Abstract
Mnt is a repressor from phage P22 that belongs to the ribbon–helix–helix family of DNA binding factors. Four amino acids from the N-terminus of the protein, Arg2, His6, Asn8 and Arg10, interact with the base pairs of the DNA to provide the sequence specificity. Raumann et al. (Nature Struct. Biol., 2, 1115–1122) identified position 6 as a ‘master residue’ that controls the specificity of the protein. Models for the interaction have residue 6 of Mnt interacting directly with position 5 of the operator. In vivo selections demonstrated that protein variants at residue 6 bound specifically to operator mutations at that position. Operators in which the wild-type G at position 5 was replaced by T specifically bound to several different protein variants, primarily hydrophobic residues. The obtained protein variants, plus some others, were used in in vitro selections to determine their preferred binding sites. The results showed that the residue at position 6 influenced the preference for binding site bases predominantly at position 5, but that the effects of altering it can extend over longer distances, consistent with its designation as a ‘master residue’. The similarities of binding sites for different residues do not correlate strongly with common measures of amino acid similarities.
INTRODUCTION
The Mnt repressor of bacteriophage P22 is an 82-residue protein that exists as a tetramer in solution (1). It binds specifically to a 17 bp operator site that is symmetric about a central base pair. Each half of the operator is probably recognized by symmetrically related dimers of the Mnt tetramer (2,3). The Mnt residues that are functionally important for DNA binding are located at the N-terminus of the protein (4,5). Substitutions of Ala for each residue throughout the N-terminal region showed that the side chains of Arg at position 2, His at position 6, Asn at position 8 and Arg at position 10, are critical for high affinity binding to the wild-type operator DNA. An Mnt mutant bearing a His to Pro substitution at position 6 was able to bind with wild-type affinity to a mutated Mnt operator, with symmetric G-C to A-T changes at base pairs 5 and 17 (6). In addition, protection and interference experiments (2,3) support the model that the His residues at position 6 of the Mnt tetramer contact base pairs 5 and 17 in the operator sequence. Knight and Sauer (4) also showed that the hybrid repressor in which the N-terminal six residues of Mnt are replaced by the corresponding nine residues of the Arc protein bind specifically to the arc operator, but not to the mnt operator. The crystal structure of Arc bound to its operator revealed the N-terminal amino acids in contact with the DNA (7). Together, all of these results support the model of Mnt binding to DNA (3,8) shown in Figure 1.
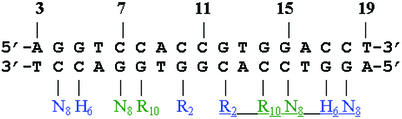
Figure 1.
Model for the Mnt–operator interaction from Knight and Sauer (3) and Raumann et al. (8). Positions 3–19 are thought to include all of the direct interactions with the protein. The amino acids modeled to interact directly with the DNA are Arg2, His6, Asn8 and Arg10. The figure shows the two dimers, one underlined the other not, that each bind to one half-site. Each dimer contains an anti-parallel β-ribbon of the N-terminal amino acids. Amino acids R2, H6 and N8 from one monomer of each dimer (in blue) and amino acids N8 and R10 of the other monomer (in green) are thought to interact with the base pairs indicated.
Raumann et al. (8) showed that changes to His6 of Mnt could dramatically alter its specificity, including changes at positions other than 5 and 17. The specificity of Mnt is determined by amino acids R2, H6, N8 and R10. The homologous Arc repressor has a longer N-terminal tail, so its equivalent specificity determining amino acids are S5, Q9, N11 and R13. The crystal structure of the Arc–DNA complex (7) shows that S5 interacts with the backbone only and so should not contribute significantly to the specificity of Arc. A hybrid protein, with most of the residues from Mnt to allow for tetramer formation but the N-terminal residues from Arc, binds to the arc operator with high affinity, but not to the mnt operator (8). Replacements S5R and Q9H (or even just Q9H alone) shows high affinity to the mnt, but not to the arc, operator. The most surprising result was that a hybrid protein with the replacement of S5R alone, still containing Q9, was able to bind to both operators with high affinity, despite the fact that the two operators have almost no base pairs in common. The Arc protein contacts directly the sequence TAGA in each half-site of the arc operator, whereas the conserved positions in the mnt operator that are modeled to interact with the homologous residues are GGTCC [see Raumann et al. (8) for more details]. These authors concluded that position 6 was a ‘master residue’ that controlled how the protein recognized the DNA sequence and could alter the contacts made by the other residues. The hybrid protein also had much greater affinity for non-specific DNA than either protein alone, showing that it had lost some of its specificity. In the work presented here we show that the preferred binding site for Mnt with the H6Q replacement, as determined by in vitro selections, is a hybrid operator with features from each of the mnt and arc operators.
Other work has also demonstrated that the binding site positions do not contribute independently to the affinity. Fields et al. (9) measured the change in binding energy for all single base substitutions to the wild-type binding site and their results are largely consistent with the model of Knight and Sauer. However, their in vitro selections showed that positions 5 and 6 were highly correlated (9,10). Quantitative relative affinity assays to all possible 16 dinucleotides at positions 5 and 6 measured the degree of interdependence between the positions (11). The primary interaction is that the preference for T at position 6 depends on a G at position 5. If any other base occurs at position 5 the preference at position 6 changes to G. Together, all of these findings and the model for interaction suggest that Mnt residue 6, a His in the wild-type protein, is critical in determining the specificity of the protein, a ‘master residue’ in the phrase of Raumann et al. (8). For that reason we undertook the analyses described in this paper of selecting in vivo for variant proteins that specifically recognize operators with changes at position 5, and to determine from in vitro selections the preferred binding sites for several protein variants at residue 6.
MATERIALS AND METHODS
Protein selections using transcriptional interference
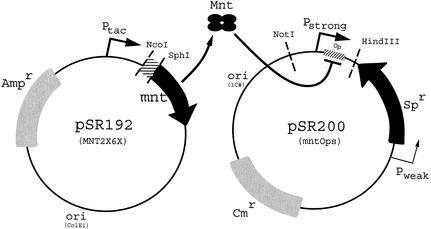
Transcriptional interference allows the selection of repressors that bind to specific operator sequences (12). Figure 2 is a diagram of the method used in this paper. Plasmid pSR192 expresses variant Mnt proteins. If the Mnt expressed in a particular cell can bind to the operator on plasmid pSR200 (Op in Fig. 2) and repress transcription from Pstrong, then the cells will be resistant to spectinomycin (Sp). Otherwise, transcription from Pstrong prevents adequate expression from Pweak and the cells are sensitive to Sp.
Figure 2.
