Abstract
Homologous recombination is a fundamental process for genome maintenance and evolution. Various proteins capable of performing homology recognition and pairing of DNA strands have been isolated from many organisms. The RecA family of proteins exhibits a number of biochemical properties that are considered hallmarks of homology recognition. Here, we investigated whether the unrelated Escherichia coli RecT protein, which mediates homologous pairing and strand exchange, also exhibits such properties. We found that, like RecA and known RecA homologs: (i) RecT promotes the co-aggregation of ssDNA with duplex DNA, which is known to facilitate homologous contacts; (ii) RecT binding to ssDNA mediates unstacking of the bases, a key step in homology recognition; (iii) RecT mediates the formation of a three-strand synaptic intermediate where pairing is facilitated by local helix destabilization, and the preferential switching of A:T base pairs mediates recognition of homology; and (iv) RecT-mediated pairing occurs from both 3′- and 5′-single-stranded ends. Taken together, our results show that RecT shares fundamental homology-recognition properties with the RecA homologs, and provide new insights on an underlying universal mechanism of homologous recognition.
Keywords: DNA base unstacking/DNA co-aggregation/DNA unwinding/homologous pairing/RecT
Introduction
In Escherichia coli, the major recombination pathway operating in wild-type cells requires the function of RecA and RecBCD proteins (Kowalczykowski et al., 1994). In the absence of a functional RecBCD complex, cells are deficient for recombination and some types of repair. Two types of suppressor mutations, sbcA and sbcB(sbcC), which restore recombination proficiency to recBC mutants, have been isolated (Clark and Sandler, 1994). The sbcA mutations map in the cryptic Rac prophage and induce the expression of the RecE and RecT proteins (for a review, see Kolodner et al., 1994). Remarkably, when RecE and RecT are expressed, many types of recombination events can occur in the absence of RecA (Fishel et al., 1981; Gillen et al., 1981; Symington et al., 1985; Luisi-DeLuca and Kolodner, 1992; Takahashi et al., 1993). These recombination events include plasmid recombination, double-strand break (DSB) and gap repair, and recombination of λ red mutant bacteriophages. Mechanistic studies have suggested that some RecET-mediated recombination events involve the formation of strand-exchange intermediates (Kobayashi and Takahashi, 1988; Takahashi et al., 1997). In addition, the RecET system can promote recombination between short homologous sequences such as occurs in PCR construct-mediated gene targeting events, recombination events that are apparently not promoted by RecA-dependent recombination pathways (Zhang et al., 1998; Muyrers et al., 2000).
The recE gene product, also called exonuclease VIII (Kushner et al., 1971), is an ATP-independent exonuclease that preferentially degrades linear double-stranded DNA (dsDNA) in the 5′ to 3′ direction and also degrades single-stranded DNA (ssDNA) at low rates (Joseph and Kolodner, 1983). The recT gene product binds to ssDNA and promotes the renaturation of ssDNA (Hall et al., 1993). RecT, in the absence of ATP, promotes homologous pairing and strand exchange between a circular ssDNA and a linear dsDNA containing short single-strand tails by a mechanism that results in strand displacement and the formation of heteroduplex joints as long as 6 kb (Hall and Kolodner, 1994). RecT in combination with the RecE exonuclease will also promote a similar reaction between a circular ssDNA and a linear dsDNA with a blunt end. In addition, RecT promotes D-loop formation between a supercoiled dsDNA and an homologous ssDNA, or the 3′-single-stranded ends of a linear duplex, the latter being a substrate similar to that generated by the action of the RecE exonuclease. This DNA strand-transfer reaction is mediated by RecT–ssDNA nucleoprotein complexes, and does not require a high-energy cofactor (Noirot and Kolodner, 1998). In many regards, the homologous pairing and strand-exchange reactions promoted by RecT are similar to those promoted by RecA in the presence of non-hydrolyzable ATP analogs like ATPγS (Menetski et al., 1990; Rosselli and Stasiak, 1990; Kowalczykowski and Krupp, 1995). Similarly, the RecA family members Rad51 and Dmc1 can promote homologous-pairing reactions in the absence of ATP hydrolysis (Sung and Stratton, 1996; Hong et al., 2001). Based on these observations, it has been suggested that RecT is a homologous-pairing protein that shares some properties with RecA even though RecT and RecA do not share any sequence homology (Hall and Kolodner, 1994; Noirot and Kolodner, 1998).
Many functional similarities between RecE/RecT- and the bacteriophage λ Redα/Redβ-mediated recombination have been established (Kolodner et al., 1994). Recently, it has been shown that RecE/RecT and Redα/Redβ promote the RecA-independent integration of linear DNA fragments flanked by short homologous arms into the E.coli chromosome or into endogenous plasmids (Zhang et al., 1998; Datsenko and Wanner, 2000; Yu et al., 2000). In addition, a specific protein–protein interaction between the exonuclease (Redα or RecE) and its cognate recombinase (Redβ or RecT) is required for recombination (Muyrers et al., 2000). Moreover, RecT and Redβ are evolutionarily related, and define a distinct superfamily of recombination proteins (Iyer et al., 2002). Thus, RecE/RecT- and Redα/Redβ-mediated recombination appear to be mechanistically equivalent. Consistent with this, the RecET system will also promote recombination of bacteriophage λ red mutants (Kolodner et al., 1994). Recombination could take place by a single strand annealing (SSA) mechanism, first proposed for Redα/Redβ-mediated recombination (Cassuto et al., 1971), and which contributes to the majority of the recombination events in non-replicating λ chromosomes (Stahl et al., 1997), because RecT and Redβ promote annealing of complementary single strands and strand exchange in vitro (Hall et al., 1993; Hall and Kolodner, 1994; Li et al., 1998). However, it is possible also that RecET-mediated recombination of bacteriophage λ red mutants might occur by a DNA strand invasion mechanism (Noirot and Kolodner, 1998), as proposed to account for DSB and gap repair (Symington et al., 1985; Kobayashi and Takahashi, 1988; Luisi-DeLuca and Kolodner, 1992; Takahashi et al., 1997).
In this study, we report that RecT, despite its genetic unrelatedness to RecA and its independence of ATP, exhibits some of the hallmarks of the reactions by which members of the RecA family of proteins promote the recognition of homology between a single-strand and duplex DNA. Our findings may provide new insights on an underlying universal mechanism of homologous recognition.
Results
RecT binding promotes aggregation of ssDNA but not of dsDNA
DNA binding assays were performed by incubating 7.2 kb M13mp19 ss- and dsDNA with different concentrations of RecT protein, and the formation of RecT–DNA complexes was monitored by agarose gel electrophoresis (Figure 1). RecT protein modestly retarded the migration of dsDNA up to 5 µM RecT, and at higher concentrations the RecT–dsDNA complexes migrated as a smear. These results suggest that the RecT–dsDNA complexes might not be stable under the electrophoresis conditions used here. Indeed, upon fixation of the protein–DNA complexes with glutaraldehyde (0.2% final concentration, 10 min at 37°C) prior to electrophoresis, much larger RecT–dsDNA complexes, which did not enter the gel, were observed at all RecT concentrations. Saturation of the dsDNA was reached at a ratio of 1 RecT monomer/7.5 base pairs (data not shown). This confirms previous observations that RecT binds to dsDNA with a high affinity (Noirot and Kolodner, 1998). However, the formation of aggregates after fixation cannot be interpreted as an indication that RecT causes dsDNA aggregation because glutaraldehyde generates DNA–protein and protein–protein cross-links.
Fig. 1. Aggregation of ssDNA but not dsDNA upon RecT binding. Reaction mixtures (30 µl) containing the indicated RecT concentrations were incubated in buffer T with 25 µM 32P-labeled M13mp19 linear dsDNA (lanes a–i) or with 12.5 µM 32P-labeled M13mp19 ssDNA (lanes j–r) for 20 min at 37°C. The reaction products were resolved by electrophoresis in a 0.6% agarose gel. Aggregated material remained at the gel origin. The positions of linear dsDNA and ssDNA are indicated by the arrows.
In contrast, RecT binding to ssDNA produced large aggregates that failed to enter the gel without fixation. These aggregates could also be pelleted by centrifugation in a microcentrifuge (data not shown). The mobility of these complexes remained unchanged upon fixation (data not shown), indicating that RecT–ssDNA complexes were stable under the gel conditions used. Unbound ssDNA was observed at sub-saturating concentrations of RecT, indicative of cooperative binding. Saturation of the ssDNA was observed at 1 RecT monomer/3 bases, which is near the optimum ratio for D-loop formation (Noirot and Kolodner, 1998).
