Abstract
Alterations in the activity of the centrosomal kinase, Aurora-A/STK15, have been implicated in centrosome amplification, genome instability and cellular transformation. How STK15 participates in all of these processes remains largely mysterious. The activity of STK15 is regulated by phosphorylation and ubiquitin-mediated degradation, and physically interacts with protein phosphatase 1 (PP1) and CDC20. However, the precise roles of these modifications and interactions have yet to be fully appreciated. Here we show that STK15 associates with a putative tumor and metastasis suppressor, NM23-H1. STK15 and NM23 were initially found to interact in yeast in a two-hybrid assay. Association of these proteins in human cells was confirmed by co-immunoprecipitation from cell lysates and biochemical fractionation indicating that STK15 and NM23-H1 are present in a stable, physical complex. Notably, SKT15 and NM23 both localize to centrosomes throughout the cell cycle irrespective of the integrity of the microtubule network in normal human fibroblasts.
INTRODUCTION
Correct partitioning of the genome during mitosis depends upon the tightly regulated function of the mitotic spindle, which is composed of centrosomes, microtubules, molecular motors, chromosomes and kinetochores. The centrosome is the major microtubule-organizing center in mammalian cells and the counterpart to the spindle pole body of the yeast Saccharomyces cerevisiae. The centrosome in mammalian cells is composed of two perpendicularly positioned centrioles and the surrounding amorphous pericentriolar material. γ-Tubulin and pericentrins are constitutive components of the centrosome, while other proteins, such as p53, pRB, BRCA1, BRCA2 and CDK2, accumulate at centrosomes in a cell cycle-dependent manner (1–4).
The centrosome normally duplicates once per cell cycle. This process is initiated during G1 after cells pass the restriction point and is completed during G2. Centrosomes separate at G2/M, migrating to opposite poles of the cell to establish the microtubule network that is required to separate condensed chromosomes during M phase. Centrosomes play a vital role in establishing spindle bipolarity, in assembling spindle microtubules and in determining the plane of cytokinesis (for reviews see 5–10). More recently, accumulating evidence suggests a more direct role than had previously been appreciated for the centrosome in cytokinesis and cell cycle progression during the following G1 and S phases (11–13).
Centrosome abnormalities, including morphological alterations, supernumerary centrosomes and acentriolar centrosomes, have been demonstrated in most human cancer cells, including those derived from breast, prostate, lung, colon and brain (for reviews see 14,15; see also 2,16–18). Several oncogenes, tumor suppressor genes, cell division cycle and mitotic checkpoint genes are required for or involved in centrosome duplication, such as Ras, BRCA1 and BRCA2, CDK2, cyclin A and ATR (3,19–23). Like other cellular processes, protein kinases play critical roles in the centrosome duplication process. Among the centrosome-associated kinases, Aurora-A/AIK1/BTAK/STK15 kinase has been identified as a candidate oncogene with connections to the centrosome cycle (24–27). Aurora-A/STK15 is overexpressed at both the mRNA and protein levels in a number of cancer cell lines, including breast, ovarian and prostate (25,27), and also in breast cancer tissues (28–30). Its kinase activity peaks at the G2/M phase of the cell cycle (25). A mutant, kinase-inactive STK15 (Stk15 K162M) abolishes the oncogenic activity of STK15 in Rat1 fibroblasts, while a mutation conferring constitutive activation (Stk15 T288D) increases the kinase activity and enhances transforming potential (25). These results strongly suggest that the kinase activity of STK15 is essential for STK15 function in vivo. STK15 is regulated by phosphorylation and ubiquitin-mediated degradation and interacts with CDC20 and protein phosphatase 1 (PP1) (31–35).
As a putative tumor suppressor, the nm23 gene was discovered on the basis of its reduced expression in highly metastatic cell lines (36). Several studies have shown that NM23 overexpression can reduce the metastatic potential of melanoma and breast carcinoma cells in vivo (37–39). Furthermore, nm23 expression, at both the protein and mRNA levels, inversely correlates with high metastatic potential in numerous human cancers, including breast, gastric, cervical and ovarian carcinoma and melanoma (for a review see 40).
The nm23 genes encode a protein family with eight subfamilies in human (41). The well characterized biochemical activities of these proteins include NDP kinase (42), protein histidine kinase, histidine-dependent protein phosphotransferase (43–45) and serine autophosphorylation (46,47). Data suggest that it is the level of autophosphorylation that correlates with tumor suppression by NM23 in melanoma cells (46). NM23 associates with the cytoskeleton through an interaction with β-tubulin (48). Awd, the fly homolog of NM23, co-localizes with microtubules in Drosophila cells (49) and more recent data have indicated that NM23 may also be a component of the centrosome (50).
Despite the strong evidence that STK15/Aurora-A/BTAK regulates centrosome duplication, cellular transformation and aneuploidy, little evidence directly connects this kinase with known oncogenes or tumor suppressors. Here we provide the data that STK15 interacts with the tumor suppressor NM23, both in yeast and in normal human fibroblasts. NM23 and STK15 co-fractionate in a high molecular weight complex and co-localize at centrosomes throughout the cell cycle.
MATERIALS AND METHODS
Plasmids and primers
Full-length STK15 was obtained by PCR from plasmid pcDNA3-STK15/BTAK (a kind gift from Dr S. Sen; 27) using the primers STK15-CHIS5 (CGGGATCCCGG GGATGGACCGATCTAAAGAAACTGC) and STK15-3XhoI (CCGCTCGAGCTAAGACTGTTTGCTAGCTGA TTC). STK15-GBT8 was constructed by cloning into the BamHI and XhoI sites of pGBT8. A HeLa cDNA two-hybrid library was used in the two-hybrid screen (51). Full-length nm23-H1 cDNA was obtained by PCR of EST clones ordered from Genome Systems Inc. (EST clones 590228 and 3454119) using the primers NM23GAD-5EcoRI (CGGAATTCCA TGGCCAACTGTGA-GCG) and NM23-3XhoI (CCGCTC GAGTCATTCATAGATCCAG). pGADGH-NM23-H1 was constructed by cloning the the PCR product into the EcoRI and XhoI sites of pGADGH (51).
