Abstract
Telomerase is a reverse transcriptase that maintains chromosome ends. The N-terminal half of the catalytic protein subunit (TERT) contains three functional domains (I, II, and III) that are conserved among TERTs but not found in other reverse transcriptases. Guided by an amino acid sequence alignment of nine TERT proteins, mutations were introduced into yeast TERT (Est2p). In support of the proposed alignment, mutation of virtually all conserved residues resulted in loss-of-function or temperature sensitivity, accompanied by telomere shortening. Overexpression of telomerase component Est3p led to allele-specific suppression of the temperature-sensitive mutations in region I, suggesting that Est3p interacts with this protein domain. As predicted by the genetic results, a lethal mutation in region I resulted in loss of Est3p from the telomerase complex. We conclude that Est2p region I is required for the recruitment of Est3p to yeast telomerase. Given the phylogenetic conservation of region I of TERT, this protein domain may provide the equivalent function in all telomerases.
INTRODUCTION
Telomeres, the DNA-protein complexes located at the ends of most linear, eukaryotic chromosomes, protect chromosome ends from nucleolytic digestion, distinguish normal chromosome ends from broken ends, and facilitate complete replication of the genome. This last function is mediated by telomerase, a ribonucleoprotein enzyme that uses its intrinsic RNA subunit as a template for addition of telomeric DNA to the end of the chromosome (Greider and Blackburn, 1987, 1989). The reverse transcription reaction is catalyzed by a protein subunit of telomerase (TERT; TElomerase Reverse Transcriptase; Lingner et al., 1997a), homologues of which have been identified in a number of organisms (Harrington et al., 1997; Kilian et al., 1997; Meyerson et al., 1997; Nakamura et al., 1997; Bryan et al., 1998; Collins and Gandhi, 1998; Greenberg et al., 1998; Fitzgerald et al., 1999; Oguchi et al., 1999; Malik et al., 2000).
The TERT proteins share a central domain (RT domain) that is homologous to the catalytic domain of other reverse transcriptases, including a triad of invariant and essential aspartate residues (Nakamura et al., 1997; Lingner et al., 1997a). Saccharomyces cerevisiae TERT is encoded by the gene EST2, first identified in a screen for mutations that result in Ever Shorter Telomeres (Lendvay et al., 1996). Although Est2p and the telomerase RNA (TLC1 RNA; Singer and Gottschling, 1994) are thought to form the catalytic core of telomerase, additional proteins of the telomerase holoenzyme are required for telomerase activity in vivo. Deletion of EST1 or EST3 results in a telomere-loss phenotype identical to disruption of EST2 (Lundblad and Szostak, 1989; Lendvay et al., 1996). However, these proteins are not required for the in vitro activity of telomerase (Cohn and Blackburn, 1995; Lingner et al., 1997b). Based on this observation, Est1p and Est3p have been proposed to play essential regulatory roles in vivo (Nugent and Lundblad, 1998). Indeed, Est1p directly interacts with Cdc13p, a protein that binds single-stranded telomeric DNA, and this interaction recruits or activates telomerase at the telomere (Evans and Lundblad, 1999; Qi and Zakian, 2000; Pennock et al., 2001; Taggart et al., 2002). The role of Est3p in the telomerase complex is unknown.
Yeast Est2p contributes to the catalytic activity of telomerase and to other aspects of telomerase assembly and function. For example, coimmunoprecipitation of Est3p with the telomerase RNA is dependent on Est2p (Hughes et al., 2000). Some of these additional functions of Est2p may be mediated by the N-terminal region that precedes the RT domain, a region found in all TERTs but not in other RTs. We previously undertook wide-scale mutagenesis (Unigenic Evolution; Deminoff et al., 1995) and identified three novel regions (I–III) in the N-terminal half of Est2p that are essential for telomerase function in vivo (Friedman and Cech, 1999). Region III contributes to binding of TLC1 RNA (Friedman and Cech, 1999). Indeed, similar regions of Tetrahymena thermophila and human TERT also bind telomerase RNA (Beattie et al., 2000; Bryan et al., 2000; Lai et al., 2001), suggesting that region III of Est2p may correspond to a protein domain with similar functions in all TERT homologues. The roles of regions I and II have been less clear. We previously identified a mutation in region I (est2-ala1) that is defective for telomere maintenance in vivo, but has minimal effects on telomerase activity in vitro (Friedman and Cech, 1999). This phenotype is reminiscent of that resulting from deletion of EST1 or EST3 and suggests that residues in region I might mediate interactions with telomerase accessory proteins. Intriguingly, mutations within a subdomain of region I of human TERT (the DAT domain) also destroy the in vivo activity of telomerase, but leave catalytic activity intact (Armbruster et al., 2001).
In this study, we use an alignment of TERT sequences to guide site-directed mutagenesis of conserved residues within essential regions I–III of Est2p. Single mutations of the most highly conserved amino acids either destroy Est2p function or confer a temperature-sensitive (ts) phenotype on strains carrying those alleles. Strikingly, overexpression of Est3p specifically suppresses the ts growth of mutants in region I of Est2p, but has little or no effect on the growth of mutants in regions II or III, suggesting that Est2p region I recruits Est3p to the telomerase complex. Indeed, both Est3p and Est1p fail to coimmunoprecipitate with Est2p bearing a lethal mutation in region I (est2-ala1). Therefore, we propose that one essential role of Est2p region I is to stabilize the association of these accessory proteins with the telomerase complex.
MATERIALS AND METHODS
Alignment of TERT Sequences
TERT sequences were obtained from GenBank and aligned using Clustal W (http://www.ebi.ac.uk/clustalw/). Refinement of the alignment was done manually.
Site-directed Mutagenesis of EST2
Plasmid pKF410 was used as the base for mutagenesis of EST2. This plasmid is derived from pKF409 (Friedman and Cech, 1999) by cloning of a KpnI-SacI fragment containing ProA-tagged EST2 into the corresponding sites of pRS316 (URA3, CEN; Sikorski and Hieter, 1989). Mutants were created by site-directed mutagenesis (Deng and Nickoloff, 1987) to change the desired amino acids to alanine (GCA) or aspartic acid (GAT). Two independent isolates of each mutant were created and tested for Est2p function. N80D is an AAC-to-GAC mutation recovered during Unigenic Evolution (Friedman and Cech, 1999). Function of mutant est2 alleles was monitored by transformation into YKF102 (est2::HIS3 rad52::LEU2) as previously described (Friedman and Cech, 1999).
Integration of est2 Alleles
Functional est2 alleles were integrated into strain YKF101 (est2::HIS3; Friedman and Cech, 1999) by standard two-step integration. Mutant alleles created in pKF410 (described above) were subcloned into the integrating vector pRS306 (URA3; Sikorski and Hieter, 1989) by cleavage with AlwNI. Resulting plasmids (pKF409 series) were cleaved with ClaI to target integration 5′ of the EST2 coding region. Candidate strains selected for 5-FOA resistance were screened for loss of the HIS3 marker as an indication of retention of the mutant est2 allele. All strains were screened by Southern analysis, and a representative subset was sequenced to confirm presence of the mutation. Note that the use of an est2::HIS3 strain as the recipient of the integration construct directs recombination downstream of EST2, resulting in integration of the entire plasmid-borne coding region of EST2. As a result, all integrated strains contain the C terminus of EST2 derived from plasmid pKF409.
