Abstract
Lignin content and composition are two important agronomic traits for the utilization of agricultural residues. Rice (Oryza sativa) gold hull and internode phenotype is a classical morphological marker trait that has long been applied to breeding and genetics study. In this study, we have cloned the GOLD HULL AND INTERNODE2 (GH2) gene in rice using a map-based cloning approach. The result shows that the gh2 mutant is a lignin-deficient mutant, and GH2 encodes a cinnamyl-alcohol dehydrogenase (CAD). Consistent with this finding, extracts from roots, internodes, hulls, and panicles of the gh2 plants exhibited drastically reduced CAD activity and undetectable sinapyl alcohol dehydrogenase activity. When expressed in Escherichia coli, purified recombinant GH2 was found to exhibit strong catalytic ability toward coniferaldehyde and sinapaldehyde, while the mutant protein gh2 completely lost the corresponding CAD and sinapyl alcohol dehydrogenase activities. Further phenotypic analysis of the gh2 mutant plants revealed that the p-hydroxyphenyl, guaiacyl, and sinapyl monomers were reduced in almost the same ratio compared to the wild type. Our results suggest GH2 acts as a primarily multifunctional CAD to synthesize coniferyl and sinapyl alcohol precursors in rice lignin biosynthesis.
In addition to cellulose, lignin is the second major biopolymer component of the plant cell wall. Lignin cross-links with carbohydrates, such as cellulose and hemicellulose, to play an important role of the cell walls, enhancing the rigidity, conferring resistance to pathogens and mechanical stress, and enabling solute transport in the xylem (Brill et al., 1999; Chabannes et al., 2001; Jones et al., 2001). Lignin is a complex phenolic polymer that is mainly derived from the oxidative polymerization of p-coumaryl, coniferyl, and sinapyl alcohol precursors as the principal monomers, and the monomers are defined as p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units. The types and amount of lignin units vary depending on plant species, tissues, cell types, and cell wall layers and are influenced by developmental and environmental cues (Campbell and Sederoff, 1996; Donaldson, 2001).
Apart from its critical role in plant development, lignin has great economical and environmental value. Considerable monetary and environmental costs are incurred by industry in removing lignin from cellulose during production of pulp and paper (Hu et al., 1999). Similarly, lignin limits the digestibility and energy yield of forage crops (Jung and Vogel, 1986; Cherney et al., 1990). Studies on the lignin metabolic pathway help to understand plant development and modify plants for meeting the industrial and livestock needs (Baucher et al., 1996; Halpin et al., 1998). Many investigations were conducted on a series of maize (Zea mays) brown midrib (bm) mutants, which have been used as a model for digestibility and lignin studies (for review, see Barrière et al., 2004). bm mutants have a reddish-brown pigmentation in the leaf midrib and stalk pith, which is associated with lignified tissues. Researches show that these bm mutants are deficient in the enzymes that control the lignin content or composition in lignin biosynthesis (Vignols et al., 1995; Halpin et al., 1998; Piquemal et al., 2002). The mutants have aroused considerable agronomic interest in past decades because the changes in lignin content and composition may affect intake digestion and milk production by dairy cows (Ebling and Kung, 2004; Taylor and Allen, 2005). Rice (Oryza sativa) is one of the most important crops in the world, feeding more than half of the world's population, so identification of the corresponding lignin mutants will have important implications for agriculture.
Cinnamyl-alcohol dehydrogenase (CAD) was one of the first enzymes studied in lignin biosynthetic pathway, and it catalyzes the final reduction of the hydroxyl-cinnamaldehydes to the corresponding alcohols (Mansell et al., 1974). For the important role in regulation of the lignin content and composition, many CAD homologs have been isolated from different plant species and used to modify plants by transgenic approach (Halpin et al., 1994; Baucher et al., 1996, 1999; Ralph et al., 1997). CAD was initially believed to be multifunctional to catalyze the reduction of three hydroxyl-cinnamaldehydes (p-coumaryl, coniferyl, and sinapyl aldehydes) to the corresponding alcohols (for review, see Boerjan et al., 2003). A sinapyl alcohol dehydrogenase (SAD) has previously been isolated from aspen (Populus tremuloides) and proven to be specifically involved in the reduction of sinapaldehyde, while CAD from aspen has been proven to be specifically involved in the reduction of coniferaldehyde (Li et al., 2001). The result supports a hypothesis that CAD is involved only in the reduction of coniferylaldehyde, while SAD is specifically involved in the reduction of sinapaldehyde in angiosperm plants. The genomes of Arabidopsis (Arabidopsis thaliana) and rice have been fully sequenced and annotated (Arabidopsis Genome Initiative, 2000; International Rice Genome Sequencing Project, 2005), which provides a good opportunity to conduct genome-wide analyses of CAD family members. So far, nine Arabidopsis CAD members have been annotated as the bona fide CAD homologs, and their biochemical properties have been well studied (Kim et al., 2004). AtCADC and AtCADD of the family have been studied in null mutants and double mutants (Sibout et al., 2003, 2005) and proven to be the primary CAD genes in Arabidopsis lignification. Tobias and Chow (2005) have recently shown that there are 12 CAD family members in rice based on bioinformatics criteria, but the corresponding biochemical properties and physiological functions remain unknown.
The rice gold hull and internode (gh) mutant was first described as early as 1917 (http://www.gramene.org/db/mutant/search_mutant?id=GR:0060365). It exhibits a reddish-brown pigmentation in the hull and internode, instead of in the midrib. The hull and internode become golden yellow at maturation, so the mutant is named as gh rice. Based on the morphological and genetic descriptions, a series of gh mutates (gh1, gh2, gh3, and gh4) have been identified. Among them, gh1 was located on chromosome 5 (Nagao and Takahashi, 1963), and gh2 and gh3 were both located on chromosome 2 (Iwata and Omura, 1971, 1977). Despite the fact that these mutants have long been used as marker genes in rice breeding and genetics study, the mechanism underlying the phenotype is still unknown. In this study, the gh2 was identified to be a lignin-deficient mutant by a map-based cloning approach, and the GH2 gene codes a CAD. Both native and recombinant GH2 protein exhibit CAD activity and SAD activity, but native and recombinant gh2 protein lost the corresponding activities. Additionally, the H, G, and S monomers were similarly reduced in the gh2 mutant plants compared with the wild type. Our data suggest that coniferyl alcohol precursors and sinapyl alcohol precursors are both derived from the GH2-catalyzing reduction of coniferaldehyde and sinapaldehyde in the rice hull and internode.
RESULTS
Morphological Characterization of the gh2 Mutant
We evaluated the morphology of the gh2 mutant and compared it with the wild-type variety, Zhefu802. gh2 exhibits an obvious reddish-brown pigment in the panicle (hull), internode, and basal leaf sheath at the heading stage (Fig. 1, A and B). The reddish-brown pigments gradually become intensive starting from the basal internode to the apical internode. During plant maturing, seed coloration becomes darker and finally changes to golden yellow at maturation (Fig. 1C). Except the reddish-brown coloration in the above specific tissues, the gh2 mutant plants show similar development with the wild type under normal cultivated condition.
Figure 1.
Phenotype of wild type and gh2 mutant. A, The panicles of wild type (left) and gh2 mutant (right) at the heading stage. The wild-type panicle is green, while the gh2 panicle has turned reddish-brown. B, The internodes of the wild type and gh2 mutant at the heading stage. The wild-type internode is green (left), while the gh2 internode is reddish-brown (right). C, The seeds of wild type and gh2 at maturation. The wild-type seeds are light yellow (left), while the gh2 seeds are golden yellow (right).