Cell culture
Human IMR90 cells were purchased from ATCC (CCL-186). These were immortalized by infection with a hTERT retrovirus at passage 25 (52). The cells were cultured at 37°C in a 5% CO2 incubator in Dulbecco’s modified medium (DMEM) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS), 10% (v/v) non-essential amino acids (NEAA) and penicillin/streptomycin (100 IU/ml and 100 µg/ml, respectively). Nocodazole treatment was done by addition of 10 µg/ml nocodazole to the culture medium followed by incubation for 10 min at 37°C.
Two-hybrid screening
To identify proteins interacting with STK15, we screened a plasmid library of fusions between the GAL4 activation domain (GAD, residues 768–881) and HeLa cell cDNA fragments. The Gal4 DNA-binding domain (GBD) is fused with STK15 in the pSTK15-GBT8 construct. The screen was done in a S.cerevisiae reporter strain (pJ64-4a, W303MATa trp9-901, 112 ura3-52 his3-200 gal4 gal80 GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-LacZ, a kind gift from R. Rothstein). A total of 1.2 × 106 transformants were assayed on synthetic drop-out medium (without leucine, histidine and tryptophan, SC –HLW) plates. A total of 104 colonies turned blue on X-gal plates, and the plasmids were recovered from 54 colonies. Retransformation of the plasmids into the test strain confirmed that 20 plasmids retained the ability to activate the β-galactosidase reporter. Sequencing analysis revealed four plasmids contained coding sequences from nm23-H1 genes. The four plasmids were derived from two independent cDNAs comprising nucleotides 25–330 and 4–159 of the nm23-H1 coding region.
Antibodies
A polyclonal antiserum against STK15 (anti-STK15) was raised in rabbits by presenting a KLH-conjugated nine amino acid peptide (synthesized by Research Genetics, Huntsville, AL) from the C-terminus of STK15 (NKESASKQS). The serum was affinity purified using a resin prepared from the synthesized peptide (53). The antibody recognizes a 46 kDa band in whole cell lysates. For some studies, the affinity-purified antibody was labeled with Alexa Fluor 647 dye (Molecular Probes, Eugene, OR), for example for examination of cells triple labeled by anti-β-tubulin-FITC, anti-NM23-H1-TRITC and anti-STK15-AF647 antibodies. Affinity-purified anti-NM23 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) (rabbit polyclonal sc-343 and sc-343-TRITC for immunofluorescence and mouse monoclonal sc-465 for immunoprecipitation). Mouse monoclonal anti-centrin2 antibody was a kind gift from Dr J. L. Salisbury (Mayo Clinic, Rochester, MN). Monoclonal anti-γ-tubulin (T-3195) and FITC-conjugated mouse monoclonal anti-β-tubulin were obtained from Sigma (St Louis, MO). Secondary antibodies were obtained from Pierce (Rockford, IL), Jackson Immunoresearch Laboratory Inc. (West Grove, PA) and Molecular Probes Inc. (Eugene, OR).
Immunoblotting, immunoprecipitation and fluorescence microscopy
IMR90 cells were lysed in IP buffer [125 mM NaCl, 1 mM Mg(OAc)2, 1 mM CaCl2, 5 mM EGTA, 20 mM HEPES (pH 7.6), 1% (v/v) NP-40, with freshly dissolved 2 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride and protease inhibitors (Roche, Mannheim, Germany)] on ice for 10–15 min. Protein concentrations were determined by Bradford assay (Bio-Rad). A total of 10–50 µg protein was analyzed by 10 or 12% (v/v) SDS–PAGE (54).
Immunoprecipitation was performed as described (55) with the following modifications. IMR90 cells were lysed in IP buffer at 4°C for 10 min. The cell lysate was cleared by centrifugation at 10 000 g for 10 min at 4°C. The supernatant was pre-cleared by incubation for 30 min with agarose–protein A or agarose–protein G beads (Pierce). The supernatant was further incubated with 5 µg antibodies for 3–4 h at 4°C with agitation, after which agarose–protein A or A/G mixture (pre-blocked by incubation with 5% BSA) was added for 1 h at 4°C. The beads were then washed three times with IP buffer. For release of STK15 and associated proteins, beads were incubated with 1 mg/ml synthetic antigen in phosphate-buffered saline (PBS) for 1 h at 4°C with agitation.
For indirect immunofluorescence, cells were grown on acid-washed 13 mm square glass coverslips (Fisher Scientific, Pittsburgh, PA). Cells were first fixed with 0.5% (w/v) formaldehyde in PBS (pH 7.2) for 15 min at room temperature and then washed four times with PBS. Fixed cells were permeabilized by 0.1% (v/v) Triton X-100 in PBS with 1% (v/v) normal goat serum (NGS) and then washed with PBS with 1% (v/v) NGS (Gibco-BRL). The primary antibodies used were anti-STK15 1:100, anti-NM23 1:200, anti-centrin2 1:200 and anti-β-tubulin-FITC 1:25, at room temperature for 1 h. Secondary antibodies were diluted in the blocking buffer at 1:100 or 1:200 and incubated for 1 h at room temperature in the dark. The first wash after secondary antibody included 1 µg/ml Hoechst 33342 (Sigma) for 15 min at room temperature. For triple labeling, the primary antibodies used were as follows: anti-β-tubulin-FITC 1:25, anti-NM23-TRITC 1:50 and anti-STK15-AF647 1:50. Coverslips were mounted on glass slides with mounting medium. Microscopy was carried out on Nikon or Zeiss microscopes with 63× or 100× oil immersion lens. The images were captured with Openlab (Lexington, MA) software and saved as TIFF files.
Biochemical fractionation of STK15 and NM23
For fractionation of STK15 and NM23, IMR90 cells were lysed as above and the lysates were cleared by centrifugation at 10 000 g for 10 min at 4°C. The supernatants were cleared by passage through a 0.2 µm filter and fractionated through three chromatographic steps, Mono S, Mono Q and Superose 6 HR10/30 (Amersham-Pharmacia Biotech, Piscataway, NJ), in an AKAT FPLC system. The cell lysates were loaded first on Mono S and eluted with a linear gradient of from 100 to 1000 mM potassium chloride in MonoS buffer [20 mM HEPES (pH 7.6), 10% (v/v) glycerol, 1 mM DTT, 1 mM EDTA, 0.01% (v/v) NP40]. Fractions were collected (0.5 ml) and those containing the peak of STK15 1.5 ml (from 280 to 370 mM) were collected and dialyzed against MonoQ buffer [20 mM HEPES (pH 8.0), 10% (v/v) glycerol, 1 mM DTT, 1 mM EDTA, 0.01% (v/v) NP40, containing 100 mM NaCl]. The dialyzed eluate was then loaded onto Mono Q and was eluted with a linear gradient from 0 to 1000 mM potassium chloride in MonoQ buffer. The peak fractions for STK15 (from 350 to 450 mM) were collected and pooled before 0.5 ml was loaded onto Superose 6. The column was run at 0.4 ml/min with Super6 buffer [20 mM HEPES (pH 7.6), 10% (v/v) glycerol, 1 mM DTT, 1 mM EDTA, 0.01% NP40 and 300 mM NaCl]. Fractions of 1 ml were collected and proteins were precipitated with TCA. The pellets were then re-dissolved in 0.1 N NaOH and loaded onto SDS–PAGE gels for immunoblot analysis.