As described in the RESULTS section, the original EST2 clone (Lendvay et al., 1996) contains a four-nucleotide insertion in a TA-dinucleotide tract at the extreme C terminus of EST2 compared with the EST2 gene in our laboratory strain (TVL268). As a result, the TVL268-specific allele ends in the sequence YIYIYI, whereas the published allele terminates with the sequence YIYIYIHIVN. In the course of our site-directed mutagenesis, the TA-dinucleotide repeat incurred an additional six-nucleotide insertion to result in the predicted protein YIYIYIHIHIVN. The method used to integrate est2 alleles results in preservation of this alternative C terminus (see above).
Plasmid Construction
Overexpression plasmids were constructed as follows. Plasmid pVL249 [Yeplac 112 (TRP1, 2 μ) containing EST1 with ADH promoter and terminator sequences] was obtained from V. Lundblad. The SphI fragment containing EST1 was cloned from pVL249 into the BamHI site of pRS423 (HIS3, 2 μ; Christianson et al., 1992) after blunting of DNA ends with T4 DNA polymerase. Plasmid pVL325 [Yeplac351 (LEU2, 2 μ) containing EST3] was obtained from V. Lundblad. The EcoRI-SalI fragment containing EST3 was cloned into the same sites in the polylinker of pRS423. To create a plasmid for overexpression of SMD3, a BamHI fragment containing HA-SMD3 from plasmid pAS503 (Seto et al., 1999) was cloned into the BamHI site of pRS423.
Plasmids for expression of EST2 and ProA-EST2 were constructed as follows. PvuII fragments containing EST2 or ProA-EST2 were obtained by cleavage of pKF404 or pKF409, respectively (Friedman and Cech, 1999). These fragments were cloned into the corresponding sites of pRS315 (LEU2 CEN; Sikorski and Hieter, 1989) to create pKF417 (EST2) and pKF416 (ProA-EST2). A plasmid for overexpression of ProA-est2-ala1 was obtained by cloning the KpnI-SacI fragment from pKF409-Ala1 into pRS425 (LEU2, 2 μ; Christianson et al., 1992) to yield pRS425 est2-ala1.
Southern Blot Analysis
Yeast DNA was prepared using the DNA-Pure Yeast Genomic Kit (CPG Inc., Fairfield, NJ) and cleaved with XhoI. The DNA was separated on a 1.1% agarose gel, transferred to Hybond N+ membrane (Amersham Biosciences, Piscataway, NJ), and probed with a random-primed yeast telomere sequence probe. A fragment from chromosome IV serves as a loading control (Friedman and Cech, 1999).
Growth Curve Analysis
Cells containing either pRS423 (HIS3, 2 μ) or pRS423-EST3 (see Plasmid Construction) were grown to saturation at 25°C in defined media lacking histidine. Cell density was determined by hemocytometer, and the cells were diluted to 5 × 105 cells per ml in 10 ml of media. The cultures were grown at 35°C with agitation in 50 ml conical tubes (Corning 430291, Corning, NY). Every 24 h, cell density was determined as above, and the cells were diluted to 5 × 105 cell/ml. The cell density achieved during every 24 h of growth was recorded as shown in Figure 6.
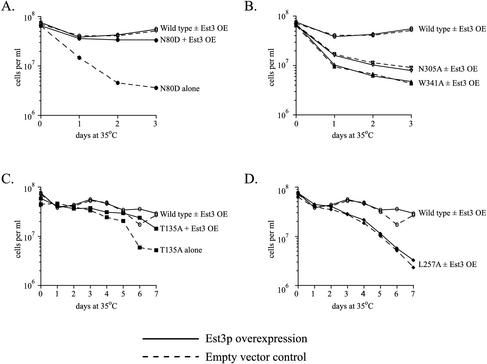
Figure 6.
Quantitation of suppression by Est3p overexpression in liquid culture. The growth of wild-type and integrated est2 mutant strains at 35°C is shown in the presence of empty vector (dashed lines) or 2 μ EST3 (solid lines). OE, overexpression. The cell density plotted represents the amount each culture grew over a 24-h period after dilution to 5 × 105 cell/ml. Note that the time scale on the X-axis is significantly different for alleles with strong temperature-sensitive phenotypes (A and B) and weak temperature-sensitive phenotypes (C and D). Growth of the wild-type strain is shown in each graph for comparison (○). (A) Region I mutant N80D (●). (B) Region III mutants W341A (▴) and N305A (▾). (C) Region I mutant T135A (▪). (D) Region II mutant L257A (♦).
Coimmunoprecipitation Assay and Western Blotting
The EST2 coding region was replaced with TRP1 in strains pVL288 (HA3-EST1; Hughes et al., 2000) and pVL293 (EST3-HA3-GST; Hughes et al., 2000). After minimal growth, these strains were transformed with a URA3 centromere plasmid expressing untagged EST2 (pKF404). These strains were subsequently transformed with pKF416, pKF417, or pRS425 est2-ala1. Strains were grown at 30°C in YPD to OD600 ∼1. Extracts were prepared by glass bead lysis in TMG (10 mM Tris-Cl pH 8, 1 mM MgCl2, 10% glycerol, 0.1 mM DTT) with 200 mM NaCl. One Complete, Mini protease tablet (Roche Molecular Biochemicals, Indianapolis, IN) was added for each 10 ml of TMG. RNasin (Promega, Madison, WI) was added at a concentration of 1 μl per 500 μl extract. For immunoprecipitation of ProA-Est2p, extract was adjusted to 0.5% Tween-20 and incubated for 3 h at 4°C with 60 μl IgG Sepharose beads (Amersham Biosciences) equilibrated with TMG (plus 200 mM NaCl, 0.5% Tween-20). After incubation, beads were washed three times with TMG (plus 200 mM NaCl, 0.5% Tween-20), once with TMG (plus 50 mM NaCl) and resuspended in 20 μl TMG (plus 50 mM NaCl, 0.5 mM DTT). To visualize coimmunoprecipitation of HA3-Est3p, ProA-Est2p was immunoprecipitated under nonreducing conditions. Extract was prepared with DTT as described above. However, all solutions used for the immunoprecipitation were prepared without DTT and RNasin.
For immunoprecipitation of HA3-tagged proteins, 500 μl extract was adjust to 0.5% Tween-20 and 1 μl RNasin was added. Adjusted extract was incubated with 20 μl rabbit polyclonal HA antibody (Y11, Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at 4°C. Protein G–conjugated beads (60 μl; Amersham Biosciences) equilibrated with TMG (plus 200 mM NaCl, 0.5% Tween-20) were added, and the incubation was continued for 6 h at 4°C. Beads were washed as described above.
Immunoprecipitation beads (10 μl) were mixed with an equal volume of 2× Laemmli loading buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 0.05% bromophenol blue, 20% glycerol, 5% β-mercaptoethanol) and heated to 98°C for 5 min, and the supernatant was subjected to electrophoresis either in a 6% Tris-glycine Novex gel (Invitrogen, Carlsbad, CA; for detection of ProA-Est2p) or in a 4–20% Tris-glycine Novex gel (Invitrogen; for detection of HA3-Est1p and HA3-Est3p). IgG beads prepared from the HA3-EST3 strain were loaded in nonreducing Laemmli loading buffer (same as above without β-mercaptoethanol). For detection of HA3-tagged proteins, protein was transferred to PVDF membrane (Amersham Biosciences) by wet transfer in 20 mM phosphate buffer, pH 6.8. For detection of protein A–tagged Est2p, protein was transferred to Zetabind membrane (Cuno Laboratory Products, Meriden, CT) as above. HA3-tagged proteins were detected with monoclonal HA.11 antibody (Babco, Richmond, CA). Secondary was peroxidase-conjugated goat anti-mouse antibody (Chemicon, Temecula, CA). Protein A–tagged Est2p was directly detected with HRP-conjugated F-7 HA antibody (Santa Cruz Biotechnology) that cross-reacts efficiently with protein A.
