Abstract
Gene regulatory pathways converge at the level of transcription, where interactions among regulatory genes and between regulators and target genes result in the establishment of spatiotemporal patterns of gene expression. The growing identification of direct target genes for key transcription factors (TFs) through traditional and high-throughput experimental approaches has facilitated the elucidation of regulatory networks at the genome level. To integrate this information into a Web-based knowledgebase, we have developed the Arabidopsis Gene Regulatory Information Server (AGRIS). AGRIS, which contains all Arabidopsis (Arabidopsis thaliana) promoter sequences, TFs, and their target genes and functions, provides the scientific community with a platform to establish regulatory networks. AGRIS currently houses three linked databases: AtcisDB (Arabidopsis thaliana cis-regulatory database), AtTFDB (Arabidopsis thaliana transcription factor database), and AtRegNet (Arabidopsis thaliana regulatory network). AtTFDB contains 1,690 Arabidopsis TFs and their sequences (protein and DNA) grouped into 50 (October 2005) families with information on available mutants in the corresponding genes. AtcisDB consists of 25,806 (September 2005) promoter sequences of annotated Arabidopsis genes with a description of putative cis-regulatory elements. AtRegNet links, in direct interactions, several hundred genes with the TFs that control their expression. The current release of AtRegNet contains a total of 187 (September 2005) direct targets for 66 TFs. AGRIS can be accessed at http://Arabidopsis.med.ohio-state.edu.
Genome-wide gene regulatory networks govern the phenotypic states of different cell types, tissues, and developmental stages in eukaryotic organisms (Wellmer and Riechmann, 2005). The gene regulatory networks converge at the level of transcription, where the DNA-binding transcription factors (TFs) recognize cis-regulatory elements in the promoter regions of target genes. It is estimated that approximately 5% of the genes in the genome of a eukaryotic organism encode TFs (Riechmann and Ratcliffe, 2000; Riechmann et al., 2000). TFs and the cis-regulatory elements to which they bind represent the protein-DNA interactions and act as the nodal points of gene regulatory networks (Blais and Dynlacht, 2005; Xing and van der Laan, 2005).
Over the last 50 years, molecular biology has provided key insights into the stunning array of gene-regulatory network components. The advent of high-throughput experimental technologies, such as ChIP-chip, gene expression arrays, and yeast two-hybrid, has generated vast amounts of data that describe protein-protein and protein-DNA interactions (Harmer et al., 2000; Gao et al., 2004), which are the key features of the gene regulatory networks. A first toward understanding the gene regulatory network architecture and its associated biological meaning is to integrate and organize the expanding information into databases, and to present the information in a visual form that is accessible for analysis by the experimentalist.
There has been a dramatic increase in the number of large-scale comprehensive databases that provide useful resources to the community on, for example, biochemical pathways (e.g. the Kyoto Encyclopedia of Genes and Genomes [Kanehisa and Goto, 2000], AraCyc [Mueller et al., 2003], and MapMan [Thimm et al., 2004]), protein-protein interactions (e.g. the Biomolecular Interaction Network Database), or systems like Dragon Plant Biology Explorer and Pathway Miner for integrating associations in metabolic networks and ontologies. Other databases, such as Regulon DB (Huerta et al., 1998), PlantCARE (Rombauts et al., 1999), PLACE (Higo et al., 1999), Eukaryotic Promoter Database (Perier et al., 2000), Transcription Regulatory Regions Database (Kolchanov et al., 2002), AthaMap (Steffens et al., 2004), and TRANSFAC (Wingender et al., 2000) store information related to transcriptional regulation. We previously developed AtcisDB (Arabidopsis thaliana cis-regulatory database) and AtTFDB (Arabidopsis thaliana transcription factor database) as part of the Arabidopsis Gene Regulatory Information Server (AGRIS; Davuluri et al., 2003) to annotate Arabidopsis (Arabidopsis thaliana) promoter sequences, the corresponding cis-regulatory elements, and TFs. The development of an enhanced Web-based portal with additional features and functionalities resulted in a significant expansion of both AtcisDB and AtTFDB, as well as in the addition of a new database (AtRegNet, for the Arabidopsis thaliana regulatory network). AtRegNet visualizes regulatory networks and allows quick access and download of any Arabidopsis sequences with information on direct targets and their interactions, providing a significant new resource to the Arabidopsis research community. With all the above-mentioned information integrated as one resource, AGRIS has become a primary resource for establishing regulatory networks and expression for all Arabidopsis genes.
RESULTS AND DISCUSSION
AGRIS is a user-friendly online database tool conceived as a resource for retrieving information regarding Arabidopsis promoters, cis-regulatory elements, TFs, and their interactions into regulatory networks. AGRIS currently consists of three integrated databases, AtcisDB, AtTFDB, and AtRegNet, which talk to each other in multiple ways. These three databases, used in combination, provide a powerful resource for research related to TFs, cis-regulatory elements, and the interactions between them. The Web site is updated once every 3 months by semiautomated and manual search processes, based on new results obtained from the literature for TF families, the insightful comments of curators for specific TF families, novel binding-site motifs, and the identification of new direct and indirect targets for TFs.