The GH2 Gene Encodes a CAD
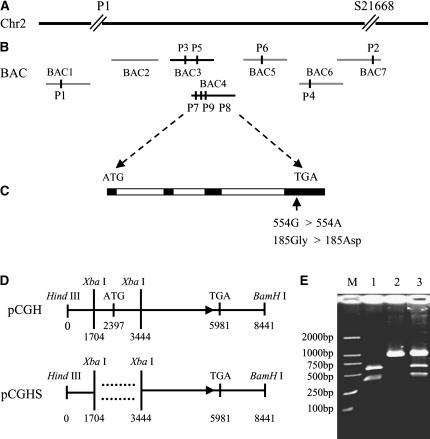
The gh2 gene has been genetically mapped to the region around 20 cM on chromosome 2 by the primary trisomics (Sanchez and Khush, 1994). To achieve a more precise map for the gh2 locus, we generated a large F2 mapping population between gh2 and Nipponbare, a japonica rice variety that has been fully sequenced. Of the approximately 13,000 F2 plants, 3,256 segregants showing the gh2 mutant phenotype were further used for fine mapping. Based on this, the gh2 gene was further located to a genomic region demarcated by sequence tag site (STS) markers P1 and S21668 (Fig. 2A). Between the two markers, we developed six additional STS markers and two cleaved amplified polymorphic sequence (CAPS) markers (Table I), leading to the mapping of this gene to a narrow region between two CAPS markers (P7 and P8; Fig. 2B). No recombinant near the STS marker P9 was found. The candidate genes near the P9 marker in the 30-kb region were chosen to be sequenced, and a gene encoding a CAD was found to be mutated by one nucleotide substitution G to A (554G > 554A) in the fourth extron, which substituted a zero-charge Gly to an electronegative Asp (Fig. 2C).
Figure 2.
Cloning and confirmation of the GH2 gene. A, The GH2 locus was mapped to a region between markers P1 and S21668 in the short arm of rice chromosome 2 (Chr2). B, Fine mapping of the GH2 locus using primers developed from seven bacterial artificial chromosomes (BACs): BAC1, OJ1297_C09; BAC2, OJ1572_F02; BAC3, OJ1073_F05; BAC4, OJ1145_F01; BAC5, OSJNBb0031B09; BAC6, OJ1136_D07; and BAC7, J1225_F07. P1 to P9 are the developed markers in the work (Table I). C, GH2 gene structure, showing the mutation site of the gh2. The start codon (ATG) and the stop codon (TGA) are indicated. Black and white boxes indicate the extron and intron sequences, respectively. The G to A substitution in the gh2 converted a zero-charge Gly to an electronegative Asp. D, Complementation constructs. The construct pCGH contains the entire GH2 gene, plus a 2,397-bp upstream region and 2,460-bp downstream region. The plasmid pCGHS contains a partial coding region of the ORF. E, Identification of transgenic plants. The G to A substitution in the gh2 creates an Aat II site that is used for identifying the mutant line, the complement line, and the wild-type line. Lane M, DL2000 marker; lane 1, gh2; lane 2, wild type; lane 3, the pCGH-transformed rice line 1.
Table I.
List of the PCR-based molecular markers developed in this study
| Marker | Primer Pairsa | Fragment Sizeb | Restriction Enzyme |
|---|---|---|---|
| P1 | F 5′-TATGTTTTCTAACGCAGGCT-3′; R 5′-AGAACACCCCAATATTACAT-3′ | N = 167, Z = 173 | |
| P2 | F 5′-ATTCTGCAATCCATAGTCAT-3′; R 5′-AATTGAAATCCTTAGGTGGA-3′ | N = 99, Z = 83 | |
| P3 | F 5′-CTCTGCGTTCACAAGTAGAT-3′; R 5′-TACCTATCTTGGCGGTGTAT-3′ | N = 105, Z = 89 | |
| P4 | F 5′-GCATAAGTCACGCTTAAA AT-3′; R 5′-TAAGTCGTCATGGTTACATT-3′ | N = 106, Z = 92 | |
| P5 | F 5′-AGAACGTGAGAAGTATGTGT-3′; R 5′-GCATGAATAACCACAGCAGA-3′ | N = 95, Z = 82 | |
| P6 | F 5′-TGCTGGAGAGGAACTATGCT-3′; R 5′-GCATGAATAACCACAGCAGA-3′ | N = 156, Z = 130 | |
| P7 | F 5′-ACGATTGCATGGTCTCCTAA-3′; R 5′-CATTCATCACATCACGCTCA-3′ | N = Z = 750 | XbaI |
| P8 | F 5′-TAGAAGGGCGATGGAGAAGC-3′; R 5′-GTATCCCACAAACGCGAAGC-3′ | N = I = 1,049 | XbaI |
| P9 | F 5′-GCAACAGTAGTAGTAGTAGA-3′; R 5′-CACTGATTACATGTGCCTCA-3′ | N = 200, Z = 244 |
F, Forward primer; R, reverse primer.
Numbers indicate the size (in bp) of amplified fragments; N, Nipponbare; Z, Zhefu802.
The identity of GH2 was confirmed by genetic complementation experiments. The plasmid pCGH, containing the entire open reading frame (ORF), 2,397-bp upstream region and 2,460-bp downstream region, and pCGHS, containing a partial coding region of the ORF (Fig. 2D), were introduced into the recessive mutants from the F3 generation of the mapping population, then 12 and eight independent transgenic lines were obtained from the two constructs, respectively. All 12 lines of pCGH showed complete complementation of the gh2 phenotype, whereas all eight lines of pCGHS failed to rescue the gh2 mutant. Additionally, the mutation in the gh2 produces an Aat II site in the genomic DNA, which can be used as a CAPS marker to determine the gh2 mutant background in the complementation test (Fig. 2E). Therefore, we conclude that the candidate gene controls the gh2 phenotype in rice.
Phylogenetic Analysis of GH2
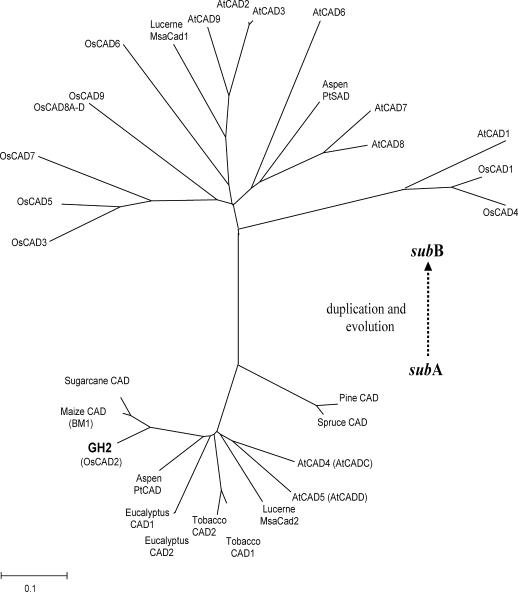
To determine the evolutionary relationship between GH2 and CAD family members from the rice and Arabidopsis CAD families as well as other representative identified CAD homologs, an unrooted tree was built using the neighbor-joining method based on full-length protein sequences (Fig. 3). The result indicates that all the CAD protein sequences are divided into two subfamilies: subA (GH2, sugarcane [Saccharum officinarum] CAD, maize CAD, aspen PtCAD, eucalyptus [Eucalyptus globules] CAD, tobacco [Nicotiana tabacum] CAD1, tobacco CAD2, Lucerne [Medicago sativa] MsaCad2, Arabidopsis CADC, Arabidopsis CADD, pine [Pinus taeda] CAD, and spruce [Picea abies] CAD) and subB (Lucerne MsaCad1, aspen PtSAD, OsCAD1, OsCAD3, OsCAD4, OsCAD5, OsCAD6, OsCAD7, OsCAD8A-D, OsCAD9, AtCAD1, AtCAD2, AtCAD3, AtCAD6, AtCAD7, AtCAD8, and AtCAD9).
Figure 3.