For the Western blot shown in Figure 2B, extract from each of the cultures was prepared as described above, and the protein concentration was adjusted to 20 mg/ml. ProA-Est2p was immunoprecipitated as described above. The indicated volume of beads were mixed with loading buffer and heated for 10 min at 70°C, and the supernatant was loaded on a 7% Tris acetate NuPAGE gel (Invitrogen). Proteins were transferred by wet transfer to PVDF membrane (Amersham). Primary antibody was monoclonal anti–protein-A antibody (clone SPA-27; Sigma, St. Louis, MO). Secondary was peroxidase-conjugated goat anti-mouse antibody (Chemicon).
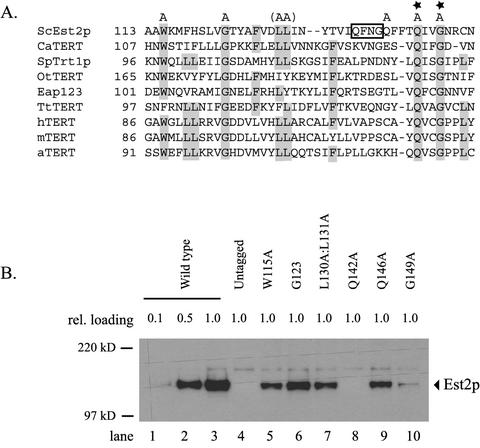
Figure 2.
Lethal mutations in Est2p do not eliminate protein expression. (A) Alignment of TERT proteins in a short segment of region I. The proposed alignment of 9 TERT proteins is shown. Conserved amino acids are highlighted. Note the invariant glutamine (Q) and glycine (G) motif (starred). A neighboring QxxG motif is boxed. The positions of amino acids that were mutated to alanine and that failed to complement an est2 deletion are indicated by an “A” above the alignment. L130 and L131 were simultaneously mutated to create a single allele (L130A:L131A). ScEst2p, S. cerevisiae; CaTERT, Candida albicans; SpTRT1p, Schizosaccharomyces pombe; OtTERT, Oxytricha trifallax; Eap123, Euplotes aediculatus; TtTERT, Tetrahymena thermophila; hTERT, human; mTERT, mouse; aTERT, Arabidopsis thaliana. (B) Expression of ProA-Est2p in strains carrying lethal est2 mutations. A yeast strain containing a chromosomal deletion of est2 (est2::HIS3) and carrying a URA3 CEN plasmid bearing wild-type, untagged EST2 was transformed with a LEU2 CEN plasmid encoding the indicated mutant alleles of ProA-est2. Loss of the complementing plasmid was selected on 5-FOA and the resulting senescent cultures were grown to midlog phase. Cell extracts were adjusted to 20 mg/ml total protein and ProA-Est2p was immunoprecipitated on IgG-conjugated beads. Relative loading is indicated. “Untagged” indicates a strain expressing Est2p lacking the protein A tag.
RESULTS
Mutational Analysis of Conserved Residues in Yeast
As a step toward understanding the function of the three essential N-terminal domains of Est2p, nine predicted TERT amino acid sequences (human, mouse, Oxytricha trifallax, T. thermophila, Euplotes aediculatus, S. cerevisiae, Schizosaccharomyces pombe, Candida albicans, and Arabidopsis thaliana) were aligned using Clustal wPCC (Supplementary Data) to identify highly conserved residues. The alignment presented here is similar, but not identical, to several complete or partial alignments that have been recently proposed (Malik et al., 2000; Miller et al., 2000; Xia et al., 2000). The amino acid sequence of the N-terminal 370 amino acids of Est2p (yeast TERT) is shown in Figure 1A. Those residues that are conserved among TERT family members are indicated. The regions showing the highest levels of amino acid conservation among all TERT proteins are coincident with essential regions I, II, and III of yeast TERT (Est2p) identified by Unigenic Evolution (Figure 1A). These three regions are also essential for human telomerase activity as demonstrated by mutational analysis (Armbruster et al., 2001), supporting the high level of functional conservation between the TERT proteins.
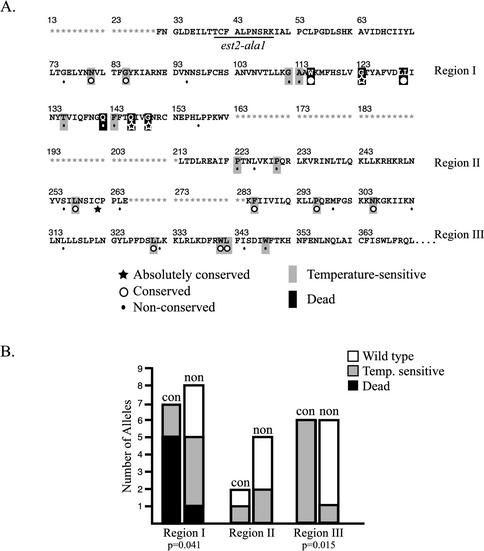
Figure 1.
Mutations in conserved residues of yeast TERT affect protein function. (A) Summary of individual point mutations created in yeast TERT (Est2p). The amino acid sequence is shown by single amino acid code for residues within essential regions I, II, and III. Residues within nonessential regions are represented by an asterisk (*). Positions that were mutated are indicated (★, conserved position containing an identical amino acid in all 9 TERT proteins; ○, conserved position containing an identical amino acid in at least 6 of 9 TERT proteins; ●, nonconserved position containing an identical amino acid in 4 or fewer TERT proteins). In two cases, an amino acid was mutated in S. cerevisiae TERT that corresponded to a conserved amino acid in most family members but was not conserved in yeast Est2p (G112A and T135A). These positions are classified as nonconserved. Two residues (N80 and L257) are conserved in 5 of 9 TERT homologues and were considered conserved for the purposes of this study. All mutations were to alanine with the exception of N80D and A113D. The N80D mutant was fortuitously isolated in the course of the Unigenic Evolution analysis. L130 and L131 were simultaneously mutated to alanine. The position of the est2-ala1 mutation is shown (residues 40–49 changed to alanine). The phenotype of each mutant is indicated by shading: Light shading, temperature-sensitive growth; dark shading, senescence at all temperatures. (B) Bar graph summarizing phenotypes of region I, II, and III mutants. The height of each bar represents the total number of mutants, whereas the shading corresponds to the mutant phenotype as indicated in the legend. Conserved (con) and nonconserved (non) positions are graphed separately. Mutant alleles were classified as “wild-type” if they supported colony growth at 25 and 35°C that was qualitatively similar to the wild-type control strain. Mutant alleles were classified as “temperature sensitive” if they showed senescence at 35°C that was accelerated compared with the wild-type strain. Mutant alleles classified as “dead” failed to rescue an est2 rad52 strain at 30°C. p value shown for region I indicates a significant difference between the proportion of lethal mutations in conserved vs. nonconserved positions as calculated by Fisher's exact test. p value shown for region III indicates a significant difference between the proportion of temperature-sensitive mutations in conserved vs. nonconserved positions.