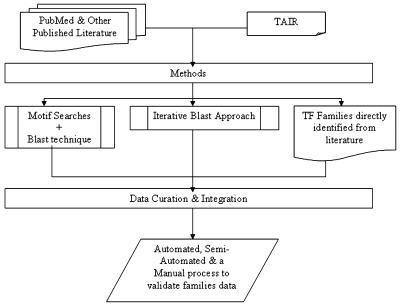
AtTFDB
AtTFDB contains information on TFs. TFs are grouped into families based on the presence of conserved domains and following prior classifications of plant TFs (Riechmann et al., 2000). There are differences between the previously estimated numbers of TFs per family (Riechmann and Ratcliffe, 2000; Riechmann et al., 2000) and those listed on AGRIS. This is due to continuous refinement of genome annotations since the first publication in December 2000, and the increasing experimental data that result in the addition of several new TFs each quarter. Several families were identified directly from published literature through a manual curation process (Fig. 1). Expert curators for specific families and personnel in our labs are constantly making sure that our data are accurate and up to date. The current version of AtTFDB contains 50 families and 1,690 TFs. Since the previous version of AtTFDB (Davuluri et al., 2003), 16 new families and 315 new TFs have been identified and added to the database. As an additional new feature, links are provided for the binding sites of TFs, when known. The breakdown of TFs into families and the queries used for their identification are shown in Table I. AtTFDB provides hidden Markov models, ClustalW alignments, references, and protein and nucleotide sequences for each family listed and links to the databases Munich Information Center for Protein Sequences (MIPS) Arabidopsis thaliana Database (Schoof et al., 2004), SALK (Borevitz and Ecker, 2004), The Arabidopsis Information Resource (TAIR; Rhee et al., 2003), and The Institute for Genomic Research (TIGR; Wortman et al., 2003) for each gene in the family.
Figure 1.
TF family identification process.
Table I.
TF families in AtTFDB
The main queries used to initially identify and classify TFs into different families are mentioned in the third column. Subsequent addition of members was done from the literature. Zn, Zinc.
| TF Family | No. of Members in AtTFDB | Members Initially Identified by: |
|---|---|---|
| AB13VP1 | 20 | Identify domain and BLAST |
| Alfin-like | 7 | Iterative BLAST |
| AP2-EREBP | 128 | Identify domain and BLAST |
| ARF | 24 | Identify domain and BLAST |
| ARID | 9 | Identified in literature |
| ARR-B | 15 | Identify domain and BLAST |
| AtRKD | 5 | Identified in literature |
| BBR/BPC | 7 | Identified in literature |
| bHLH | 162 | Iterative BLAST |
| bZIP | 73 | TAIR Web site |
| BZR | 6 | Identified in literature |
| C2C2 (Zn) CO-like | 30 | Identify domain and BLAST |
| C2C2 (Zn) Dof | 36 | Identify domain and BLAST |
| C2C2 (Zn) GATA | 29 | Identify domain and BLAST |
| C2C2 (Zn) YABBY | 6 | Identify domain and BLAST |
| C2H2 | 107 | Iterative BLAST |
| C3H | 163 | Iterative BLAST |
| CAMTA | 6 | Identified in literature |
| CCAAT-Dr1 | 2 | Identified in literature |
| CCAAT-HAP2 | 10 | Identify representative protein and BLAST |
| CCAAT-HAP3 | 10 | Identify representative protein and BLAST |
| CCAAT-HAP5 | 13 | Identify representative protein and BLAST |
| CPP (Zn) | 8 | Iterative BLAST |
| E2F-DP | 8 | Identified in literature |
| EIL | 6 | Iterative BLAST |
| G2-like | 40 | Identify domain and BLAST |
| GeBP | 16 | Identify representative protein and BLAST |
| GRAS | 33 | Identify domain and BLAST |
| GRF | 9 | Identify representative protein and BLAST |
| Homeobox | 89 | Identify domain and BLAST |
| HRT | 3 | Identified in literature |
| HSF | 21 | Identify domain and BLAST |
| JUMONJI | 5 | DATF and TAIR Web sites |
| MADS | 110 | Iterative BLAST |
| MYB | 138 | TAIR Web site |
| MYB related | 21 | Iterative BLAST |
| NAC | 90 | Identify domain and BLAST |
| NLP | 7 | Identified in literature |
| Orphans | 2 | Documentation |
| PHD | 11 | Identified in literature |
| RAV | 11 | Identified in literature |
| REM | 21 | Identified in literature |
| SBP | 16 | Identify domain and BLAST |
| TCP | 26 | Iterative BLAST |
| Trihelix | 29 | Iterative BLAST |
| TUB | 10 | Identify representative protein and BLAST |
| VOZ-9 | 2 | Identified in literature |
| WHIRLY | 3 | Identified in literature |
| WRKY | 72 | TAIR Web site |
| ZF-HD | 15 | Identified in literature |
AtcisDB
AtcisDB is an integrated database of 25,806 promoter sequences (September 2005) with annotations of cis-regulatory elements. The current version of AGRIS contains only 5′ promoter regions; regulatory sequences in introns will be incorporated as they continue to be identified. The coding sequences (ftp://ftp.arabidopsis.org) based on the current genome annotation were mapped to the chromosomal sequences by BLAT (Kent, 2002). Then, for each gene, if the upstream intergenic region is greater than 3 kb, the sequence upstream of the ATG of length 3 kb was retrieved. Otherwise, we consider that intergenic region as the promoter of the downstream gene, to exclude any coding region of upstream genes (Davuluri et al., 2003). Subsequently, with the increasing accumulation of full-length (FL) cDNAs, these gene upstream regions were curated to represent bona fide 5′ regulatory sequences, in every case where this was possible. Currently, the promoters in AtcisDB can be divided into three classes: (1) curated promoters (12,500 total), established based on the availability of FL cDNAs (Molina and Grotewold, 2005); (2) upstream sequences (13,069 total), corresponding to regions upstream of the annotated ATG and including promoters and 5′ untranslated regions, but without the support provided by FL cDNAs that confirms the position of the transcription start site; and (3) manually annotated (237 total), based on reports from the literature or personal communications (e.g. Schuler and Werck-Reichhart, 2003). The breakdown of number of promoters acquired through each process is shown in Table II.