Phylogenetic analysis of GH2. A neighbor-joining tree was built on the full-length protein sequences of rice CAD family members, Arabidopsis CAD family members, and other representative identified CAD homologs. The scale bar is an indicator of genetic distance based on branch length. The subA subfamily includes rice GH2, sugarcane CAD, maize CAD, aspen PtCAD, eucalyptus CAD, tobacco CAD1, tobacco CAD2, Lucerne MsaCad2, Arabidopsis CADC, Arabidopsis CADD, pine CAD, and spruce CAD. The subB subfamily includes Lucerne MsaCad1, aspen PtSAD, rice OsCAD1, OsCAD3, OsCAD4, OsCAD5, OsCAD6, OsCAD7, OsCAD8A, OsCAD8B, OsCAD8C, OsCAD8D, OsCAD9, Arabidopsis AtCAD1, AtCAD2, AtCAD3, AtCAD6, AtCAD7, AtCAD8, and AtCAD9. Accession numbers of the sequences used to build the tree are as follows: AY288079 (AtCAD1), AY302077 (AtCAD2), AY302078 (AtCAD3), AY302081 (AtCAD4), AY302082 (AtCAD5), AY302075 (AtCAD6), AY302079 (AtCAD7), AY302080 (AtCAD8), AY302076 (AtCAD9), OsCAD1 (AAN09864), GH2 (DQ234272), OsCAD3 (AAP53892), OsCAD4 (BK003970), OsCAD5 (BK003971), OsCAD6 (CAD39907), OsCAD7 (CAE05207), OsCAD8A to D (BK003972), OsCAD9 (AAN05338), sugarcane CAD (AJ231135), maize CAD (AJ005702), spruce CAD (X72675), aspen CAD (AF217957), aspen SAD (AF273256), eucalyptus (AF038561), tobacco CAD1 (X62343), tobacco CAD2 (X62344), pine (Z37992), Lucerne MsaCAD1 (AAC35846), and Lucerne MsaCAD2 (AAC35845).
SubA members are evolutionally conserved CAD genes in gymnosperm and angiosperm, which has been regarded as the true CAD clade before angiosperm and gymnosperm split (Raes et al., 2003; Peter and Neale, 2004). In the complete CAD family of Arabidopsis, two CAD members (AtCADC and AtCADD) belong to subA subfamily. Only double mutation of the two subA CAD genes (AtCADC and AtCADD) can cause the lignin-deficient phenotype. Although rice has 12 CAD members, only GH2 belongs to the subA CAD subfamily. The fact that the point mutation of GH2 in rice has led to the lignin-deficient phenotype demonstrates that GH2 is the only primary enzyme that catalyzes the last step of lignin precursor biosynthesis. SubB is an angiosperm-specific subfamily, which can be regarded as the duplication of subA, and the members are likely to gain new functions in evolution. So far, Lucerne MsaCAD1 and Arabidopsis AtCAD7 in SubB have been proven to participate in plant defense, aspen PtSAD has been proven to catalyze sinapyl alcohol biosynthesis (Kiedrowski et al., 1992; Brill et al., 1999; Li et al., 2001), but the functions of rice subB subfamily remain to be elucidated. Based on the role of subB members in other plants, we predict that the subB CAD members in rice may function in enhancing the cell wall for plant defense or act in assistant roles of lignification.
GH2 Expression Pattern and CAD Activities of Total Proteins
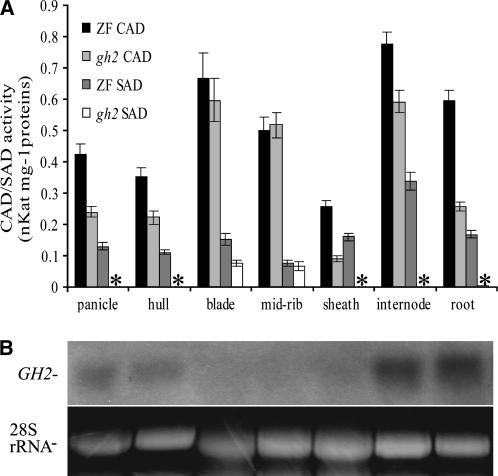
Since the mutation is a point mutation, GH2 gene expression was detected in the wild-type plants only at the heading stage (Fig. 4B). We detected GH2 gene transcription in all seven tissues, including panicles, hulls, blades, midribs, leaf sheaths, internodes, and young roots, with a 217-bp 3′ untranscriptional sequence as the specific probe. The results showed GH2 gene was expressed in the lignified tissues, including panicles, hulls, internodes, young roots, and leaf sheaths. In the leaves (blade and midrib), which are slightly lignified, the GH2 transcripts cannot be obviously detected by northern blot. Furthermore, GH2 gene expressed stronger in the first internode than in the second internode, suggesting that the gene expressed gradually stronger starting from the basal internode to the apical internode (data not shown). Consequently, the GH2 expression pattern is closely related to the rice lignification pattern and the localization of reddish-brown pigments.
Figure 4.
CAD activities of total proteins and GH2 expression pattern. A, CAD activity and SAD activity of the total proteins from different tissues of the wild-type and gh2 plants at the heading stage. The se bars were obtained from triplicate independent measurements. *, Not detectable. B, Northern-blot analysis of GH2 expression in different tissues of the wild type at the heading stage. Ethidium bromide staining of the 28S rRNA band is shown as the loading control. For both assays, the tissues include panicles, hulls, leaf blades, midribs, sheaths, the first two internodes, and young roots. The leaves eliminated of midrib (blades), midribs, and sheaths were all derived from flag leaves.
To study the catalytic ability of native GH2 in rice, CAD activity and SAD activity of the total proteins were assayed in all tissues of wild-type and gh2 plants at the heading stage. Strong CAD activity and weak SAD activity were detected in all the tissues of the wild type (Fig. 4A). In the gh2 sheaths, roots, panicles, hulls, and internodes in which GH2 gene was strongly or relatively strongly expressed, the CAD activity was drastically reduced and the SAD activity was not detectable, whereas, in the leaves (blade and midrib) in which GH2 gene was weakly expressed, the CAD activity and SAD activity were slightly reduced. The reductions are consistent with the expression pattern of GH2 described above (Fig. 4B). Thus, the result shows that GH2 protein has CAD activity and SAD activity, and the CAD activity and SAD activity were drastically reduced in the gh2 plants. In the leaves in which the GH2 gene is slightly expressed, the total proteins of the gh2 mutant still exhibit high CAD activity and weak SAD activity. Additionally, the reduction ratios of CAD activity in the different tissues are unequal. So we speculate it is possible that additional CAD isoenzymes with CAD activity also unevenly exist in different tissues of the plants. Most importantly, the fact that the SAD activity was completely abolished due to the defective GH2 protein in panicles, hulls, internodes, and sheaths shows that no other SAD homolog with obvious SAD activity exists in these tissues.