RESULTS AND DISCUSSION
NM23 and STK15 interact
To search for proteins that interact with STK15 we conducted a yeast two-hybrid screen using the full-length STK15 coding sequence as a bait (see Materials and Methods). Among the positives, we found four plasmids containing the human nm23-H1 coding sequence. Two different fusions were represented, which cover a common N-terminal region of NM23-H1 (three represented fusions of Gal4 to amino acids 9–110, and another represented amino acids 2–53). This suggests that the region of NM23-H1 between amino acids 2 and 53 binds directly to STK15. We inserted the full-length nm23-H1 coding sequence into pGADGH (pGADGH-NM23-H1) and confirmed that this fusion protein also interacts specifically with STK15 (Fig. 1). Only in combination with STK15 did colonies carrying NM23-H1 show significant β-galactosidase expression. Neither NM23-H1 alone nor NM23 combined with other Gal4 fusion proteins (e.g. ras) induced β-galactosidase expression. However, at least in yeast, the degree of X-gal staining indicated that the interaction between STK15 and Nm-23H1 was relatively weak, as compared to the positive control (pGBT8-Ras and pGADGH-Raf in this case). It was therefore essential to investigate whether these proteins physically interacted in mammalian cells.
Figure 1.
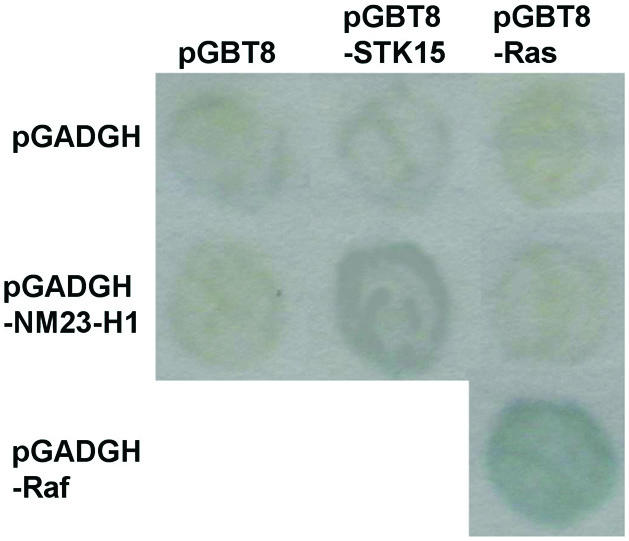
STK15 and NM23 interact specifically in the two-hybrid assay. Transformants of S.cerevisiae strain pJ64-4a were grown and re-patched on synthetic complete plates lacking Leu and Trp to select for both pGBT and pGADGH plasmid derivatives. The patches were transferred to Whatman No. 1 filter paper for β-galactosidase assays and the color was developed at 37°C for 1 h. A combination of pGBT8-Ras and pGADGH-Raf served as the positive control.
To test whether STK15 and NM23 also interact in vivo in human cells, proteins were immunoprecipitated from IMR90 cell lysates using anti-STK15 and anti-NM23 antibodies (see Materials and Methods). The presence of NM23 and STK15 in the precipitate was examined by western blotting (Fig. 2). Anti-STK15 specifically co-immunoprecipitates STK15 and NM23. Pre-incubation of the STK15 antibody with its cognate synthetic antigen coordinately abolished immunoprecipitation of both STK15 and NM23 (Fig. 2, lanes 1 and 2). The NM23 antiserum also immunoprecipitated NM23 and STK15, while normal mouse IgG did not bring down significant amounts of either NM23 or STK15 (Fig. 2, lanes 3 and 4). We did detect some non-specific binding of NM23 to protein A/G beads; however, this is not unexpected considering that NM23 is a relatively abundant protein (lane 3). The foregoing results indicate that STK15 and NM23 associate with each other in human cells. Comparing the total amount of NM23 present in the cell and the amount precipitated by anti-NM23-H1 antibody (Fig. 2, lane 4, and western data not shown) to the amount of NM23 immunoprecipitated by the STK15 antibody (Fig. 2, lane 2; both lanes 2 and 4 were from the same amount of cell lysate) suggests that a relatively small percentage of NM23-H1 exists in a stable, physical complex with STK15. In western blots of the anti-STK15 immunoprecipitations, we also detected another protein of ∼54 kDa, in addition to the 46 kDa protein that is predicted to be an STK15 protein derived from an alternatively spliced mRNA. We have not definitively determined the identity of the 54 kDa protein; however, the human genome sequencing project has predicted an alternatively spliced STK15 transcript that is predicted to generate a protein of this size (accession no. XP_009546). In contrast, we have confirmed that the ∼46 kDa band is STK15 by MALDI-TOF mass spectrometry.
Figure 2.