To generate Est2p alleles suitable for functional and genetic analysis, a number of single amino acid changes (and one double mutation, L130A:L131A) were introduced into protein A–tagged Est2p (ProA-Est2p) encoded by a low-copy-number centromere plasmid. This epitope-tagged version of Est2p supports virtually normal telomerase activity as assayed by the ability of the ProA-EST2 strain to maintain telomere length <20 base pairs shorter than wild type (Friedman and Cech, 1999). All positions in the N-terminal 400 amino acids of Est2p that were conserved in at least 8 of the 9 TERT family members were mutated to alanine. Most positions that were conserved in at least 7 of 9 family members were mutated as well, provided that S. cerevisiae TERT contained the conserved residue at that position. Because the amino acid at position 113 was alanine, this residue was mutated to aspartic acid. For comparison, 17 nonconserved positions (an identical amino acid in four or fewer TERT proteins without similarity among the remaining family members) were mutated (Figure 1A).
Multiple isolates of the centromere plasmid-borne est2 alleles were tested for complementation in an est2 rad52 strain at 30°C. The rad52 mutation prevents survival in the absence of telomerase by inhibiting the maintenance of telomeres by a Rad52p-dependent recombination mechanism (Lundblad and Blackburn, 1993). The ability of each of the mutated alleles of Est2p to rescue the growth of senescing yeast is summarized in Figure 1. Strikingly, all but two of the conserved positions in region I are essential for telomerase activity as measured in this assay, whereas only one nonconserved residue is essential. Indeed, the two mutations in conserved residues that are viable (G85A and N80D) result in severely reduced telomere length (>150 base pairs shorter than wild type; unpublished data). This difference in the effect of mutations at conserved and nonconserved positions is statistically significant (p = 0.041) and supports the proposed sequence alignment in region I.
This result is comparable to that reported by Xia et al. (2000) in which a similar alignment of TERT homologues was used to guide mutagenesis within region I of Est2p. Two of the mutations that we created in EST2 (G85A and W115A) are identical to those previously reported (Xia et al., 2000). Consistent with their results, we find that the W115A mutant is inviable, whereas the G85A mutant is viable but results in severely reduced telomere length. On the other hand, the alignment that we propose differs slightly from that of Xia et al. (2000). Most significantly, an absolutely conserved glutamine and glycine pair separated by two amino acids (QxxG) within region I of yeast TERT is aligned differently with the other TERT proteins. Figure 2A shows the detailed protein alignment in this region. Est2p contains two closely juxtaposed QxxG motifs. We propose that the more C-terminal of these sequences provides the correct alignment with the other TERT proteins (see also the alignments proposed by Malik et al., 2000 and Miller et al., 2000). Mutation of either Q146 or G149 to alanine is lethal (Figure 1A), consistent with the absolute conservation of these amino acids in the other TERT proteins. In contrast, mutations in the alternative QxxG sequence (Q138 and G141) reduce telomere length but do not eliminate protein function (Xia et al., 2000).
Protein levels were examined in strains carrying each of the lethal alleles within region I (Figure 2). An est2::HIS3 strain containing untagged EST2 on a URA3 CEN plasmid was transformed with each protein A–tagged est2 mutant allele on a LEU2 CEN plasmid (pKF416; see MATERIAL AND METHODS). The URA3 plasmid was subsequently evicted by growth of the strains on plates containing 5-fluoroorotic acid (5-FOA). ProA-Est2 protein levels were assessed by immunoprecipitation on IgG-conjugated beads and Western blotting. As shown in Figure 2B, the W115A, G123A, L130A:L131A, and Q146A mutations reduce Est2 protein levels to approximately half of the wild-type level. Decreases in Est2p levels of this magnitude do not by themselves inhibit cell growth (Friedman and Cech, 1999). The G149A mutation has a larger effect, resulting in a nearly 10-fold decrease in protein (Figure 2B, lane 10). The Q142A protein is extremely unstable and cannot be detected (Figure 2B, lane 8).
Temperature Sensitivity of est2 Alleles
Somewhat surprisingly, none of the mutations to alanine in conserved residues of region II or III eliminated protein function at 30°C. Therefore, we were unable to test the validity of the sequence alignment through region II and III using viability as an assay. To provide a more sensitive measure of protein function, all proA-est2 alleles that were able to rescue the est2 rad52 strain at 30°C were integrated into the genome at the endogenous EST2 locus. As predicted by the ability of these alleles to complement an est2 mutation when plasmid-borne, strains containing integrated versions of these alleles grew well at 25°C over multiple streaks. However, these strains showed large differences in growth at 35°C (Figure 3). Because senescence is a delayed lethal phenotype manifested as telomeres reach a critically short length, each strain was successively streaked on plates at 35°C to assess temperature sensitivity, with each streak of a single colony representing ∼25 generations of growth.
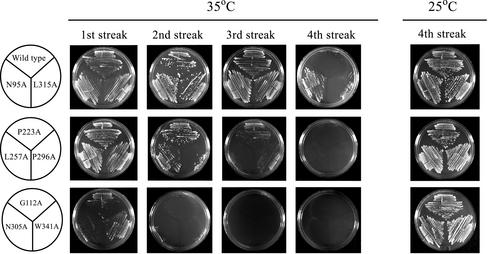
Figure 3.
Representative examples of temperature-sensitive alleles of est2. Strains bearing integrated versions of the indicated est2 alleles were restreaked multiple times at 35 or 25°C. Each successive restreak is shown for the plates at 35°C, whereas only the fourth restreak is shown for plates grown at 25°C. The wild-type strain and two mutants classified as wild-type are shown in the top row. The middle row represents strains with a weak temperature-sensitive phenotype, whereas the bottom row shows examples of severely temperature-sensitive strains. Both the intermediate and severe ts mutants were combined for the summary in Figure 1.
As shown in Figure 3, the parental ProA-EST2 strain used in these studies is moderately temperature sensitive (by streak 4). This temperature sensitivity is not due to the addition of the protein A tag to EST2, but rather arises from a remarkable polymorphism at the C terminus of Est2p. During the course of these studies, we discovered that our plasmid-borne EST2 allele (derived from the initial cloning of EST2; Lendvay et al., 1996) has a four-nucleotide duplication in a TA-dinucleotide tract at the extreme C terminus of EST2 compared with the EST2 gene in our laboratory strain (TVL268). As a result, the TVL268-specific allele ends in the sequence YIYIYI, whereas the allele on the plasmid (and in the database) terminates with the sequence YIYIYIHIVN. Early in our manipulations of plasmid-borne EST2, the TA-dinucleotide repeat incurred an additional six-nucleotide insertion to result in the predicted protein YIYIYIHIHIVN. Cells expressing ProA-tagged Est2p with the YIYIYI terminus grow robustly for at least 200 generations at the restrictive temperature (unpublished data), whereas cells expressing ProA-tagged Est2p with either of the extended C-termini (YIYIYIHIVN or YIYIYIHIHIVN) grow poorly at 35°C. The integration strategy that was used to create the wild-type and mutant ProA-EST2 strains analyzed in this work resulted in the incorporation of the plasmid (YIYIYIHIHIVN) C terminus into the chromosome (see MATERIALS AND METHODS). Subsequent to the work described here, all of the temperature-sensitive est2 mutants were integrated into the chromosome using a strategy that maintained the more robust (YIYIYI) C-terminal sequence. Each of these mutants strains senesces at the restrictive temperature, although this senescence is delayed by ∼50 generations compared with an isogenic strain containing the YIYIYIHIHIVN C terminus (unpublished data). Because of this C-terminal polymorphism, all of the strains described in this study are sensitized to high-temperature growth, and all of the following mutant phenotypes are described in comparison to the otherwise isogenic parental strain.