Table II.
Different methods through which promoter sequences are acquired for AtcisDB
The number of promoters acquired for each method may change based on periodic updates that take place in genome annotation.
| Promoter Acquisition Process | No. of Promoters | Promoter Type |
|---|---|---|
| Annotated genomic sequences without FL cDNAs | 13,069 | Predicted |
| Annotated genomic sequences with FL cDNAs | 12,500 | Predicted and curated |
| Manually annotated from literature or communication | 237 | Experimentally curated |
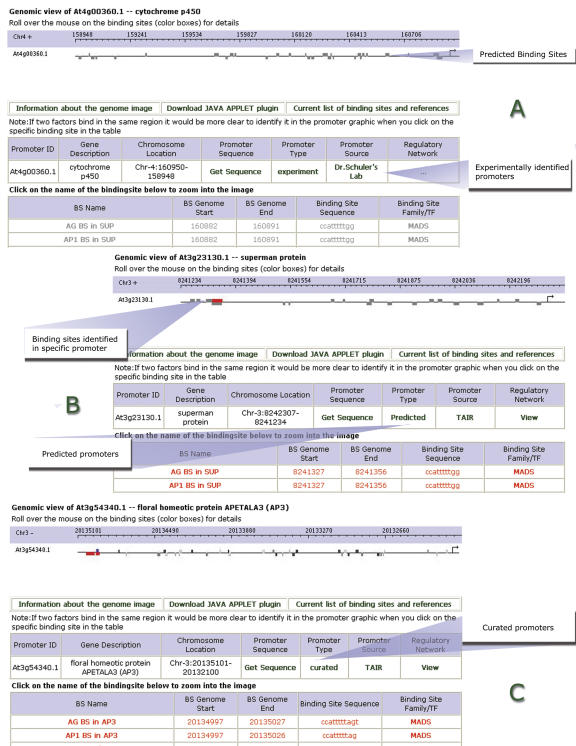
For prediction of the cis-regulatory elements currently available through the AtcisDB (Fig. 2), consensus-binding sequences for known TFs have been collected from the literature, and that information is housed in AGRIS. The prediction of cis-regulatory elements was done primarily by scanning promoter sequences for known TF consensus binding sites obtained from AtTFDB (Table III). A complete list of binding sites and its references is provided online (Supplemental Table I). Experimentally validated binding sites are represented as red color-coded rectangular boxes in the genomic image view of the promoter (explained in the “Data Visualization and User Interface” section below).
Figure 2.
Display of promoters identified in AtcisDB. A represents experimental promoters, B represents predicted promoters, and C represents curated promoters with binding sites in red indicating them to be present in that specific promoter.
Table III.
List of TFs by families for which cis-binding elements have been known
| TF Family | No. of cis-Elements Identified |
|---|---|
| AP2-EREBP | 4 |
| ARF | 2 |
| bHLH | 1 |
| bZIP | 14 |
| C2C2-Dof | 4 |
| E2F-DP | 3 |
| EIL | 4 |
| Homeobox | 8 |
| HSF | 1 |
| MADS | 13 |
| MYB | 6 |
| MYB related | 3 |
| Orphan (LFY) | 1 |
| RAV | 2 |
| VOZ-9 | 1 |
The data can be viewed by an interactive browser developed using the Genome Data Visualization Toolkit (GDVTK; Sun and Davuluri, 2004). Random manual error checking is done routinely to avoid errors in the data presented, and data analysis is performed by automated tools developed in Perl.