Enzyme Kinetics of the Recombinant GH2 and gh2
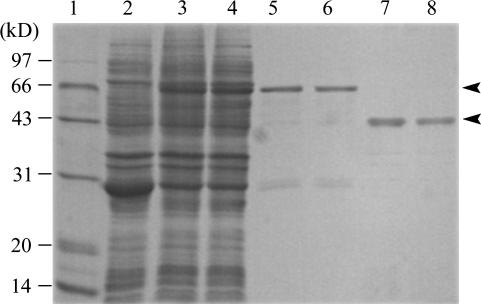
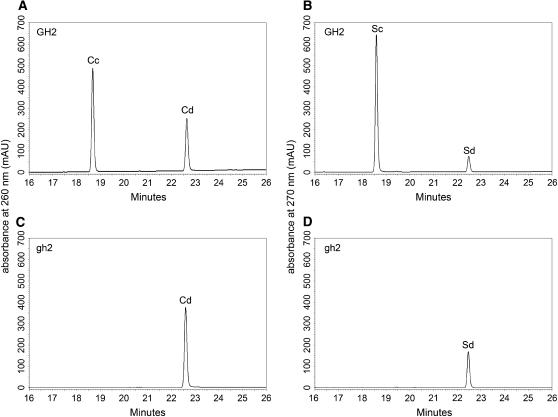
To evaluate the way in which the G to A nucleotide substitution impaired enzymatic function in the gh2 mutant plant, we expressed the wild-type protein GH2 and the point-mutated protein gh2 (G185D) in Escherichia coli. The recombinant proteins were purified by glutathione-Sepharose 4B affinity chromatography and cleaved by PreScission Protease on column (Fig. 5). The enzyme activities of the cleaved recombinant proteins were assayed toward the substrates coniferaldehyde and sinapaldehyde. GH2 exhibited strong CAD and SAD activities, while gh2 completely lost the CAD and SAD activities (Fig. 6; Table II). The result shows the point mutation G185D completely abolished the enzyme activities of GH2 for substrates coniferaldehyde and sinapaldehyde. Furthermore, the result confirms that native GH2 also completely lost the enzyme activities, and additional CAD isoenzymes with CAD activity exist in different tissues of rice.
Figure 5.
Expression and purification of recombinant GH2 and gh2 proteins in E. coli. Lane 1, The molecular mass ladder; lanes 2, 3, and 4, the crude extracts from induced recombinant E. coli of pGEX6P-1, pGEXGH2, and pGEXgh2; lanes 5 and 6, the purified glutathione S-transferase (GST)-GH2 and GST-gh2 (about 66 kD); lanes 7 and 8, the cleaved fusion proteins GH2 and gh2 (about 40 kD). The arrowheads point to the GST fusion proteins and the cleaved fusion proteins. Proteins were visualized by Coomassie Blue R-250 staining.
Figure 6.
CAD and SAD activities of purified recombinant GH2 and gh2. A and B, HPLC chromatograms of GH2-catalyzing coniferaldehyde and sinapaldehyde to produce coniferyl alcohol and sinapyl alcohol, showing GH2 exhibits strong CAD activity and SAD activity; C and D, HPLC chromatograms of gh2-catalyzing coniferaldehyde and sinapaldehyde to produce coniferyl alcohol and sinapyl alcohol, showing gh2 completely lost the CAD activity and SAD activity. AU, Absorbance units; Cd, coniferaldehyde; Cc, coniferyl alcohol; Sd, sinapaldehyde; Sc, sinapyl alcohol.
Table II.
Kinetics properties of the recombinant GH2 proteina
| Substrate | Km | Vmax | Kcat | Kenz | Specific Activity |
|---|---|---|---|---|---|
| μm | pKat μg−1 | s−1 | m−1 s−1 | pKat μg−1 | |
| Coniferaldehyde | 4.4 ± 0.2 | 32.9 ± 1.7 | 1.3 ± 0.1 | 288,640 | 23.3 ± 1.0 |
| Sinapaldehyde | 20.8 ± 2.3 | 87.0 ± 8.1 | 3.4 ± 0.3 | 161,540 | 59.5 ± 0.7 |
Values are means ± se for three independent assays.
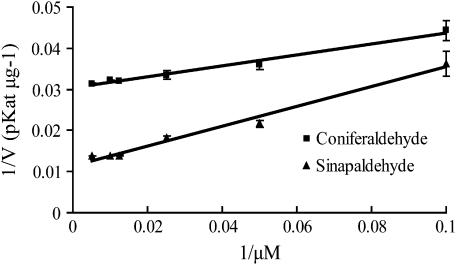
To further know the substrate specificity of GH2 in vitro, we study the kinetics of the purified recombinant GH2 toward the substrates coniferaldehyde and sinapaldehyde by HPLC analysis. The overall catalytic properties were revealed by Km, Vmax, Kcat, and Kenz (calculated by dividing Kcat by Km) values (Table II) calculated on the basis of the Lineweaver-Burk plots (Fig. 7). The Km value of GH2 for sinapaldehyde is 4.7-fold higher than that for coniferaldehyde, showing GH2 has higher affinity for coniferaldehyde than sinapylaldehyde. The Vmax value of GH2 for sinapylaldehyde is 2.6-fold higher than that for coniferaldehyde, indicating that the GH2 exhibits higher catalytic ability for coniferaldehyde than sinapylaldehyde. Based on the above, the specific constant Kenz (Kcat/Km) for coniferylaldehyde is 1.8-fold higher than that for sinapylaldehyde. The results suggest that coniferaldehyde and sinapaldehyde both are the favored substrates of GH2, but coniferaldehyde is the main substrate and sinapaldehyde is the secondary substrate.
Figure 7.
Lineweaver-Burk plots of GH2 for the substrates coniferaldehyde and sinapaldehyde: ▪, coniferaldehyde; ▴, sinapaldehyde. The se bars were obtained from triplicate assays.
Phloroglucinol and Mäule Staining of the Wild Type and gh2 Mutant
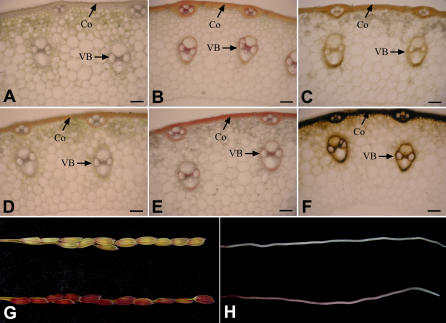
Phloroglucinol, a reagent traditionally used to detect cinnamyl aldehyde and lignin (Speer, 1987), appears to be more sensitive for cinnamyl aldehyde detection than other methods (Halpin et al., 1998). So the accumulated cinnamyl aldehydes in the internode sections, young roots, and young branch panicles were detected with phloroglucinol reagent. Obvious differences of staining patterns between wild type and gh2 were observed. In the wild-type plants, little phloroglucinol staining was evident, whereas in the mutant plants strong staining occurred in the internode sections, young branch panicles, and roots (Fig. 8, B, D, G, and H). When the fresh hand-cut internode sections were observed under light microscopy, the reddish-brown coloration and phloroglucinol staining are found in the same cortex (Co) region and vascular bundle (VB) region (Fig. 8, D and E). The results suggest that accumulated hydroxyl-cinnamaldehydes deposited in the high-lignified tissues or regions of the gh2 plants due to the deficient GH2 protein. Coniferaldehyde residues had been proven to be responsible for the reddish-brown color in woody internodes of caffeic acid O-methyltransferase (COMT) down-regulated transgenic plants (Tsai et al., 1998). Considering the same localization of the reddish-brown pigments and the hydroxyl-cinnamaldehydes, we speculate the phenotype of gh2 is caused by the polymerization of accumulated aldehydes into lignin.
Figure 8.
Phloroglucinol and Mäule staining of wild type and gh2. A and D, Transverse internode sections of wild type (A) and gh2 (D). The reddish-brown coloration deposited in the Co and VB regions. B and E, Transverse internode sections of the wild type and gh2 after phloroglucinol staining. C and F, Transverse internode sections of wild type (C) and gh2 (F) after Mäule staining. G, Young branch panicles of the wild type (top) and gh2 (bottom) after phloroglucinol staining. H, Young roots of wild type (top) and gh2 (bottom) after phloroglucinol staining. Bars = 100 μm.
