report that the rate of ATPase activity in the spike is very sensitive to the monovalent cation concentration. Furthermore, they report unpublished data of similar hyperstimulation for yeast topo II, yet Lindsley’s group have never reported this phenomenon in their extensive studies on the same enzyme. The cation concentration used in this study was 50 mM, while those used by Maxwell’s group were 125 (10) and 10 mM (38). Olland and Wang (41) demonstrated a spike in ATPase activity for an N-terminal fragment of yeast topo II at 50 mM KCl but not at 100 mM KCl. They postulate that the monomer–dimer equilibrium of this fragment may have influenced these results. Olland and Wang also suggest that the hyperstimulation previously observed for full-length topo IIα may arise from a lower rate of ATPase activity on certain weak DNA-binding sites. In conclusion, it is not clear whether the absence of DNA hyperstimulation for topo IIβ reflects a true difference from the α isoform or simply a difference in reaction conditions, and this determination will await testing of the two isoforms under identical laboratory conditions.
DNA binding and DNA cleavage in the absence of ATP appear to be unaffected by the S165R mutation. ATP stimulates DNA cleavage by wild-type topo IIβ by ∼2-fold; however, no ATP stimulation of cleavage was seen with βS165R. Previous studies have shown that etoposide, another topoisomerase II poison, inhibits DNA religation after hydrolysis of the first ATP molecule and the release of its products (18,42). Hydrolysis of the second ATP molecule is inhibited by etoposide. Our data are consistent with this scheme, with the reduced rate of ATP binding and hydrolysis in S165R resulting in an absence of ATP-stimulated cleavage.
The reduction in ATPase activity is reflected in a 5-fold reduction in strand passage activity in both relaxation and decatenation assays. Consequently, βS165R is unable to complement the functions of ts yeast topo II at the non-permissive temperature in vivo. These results show that efficient ATP hydrolysis is crucial to topo II function in vivo. The relaxation assays also indicate that βS165R is more processive than wild-type topo IIβ. As the S165R mutation does not affect DNA binding, the more distributive relaxation pattern of βS165R is probably due to its reduced strand passage activity, allowing more time to dissociate and reassociate with the substrate DNA between reaction cycles.
In the kinetic scheme proposed by Lindsley and co-workers for ATP hydrolysis by topo II, kcat/Kmapp includes only terms involved in ATP binding. While we find that Kmapp is unchanged by the S165R mutation, kcat/Kmapp is reduced by 10-fold, suggesting that some aspect of ATP binding has been altered. To investigate how the mutation might affect the ATP binding pocket, we used computer modelling to predict the structure of the topo IIβ ATPase domain, based on the DNA gyrase B structure (28). The arrangement of key residues at the active site is predicted to be very similar to that in DNA gyrase B. The residue adjacent to Ser165, Asn166, aligns with the gyrase B residue Lys103, which forms a salt bridge with the β-phosphate of the bound ATP (28). In topo IIβ, Asn166 appears to be in a suitable position to form a hydrogen bond with ATP. The adjacent Ser165 may also interact with the bound ATP, or it could help to position Asn166 correctly. Replacement of serine with arginine could thus remove contacts that are important for ATP binding. The positively charged side chain of the introduced arginine residue could also influence the binding and positioning of ATP within the binding pocket, as it is predicted to lie close to the ribose and adenine rings.
The S165R mutation caused a 7-fold reduction in kcat, which reflects the rate of ATP hydrolysis and product release. In the presence of saturating ATP concentrations, release of the ADP and Pi products is rate determining. The proximity of the Arg165 side chain to the ribose and adenine rings of the ATP leads us to speculate that the reduction in kcat may be due to a reduction in the dissociation rate of ADP, rather than a reduction in the rate of substrate to product conversion. However, more detailed kinetic analyses are necessary to test this hypothesis.
In summary, the S165R mutation is the first reported mutation in the ATPase domain of human DNA topo IIβ. Our data show that this mutation affects the ATPase activity of the enzyme, reducing the kcat for ATP hydrolysis by 7-fold, and reducing the kcat/Kmapp by 10-fold. DNA binding and DNA cleavage in the absence of ATP appear to be unaffected.
The reduction in ATPase activity is reflected in a 5-fold reduction in relaxation activity. Consequently, βS165R is unable to complement the functions of ts yeast topo II at the non-permissive temperature in vivo. These results show that efficient ATP hydrolysis is crucial to topo II function in vivo and also indicate that ATP hydrolysis is not required for DNA binding or cleavage.
Acknowledgments
ACKNOWLEDGEMENTS
The coordinates for the X-ray crystal structure of the E.coli DNA gyrase B fragment were kindly provided by D. B. Wigley. We thank Dr R. Virden for helpful discussions. This work was supported by grants from the BBSRC, NECCRF and CRC.
