Abstract
The frequency (frq) gene, the central component of the frq-based circadian negative feedback loop, regulates various aspects of the circadian clock in Neurospora. However, the biochemical function of its protein products, FRQ, is poorly understood. In this study, we demonstrated that the most conserved region of FRQ forms a coiled-coil domain. FRQ interacts with itself in vivo, and the deletion of the coiled-coil region results in loss of the interaction. Point mutations, which are designed to disrupt the coiled-coil structure, weaken or completely abolish the FRQ self-association and lead to the arrhythmicity of the overt rhythm. Mutations of the FRQ coiled-coil that inhibit self-association also prevent its interaction with two other key components of the Neurospora circadian clock, namely WC-1 and WC-2, the two PAS domain-containing transcription factors. Taken together, these data strongly suggest that the formation of the FRQ–FRQ and FRQ–WC complexes is essential for the function of the Neurospora clock.
Keywords: circadian/clock/frequency/wc-1/wc-2
Introduction
Circadian clocks (period ∼24 h) are found in almost all groups of organisms and they control a wide variety of daily endogenous (circadian) rhythms of molecular, physiological and behavioral activities in these organisms (Bünning, 1973; Dunlap, 1999). The identification of clock genes in several model systems and the understanding of how these genes are regulated have started to unveil how circadian clocks work at the molecular level (Hall, 1998; Dunlap, 1999; Johnson and Golden, 1999; Liu et al., 1999; Young, 1999; King and Takahashi, 2000; Somers et al., 2000). From these studies, a common mechanistic theme has emerged: clocks consist of a network of positive and negative interactors that form the core of the oscillators that establish the negative feedback loops generating the basic circadian rhythm (Aronson et al., 1994a; Crosthwaite et al., 1997; King et al., 1997; Allada et al., 1998; Darlington et al., 1998; Hardin, 1998; Ishiura et al., 1998; Rutila et al., 1998; Wang and Tobin, 1998; Glossop et al., 1999; Shearman et al., 2000).
In Neurospora, there is at least one circadian autoregulatory negative feedback loop (Dunlap, 1999; Merrow et al., 1999; Lakin-Thomas and Brody, 2000; Lee et al., 2000) with frq mRNA and FRQ proteins as the central components (Aronson et al., 1994a). The deletion of the frq locus leads to the loss of normal circadian rhythmicity (Aronson, 1994b). Mutations of frq result in long and short period length (from 16 to 35 h), arrhythmia, and loss of temperature and nutritional compensation of the clock (Aronson et al., 1994b; Liu et al., 2000). Levels of both frq mRNA and FRQ protein cycle with the same period as the overt rhythms, and FRQ protein acts negatively to repress its own transcription (Aronson et al., 1994a). Importantly, the rhythmic expression of frq is essential for the negative feedback loop because constitutive expression of frq results in the loss of the overt rhythm, and step changes in frq expression reset the phase of the clock (Aronson et al., 1994a; Garceau et al., 1997). Light and temperature, two of the most important environmental signals, reset the Neurospora clock by changing the levels of frq mRNA and FRQ protein (Crosthwaite et al., 1995; Liu et al., 1998).
Although transcriptional regulation of frq is important for rhythm generation, post-transcriptional regulation of frq also plays a major role in regulating the clock. First, temperature-regulated alternative initiation of FRQ protein translation produces two forms of FRQ: a long form of 989 amino acids (LFRQ) and a shorter form of 890 amino acids (SFRQ) (Garceau et al., 1997). This mechanism allow the Neurospora clock to function optimally over a wide range of temperatures (Liu et al., 1997). Phosphorylation of FRQ is another important step of regulation. FRQ protein is progressively phosphorylated over time and its level decreases after it is extensively phosphorylated (Garceau et al., 1997). Although the identity of the FRQ kinase(s) is not known, one important function of phosphorylation is to regulate the protein stability of FRQ and affect the period length of the clock (Liu et al., 2000).
WC-1 and WC-2, two PAS-domain-containing transcription factors, are the other two known components of the Neurospora clock. Not only are they two of the essential components of the light signal transduction pathway of the clock (Crosthwaite et al., 1997; Talora et al., 1999), but they are also required for normal circadian rhythmicity in constant darkness. In strains with lesions of either wc-1 or wc-2, the levels of frq mRNA and FRQ are extremely low, and these low levels of frq and FRQ expression are unable to support the overt rhythmicity (Crosthwaite et al., 1997). Like the PAS-domain-containing transcription factors BMAL1 and CLOCK in mammals, and dCLOCK and CYC in Drosophila, WC-1 and WC-2 form heterodimers in vivo (Dunlap, 1999; Talora et al., 1999). Recently, it was reported that FRQ interacts with WC-1 and WC-2 in vivo, suggesting that FRQ might negatively regulate its own expression by inhibiting the transcriptional activation of the WC proteins (Denault et al., 2000). Together, these data suggest that the WC proteins might function as the transcription activators of frq and behave as the positive limb of the Neurospora circadian negative feedback loop (Dunlap, 1999).
Despite the importance of FRQ protein in the Neurospora clock, how it works in the FRQ–WC based circadian negative feedback loop is poorly understood. In this study, we identified the most conserved region of FRQ as a coiled-coil domain. We have shown that FRQ interacts with itself in vivo, and such interaction depends on the presence of the coiled-coil region. Point mutations in this region, designed to disrupt the coiled-coil structure, weaken or completely abolish the FRQ self-interaction, and lead to the arrhythmicity of the overt rhythm. In addition, mutant forms of FRQ that fail to self-associate, also fail to interact with WC-1 and WC-2, the two PAS-domain-containing transcription factors. Taken together, these data demonstrate that the formation of the FRQ–FRQ and FRQ–WC complexes are essential for the function of the Neurospora clock.