Direct Targets of TFs Link AtcisDB and AtTFDB
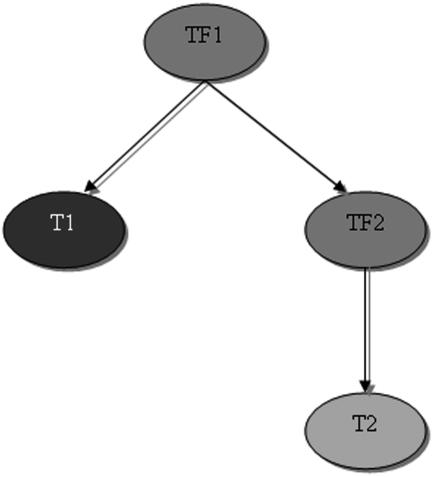
As more is discovered about how TFs function, it is increasingly recognized that they act as part of complex networks. Within these networks, TFs often regulate other TFs, forming “branched” networks. TFs also control the expression of other non-TF-encoding genes directly participating in cell differentiation and responses to biotic or abiotic stimuli. A single TF may regulate, directly or indirectly, hundreds of genes. We define here as a direct target of a TF a gene that is directly recognized by the TF, for example, through binding to its cis-regulatory elements, and that may result in the activation or repression of the gene. For example, T1 is a direct target of TF1, and T2 is an indirect target of TF1 as it requires the prior activation (or repression) of TF2 (Fig. 3). While there are many ways in which the direct interaction of a TF to a target gene can be demonstrated, two approaches allow the simultaneous identification of many direct targets for a given TF. The first method is based on chromatin immunoprecipitation (ChIP) followed by hybridization of microarrays (chip), thus called ChIP-chip (e.g. Lee et al., 2002). The second method is based on the posttranscriptional control of a TF fused to the hormone-binding domain of the glucocorticoid receptor (GR), followed by induction with a steroid hormone (e.g. dexamethasone [DEX]) in the absence and presence of protein synthesis inhibitors, such as cycloheximide (CHX). Fusion of TFs to the GR domain has been demonstrated to cause the fusion protein to be retained in the cytoplasm in the absence of a steroid hormone (Aoyama and Chua, 1997). Upon application of the synthetic hormone DEX, the protein moves into the nucleus and activates/represses transcription. Direct targets are those that are either induced or repressed by a given TF-GR upon DEX induction, even in the presence of the translation inhibitor CHX. For example, in Figure 3, TF1-GR and DEX would activate T1, TF2, and T2, but in the presence of CHX only T1 and TF2 would remain activated. While the ChIP-chip method (Wang et al., 2002) has been used in only a few instances to identify direct targets for Arabidopsis TFs, TF-GR fusions have proven to be very useful in the identification of many TF direct targets (Sablowski and Meyerowitz, 1998; Zik and Irish, 2003a; William et al., 2004).
Figure 3.
T1 and TF2 are direct targets of TF1. T2 is an indirect target of TF1. T1 is a direct target of TF1, and T2 is an indirect target of TF1, as it requires the prior activation (or repression) of TF2.
Based on the analysis of the published literature, we have defined two groups of direct targets: confirmed and unconfirmed. A confirmed direct target has been defined as a gene that responds to a given TF according to the following criteria. (1) The TF binds directly to the regulatory region of the target gene, as shown by electrophoretic mobility shift assay, yeast one-hybrid analysis, and/or ChIP; or (2) the TF directly regulates the target gene, based on use of transgenic plants expressing an inducible TF-GR fusion protein, and the effect of CHX on the DEX activated/repressed genes; and (3) in vivo evidence of regulation showing expression of the target gene is affected by either loss-of-function mutations in the TF or ectopic expression of that TF in the plant.
An unconfirmed direct target is one that has been suggested to be a direct target of a given TF by one of the approaches described above.
AtRegNet
To understand the function of any TF, the complex networks in which these TFs are involved must be recognized and visualized. AtRegNet was developed as a tool used to display the regulatory networks of Arabidopsis. It provides a fast and easy way for researchers to visualize the network of interactions in this plant.
AtRegNet documents and visualizes networks formed by TFs and their direct target genes. This means that many interactions that have only been defined based on genetic data are not coded for in AtRegNet. This provides a fundamental difference with other similar efforts that have tried to integrate TF and regulated genes into networks (e.g. Espinosa-Soto et al., 2004). As more data becomes available to support direct interactions, these will be added to the database. Information presented in this database is taken from data validated by peer-reviewed publications.
Once it has been documented that a given gene is a direct target of a TF found in AtTFDB, it is entered into the AtRegNet database. Unpublished data on any TFs (either generated in-house by our group or communicated to us by others) will be added to the database as unconfirmed direct targets. Once the data have been published, the interactions are updated. The current version of AtRegNet (September 2005) has 187 direct targets identified, of which 66 have been identified to target TFs (Table IV).
Table IV.
List of TFs by families for which direct targets have been identified
Confirmed targets are denoted in parentheses. Total targets equals 187, of which 79 are confirmed and 108 are unconfirmed.
| TF Family | Direct Targets (Confirmed) |
|---|---|
| ABI3VP1 | 2 (2) |
| BBR/BPC | 3 (3) |
| bHLH | 1 (1) |
| bZIP | 49 (4) |
| BZR | 2 (2) |
| C2C2-CO-like | 2 (2) |
| C2H2 | 1 (1) |
| EIL | 3 (3) |
| GeBP | 1 (1) |
| Homeobox | 5 (5) |
| HSF | 1 (1) |
| MADS | 24 (16) |
| MYB | 4 (1) |
| MYB related | 2 (2) |
| NAC | 3 (3) |
| Orphan (LFY) | 20 (20) |
| WRKY | 64 (12) |
Implementing AtRegNet with a Flower Developmental Network
Understanding the mechanisms that underlie development of a cell type or organ requires knowledge of the gene regulatory network controlling those mechanisms. Flower development is one of the better-understood developmental processes in plants and is coordinated by an integrated network of many genetic interactions or pathways in Arabidopsis. A large number of genes that control different steps of flower development, from control of flowering time (Komeda, 2004; Simpson, 2004) to differentiation of specific cell types within the flower (e.g. Chalfun-Junior et al., 2005; Nakayama et al., 2005), have now been identified, revealing some of the regulatory interactions at work. Flower development can be understood in the framework of the currently well known ABC model (Coen and Meyerowitz, 1991), recently modified by the identification of the redundant SEPALLATA (SEP) genes (Pelaz et al., 2000; Jack, 2001; Ditta et al., 2004). In this model, there are four gene functions, the A, B, and C, and SEP floral homeotic genes. They act combinatorially to specify identity of each organ type within the flower. The A group genes APETALA1 and APETALA2 (AP2) function in the first and second whorl; B group genes APETALA3 (AP3) and PISTILLATA (PI) function in the second and third whorls; and the C group gene AGAMOUS (AG) functions in the third and fourth whorls. The SEP1, 2, 3, and 4 genes function redundantly in all four whorls. Thus, for example, A + SEP in the first whorl specifies sepals, and A + B + SEP in the second whorl specifies petals. All these genes encode TFs, which serve as master regulatory switches (Jack, 2001). Recently, it has been found that these genes act in larger order protein complexes, with the SEP proteins serving as molecular bridges (Goto, 2001; Pelaz et al., 2001).