Plasmid designs for interference selections. Plasmid pSR192 contains the Mnt protein that is randomized at codons 2 and 6. Plasmid pSR200 contains one of the selection operators where Mnt binding will repress transcription from Pstrong and allow the cells to be Sp resistant.
pSR192 is derived from pTM201, a gift from Knight and Sauer (5). The engineered NcoI and SphI sites flank the N-terminal DNA-binding domain of Mnt, and were used to remove and replace that N-terminal region. The Mnt library, where amino acids 2 and 6 are randomized, was produced by subcloning a synthetic oligonucleotide, purchased from Gibco BRL, of sequence: d(CATGGCANNNGATGATCCGNN NTTCAACTTCCGCATG) (referred to here as Mnt2X6X) into pSR192 (Fig. 2). In Mnt2X6X, the Ns are the random ized bases where N = ‘A or C or G or T’. The synthetic single-stranded oligonucleotides were subcloned into double-stranded plasmids by a procedure called ‘notch’ cloning (13,14). The linearized vector is flanked by a 5′ overhang from the NcoI site and a 3′ overhang from the SphI site such that a single-stranded oligonucleotide can anneal to both overhangs. The oligonucleotide is phosphorylated at the 5′ end so it can be ligated at both ends. Filling in the single-stranded region in vitro prior to transformation was not observed to improve the efficiency. Therefore, plasmids containing a short ssDNA region were used in the transformations.
Plasmid pSR200 is derived from plasmid pNN388, a gift from Elledge and Davis (12), and contains the aadA gene (for Sp resistance) transcribed from a weak promoter. Operator mutants were synthesized as 17-base oligonucleotides which, because they are palindromes, anneal to themselves creating a blunt-ended fragment. The double-stranded operator fragment was then blunt-cloned into pSR200 cut with SmaI, at a site near the transcription start base. Individual specific operators were made with the following sequences: d(AGNXTCCANYSNY′TGGANX′CT).
Because the mnt operator is a palindrome, only symmetric oligos were synthesized; whatever base is at operator position 5 (NX) its compliment will be found at position 17 (NX′). Likewise, whatever base is found at position 10 (NY), its compliment will be found at position 12 (NY′). Position 11 contains an S (G or C) at the palindromic axis to allow oligos to anneal without a mismatch. If a central mismatch does occur, it will be repaired in vivo or resolved during replication. Although symmetric mutations were made in the operator, for simplicity we will refer only to one operator half-site. Note that position 3 in the oligo represents the fifth operator position in the conventional numbering of the mnt operator as 21 bases long (3) (see Fig. 1). Only the central 17 positions are perfectly palindromic (except for the central base) and are the only positions thought to contribute to the specificity of the interaction. We use that convention throughout, so when we refer to operator position 5, we are also referring to oligonucleotide position 3. Each operator variant was verified by DNA sequencing. All transformations and selections were performed with the JM107 Escherichia coli strain, which was previously shown to support transcriptional interference selection.
The library size for all the selections was determined to be between 50 000 and 100 000, a sufficient size to con tain all combinations of the two randomized codons. Cells transformed with both plasmids were selected on LB agar plates with chloramphenicol (Cm) and a Sp concentration in the range of 80–200 µg/ml. The number of colonies visible on the Sp/Cm plates was generally far more than the number of colonies actually picked, therefore results of the selections should not be interpreted as the total number of possible functional pairings of operator bases and Mnt side chains. Advantageous mutations in either the chromosome or the interference plasmid overtook other colonies as the incubation time increased. It was not always possible to identify these false positives by eye on a plate. Consequently, for some promoters, up to half of all selected colonies were false positives. False positives were identified and eliminated by transferring the Mnt containing plasmid to a new cell containing the interference plasmid. If the cell containing the transferred plasmid is sensitive to the Sp, then we can assume that it was a false positive due to a genomic or interference plasmid mutation. For each operator sequence several true positive selectants were picked and the sequence of the Mnt N-terminus was determined by DNA sequencing. No variations were observed other than in the randomized positions.
In vitro selections of binding sites for each protein variant
Binding sites for each protein were selected using the SELEX procedure as previously applied to Mnt (9). The oligonucletide pool, from which the binding sites were selected, contains an 80-base template strand with 20 bases of randomized sequence (20N) between two fixed stretches of DNA with HindIII and BamHI recognition sites: d(CCCAAGCTTAATACGACG CACTATAGGGAGGAT-20N-TTGCAGCATCGTGAACT AGGATCCGG).
The randomized DNA was amplified by PCR. Twenty reactions were pooled and the PCR products were phenol/chloroform treated and separated in 10% polyacrylamide gel (20:1, mono:bis) to create a pool of 80 bp double-stranded DNA containing the random 20 bp sequence. This desired 80 bp PCR product was sliced from the gel and purified by the freeze–squeezing method as described by Beutel and Gold (15).
To facilitate purification of the Mnt proteins and the SELEX procedure, the genes were recloned into the pET-24 vector (Novagen) so as to include six histidines (His-tag) on the C-terminus. Each protein obtained in the interference selections was amplified by PCR using the following primers: Mnt forward primer, d(GGGGAATTCAAGGAGATATACCCA TGGCTAGAGATGATCCG); Mnt reverse primer, d(CCC CCTCGAGGGTGGTTTTTTTGTA).
The forward primer contains the sequence for the first six amino acids of Mnt preceded by the Shine–Dalgarno sequence and an EcoRI site for cloning into pET-24. The reverse primer contains the sequence of the last five residues and an XhoI site for cloning into pET-24 such that six His residues are added to the C-terminus of the protein.
In addition to those proteins obtained in the transcriptional interference selection, four additional Mnt variants with mutations at residue 6 were generated. Mnt mutants H6R, H6L, H6N and H6Q were obtained from the wild-type clone using the Mnt reverse primer and variants of the Mnt forward primer that substituted the codons for the desired amino acid for the His residue at position 6. We also created two truncated Mnt genes which lack three (Mnt79) and four (Mnt78) amino acids of the C-terminus by using the Mnt forward primer and appropriate variants of the Mnt reverse primer. All of the Mnt protein variants were verified by DNA sequencing of the modified regions.
All of the Mnt plasmids were transformed into BL21 E.coli (Novagen). Cells containing each expression vector were incubated in 400 ml of LB media containing 25 µg/ml kanamycin, then induced for 5 h with 2 mM IPTG. The cells were harvested and the pellet resuspended into buffer A (6 M GuHCl, 0.1 M Na-phosphate, 0.01 M Tris–HCl, pH 8.0) for 1 h at room temperature. The lysate was centrifuged at 10 000 g for 15 min at 4°C, and the supernatant transferred into a new tube with 4 ml of 50% slurry nitrilo-tri-acetic acid (Ni-NTA) resin (QIAGEN) and incubated at room temperature for 45 min with gentle shaking. The solution was transferred to a 1.6 cm diameter column, and washed with 100 ml of buffer B (8 M urea, 0.1 M Na-phosphate, 0.01 M Tris–HCl pH 8.0) followed by 100 ml of buffer C (8 M urea, 0.1 M Na-phosphate, 0.01 M Tris–HCl, pH 6.3). The protein was eluted by washing the column with buffer C with increasing concentration of imidazole (50–300 mM) and 1.5 ml fractions were collected and analyzed by 15% SDS–PAGE. The different fractions with lowest contamination were pooled together and the protein was dialyzed gradually against decreasing amounts of urea (4–0.2 M) in 50 mM Tris pH 7.5, 0.1 mM EDTA, 5% glycerol, 200 mM KCl, and finally against binding buffer (BB: 50 mM Tris pH 7.5, 200 mM KCl, 10 mM MgCl2, 5% glycerol). The protein concentrations were determined by the Bio-Rad Protein Assay method using gamma globulin as a standard. All proteins were kept at –80°C for further investigation. The proteins appeared over 95% pure by SDS–PAGE (data not shown). Binding activity of each protein was verified by EMSA (‘band-shift’ assay) on different operator sequences (data not shown). Protein H6P was not recovered with sufficient activity to be used in the SELEX experiments, but all other proteins were recovered.