Results
FRQ interacts with itself
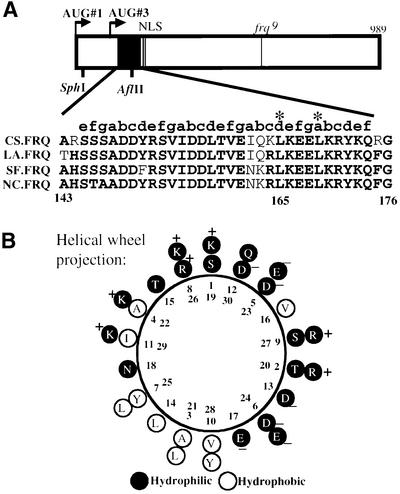
Analysis of the FRQ open reading frame (ORF) revealed that a 30 amino acid region (aa 145–174) near the N-terminus of the protein has potential to form a coiled-coil structure. MultiCoil and Paircoil scores indicate that this region has >90% probability of forming a dimer (Berger et al., 1995; Wolf et al., 1997). Previously, sequence comparison among different FRQ homologs has shown that this region is the most conserved region (Lewis et al., 1997) (Figure 1A), suggesting its important role for the function of FRQ. Coiled-coil domain is one of the common structural motifs mediating protein–protein interactions. It is a structure of heptad repeats (amino acid positions are labeled a–g), positions a and d are hydrophobic and form the helix interface that mediates protein–protein interactions, while other positions (b, c, e, f and g) are typically hydrophilic and form the solvent-exposed part of the structure (Lupas, 1996). The computer analysis reveals that this domain of FRQ consists of four heptad repeats. The helical wheel projection of this region of FRQ shows that the a and d positions are mostly hydrophobic and have the potential of forming the helix interface, while the rest of the positions are more hydrophilic (Figure 1B).
Fig. 1. The putative FRQ coiled-coil domain. (A) Schematic diagram of the FRQ ORF and sequencing alignments of the putative FRQ coiled-coil domain from four different fungal species. The open box represents the FRQ ORF. Arrows labeled AUG#1 and AUG#3 are the two alternative protein initiation sites that produce LFRQ and SFRQ. The filled black box represents the putative FRQ coiled-coil domain. NLS, nuclear localization signal. The line in the middle represents the location of the frq9 mutation. In the sequence alignments, the bold letters are the conserved amino acids in different frq homologs. On the top of the alignment, letters a–g label the position of each amino acid within the helical heptad repeats. Asterisks mark the two amino acids (Leu165 and Leu169) that are mutated in this study. CS, Chromocrea spinlosa; LA, Leptosphaeria australiensis; SF, Sordaria fimicola; NC, Neurospora crassa. (B) Helical wheel projection of the FRQ coiled-coil domain. In the coiled-coils, seven residues allow the helix to make two turns to show a heptad repeat. The Ser145 denotes as position ‘1’ in the helical wheel projection. The charge and the hydrophobicity of each residue are labeled.
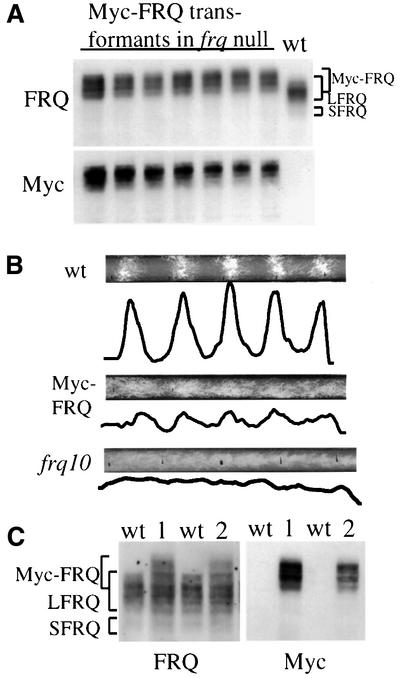
To demonstrate the function of this region in protein– protein interactions, we first examined its role in mediating FRQ self-association. We made a construct in which five c-Myc epitopes were inserted into the N-terminus (SphI site; Figure 1A) of FRQ such that only the large FRQ form (LFRQ) is tagged. This Myc-tagged FRQ was expressed at a level comparable to that of the wild-type FRQ and it was also phosphorylated normally (Figure 2A, the various bands are the results of protein phosphorylation) (Garceau et al., 1997). The Myc-FRQ bands migrate more slowly than the wild-type FRQ in SDS–PAGE gel because of the addition of the five c-Myc epitopes (79 amino acids), and they are specifically recognized by a monoclonal c-Myc antibody (Figure 2A and C). However, this construct failed to express the small FRQ form (SFRQ, not tagged by c-Myc), suggesting that the region where the epitopes were inserted is important for the expression of SFRQ. The Myc-tagged FRQ is functional because it can rescue the circadian rhythm of conidiation banding of the frq null strain (frq10) (Aronson et al., 1994b), albeit with a slightly shorter period (∼21 h) than that of the wild-type strain (∼22 h) (Figure 2B). The low amplitude and the short period rhythms of the Myc-FRQ transformants are likely to be due to the lack of expression of SFRQ, since both FRQ forms are required for optimal clock function (Liu et al., 1997).
Fig. 2. The expression of Myc-FRQ in Neurospora and the rescue of the conidiation banding rhythm of the frq null strain by Myc-FRQ. (A) Western blot analysis showing the expression profile of Myc-FRQ in Neurospora. pKAJ120⋅Myc-FRQ was transformed into 93–4 (frq null strain) and seven independent transformants were selected for protein analysis. Cultures were grown and harvested in constant light (LL). The protein blot was first probed with FRQ antiserum (top) and then stripped and reprobed with a monoclonal c-Myc antibody (bottom). Note the slower mobility of Myc-FRQ bands relative to the wild-type FRQ. (B) The rescue of the circadian conidiation banding rhythm of frq null strain by Myc-FRQ. Both the original image of the race tubes and the densitometric analysis of the race tubes are shown. frq10 is the frq null strain. The race tubes shown are representative samples from six replicate tubes. (C) Expression of Myc-FRQ in an frq+ strain. pKAJ120⋅Myc-FRQ was transformed into 87–12 (frq+) and two independent transformants (1 and 2) are analyzed along with the wild-type strain. The blot was probed with either FRQ (upper panel) or c-Myc antibody (lower panel).