Formation of ternary complexes ssDNA–RecT–dsDNA
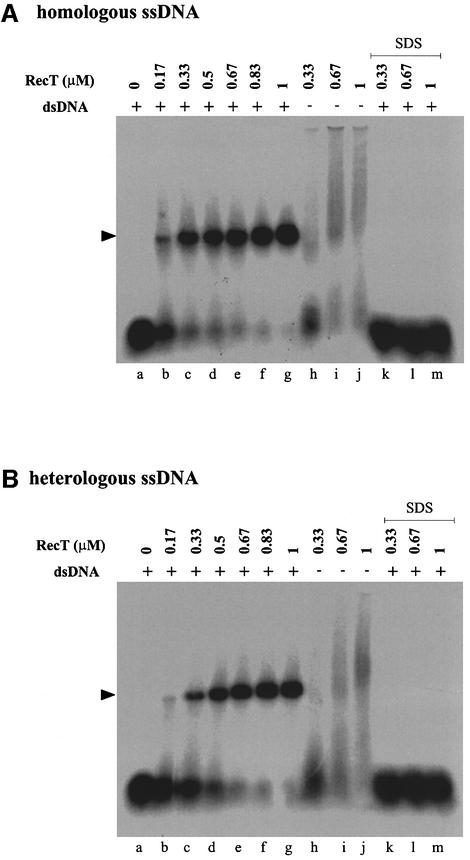
To examine the ability of RecT–ssDNA complexes to promote co-aggregation with dsDNA, the formation of a complex between either a homologous or heterologous 32P-labeled ssDNA (50 nucleotides) and a linear dsDNA was monitored. The ssDNA was pre-incubated with RecT to form complexes capable of migrating as a smear in an agarose gel (Figure 2, lanes h–j). The reaction was initiated by adding the linear dsDNA, and the formation of a ssDNA–dsDNA complex was monitored by measuring the co-migration of radioactivity with the linear dsDNA during electrophoresis through agarose gels (Figure 2, lanes a–g). The amount of complex formed increased with increasing RecT concentration until all the ssDNA was present in complexes. Complex formation did not occur if either RecT or dsDNA was omitted from the reaction, indicating that a ternary complex ssDNA–RecT–dsDNA was formed. Ternary complex formation occurred with similar efficiency when either homologous ssDNA (Figure 2A) or heterologous ssDNA (Figure 2B) was present in the reaction. Upon deproteinization, only the two parental DNA molecules were observed, indicating that stable strand exchange with linear dsDNA did not occur under the reaction conditions used. Taken together, these results indicate that RecT promotes a homology-independent co-aggregation of ssDNA and dsDNA.
Fig. 2. RecT promotes co-aggregation of ssDNA with dsDNA. The indicated RecT concentrations were pre-incubated with 0.83 µM 32P-labeled ssDNA in buffer T for 10 min at 25°C. Unlabeled linear pUC18 DNA (30 µM) was added (lanes a–g and lanes k–m), or omitted in controls (lanes h–j), to the reaction mixtures (30 µl) and incubated for another 40 min at 37°C. Untreated (lanes a–j) and deproteinized (lanes k–m) reaction products were resolved by electrophoresis in a 0.9% agarose gel. (A) Homologous ssDNA (O34). (B) Heterologous ssDNA (O26). The position of linear pUC18 DNA is indicated by the arrow.
RecT binding results in unstacking of ssDNA bases
If the RecT–ssDNA complexes are proficient for homologous pairing, RecT binding might be expected to promote unstacking of the ssDNA bases. This property is a hallmark of homology recognition by RecA and RecA-like proteins (Voloshin et al., 1996; Nishinaka et al., 1997). Unstacked DNA bases are more susceptible to oxidation by potassium permanganate (PP) than stacked bases. Potassium permanganate reacts mostly with thymine residues and creates a lesion that becomes a strand cleavage site after treatment with piperidine (Voloshin et al., 1996). Incubation of RecA with ssDNA thus generally sensitizes the T residues in the ssDNA to modification by PP (Voloshin et al., 1996). In contrast, incubation of E.coli single-strand binding protein (SSB) with ssDNA appears to decrease the accessibility of the bases to PP in single-strand regions without secondary structures, and as expected, to make the bases within the double-strand regions of secondary structures accessible to PP because SSB denatures these regions (Kumar and Varshney, 1997; O.Voloshin and D.Camerini-Otero, personal communication).
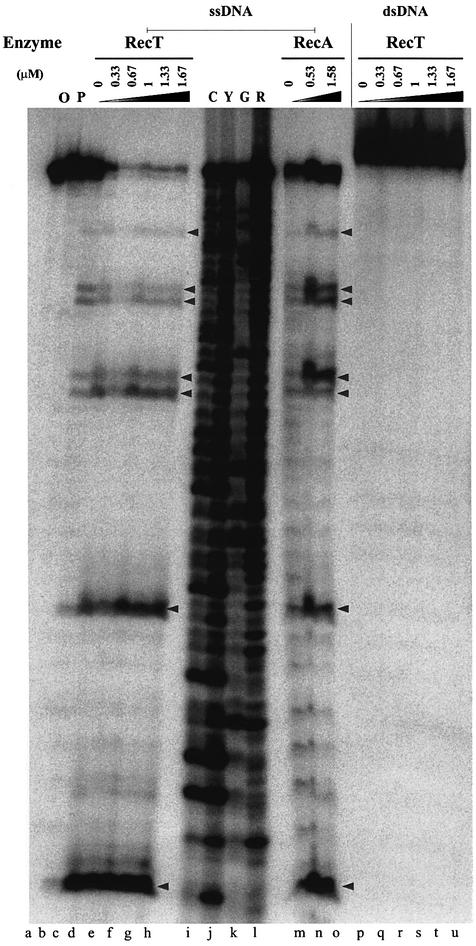
To analyze RecT-promoted unstacking, varying amounts of RecT or RecA proteins were incubated with 5′ 32P-labeled ssDNA prior to PP treatment (Figure 3). ssDNA was weakly sensitive to modification by PP under the conditions used. Upon RecT binding, the sensitivity to PP increased dramatically. As a result, most of the radioactivity was present in the shorter cleavage products and very little intact ssDNA remained, suggesting modification occurred at multiple thymine residues. Maximum modification of the ssDNA was obtained at about 1 RecT monomer/3 bases, consistent with the saturation level observed in the ssDNA binding experiments (Figure 1). RecA binding to the same ssDNA was found to cause a similar PP-sensitivity pattern as RecT, although with a somewhat reduced efficiency. This effect is likely to be caused by the DTT present at low concentration in the RecA preparation, which may partially quench the PP modification reaction. These results show that RecT unstacks ssDNA upon binding.
Fig. 3. RecT binding entails unstacking of ssDNA but not dsDNA. Sensitivity of thymine residues to KMnO4 in ssDNA (lanes c–h and m–o) or dsDNA (lanes p–u) was monitored as a function of RecT (lanes c–h and p–u) or RecA concentrations (lanes m–o). The indicated enzyme concentrations were incubated with 1.7 µM 32P-labeled ssDNA or 3.4 µM 32P-labeled dsDNA in 75 µl of reaction mixtures, for 20 min at 37°C and 10 min at 25°C. The protein–DNA complexes were reacted with 0.6 mM KMnO4 for 1 min at 25 °C, treated with piperidine as described in Materials and methods, and the DNA products were separated in a 20% urea–polyacrylamide gel. KMnO4-untreated controls: ssDNA (lane O) and ssDNA reacted with piperidine (lane P). Lanes C, Y, G and R are standard Maxam–Gilbert sequencing reactions. Arrows indicate the positions of thymines.
The PP-modification assay was also used to monitor the effect of RecT binding to dsDNA. The 5′ 32P-labeled oligonucleotide used above as ssDNA was annealed with its complementary unlabeled oligonucleotide and used as a substrate in reactions with PP. dsDNA did not exhibit any detectable sensitivity to PP upon RecT binding, even at saturating RecT concentrations (Figure 3). The same results were obtained when dsDNA was modified with 1.6 mM PP instead of 0.6 mM PP (data not shown). These results indicate that RecT binding to dsDNA does not result in unstacking of the bases and does not promote formation of single-stranded regions in the dsDNA.
RecT binding promotes unwinding of dsDNA
A topoisomerase I-relaxation assay was used to investigate whether RecT binding to dsDNA induces a conformation change in the dsDNA (Rould et al., 1992). If binding of RecT to a circular dsDNA molecule causes conformation changes, compensatory changes will be generated elsewhere in the dsDNA molecule. These compensatory changes can be relaxed by a topoisomerase I, which is capable of relaxing positive and negative supercoiling. Upon deproteinization, the protein–DNA contacts maintaining the initial conformation changes will be lost, leaving the dsDNA molecule in a superhelical state. The extent and the sign of this superhelicity will reflect the extent to which the conformation of the dsDNA molecule has been altered by the binding of RecT.