The lignin composition of the wild-type and gh2 plants was also estimated by Mäule regent, which can stain G residues yellow and S residues red (Meyer et al., 1998). In the Mäule staining, the fresh hand-cut internode sections of wild type were stained reddish yellow in the Co region and purely yellow in the VB region, suggesting that the rice Co region contains abundant G monomers and a small amount of S monomer while the VB region mainly contains G monomers (Fig. 8C). In the corresponding tissues of the gh2 plants, the Co region and VB region were stained black-yellow by Mäule reagent (Fig. 8F). The phenomenon showed that the lignin monomers of the Co and VB regions were both altered in the gh2 mutant for the deficient GH2 protein, but the reason the tissues were stained black-yellow is unknown. The lignin properties of the gh2 mutants will be studied to explain the phenomenon.
gh2 Plants Have Altered Lignin Content and Composition
To further detect the lignin deficiency of the gh2 mutant plants, lignin content and composition were estimated in the wild-type and gh2 plants. Cell wall residue (CWR) was prepared from the dried mature hulls and internodes. Klason lignin analysis of the CWR showed that the total lignin content of the gh2 plants was reduced by 5% to 6% compared with the wild-type plants (Table III). The result demonstrates that GH2 is a critical enzyme that regulates lignin content in rice. Lignin composition of the wild-type and gh2 plants was also detected in the hulls and internodes by the derivatization followed by reductive cleavage (DFRC) method. The results show rice lignin consists of the three kinds of basal monomers, the H, G, and S monomers (Table III). In wild-type rice, G monomer is the major monomer and the G:S ratio is 60.66 in the hulls and 6.05 in the internodes, which is consistent with the Mäule staining (Fig. 8C). In the gh2, the three monomers are all reduced by approximately 35% in the hulls and approximately 25% in the internodes compared with the wild type. Although the reduction ratios are different in different tissues (hull and internode), the reductions of H, G, and S monomers in the same tissue are almost similar in degree. The results show that the GH2 enzyme takes the role of synthesizing H, G, and S monomer precursors in the hulls and internodes. The unequal reductions in hulls and internodes may be caused by the different lignification pattern between the hulls and internodes. Such analogous differences have been observed among Arabidopsis floral stems, roots, and hypocotyls (Sibout et al., 2003, 2005). Compared with the dramatic reductions in lignin monomers (25%–35%), the total lignin content reductions are quite low (only 5%–6%; Table III). The difference shows there are other substrates besides the three basal monomers participating in lignin biosynthesis.
Table III.
Klason and DFRC analysis of the wild type and gh2 mutanta
| Lignin Contentb
|
Lignin Monomer Composition
|
||||
|---|---|---|---|---|---|
| Total H Monomers | Total G Monomers | Total S Monomers | G:S | ||
| % CWR | μmol g−1 Klason lignin | ||||
| Wild-type hull | 34.6 ± 0.1 | 41.61 ± 6.35 | 688.44 ± 48.30 | 11.35 ± 1.28 | 60.66 |
| gh2 hull | 32.5 ± 0.1 | 29.20 ± 2.16 | 444.27 ± 14.26 | 7.63 ± 0.83 | 58.23 |
| Wild-type internode | 15.0 ± 0.1 | 54.36 ± 8.62 | 233.56 ± 46.66 | 38.60 ± 8.62 | 6.05 |
| gh2 internode | 14.2 ± 0.1 | 41.22 ± 3.70 | 164.95 ± 25.91 | 27.87 ± 4.37 | 5.91 |
The data correspond to the means and ses from triplicate analyses.
Klason lignin content (percentage of weight of the extract-free dried cell wall from the hull and internode).
DISCUSSION
The gh2 Plant Is Defective in Lignin Biosynthesis
gh mutants have long been used as morphological markers and classified into coloration mutant categories (Zeng et al., 2003; Kurata et al., 2005). We found the GH2 gene encodes a CAD by map-based cloning, which shows the gh2 mutant is a lignin-deficient mutant. Maize bm lignin mutants have been used as a model to study crop lignin digestibility. Following the first bm mutant documented in maize in 1924 (Barrière et al., 2004), bm phenotype has also been identified in sorghum (Sorghum bicolor) and millet (Panicum miliaceum), which are all diploid C4 plants (Cherney et al., 1991). However, no corresponding bm mutant has been found in rice, to our knowledge. In this study, the gh2 mutant has been proven to be the so-called rice bm mutant.
Comparing the maize CAD mutant bm1 with the rice gh2 mutant in our study, the phenotype difference between bm1 and gh2 is that maize bm1 mutant plants have reddish-brown pigments in the leaf midrib and stalk pith, whereas rice gh2 mutant plants have the pigments in the hull, internode, and basal leaf sheath. In truth, the reddish-brown pigments in the two mutants are all associated with the high-lignified tissues. When the expression pattern of CAD genes was investigated, maize CAD gene was expressed strongly in leaves but GH2 gene was slightly expressed in the same tissue (Fig. 4B). So it is likely that the phenotype difference may be caused by the expression pattern or loss-of-function extent of the CAD genes between bm1 and gh2. The identification of the gh phenotype provides an important clue for finding lignin mutants in grass crops such as wheat (Triticum aestivum) and barley (Hordeum vulgare).
Rice is the first crop plant whose genome has been sequenced (International Rice Genome Sequencing Project, 2005). Many characterized genes in maize have homologs in rice (Gaut and Doebley, 1997; Devos and Gale, 2000). With the advantages of a fully sequenced genome and an effective transformation system, rice seems to be the best model in the grass family to study lignin biosynthesis. Therefore, cloning and characterization of the other rice GH genes will be critical for us to get a deep understanding of lignin biosynthesis in grasses.
GH2 Is a Multifunctional Enzyme
We conclude GH2 is the primary CAD that acts as a multifunctional CAD enzyme to synthesize coniferyl alcohol and sinapyl alcohol precursors in lignified tissues of rice for the following reasons. First, in the rice CAD family, GH2 is the only one CAD belonging to the subA subfamily, which is widely proven to participate in constitutive lignin biosynthesis (Fig. 3). Second, when the GH2 gene was mutated, the CAD and SAD activities of both the native and purified recombinant GH2 protein were drastically reduced relative to the wild type (Figs. 4A and 6). Third, the kinetics properties of recombinant GH2 on the substrates of coniferaldehyde and sinapaldehyde show GH2 exhibits strong catalytic ability toward coniferaldehyde and sinapaldehyde (Table II). Finally, the G monomers and S monomers were reduced to a similar degree in the gh2 mutant plants compared with the wild type (Table III).
The difference between gymnosperm and angiosperm lignin is that gymnosperm plants contain mainly G monomers and monogenic CAD gene, whereas angiosperm plants consist of G monomers, S monomers, and one CAD family (Galliano et al., 1993; MacKay et al., 1997; Baucher et al., 1999). Consistent with this, CAD enzymes extracted from gymnosperm plants are quite specific for the reduction of coniferaldehyde (Kutsuki et al., 1982; Galliano et al., 1993), whereas CAD enzymes extracted from the gymnosperm plants have a significant affinity for both coniferaldehyde and sinapaldehyde (Mansell et al., 1974; Hawkins and Boudet, 1994; Brill et al., 1999). The relations between lignin composition and CAD enzyme activities in different plants have led to a hypothesis that a specialized CAD protein is involved in either sinapyl alcohol or coniferyl alcohol biosynthesis (Hawkins and Boudet, 1994). This hypothesis is supported by the identification of aspen PtCAD and PtSAD, which can specifically synthesize coniferyl alcohol and sinapyl alcohol, respectively (Li et al., 2001). On the contrary, Sibout et al. (2005) have recently proven that AtCADC and AtCADD are the primary CAD genes participating in lignin biosynthesis of the floral stems in Arabidopsis and speculated that no specific SAD is needed to synthesize sinapyl alcohol. Furthermore, aspen PtCAD cDNA has been found to be able to complement the cad-c cad-d double mutant (Sibout et al., 2005), completely restoring the level of conventional G and S units, while PtSAD cDNA only partially complemented the S monomer reduction. In the study, GH2 protein has been proven to have strong CAD activity and SAD activity (Fig. 6; Table II), and no additional SAD with obvious SAD activity was detectable in the highly lignified tissues, such as hull, sheath, and internode (Fig. 4A). Our results suggest GH2 exhibits multifunctional activities and no additional specific SAD exists in rice highly lignified tissues.