REFERENCES
- 1.DiNardo S., Voelkel,K. and Sternglanz,R. (1984) DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc. Natl Acad. Sci. USA, 81, 2616–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holm C., Goto,T., Wang,J.C. and Botstein,D. (1985) DNA topoisomerase II is required at the time of mitosis in yeast. Cell, 41, 553–563. [DOI] [PubMed] [Google Scholar]
- 3.Uemura T., Ohkura,H., Adachi,Y., Morino,K., Shiozaki,K. and Yanagida,M. (1987) DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell, 50, 917–925. [DOI] [PubMed] [Google Scholar]
- 4.Wang J.C. (1996) DNA topoisomerases. Annu. Rev. Biochem., 65, 635–692. [DOI] [PubMed] [Google Scholar]
- 5.Burden D.A. and Osheroff,N. (1998) Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim. Biophys. Acta, 1400, 139–154. [DOI] [PubMed] [Google Scholar]
- 6.Chen A.Y. and Liu,L.F. (1994) DNA topoisomerases: essential enzymes and lethal targets. Annu. Rev. Pharmacol. Toxicol., 34, 191–218. [DOI] [PubMed] [Google Scholar]
- 7.Maxwell A. and Gellert,M. (1984) The DNA dependence of the ATPase activity of DNA gyrase. J. Biol. Chem., 259, 14472–14480. [PubMed] [Google Scholar]
- 8.Halligan B.D., Edwards,K.A. and Liu,L.F. (1985) Purification and characterization of a type II DNA topoisomerase from bovine calf thymus. J. Biol. Chem., 260, 2475–2482. [PubMed] [Google Scholar]
- 9.Lindsley J.E. and Wang,J.C. (1993) On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. J. Biol. Chem., 268, 8096–8104. [PubMed] [Google Scholar]
- 10.Hammonds T.R. and Maxwell,A. (1997) The DNA dependence of the ATPase activity of human DNA topoisomerase II alpha. J. Biol. Chem., 272, 32696–32703. [DOI] [PubMed] [Google Scholar]
- 11.Roca J. and Wang,J.C. (1992) The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell, 71, 833–840. [DOI] [PubMed] [Google Scholar]
- 12.Roca J. and Wang,J.C. (1994) DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism Cell, 77, 609–616. [DOI] [PubMed] [Google Scholar]
- 13.Harkins T.T., Lewis,T.J. and Lindsley,J.E. (1998) Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 2. Kinetic mechanism for the sequential hydrolysis of two ATP. Biochemistry, 37, 7299–7312. [DOI] [PubMed] [Google Scholar]
- 14.Harkins T.T. and Lindsley,J.E. (1998) Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 1. A DNA-dependent burst in ATP hydrolysis. Biochemistry, 37, 7292–7298. [DOI] [PubMed] [Google Scholar]
- 15.Morris S.K., Harkins,T.T., Tennyson,R.B. and Lindsley,J.E. (1999) Kinetic and thermodynamic analysis of mutant type II DNA topoisomerases that cannot covalently cleave DNA. J. Biol. Chem., 274, 3446–3452. [DOI] [PubMed] [Google Scholar]
- 16.Lindsley J.E. (1996) Intradimerically tethered DNA topoisomerase II is catalytically active in DNA transport. Proc. Natl Acad. Sci. USA, 93, 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baird C.L., Harkins,T.T., Morris,S.K. and Lindsley,J.E. (1999) Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl Acad. Sci. USA, 96, 13685–13690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baird C.L., Gordon,M.S., Andrenyak,D.M., Marecek,J.F. and Lindsley,J.E. (2001) The ATPase reaction cycle of yeast DNA topoisomerase II. Slow rates of ATP resynthesis and P(i) release. J. Biol. Chem., 276, 27893–27898. [DOI] [PubMed] [Google Scholar]
- 19.Austin C.A. and Marsh,K.L. (1998) Eukaryotic DNA topoisomerase IIβ. Bioessays, 20, 215–226. [DOI] [PubMed] [Google Scholar]
- 20.Meczes E.L., Marsh,K.L., Fisher,L.M., Rogers,M.P. and Austin,C.A. (1997) Complementation of temperature-sensitive topoisomerase II mutations in Saccharomyces cerevisiae by a human TOP2beta construct allows the study of topoisomerase IIbeta inhibitors in yeast. Cancer Chemother. Pharmacol., 39, 367–375. [DOI] [PubMed] [Google Scholar]
- 21.Austin C.A., Marsh,K.L., Wasserman,R.A., Willmore,E., Sayer,P.J., Wang,J.C. and Fisher,L.M. (1995) Expression, domain structure and enzymatic properties of an active recombinant human DNA topoisomerase IIβ. J. Biol. Chem., 270, 15739–15746. [DOI] [PubMed] [Google Scholar]
- 22.Nitiss J. and Wang,J.C. (1988) DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc. Natl Acad. Sci. USA, 85, 7501–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West K.L. and Austin,C.A. (1999) Human DNA topoisomerase II beta binds and cleaves four-way junction DNA in vitro. Nucleic Acids Res., 27, 984–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sali A. and Blundell,T.L. (1993) Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 25.Caron P.R. and Wang,J.C. (1993) DNA topoisomerase as targets of therapeutics: a structural overview. In Andoh,T., Ikeda,H. and Oguro,M. (eds), Molecular Biology of DNA Topoisomerases. CRC Press, Boca Raton, FL, pp. 243–263.