As shown in Figure 3, the mutants fall into several classes based on their growth at elevated temperature. Several of the strains (illustrated by N95A and L315A) grow well for up to three streaks at 35°C. Like the wild-type strain, these strains show hallmarks of yeast senescence—small and irregular colony size—by the fourth streak at 35°C (Figure 3). These mutants are classified as “wild type” in the summary in Figure 1B. A second class of mutants (P223A, L257A, and P296A) has an intermediate phenotype. A third class of mutants (G112A, N305A, and W341A) displays a severe high-temperature growth defect. These mutants grow poorly during the first streak at 35°C, and form microcolonies after a second streak.
Multiple temperature-sensitive mutations were identified in each of the three regions of the Est2p N-terminus. The hierarchy of temperature sensitivity remained identical among the alleles, regardless of the sequence that was present at the extreme C terminus of the gene (unpublished data), suggesting that there are no allele-specific interactions between the C-terminal polymorphism and the upstream mutations described here. Each of the est2 temperature-sensitive (est2ts) strains was transformed with a centromere plasmid encoding wild-type ProA-Est2p. This low-level expression of wild-type Est2p rescued the temperature-sensitive growth defect of all est2ts strains (unpublished data), suggesting that temperature sensitivity is due in each case to the introduced mutation in Est2p and that none of the mutations is strongly dominant to wild type.
To monitor any change in telomere length in these mutant strains at the restrictive temperature, liquid cultures of each strain were grown at 25°C to saturation. The cultures were diluted 100-fold, placed at 35°C, and allowed to regrow for 24 h. These cultures were subsequently diluted 100-fold every 24 h to allow multiple generations of growth to occur at the restrictive temperature. As shown in Figure 4 (lanes 1–7), the telomeric XhoI restriction fragment undergoes significant shortening at the restrictive temperature in the wild-type strain before reaching a stable length that is maintained over many generations (a total of 22 successive dilutions in this experiment). These results are consistent with the observation that the wild-type strain experiences a decline in colony size after successive restreaks at high temperature (Figure 3). By the fifth streak, the wild-type strain adapts to high temperature growth and can be cultured indefinitely (unpublished data). The N95A and L315A strains exhibit telomere loss at the restrictive temperature similar to that of the wild-type strain (Figure 4, lanes 8–21). Note that the N95A mutant has slightly elongated telomeres at permissive temperature (Figure 4, compare lanes 1 and 8), a phenotype that is presumably responsible for the more robust growth of this mutant on plates at the restrictive temperature (Figure 3).
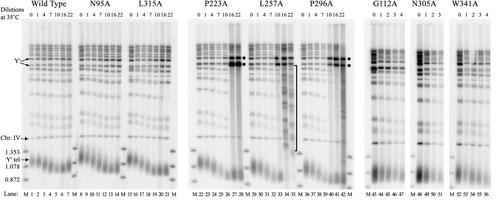
Figure 4.
Telomere length decreases in mutant strains grown at the restrictive temperature. Strains containing either wild-type ProA-Est2p or the indicated mutant versions of chromosomal ProA-Est2p were grown to saturation in rich media at 25°C to produce the samples shown in lanes marked “0 dilutions at 35°C.” Cultures were diluted 100-fold, placed at 35°C, and allowed to regrow. An identical dilution regimen was repeated for the indicated number of dilutions (cultures were typically diluted every 24 h; see RESULTS for details). DNA was harvested from these cells at the indicated times, cleaved with XhoI, separated in a 1.1% agarose gel, transferred to nylon membrane, and simultaneously probed with DNA fragments corresponding to the yeast telomeric repeat and to a 1.6-kb internal fragment from chromosome IV (Chr. IV) that serves as a loading control. Chromosomes containing a Y′ element yield a heterogeneous telomere band (Y′ tel). Note that successive dilutions are shown for the severely temperature-sensitive mutants G112A, N305A, and W341A. A subset of dilutions is shown for the remaining mutants and wild type. The sizes of marker fragments (M) are shown. Bands of 6.7 and 5.2 kb that result from XhoI cleavage within tandemly repeated Y′ elements are indicated (Y′). These fragments become amplified in type I survivors (★). The bracket adjacent to lane 35 highlights bands that result from telomere amplification in type II survivors (see text for details).
In contrast to the N95A and L315A mutants, the P223A, L257A, and P296A strains exhibit an extreme growth lag at the restrictive temperature. Strains L257A and P296A begin to grow slowly by the sixth dilution at restrictive temperature and require 48 h between dilutions for regrowth of the culture. However, normal growth resumes by dilution 10. The P223A strain exhibits a similar growth delay between dilutions 8 and 12. Strikingly, this period of slow growth corresponds to the appearance of hybridizing bands in the telomere restriction pattern that are indicative of telomere-telomere recombination. Yeast strains with critically short telomeres are able to survive in the absence of telomerase activity by utilizing telomerase-independent pathways for telomere maintenance that are dependent on the Rad52 protein (Lundblad and Blackburn, 1993). Two types of survivors have been described: Type I survivors show amplification of the Y′ element, resulting in overrepresentation of XhoI fragments of 6.7 and 5.2 kb (see starred bands in Figure 4, lanes 27, 28, 41, and 42; Lundblad and Blackburn, 1993); type II survivors show sudden increases in telomere length resulting from telomerase-independent mechanisms (brackets in Figure 4, lanes 34 and 35; Lundblad and Blackburn, 1993; Teng and Zakian, 1999). The appearance of banding patterns characteristic of survivor strains in these temperature-sensitive mutants after prolonged growth at the high temperature attests to the inability of these mutant telomerases to function adequately at the restrictive temperature and clearly distinguishes these mutants from the wild-type strain.
The three mutants that display a strong temperature-sensitive growth defect at the high temperature on plates (Figure 3; G112A, N305A, W341A) were also examined for telomere length after growth at the high temperature. Each of these mutants has a telomere length at the permissive temperature that is significantly reduced from wild type (compare Figure 4, lanes 1, 43, 48, and 52). In each case, these mutants were unable to proliferate more than 4 days at 35°C. During that time, the smear corresponding to the Y′ terminal fragment (centered around 1 kb) became less broad, corresponding to progressive shortening of the longer telomeres in the population. Interestingly, these mutants failed to form survivor strains at the restrictive temperature, even after the final diluted culture was incubated for multiple days at the restrictive temperature. This failure to grow was not due to cell death because returning these cultures to the permissive temperature allowed the cultures to resume growth. Despite our failure to detect the typical changes that accompany a switch to recombination-dependent telomere maintenance, the growth of these mutants at restrictive temperature appears to be facilitated by RAD52, because deletion of RAD52 in these temperature-sensitive strains reproducibly increases the rate of senescence at the high temperature (A. Brown and K. Friedman, unpublished data).
The temperature-sensitive phenotypes of the mutant strains are summarized in Figure 1B. The growth of these strains at high temperature provides a sensitive indicator of the degree to which Est2p function is compromised. Using this criterion, there is a striking tendency for mutations in conserved positions of region III to result in temperature sensitivity, whereas mutations at the majority of nonconserved positions have no effect on growth at 35°C. The difference between conserved and nonconserved positions is statistically significant (p = 0.015) and suggests that the sequence alignment proposed here correctly identifies important residues in region III of TERT. Because region II contains a small number of conserved residues, we were not able to statistically analyze the effect of mutations in conserved residues of this small region.