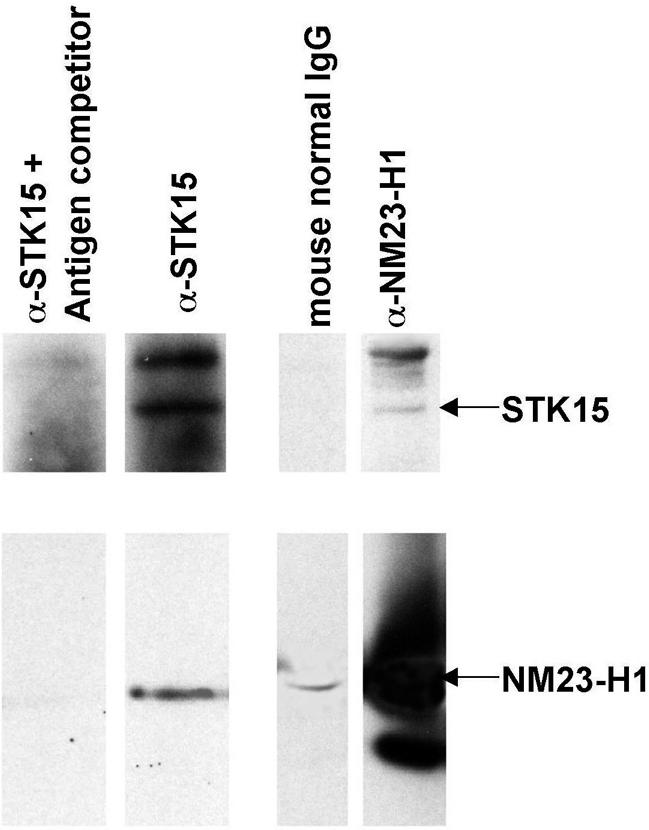
STK15 and NM23 co-immunoprecipitate. Protein lysates from immortalized IMR90 cells were cleared by centrifugation and immunoprecipitated with anti-STK15 plus STK15 polypeptide antigen competitor (lane 1), anti-STK15 alone (lane 2), normal mouse IgG (lane 3) and anti-NM23-H1 (lane 4). The immunoprecipited proteins were fractionated by SDS–PAGE and transferred to nitrocellulose membrane for western blotting. STK15 and NM23-H1 are indicated by arrows.
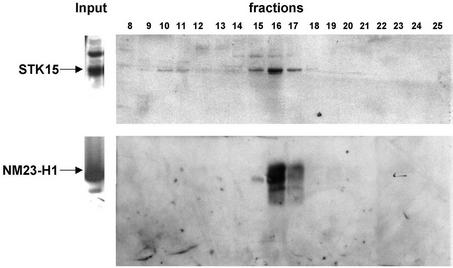
To verify the interaction between STK15 and NM23-H1, protein extracts from IMR90 cells were fractionated by FPLC. Throughout the purification, we followed STK15 by western blotting. Western analysis of the purification through Mono S, Mono Q and Superose 6 columns revealed that STK15 is present in two distinct forms, a high molecular weight protein complex (Fig. 3, fractions 10 and 11, ∼2 MDa) and a relatively low molecular weight complex (fractions 15–17, ∼350 KDa). NM23-H1 also shows a peak around fractions 15–17, which exactly co-migrates with STK15 in the ∼350 kDa complex (Fig. 3). This result supports the notion that STK15 and NM23 coexist in a protein complex in vivo.
Figure 3.
STK15 and NM23-H1 co-fractionate. Protein lysates from immortalized IMR90 cells were fractionated on Mono S, Mono Q and Superose 6 columns. Fractions 8–25 of 1 ml volume were collected after the final Superose 6 gel filtration column and constituent proteins were separated by SDS–PAGE for western blotting. STK15 and NM23-H1 are indicated by arrows.
STK15 and NM23 co-localize at centrosomes
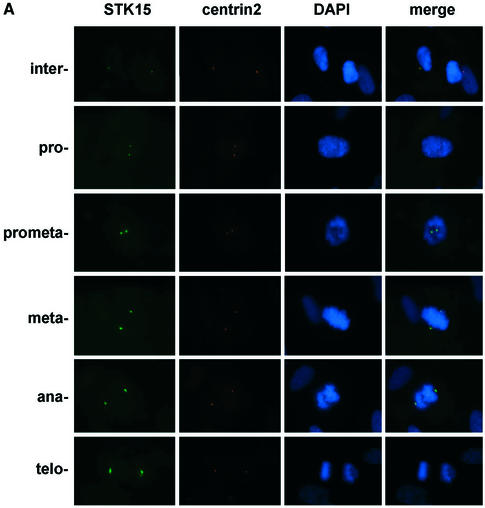
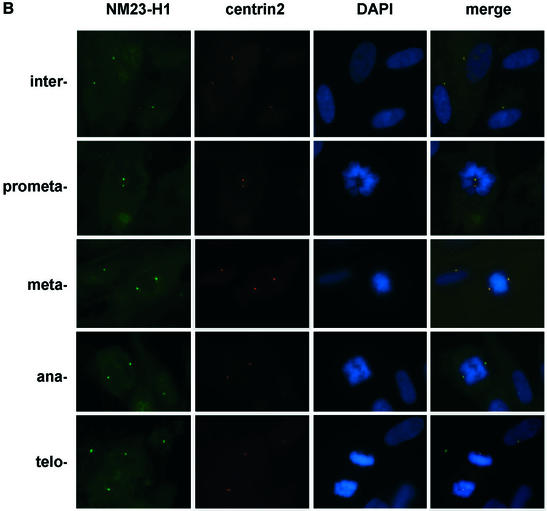
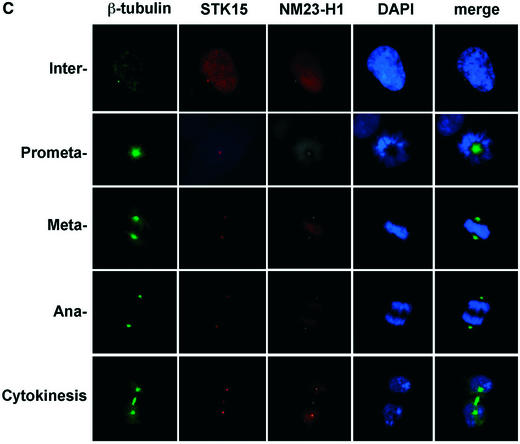
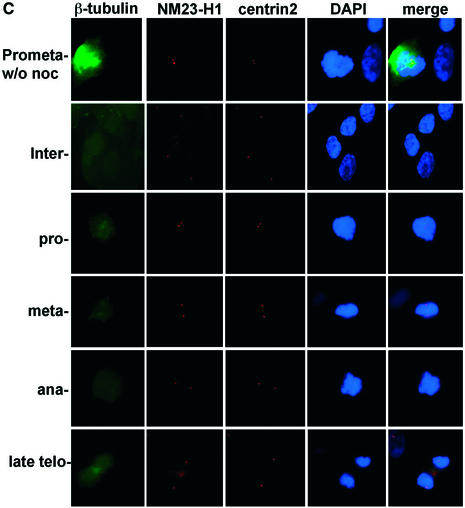
It has been shown that STK15 localizes to centrosomes in mitotic cells (25) (Fig. 4A). By fixing IMR90 cells in 0.5% (w/v) formaldehyde, we can also detect STK15 at centrosomes during interphase in immortalized human IMR90 cells. Centrosomes were visualized by simultaneous staining with an anti-centrin2 antiserum, which locates the two centrioles in the centrosome (Fig. 4). The amount of STK15 at centrosomes increases as cells move from interphase to prophase, and SKT15 also becomes apparent on the mitotic spindle. These results suggest that STK15 is a centrosome component throughout the cell cycle (Fig. 4A), but becomes enriched at centrosomes during mitosis. NM23-H1 also localizes to centrosomes in interphase cells (Fig. 4B) (50). At the beginning of prophase NM23-H1 accumulates at the centrosome, as judged from the increased intensity of staining (Fig. 4B, prometaphase) (50). The association of NM23 with centrosomes persists through the mitotic phase and NM23-H1 also distributes somewhat to the microtubule spindles adjacent to the centrosome from metaphase to telophase (Fig. 4B) (50). From late telophase to cytokinesis, NM23-H1 also accumulates at the newly forming midbody microtubules (Figs 4C and 5C in late telophase and cytokinesis cells). Interestingly, an identical distribution pattern at the midbody microtubules is also observed for STK15 (Figs 4C and 5B in late telophase and cytokinesis cells). The accumulation of NM23-H1 on centrosomes coincides with the enrichment of STK15 at the centrosome and with increased STK15 kinase activity at the beginning of mitosis (25).