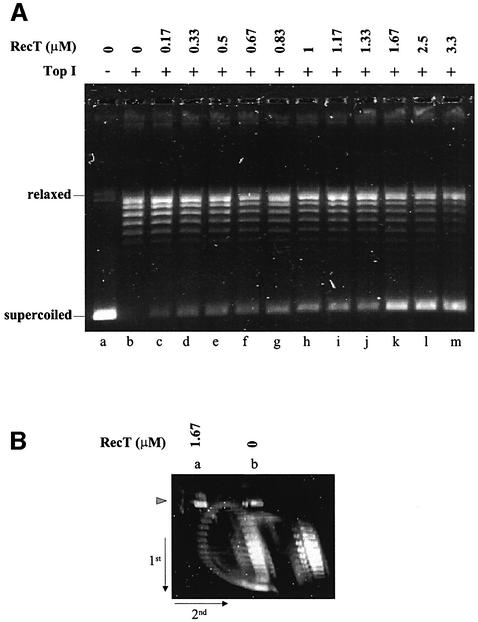
Supercoiled DNA was fully relaxed by treatment with an excess of wheat germ topoisomerase I. This relaxation eliminated the single-strand regions that could initially be present in the DNA. Indeed, it has been shown previously that E.coli SSB can interact with regions that have single-stranded character present in supercoiled circular DNA but cannot interact with relaxed circular DNA (Glikin et al., 1983). Then, the topoisomerase I–DNA mixture was further incubated in the presence of increasing amounts of RecT. Wheat germ topoisomerase I is fully active in RecT binding buffer (no Mg++). After a 45 min incubation at 37°C, the reaction mixture was deproteinized and DNA products were separated by electrophoresis through an agarose gel (Figure 4A). Addition of RecT provoked the appearance of a fast-migrating band that co-migrated with supercoiled DNA. The amount of material in this band increased with RecT concentration to reach a plateau at 1.67 µM, where it represented ∼65% of the total DNA. The DNA in this band corresponded to highly-negatively supercoiled DNA, as demonstrated by two-dimensional chloroquine gel electrophoresis (Figure 4B). From these results, we conclude that RecT binding induces unwinding of the dsDNA. Interestingly, the unwinding promoted by RecT parallels that promoted by RecA in the presence of ATPγS, under conditions where extensive unwinding of duplex DNA is observed even in the absence of ssDNA (Stasiak and Di Capua, 1982). When RecT was added in the reaction as a homologous ssDNA–RecT complex, the extent of unwinding observed was similar to that caused by free RecT (data not shown). These results suggest that either ssDNA–RecT complexes can unwind dsDNA directly or RecT can transfer from the ssDNA–RecT complexes to the dsDNA.
Fig. 4. RecT binding promotes unwinding of dsDNA. (A) Supercoiled pHV2900OB DNA (30 µM) was relaxed by incubation with 4 U of wheat germ topoisomerase I for 20 min at 37°C. Then, RecT was added to the reaction mixtures (30 µl) at the concentrations indicated and incubated for a further 45 min at 37°C in buffer T. The reaction products were deproteinized, separated by electrophoresis in a 1% agarose gel and revealed by SYBR green staining. Unreacted supercoiled pHV2900OB DNA is in lane a. The positions of supercoiled and relaxed DNA molecules are indicated. (B) pHV2900OB DNA treated exactly as described above, with 1.67 µM RecT (lane a) or no RecT (lane b). The deproteinized reaction products were subjected to two- dimensional chloroquine gel electrophoresis. Chloroquine concentrations used were 0.66 µM and 2 µM in the first and second dimension, respectively. Position of the nicked circle is indicated by an arrow-head.
Formation of synaptic intermediates by RecT
The ability of RecT to pair two homologous DNAs and exchange strands between the paired DNAs was studied by using assays based on fluorescence resonance energy transfer (FRET). The fluorometric assays are non-disruptive and allow one to observe the reaction as it occurs in solution (Bazemore et al., 1997b). In the pairing assay, fluorescein is located on the single-stranded oligonucleotide, and rhodamine is located on its complementary strand in the duplex substrate. In the assay for strand exchange, the two dyes are located initially on the two strands of the duplex DNA. These assays have been used previously to investigate the mechanism of homologous pairing and strand exchange by E.coli RecA and its human homologs, Rad51 and Dmc1 (Bazemore et al., 1997a,b; Gupta et al., 1997, 1999a, 2001).
In the pairing assay, strands labeled with fluorescein and rhodamine come into proximity in a three-stranded intermediate, and they remain in proximity in subsequent products. In that state, the fluorescent dyes can participate in fluorescence resonance energy transfer: fluorescein excited by light at its absorption maximum transfers energy non-radiatively to rhodamine, which results in enhanced or sensitized emission of light from rhodamine. Measurement of the sensitized emission is essential to demonstrate homology-dependent interaction of strands of DNA (Bazemore et al., 1997a,b; Gupta et al., 1997, 1999a, 2001)
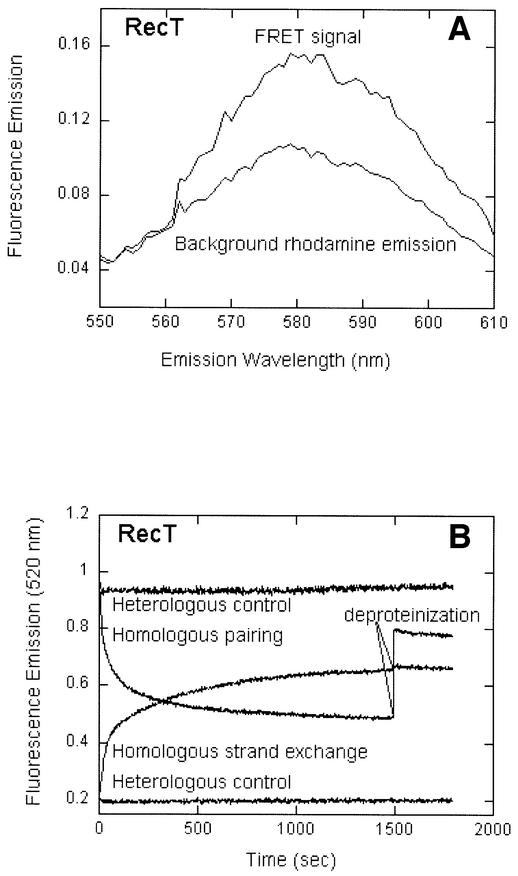
RecT-mediated pairing and strand-exchange reactions were studied with the 83mer oligonucleotide G16 (16% GC content) containing fluorescein and rhodamine fluorophores. In pairing assays carried out with RecT and homologous substrates, the amplitude of the sensitized emission from rhodamine was 50% greater than the background non-sensitized emission from dyes that were not in proximity (Figure 5A), whereas the emission from heterologous substrates was indistinguishable from the background emission (data not shown). The amplitude of the sensitized emission measured for reactions of RecT was similar to that previously observed for human Dmc1 (Gupta et al., 2001). The demonstration of energy transfer shows that the dye-labeled homologous strands have been brought into close proximity.
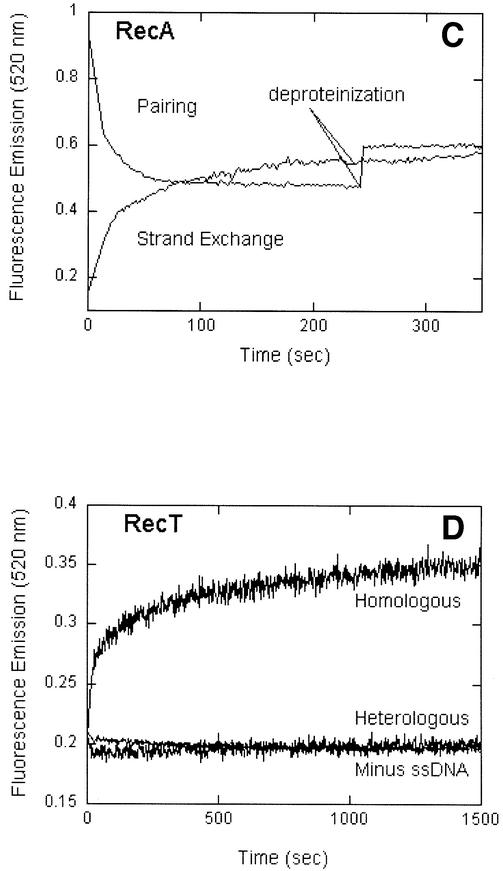
Fig. 5. RecT forms protein-dependent synaptic complexes. (A) The FRET signal or sensitized emission for homologous pairing promoted by RecT. The background rhodamine emission represents the signal obtained in a control experiment in which the ssDNA was devoid of fluorescein fluorophore. (B and C) Pairing and strand-exchange reactions were performed with oligonucleotide G16 (16% GC) by fluorometric assays as described in Materials and methods. The fluorescein emission was recorded every 2 sec at 520 nm on excitation at 493 nm. The reaction was deproteinized by the addition of SDS (final concentration, 0.3%) at 1500 s for RecT (B) and at 240 s for RecA (C). Heterologous controls for pairing and strand exchange (B) contained G26(–)ssDNA and G16 duplex substrates. (D) A strand-exchange reaction by RecT with G16 substrates in which a control lacking any ssDNA shows that the strand-exchange signal did not result from helicase activity or any other adventitious separation of the strands of duplex DNA.