Rescue of the est2ts Alleles at High Temperature by Overexpression of Est1p and Est3p
To address the specific defects imparted by mutations in regions I, II, and III, the effect of overexpression of several known telomerase components on growth of the est2ts strains was measured. Two micron plasmids containing inserts encoding EST1, EST3, and SMD3 were transformed into each of the est2ts strains described above. EST1 was expressed from the constitutive high-level Adh promoter, whereas EST3 and SMD3 were expressed from their endogenous promoters. Est1p and Est3p are known components of the telomerase holoenzyme (Lendvay et al., 1996; Evans and Lundblad, 1999; Hughes et al., 2000). Smd3p is one of seven Sm proteins that bind a site near the 3′ end of TLC1 RNA; these proteins mediate TLC1 RNA stability and are contained in the mature telomerase complex (Seto et al., 1999). Mutant strains containing pRS423 (empty vector) or 2 μ SMD3 grew equivalently over four streaks at 35°C, showing senescence in both cases. Somewhat surprisingly, transformation with 2 μ EST1 rescued growth of all region I, II, and III est2ts mutants at the restrictive temperature (unpublished data). In contrast, overexpression of Est3p rescued a specific subset of the est2ts mutants. Region I est2ts mutants transformed with 2 μ EST3 continued to grow for multiple generations on plates at 35°C, whereas the same strains transformed with empty vector underwent senescence (Figure 5, top). In contrast, 2 μ EST3 had minimal effect on the growth of mutants in regions II and III (Figure 5, middle and bottom).
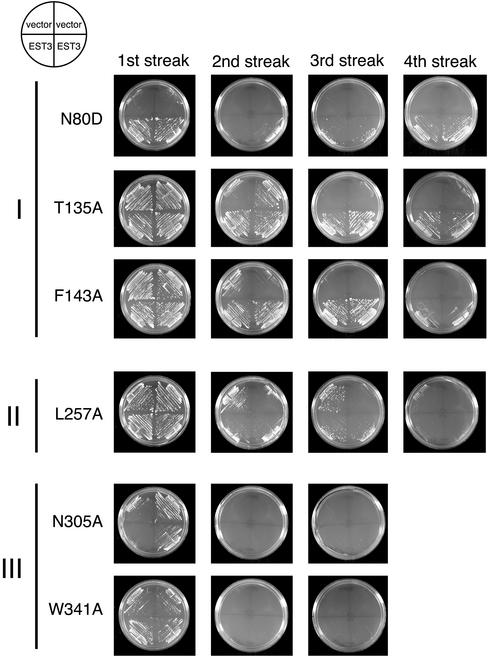
Figure 5.
Est3p overexpression specifically suppresses mutations in region I of EST2. Yeast strains carrying integrated est2ts mutations located in regions I, II, or III were transformed with pRS423 (empty vector) or pRS423-EST3 (2-μm, EST3 plasmid) at 25°C. Two independent transformants of each were restreaked four successive times at the restrictive temperature of 35°C. As indicated, the two strains transformed with pRS423-EST3 were streaked in the bottom two quadrants of the plate, whereas the two strains transformed with empty vector were streaked in the top two quadrants. The position of each mutation is indicated.
Because the ability of the cells to grow on plates provides only a qualitative assay of cell viability, the effect of Est3p overexpression was quantitated by monitoring the long-term growth of est2ts strains expressing empty vector or 2 μ EST3 in liquid culture at the restrictive temperature (35°C). Every 24 h, cell density was determined and the cultures were diluted to 5 × 105 cells per milliliter. Senescence of the est2ts strains carrying empty vector was manifested as a gradual reduction in the growth rate over each 24-h period, reflected in the inability of the strain to regrow an equivalent amount each day (Figure 6A-D, dashed lines). The strength of this effect correlated well with that seen for each mutant on solid media. Growth of the strong mutants N80D, N305A, and W341A expressing empty vector declined rapidly at 35°C (Figure 6, A and B). In contrast, the weaker mutants T135A and L257A showed a clear but gradual decline in growth rate at the restrictive temperature (Figure 6, C and D). When the est2ts strains were transformed with 2 μ EST3 (solid lines), no change in the growth of region II or III mutants was detectable (Figure 6, B and D). In contrast, the region I mutants overexpressing Est3p showed growth at the restrictive temperature that was restored nearly to wild-type levels (Figure 6, A and C). Significantly, the region I–specific pattern of rescue by Est3p overexpression was true of both strong mutants (compare N80D with N305A and W341A) and weak mutants (compare T135A and L257A), indicating that this rescue is dependent on the location of the mutation within EST2 and not on the strength of the mutant phenotype. Growth of all six region I ts mutants was rescued by Est3p overexpression (Figures 5 and 6 and unpublished data). The allele specificity of suppression by Est3p overexpression suggests that Est3p interacts with Est2p through region I.
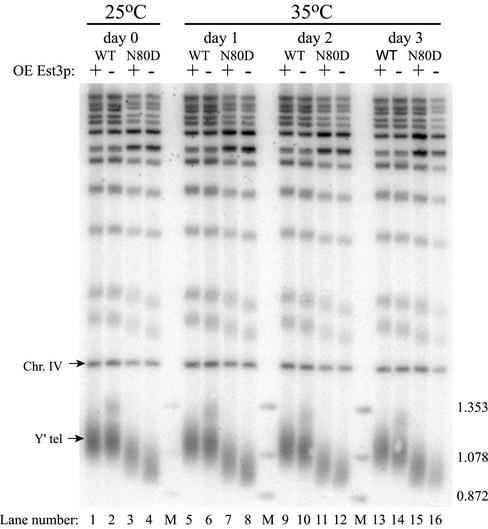
To examine the effect of Est3p overexpression on telomere length, cells were harvested and DNA was prepared from wild-type and N80D cells during the growth curve experiment shown in Figure 6A. Telomere length was examined by cleavage of the DNA with XhoI and hybridization to a labeled telomeric DNA probe. Overexpression of Est3p from a high copy number plasmid resulted in a small increase in telomere length in the N80D mutant strain at 25°C (Figure 7, compare lanes 3 and 4) while having no demonstrable effect on telomere length in the wild-type strain (lanes 1 and 2). After 3 days of growth at 35°C, telomere lengths in the N80D strain overexpressing Est3p remained slightly longer than those of the N80D strain containing empty vector. Thus, overexpression of Est3p does not restore normal telomere length in the mutant strain, but rather may allow telomeres to be maintained at a length just above a threshold required for continued growth. A similar effect on telomere length in the T135A mutant strain was also observed upon Est3p overexpression (unpublished data). Two pieces of evidence suggest that the increased survival of the region I mutants upon Est3p overexpression is not due to an increase in the rate of survivor formation. First, strains grown at high temperature show no sign of telomeric or subtelomeric repeat amplification (Figure 7, lane 15). Second, overexpression of Est3p rescues the high temperature growth of both the N80D and G85A mutants in the absence of Rad52p (A. Brown and K. Friedman, unpublished data).
Figure 7.
Overexpression of Est3p causes slight telomere lengthening in an Est2p region I mutant. Cultures corresponding to each of the time points in Figure 6A were harvested and DNA was extracted, cleaved with XhoI, separated in a 1.1% agarose gel, and Southern blotted. The blot was hybridized with fragments corresponding to the yeast telomere repeat and to a 1.6-kb fragment of chromosome IV (Chr. IV). Chromosomes containing a Y′ element yield a heterogeneous telomere band (Y′ tel). The sizes of marker bands (M) are indicated.