Figure 4.
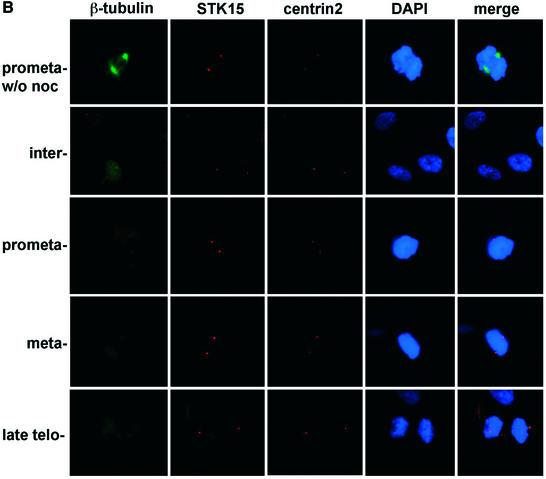
STK15 and NM23-H1 localize to centrosomes throughout the cell cycle. IMR90 cells were plated on glass coverslips and cultured in growth medium for 24 h before fixation with paraformaldehyde. The centrosome was visualized with anti-centrin2 and Texas red-labeled secondary antibodies. (A) STK15 and (B) NM23-H1 were visualized with anti-STK15 and anti-NM23-H1, respectively, and FITC-labeled secondary antibodies. DNA was labeled by Hoechst 33342 or DAPI staining. The composite images are STK15 or NM23 merged with centrin2 and DAPI. The green NM23 or STK15 merged with red centrin2 produces yellow spots in the composite image, indicating that NM23 and STK15 localize to the centrosomes. (C) Cells were labeled with anti-β-tubulin-FITC, anti-NM23-TRITC and anti-STK15-AF647 (Cy5) simultaneously and visualized on the microscope with FITC, TRITC, Cy5 and DAPI filters, respectively. Cell cycle stages are indicated on the left.
Figure 5.
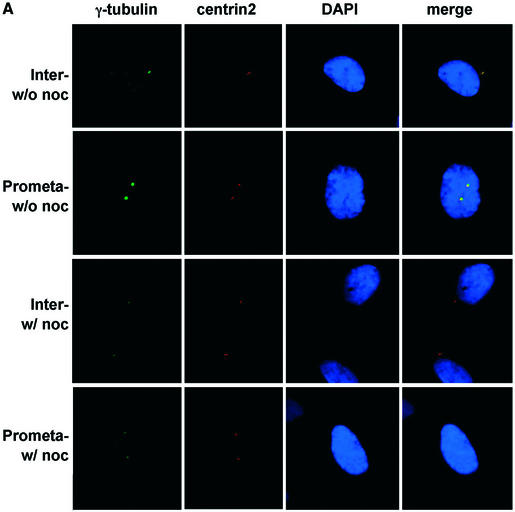
(Previous two pages and above) The centrosome localization of STK15 and NM23-H1 is not microtubule dependent. (A) IMR90 cells grown on coverslips were treated with or without nocodazole and fixed for immunofluorescence. The centrosome was visualized by staining with anti-centrin2 and anti-γ-tubulin antibodies. As predicted, the signal representing centrosome-associated γ-tubulin becomes significantly weakened, especially in mitotic cells (prometaphase cells here), upon nocodazole treatment (labeled w/ noc on the left), as compared to that seen in cells without nocodazole (labeled w/o noc on the left). (B) STK15 and (C) NM23-H1 still localize to centrosome in nocodazole-treated cells throughout the cell cycle. IMR90 cells treated with or without nocodazole (w/o noc) were fixed and triple labeled for β-tubulin (FITC), STK15/NM23-H1 (Cy5), centrin2 (Texas red) and DNA (DAPI). The nocodazole treatment totally dissipates the microtubule spindles so the cells with nocodazole fail to show the characteristic β-tubulin staining pattern (the β-tubulin columns). Cell cycle stages are indicated on the left.
In order to test whether STK15 and NM23 co-localize, we simultaneously labeled IMR90 cells with β-tubulin-FITC, NM23-TRITC and STK15-AF647 (Cy5) and visualized the localization of each protein (Fig. 4C). β-Tubulin immunofluorescence reveals interphase microtubule filaments and centrosomes. In mitotic phase, β-tubulin localizes both to the mitotic spindles and to the centrosomes (Fig. 4C). NM23 and STK15 fluorescence is present together at the centrosome at all cell cycle phases (Fig. 4C, the right side merged lane). These results confirm that STK15 and at least some fraction of NM23 co-localize to centrosomes throughout the cell cycle.