In the first round of SELEX, ∼2 × 10–7 M of each Mnt protein was incubated with 5 × 10–7 M of double-stranded 80mer template for 2 h in binding buffer BB. In order to separate the Mnt–DNA complex from the free DNA, the mixture was passed through 30 ml of a 50% slurry of Ni-NTA pre-washed with BB. The column was washed with 200 µl of BB including 100 µg/ml BSA, 50 µg/ml herring sperm DNA, and finally with 200 µl BB. The volumes and rate of washing were optimized using His-tagged Mnt and radiolabled wild-type operator DNA.
The protein–DNA complex was eluted with 500 mM imidazole (Sigma) in BB. Different volumes of the eluted complex were amplified by PCR for 10–15 cycles. The amplified DNA reactions were monitored after various cycles by PAGE. The PCR conditions, DNA amount and number of PCR cycles were selected for further amplification for each DNA (five tubes of each).The pooled PCR DNA product at 80 bp size was sliced from the gel and purified by the freeze–squeezing method for the next round of selection. With the progress of the enrichment, the protein concentrations were lowered to 10–9 M and were kept higher than the concentration of the DNA. SELEX steps were repeated until the EMSA showed a significant binding of the different proteins to their corresponding SELEX products (mainly three to five rounds of SELEX depending on washing conditions).
DNA purified from different rounds of SELEX was digested by HindIII and BamHI restriction enzymes and run into 2% LMA for further purification. The DNA fragment with the appropriate size was ligated into pBluescript KS II (Stratagene) and transformed into DH5 competent cells. Recombinant clones were selected and the operator DNAs sequenced.
RESULTS
Selections in vivo for Mnt proteins with altered specificity
Although the structure for the Mnt–DNA complex has not been solved crystallographically, a model for the interaction has been proposed (see Fig. 1) that is consistent with a large body of evidence, including a comparison with the homologous Arc protein, for which a protein–DNA complex structure has been solved (7). We tested whether Mnt variants at positions 2 and 6 could be selected that would bind with high affinity and specificity to operators modified at positions 5 and 10 (with symmetry maintained at positions 12 and 17). Originally we planned to test all 16 combinations of bases at those positions, but initial experiments showed that no mutant proteins could be selected if position 10 were changed to either A or G. So, in subsequent experiments only position 5 (and 17 to maintain symmetry) was modified. The complete list of all operators tested is provided in Supplementary Material, Table S1. In every selection both positions 2 and 6 of the protein were completely randomized, but only R2 was ever selected. The fact that many different codons for R2 were obtained showed that the randomization procedure worked, but only R2 provided sufficient affinity and specificity to be selected in our experiments.
For operators modified at position 5, G (the wild-type base), A and T all were successful in selecting specific proteins, more than one in each case. A C at position 5 did not select any proteins from the randomized pool, suggesting that no Mnt variant at amino acid 6 is capable of making high affinity and specific contacts with a C-G base pair at position 5 of the operator. All of the selected proteins for each operator are listed in Supplementary Material, Tables S2–S4. The successful selections are summarized in Table 1. The wild-type operator, G at position 5, selected the wild-type protein, H6, in 9 of 11 sequenced clones. Ser and Thr (H6S and H6T) were each selected once. The operator with an A at position 5 is modified in vivo by the E.coli dam methylase so that the As on both strands of the GATC sequence (operator positions 5, 6, 16 and 17) are methylated. This operator selected primarily H6P proteins (six out of seven), as reported previously (6). One example of H6T was also obtained. Operators with T at position 5 selected a variety of amino acids at residue 6, primarily but not exclusively hydrophobic. Seven different proteins were obtained in 12 sequenced clones, and the number of each is shown in Table 1.
Table 1. Selected Mnt proteins for each operator.
| Base ↓ residue → | H | S | T | P | I | V | A | G | M |
|---|---|---|---|---|---|---|---|---|---|
| G(wt) | 9 | 1 | 1 | ||||||
| A* | 1 | 6 | |||||||
| T | 2 | 3 | 2 | 2 | 1 | 1 | 1 |
The base at position 5 of the operator is listed in the first column. G is the wild-type base, and A* creates a dam methylation site. The number of times each protein variant at position 6 was obtained are shown.
Selections in vitro for binding sites for variant proteins
All of the proteins that were obtained in the in vivo studies except H6P, which we were unable to purify in sufficient, active quantity, were used in in vitro SELEX experiments (see Materials and Methods) to determine their specificity. Four additional proteins, H6L, H6N, H6Q and H6R, were also included in the SELEX procedure. To simplify the purification, these proteins were cloned so as to include six His residues as the C-terminus of the protein. The His-tag is not expected to change the specificity of the protein but might reduce the affinity. To test if there were differences in specificity we selected binding sites for three variants of the His-tagged wild-type protein: full length and having three (Mnt79) and four (Mnt78) amino acids deleted from the C-terminus. Previous work had shown that deleting K79 significantly reduced the affinity of Mnt for operator DNA (16), but changes in specificity were not tested. By comparing the SELEX products from each of those proteins with our previous SELEX experiments using the wild-type protein without the His-tag (9) we could determine if changes in the specificity accompanied the addition of the His-tag or the reduction in affinity.