Coimmunoprecipitation of Telomerase Proteins with Mutant Est2p
To test whether mutations in region I affect the ability of Est3p to associate with the telomerase complex, we turned to a well-characterized mutant in region I. The est2-ala1 mutant has 10 consecutive amino acids changed to alanine (residues 40–49; Figure 1A). This mutation is lethal: strains carrying est2-ala1 undergo progressive telomere shortening and senescence. However, we have previously shown that this mutant retains the ability to bind to telomerase RNA and to exhibit in vitro telomerase activity (Friedman and Cech, 1999). Although protein levels are reduced (one tenth of wild-type), expression of ProA-est2-ala1 on a 2 μ plasmid to levels that approximate those of wild-type Est2p does not rescue cell growth. On the basis of these phenotypes, we previously proposed that this mutant is defective for interactions with one or more accessory proteins of telomerase.
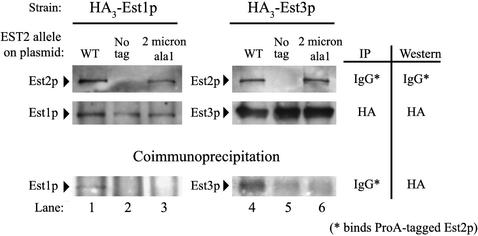
To address this possibility, strains were constructed that contain HA3-tagged versions of EST1 or EST3 integrated at the endogenous loci. These strains are est2::TRP1 and are maintained by an untagged version of Est2p carried on a URA3 centromere plasmid. The HA3-EST1 est2::TRP1 and HA3-EST3 est2::TRP1 strains were transformed with LEU2 centromere plasmids bearing untagged EST2 or ProA-EST2, and the URA3 complementing plasmid was subsequently lost by selection on plates containing 5-FOA. As shown in Figure 8, immunoprecipitation of ProA-Est2p results in coimmunoprecipitation of both Est1p and Est3p (lanes 1 and 4).
Figure 8.
Est1p and Est3p fail to coimmunoprecipitate with Est2-ala1p. Western analysis is shown for an HA3-EST1 est2:: HIS3 strain (lanes 1–3) or HA3-EST3 est2:: HIS3 strain (lanes 4–6). Strains contained the following plasmids: WT (lanes 1 and 4), ProA-EST2 on a centromere plasmid; No tag (lanes 2 and 5), untagged EST2 on a centromere plasmid; 2 micron ala1, ProA-est2-ala1 on a 2 μ plasmid and untagged EST2 on a centromere plasmid. Top row: Protein A–tagged Est2 protein immunoprecipitated on IgG-conjugated beads and detected by Western analysis. Middle row: HA3-tagged protein immunoprecipitated on Protein G beads bound to polyclonal HA antibody and detected by Western analysis. Bottom row: HA3-tagged proteins coimmunoprecipitated with ProA-Est2p on IgG beads (identical to those analyzed in the top row) and detected by Western analysis. The background signal in lanes 5 and 6 (bottom row) derives from IgG heavy chain that comigrates with HA3-Est3p.
To test the effect of the est2-ala1 mutation on incorporation of Est1p and Est3p into the telomerase complex, 2 μ LEU2 plasmids bearing protein A–tagged est2-ala1 were transformed into the HA3-tagged strains. To allow robust growth of these strains, the complementing plasmid expressing untagged wild-type EST2 was retained. Mutant telomerase complex was specifically assayed by immunoprecipitation of the tagged mutant protein. Although expression of ProA-est2-ala1 from the 2 μ plasmid approximates that of wild-type EST2 and levels of HA3-Est1p and HA3-Est3p are equivalent in all strains, neither Est3p or Est1p was detectably coimmunoprecipitated with protein A–tagged Est2-ala1p (Figure 8, lanes 3 and 6). A small amount of IgG heavy chain, which comigrates with the 54-kDa tagged Est3p, accounts for the background seen in lanes 5 and 6 (Figure 8, bottom). We repeated these experiments in senescing cells expressing only ProA-est2-ala1 (after selection for loss of the complementing plasmid on plates containing 5-FOA) and obtained identical results (unpublished data). To verify that the ProA-est2-ala1 mutant allele retains functional association with the telomerase RNA, we confirmed that the immunoprecipitation beads assayed in the Western analysis shown in Figure 8, lanes 1 and 3, are capable of supporting in vitro telomerase activity (unpublished data). Taken together, these results indicate that ProA-est2-ala1 does not destroy the integrity of the core telomerase complex but rather results in loss of telomerase accessory proteins.
DISCUSSION
Conserved Sequence Motifs of Regions I and III
The mutational analysis presented here provides evidence for the functional importance of conserved sequence motifs in regions I and III of TERT. In agreement with previous reports (Malik et al., 2000; Miller et al., 2000; Xia et al., 2000), mutations in conserved residues of region I are more likely to result in lethality than those in nonconserved residues, suggesting that the sequence alignment has correctly identified critical residues. Although our sequence alignment is in general agreement with others, we do find a discrepancy in the C-terminal portion of region I. Here, the S. cerevisiae sequence can be aligned in two different registers with the other TERT proteins because there are two QxxG motifs in close proximity. Mutations at Q146 and G149 result in loss of Est2p function, suggesting that these residues comprise a QxxG motif that is absolutely conserved among the TERT family members.
Lethality of mutations in the conserved residues of region I could result either from destabilization of the protein or through mutation of a functionally important motif. Our Western analysis shows that mutations at conserved residues W115, G123, L130:L131, and Q146A reduce protein levels only about twofold from wild type. We have previously observed that Est2p levels as little as one tenth of the wild-type amount can support normal cell growth (Friedman and Cech, 1999). Therefore, the lethal phenotypes of the W115A, G123A, L130A:L131A, and Q146A mutations are unlikely to result solely from the twofold reduction in protein levels but rather may reflect essential roles of these conserved amino acids. Interestingly, the only nonconserved amino acid within region I that results in lethality when mutated (Q142A) eliminates detectable Est2 protein. Therefore, the functional significance of this amino acid position cannot be determined.
Attempts to validate the sequence alignment in region III have been previously hampered by an inability to identify lethal mutations in this region of the protein. Despite identification of numerous residues that are conserved among the TERT proteins, none of these positions is essential for the function of yeast Est2p. Here we have assessed the temperature-sensitive growth of strains carrying these Est2p alleles in order to gain a sensitive measure of Est2p activity. The rationale is that mutations that perturb, but do not abrogate, telomerase activity may be exacerbated by high temperature. The version of Est2p used as the template for the mutations described in this study contains a polymorphism at the C terminus that results in modest temperature sensitivity of the wild-type protein. Therefore, this strain is optimally suited to identify additional functional defects at high temperature. Indeed, mutations in all six of the highly conserved residues of region III resulted in temperature sensitivity, whereas only one of six mutations in nonconserved residues affected high temperature growth to a larger extent than with the wild-type gene. These results support the validity of the sequence alignment in region III. Furthermore, the differential response of Est2p to mutations in region I and III may reflect the different roles of these protein regions in the telomerase complex. Region III has been implicated in binding the telomerase RNA both in yeast (Friedman and Cech, 1999) and in other organisms (Beattie et al., 2000; Bryan et al., 2000; Lai et al., 2001). If the conserved residues in region III are involved in binding to the telomerase RNA, our results suggest that each residue makes only a partial contribution to RNA binding, the effect of which can only be observed at high temperature.