STK15 and NM23 centrosomal localization is microtubule-independent
There is some evidence indicating that NM23-H1 is β-tubulin associated and localized through γ-tubulin to centrosomes (48,50). In order to clarify that STK15 and NM23 do not associate indirectly at centrosomes through their separate interactions with microtubules, we treated IMR90 cells with nocodazole (see Materials and Methods), which rapidly dissipates the microtubule network in cells (Fig. 5B and C, β-tubulin column).
There are several centrosome-associated proteins that localize to this structure in a spindle-dependent fashion. Of these, perhaps the most well studied is γ-tubulin (56). Upon nocodazole treatment of mammalian cells, centrosomal γ-tubulin is reduced by ∼2-fold in interphase cells and by at least 4-fold in mitotic cells (56). Using β- and γ-tubulin antisera, we confirmed that our nocodazole treatment dissipates the microtubule network and dramatically reduces the amount of γ-tubulin associated with the centrosome (Fig. 5A–C). In such cells, STK15 and NM23-H1 still localize to the centrosome with the same pattern and intensity of staining seen in untreated cells (Fig. 5B and C). The result suggests that the interaction between STK15 and NM23 and their centrosome localization are microtubule-independent.
A previous study (57) showed that the centrosomal localization of a Xenopus laevis homolog of STK15, Aurora-A, is microtubule-dependent. This result was obtained using a construct consisting of the only N-terminal domain of Aurora-A (57). However, microtubule-independent localization of Aurora-A to the centrosome was observed using a full-length Aurora-A construct (57). Here, we also observed that the localization of full-length STK15 to the centrosome is not dependent on microtubules (Fig. 5B).
A previous study showed that STK15 associates with centrosomes only at onset of mitosis (25). However, our immunofluorescence studies indicate that STK15 associates with centrosomes throughout the cell cycle. This discrepancy could arise because of the different cell types used in these studies. Here we examined STK15 in normal or immortalized human fibroblasts cells while previous studies have used human cancer cells and rodent fibroblast cells. Furthermore, different antisera and fixation conditions were used in this and in previous studies. We do, however, observe an increase in the abundance of STK15 at centrosomes in mitotic cells.
STK15 is a centrosome-associated kinase, overexpression of which in rodent or human cancer cells causes improper centrosome duplication, aneuploidy and cellular transformation (25,27). The abundance of STK15 is controlled by the ubiquitin pathway (32,33) and has been linked to CDC20, an APC activator (31). STK15 kinase activity is also controlled by PP1, a protein phosphatase (34). Here we provide evidence that STK15 is associated with a putative tumor and metastasis suppressor protein, NM23-H1. These proteins physically interact and co-localize to centrosomes throughout the cell cycle in human IMR90 cells. NM23-H1 plays important roles in cell proliferation, differentiation, tumorigenesis and metastasis.
The interaction between NM23 and STK15 might potentially modulate either of their activities through a number of mechanisms, ranging from allosteric regulation to induced localization to enzymatic modification. We tested the latter possibility using STK15 and NM23-H1, purified from Escherichia coli. Purified STK15 was enzymatically active, as judged by its ability to phosporylate a model substrate, MBP. NM23 also displayed activity as judged by auto-phosphorylation and NDP kinase activities (data not shown; 46,58). Neither of the purified proteins was capable of chemically modifying the other nor did mixing these proteins affect their activities towards model substrates in vitro. Therefore, the exact nature of the functional relationship between NM23 and SKT15 remains unknown.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank J. L. Salisbury (Mayo Clinic, Rochester, MN) for the kind gift of the anti-centrin2 antiserum, S. Sen (University of Texas M.D. Anderson Cancer Center, Houston, TX) for the STK15 plasmid, R. Rothstein (Columbia University) for the yeast two-hybrid strain pJ64-4a and Y. Seger (Cold Spring Harbor) for the hTERT retroviral plasmid. The authors would also like to thank D. Conklin, M. Carmell, S. Hammond, A. Caudy and other members in the laboratory for technical support and helpful discussions. We are thankful to Stephen Hearn and Gayle Lark at the core microscopy facility and Jim Duffy in the art department. J.D. is supported by a post-doctoral fellowship (PC001528) from the Department of Defense Prostate Cancer Research Program (PCRP). This work was supported by a grant from the NIH (P01-CA13106) to G.J.H.
REFERENCES
- 1.Zheng Y., Jung,M.K. and Oakley,B.R. (1991) γ-Tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell, 65, 817–823. [DOI] [PubMed] [Google Scholar]
- 2.Carroll P.E., Okuda,M., Horn,H.F., Biddinger,P., Stambrook,P.J., Gleich,L.L., Li,Y.,Q., Tarapore,P. and Fukasawa,K. (1999) Centrosome hyperamplification in human cancer: chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene, 18, 1935–1944. [DOI] [PubMed] [Google Scholar]
- 3.Hsu L.C. and White,R.L. (1998) BRAC1 is associated with the centrosome during mitosis. Proc. Natl Acad. Sci. USA, 95, 12983–12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsumoto Y., Hayashi,K. and Nishida,E. (1999) Cyclin dependent kinase 2 (CDK2) is required for centrosome duplication in mammalian cells. Curr. Biol., 9, 429–432. [DOI] [PubMed] [Google Scholar]