Although the above-mentioned genes have well documented roles in floral organ specification, only recently have targets of these TFs been identified, with only a few of these targets known to be direct targets (Zik and Irish, 2003b; Ito et al., 2004; Wellmer et al., 2004; Gomez-Mena et al., 2005). This information is beginning to reveal a complex regulatory network among the ABC and SEP genes themselves. It is still unclear how later steps in tissue differentiation are regulated by these TFs. In addition, information about the direct control of expression of the ABC and SEP genes, while incomplete, is also revealing a complex network of TF interactions (e.g. Wellmer and Riechmann, 2005).
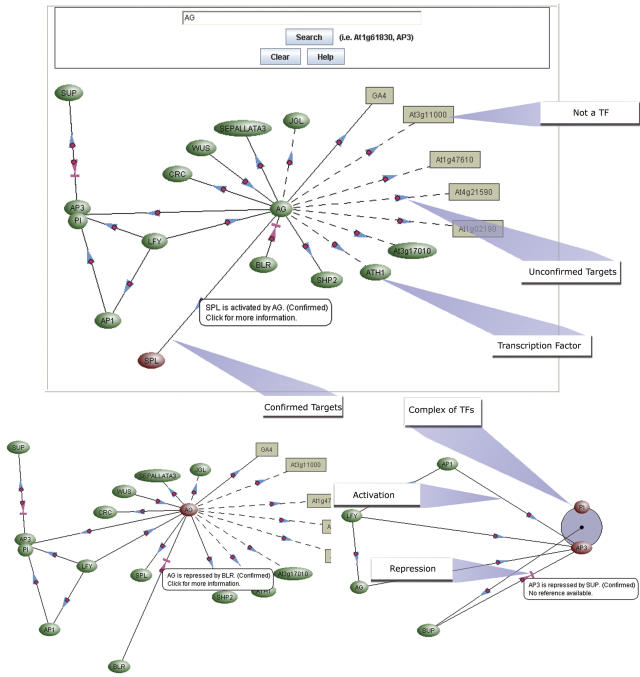
To understand the entire process of flower formation and to view its complexity all at once, it becomes imperative to have a resource that houses all these data. AtRegNet acts as an information portal in which users can infer networks that regulate gene expression, based on the experimental data from microarray experiments, and also as a platform to identify the components and determine how they interact with one another. To see how AtRegNet can help visualize a network such as that involved in flower development, take the C group gene AG as an example (Fig. 4). AG has been shown to directly regulate a number of genes, including other floral homeotic genes (AP3, PI, SEP3; Gomez-Mena et al., 2005), other TFs involved in flower development (e.g. CRABS CLAW [Gomez-Mena et al., 2005] and WUSCHEL [Lenhard et al., 2001; Lohmann et al., 2001]), and other non-TF genes (e.g. GA4; Gomez-Mena et al., 2005). In turn, AG has been shown to be regulated by other TFs, such as the floral meristem identity LEAFY (LFY) TF (Lenhard et al., 2001; Lohmann et al., 2001). This places AG at the center of a complex network of interactions, experimentally determined by a variety of groups and published at different times. These interactions can be easily visualized simultaneously using AtRegNet, and the network can be built out from AG.
Figure 4.
A screen shot of the AtRegNet database and its search results for AG. AG has been shown to directly regulate a number of genes, including other floral homeotic genes.
AGRIS and Other Similar Resources
There are other currently available databases that partially overlap in function with AGRIS. PlantCARE, a cis-regulatory elements database, contains very limited information on TF-binding sites and associated promoters. AthaMap (Steffens et al., 2005) also is a useful resource that displays TF-binding sites at the genome level, thus complementing some features of AtcisDB. However, to our knowledge, AtcisDB is the only database that provides an annotation of both computationally predicted and experimentally validated cis-regulatory elements for all promoter sequences.
The Database of Arabidopsis Transcription Factors (DATF; http://datf.cbi.pku.edu.cn/) was recently described (Guo et al., 2005) and contains significant overlap in the TF information with the initial description of AGRIS (Davuluri et al., 2003). However, DATF has significant differences with AGRIS. One major difference is that DATF contains so far only information on TFs and thus can only be compared to AtTFDB, as it does not contain promoter sequences (as found in AtcisDB) or regulatory networks (as found in AtRegNet). There are some small differences in the number (50 in AGRIS versus 56 in DATF) and names of the TF families between the two databases (see supplemental data for a more detailed description of the differences).