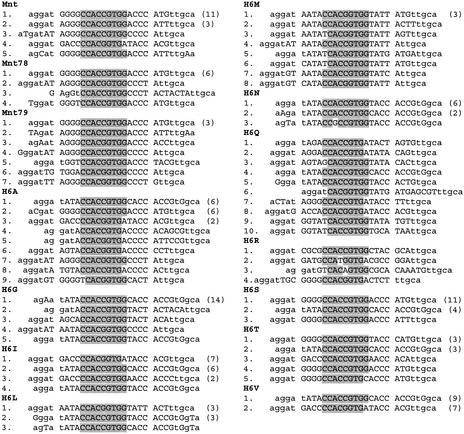
Figure 3 shows the selected binding sites for each protein. The central nine bases of the binding site, CCACSGTGG (shaded in Fig. 3), are highly conserved as expected because all of the amino acids that interact with those positions are unchanged in the variant proteins (Fig. 1). This makes it easy to align the sites, as the core region is conserved with no more than one mismatch, except for H6R where two of the sites contain two mismatches. A few of the selected sequences did not contain a recognizable core binding site. In each case there is still a recognizable half-site, consisting of at least the sequence CCA (on either strand). Binding to operator half-sites is much weaker than to full sites (17) but is thought to be on the pathway to the complete, cooperative binding (18) so it is not too surprising to see a few of those sequences remain after a few rounds of SELEX. Only the sites with complete core regions, shown in Figure 3, are considered in further analysis. The complete list of all selected sites is available in Supplementary Material, Figure S1. A few sites have a mutation in the right fixed sequence that change it from TTGCA to TGGCA. In the right context this can create two core regions in the same selected sequence, such as in H6A:1 and H6G:1, which may contribute to their affinity. Even though the two core regions overlap, and would presumably be mutually exclusive for binding, the fact that two of them occur on the same oligo could at least increase the on-rate for complex formation, and therefore the affinity. In the following analyses only the sites shown aligned in Figure 3 are considered, and those secondary ones are not.
Figure 3.
Selected binding sites for each Mnt variant obtained in the SELEX experiments. The binding sites, which contain the conserved core operator sequence CCACSGTGG (shaded), are aligned between the spaces in the sequences. Upper case letters are from the randomized regions and lower case letters are from the fixed regions. Sometimes a fixed region obtained a mutation, and those have been capitalized (e.g. H6A:1). The number in parentheses following some of the sequences are the number of times that sequence appeared in the selected collection.
Table 2 summarizes the results from Figure 3. Only the outer positions, 3–6 and 16–19, are included because the core region is highly conserved and nearly all of the variation is confined to those positions. Since each binding site is composed of two half-sites, they have been merged into a single half-site list in the table which includes the tetramers from positions 3–6 and the complements of positions 19–16 from Figure 3. Table 3 further reduces the information in Table 2 to show, for each protein, the composition of selected binding sites at each position. If the positions interacted independently with the protein then the tetramer frequencies (Table 2) could be well predicted from the monomer frequencies (Table 3). Clear preferences show up in the monomers which reveal important interactions between particular amino acids at position 6 and the binding site bases, but there are also significant interactions between positions (see Discussion).
Table 2. Operator positions 3–6 and 16–19 for each Mnt protein variant.
| H(wt) | A | G | I | L | M | N | Q | R | S | T | V | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TATA | 0 | 6 | 15 | 7 | 4 | 1 | 9 | 5 | 0 | 4 | 3 | 9 | 63 |
| GGGG | 24 | 11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 12 | 5 | 0 | 54 |
| GGGT | 29 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 2 | 0 | 49 |
| GGTG | 0 | 6 | 14 | 6 | 1 | 0 | 2 | 1 | 0 | 4 | 3 | 9 | 46 |
| GGTA | 1 | 2 | 1 | 8 | 3 | 0 | 7 | 5 | 0 | 0 | 3 | 7 | 37 |
| AATA | 0 | 0 | 1 | 0 | 6 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 22 |
| GACC | 1 | 2 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 7 | 20 |
| AGGG | 12 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 15 |
| AGTA | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 5 |
| CATA | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 4 |
| GATA | 0 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| GGTT | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 |
| AGGA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 |
| GATG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 |
| ACCA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| AGAG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| AGCA | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| AGGT | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| AGTG | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| CGCG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| GGCG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| GTAG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| TAGA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| TGCA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| TGCG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| TGGA | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| TGGT | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| TGTA | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Total | 70 | 40 | 36 | 32 | 14 | 20 | 18 | 20 | 8 | 32 | 18 | 32 | 340 |
Combining the two half-sites of each operator, the outer four positions are listed for all of the selected binding sites. For each protein variant in position 6 (listed across the top) is shown the number of times each operator was obtained.
Table 3. Binding site base frequencies at positions 3–6 and 16–19 for each Mnt variant.
| Amino acid | A | G | H | I | L | M | N | Q | R | S | T | V | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Base | ||||||||||||
| 3 | A | 4 | 4 | 13 | 0 | 6 | 15 | 0 | 6 | 1 | 0 | 0 | 0 |
| C | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | |
| G | 29 | 17 | 55 | 25 | 4 | 1 | 9 | 6 | 5 | 28 | 15 | 23 | |
| T | 7 | 15 | 2 | 7 | 4 | 1 | 9 | 7 | 1 | 4 | 3 | 9 | |
| 4 | A | 10 | 17 | 1 | 16 | 10 | 20 | 9 | 7 | 2 | 4 | 4 | 16 |
| C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | |
| G | 30 | 19 | 69 | 16 | 4 | 0 | 9 | 12 | 4 | 28 | 14 | 16 | |
| T | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| 5 | A | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| C | 2 | 1 | 1 | 9 | 0 | 0 | 0 | 2 | 3 | 0 | 1 | 7 | |
| G | 19 | 1 | 68 | 0 | 0 | 0 | 0 | 4 | 1 | 24 | 7 | 0 | |
| T | 19 | 34 | 1 | 23 | 14 | 20 | 18 | 14 | 2 | 8 | 10 | 25 | |
| 6 | A | 12 | 21 | 2 | 15 | 13 | 20 | 16 | 18 | 0 | 4 | 6 | 16 |
| C | 2 | 0 | 1 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 7 | |
| G | 20 | 15 | 36 | 6 | 1 | 0 | 2 | 2 | 8 | 16 | 8 | 9 | |
| T | 6 | 0 | 31 | 2 | 0 | 0 | 0 | 0 | 0 | 12 | 3 | 0 |
Taken from the data of Table 2, but now the total number of times each base occurs at each position in the sites for each protein are shown.
Table 4 shows the correlations between the amino acid vectors using both the mono- and tetra-nucleotide occurrences (Tables 3 and 2, respectively). Most of the tetra-nucleotide correlations are lower than their corresponding mono-nucleotides. This is not surprising since there are some features that are shared by the binding sites of all proteins, such as a paucity of Cs at all positions and lack of A at position 5, that give them some similarity when compared to each other. In fact, only five of the 66 correlations are negative for the mono-nucleotides. In contrast, 23 of the 66 correlations for tetra-nucleotides are negative, indicating much more diversity at that level of comparison, which also highlights the non-independence of the binding site positions. Table 5 shows the correlations between the outer tetra-nucleotides for the eight most common ones. These eight tetra-nucleotides each occur at least 15 times, and account for 90% of them all, whereas the remaining ones occur five times or less. The sequence AATA is negatively correlated with all of the other sequences, primarily because it is selected by only two proteins, H6M and H6L. The remaining seven tetra-nucleotides fall into two classes, where the correlations within a class are all positive and the correlations between classes are all negative. The classes are distinguished by the base at position 5, where one class (shown in bold) has a G and the other (in italics) a T or C. Since position 5 is modeled to interact directly with amino acid 6, it is not surprising that it should be the dominant determinant of binding site class. The upper right half of Table 5 shows the number of identical bases between each pair of operators. There are examples of high similarity with negative correlation and also low similarity with high correlation, indicating that the contributions of each position to the binding are not additive.