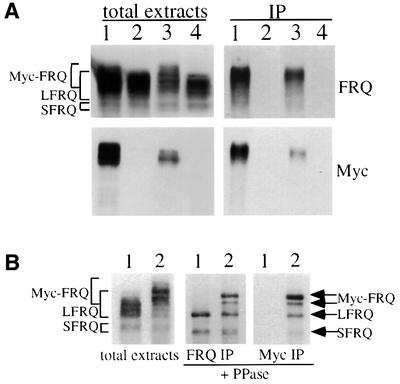
To examine whether FRQ interacts with itself, the Myc-FRQ construct was transformed into a wild-type Neurospora strain. As can be seen from Figure 2C, all three forms of FRQ (endogenous LFRQ and SFRQ, and MYC-FRQ) are expressed in these transformants. Protein extracts from cultures grown in constant light (LL) or dark (DD20: 20 h after light/dark transition) were prepared. Immunoprecipitation was performed using the monoclonal c-Myc antibody. The smear of lower molecular weight FRQ signals in Figure 3A (upper right panel) suggests that the endogenous LFRQ and SFRQ were pulled down by Myc-FRQ. To obtain visually clearer results, the immunoprecipitates using either FRQ or Myc antibodies were treated with lambda phosphatase to dephosphorylate different FRQ forms. As shown in the center panel of Figure 3B, immunoprecipitation using FRQ antiserum pulled down both LFRQ and SFRQ in the wild-type strain, while in the Myc-FRQ strain, in addition to the two endogenous wild-type FRQ forms, two Myc-FRQ forms are also pulled down (Figure 3B, center panel). The expression of the smaller Myc-FRQ form is very likely to be the result of internal protein initiation from one of the six methionines within the five Myc epitope region. When c-Myc antibody was used to perform immunoprecipitation, as expected, the endogenous LFRQ and SFRQ are pulled down along with the Myc-FRQ forms, indicating the self-association of FRQ (Figure 3B). Comparison of the relative amounts of Myc-FRQ and the endogenous FRQ forms in the FRQ immunoprecipitation (IP) and the Myc IP samples, we estimate that there is ∼50% of FRQ associated with each other to form complexes (Figures 3B and 4B). Since this comparison can only estimate the amount of FRQ that forms heterodimer and the immunoprecipitation procedures might also disrupt the complexes, the actual number in vivo should be >50%. In addition, yeast two-hybrid assays using FRQ regions containing this coiled-coil domain also showed that FRQ interacts with itself in yeast (data not shown). Together, these results demonstrate that both LFRQ and SFRQ interact with each other to form complexes in vivo.
Fig. 3. FRQ interacts with itself in vivo. (A) Western blot analyses (probed with FRQ or c-Myc antibody) showing the expression of various FRQ forms in total extracts (left two panels) and in immunoprecipitates (right two panels). Immunoprecipitation was performed using a c-Myc monoclonal antibody. Note the lack of low molecular weight smear in the right bottom blot probed with c-Myc antibody. Lane 1, frq+, Myc-FRQ; LL; lane 2, wild type, LL; lane 3, frq+, Myc-FRQ; DD20; lane 4, wild type, DD20. (B) Phosphatase treatment after immunoprecipitation shows that Myc-FRQ interacts with the endogenous FRQ forms. Before phosphatase treatment, the protein extracts were immunoprecipitated with either FRQ (center panel) or c-Myc (right panel) antibodies. The cultures were grown and harvested in LL. Lane 1, wild type; lane 2, frq+, Myc-FRQ.
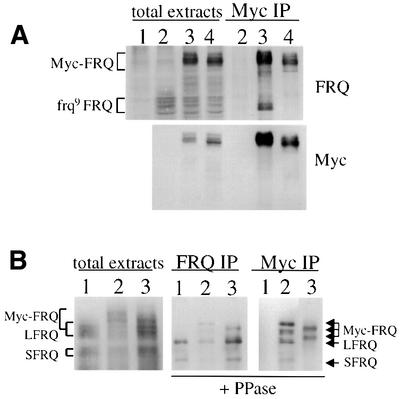
Fig. 4. Deletion of the putative coiled-coil domain abolishes FRQ–FRQ interaction. (A) The interaction between Myc-FRQ and the truncated frq9 FRQ. The cultures were grown in LL. Lane 1, frq10 (frq null); lane 2, frq9; lane 3, frq9, Myc-FRQ; lane 4, frq9, Myc-FRQ1 (deletion of the coiled-coil region). The left four lanes are total protein extracts and the three lanes on the right are IP products with c-Myc antibody. The protein blots were probed with FRQ or c-Myc antibody. Note the disappearance of the frq9 FRQ bands in lane 4 of the IP samples. (B) Myc-FRQ with the deletion of the coiled-coil domain can no longer interact with the endogenous FRQ forms. Lane 1, wild type; lane 2, frq+, Myc-FRQ; lane 3, frq+, Myc-FRQ1. The protein extracts were immunoprecipitated with FRQ (center) or c-Myc (right) antibody and then treated with phosphatase to dephosphorylate various FRQ forms. The protein blots were probed with FRQ antiserum. Note the disappearance of the LFRQ and SFRQ bands in lane 3 of the Myc IP panel.
Deletion of the coiled-coil domain eliminates the FRQ–FRQ interaction
To locate the region responsible for the FRQ–FRQ interaction, the Myc-FRQ construct was transformed into an frq9 strain. In the frq9 strain, a one nucleotide frame-shift deletion within the FRQ ORF results in the translation of a truncated FRQ protein of ∼75 kDa (Aronson et al., 1994b). Although the truncated FRQ in frq9 is only marginally stable, it can be detected by western blot analysis (Figure 4A, top panel). The various bands of frq9 FRQ suggest that it is also phosphorylated. In the Myc-FRQ, frq9 strain, both Myc-FRQ and frq9 FRQ are expressed (Figure 4A, lane 3, left). When using Myc antibody to perform immunoprecipitation, the smaller frq9 FRQ can be pulled down by the Myc-FRQ forms (Figure 4A, lane 3, right), but not in the negative control (frq9 strain; Figure 4A, lane 2). These data further indicate that FRQ self-associates and that the region responsible for the FRQ–FRQ interaction is in the region upstream of the frq9 mutation.
