Abstract
Bni4 is a scaffold protein in the yeast Saccharomyces cerevisiae that tethers chitin synthase III to the bud neck by interacting with septin neck filaments and with Chs4, a regulatory subunit of chitin synthase III. We show herein that Bni4 is also a limiting determinant for the targeting of the type 1 serine/threonine phosphatase (Glc7) to the bud neck. Yeast cells containing a Bni4 variant that fails to associate with Glc7 fail to tether Chs4 to the neck, due in part to the failure of Bni4V831A/F833A to localize properly. Conversely, the Glc7-129 mutant protein fails to bind Bni4 properly and glc7-129 mutants exhibit reduced levels of Bni4 at the bud neck. Bni4 is phosphorylated in a cell cycle-dependent manner and Bni4V831A/F833A is both hyperphosphorylated and mislocalized in vivo. Yeast cells lacking the protein kinase Hsl1 exhibit increased levels of Bni4-GFP at the bud neck. GFP-Chs4 does not accumulate at the incipient bud site in either a bni4::TRP1 or a bni4V831A/F833A mutant but does mobilize to the neck at cytokinesis. Together, these results indicate that the formation of the Bni4-Glc7 complex is required for localization to the site of bud emergence and for subsequent targeting of chitin synthase.
INTRODUCTION
Chitin, an N-acelylglucosamine polymer, is a minor yet essential component of the yeast cell wall (Bulawa, 1993). In vegetative cells, chitin is deposited in a ring that marks the site of the bud emergence in late G1, and it forms the primary septum at cytokinesis. Three enzymes are responsible for the synthesis of chitin (Smits et al., 2001). Chitin synthase III (Chs3), which produces the majority of chitin, lays down the polymer in the lateral wall, and forms the chitin ring at bud emergence (Choi et al., 1994b; Chuang and Schekman, 1996; Cos et al., 1998). Chs3 is also responsible for the synthesis of chitin during mating and sporulation (Bulawa, 1992; Pammer et al., 1992). Chitin synthase II (Chs2) synthesizes chitin that constitutes the primary septum (Sburlati and Cabib, 1986; Silverman et al., 1988). Chs1 is a repair enzyme that counteracts hydrolysis of chitin caused by chitinase (Cabib et al., 1989). Chs1 and Chs2 are regulated primarily at the transcriptional level (Choi et al., 1994a), whereas Chs3 is metabolically stable and is regulated posttranscriptionally (Chuang and Schekman, 1996), by periodic deposition at the cell surface and recruitment to the incipient bud site in G1 followed by endocytosis later in the cell cycle and redistribution to the mother-bud neck at cytokinesis. Chs3 protein resides in endosomal-like compartments referred to as chitosomes (Ziman et al., 1996, 1998).
A number of proteins have been identified that are necessary for the proper activity of Chs3. Chs7 is involved in the export of Chs3 from the ER (Trilla et al., 1999), whereas Chs5 and Chs6 are required for the proper delivery of Chs3 to the plasma membrane (Santos and Snyder, 1997; Ziman et al., 1998; Valdivia et al., 2002). Chs4 is a regulatory subunit essential for chitin synthase III activity (Trilla et al., 1997; Ono et al., 2000) and is also responsible for the deposition of Chs3 at the incipient bud site by physically linking Chs3 to the septin ring through the interaction with the scaffolding protein Bni4 (DeMarini et al., 1997). Bni4 first appears at the site of bud emergence before budding and remains on the mother side of the bud until late in the cell cycle. Its accumulation at the bud neck is dependent upon septins, a family of GTP binding proteins that form the bud neck filaments (Longtine et al., 1996). Chitin deposition is delocalized in bni4 null mutants, due in part to a failure to tether Chs3 and Chs4 to the bud neck. Hence, the deposition of chitin at the bud neck is regulated not only by the controlled delivery of Chs3 to the plasma membrane from chitosomes but also through Bni4-dependent targeting of chitin synthase to the septin filaments at the bud neck.