CONCLUSION
AGRIS provides a valuable resource regarding Arabidopsis TFs, promoters, and the interactions between them. The three databases present in AGRIS are actively linked with each other and provide links to other resources widely used by the Arabidopsis community. All the data present in AGRIS are downloadable and freely available to the community. AtRegNet provides a powerful tool that enables investigators to create their own regulatory motifs, either from existing nodes or de novo.
MATERIALS AND METHODS
AtTFDB
The development of AtTFDB was based on searches of the TAIR Web site http://www.arabidopsis.org resulting in few TF sequences retrieved searching with the key words “transcription factor” or “regulatory protein.” Thus, to identify TFs, several different approaches were used, and many families were identified through a domain search and BLAST technique. Publications were found through PubMed, and the conserved domain motif that characterizes each TF family was identified. Using the motif, a BLAST was conducted on the TAIR Web site, and the resultant sequences were then aligned and mismatches were discarded. Another approach, especially for large families where very few TFs had been identified, was an iterative BLAST approach. A few representative proteins were used to perform a BLAST on the TAIR Web site (Fig. 1). In addition, published data were used to manually curate and update the TFs included in this database.
Design and Web Implementation of AGRIS
A Web interface for AGRIS was developed using the J2EE technology (JSP and Servlet) because of its rapid development and easy maintainability qualities in development of dynamically generated Web pages and because it takes advantage of the java technology provided by the Apache Tomcat server http://jakarta.apache.org/tomcat. The databases AtTFDB, AtcisDB, and AtRegNet were developed using MySql (http://www.mysql.com). Supplemental Figure 1 contains a simplified diagram revealing the interaction of the different components behind the AGRIS online resource.
Data Visualization and User Interface
AtcisDB
All cis-regulatory element data are presented using GDVTK, which provides a graphical view of the annotations. A line with a small arrow indicates the position of the predicted or experimentally demonstrated transcription start site. A set of small colored squares represents the TF-binding sites. When the user moves the mouse on these colored squares, a contextual menu will pop out and show the detailed information for that binding site. The user can zoom in or out of the promoter annotations.
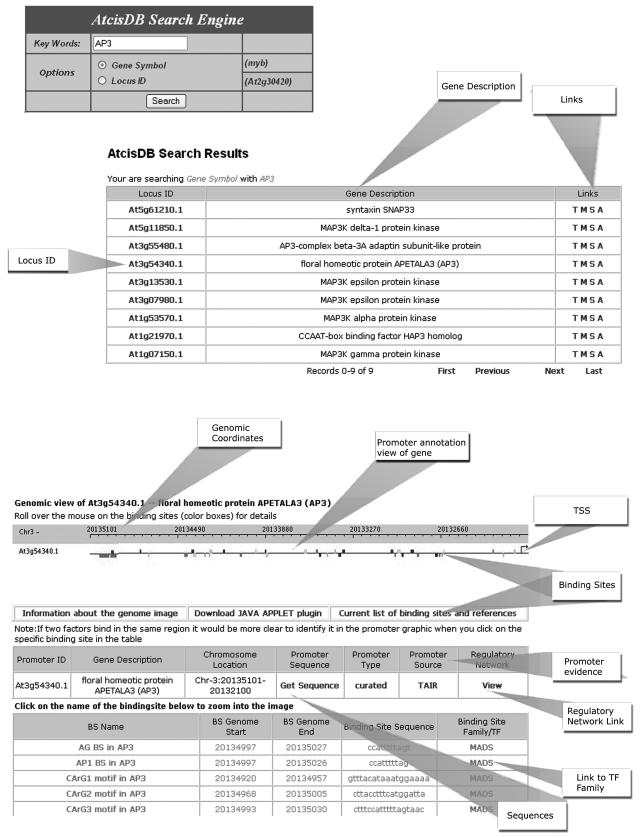
The AtcisDB database can be searched by Gene Name or Symbol (NAC, MYB), or Locus ID (At1g01010). The search results will include the Locus ID, the Gene Description that represents that Locus, and cross-reference for gene annotation from MIPS, TIGR, TAIR, and SALK. The cis-regulatory element annotation for each gene can also be obtained. Figure 5 shows a sample screen shot of the AtcisDB Web page.
Figure 5.
A screen shot of the AtcisDB Web search engine and its corresponding search result pages.
AtTFDB
AtTFDB is a comprehensive and public Arabidopsis (Arabidopsis thaliana) TF database. From this page, users may search the database for TFs in multiple ways.