Since the putative coiled-coil domain is included in the frq9 FRQ, we made a construct (pMyc-FRQ1) in which the coiled-coil domain is deleted (FRQ aa 107–178) from Myc-FRQ to see whether the coiled-coil domain is necessary for the FRQ–FRQ interaction. This construct was transformed into either frq9 (Figure 4A) or the wild-type strain (Figure 4B). As predicted, Myc-FRQ1 did not co-precipitate with frq9 FRQ (Figure 4A, lane 4, right) or the endogenous LFRQ and SFRQ forms (Figure 4B, lane 3, right panel). The slightly faster mobility of the Myc-FRQ1 protein species is due to the deletion of the coiled-coil domain. These data demonstrate that this putative coiled-coil domain region is required for the FRQ–FRQ interaction. In addition, when this region is deleted from the wild-type FRQ, it no longer rescues the circadian banding rhythm of the frq null strain (data not shown), suggesting that this region and the FRQ–FRQ interaction mediated by this region are essential for the clock function of FRQ.
Mutations of the coiled-coil domain disrupt the FRQ–FRQ interaction and eliminate its clock function
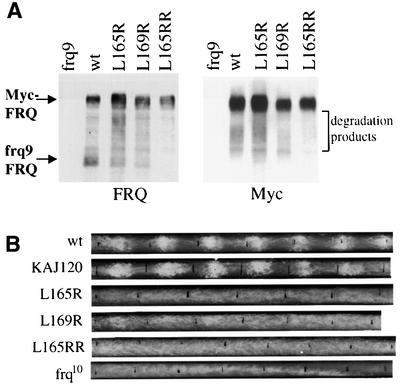
To access the molecular surface involved in FRQ–FRQ interaction, we introduced point mutations at the critical a and d positions in the predicted coiled-coil structure (Figure 1B) that form the hydrophobic helix interface. Therefore, mutation of these amino acids to charged hydrophilic residues would be predicted to weaken or prevent the protein–protein interaction. Point mutations (to arginine) were introduced at one or both of the conserved leucines at position 165 and 169 (Figure 1), and the Myc-FRQ constructs with these mutations were transformed into the frq9 strain. Such mutations were predicted by the MultiCoil and Paircoil programs to reduce significantly the probability of forming coiled-coil structure (Berger et al., 1995; Wolf et al., 1997). As predicted, the single site mutations (Leu165Arg and Leu169Arg) significantly weakened the interaction between Myc-FRQ and frq9 FRQ, while in the double mutant (Leu165ArgArg: Leu165Arg and Leu169Arg), such interaction was completely abolished (Figure 5A). These data further support the role of the coiled-coil structure in the formation of the FRQ–FRQ complexes.
Fig. 5. Point mutations of the coiled-coil domain lead to the weakening or complete loss of the FRQ–FRQ interaction and loss of rescue of the conidiation banding rhythm in the frq null strain. (A) Leu165 and Leu169 are important residues for FRQ–FRQ interaction. Single (Leu165Arg and Leu169Arg) or double (Leu165ArgArg) mutations were introduced into pKAJ120⋅Myc-FRQ. Mutation constructs were transformed into 94–1 (frq9, his-3), so that both Myc-FRQ and frq9 FRQ are expressed in these transformants. Various Myc-FRQ forms were immunoprecipitated down using c-Myc antibody. The protein blots were probed with FRQ or Myc antibody. Note the decrease of the frq9 FRQ signals in Leu165Arg and Leu169Arg samples, and its complete disappearance in 165ArgArg sample. The smearing signals between Myc-FRQ and frq9 FRQ are degradation products of Myc-FRQ. (B) Race tube results showing the arrhythmicity in constant darkness for the FRQ coiled-coil mutant strains. Leu165Arg, Leu169Arg and Leu165ArgArg mutations were introduced into pKAJ120. The mutations constructs were then transformed into the frq null strain (frq10). The resulting transformants were first screened by western blot analysis for FRQ expression before being examined by race tube assays. At least 10 positive transformants were examined by race tube assays. The wild-type strain and the pKAJ120 transformants were used as controls. The race tubes shown are representative samples from six replicate tubes.
The ability of FRQ containing these mutations to rescue the circadian rhythm of conidiation of the frq null strain (frq10) is a stringent test of the functional significance of the FRQ–FRQ interaction. These mutations were introduced into the wild-type frq construct (pKAJ120) and transformed into frq10. None of the constructs, even the single mutation, was able to rescue to the normal circadian banding rhythm of frq10 (Figure 5B). The occasional very broad conidiation peaks in the Leu165Arg and Leu169Arg mutants suggest that the clock might be running with a very low amplitude in these mutant strains. The circadian phenotype of the Leu165ArgArg mutant strains is indistinguishable from that of the frq null strain. These data indicate that the coiled-coil domain-mediated FRQ–FRQ interaction is essential for the circadian function of FRQ in Neurospora, and even the weakening of the FRQ–FRQ interaction disrupts the circadian clock.
FRQ–FRQ interaction is required for its interactions with the WC proteins
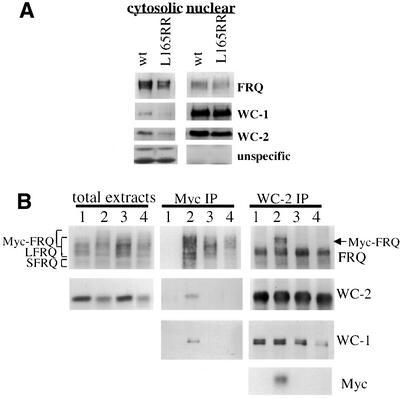
Since the formation of the FRQ–FRQ complexes is required for the function of FRQ, we wondered what the biochemical role of such complexes is. Because the coiled-coil domain is proximal to the nuclear localization signal (NLS) of FRQ (Luo et al., 1998), it is possible that the formation of the FRQ–FRQ complex is important for the nuclear localization of FRQ. To examine this possibility, nuclei preparations from both the wild-type strain and the L165RR (pKAJ120⋅165RR transformed into frq10) strain were made and the presence of FRQ and several other proteins was examined. As seen in Figure 6A, the mutant FRQ, like wild type, is found in the nucleus (Luo et al., 1998). In agreement with previous studies (Talora et al., 1999; Schwerdtfeger and Linden, 2000), both WC-1 and WC-2 are greatly enriched in the nuclear fractions. Thus, the formation of the FRQ–FRQ complex is not required for the nuclear localization of FRQ. The different nuclear and cytoplasmic distributions of FRQ and WCs suggest that FRQ nuclear localization is regulated by some unknown mechanisms.