Table 4. Correlations of operator sequences between Mnt variants.
| Amino acid | A | G | H | I | L | M | N | Q | R | S | T | V |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 100 | 64 | 81 | 59 | 43 | 13 | 53 | 57 | 62 | 91 | 91 | 64 |
| G | 52 | 100 | 23 | 78 | 93 | 75 | 90 | 96 | 15 | 38 | 46 | 79 |
| H | 71 | –9 | 100 | 13 | –5 | –24 | 4 | 21 | 41 | 88 | 79 | 14 |
| I | 34 | 57 | –12 | 100 | 77 | 49 | 87 | 71 | 26 | 49 | 64 | 99 |
| L | 16 | 45 | –12 | 4 | 100 | 87 | 93 | 89 | 3 | 19 | 26 | 77 |
| M | –10 | 2 | –9 | –8 | 76 | 100 | 66 | 70 | –7 | –14 | –7 | 48 |
| N | 36 | 68 | –9 | 71 | 61 | –2 | 100 | 90 | 12 | 36 | 44 | 87 |
| Q | 22 | 52 | –13 | 59 | 50 | –6 | 89 | 100 | 1 | 34 | 39 | 71 |
| R | –1 | –17 | 4 | –23 | –19 | –14 | –17 | –29 | 100 | 55 | 55 | 37 |
| S | 89 | 26 | 89 | 7 | 3 | –8 | 13 | 1 | 6 | 100 | 97 | 50 |
| T | 91 | 54 | 54 | 56 | 30 | –10 | 56 | 42 | –1 | 76 | 100 | 64 |
| V | 44 | 77 | –12 | 95 | 45 | –6 | 77 | 63 | –22 | 14 | 60 | 100 |
Table 5. Correlations of protein sequences for the most commonly selected operators.
| GGGG | GGGT | AGGG | TATA | GGTG | GGTA | GACC | AATA | |
|---|---|---|---|---|---|---|---|---|
| GGGG | 3 | 3 | 0 | 3 | 2 | 1 | 0 | |
| GGGT | 0.97 | 2 | 0 | 2 | 2 | 1 | 0 | |
| AGGG | 0.85 | 0.91 | 0 | 2 | 1 | 0 | 1 | |
| TATA | –0.39 | –0.40 | –0.37 | 1 | 2 | 1 | 3 | |
| GGTG | –0.18 | –0.24 | –0.27 | 0.87 | 3 | 0 | 1 | |
| GGTA | –0.42 | –0.37 | –0.22 | 0.43 | 0.19 | 1 | 2 | |
| GACC | –0.15 | –0.13 | –0.08 | 0.25 | 0.38 | 0.68 | 1 | |
| AATA | –0.26 | –0.21 | –0.16 | –0.29 | –0.30 | –0.33 | –0.24 |
The lower-left half contains the correlation coefficients (times 100) for the top eight rows of Table 2. The upper-right half contains the number of identical bases in each pair of operators.
DISCUSSION
Figure 1 shows the model of the Mnt–DNA interaction first proposed by Knight and Sauer (3) and reinforced by Raumann et al. (7,8) after determining the structure of the Arc–DNA complex and comparing the affinities of several hybrid proteins to both the mnt and arc operators. H6 and R2 of the Mnt repressor are proposed to make hydrogen bond contacts with operator base pairs 5 and 10, respectively (and 17 and 12 on the other half-site). The fact that every protein selected to operators with C at position 10 and G at position 12 contained R2, encoded by all six Arg codons, leads us to conclude that no other amino acid can provide the affinity and specificity necessary for the in vivo selection. In particular, we never obtained R2S even though Raumann et al. (8) determined that S2 had essentially the same affinity to the mnt operator as R2 in that position in a hybrid protein. However, S2 contributes to affinity through a hydrogen bond to the backbone, whereas R2 is modeled to interact with the base pair such that it can contribute the specificity required for proper function in vivo. In addition, no proteins were selected on operators that contained bases other than C at position 10, suggesting that the particular interaction of R2 with the C10 and G12 base pairs is required for Mnt to achieve the necessary specificity for in vivo function. This is reinforced by the fact that of the 340 half-sites in Figure 3, all selected by proteins with R2, only one is missing the C10 base pair. Equally impressive is that of the 340 half-sites, only one is missing the A9 (and T13) base pair, even though a direct contact with that position is not included in the model. Previous measurements of relative affinity for variants of position 9 also showed its important contribution to Mnt binding (3,9). We suspect that R2 also interacts with it, probably through a hydrophobic interaction between the Arg side chain and the 5-methyl group of T. The combination of all those contacts, R2 from two of the protein monomers with A9, C10, G12 and T13, may be necessary for the specificity required in vivo.
The fact that no proteins were selected for operators with C5 also appears significant given that multiple proteins were selected for each of the other position 5 variants. In addition, since the only C5 sites selected in vitro included variations at adjacent positions as well (Table 2), it appears that no amino acid is capable of interacting with the C5 base pair, at least in the context of the normal mnt operator, with sufficient specificity to function in vivo.
The successful in vivo selections, summarized in Table 1, provide further evidence for the model because changes in operator position 5 can be complemented by changes in Mnt position 6. Furthermore, the exact pattern of combinations obtained provides additional constraints on this interaction and more information about protein–DNA interactions in general. In a compilation of many protein–DNA complexes determined by X-ray crystallography, it was found that a hydrogen bond between a His and a G is a common interaction, but not as common as Arg or Lys with G (19). In an analysis of zinc-finger proteins selected to bind particular sites, it was also found that His interaction with G is common, but Arg and Lys are used more frequently (20). However, a more detailed analysis of the data from zinc-finger proteins shows that the preferences for His, Arg and Lys are distinct, depending on the position within the binding site (21–23). Arg and Lys are the preferred amino acids when the interacting G is at either end of the 3-long binding site, but when a G is in the middle position it is preferentially bound by His. This change in preference is due to the particular geometry and proximity of the protein α-helix with the DNA major groove. The fact that we selected mostly His, and no Arg or Lys, suggests that the orientation of the contact in Mnt is similar to the central positions of zinc-finger interactions.
The selection of Pro with operator A5 is something of a special case due to the dam methylation of both A5 and A6′. The fact the we obtained Pro primarily, and that this was the only mutation obtained by Youderian et al. (6) in a different selection with the same operator sequence, indicates very high specificity for this particular combination. Methylation of the dam sites is required for the high affinity interaction with H6P Mnt (24), suggesting that hydrophobic interactions are involved. However, the specificity cannot be solely due to hydrophobic interactions, or we would expect to obtain other amino acids as well, as we did with the T5 operator. As suggested previously (6), it is likely that specific van der Waals contacts occur between the Pro and the methylated bases, and that other amino acids cannot make the same set of contacts. A distortion of the β-ribbon by the Pro may also contribute to the specific interaction and the fact that other amino acids cannot serve as well.