- 26.Jackson A.P. and Maxwell,A. (1993) Identifying the catalytic residue of the ATPase reaction of DNA gyrase. Proc. Natl Acad. Sci. USA, 90, 11232–11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saraste M., Sibbald,P.R. and Wittinghofer,A. (1990) The P-loop— a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci., 15, 430–434. [DOI] [PubMed] [Google Scholar]
- 28.Wigley D.B., Davies,G.J., Dodson,E.J., Maxwell,A. and Dodson,G. (1991) Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature, 351, 624–629. [DOI] [PubMed] [Google Scholar]
- 29.Tamura J.K. and Gellert,M. (1990) Characterization of the ATP binding site on Escherichia coli DNA gyrase. Affinity labelling of Lys-103 and Lys-110 of the B subunit by pyridoxal 5′-diphospho-5′-adenosine. J. Biol. Chem., 265, 21342–21349. [PubMed] [Google Scholar]
- 30.O’Dea M.H., Tamura,J.K. and Gellert,M. (1996) Mutations in the B subunit of Escherichia coli DNA gyrase that affect ATP-dependent reactions. J. Biol. Chem., 271, 9723–9729. [DOI] [PubMed] [Google Scholar]
- 31.Smith C.V. and Maxwell,A. (1998) Identification of a residue involved in transition-state stabilization in the ATPase reaction of DNA gyrase. Biochemistry, 37, 9658–9667. [DOI] [PubMed] [Google Scholar]
- 32.Hu T., Chang,S. and Hsieh,T. (1998) Identifying Lys(359) as a critical residue for the ATP dependent reactions of Drosophila DNA topoisomerase II. J. Biol. Chem., 273, 9586–9592. [DOI] [PubMed] [Google Scholar]
- 33.West K.L., Meczes,E.L., Thorn,R., Turnbull,R.M., Marshall,R. and Austin,C.A. (2000) Mutagenesis of E477 or K505 in the B′ domain of human topoisomerase IIβ increases the requirement for magnesium ions during strand passage. Biochemistry, 39, 1223–1233. [DOI] [PubMed] [Google Scholar]
- 34.Roca J., Ishida,R., Berger,J.M., Andoh,T. and Wang,J.C. (1994) Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc. Natl Acad. Sci. USA, 91, 1781–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corbett A.H., DeVore,R.F. and Osheroff,N. (1992) Effect of casein kinase II-mediated phosphorylation on the catalytic cycle of topoisomerase II. Regulation of enzyme activity by enhancement of ATP hydrolysis. J. Biol. Chem., 267, 20513–20518. [PubMed] [Google Scholar]
- 36.Corbett A.H., Fernald,A.W. and Osheroff,N. (1993) Protein kinase C modulates the catalytic activity of topoisomerase II by enhancing the rate of ATP hydrolysis: evidence for a common mechanism of regulation by phosphorylation. Biochemistry, 32, 2090–2107. [DOI] [PubMed] [Google Scholar]
- 37.Hu T., Sage,H. and Hsieh,T.S. (2002) ATPase domain of eukaryotic DNA topoisomerase II: inhibition of ATPase activity by the anticancer drug bisdioxopiperazine and ATP/ADP-induced dimerization. J. Biol. Chem., 277, 5944–5951. [DOI] [PubMed] [Google Scholar]
- 38.Campbell S. and Maxwell,A. (2002) The ATP-operated clamp of human DNA topoisomerase IIα: hyperstimulation of ATPase by “Piggy-back” binding. J. Mol. Biol., 320, 171–188. [DOI] [PubMed] [Google Scholar]
- 39.Niimi A., Suka,N., Harata,M., Kikuchi,A. and Mizuno,S. (2001) Co-localization of chicken DNA topoisomerase IIalpha, but not beta, with sites of DNA replication and possible involvement of a C-terminal region of alpha through its binding to PCNA. Chromosoma, 110, 102–114. [DOI] [PubMed] [Google Scholar]
- 40.Christensen M.O., Larsen,M.K., Barthelmes,H.U., Hock,R., Andersen,C.L., Kjeldsen,E., Knudsen,B.R., Westergaard,O., Boege,F. and Mielke,C. (2002) Dynamics of human DNA topoisomerases IIalpha and IIbeta in living cells. J. Cell Biol., 157, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olland S. and Wang,J.C. (1999) Catalysis of ATP hydrolysis by two NH(2)-terminal fragments of yeast DNA topoisomerase II. J. Biol. Chem., 274, 21688–21694. [DOI] [PubMed] [Google Scholar]
- 42.Morris S.K. and Lindsley,J.E. (1999)Yeast topoisomerase II is inhibited by etoposide after hydrolyzing the first ATP and before releasing the second ADP. J. Biol. Chem., 274, 30690–30696. [DOI] [PubMed] [Google Scholar]