Fig. 6. The FRQ coiled-coil domain is not required for the nuclear localization of FRQ but it is necessary for the FRQ–WC interactions. (A) Western blot analysis of the nuclear and cytosolic preparations shows that the coiled-coil mutation does not affect the nuclear localization of FRQ. Wild-type strain and Leu165ArgArg mutant strain (frq10, pKAJ120⋅L165RR) were grown and harvested in LL. The protein blots were probed with either FRQ, WC-1 or WC-2 antiserum. The two non-specific bands (bottom panels), recognized by the WC-2 antiserum, can only be found in the cytosolic fractions, suggesting that the nuclei preparations are free of cytosolic contamination. (B) The formation of the FRQ coiled-coil structure is required for the interaction between FRQ and the WC proteins. Total protein extracts from different strains grown in LL were immunoprecipitated by c-Myc or WC-2 antibody, and protein blots were probed with FRQ, WC-1, WC-2 or c-Myc antibody. Lane 1, frq+; lane 2, frq+, Myc-FRQ; lane 3, frq+, Myc-FRQ1; lane 4, frq+, FRQ-165RR.
Recently, it was reported that FRQ interacts with two PAS-domain-containing transcriptional factors, WC-1 and WC-2 (Denault et al., 2000), probably leading to the negative repression of frq transcription. To test the possibility that the formation of the FRQ–FRQ complex is important for the interaction between FRQ and the WC proteins, we examined the interactions between FRQ and the WC proteins. Wild-type strains containing Myc-FRQ with or without the coiled-coil mutations (FRQ1 or Leu165ArgArg) allowed the endogenous wild-type FRQ to be used as an internal control in the co-immunoprecipitation experiments. As expected, both WC-1 and WC-2 were specifically co-immunoprecipitated with the wild-type Myc-FRQ (Figure 6B, center panels, lane 2) when Myc antibody was used to perform immunoprecipitation. In contrast, Myc-FRQ containing the coiled-coil mutations (Figure 6B, center panels, lanes 3 and 4) failed to pull down either WC protein.
To confirm these results, the same extracts were immunoprecipitated with the WC-2 antiserum. As expected, the endogenous LFRQ and SFRQ co-immunoprecipitated with WC-2 in all strains. WC-2 also pulled down the wild-type Myc-FRQ (Figure 6B, right panels, lane 2), but it failed to pull down the Myc-FRQ containing the coiled-coil mutations (Figure 6B, right panels, lanes 3 and 4). In agreement with a previous report (Talora et al., 1999), WC-2 also interacts with WC-1 to form a heterodimer (Figure 6B, right panels). These data demonstrate that the formation of the FRQ coiled-coil structure is required for the interaction between FRQ and the WC proteins, and it is possible that this latter interaction is essential for FRQ to carry out its biochemical role in the circadian clock of Neurospora.
Discussion
Previous studies have shown that the frq gene is one of the central components of a Neurospora circadian negative feedback loop (Dunlap, 1999). In this study, we have investigated the role of FRQ protein–protein interactions. We have shown that FRQ interacts with itself in vivo through a coiled-coil domain. The data presented demonstrate that the coiled-coil-mediated FRQ–FRQ interaction is essential for its function in the Neurospora clock. We showed that point mutations of the coiled-coil structure weaken or prevent the FRQ self-association, and result in the arrhythmicity of the conidiation rhythm. Furthermore, we have also shown that the formation of the FRQ coiled-coil structure is required for the interaction between FRQ and the WC proteins. Taken together, these data indicate that the formation of the FRQ–FRQ and FRQ–WC complexes are essential for the function of the Neurospora clock.
Coiled-coils are one of the most commonly used protein structure motifs mediating protein–protein interactions (Lupas, 1996). The leucine zipper domain found in some transcription factors is the best known example (Landschulz et al., 1988; McKnight, 1991; Ellenberger et al., 1992). Previously, this region of FRQ was proposed to be a helix–turn–helix (HTH) DNA binding motif by computer analysis; however, the scores obtained for FRQ are low, and there is no experimental support for the hypothesis (Lewis et al., 1997). The evidence presented in this study strongly supports the formation of a coiled-coil structure by this region. First, we have shown that this region of FRQ mediates FRQ–FRQ interactions in vivo. Secondly, mutations that are designed to disrupt the coiled-coil hydrophobic interface weaken or completely abolish such protein–protein interaction. Thirdly, the interaction between FRQ and the WC complex and the loss of function for the mutant monomeric FRQ forms suggest that FRQ might function as a co-factor for the WC proteins. Although we do not know the orientation of the FRQ–FRQ coiled-coil complexes, the opposite charged residues flanking the hydrophobic core in the proposed coiled-coil structure (Figure 1B) suggest that FRQ might form anti-parallel coiled-coil complexes.
Previously, the sucrose gradient experiments suggested that FRQ existed mostly as a monomer although the results of the gel filtration experiments indicated that FRQ was in a large complex (Garceau et al., 1997). The failure to detect the FRQ–FRQ complexes in the sucrose gradient experiments might be due to limited resolution of the method, weak interactions among FRQ and the WCs, and the large size and various forms of FRQ. From the results of the co-immunoprecipitation experiments presented here (Figures 3 and 4), we estimate that most of the FRQ proteins form complexes with each other. We have formally shown the functional significance of the FRQ coiled-coil domain, and the simplest interpretation of our results is that FRQ proteins form dimers before interacting with the WC proteins. If this is the case, it suggests that another region of FRQ is responsible for the FRQ–WC interactions, and the formation of the FRQ–FRQ complex allows that to happen. However, it is also possible that the FRQ–FRQ and FRQ–WC complexes are mutually exclusive and FRQ interacts with the WCs through its coiled-coil domain. It is also not known which of the WC proteins FRQ interacts with and whether both WCs are required for such interactions. Examining these possibilities is the focus of our current research and it should provide us with better understanding on how these proteins work.