In the pairing assay, as a consequence of the non-radiative transfer of energy from fluorescein to rhodamine, the fluorescence emission from fluorescein is quenched. This aspect of FRET is useful for studying the kinetics of homologous pairing (Bazemore et al., 1997b,a; Gupta et al., 2001). In observations on the time course of homologous pairing of the G16 substrate promoted by RecT, the fluorescence emission from fluorescein was quenched, whereas no quenching was observed with a heterologous substrate (Figure 5B).
In the assay for strand exchange, fluorescein and rhodamine are initially in proximity and participate in FRET until strand exchange occurs, at which time the dyes separate, the FRET signal is lost and the fluorescence emission from fluorescein is correspondingly enhanced. Fluorescence enhancement was observed in reactions of homologous substrates promoted by RecT, whereas no enhancement was observed when heterologous substrates were tested (Figure 5B) or when the single-stranded substrate was omitted (Figure 5D). These two controls show that fluorescence enhancement was not attributable to helicase activity or to melting of the duplex substrate for other adventitious reasons. Thus, acting on oligonucleotide substrates, RecT promoted strand exchange as well as homologous pairing.
In similar experiments on human Dmc1, the effects of the deproteinization of reactions that have reached equilibrium have been investigated (Gupta et al., 1997, 2001). In the pairing assay, deproteinization leads to partial reversal of the quenching of fluorescein because the disruption of the synaptic complex has the potential to produce some of the original substrates, thereby in part reversing FRET. In contrast, in the assay for strand exchange, deproteinization has no effect on the enhanced emission from fluorescein because removing the protein cannot be expected to lead to the reunion of the separated dye-bearing strands. In the case of RecT, when the pairing reaction at equilibrium was deproteinized by the addition of SDS, ∼65% of the signal was reversed, whereas the strand-exchange signal was not affected (Figure 5B). For comparison, we did the same experiment with RecA protein: with substrates labeled for the pairing assay, deproteinization at equilibrium caused a partial, but smaller reversal of the fluorescence signal, whereas deproteinization of the strand-exchange reaction again had no observable effect (Figure 5C). Thus, the effects of deproteinization suggest that RecT, like RecA and Dmc1, forms a synaptic intermediate, the disruption of which can yield either the precursor substrates or products that have undergone strand exchange.
Mechanism of recognition of homology
The data indicating that RecT promotes the formation of a three-stranded synaptic intermediate prompted us to explore how RecT promotes the recognition of homology. Previous studies have shown that homologous recognition mediated by human Rad51 and Dmc1, respectively, requires the preferential breathing of A:T base pairs (Gupta et al., 1999a, 2001). We used similar methods to investigate the mechanism of homologous pairing catalyzed by RecT.
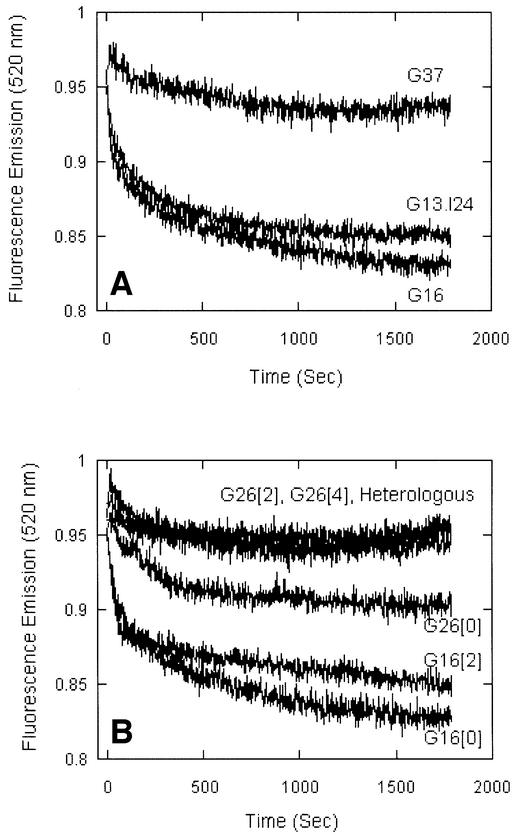
The pairing reaction promoted by RecT was found to be inversely proportional to the GC content of the DNA substrates as is the case for RecA, human Rad51 and human Dmc1 (Gupta et al., 1999a, 1999b, 2001). At 37% GC, as compared with 16% GC, the pairing reaction promoted by RecT was completely eliminated (Figure 6A), but when in the same duplex DNA substrate, inosine was partially substituted for guanine (G13.I24), the reaction was restored. Since I:C base pairs are less stable than G:C base pairs, the stimulation of pairing by inosine substitution suggested that the recognition of homology by RecT is related to helix stability.
Fig. 6. Effects of inosine substitution and mismatches on RecT-promoted pairing. (A) Homologous-pairing reactions promoted by RecT were done with three different pairs of single-stranded and duplex oligonucleotides: G16 (16% GC), G37 (37% GC) and inosine- substituted G37 (13% GC and 24% IC). The inosine residues were substituted in G37 in the place of guanines. (B) Two, four or six mismatches at the sites of A:T base pairs were created by transversions in the duplex substrates, G16 and G26 (28). The numbers in square brackets represents the number of transversions, i.e. mismatches present.
The specific role of A:T base pairs in homology recognition was analyzed in an experiment in which the effect of mismatches on joint molecule formation was tested. As in previous studies (Gupta et al., 1999a), mismatches were introduced by inverting base pairs in the duplex substrate. This experimental design maintained the same base composition as the unaltered substrate and avoided the introduction of deformations in the structure of the substrates. If A:T base pairs play a rate limiting or critical role in recognition of homology, mismatches opposite A:T in relatively AT-rich DNA should have a smaller effect on recognition of homology than the same mismatches in relatively GC-rich DNA.
Two, four or six A:T transversions were made in G16 (16% GC) and G26 (26% GC) to construct mismatched duplexes (Gupta et al., 1999a). As can be seen in Figure 6B, two or more mismatches in G26, the relatively less AT-rich substrate, completely inhibited homologous recognition, whereas two mismatches in G16, the relatively more AT-rich substrate, had virtually no effect on the reaction. This suggests that A:T base pairs play an important role in the RecT-promoted homology search. Together, the effect of inosine substitution and mismatches on RecT-promoted homologous pairing suggest that the breathing of DNA, and in particular the breathing of A:T base pairs, plays a critical role in the recognition of homology by RecT.
RecT mediates strand invasion by DNA with 5′-single-stranded ends
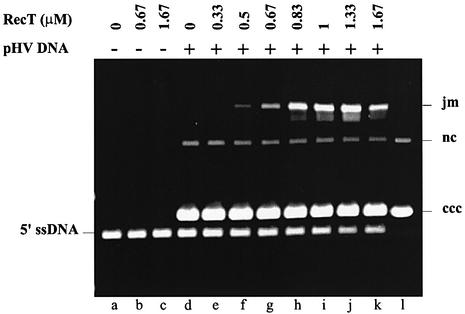
Previously, we showed that RecT could promote D-loop formation between a 3′-single-stranded end of a linear dsDNA, a DNA substrate similar to that the RecE exonuclease would generate, and a supercoiled circular DNA (Noirot and Kolodner, 1998). Here, we investigated whether RecT was capable of mediating strand transfer of 5′-ssDNA ends. Such a DNA substrate was produced by linearizing pUC18 DNA and partially resecting the DNA ends with exonuclease III, which degrades the 3′-strands. The 5′-ssDNA was then pre-incubated with varying amounts of RecT, and supercoiled DNA was added to initiate the reaction. The formation of joint molecules (Figure 7) increased with increasing amounts of RecT and reached a plateau at 1.33 µM RecT where ∼50% of 5′-ssDNA molecules were present in joint molecules. Increasing the RecT concentration further diminished the yield of joint molecules, consistent with the observation that an excess of RecT inhibited pairing (Noirot and Kolodner, 1998). Joint molecules were not formed if the supercoiled DNA was omitted, showing that the joint molecules did not result from stable pairing of 5′-ssDNA molecules with each other. These results show that RecT promotes the pairing of supercoiled DNA with a linear dsDNA having 5′ homologous single-stranded tails.