The available data lead to a paradox that questions whether there is a specific SAD in angiosperm plants. We think the conflicting data may be interpreted by elucidating the source of the S monomers. The proportion of S monomers in the total lignin monomers changes greatly among various kinds of species (Sederoff et al., 1999). In detail, S monomers exist in the wood angiosperm plants with high proportion, in the herbaceous angiosperm plants with relatively low proportion, and cannot be detected in the gymnosperm plants (Baucher et al., 1996; Campbell and Sederoff, 1996; Sibout et al., 2005). At present, F5H, COMT, and SAD (including the SAD activity of CAD) are regarded as the three factors that can affect the biosynthesis of S monomers (Boerjan et al., 2003). The alterations of the expression pattern of the three kinds of genes in plants have been found to be able to influence the proportion of S lignin monomers in lignin (Vignols et al., 1995; Meyer et al., 1998; Piquemal et al., 2002). Among different species, especially between wood angiosperm and herbaceous angiosperm, the different proportions of S monomers in total monomers may be caused by the diversities of the three kinds of genes, including gene copy members, expression patterns, or protein biochemical properties. So in some species such as aspen, it is also possible for some subfamily members to gain SAD function to act in an assistant role in maintaining enough sinapyl alcohol concentration for lignin polymerization in specific development phage or tissue. Although GH2 has been proven to be the primary CAD gene in rice lignin biosynthesis, the reason rice lignin consists of a low proportion of S monomers remains unknown. Maize is a grass plant that has a relatively high proportion of S monomers (Halpin et al., 1998). Further study on the three factors of F5H, COMT, and CAD genes in rice and maize, which is under way, will be very useful for understanding the mechanism of low S monomer composition in rice.
Potential Application of the gh2 Mutant in Agriculture
Rice is an important crop that feeds more than half of the world's population. Effective utilization of rice by-products will definitely increase the income of rice producers. Rice residue has the potential to be used for bio-oil extraction by pyrolysis, for mushroom growth, and even for dietary fiber preparation (Kong et al., 2005). However, the most important applications are for livestock feeding, pulp, and paper making (Reddy and Yang, 2005). Lignin content and composition are the most important traits that limit the above applications. Point mutation of gh2 has been proven to affect lignin content and composition without influencing plant development under normal cultivar conditions, and further study on the mutant may improve the qualities of the rice residues. The molecular cloning of other gh mutants will help to discover new genes controlling lignin content and composition. Just like the wide application of maize bm mutants in agricultural production, cloning of GH genes and their application in rice will arouse many agricultural interests in the future.
MATERIALS AND METHODS
Materials
The original rice (Oryza sativa) gh2 mutant (RGS no. 6) was kindly provided by Dr. Khush at the International Rice Research Institute, The Philippines. The gh2 gene was transferred to Zhefu802 (spp. indica) by 10 rounds of backcrosses with Zhefu802 (Zeng et al., 2003). We obtained the isogenic line with the gh2 gene and also named it as gh2, while Zhefu802 represents the wild type. The gh2 was crossed with a japonica rice variety, Nipponbare, to construct the F2 mapping population.
Genetic Analysis and Marker Development
The primary locus of GH2 locus was determined by newly developed marker P1 and reported STS marker S21668. To fine map the gh2, seven STS markers and two CAPS markers (Table I) were developed on comparisons of original or CAPS length between indica var. 9311 and japonica var. Nipponbare according to the data published in http://www.ncbi.nlm.nih.gov.
Complementation Test
A 8.44-kb genomic DNA fragment containing the entire GH2 coding region, the 2,397-bp upstream sequence, and the 2,460-bp downstream sequence was inserted into the binary vector pCAMBIA1300 to generate the transformation plasmid pCGH for complementation test. A control plasmid, pCGHS, containing truncated GH2 gene also was constructed according to the strategy described previously (Li et al., 2003a). The two binary plasmids were introduced into Agrobacterium tumefaciens EHA105 by electroporation, and transformed to rice for complementation testing according to a published method (Hiei et al., 1994).
Phylogenetic Analyses
The full-length CAD protein sequences were retrieved from GenBank and used for phylogenetic analyses according to the methods described by Li et al. (2003b). Multiple sequence alignments were conducted using ClustalX version 8.0 with the PAM matrix. A neighbor-joining tree was built using MEGA version 2.1 adopting Poisson correction distance and was presented using radiation TreeView. Support for the tree obtained was assessed using the bootstrap method with 1,000 replicates. The Arabidopsis (Arabidopsis thaliana) CAD family was named as described by Kim et al. (2004) and Sibout et al. (2003). The rice CAD family was named as described by Tobias and Chow (2005) except that OsCAD2 was renamed as GH2.
RNA Extraction and Gel Blot
Total RNA was extracted from tissues of wild-type plants at the heading stage with TRIZOL reagent (Invitrogen). Twenty micrograms of total RNA per lane was electrophoresed on 1% agarose-formaldehyde gels and capillary blotted onto Hybond-N+ membranes (Amersham). The 217-bp 3′-terminal untranscriptional fragment of the GH2 gene was labeled with α-32p-dCTP using the primer-a-gene labeling system (Promega) as the specific probe, and ethidium bromide staining of the 28S ribosomal RNA band was used as the loading control. The membrane was incubated in standard prehybridization solution at 65°C for 6 h and then hybridized with the α-32P-labeled probe at 65°C for 16 h. Following hybridization and sequential washing, the radioactive membrane was then exposed to x-ray film.
Plant Protein Extraction and Enzyme Assay
Plant total proteins were extracted as the method described by Halpin et al. (1998) with a little modification. Rice fresh tissues (about 300 mg) at the heading stage were separately milled to fine power in liquid N2, then extracted with 1,000 μL extraction buffer (100 mm Tris-HCl, pH 7.5; 2% polyethylene glycol 6000, 5 mm dithiothreitol, 2% polyvinylpolypyrrolidone) for 2.5 h at 4°C. The supernatant was decanted and about 100 μg total proteins were used for CAD activity and SAD activity assay. Assays of crude enzyme were carried out using coniferyl alcohol and sinapyl alcohol as the substrates, and the formation of hydroxy-cinnamaldehydes was monitored spectrophotometrically (TU-1201, GENERAL) at 400 nm using the following molar extinction coefficients: coniferaldehyde 2.10 × 104 m−1 cm−1 and sinapaldehyde 1.68 × 104 m−1 cm−1, as described by Sibout et al. (2003). Assays were carried out at 30°C for 10 min in 500 μL of 100 mm Tris-HCl, pH 8.8, 100 μm NADP, and 250 μm of coniferyl alcohol or sinapyl alcohol. The enzyme reactions were initiated by enzyme addition and stopped by holding at 85°C for 10 min. An assay without NADP was used as the control. Protein concentration was determined using the Bradford reagent. Resulting units are defined as the amount of activity that converts 1 nmol of hydroxy-cinnamyl alcohol into the corresponding aldehyde per second (1 nKatal) per microgram of crude protein extract.