Perhaps the most interesting results are the selection of several different amino acids with the operator T5. These are predominantly hydrophobic; in fact, every aliphatic amino acid was obtained except Leu, and that might have been obtained had we sequenced more selected colonies. However, the preference for Ile and Val over Leu and Met is consistent with the comparison to zinc-finger proteins described above. Leu and Met are preferred as contacts to T in the outer positions of the binding sites, whereas Ile and Val are preferred at the center position (22). This again suggests that the orientation and proximity of amino acid 6 with respect to the base pair at position 5 is similar to the central position of a zinc-finger interaction. The large number of different amino acids selected suggests that the protein–DNA complex is driven by the hydrophobic effect, where desolvation of the methyl group of T5 and the amino acid side chains provide the free energy of interaction (25–28). While specific van der Waals contacts may contribute too, at least in some cases, that would not seem to be the primary driving force.
There are many previous examples of protein changes involving hydrophobic amino acids being correlated with changes in T-A base pairs in the binding sites. In at least three different proteins, changing Gln to Ala results in the binding site changing from A to T (i.e. the base pair is ‘flipped’ from an A-T to a T-A) (29–31). In another example, a Thr to Ala change results in a change of the binding site base pair from a C-G to a T-A (32). In the lac repressor, changing Gln18 to any of Ala, Thr, Val or Met, changes the binding site base pair from G-C to either A-T or T-A (33). There are also examples in the opposite direction, where the wild-type protein has a hydrophobic amino acid interacting with a T-A base pair. For example, in the Trp repressor, changing Ile79 to Lys alters the preferred base pair from T-A to G-C (34). In addition, in the yeast zinc-finger protein Adr1p, changing Leu146 to His alters the preferred binding site from T-A to G-C at two adjacent positions (35).
The nine-base central core of the Mnt operator is conserved almost completely in the SELEX products for all of the protein variants. This is consistent with the model for the Mnt–DNA interaction since positions 7–15 of the operator are proposed to interact with Mnt amino acids R2, N8 and R10, which are conserved in all of the variant proteins. The results also verify that Mnt proteins with His-tags, even with up to four amino acids deleted from the C-terminus, bind to the full operator with presumably the same cooperative interactions between the dimers as observed for the wild-type protein (18).
There is no significant difference in the base preferences between the full-length His-tagged wild-type protein and the shorter Mnt78 and Mnt79 variants, so their binding sites have all been combined in the ‘H’ columns of Tables 2 and 3. However, there is a subtle but consistent difference between the sites selected with these proteins from those obtained previously using wild-type Mnt without the His-tag (9). As seen before, there is a preference for G over the wild-type A at position 3. However, previously there was a strong preference for T at position 6, with 93 of the 124 sites containing T and 19 containing G (with the remaining 12 being 9 A and 3 C) (9). In the current selections there is a slight preference for G over T, 36 to 31, at position 6. More importantly, there is an asymmetry that did not appear with the wild-type protein. The nearly symmetric operator has a central base that can used to define an orientation. If we choose the strand with a C at position 11 as the ‘top strand’, as in Figure 1, then we can ask whether specific variations are more likely to occur on one side of the operator or the other. In the previous selections with wild-type Mnt without a His-tag (9), the ratio of T to G on the left side (position 6) was 42 to 9, and on the right side (position 16) was 46 to 7. In the selections reported here for all of the Mnt proteins with wild-type H6, the left ratio of T to G is 4 to 30, and on the right side is 27 to 6. The right side now behaves as both sides did previously, with a strong bias towards T over G, but the left side has switched to a strong preference for G over T. The difference between the proteins is only in the C-terminus and, excluding the shortened proteins, is only the presence of the His-tag in the current selections. Berggrun and Sauer (18) measured a strain incurred upon cooperative binding of the full operator, and postulate this could be due to the specific geometry of the Mnt tetramers created by the interactions within the C-terminal tetramerization domains. Those measurements were performed on Mnt proteins containing His-tags and binding to wild-type operators. The T6G mutant operator is preferred by the His-tagged protein, but not the wild-type protein, possibly because the His-tag modifies the interactions between the C-terminal domains and alters the preferred orientation of the N-terminal domains with the DNA. In the study of non-independence of positions 5 and 6 (11) it was shown that the preference for T at position 6 depended on having a G at position 5, and otherwise a G was preferred at position 6. One explanation for the results in this paper is that the His-tagged protein does not allow for the optimal interaction between residue His6 and G5 of the operator, thereby allowing the preference at position 6 to behave as if G was not the adjacent base. All of the other base/position preferences are essentially the same between the His-tagged and non-tagged wild-type proteins.
The most important change in the binding sites that accompanies changes in residue 6 are at operator position 5, consistent with the model for the Mnt–DNA interaction shown in Figure 1. This can be seen in the classes of operator sites observed in Table 5. It can also be seen in the mono-nucleotide preferences for each protein shown in Table 3, but it is also clear that the base preferences at other positions also change. Most other amino acids prefer an A at position 6, even though the model postulates no interaction between the protein and that operator position. This is consistent with previous work showing that even the wild-type protein interacts with both positions 5 and 6 in a non-additive manner (9,11). H6A provides a good example of non-independence between positions 5 and 6. H6A prefers either G or T at position 5 and A or G at position 6, but the combination GA never occurs, with TA and GG being the preferred di-nucleotides. TA is the sequence at the equivalent positions in the arc operator and, not surprisingly, is the preferred sequence of H6Q, which has the Arc substitution at residue 6. Several other proteins also prefer the TA combination at positions 5 and 6. Mnt prefers the GG combination and H6A appears to be able to bind to both sequences well, binding in either an Arc-like or Mnt-like mode.
Most surprising are the changes in base preferences at positions 3 and 4. Mnt selected almost exclusively G at position 4 while most other proteins prefer either G or A at position 4, with H6L and H6M strongly preferring A. In the model, N8 interacts with position 4 and it is the same in all of the proteins, but Asn has been observed interacting with each of the possible base pairs in different protein–DNA complexes (19). Small changes in its orientation, created by substitutions for H6, leading to changes in the base preferences at position 4 are therefore consistent with the interaction of N8 with that position.
For most proteins the specificity at position 3 is weaker than at the others, also consistent with the model which includes no direct interaction with that position. The wild-type protein prefers A or G. Other proteins can prefer A or G or T, but never C, and in most cases more than one base is often selected with a fairly high frequency. This effect can also be explained by small differences in the orientation, or distance, of the protein relative to the outer positions in the operator which are caused by changes at residue 6. These results are all consistent with Mnt residue 6 being a ‘master residue’ that both interacts with DNA directly and influences how other amino acids interact (8). However, in our examples, the influence is only on the outer positions, 3–6, and not on the inner positions, 7–10. In the context of the Mnt N-terminus, all of the different proteins select the same wild-type core sequence at positions 7–15, but the operator positions 3–6 and 16–19 vary considerably depending on amino acid 6.