Is it possible that the monomeric FRQ is also functional? Several lines of evidence suggest that this is unlikely. First, the coiled-coil domain is the most conserved region in the FRQ ORF, suggesting that the protein–protein interaction mediated by this region is important for the function of FRQ. In this regard, the mutant strains with only the monomeric mutant FRQ is not functional (Figure 5B). In addition, we have shown that the mutant FRQ forms that failed self-association can no longer interact with the WC proteins, the positive elements in the circadian feedback loop.
Protein interactions of clock components are important regulatory steps for circadian negative feedback loops. In Drosophila, the formation of the PER–TIM complex is essential for the nuclear localization of both proteins (Vosshall et al., 1994; Saez and Young, 1996). Although Drosophila PER can interact to form a homodimer through the PAS domains, the functional significance of such interactions is unclear (Huang et al., 1993; Zeng et al., 1996). In mammals, three mPer genes (mPer1, mPer2 and mPer3) are part of the circadian oscillator. The interactions among the three mPERs and mCRYs appear to be important for the nuclear localization of mPER proteins (Kume et al., 1999; Field et al., 2000; Yagita et al., 2000). Once in the nucleus, PER proteins, in both Drosophila and mammals, can then interact with the protein complex formed by two PAS-domain-containing transcription factors (dCLOCK–CYC in Drosophila, BMAL1–CLOCK in mammals) to inhibit their transcriptional activation of per and some clock-controlled genes (Darlington et al., 1998; Gekakis et al., 1998; Lee et al., 1999), thus closing the negative feedback loop in each system.
In contrast, the formation of the FRQ–FRQ complex does not affect the nuclear localization of FRQ (Figure 6A). Instead, we found that mutant forms of FRQ, which fail to interact with each other, also do not interact with WC-1 and WC-2 (Figures 5 and 6). Since WC-1 and WC-2 are thought to be the positive elements in the Neurospora negative feedback loop (Dunlap, 1999), the failure of FRQ to interact with the WC complex may break the feedback loop and cause the loss of rhythmicity in these mutants (Figure 5). If this is the case, the interactions between FRQ and the WC complex play an essential role in closing the negative feedback loop in Neurospora.
In conclusion, our studies demonstrated that FRQ associations with itself and the WC proteins are essential for the function of the Neurospora circadian clock. Our data are consistent with the negative feedback model in which FRQ is the negative element in the loop, and the WC proteins are the positive elements of the loop.
Materials and methods
Strains, culture conditions and race tube assay
Neurospora strains used in this study are bdA (clock wild type), 87–12 (bd; A; his-3), 93–4 (bd; frq10; his-3), 94–1 (bd; frq9; his-3) (Aronson et al., 1994a,b) and different frq mutant strains created in this study. Strains 87–12 (frq wild-type strain), 93–4 (frq null strain) and 94–1 (frq9, making truncated FRQ) are the host strains for various frq mutation constructs described. Culture conditions were as described previously (Aronson et al., 1994a; Crosthwaite et al., 1995). Race tube assay media contains 1× Vogel’s, 0.1% glucose, 0.17% arginine and 50 ng/ml biotin. Densitometric analyses of race tubes and calculations of period length were performed as described previously (Liu et al., 1997) using Chrono II version 11.1 (Dr Till Roenneberg, Ludwigs-Maximillian University, Munich, Germany).
Plasmid constructs and Neurospora transformation
pKAJ120 (containing the entire frq gene including its promoter and a his-3 targeting sequence) is the parental plasmid for all frq constructs described in this study. To create the Myc-FRQ construct (pKAJ120⋅Myc-FRQ), a PCR fragment (using pCS2-Myc as the template, generously provided by Dr Hongtao Yu) containing five tandem repeats of c-Myc epitope (79 amino acids) was inserted into the SphI site of pKAJ120 (Figure 1A). All the deletion and point mutations of the FRQ ORF were constructed using the Transformer Site-Directed Mutagenesis kit (Clontech Laboratories, Inc., Palo Alto, CA), and pUC19Mfrq was used as the in vitro mutagenic template (Liu et al., 2000). The mutagenic primers used were FRQ1 (to delete FRQ aa 107–178), 165Arg (to mutate Leu165 to Arg), 169Arg (to mutate Leu169 to Arg) and 165ArgArg (to mutate both Leu165 and Leu169 to Arg). After mutagenesis, the region containing the mutation was subcloned into either pKAJ120 (for rescuing the circadian conidiation rhythm of frq null strain) or pKAJ120⋅Myc-FRQ (for immunoprecipitation assay). All constructs were confirmed by DNA sequencing, and they were targeted by transformation to the his-3 locus of the host strains as previously described (Bell-Pedersen et al., 1996). For protein analyses, at least three or four independent transformants were examined. For race tube analysis, at least 10 independent positive transformants were examined.
Computer analysis of the FRQ coiled-coil structure
Web-based MultiCoil (http://nightingale.lcs.mit.edu/cgi-bin/multicoil) and Paircoil (http://nightingale.lcs.mit.edu/cgi-bin/score) programs were used to predicted the probability of coiled-coil formation (Berger et al., 1995; Wolf et al., 1997). They were also used to help design mutations to be introduced into the FRQ coiled-coil domain.