GH2 and gh2 Expression in Escherichia coli
Both GH2 and gh2 full-length cDNAs were isolated by reverse transcriptase-PCR from the total RNA from gh2 and wild-type plant roots with RT-PCR system (Invitrogen) using primer 1 (5′-CTTCTTGTTCTTGTTCTCTT-3′) and primer 2 (5′-GCCGAATTTATACCGGAAAG-3′). The PCR products were inserted into pGEM-T vectors and sequenced to obtain the correct clones pGEMGH2 and pGEMgh2. The ORF sequences of GH2 and gh2 were amplified from cDNA clones pGEMGH2 and pGEMgh2 using primer 3 (5′-GCGGGATCCATGGGCAGCCTCGCCGCCGA-3′) and primer 4 (5′-GCGGAATTCCTAGCGGACGTCGTTGCGCT-3′). The primers incorporated a BamH site at the N-terminal end and an EcoRI site at the C-terminal end of the ORF. The PCR products were digested and cloned into the corresponding site of pGEX-6P-1 to generate pGEXGH2 and pGEXgh2, sequenced to assure GH2 and gh2 sequence, and then introduced into E. coli BL21 for protein expression. The bacteria containing pGEXGH2 or pGEXgh2 were cultured overnight at 37°C, diluted to 400 times, and grown at 37°C for about 3 h to an OD600 of 0.6, then transferred to 18°C for 16 h after adding isopropyl-β-d-thiogalactopyranoside to 0.5 mm. The cells were collected, and lysed by moderate ultrasonic. The recombinant GH2 and gh2 protein were purified by glutathione-Sepharose 4B affinity chromatography and cleaved by PreScission Protease on a column according to the manufacturer's instructions (Pharmacia Biotech). The purified proteins were quantified by the bicinchoninic acid method and used for the enzyme kinetics analysis.
HPLC Analysis of Enzyme Functions and Reaction Kinetics
For the substrate-specific activity test, the enzyme reaction mixture contained 100 mm Tris-HCl, pH 8.8, buffer, 200 μm NADPH, 200 μm aldehyde substrate in a final volume of 250 μL, and 1.2 μg of purified recombinant GH2 or gh2 protein were used. The reactions were for 10 min at 30°C and stopped by adding glacial acetic acid (10 μL). For the kinetic analyses, the enzyme reaction mixture contained 100 mm Tris-HCl, pH 8.8, buffer, 200 μm NADPH, 0.5 μg purified recombinant protein, and varying concentrations (10–200 μm) of coniferaldehyde (Aldrich, High Purity) or sinapaldehyde (Apin Chemicals, High Purity) in a final volume of 250 μL. Enzymatic reactions were initiated by cinnamyl aldehyde addition and, after 4 min incubation at 30°C, were stopped by adding glacial acetic acid (10 μL). All the reactions were carried out with NADPH-free mixture as the control. An aliquot (50 μL) of each assay mixture was subjected to reverse-phase HPLC analysis with UV detection for the product identification. Authentic coniferyl alcohol (Aldrich, High Purity) and sinapyl alcohol (Aldrich, technical grade) were used as the standards for qualifying and quantifying the hydroxy-cinnamyl alcohol products. The kinetics values, Km, Vmax, Kcat, and Kenz, were defined as described by Kim et al. (2004). Reverse-phase HPLC (SCL-10AVP SHIMADZU) analysis was carried out using a 5μ C18 column (250 × 4.60 mm, SUPELCOSIL LC-18, SUPELCO) maintained at 30°C. A linear gradient of acetonitrile in water (all solution contained 0.1% trifluoroacetic acid) at 1 mL/min for 30 min was used to separate the substrates and products. The HPLC methods were as follows: coniferaldehyde and sinapaldehyde, λ 340 nm, 10% to 47% acetonitrile; coniferyl alcohol, λ 260 nm, 10% to 30% acetonitrile; and sinapyl alcohol, λ 270 nm, 10% to 25% acetonitrile.
Lignin Analysis
Histochemical Staining
For histochemical localization of the accumulated cinnamyl aldehydes, phloroglucinol staining was performed according to a standard protocol (Halpin et al., 1998) with a little modification. Fresh hand-cut sections from rice second internode, young roots, and branch panicles at the heading stage were incubated for 10 min in phloroglucinol solution (1% in ethanol:water [70:30]), the phloroglucinol was removed and treated with 18% HCl for 5 min, then photographed using a color digital camera directly or under light microscope (Olympus BX51). Mälue staining was according to Chapple's method (Chapple et al., 1992) with a few modifications. Fresh hand-cut sections of the second internode were treated with 0.5% KMnO4 for 10 min, rinsed with water, treated with 10% HCl for 2 min, rinsed with water again, then mounted in concentrated NH3·H2O and examined immediately under light microscopy and photographed as above.
Lignin Content and Composition Analysis
Dried mature internodes and hulls were collected after the removal of leaves, leaf sheaths, and the seeds. The milled hull or internode power was sieved through a 0.5-mm screen, washed in phosphate buffer (50 mm, pH 7.2) three times, and extracted with 70% ethanol twice at 70°C for 1 h. The CWR was dried in a vacuum and used to measure lignin content and composition. Lignin content was quantified according to Li et al. (2003b). Lignin compositions were assayed using the DFRC protocol (Lu and Ralph, 1997).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number DQ234272 (GH2).
Acknowledgments
We thank Dr. John Ralph (University of Wisconsin, Madison, WI) for supplying authentic standards for DFRC analysis, Dr. Khush (International Rice Research Institute, The Philippines) for providing the original gh2 mutant, Dr. Mu Zhang (Institute of Genetics and Developmetal Biology, Chinese Academy of Sciences) for assistance in gas chromatography-mass spectrometry analysis. We also thank the two anonymous reviewers and the editor for critical comments on the manuscript.
This work was supported by the Ministry of Sciences and Technology of China (grant no. 2005CB120805), the Chinese Academy of Sciences, and the National Natural Science Foundation of China (grant nos. 30325008 and 30530070).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Zhukuan Cheng (zkcheng@genetics.ac.cn).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.073007.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Barrière Y, Ralph J, Mechin V, Guillaumie S, Grabber JH, Argillier O, Chabbert B, Lapierre C (2004) Genetic and molecular basis of grass cell wall biosynthesis and degradability. II. Lessons from brown-midrib mutants. C R Biol 327: 847–860 [DOI] [PubMed] [Google Scholar]
- Baucher M, Bernard-Vailhé MA, Chabbert B, Besle JM, Opsomer C, Van Montagu M, Botterman J (1999) Down-regulation of cinnamyl alcohol dehydrogenase in transgenic alfalfa (Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Mol Biol 39: 437–447 [DOI] [PubMed] [Google Scholar]
- Baucher M, Chabbert B, Pilate G, Van Doorsselaere J, Tollier MT, Petit-Conil M, Cornu D, Monties B, Van Montagu M, Inze D, et al (1996) Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol 112: 1479–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Brill EM, Abrahams S, Hayes CM, Jenkins CL, Watson JM (1999) Molecular characterisation and expression of a wound-inducible cDNA encoding a novel cinnamyl-alcohol dehydrogenase enzyme in lucerne (Medicago sativa L.). Plant Mol Biol 41: 279–291 [DOI] [PubMed] [Google Scholar]
- Campbell MM, Sederoff RR (1996) Variation in lignin content and composition: mechanisms of control and implications for the genetic improvement of plants. Plant Physiol 110: 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabannes M, Ruel K, Yoshinaga A, Chabbert B, Jauneau A, Joseleau JP, Boudet AM (2001) In situ analysis of lignins in transgenic tobacco reveals a differential impact of individual transformations on the spatial patterns of lignin deposition at the cellular and subcellular levels. Plant J 28: 271–282 [DOI] [PubMed] [Google Scholar]
- Chapple CC, Vogt T, Ellis BE, Somerville CR (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherney DJ, Patterson JA, Johnson KD (1990) Digestibility and feeding value of pearl millet as influenced by the brown-midrib, low-lignin trait. J Anim Sci 68: 4345–4351 [DOI] [PubMed] [Google Scholar]