Fig. 7. D-loop formation by DNA with 5′-single-stranded ends mediated by RecT. pUC18 DNA was linearized by ScaI and partially degraded with exonuclease III, as described in Materials and methods, to generate a resected duplex carrying 5′-single-stranded tails about 100 bases long, designated 5′-ssDNA. 5′-ssDNA (20 µM) was incubated with the indicated amounts of RecT for 20 min at 25°C. Supercoiled pHV2900OB DNA (40 µM) was added to initiate the reactions (lanes d–k) or was omitted in controls (lanes a–c). The reaction mixtures were incubated for 50 min at 37°C, deproteinized and the products resolved by electrophoresis in a 0.9% agarose gel revealed by SYBR green staining. Unreacted supercoiled pHV2900OB DNA is in lane l. jm, joint molecules; nc, nicked circular duplex; ccc, supercoiled pHV2900OB.
Discussion
The E.coli RecT homologous-pairing protein was discovered through studies of the RecE recombination pathway, which can substitute for the RecA–RecBCD-dependent recombination pathway. To understand how the RecT protein can promote homologous pairing, we investigated whether RecT mediates key biochemical reactions thought to be critical for the unrelated RecA family of recombination proteins to promote homologous pairing and strand exchange. We found that RecT exhibited cooperative binding to ssDNA and formed large, stable protein–DNA aggregates. RecT also bound to dsDNA but fixation was required to observe the formation of large protein–DNA aggregates, suggesting that RecT–dsDNA complexes had a lower stability than RecT–ssDNA complexes. Binding of RecT to ssDNA resulted in unstacking of the bases whereas binding of RecT to dsDNA resulted in unwinding of the dsDNA, as defined operationally by a change in linkage number, but did not result in unstacking of the bases. The RecT–ssDNA complexes were found to co-aggregate dsDNA in a homology-independent manner. Under conditions where RecT promotes unstacking of bases in ssDNA, unwinding of dsDNA and co-aggregation of ssDNA with dsDNA, RecT was found to promote D-loop formation with both 5′- and 3′-single-stranded ends. These results indicate that RecT promotes most if not all of the key biochemical reactions thought to be hallmarks of RecA-mediated homologous pairing even though RecT and RecA are genetically unrelated proteins.
The E.coli RecA protein can bind to both ssDNA and dsDNA, although in the presence of ATP, there is a kinetic barrier that strongly favors the formation of nucleoprotein filaments on single strands (Pugh and Cox, 1987). These filaments then synapse with naked dsDNA (Kowalczykowski et al., 1994). Similarly, RecT-mediated pairing involves a ssDNA nucleoprotein complex that synapses with naked dsDNA (Thresher et al., 1995; Noirot and Kolodner, 1998). However, the order of addition was important because D-loops were formed when RecT was pre-incubated with ssDNA but not when pre-incubated with dsDNA, and the yield of D-loops was much reduced when RecT was added to a mixture of ssDNA and dsDNA. Those observations are consistent with the apparent higher affinity of RecT for dsDNA (Noirot and Kolodner, 1998). Thus, there may be other factors in E.coli that can overcome the need for a prescribed order of addition in vitro. Related observations reveal that eukaryotic RecA-like proteins, such as Saccharomyces cerevisiae Rad51 (ScRad51) and human Rad51 (HsRad51) also form nucleoprotein filaments with both ssDNA and dsDNA, under standard reaction conditions (Sung and Robberson, 1995; Baumann et al., 1996).
Under strand-exchange conditions, RecT promoted the co-aggregation of ssDNA with dsDNA in an homology-independent fashion. The RecT-promoted co-aggregates were detected without fixation, suggesting that the stable RecT–ssDNA aggregates were the active species mediating the co-aggregation of dsDNA. Co-aggregation was inhibited by addition of 5 mM Mg++ (P.Noirot, unpublished results) as was the RecT-mediated homologous pairing (Noirot and Kolodner, 1998), supporting the view that co-aggregation may be required for homologous pairing; note that both processes do occur at lower Mg++ concentrations. Co-aggregation is also promoted by RecA, ScRad51 and HsRad51 under reaction conditions where homologous pairing occurs (Tsang et al., 1985; Sung and Robberson, 1995; Baumann et al., 1996). In the case of RecA, co-aggregation was shown to facilitate the establishment of homologous contacts (Chow and Radding, 1985; Tsang et al., 1985).
Under conditions where RecA promotes homologous pairing, RecA–ssDNA filaments are formed in which the ssDNA within the filament is stretched resulting in unstacking of the DNA bases, a reaction that is also promoted by T4 UvsX, ScRad51 and HsRad51 (Stasiak and Di Capua, 1982; Ogawa et al., 1993; Yu and Egelman, 1993; Benson et al., 1994; Voloshin et al., 1996; Nishinaka et al., 1997, 1998). Furthermore, an oligopeptide derived from RecA that retains the ability to unstack the ssDNA bases promotes limited homologous-pairing reactions (Voloshin et al., 1996). This led to the proposal that formation of a well-defined nucleoprotein filament within which ssDNA is extended and ssDNA bases are unstacked is the hallmark of homology recognition mediated by the RecA/Rad51 family of proteins (Voloshin et al., 1996). RecT does not belong to the RecA/Rad51 family based on the lack of homology and on the prominent functional difference that RecT-mediated strand transfer reaction does not require ATP or another nucleotide cofactor (Noirot and Kolodner, 1998). It should, however, be noted that the homologous pairing and strand-exchange reactions between a linear duplex and a ssDNA that are promoted by RecT (this work; Hall and Kolodner, 1994) are similar to those promoted by RecA, Rad51 and Dmc1 in the presence of non-hydrolyzable ATP analogs like ATPγS (Menetski et al., 1990; Rosselli and Stasiak, 1990; Sung and Stratton, 1996; Hong et al., 2001). Furthermore, RecT does not form extended helical filaments on ssDNA like the RecA family of proteins (Thresher et al., 1995). However, the results presented here show that RecT binding to ssDNA, like RecA binding, dramatically increases the sensitivity of thymine residues in the ssDNA to PP modification and thus unstacks the ssDNA bases. The conservation of ssDNA unstacking in such distantly related pairing proteins suggests that unstacking of DNA bases might be a basic property common to all recombination proteins that promote homology-recognition reactions.
We observed that RecT promoted joint molecule formation between a supercoiled DNA and the homologous 5′-single-stranded tails of a linear dsDNA. Joint molecule formation was previously shown to occur with homologous 3′-single-stranded tails (Noirot and Kolodner, 1998). The optimum yields of joint molecule formation with each ssDNA substrate, and under the same reaction conditions, were reached within a 2-fold range of RecT concentration (0.67–1.33 µM). This observation suggests that RecT mediates DNA strand invasion using either 5′- or 3′-single-stranded tails. Interestingly, ScRad51 protein can also promote strand exchange using 3′- or 5′-ssDNA ends (Namsaraev and Berg, 1998; Mazin et al., 2000). In contrast, RecA exhibits a marked preference for 3′-single-stranded tails in strand-exchange reactions because while both 5′- or 3′-single-stranded tails are used as substrates for strand exchange, only 3′-single-stranded tails lead to stable products (Konforti and Davis, 1990, 1992).
In a previous study, the inhibitory effect of increasing GC content on reactions of RecA and human Rad51 proteins provided the initial clues to the importance of A:T base pairs to the recognition of homology (Gupta et al., 1999b). Investigation of this effect showed that as the GC content of substrates was increased, the rate constant for strand exchange was driven toward zero, before the rate constant for homologous pairing was reduced (Gupta et al., 1999a). This led to the hypothesis that the switching of A:T base pairs plays a special role in the recognition of homology, and the corollary that the greater stability of G:C base pairs impedes the completion of strand exchange. If we grant the validity of that hypothesis, which survived a number of tests, it means that recognition of homology and at least the initiation of strand exchange are the same event, based on the disruption of parental base pairs and the formation of new heteroduplex base pairs. Such observations made originally on RecA and human Rad51 were extended to human Dmc1 (Gupta et al., 2001), and in this paper, to RecT. Of these four proteins, Dmc1 and RecT were the most sensitive to GC content: reactions were undetectable for substrates whose GC content exceeded 25%. In the case of Dmc1, pairing occurred when the substrate was 25% GC, but strand exchange was undetectable.