Generation of antisera against WC-1 and WC-2
Glutathione S-transferase (GST)–WC-1 (containing WC-1 aa 288–709) and GST–WC-2 (containing the entire WC-2 ORF) fusion proteins were expressed in BL21 cells and the inclusion bodies containing the recombinant proteins were purified. To purify the inclusion bodies, the pellets of 500 ml of Escherichia coli cultures (after 2 h of 1 mM isopropyl-β-d-thiogalactopyranoside induction) was resuspended in 40 ml of buffer A (50 mM Tris–HCl pH 8.3, 1 mM EDTA, 2 mM 2-mercaptoethanol) containing freshly added 0.1% lysozyme. The resuspensions were kept at room temperature for 40 min before the cells were broken by sonication (5′′ × 30, Sonifier 450, VWR Scientific). The pellets were collected by centrifugation (9000 r.p.m. × 25′) and washed in 20 ml of buffer C (Tris–HCl pH 8.3, 2 mM EDTA, 2 mM 2-mercaptoethanol, 2 M urea, 0.5% Triton X-100). The resulting pellets were then resuspended in 8 M urea and the GST fusion proteins were then purified by electroelution after running on preparative 7.5% SDS–PAGE gels. WC-1 and WC-2 antisera were generated by immunizing New Zealand white rabbits with the purified recombinant proteins according to standard protocols. WC-2 antiserum was used for western blot analysis and immunoprecipitation assays without further purification. For WC-1 antiserum, it was affinity purified using nitrocellulose membranes blotted with purified GST–WC-1 protein.
Protein analyses
Protein extraction, quantification and western blot analysis are as described previously (Garceau et al., 1997). Equal amounts of total protein (∼100 µg) were loaded in each protein lane and after the blots were developed by chemiluminescence (ECL, Amersham Pharmacia, Piscataway, NJ), they were stained by amido black to verify equal loading of protein (Liu et al., 1997). The purification of Neurospora nuclei was performed as described previously (Luo et al., 1998).
For IP, 0.5 mg of soluble protein extracts in extraction buffer (Garceau et al., 1997) were incubated at 4°C for 2 h with either the monoclonal c-Myc antibody (2 µg; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or WC-2 antiserum (1:250 dilution). Then protein G–agarose beads (10 µl) were added and incubated for 1–2 h at 4°C, washed four times with ice-cold extraction buffer before the IP products were resuspended in protein loading buffer or 50 µl of phosphatase buffer.
For phosphatase treatments, 1000 U of lambda PPase (New England Biolabs, Beverly, MA) were added to the IP complexes resuspended in the phosphatase buffer and incubated at 30°C for 30 min before loading on SDS–PAGE gels (Garceau et al., 1997).
Acknowledgments
Acknowledgements
We are grateful to Drs Jay Dunlap and Jennifer Loros for early support (R37-GM 34985 to J.C.D. and MH44651 to J.C.D. and J.J.L.). We thank Drs Philip Thomas, Jim Stull, Stephen Hammes, Helen Yin and Kevin Gardner for helpful discussion and comments to the paper, Cindy Patterson for administrative assistance, Drs Mingsheng Zhu and Huiqiao Sun for technical help in antisera generations, Leng Wen for providing c-Myc antibody, and Allan C.Froehlich for advice on nuclei preparation. Y.L. is a Louise W.Kahn endowed scholar in Biomedical Research at UT Southwestern Medical Center.
References
- Allada R., White,N.E., So,W.V., Hall,J.C. and Rosbash,M. (1998) A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell, 93, 791–804. [DOI] [PubMed] [Google Scholar]
- Aronson B., Johnson,K., Loros,J.J. and Dunlap,J.C. (1994a) Negative feedback defining a circadian clock: autoregulation in the clock gene frequency. Science, 263, 1578–1584. [DOI] [PubMed] [Google Scholar]
- Aronson B.D., Johnson,K.A. and Dunlap,J.C. (1994b) The circadian clock locus frequency: A single ORF defines period length and temperature compensation. Proc. Natl Acad. Sci. USA, 91, 7683–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D., Shinohara,M., Loros,J. and Dunlap,J.C. (1996) Circadian clock-controlled genes isolated from Neurospora crassa are late night to early morning specific. Proc. Natl Acad. Sci. USA, 93, 13096–13101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B., Wilson,D.B., Wolf,E., Tonchev,T., Milla,M. and Kim,P.S. (1995) Predicting coiled coils by use of pairwise residue correlations. Proc. Natl Acad. Sci. USA, 92, 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bünning E. (1973) The Physiological Clock. Springer-Verlag, New York, NY.
- Crosthwaite S.C., Loros,J.J. and Dunlap,J.C. (1995) Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell, 81, 1003–1012. [DOI] [PubMed] [Google Scholar]
- Crosthwaite S.C., Dunlap,J.C. and Loros,J.J. (1997) Neurospora wc-1 and wc-2: Transcription, photoresponses and the origins of circadian rhythmicity. Science, 276, 763–769. [DOI] [PubMed] [Google Scholar]
- Darlington T.K., Wager-Smith,K., Ceriani,M.F., Stankis,D., Gekakis,N., Steeves,T., Weitz,C.J., Takahashi,J. and Kay,S.A. (1998) Closing the circadian loop: CLOCK induced transcription of its own inhibitors, per and tim. Science, 280, 1599–1603. [DOI] [PubMed] [Google Scholar]
- Denault D.L., Dunlap,D.C. and Loros,J.J. (2000) Biochemical interactions between FRQ and WC-2: critical clock proteins required for the normal operation of the Neurospora. The NEUROSPORA 2000 Meeting. Abstract 26.