- Cherney JH, Cherney DJR, Akin DE, Axtell JD (1991) Potential of brown-midrib, low-lignin mutants for improving forage quality. Adv Agron 46: 157–198 [Google Scholar]
- Devos KM, Gale MD (2000) Genome relationships: the grass model in current research. Plant Cell 12: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson LA (2001) Lignification and lignin topochemistry: an ultrastructural view. Phytochemistry 57: 859–873 [DOI] [PubMed] [Google Scholar]
- Ebling TL, Kung L Jr (2004) A comparison of processed conventional corn silage to unprocessed and processed brown midrib corn silage on intake, digestion, and milk production by dairy cows. J Dairy Sci 87: 2519–2526 [DOI] [PubMed] [Google Scholar]
- Galliano H, Cabane M, Eckerskorn C, Lottspeich F, Sandermann H Jr, Ernst D (1993) Molecular cloning, sequence analysis and elicitor-/ozone-induced accumulation of cinnamyl alcohol dehydrogenase from Norway spruce (Picea abies L.). Plant Mol Biol 23: 145–156 [DOI] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF (1997) DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA 94: 6809–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin C, Holt K, Chojecki J, Oliver D, Chabbert B, Monties B, Edwards K, Barakate A, Foxon GA (1998) Brown-midrib maize (bm1): a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J 14: 545–553 [DOI] [PubMed] [Google Scholar]
- Halpin C, Knight ME, Foxon GA, Campbell MM, Boudet AM, Boon JJ, Chabbert B, Tollier MT, Schuch W (1994) Manipulation of lignin quality by downregulation of cinnamyl alcohol dehydrogenase. Plant J 6: 339–350 [Google Scholar]
- Hawkins SW, Boudet AM (1994) Purification and characterization of cinnamyl alcohol dehydrogenase isoforms from the periderm of eucalyptus gunnii hook. Plant Physiol 104: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, Tsai CJ, Chiang VL (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 17: 808–812 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Iwata N, Omura T (1971) Linkage analysis by reciprocal translocation method in rice plants (Oryza sativa L.). II. Linkage groups corresponding to the chromosomes 5, 6, 8, 9, 10 and 11. Sci Bull Fac Agric Kyushu Univ 25: 137–153 [Google Scholar]
- Iwata N, Omura T (1977) Linkage studies in rice (Oryza sativa L.) on some mutants derived from chronic gamma irradiation. J Fac Agric Kyushu Univ 21: 117–127 [Google Scholar]
- Jones L, Ennos AR, Turner SR (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J 26: 205–216 [DOI] [PubMed] [Google Scholar]
- Jung HG, Vogel KP (1986) Influence of lignin on digestibility of forage cell wall material. J Anim Sci 62: 1703–1712 [DOI] [PubMed] [Google Scholar]
- Kiedrowski S, Kawalleck P, Hahlbrock K, Somssich IE, Dangl JL (1992) Rapid activation of a novel plant defense gene is strictly dependent on the Arabidopsis RPM1 disease resistance locus. EMBO J 11: 4677–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Kim MR, Bedgar DL, Moinuddin SG, Cardenas CL, Davin LB, Kang C, Lewis NG (2004) Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc Natl Acad Sci USA 101: 1455–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Xie J, Wu X, Huang Y, Bao J (2005) Rapid prediction of acid detergent fiber, neutral detergent fiber, and acid detergent lignin of rice materials by near-infrared spectroscopy. J Agric Food Chem 53: 2843–2848 [DOI] [PubMed] [Google Scholar]
- Kurata N, Miyoshi K, Nonomura K, Yamazaki Y, Ito Y (2005) Rice mutants and genes related to organ development, morphogenesis and physiological traits. Plant Cell Physiol 46: 48–62 [DOI] [PubMed] [Google Scholar]
- Kutsuki H, Shimada M, Higuchi T (1982) Regulatory role of cinnamyl alcohol dehydrogenase in the formation of guaiacyl and syringyl lignins. Phytochemistry 21: 19–23 [Google Scholar]
- Li L, Cheng XF, Leshkevich J, Umezawa T, Harding SA, Chiang VL (2001) The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell 13: 1567–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D, Wang X, Liu X, Teng S, Hiroshi F, et al (2003. a) Control of tillering in rice. Nature 422: 618–621 [DOI] [PubMed] [Google Scholar]
- Li Y, Qian Q, Zhou Y, Yan M, Sun L, Zhang M, Fu Z, Wang Y, Han B, Pang X, et al (2003. b) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15: 2020–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu FC, Ralph J (1997) Derivatization followed by reductive cleavage (DFRC Method), a new method for lignin analysis: protocol for analysis of DFRC monomers. J Agric Food Chem 45: 2590–2592 [Google Scholar]
- MacKay JJ, O'Malley DM, Presnell T, Booker FL, Campbell MM, Whetten RW, Sederoff RR (1997) Inheritance, gene expression, and lignin characterization in a mutant pine deficient in cinnamyl alcohol dehydrogenase. Proc Natl Acad Sci USA 94: 8255–8260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansell RL, Gross GG, Stockigt J, Franke H, Zenk MH (1974) Purification and properties of cinnamyl alcohol dehydrogenase from higher plants involved in lignin biosynthesis. Phytochemistry 13: 2427–2435 [Google Scholar]
- Meyer K, Shirley AM, Cusumano JC, Bell-Lelong DA, Chapple C (1998) Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc Natl Acad Sci USA 95: 6619–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao S, Takahashi M (1963) Trial construction of twelve linkage groups in Japanese rice: genetical studies on rice plant, XXVII. J Fac Agric Hokkaido Univ 53: 72–130 [Google Scholar]
- Peter G, Neale D (2004) Molecular basis for the evolution of xylem lignification. Curr Opin Plant Biol 7: 737–742 [DOI] [PubMed] [Google Scholar]
- Piquemal J, Chamayou S, Nadaud I, Beckert M, Barriere Y, Mila I, Lapierre C, Rigau J, Puigdomenech P, Jauneau A, et al (2002) Down-regulation of caffeic acid O-methyltransferase in maize revisited using a transgenic approach. Plant Physiol 130: 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, MacKay JJ, Hatfield RD, O'Malley DM, Whetten RW, Sederoff RR (1997) Abnormal lignin in a loblolly pine mutant. Science 277: 235–239 [DOI] [PubMed] [Google Scholar]
- Reddy N, Yang Y (2005) Biofibers from agricultural byproducts for industrial applications. Trends Biotechnol 23: 22–27 [DOI] [PubMed] [Google Scholar]
- Sanchez AC, Khush GS (1994) Chromosomal location of some marker genes in rice using the primary trisomics. J Hered 85: 297–300 [Google Scholar]
- Sederoff RR, MacKay JJ, Ralph J, Hatfield RD (1999) Unexpected variation in lignin. Curr Opin Plant Biol 2: 145–152 [DOI] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Seguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Pollet B, Goujon T, Mila I, Granier F, Seguin A, Lapierre C, Jouanin L (2003) Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis: isolation and characterization of the corresponding mutants. Plant Physiol 132: 848–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer EO (1987) A method of retaining phloroglucinol proof of lignin. Stain Technol 62: 279–280 [DOI] [PubMed] [Google Scholar]
- Taylor CC, Allen MS (2005) Corn grain endosperm type and brown midrib3 corn silage: site of digestion and ruminal digestion kinetics in lactating cows. J Dairy Sci 88: 1413–1424 [DOI] [PubMed] [Google Scholar]
- Tobias CM, Chow EK (2005) Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification. Planta 220: 678–688 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Popko JL, Mielke MR, Hu WJ, Podila GK, Chiang VL (1998) Suppression of O-methyltransferase gene by homologous sense transgene in quaking aspen causes red-brown wood phenotypes. Plant Physiol 117: 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignols F, Rigau J, Torres MA, Capellades M, Puigdomenech P (1995) The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng DL, Qian Q, Dong GJ, Zhu XD, Dong FG, Teng S, Guo LB, Cao LY, Cheng SH, Xiong ZM (2003) Development of isogenic lines of morphological markers in indica rice. Acta Bot Sin 45: 1116–1120 [Google Scholar]