Table 4 shows the correlation between the operators selected by each amino acid for both the mono- and tetra-nucleotide compositions (i.e. correlations between the columns of Tables 2 and 3). Amino acid pairs with high correlation impose similar constraints on the binding sites. Using the mono-nucleotide correlations (the upper half of Table 4) and grouping together amino acids with a correlation above 0.8 for any pair (i.e. single-link clustering) results in three groups: group 1 = {A, H, S, T}; group 2 = {G, I, L, M, N, Q, V}; group 3 = {R}.
Using the tetra-nucleotide compositions and a threshold of 0.75 results in a similar grouping, except that group 2 splits into two: group 2a = {G, I, N, Q, V}; group 2b = {L, M}.
At a lower threshold of 0.6, required to merge groups 2a and 2b, group 1 also merges. The difference between the groupings at the two levels is due to the operator sequence AATA which is preferred exclusively by H6L and H6M. At the mono-nucleotide level the operators for those two proteins are fairly similar to those from group 2a and so form the entire group 2.
Similar groups are obtained if one clusters the operator sequences rather than the amino acids. Table 5 shows the correlations between the rows of Table 2, using only the eight most frequent operator sequences. The correlations between operators fall into three distinct classes, where within each class the correlations are all positive and between classes are all negative. One class is just the operator AATA which has a negative correlation with all of the others. As stated above it is preferred only by H6L and H6M and clusters those two proteins as group2b. The other classes of operators are distinguished primarily by the base at position 5. Those with a G there (GGGG, GGGT and AGGG) are all positively correlated and they are primarily the preferred operators for proteins in group 1. The other class of operators has T or C at position 5, and they are primarily preferred by proteins in group 2a. The protein group 3 is occupied only by the protein H6R which is a unique case. It has essentially no preference for bases at positions 3–5, but requires G at position 6. This is consistent with the comparison made above with zinc-finger proteins. When zinc-finger binding sites have G in the center position, His is the preferred amino acid, but for Gs in the outer positions, Arg and Lys are preferred. These differences are due to the sizes of the different amino acids and the distance between the protein and the base pairs in those two positions. The interaction of wild-type H6 of Mnt with G at operator position 5 suggests a close interaction, but with the H6R replacement the preference changes to G at the neighboring position 6, presumably because the distance to that base pair is appropriate for the Arg contact. In doing so, the orientation or distance of the N8 contact with position 4 is apparently lost because H6R has little preference for the outer operator positions.
The amino acids in groups 1 and 2a are not intuitive; they certainly do not correspond to typical amino acid similarity tables such as PAM or BLOSUM matrices (36,37). That may not be too surprising because the criteria here for similarity is what bases are preferred in the binding sites, which has to do with specific interaction potentials rather than overall similarities of amino acid properties. Different amino acids may select the same base pair for different reasons. For example, the hydrophobic amino acids may prefer the T at position 5 because of its methyl group, whereas N and Q can form hydrogen bonds with A-T base pairs. The Mnt interaction with DNA is also complicated by the fact that both strands of the β-ribbon are in, or near, the major groove of the DNA, but the model has only one of the H6 and R10 amino acids interacting directly with the base pairs. In the Arc–DNA interaction, Q9 and R13 (the homologous positions) from both monomers interact with base pairs in the operator (7,8). Perhaps the Mnt variants allow the amino acid on the other strand, or both amino acids, to interact directly with the DNA. Such a rearrangement could easily propagate outwards and modify the contacts between those bases and residues.
Substitutions of H6 of the Mnt repressor show the characteristics of a ‘master residue’ as defined by Raumann et al. (8). It interacts directly with DNA at operator position 5 where mutations can be suppressed in vivo by altered residues in the protein. Proteins with a variety of different substitutions for H6 primarily affect the base preferences of selected sites at operator position 5, but such variants also affect the base preferences at other operator positions, indicating that the interaction between amino acid 6 and DNA can influence the interactions of other residues and base pairs. Because the β-ribbon structure of the Mnt-binding domains positions two protein strands near the DNA it is possible that both of them interact directly with the base pairs, as in Arc (7,8), at least in some of the mutant proteins. Changes in the interaction between residue 6 and operator position 5 do not significantly affect the contacts with the central core of the operator site, as it is highly conserved in all protein variants. However, changes in base preferences for all positions from 3 to 6 are evident. Those operator regions can be grouped into three classes with high correlations within the classes and negative correlations between them. Distinct sets of amino acids prefer each class of operator, but the sets do not conform to typical measures of amino acid similarity. This indicates that particular base pairs can be selected by quite different amino acids, presumably by contacting different features of those base pairs, which contributes to the degeneracy of any protein–DNA recognition code (38).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr Stephen Elledge and Dr Ronald Davis for their donation of the transcriptional interference plasmids pNN388 and pNN396. We also thank Dr Kendall Knight and Dr Robert Sauer for their donation of Mnt plasmid pTM201, and Dr Larry Gold for the donation of E.coli strain JM107. We thank Nexagen for the gift of some DNA oligos. This work was supported by NIH grant GM28755.
REFERENCES
- 1.Vershon A.K., Youderian,P., Susskind,M.M. and Sauer,R.T. (1985) The Bacteriophage P22 Arc and Mnt repressors. J. Biol. Chem., 260, 12124–12129. [PubMed] [Google Scholar]
- 2.Vershon K.A., Liao,S.-M., McClure,W.R. and Sauer,R.T. (1987) Bacteriophage P22 Mnt repressor DNA binding and effects on transcription in vitro. J. Mol. Biol., 195, 311–322. [DOI] [PubMed] [Google Scholar]
- 3.Knight K.L. and Sauer,R.T.(1992) Biochemical and genetic analysis of operator contact made by residues within the β-sheets DNA binding motif of Mnt repressor. EMBO J., 11, 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knight K.L. and Sauer,R.T.(1989) DNA binding specificity of the Arc and Mnt repressors is determined by a short region of N-terminal resirues. Proc. Natl Acad. Sci. USA, 86, 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knight K.L. and Sauer,R.T.(1989b) Identification of functionally important residues in the DNA binding region of Mnt repressor. J. Biol. Chem., 264, 13706–13710. [PubMed] [Google Scholar]
- 6.Youderian P., Vershon,A., Bouvier,S., Sauer,R.T. and Susskind,M.M. (1983) Changing the DNA-binding specificity of a repressor. Cell, 35, 777–783. [DOI] [PubMed] [Google Scholar]
- 7.Raumann B.E., Rould,M.A., Pabo,C.O. and Sauer,R.T. (1994) DNA recognition by β-sheets in the Arc repressor-operator crystal structure. Nature, 367, 754–757. [DOI] [PubMed] [Google Scholar]
- 8.Raumann B.E., Knight,K.L. and Sauer,R.T. (1995) Dramatic changes in DNA-binding specificity caused by single residue substitutions in an Arc/Mnt hybrid repressor. Nature Struct. Biol., 2, 1115–1122. [DOI] [PubMed] [Google Scholar]
- 9.Fields D.S., He,Y-Y., Al-Uzri,A.Y. and Stormo,G.D. (1997) Quantitative specificity of Mnt repressor. J. Mol. Biol., 271, 178–194. [DOI] [PubMed] [Google Scholar]
- 10.Stormo G.D. and Fields,D.S. (1998) Specificity, energy and information in DNA–protein interactions. Trends Biochem. Sci., 23, 109–113. [DOI] [PubMed] [Google Scholar]
- 11.Man T.-K. and Stormo,G.D. (2001) Non-independence of Mnt repressor-operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucleic Acids Res., 29, 2471–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elledge S.J. and Davis,R.W. (1989) Position and density effects on repression and stationary and mobile DNA-binding proteins. Genes Dev., 3, 185–197. [DOI] [PubMed] [Google Scholar]
- 13.Childs J., Villanueba,K., Barrick,D., Schneider,T., Stormo,G., Gold,L., Leitner,M. and Caruthers,M. (1985) Ribosome binding site sequences and function. In Calendar,R. and Gold,L. (eds), Sequence Specificity in Transcription and Translation. Alan R. Liss, Inc., New York, pp. 341–350.