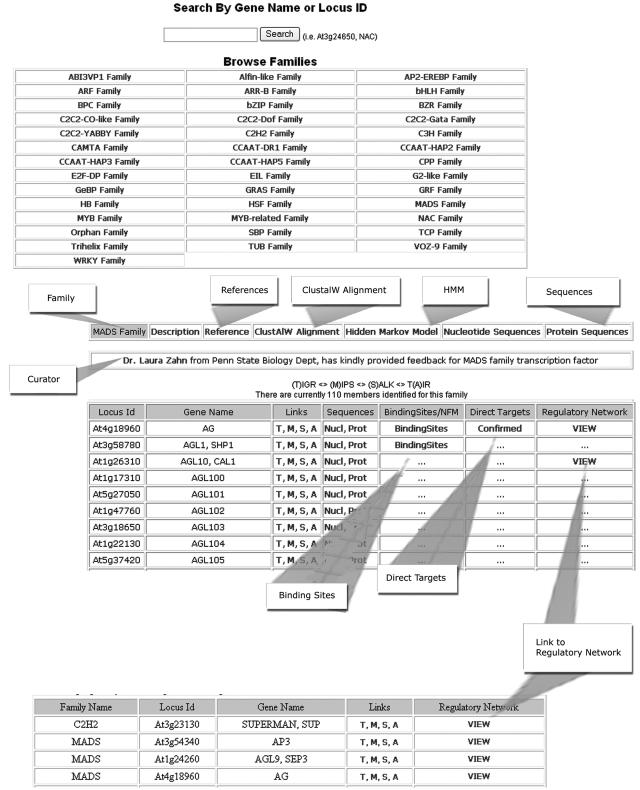
Users may search using a specific Locus ID (Arabidopsis Genome Initiative) or gene name search. The user may enter a Locus ID, such as At3g24650, or a text (such as NAC), or browse families by clicking on a link to one of the families. The results are listed in a Search Results table. The family name, gene name, and description are displayed. In addition, there are four Arabidopsis resources the user may use to retrieve additional information: (M) for MIPS (http://mips.gsf.de); (T) for TIGR (www.tigr.org); (S) for SALK (http://signal.salk.edu); and (A) for TAIR (www.arabidopsis.org). Figure 6 shows a sample screen shot of the AtTFDB search engine and its results with an additional link to binding-site information for TFs (when known).
Figure 6.
A screen shot of the AtTFDB Web search engine and its corresponding search result pages.
AtRegNet
Using java's threading and applet technology allows for quick, on-the-fly generation of the network. We developed in-house a java-based network generation module named Regulatory Information Network (S. James, S. Subramaniam, S.K. Palaniswamy, R.V. Davuluri, and E. Grotewold, unpublished data). This tool helps to create a network with different types of interactions between TFs and their functionalities. AtRegNet is easily updatable. Since it is Web based, the application can be tweaked and new features can be added to fit the needs of the scientific community without requiring the user to download updates or install patches. It was developed to provide a fast and easy way for researchers to visualize the network of interactions specifically in the Arabidopsis plant. The original format of the data is tab-delimited text. The files are parsed into tables in the AtRegNet database with a series of Perl scripts. We provide a Web link to the reference publication in PubMed.
AtRegNet pulls all the information it needs to generate a network from the database and integrates the information with AtcisDB and AtTFDB. Each record from the table contains a TF and a target for that TF as well as other data, such as type of regulation (like activation or repression).
AtRegNet can be accessed from the AGRIS Web site. In the title bar of all AGRIS pages is a link to Regulatory Networks, which will bring up a description of the AtRegNet database, and a blank workspace. A search bar for AtRegNet is provided in the interface. Another way to access AtRegNet is to search for a TF in AtTFDB and if regulatory network data are present on the TF, then a link is automatically provided to AtRegNet. When these links are used, the TF is immediately searched and the network is displayed when the AtRegNet page loads. Figure 4 shows a sample screen shot of the AtRegNet database.
TFs are represented as circles with lines connecting them to their targets. Lines with blue arrows indicate that the TF positively regulates the target, while lines with red arrows indicate the TF represses the target. Non-TF targets (which can be any gene that encodes a product that does not act as a DNA-binding TF) are represented as rounded squares. TFs are provided with links to their interactions (if known) and both AtTFDB and AtcisDB. Non-TF targets are linked to AtcisDB. Clicking on the arrows linking the TF to the target can access links to papers documenting the interactions.
Downloads and Help Pages
All the data in AGRIS can be downloaded for free after a quick registration process by the user. The download menu on the top navigation bar takes the user to the registration and download process page. The users can download promoter annotations and sequences, binding sites, TF families, references, and regulatory network interactions. The downloaded data files are provided in tab-delimited text format. The users can also read the comprehensive Help pages on how to use and interpret the data in all three databases. The help information can be accessed from http://Arabidopsis.med.ohio-state.edu/faq.jsp.
Curators for AGRIS
Currently, there are curators for some families; based on our request and their expertise, the curators help us validate data through cross-verification approach and make sure that data represented are accurate for the families they curate. The list of curators is provided on http://Arabidopsis.med.ohio-state.edu/credits.jsp.
Supplementary Material
Acknowledgments
We thank Sarat Subramanian for his help in AtTFDB, and Twyla Pohar for her comments and header design for the Web site. We also thank the AGRIS curators and the anonymous reviewers for their feedback that resulted in significant AGRIS improvements.