How should one think about pairing and strand-exchange reactions that occur only in DNA whose AT content is out of the range of most naturally occurring DNA? Perhaps short islands of AT-rich DNA suffice for rapid recognition of homology, whereas both the initiation and extension of strand exchange is impeded by G:C base pairs, which switch more slowly. This might explain the observation that RecA can promote homologous pairing and limited strand exchange in the absence of ATP hydrolysis, but requires ATP hydrolysis to drive extensive strand exchange or drive exchange through regions of non-homology (Menetski et al., 1990; Rosselli and Stasiak, 1991; Jain et al., 1994). By extension, in order to compensate for lack of ATP hydrolysis, perhaps extensive RecT-mediated strand exchange requires other proteins, like RecE. Similar suggestions have been made about the combined action of Redα and Redβ (Li et al., 1998). In the case of Dmc1 also, extensive strand exchange may depend on other factors.
The reactions catalyzed by a number of recombinases from prokaryotes and eukaryotes, including bacterial RecA, RecT, Redβ and eukaryotic Rad51, Rad52 and Dmc1 have been studied. They differ in the reaction requirements for promoting homologous pairing and strand exchange and in the structures they form on DNA. In the presence of DNA, RecT, Redβ, HsDmc1 and HsRad52 form multimeric ring structures (Thresher et al., 1995; Passy et al., 1999a,b; Stasiak et al., 2000), while HsRad51 and RecA predominantly form nucleoprotein filaments (Stasiak and Di Capua, 1982; Benson et al., 1994). RecT, Redβ and Rad52, which exhibit strikingly similar pairing activities (Passy et al., 1999a; Kagawa et al., 2001), do not bind or hydrolyze ATP. Dmc1 and Rad51 hydrolyze ATP weakly whereas RecA is a robust ATPase. However, RecA, Rad51 and Dmc1 can promote homologous pairing and strand exchange in the absence of ATP hydrolysis (Menetski et al., 1990; Rosselli and Stasiak, 1990; Sung and Stratton, 1996; Hong et al., 2001). Notwithstanding the differences in nucleoprotein structures formed by these proteins, and notwithstanding the ability or inability to use ATP to drive exchange, Rad51, Dmc1 and RecT appear to enable a single strand to recognize homology in duplex DNA by the same mechanism. We propose that the determining factor for recognition of homology is DNA structure, which forces different protein–DNA complexes to adopt the same mechanism for recognition of homology by promoting the breathing of A:T base pairs in the original duplex substrate and the unstacking of bases in the incoming ssDNA to allow the formation of new Watson–Crick base pairs. A similar conclusion on the determining role of DNA structure has been drawn from structural studies of the single strand in nucleoprotein filaments (Shibata et al., 2001).
Materials and methods
Enzymes, proteins and reagents
RecT protein was purified as described previously (Noirot and Kolodner, 1998). Restriction enzymes, T4 polynucleotide kinase, alkaline phosphatase (CIP), exonuclease III and BSA were purchased from New England Biolabs. Proteinase K and RecA were from Roche. Wheat germ topoisomerase I was from Promega. Maxam–Gilbert DNA sequencing kits were from Sigma. KMnO4 solutions were prepared as described previously in Sasse-Dwight and Gralla (1989).
Oligonucleotides
Synthesis of oligonucleotides was performed at the Molecular Biology Core Facility, Dana Farber Cancer Institute or at the Keck Biotechnology Resource Center at Yale University School of Medicine (Gupta et al., 1999a,b). Oligonucleotides used in DNA binding assays were 50 nucleotides long. O26 (5′-CACCGTCACCGACTTGAGCCATTTG GGAATTAGAGCCAGCAAAATCACCAG) corresponds to positions 2629–2579 of phage M13 wild type. O34 (5′-CTATGCGGCATCA GAGCAGATTGTACTGAGAGTGCACCATATGCGGTGTG) corresponds to positions 146–195 of pUC18. O37 (5′-AAGGAGAAAAT ACCGCATCAGGCGCCATTCGCCATTCAGGCTGCGCAACT) corresponds to positions 215–264 of pUC18 and has no thymine residue in the first 10 bases. O38 (5′-AGTTGCGCAGCCTGAATGGCGAATG GCGCCTGATGCGGTATTTTCTCCTT) is complementary to O37.
In fluorometric assays, the single-stranded 83mer oligonucleotides used to form RecT nucleoprotein filaments were designated as minus strands, while the respective complementary strands were designated as plus strands. The labeling of oligonucleotides with fluorophores and subsequent thermal annealing to form duplex substrates has been described previously (Gupta et al., 1999a). The sequences of primary sets of oligonucleotides G16 (16% GC), G26 (26% GC) and G37 (37% GC), and the location of inosine substitutions as well as base substitutions in these oligonucleotides are described elsewhere (Gupta et al., 1999a).
DNA subtrates
All DNA concentrations are expressed as moles of nucleotide residues. The 5′-end-labeling of single-stranded oligonucleotides and the preparation of double-stranded oligonucleotides were performed as described previously (Noirot and Kolodner, 1998). M13mp19 dsDNA and ssDNA, as well as plasmid DNA were prepared as described previously (Sambrook et al., 1989). The M13mp19 dsDNA was linearized by digestion with SmaI and the circular ssDNA was partially linearized by vortexing (2 min). Both ssDNA and dsDNA were dephosphorylated with CIP, and 5′-labeled by using [γ-32P]ATP and T4 polynucleotide kinase.
To generate a 3′-resected DNA duplex, pUC18 dsDNA was digested with ScaI, the linear form was purified by electrophoresis through a 5% polyacrylamide gel and the 5′-ends were labeled as described above. Then, the 3′-ends were digested with 5 U of exonuclease III per µg of DNA, for 90 s at 37°C in the recommended buffer. Under these conditions, an average of 100 bases were removed from each 3′-end, as estimated by electrophoresis through a 1.2% agarose gel along with appropriate size controls.
DNA binding and aggregation assays
Reaction mixtures (30 µl) contained 32P-labeled ssDNA at the concentrations indicated in the figure legends in buffer T (20 mM Tris–HCl pH 7.5, 25 mM NaCl, 100 µg/ml BSA, 0.5 mM DTT). Varying RecT concentrations were added and the mixtures incubated 20 min at 37°C. In the co-aggregation experiments, Tris–HCl (pH 7.5) was replaced by triethanolamine–HCl (pH 7.5) in buffer T, and the enzyme preparation used in the co-aggregation experiments was extensively dialyzed against modified buffer A (20 mM triethanolamine–HCl pH 7.5, 10 mM 2-mercaptoethanol, 0.1 mM EDTA, 60% w/v glycerol). This buffer change did not alter RecT activity in a standard D-loop formation assay. RecT was pre-incubated with 32P-labeled ssDNA for 10 min at 25°C, the reaction was started by adding the linear dsDNA and the incubation was continued at 37°C for 40 min. Samples were analyzed by electrophoresis through 0.6 or 0.9% agarose gels in TAE buffer run at 4 V/cm. Gels were then stained with ethidium bromide, fixed with TCA, and 32P-labeled DNA species were quantified as described previously (Noirot and Kolodner, 1998).
Fluorometric assays of homologous pairing and strand exchange
For RecT reactions, 3 µM 83mer oligonucleotide G16(–) was pre-incubated with 1.2 µM RecT in buffer T (25 mM Tris pH 7.4, 0.5 mM DTT, 100 µg BSA/ml) at 37°C for 5 min. G16 duplex was then added at a final concentration of 6 µM and the reaction was continued at 37°C. For reactions mediated by RecA, the mixture contained 25 mM HEPES pH 7.4, 1 mM DTT, 100 µg BSA/ml, 2 mM ATP and 1 mM MgCl2. The reaction was carried out essentially as described for RecT, except that MgCl2 was increased to 30 mM before the addition of duplex DNA.
Fluorescence resonance energy transfer (FRET) was measured as described previously (Bazemore et al., 1997b; Gupta et al., 1999a, 2001). For pairing reactions, the 3′-end of the single-stranded oligonucleotide was labeled with fluorescein, and the 5′-end of the complementary strand in the duplex oligonucleotide was labeled with rhodamine. When the strands labeled with fluorescein and rhodamine come into proximity, either in the three-stranded intermediate or in the final product of strand exchange, the fluorescent dyes can participate in FRET: fluorescein excited by light at its absorption maximum transfers energy non-radiatively to rhodamine, which results in enhanced or sensitized emission of light from rhodamine. Measurement of the sensitized emission, which requires the subtraction of emission spectra, shows that the dyes have come into proximity, and represents a first step in using FRET to demonstrate homology-dependent interaction of strands of DNA. As a consequence of the transfer of energy from fluorescein to rhodamine, the fluorescence emission from fluorescein is quenched. This aspect of FRET is useful for studying the kinetics of homologous pairing.