- 5.Lange B.M. (2002) Integration of the centrosome in cell cycle control, stress response and signal transduction pathways. Curr. Opin. Cell Biol., 14, 35–43. [DOI] [PubMed] [Google Scholar]
- 6.Bornens M. (2002) Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol., 14, 25–34. [DOI] [PubMed] [Google Scholar]
- 7.Rieder C.L., Faruki,S. and Khodjakov,A. (2001) The centrosome in vertebrates: more than a microtubule-organizing center. Trends Cell Biol., 11, 413–419. [DOI] [PubMed] [Google Scholar]
- 8.Hinchcliffe E.H. and Sluder,G. (2001) Centrosome duplication: three kinases come up a winner! Curr. Biol., 11, R698–R701. [DOI] [PubMed] [Google Scholar]
- 9.Doxsey S. (2001) Re-evaluating centrosome function. Nature Rev. Mol. Cell Biol., 2, 688–698. [DOI] [PubMed] [Google Scholar]
- 10.Stearns T. (2001) Centrosome duplication. A centriolar pas de deux. Cell, 105, 417–420. [DOI] [PubMed] [Google Scholar]
- 11.Hinchcliffe E.H., Miller,F.J., Cham,M., Khodjakov,A. and Sluder,G. (2001) Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science, 291, 1547–1550. [DOI] [PubMed] [Google Scholar]
- 12.Khodjakov A. and Rieder,C.L. (2001) Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol., 153, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piel M., Nordberg,J., Euteneuer,U. and Bornens,M. (2001) Centrosome-dependent exit of cytokinesis in animal cells. Science, 291, 1550–1553. [DOI] [PubMed] [Google Scholar]
- 14.Salisbury J.L. (2001) The contribution of epigenetic changes to abnormal centrosomes and genomic instability in breast cancer. J. Mammary Gland Biol. Neoplasia, 6, 203–212. [DOI] [PubMed] [Google Scholar]
- 15.Pihan G.A., Purohit,A., Wallace,J., Knecht,H., Woda,B., Quesenberry,P. and Doxsey,S.J. (1998) Centrosome defects and genetic instability in malignant tumors. Cancer Res., 58, 3974–3985. [PubMed] [Google Scholar]
- 16.Weber R.G., Bridger,J.M., Benner,A., Weisenberger,D., Ehemann,V., Reifenberger,G. and Lichter,P. (1998) Centrosome amplification as a possible mechanism for numerical chromosome aberrations in cerebral primitive neuroectodermal tumors with TP53 mutations. Cytogenet. Cell Genet., 83, 266–269. [DOI] [PubMed] [Google Scholar]
- 17.Gradimi B.M., Sackett,D.L., Difilippantonio,M.J., Schrock,E., Neumann,T., Jauho,A., Auer,G. and Reid,T. (2000) Centrosome amplification and instability occurs exclusively in aneuploidy, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer, 27, 183–190. [PMC free article] [PubMed] [Google Scholar]
- 18.Marx J. (2001) Do centrosome abnormalities lead to cancer? Science, 292, 426–427. [DOI] [PubMed] [Google Scholar]
- 19.Saavedra H.I., Fukasawa,K., Conn,C.W. and Stambrook,P.J. (1999) MAPK mediate ras-induced chromosome instability. J. Biol. Chem., 274, 38083–38090. [DOI] [PubMed] [Google Scholar]
- 20.Tutt A., Gabriel,A., Bertwistle,D., Connor,F., Paterson,H., Peacock,J., Ross,G. and Ashworth,A. (1999) Absence of BRCA2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr. Biol., 9, 1107–1110. [DOI] [PubMed] [Google Scholar]
- 21.Fukasawa K., Choi,T., Kuriyama,R., Rulong,S. and Vande Woude,G.F. (1996) Abnormal centrosome amplification in the absence of p53. Science, 271, 1744–1747. [DOI] [PubMed] [Google Scholar]
- 22.Hollander M.C., Sheikh,M.S., Bulavin,D.V., Lundgren,K., Augeri-Henmueller,L., Shehee,R., Molanaro,T.A., Kim,K.T., Tolosa,E., Ashwell,J.D. et al. (1999). Genomic instability in gadd45a-deficient mice. Nature Genet., 23, 176–184. [DOI] [PubMed] [Google Scholar]
- 23.Meraldi P., Lukas,J., Fry,A.M., Bartek,J. and Nigg,E.A. (1999) Centrosome duplication in mammalian somatic cells required E2F and Cdk2-Cyclin A. Nature Cell Biol., 1, 88–93. [DOI] [PubMed] [Google Scholar]
- 24.Glover D.M., Leibowitz,M.H., McLean,D.A. and Parry,H. (1995) Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell, 81, 95–105. [DOI] [PubMed] [Google Scholar]
- 25.Bischoff J.R., Anderson,L., Zhu,Y., Mossie,K., Ng,L., Souza,B., Schryver,B., Flanagan,P., Chairvoyant,F., Ginther,C. et al. (1998). A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J., 17, 3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sen S., Zhou,H. and White,R.A. (1997) A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and over expressed in human breast cancer cell lines. Oncogene, 14, 2195–2200. [DOI] [PubMed] [Google Scholar]
- 27.Zhou H., Kuang,J., Zhong,L., Kuo,W., Grey,J., Sahin,A., Brinkley,B. and Sen,S. (1998) Tumor amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nature Genet., 20, 189–193. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka T., Kimura,M., Matsunaga,K., Fukada,D., Mori,H. and Okano,Y. (1999) Centrosomal kinase AIK1 is overexpressed in invasive ductal carcinoma of the breast. Cancer Res., 59, 2041–2044. [PubMed] [Google Scholar]
- 29.Miyoshi Y., Iwao,K., Egawa,C. and Noguchi,S. (2001) Association of centrosomal kinase STK15/BTAK mRNA expression with chromosomal instability in human breast cancers. Int. J. Cancer, 92, 370–373. [DOI] [PubMed] [Google Scholar]
- 30.Shin S.O., Lee,K.H., Kim,J.H., Baek,S.H., Park,J.W., Gabrielson,E.W. and Kwon,T.K. (2000) Alternative splicing in 5′-untranslational region of STK-15 gene, encoding centrosome associated kinase, in breast cancer cell lines. Exp. Mol. Med., 32, 193–196. [DOI] [PubMed] [Google Scholar]
- 31.Farruggio D.C., Townsley,F.M. and Ruderman,J.V. (1999) Cdc20 associates with the kinase aurora2/Aik. Proc. Natl Acad. Sci. USA, 96, 7306–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honda K., Mihara,H., Kato,Y., Yamaguchi,A., Tanaka,H., Yasuda,H., Furukawa,K. and Urano,T. (2000) Degradation of human Aurora2 protein kinase by the anaphase-promoting complex-ubiquitin-proteasome pathway. Oncogene, 19, 2812–2819. [DOI] [PubMed] [Google Scholar]