This work was supported by the National Science Foundation (grant no. MCB–0418891).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ramana V. Davuluri (ramana.davuluri@osumc.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD (2005) Constructing transcriptional regulatory networks. Genes Dev 19: 1499–1511 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Ecker JR (2004) Plant genomics: the third wave. Annu Rev Genomics Hum Genet 5: 443–477 [DOI] [PubMed] [Google Scholar]
- Chalfun-Junior A, Franken J, Mes JJ, Marsch-Martinez N, Pereira A, Angenent GC (2005) ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal-distal patterning in Arabidopsis petals. Plant Mol Biol 57: 559–575 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E (2003) AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25 [DOI] [PMC free article] [PubMed]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Espinosa-Soto C, Padilla-Longoria P, Alvarez-Buylla ER (2004) A gene regulatory network model for cell-fate determination during Arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles. Plant Cell 16: 2923–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Li J, Strickland E, Hua S, Zhao H, Chen Z, Qu L, Deng XW (2004) An arabidopsis promoter microarray and its initial usage in the identification of HY5 binding targets in vitro. Plant Mol Biol 54: 683–699 [DOI] [PubMed] [Google Scholar]
- Gomez-Mena C, de Folter S, Costa MM, Angenent GC, Sablowski R (2005) Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132: 429–438 [DOI] [PubMed] [Google Scholar]
- Goto K (2001) Conversion of floral organs into leaves, leaves into floral organs: complexes of MADS transcription factors determine floral organ identity. Tanpakushitsu Kakusan Koso 46: 1830–1835 [PubMed] [Google Scholar]
- Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J (2005) DATF: a database of Arabidopsis transcription factors. Bioinformatics 21: 2568–2569 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta AM, Salgado H, Thieffry D, Collado-Vides J (1998) RegulonDB: a database on transcriptional regulation in Escherichia coli. Nucleic Acids Res 26: 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann JL, Meyerowitz EM (2004) The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430: 356–360 [DOI] [PubMed] [Google Scholar]
- Jack T (2001) Relearning our ABCs: new twists on an old model. Trends Plant Sci 6: 310–316 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolchanov NA, Ignatieva EV, Ananko EA, Podkolodnaya OA, Stepanenko IL, Merkulova TI, Pozdnyakov MA, Podkolodny NL, Naumochkin AN, Romashchenko AG (2002) Transcription Regulatory Regions Database (TRRD): its status in 2002. Nucleic Acids Res 30: 312–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y (2004) Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol 55: 521–535 [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814 [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105: 793–803 [DOI] [PubMed] [Google Scholar]
- Molina C, Grotewold E (2005) Genome wide analysis of Arabidopsis core promoters. BMC Genomics 6: 25 [DOI] [PMC free article] [PubMed]
- Mueller LA, Zhang P, Rhee SY (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N, Arroyo JM, Simorowski J, May B, Martienssen R, Irish VF (2005) Gene trap lines define domains of gene regulation in Arabidopsis petals and stamens. Plant Cell 17: 2486–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF (2001) APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26: 385–394 [DOI] [PubMed] [Google Scholar]
- Perier RC, Praz V, Junier T, Bonnard C, Bucher P (2000) The eukaryotic promoter database (EPD). Nucleic Acids Res 28: 302–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3: 423–434 [DOI] [PubMed] [Google Scholar]
- Rombauts S, Dehais P, Van Montagu M, Rouze P (1999) PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res 27: 295–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RWM, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103 [DOI] [PubMed] [Google Scholar]
- Schoof H, Ernst R, Nazarov V, Pfeifer L, Mewes HW, Mayer KF (2004) MIPS Arabidopsis thaliana Database (MAtDB): an integrated biological knowledge resource for plant genomics. Nucleic Acids Res 32: D373–D376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MA, Werck-Reichhart D (2003) Functional genomics of P450s. Annu Rev Plant Biol 54: 629–667 [DOI] [PubMed] [Google Scholar]
- Simpson GG (2004) The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr Opin Plant Biol 7: 570–574 [DOI] [PubMed] [Google Scholar]
- Steffens NO, Galuschka C, Schindler M, Bulow L, Hehl R (2004) AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Res 32: D368–D372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens NO, Galuschka C, Schindler M, Bulow L, Hehl R (2005) AthaMap web tools for database-assisted identification of combinatorial cis-regulatory elements and the display of highly conserved transcription factor binding sites in Arabidopsis thaliana. Nucleic Acids Res 33: W397–W402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Davuluri RV (2004) Java-based application framework for visualization of gene regulatory region annotations. Bioinformatics 20: 727–734 [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Wang H, Tang W, Zhu C, Perry SE (2002) A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, a MADS domain protein that preferentially accumulates in embryos. Plant J 32: 831–843 [DOI] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL (2005) Gene network analysis in plant development by genomic technologies. Int J Dev Biol 49: 745–759 [DOI] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM (2004) Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16: 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- William DA, Su Y, Smith MR, Lu M, Baldwin DA, Wagner D (2004) Genomic identification of direct target genes of LEAFY. Proc Natl Acad Sci USA 101: 1775–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res 28: 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al (2003) Annotation of the Arabidopsis genome. Plant Physiol 132: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing B, van der Laan MJ (2005) A statistical method for constructing transcriptional regulatory networks using gene expression and sequence data. J Comput Biol 12: 229–246 [DOI] [PubMed] [Google Scholar]
- Zik M, Irish VF (2003. a) Flower development: initiation, differentiation, and diversification. Annu Rev Cell Dev Biol 19: 119–140 [DOI] [PubMed] [Google Scholar]
- Zik M, Irish VF (2003. b) Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15: 207–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.