For strand exchange assays, the single-stranded substrate was unlabeled whereas the double-stranded substrate contained fluorescein at the 3′-end of the minus strand and rhodamine at the 5′-end of the plus strand. In this case, fluorescein and rhodamine are initially in proximity and participate in FRET until strand exchange occurs, at which time the dyes separate and the fluorescence emission from fluorescein is enhanced. The enhancement of the emission from fluorescein enables one to study the kinetics of strand exchange. The quenching of fluorescein emission as a result of pairing, and the enhancement of fluorescein emission as a result of strand exchange were measured at 525 nm upon excitation at 493 nm with a SLM 8000C spectrofluorometer at 2 s intervals.
Unstacking assay
Reaction mixtures (75 µl) containing 1.7 µM 32P-labeled ssDNA (O37) or 3.4 µM 32P-labeled dsDNA (O37 + O38) were incubated with varying RecT or RecA concentrations for 20 min at 37°C or 10 min at 25°C, respectively. The RecT and RecA reactions were performed in buffer T and in buffer T supplemented with 10 mM MgCl2 and 0.3 mM ATPγS, respectively. The protein–DNA complexes were then incubated with 0.6 mM KMnO4 for 1 min at 25°C. The reactions were terminated by addition of 100 µl stop solution (0.9 M β-mercaptoethanol, 0.6 M Na acetate, 250 µg/ml glycogen) followed by precipitation with ethanol. After treatment with piperidine (90°C, 30 min), the DNA products were resolved on a 20% urea–polyacrylamide gel (Sequagel, National Diagnostic). Sequencing ladders were standard Maxam–Gilbert reactions performed on O37, as described in the sequencing kit supplier’s instructions. The gel was dried on filter paper and 32P-labeled DNA was detected by autoradiography.
Unwinding assay
pHV2900OB was previously described (Noirot and Kolodner, 1998). Supercoiled pHV2900OB DNA (4 µg) was relaxed in the buffer recommended by the supplier with 52 U of wheat germ topoisomerase I for 30 min at 37°C. This reaction mixture was divided among 2.5 µl aliquots, which were added to reaction mixtures (30 µl) containing the indicated RecT concentrations in buffer T, so that each reaction contained 30 µM relaxed pHV2900OB DNA and 4 U of wheat germ topo isomerase I. Wheat germ topoisomerase I remained fully active in buffer T for at least 1 h. Mixtures were incubated for 45 min at 37°C, deproteinized by addition of EDTA, SDS and proteinase K to final concentrations of 50 mM, 0.2% and 550 µg/ml, respectively, and further incubated for 30 min at 37°C. The DNA products were analyzed by electrophoresis through 1% agarose gel in TBE buffer run at 2 V/cm for 40 h at 4°C. The gel was then stained with SYBR green and scanned with a Fluorimager (Molecular Dynamics). For analysis by two-dimensional gel electrophoresis, the DNA products were subjected to electrophoresis through a 0.8% agarose gel in TBE buffer containing 0.66 µM chloroquine in the first dimension and in 2 µM chloroquine in the second dimension. Electrophoresis was carried out at 4°C, at 4 V/cm for 20 h and 2 V/cm for 40 h in the first and second dimension, respectively. The gel was then stained with SYBR green and scanned with a Fluorimager (Molecular Dynamics).
Strand transfer assay
Reaction mixtures (30 µl) contained 20 µM linear dsDNA resected with exonuclease III (5′-ssDNA) in buffer T and RecT as indicated in figure legends. The reactions were incubated for 20 min at 25°C and supercoiled DNA (40 µM) was then added to start the reaction. After 50 min at 37°C, the reactions were deproteinized and the DNA products were analyzed as described previously (Noirot and Kolodner, 1998).
Acknowledgments
Acknowledgements
The authors would like to thank Dan Camerini-Otero, Mike Cox and Steve Kowalczykowski for helpful discussions. This work was supported by National Institutes of Health grants GM26017 (to R.D.K.) and GM33504 (to C.M.R.).
References
- Baumann P., Benson,F.E. and West,S.C. (1996) Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell, 87, 757–766. [DOI] [PubMed] [Google Scholar]
- Bazemore L.R., Folta-Stogniew,E., Takahashi,M. and Radding,C.M. (1997a) RecA tests homology at both pairing and strand exchange. Proc. Natl Acad. Sci. USA, 94, 11863–11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazemore L.R., Takahashi,M. and Radding,C.M. (1997b) Kinetic analysis of pairing and strand exchange catalyzed by RecA. Detection by fluorescence energy transfer. J. Biol. Chem., 272, 14672–14682. [DOI] [PubMed] [Google Scholar]
- Benson F.E., Stasiak,A. and West,S.C. (1994) Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J., 13, 5764–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassuto E., Lash,T., Sriprakash,K.S. and Radding,C.M. (1971) Role of exonuclease and protein of phage λ in genetic recombination. V. Recombination of λ DNA in vitro. Proc. Natl Acad. Sci. USA, 68, 1639–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow S.A. and Radding,C.M. (1985) Ionic inhibition of formation of RecA nucleoprotein networks blocks homologous pairing. Proc. Natl Acad. Sci. USA, 82, 5646–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A.J. and Sandler,S.J. (1994) Homologous genetic recombination: the pieces begin to fall into place. Crit. Rev. Microbiol., 20, 125–142. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A. and Wanner,B.L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA, 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R.A., James,A.A. and Kolodner,R. (1981) RecA-independent general genetic recombination of plasmids. Nature, 294, 184–186. [DOI] [PubMed] [Google Scholar]
- Gillen J.R., Willis,D.K. and Clark,A.J. (1981) Genetic analysis of the RecE pathway of genetic recombination in Escherichia coli K-12. J. Bacteriol., 145, 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glikin G.C., Gargiulo,G., Rena-Descalzi,L. and Worcel,A. (1983) Escherichia coli single-strand binding protein stabilizes specific denatured sites in superhelical DNA. Nature, 303, 770–774. [DOI] [PubMed] [Google Scholar]
- Gupta R.C., Bazemore,L.R., Golub,E.I. and Radding,C.M. (1997) Activities of human recombination protein Rad51. Proc. Natl Acad. Sci. USA, 94, 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.C., Folta-Stogniew,E., O’Malley,S., Takahashi,M. and Radding,C.M. (1999a) Rapid exchange of A:T base pairs is essential for recognition of DNA homology by human Rad51 recombination protein. Mol. Cell, 4, 705–714. [DOI] [PubMed] [Google Scholar]
- Gupta R.C., Folta-Stogniew,E. and Radding,C.M. (1999b) Human Rad51 protein can form homologous joints in the absence of net strand exchange. J. Biol. Chem., 274, 1248–1256. [DOI] [PubMed] [Google Scholar]
- Gupta R.C., Golub,E., Bi,B. and Radding,C.M. (2001) The synaptic activity of HsDmc1, a human recombination protein specific to meiosis. Proc Natl Acad. Sci. USA, 98, 8433–8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S.D. and Kolodner,R.D. (1994) Homologous pairing and strand exchange promoted by the Escherichia coli RecT protein. Proc. Natl Acad. Sci. USA, 91, 3205–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S.D., Kane,M.F. and Kolodner,R.D. (1993) Identification and characterization of the Escherichia coli RecT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA (published erratum appears in J. Bacteriol., 1993, 175, 1211). J. Bacteriol., 175, 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E.L., Shinohara,A. and Bishop,D.K. (2001) Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J. Biol. Chem., 276, 41906–41912. [DOI] [PubMed] [Google Scholar]
- Iyer L.M., Koonin,E.V. and Aravind,L. (2002) Classification and evolutionary history of the single-strand annealing proteins, RecT, Redβ, ERF and RAD52. BMC Genomics, 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S.K., Cox,M.M. and Inman,R.B. (1994) On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. III. Unidirectional branch migration and extensive hybrid DNA formation. J. Biol. Chem., 269, 20653–20661. [PubMed] [Google Scholar]
- Joseph J.W. and Kolodner,R. (1983) Exonuclease VIII of Escherichia coli. II. Mechanism of action. J. Biol. Chem., 258, 10418–10424. [PubMed] [Google Scholar]
- Kagawa W., Kurumizaka,H., Ikawa,S., Yokoyama,S. and Shibata,T. (2001) Homologous pairing promoted by the human Rad52 protein. J. Biol. Chem., 276, 35201–35208. [DOI] [PubMed] [Google Scholar]