- 14.Barrick D., Villanueba,K., Childs,J., Kalil,R., Schneider,D., Lawrence,C.E., Gold,L. and Stormo,G.D. (1994) Quantitative analysis of ribosome binding sites in E.coli. Nucleic Acids Res., 22, 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beutel B.A. and Gold,L. (1992) In vitro evolution of intrinsically bent DNA. J. Mol. Biol., 228, 803–812. [DOI] [PubMed] [Google Scholar]
- 16.Knight K.L. and Sauer,R.T. (1988) The Mnt repressor of bacteriophage P22: role of C-terminal residues in operator binding and tetramer formation. Biochemistry, 27, 2088–2094. [DOI] [PubMed] [Google Scholar]
- 17.Waldburger C.D. and Sauer,R.T. (1995) Domains of Mnt repressor: role in tetramer formation, protein stability and operator DNA binding. Biochemistry, 34, 13109–13116. [DOI] [PubMed] [Google Scholar]
- 18.Berggrun A. and Sauer,R.T. (2001) Contributions of distinct quaternary contacts to cooperative operator binding by Mnt repressor. Proc. Natl Acad. Sci. USA, 98, 2301–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandel-Gutfreund Y. and Margalit,H. (1998) Quantitative parameters for amino acid–base interaction: implications for prediction of protein–DNA binding sites. Nucleic Acids Res., 26, 2306–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lustig B. and Jernigan,R. (1995) Consistencies of individual DNA base-amino acid interactions in structures and sequences. Nucleic Acids Res., 23, 4707–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choo Y. and Klug,A. (1997) Physical basis of a protein–DNA recognition code. Curr. Opin. Struct. Biol., 7, 117–125. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki M. and Yagi,N. (1994) DNA recognition code of transcription factors in the helix–turn–helix, probe helix, hormone receptor and zinc finger families. Proc. Natl Acad. Sci. USA, 91, 12357–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfe S.A., Greisman,H.A., Ramm,E.I. and Pabo,C.O. (1999) Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J. Mol. Biol., 285, 1917–1934. [DOI] [PubMed] [Google Scholar]
- 24.Vershon A., Youderian,P., Weiss,M., Susskind,M. and Sauer,R. (1985) Mnt repressor–operator interactions: altered specificity requires N-6 methylation of operator DNA. In Calendar,R. and Gold,L. (eds), Sequence Specificity in Transcription and Translation. Alan R. Liss, Inc., New York, pp. 209–218.
- 25.Ivarie R. (1987) Thymine methyls and DNA–protein interactions. Nucleic Acids Res., 15, 9975–9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ha J.H., Spolar,R.S. and Record,M.T.,Jr (1989) Role of the hydrophobic effect in stability of site-specific protein–DNA complexes. J. Mol. Biol., 209, 801–816. [DOI] [PubMed] [Google Scholar]
- 27.Plaxco K. and Goddard,W.,III (1994) Contributions of the thymine methyl group to the specific recognition of poly- and mononucleotides: an analysis of the relative free energies of solvation of thymine and uracil. Biochemistry, 33, 3050–3054. [DOI] [PubMed] [Google Scholar]
- 28.Omichinski J., Clore,G., Schaad,O., Felsenfeld,G., Trainor,C., Appella,E., Stahl,S. and Gronenborn,A. (1993) NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science, 261, 438–446. [DOI] [PubMed] [Google Scholar]
- 29.Wharton R.P. and Ptashne,M. (1987) A new-specificity mutant of 434 repressor that defines an amino acid–base pair contact. Nature, 326, 888–891. [DOI] [PubMed] [Google Scholar]
- 30.Wissmann A., Baumeister,R., Muller,G., Hecht,B., Helbl,V., Pfleiderer,K. and Hillen,W. (1991) Amino acids determining operator binding specificity in the helix–turn–helix motif of Tn10 Tet repressor. EMBO J., 10, 4145–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Leeuwen H.C., Strating,M.J., Cox,M., Kaptein,R. and van der Vliet,P.C. (1995) Mutation of the Oct-1 POU-specific recognition helix leads to altered DNA binding and influences enhancement of adenovirus DNA replication. Nucleic Acids Res., 23, 3189–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschmied L., Baumeister,R. Pfleiderer,K. and Hillen,W. (1988) A threonine to alanine exchange at position 40 of Tet repressor alters the recognition of the sixth base pair of tet operator from GC to AT. EMBO J., 7, 4011–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sartorius J., Lehming,N., Kisters,B., von Wilcken-Bergmann,B. and Muller-Hill,B. (1989) lac repressor mutants with double or triple exchanges in the recognition helix bind specifically to lac operator variants with multiple exchanges. EMBO J., 8, 1265–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfau J., Arvidson,D.N., Youderian,P., Pearson,L.L. and Sigman,D.S. (1994) A site-specific endonuclease derived from a mutant Trp repressor with altered DNA-binding specificity. Biochemistry, 33, 11391–11403. [DOI] [PubMed] [Google Scholar]
- 35.Cheng C. and Young,E.T. (1995) A single amino acid substitution in zinc finger 2 of Adr1p changes its binding specificity at two positions in UAS1. J. Mol. Biol., 251, 1–8. [DOI] [PubMed] [Google Scholar]
- 36.Henikoff S. and Henikoff,J.G. (1992) Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA, 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henikoff S. and Henikoff,J.G. (1993) Performance evaluation of amino acid substitution matrices. Proteins, 17, 49–61. [DOI] [PubMed] [Google Scholar]
- 38.Benos P.V., Lapedes,A.S. and Stormo,G.D. (2002) Is there a code for protein–DNA recognition? Probab(ilistical)ly. Bioessays, 24, 466–475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.