- 33.Walter A.O., Seghezzi,W., Korver,W., Sheung,J. and Lees,E. (2000) The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene, 19, 4906–4916. [DOI] [PubMed] [Google Scholar]
- 34.Katayama H., Zhou,H., Li,Q., Tatsuka,M. and Sen,S. (2001) Interaction and feedback regulation between STK15/BTAK/Aurora-A kinase and protein phosphatase 1 through mitotic cell division cycle. J. Biol. Chem., 276, 46219–46224. [DOI] [PubMed] [Google Scholar]
- 35.Hannak E., Kirkham,M., Hyman,A.A. and Oegema,K. (2001) Aurora-1 kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol., 155, 1109–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steeg P.S., Bevilacqua,G., Kopper,L., Thorgeirsson,U.P., Talmadge,J.E., Liotta,L.A. and Sobel,M.E. (1988) Evidence for a novel gene associated with low tumor metastatic potential. J. Natl Cancer Inst., 80, 200–204. [DOI] [PubMed] [Google Scholar]
- 37.Leone A., Flatow,U., VanHoutte,K. and Steeg,P.S. (1993). Transfection of human nm23-H1 into the human MDA-MB-435 breast carcinoma cell line: effects on tumor metastatic potential, colonization and enzymatic activity. Oncogene, 8, 2325–2333. [PubMed] [Google Scholar]
- 38.Parhar R.S., Shi,Y., Zou,M., Farid,N.R., Ernst,P. and Al-Sedairy,S. (1995) Effects of cytokine-mediated modulation of nm23 expression on the invasion and metastatic behavior of B16F10 melanoma cells. Int. J. Cancer, 60, 204–210. [DOI] [PubMed] [Google Scholar]
- 39.Baba H., Urano,T. and Okada,K. (1995) Two isotypes of murine nm23/nucleoside diphosphate kinase, nm23-M1 and nm23-M2, are involved in metastatic suppression of a murine melanoma line. Cancer Res., 55, 1977–1981. [PubMed] [Google Scholar]
- 40.Freije J.M.P., MacDonald,N.J. and Steeg,P.S. (1998) Nm23 and tumor metastasis: basic and translational advances. Biochem. Soc. Symp., 63, 261–271. [PubMed] [Google Scholar]
- 41.Lacombe M., Milon,L., Munier,A. and Lambeth,D.O. (2000) The human Nm23/nucleoside diphosphate kinases. J. Bioenerg. Biomembr., 32, 247–258. [DOI] [PubMed] [Google Scholar]
- 42.Parks R.E. Jr and Agarwal,R.P. (1973) Nucleoside diphosphokinases. In Boyer,P.D. (ed.), The Enzymes, 3rd Edn. Academic Press, New York, Vol. 8, pp. 307–333.
- 43.Wagner P. and Vu,N.D. (1995) Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase. J. Biol. Chem., 270, 21758–21764. [DOI] [PubMed] [Google Scholar]
- 44.Engel M., Veron,M., Theisinger,B., Lacombe,M.L., Seib,T., Dooley,S. and Welter,C. (1995) A novel serine/threonine-specific protein phosphotransferase activity on Nm23/nucleoside-diphosphate kinase. Eur. J. Biochem., 234, 200–207. [DOI] [PubMed] [Google Scholar]
- 45.Wagner P.D., Steeg,P.S. and Vu,N.D. (1997) Two-component kinase-like activity of nm23 correlates with its motility-suppressing activity. Proc. Natl Acad. Sci. USA, 94, 9000–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacDonald N.J., De la Rosa,A., Bebedict,M.A., Frejie,J.M.P., Krutsch,H. and Steeg,P.S. (1993) A serine phosphorylation of Nm23, and not its nucleoside diphosphate kinase activity, correlates with suppression of tumor metastatic potential. J. Biol. Chem., 268, 25780–25789. [PubMed] [Google Scholar]
- 47.Hemmerich S. and Pecht,I. (1992) A cromoglycate binding protein from rat mast cells of a leukemia line is a nucleoside diphosphate kinase. Biochemistry, 31, 4580–4587. [DOI] [PubMed] [Google Scholar]
- 48.Lombardi D., Sacchi,A., D’Angostino,G. and Tibursi,G. (1995) The association of the Nm23-M1 protein and β-tubulin correlates with cell differentiation. Exp. Cell Res., 217, 267–271. [DOI] [PubMed] [Google Scholar]
- 49.Biggs J., Hersperger,E., Steeg,P.S., Liotta,L.A. and Shearn,A. (1990) A Drosophila gene that is homologous to a mammalian gene associated with tumor metastasis codes for a nucleoside diphosphate kinase. Cell, 63, 933–940. [DOI] [PubMed] [Google Scholar]
- 50.Roymans D., Vissenberg,K., De Jongle,C., Willems,R., Engler,G., Kiruma,N., Brobben,B., Claes,P., Verbelen,J.-P., Van Broeckhoven,C. et al. (2001) Identification of the tumor metastasis suppressor Nm23-H1/Nm23-R1 as a constituent of the centrosome. Exp. Cell Res., 262, 145–153. [DOI] [PubMed] [Google Scholar]
- 51.Hannon G.J., Demetrick,D. and Beach,D. (1993) Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes Dev., 7, 2378–2391. [DOI] [PubMed] [Google Scholar]
- 52.Wang J., Xie,L.Y., Allan,S., Beach,D. and Hannon,G.J. (1998) Myc activates telomerase. Genes Dev., 12, 1769–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guan K.L. and Dixon,J.E. (1991) Eukaryotic proteins expressed in E.coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem., 192, 262–267. [DOI] [PubMed] [Google Scholar]
- 54.Du J., Nasir,I., Benton,B.K., Kladde,M.P. and Laurent,B.C. (1998) Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics, 150, 987–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiong Y., Zhang,H. and Beach,D. (1993) Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation. Genes Dev., 7, 1572–1583. [DOI] [PubMed] [Google Scholar]
- 56.Vorobjev I.A., Uzbekov,R.E., Komarova,Yu.A. and Alieva,I.B. (2000) Gamma-tubulin distribution in interphase and mitotic cells upon stabilization and depolymerization of microtubules. Membr. Cell Biol., 14, 219–235. [PubMed] [Google Scholar]
- 57.Giet R. and Prigent,C. (2001) The non-catalytic domain of the Xenopus laevis aurora-A kinase localizes the protein to the centrosome. J. Cell Sci., 114, 2095–2104. [DOI] [PubMed] [Google Scholar]
- 58.Biggs J., Hersperger,E., Steeg,P.S., Ioitta,L.A. and Shearn,A. (1990) A Drosophila gene that is homologous to a mammalian gene associated with tumor metastasis codes for a nucleoside diphosphate kinase. Cell, 63, 933–940. [DOI] [PubMed] [Google Scholar]