- Kobayashi I. and Takahashi,N. (1988) Double-stranded gap repair of DNA by gene conversion in Escherichia coli. Genetics, 119, 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R., Hall,S.D. and Luisi-DeLuca,C. (1994) Homologous pairing proteins encoded by the Escherichia coli recE and recT genes. Mol. Microbiol., 11, 23–30. [DOI] [PubMed] [Google Scholar]
- Konforti B.B. and Davis,R.W. (1990) The preference for a 3′ homologous end is intrinsic to RecA-promoted strand exchange. J. Biol. Chem., 265, 6916–6920. [PubMed] [Google Scholar]
- Konforti B.B. and Davis,R.W. (1992) ATP hydrolysis and the displaced strand are two factors that determine the polarity of RecA-promoted DNA strand exchange. J. Mol. Biol., 227, 38–53. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S.C. and Krupp,R.A. (1995) DNA-strand exchange promoted by RecA protein in the absence of ATP: implications for the mechanism of energy transduction in protein-promoted nucleic acid transactions. Proc. Natl Acad. Sci. USA, 92, 3478–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski S.C., Dixon,D.A., Eggleston,A.K., Lauder,S.D. and Rehrauer,W.M. (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev., 58, 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N.V. and Varshney,U. (1997) Contrasting effects of single stranded DNA binding protein on the activity of uracil DNA glycosylase from Escherichia coli towards different DNA substrates. Nucleic Acids Res., 25, 2336–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner S.R., Nagaishi,H., Templin,A. and Clark,A.J. (1971) Genetic recombination in Escherichia coli: the role of exonuclease I. Proc. Natl Acad. Sci. USA, 68, 824–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Karakousis,G., Chiu,S.K., Reddy,G. and Radding,C.M. (1998) The β protein of phage λ promotes strand exchange. J. Mol. Biol., 276, 733–744. [DOI] [PubMed] [Google Scholar]
- Luisi-DeLuca C. and Kolodner,R.D. (1992) Effect of terminal non-homology on intramolecular recombination of linear plasmid substrates in Escherichia coli. J. Mol. Biol., 227, 72–80. [DOI] [PubMed] [Google Scholar]
- Mazin A.V., Zaitseva,E., Sung,P. and Kowalczykowski,S.C. (2000) Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J., 19, 1148–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski J.P., Bear,D.G. and Kowalczykowski,S.C. (1990) Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc. Natl Acad. Sci. USA, 87, 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyrers J.P., Zhang,Y., Buchholz,F. and Stewart,A.F. (2000) RecE/RecT and Redα/Redβ initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev., 14, 1971–1982. [PMC free article] [PubMed] [Google Scholar]
- Namsaraev E.A. and Berg,P. (1998) Branch migration during Rad51-promoted strand exchange proceeds in either direction. Proc. Natl Acad. Sci. USA, 95, 10477–10481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishinaka T., Ito,Y., Yokoyama,S. and Shibata,T. (1997) An extended DNA structure through deoxyribose-base stacking induced by RecA protein. Proc. Natl Acad. Sci. USA, 94, 6623–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishinaka T., Shinohara,A., Ito,Y., Yokoyama,S. and Shibata,T. (1998) Base pair switching by interconversion of sugar puckers in DNA extended by proteins of RecA-family: a model for homology search in homologous genetic recombination. Proc. Natl Acad. Sci. USA, 95, 11071–11076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot P. and Kolodner,R.D. (1998) DNA strand invasion promoted by Escherichia coli RecT protein. J. Biol. Chem., 273, 12274–12280. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Yu,X., Shinohara,A. and Egelman,E.H. (1993) Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science, 259, 1896–1899. [DOI] [PubMed] [Google Scholar]
- Passy S.I., Yu,X., Li,Z., Radding,C.M. and Egelman,E.H. (1999a) Rings and filaments of β protein from bacteriophage λ suggest a superfamily of recombination proteins. Proc. Natl Acad. Sci. USA, 96, 4279–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passy S.I., Yu,X., Li,Z., Radding,C.M., Masson,J.Y., West,S.C. and Egelman,E.H. (1999b) Human Dmc1 protein binds DNA as an octameric ring. Proc. Natl Acad. Sci. USA, 96, 10684–10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh B.F. and Cox,M.M. (1987) Stable binding of recA protein to duplex DNA. Unraveling a paradox. J. Biol. Chem., 262, 1326–1336. [PubMed] [Google Scholar]
- Rosselli W. and Stasiak,A. (1990) Energetics of RecA-mediated recombination reactions. Without ATP hydrolysis RecA can mediate polar strand exchange but is unable to recycle. J. Mol. Biol., 216, 335–352. [DOI] [PubMed] [Google Scholar]
- Rosselli W. and Stasiak,A. (1991) The ATPase activity of RecA is needed to push the DNA strand exchange through heterologous regions. EMBO J., 10, 4391–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rould E., Muniyappa,K. and Radding,C.M. (1992) Unwinding of heterologous DNA by RecA protein during the search for homologous sequences. J. Mol. Biol., 226, 127–139. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Sasse-Dwight S. and Gralla,J.D. (1989) KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J. Biol. Chem., 264, 8074–8081. [PubMed] [Google Scholar]
- Shibata T., Nishinaka,T., Mikawa,T., Aihara,H., Kurumizaka,H., Yokoyama,S. and Ito,Y. (2001) Homologous genetic recombination as an intrinsic dynamic property of a DNA structure induced by RecA/Rad51-family proteins: a possible advantage of DNA over RNA as genomic material. Proc. Natl Acad. Sci. USA, 98, 8425–8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M.M., Thomason,L., Poteete,A.R., Tarkowski,T., Kuzminov,A. and Stahl,F.W. (1997) Annealing versus invasion in phage λ recombination. Genetics, 147, 961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak A. and Di Capua,E. (1982) The helicity of DNA in complexes with recA protein. Nature, 299, 185–186. [DOI] [PubMed] [Google Scholar]
- Stasiak A.Z., Larquet,E., Stasiak,A., Muller,S., Engel,A., Van Dyck,E., West,S.C. and Egelman,E.H. (2000) The human Rad52 protein exists as a heptameric ring. Curr. Biol., 10, 337–340. [DOI] [PubMed] [Google Scholar]
- Sung P. and Robberson,D.L. (1995) DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell, 82, 453–461. [DOI] [PubMed] [Google Scholar]
- Sung P. and Stratton,S.A. (1996) Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem., 271, 27983–27986. [DOI] [PubMed] [Google Scholar]
- Symington L.S., Morrison,P. and Kolodner,R. (1985) Intramolecular recombination of linear DNA catalyzed by the Escherichia coli RecE recombination system. J. Mol. Biol., 186, 515–525. [DOI] [PubMed] [Google Scholar]
- Takahashi N.K., Kusano,K., Yokochi,T., Kitamura,Y., Yoshikura,H. and Kobayashi,I. (1993) Genetic analysis of double-strand break repair in Escherichia coli. J. Bacteriol., 175, 5176–5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N.K., Sakagami,K., Kusano,K., Yamamoto,K., Yoshikura,H. and Kobayashi,I. (1997) Genetic recombination through double-strand break repair: shift from two-progeny mode to one-progeny mode by heterologous inserts. Genetics, 146, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thresher R.J., Makhov,A.M., Hall,S.D., Kolodner,R. and Griffith,J.D. (1995) Electron microscopic visualization of RecT protein and its complexes with DNA. J. Mol. Biol., 254, 364–371. [DOI] [PubMed] [Google Scholar]
- Tsang S.S., Chow,S.A. and Radding,C.M. (1985) Networks of DNA and RecA protein are intermediates in homologous pairing. Biochemistry, 24, 3226–3232. [DOI] [PubMed] [Google Scholar]
- Voloshin O.N., Wang,L. and Camerini-Otero,R.D. (1996) Homologous DNA pairing promoted by a 20-amino acid peptide derived from RecA. Science, 272, 868–872. [DOI] [PubMed] [Google Scholar]
- Yu D., Ellis,H.M., Lee,E.C., Jenkins,N.A., Copeland,N.G. and Court,D.L. (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. and Egelman,E.H. (1993) DNA conformation induced by the bacteriophage T4 UvsX protein appears identical to the conformation induced by the Escherichia coli RecA protein. J. Mol. Biol., 232, 1–4. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Buchholz,F., Muyrers,J.P. and Stewart,A.F. (1998) A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet., 20, 123–128. [DOI] [PubMed] [Google Scholar]