- Dunlap J.C. (1999) Molecular bases for circadian clocks. Cell, 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Ellenberger T.E., Brandl,C.J., Struhl,K. and Harrison,S.C. (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein–DNA complex. Cell, 71, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Field M.D., Maywood,E.S., O’Brien,J.A., Weaver,D.R., Reppert,S.M. and Hastings,M.H. (2000) Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron, 25, 437–447. [DOI] [PubMed] [Google Scholar]
- Garceau N., Liu,Y., Loros,J.J. and Dunlap,J.C. (1997) Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell, 89, 469–476. [DOI] [PubMed] [Google Scholar]
- Gekakis N., Stankis,D., Nguyen,H.B., Davis,F.C., Wilsbacher,L.D., King,D.P., Takahashi,J.S. and Weitz,C.J. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science, 280, 1564–1569. [DOI] [PubMed] [Google Scholar]
- Glossop N.R., Lyons,L.C. and Hardin,P.E. (1999) Interlocked feedback loops within the Drosophila circadian oscillator. Science, 286, 766–768. [DOI] [PubMed] [Google Scholar]
- Hall J.C. (1998) Genetics of biological rhythms in Drosophila. Adv. Genet., 38, 135–184. [DOI] [PubMed] [Google Scholar]
- Hardin P.E. (1998) Activating inhibitors and inhibiting activators: a day in the life of a fly. Curr. Opin. Neurobiol., 8, 642–647. [DOI] [PubMed] [Google Scholar]
- Huang Z.J., Edery,I. and Rosbash,M. (1993) PAS is a dimerization domain common to Drosophila Period and several transcription factors. Nature, 364, 259–262. [DOI] [PubMed] [Google Scholar]
- Ishiura M., Kutsuna,S., Aoki,S., Iwasaki,H., Anderson,C.R., Tanabe,A., Golden,S.S., Johnson,C.H. and Kondo,T. (1998) Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science, 281, 1519–1523. [DOI] [PubMed] [Google Scholar]
- Johnson C.H. and Golden,S.S. (1999) Circadian programs in cyanobacteria: adaptiveness and mechanism. Annu. Rev. Microbiol., 53, 389–409. [DOI] [PubMed] [Google Scholar]
- King D.P. and Takahashi,J.S. (2000) Molecular genetics of circadian rhythms in mammals. Annu. Rev. Neurosci., 23, 713–742. [DOI] [PubMed] [Google Scholar]
- King D. et al. (1997) Positional cloning of the mouse circadian CLOCK gene. Cell, 89, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K., Zylka,M.J., Sriram,S., Shearman,L.P., Weaver,D.R., Jin,X., Maywood,E.S., Hastings,M.H. and Reppert,S.M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell, 98, 193–205. [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas P.L. and Brody,S. (2000) Circadian rhythms in Neurospora crassa: Lipid deficiencies restore robust rhythmicity to null frequency and white-collar mutants. Proc. Natl Acad. Sci. USA, 97, 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W.H., Johnson,P.F. and McKnight,S.L. (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science, 240, 1759–1764. [DOI] [PubMed] [Google Scholar]
- Lee C., Bae,K. and Edery,I. (1999) PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK–CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol. Cell. Biol., 19, 5316–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Loros,J.J. and Dunlap,J.C. (2000) Interconnected feedback loops in the Neurospora circadian system. Science, 289, 107–110. [DOI] [PubMed] [Google Scholar]
- Lewis M., Morgan,L. and Feldman,J.F. (1997) Analysis of frequency (frq) clock gene homologs: evidence for a helix–turn–helix transcription factor. Mol. Gen. Genet., 253, 401–414. [DOI] [PubMed] [Google Scholar]
- Liu Y., Garceau,N., Loros,J.J. and Dunlap,J.C. (1997) Thermally regulated translational control mediates an aspect of temperature compensation in the Neurospora circadian clock. Cell, 89, 477–486. [DOI] [PubMed] [Google Scholar]
- Liu Y., Merrow,M.M., Loros,J.J. and Dunlap,J.C. (1998) How temperature changes reset a circadian oscillator. Science, 281, 825–829. [DOI] [PubMed] [Google Scholar]
- Liu Y., Heintzen,C., Loros,J. and Dunlap,J.C. (1999) Regulation of clock genes. Cell. Mol. Life Sci., 55, 1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Loros,J. and Dunlap,J.C. (2000) Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc. Natl Acad. Sci. USA, 97, 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Loros,J.J. and Dunlap,J.C. (1998) Nuclear localization is required for function of the essential clock protein FREQUENCY. EMBO J., 17, 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. (1996) Coiled coils: new structures and new functions. Trends Biochem. Sci., 21, 375–382. [PubMed] [Google Scholar]
- McKnight S.L. (1991) Molecular zippers in gene regulation. Sci. Am., 264, 54–64. [DOI] [PubMed] [Google Scholar]
- Merrow M., Brunner,M. and Roenneberg,T. (1999) Assignment of circadian function for the Neurospora clock gene frequency. Nature, 399, 584–586. [DOI] [PubMed] [Google Scholar]
- Rutila J.E., Suri,V., Le,M., So,W.V., Rosbash,M. and Hall,J.C. (1998) CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell, 93, 805–813. [DOI] [PubMed] [Google Scholar]
- Saez L. and Young,M.W. (1996) Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron, 17, 911–920. [DOI] [PubMed] [Google Scholar]
- Schwerdtfeger C. and Linden,H. (2000) Localization and light-dependent phosphorylation of white collar 1 and 2, the two central components of blue light signaling in Neurospora crassa. Eur. J. Biochem., 267, 414–422. [DOI] [PubMed] [Google Scholar]
- Shearman L.P. et al. (2000) Interacting molecular loops in the mammalian circadian clock. Science, 288, 1013–1019. [DOI] [PubMed] [Google Scholar]
- Somers D.E., Schultz,T.F., Milnamow,M. and Kay,S.A. (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell, 101, 319–328. [DOI] [PubMed] [Google Scholar]
- Talora C., Franchi,L., Linden,H., Ballario,P. and Macino,G. (1999) Role of a white collar-1–white collar-2 complex in blue-light signal transduction. EMBO J., 18, 4961–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall L., Price,J., Sehgal,A., Saez,L. and Young,M. (1994) Specific block in nuclear localization of period protein by a second clock mutation, timeless. Science, 263, 1606–1609. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y. and Tobin,E.M. (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell, 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wolf E., Kim,P.S. and Berge,B. (1997) MultiCoil: A program for predicting two- and three-stranded coiled coils. Protein Sci., 6, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K., Yamaguchi,S., Tamanini,F., van Der Horst,G.T., Hoeijmakers,J.H., Yasui,A., Loros,J.J., Dunlap,J.C. and Okamura,H. (2000) Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev., 14, 1353–1363. [PMC free article] [PubMed] [Google Scholar]
- Young M.W. (1999) Molecular control of circadian behavioral rhythms. Recent Prog. Horm. Res., 54, 87–94. [PubMed] [Google Scholar]
- Zeng H., Qian,Z., Myers,M. and Rosbash,M. (1996) A light-entrainment mechanism for the Drosophila circadian clock. Nature, 380, 129–135. [DOI] [PubMed] [Google Scholar]