Role of Region I in Recruitment of Accessory Telomerase Proteins
Using the temperature-sensitive mutations identified through the phylogenetic analysis described above, we found that overexpression of Est3p specifically rescues the high temperature growth defect of temperature-sensitive mutants in region I. This pronounced allele specificity suggests that Est3p interacts directly or indirectly with residues in region I. Interestingly, overexpression of Est1p suppresses all of the temperature-sensitive mutants in regions I, II, and III. Overexpression of Est1p causes slight telomere lengthening in these strains. This nonspecific effect may partially counteract telomere loss at the high temperature. Alternatively, the temperature-sensitive Est2p mutants may result in generally destabilized complexes and overexpression of Est1p may compensate for that instability by mass action. Because Est1p overexpression rescues the high temperature growth of temperature-sensitive mutants located throughout the N-terminal half of Est2p, these genetic results do not highlight any particular region of Est2p with which Est1p might interact.
Surprisingly, analysis of telomerase complex composition in the est2-ala1 mutant strain by coimmunoprecipitation indicates that the association of both Est1p and Est3p with the telomerase complex is destabilized upon mutation of region I. The failure of Est1p and Est3p to coimmunoprecipitate with Est2p does not reflect some sort of global denaturation of Est2p because the est2-ala1 mutant retains the ability to associate with the telomerase RNA and to catalyze in vitro telomerase activity (Friedman and Cech, 1999). Xia et al. (2000) describe several mutations in region I (D66A and N104A:V105A) that severely reduce in vivo telomerase activity, while having minimal effects on in vitro telomerase activity. These phenotypes are consistent with the hypothesis that a substantial portion of region I contributes to recruitment of telomerase accessory proteins. Intriguingly, recent results show that a portion of region I in human TERT (the DAT domain) also contributes to the in vivo activity of telomerase, while being dispensable for catalytic activity (Armbruster et al., 2001). This portion of region I corresponds to residues 51–112 of the S. cerevisiae sequence (using the alignment shown in Supplementary Data). This portion of region I encompasses many (but not all) of the region I temperature-sensitive mutants used in our genetic analysis and is located immediately C-terminal to the est2-ala1 mutation used in the immunoprecipitation studies. Therefore, the role of TERT region I in the recruitment of telomerase accessory proteins may be evolutionarily conserved.
Although our genetic results implicate region I as contributing to Est1p and Est3p recruitment, such contribution is not necessarily through direct protein binding. As one possibility, Est2p may directly bind Est3p, which could in turn recruit Est1p to the telomerase complex. Est1p binds the telomerase RNA (Zhou et al., 2000; Livengood et al., 2002) and deletion of Est3p does not disrupt that association (Hughes et al., 2000). However, these studies did not directly address whether Est1p associates with Est2p in the absence of Est3p.
Dimerization of Yeast Telomerase?
It is interesting to note that immunoprecipitation of ProA-Est2-ala1p failed to coimmunoprecipitate Est1p and Est3p whether or not wild-type untagged Est2p was present in the strain. Because yeast telomerase has been proposed to function as a dimer (Prescott and Blackburn, 1997), this result is somewhat surprising. Expression of the wild-type and mutant proteins at equivalent levels should result in one half of telomerase dimers containing both mutant and wild-type Est2p. In this case, the ability of the wild-type Est2p-RNA complex to bind Est1p and Est3p would be expected to give detectable coimmunoprecipitation of these proteins with tagged, mutant Est2p. There are several possible explanations for this observation. Yeast telomerase may have the ability to dimerize but not exist as a dimer under most conditions (see Livengood et al., 2002). Alternatively, because the stoichiometry of the telomerase complex is unknown, dimerization may involve only a subset of the subunits of the telomerase complex. For example, a single TERT molecule may be complexed with more than one RNA molecule. Finally, it is possible that the est2-ala1 mutation disrupts the ability of Est2p to dimerize. Additional coimmunoprecipitation experiments may distinguish among these possibilities.
Implications for Other Telomerase Complexes
The results presented here support the conclusion that region I of TERT is phylogenetically conserved across species. Residues that are highly conserved are functionally important for yeast telomerase, raising the possibility that these residues fulfill similar functions in other organisms. The strong genetic system provided by S. cerevisiae has allowed us to uncover a role for region I in the recruitment of Est3p to the telomerase complex. We propose that homologous accessory proteins exist and contribute to conserved functions in all telomerases.
Supplementary Material
ACKNOWLEDGMENTS
We thank V. Lundblad for generous gifts of strains and plasmids, Y. Han for automated DNA sequencing, P. Baumann for critical reading of the manuscript, and M. Winey and colleagues for helpful suggestions and reagents. K.L.F. was an Associate of the Howard Hughes Medical Institute.
Footnotes
Online version of this article contains dataset material. Online version is available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–06–0327. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–06–0327.
REFERENCES
- Armbruster BN, Banik SS, Guo C, Smith AC, Counter CM. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol Cell Biol. 2001;21:7775–7786. doi: 10.1128/MCB.21.22.7775-7786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie TL, Zhou W, Robinson MO, Harrington L. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol Biol Cell. 2000;11:3329–3340. doi: 10.1091/mbc.11.10.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Goodrich KJ, Cech TR. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol Cell. 2000;6:493–499. doi: 10.1016/s1097-2765(00)00048-4. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Sperger JM, Chapman KB, Cech TR. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc Natl Acad Sci USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deminoff SJ, Tornow J, Santangelo GM. Unigenic evolution: a novel genetics method localizes a putative leucine zipper that mediates dimerization of the Saccharomyces cerevisiae regulator Gcr1p. Genetics. 1995;141:1263–1274. doi: 10.1093/genetics/141.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WP, Nickoloff JA. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1987;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, Shippen DE. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman KL, Cech TR. Essential functions of N-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Harrington L, Zhou W, McPhail T, Oulton R, Yeung DS, Mar V, Bass MB, Robinson MO. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- Kilian A, Bowtell DD, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lund-blad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci USA. 1997b;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997a;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Livengood AJ, Zaug AJ, Cech TR. Essential regions of Saccharomyces cerevisiae telomerase RNA. Separate elements for Est1p and Est2p interaction. Mol Cell Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. Putative telomerase catalytic subunits from Giardia lamblia and Caenorhabditis elegans. Gene. 2000;251:101–108. doi: 10.1016/s0378-1119(00)00207-9. [DOI] [PubMed] [Google Scholar]
- Meyerson M, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Miller MC, Liu JK, Collins K. Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J. 2000;19:4412–4422. doi: 10.1093/emboj/19.16.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- Oguchi K, Liu H, Tamura K, Takahashi H. Molecular cloning and characterization of AtTERT, a telomerase reverse transcriptase homolog in Arabidopsis thaliana. FEBS Lett. 1999;457:465–469. doi: 10.1016/s0014-5793(99)01083-2. [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Prescott J, Blackburn EH. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated Est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sabel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Taggart AP, Teng S-C, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- Teng S-C, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Peng Y, Mian IS, Lue NF. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol Cell Biol. 2000;20:5196–5207. doi: 10.1128/mcb.20.14.5196-5207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Hidaka K, Futcher B. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol Cell Biol. 2000;20:1947–1955. doi: 10.1128/mcb.20.6.1947-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.