Bni4 has also been found to associate with Glc7, the catalytic subunit of protein phosphatase type 1 (PP1), in two-hybrid screens (Tu et al., 1996; Uetz et al., 2000) and in a pull-down assay (Walsh et al., 2002). PP1 is a highly conserved eukaryotic serine-threonine phosphatase that is involved in a wide range of physiological processes ranging from glycogen metabolism to protein synthesis (reviewed in Shenolikar, 1994; Stark, 1996). Its location through the cell cycle is dynamic (Andreassen et al., 1998; Bloecher and Tatchell, 2000). Glc7 is found in the nucleus throughout the cell cycle and is most abundant in the nucleolus. At the start of anaphase, Glc7 accumulates at either the spindle pole bodies or the kinetochores and transiently appears at the actomyosin ring during cytokinesis. Glc7 also accumulates in a ring at the site of bud formation before bud emergence and remains largely on the mother side of the bud until late in the cell cycle (Bloecher and Tatchell, 2000).
The specificity of PP1 is largely determined by regulatory subunits that either target the catalytic subunit of PP1 to the specified substrate or alter its activity toward a specific substrate (Bollen and Stalmans, 1992). Many of these subunits contain the short consensus sequence R/K-V/I-X-F, referred to as the RVXF motif for the conserved valine/isoleucine and phenyalanine residues, that associates with a hydrophobic groove on PP1c (Egloff et al., 1997). Missense mutations in the RVXF motif result in loss of PP1c-binding activity in mammals (Egloff et al., 1997; Kwon et al., 1997; He et al., 1998; Beullens et al., 1999; Hsieh-Wilson et al., 1999; Huang et al., 1999) and yeast (Alms et al., 1999; Dombek et al., 1999; Wu et al., 2001). A common PP1c-binding motif on many targeting subunits ensures competitive binding between subunits. However, it is not known how the distribution between different targeting subunits is regulated and in most cases it is not known whether the dynamic changes in location of PP1c are controlled by targeting subunits. The association of Glc7 and Bni4 in two-hybrid and pull-down assays and the apparent colocalization of both at the bud neck led us to hypothesize that Bni4 may act as a scaffold to tether Glc7 to the bud neck. We report herein that Bni4 is indeed limiting for the association of Glc7 to the bud neck and may have a codependent role with Glc7 in associating with the septin ring. We also extend the analysis of Bni4, showing that it is necessary for the mobilization of Chs4 to the incipient bud site but that it is not required to mobilize Chs4 to the neck at cytokinesis.
MATERIALS AND METHODS
Yeast Strains, Media, and General Methods
The yeast strains used in this work are listed in Table 1 and are congenic to KT1112 (MATa ura3-52 leu2 his3; Stuart et al., 1994) or to KT1358 (MATα ura3 leu2 his3 trp1), with the exception of DLY222 (Bose et al., 2001) and PJ69-4A (James et al., 1996). Yeast strains were grown on YPD medium (2% Bacto peptone, 1% yeast extract, 2% glucose) at 30°C, except where noted. Strains were sporulated at 24°C on medium containing 2% Bacto peptone, 1% yeast extract, and 2% potassium acetate. Synthetic complete medium and media lacking specific amino acids were made as described previously (Sherman et al., 1986). Yeast transformations, manipulation of Escherichia coli and the preparation of bacterial growth media were performed as described previously (Maniatis et al., 1989; Kaiser et al., 1994). The G1 cyclin deprivation arrest and release assays were performed as described previously (Cross and Tinkelenberg, 1991). Strains YLK74 and YLK76 were created by ectopic expression of Bni4 and Bni4V831A F833A (Bni4V-A/F-A) at the URA3 locus. Sequences encoding Bni4 and Bni4V-A/F-A were removed from p366 and pAR17 (DeMarini et al., 1997) and ligated to XhoI, SpeI-cut pRS306, thus creating pLK8 and pLK9. Strain KT1918 was transformed with StuI-digested pLK8 or pLK9 and colonies were selected on SC-Ura plates. To create YLK78, strain KT1918 was transformed with StuI-digested pRS306.
Table 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| DLY222 | MATa cln1 cln2 cln3 GAL1p-CLN3∷TRP1 | (Bose et al., 2001) |
| PJ69-4A | MATa ura3 leu2 his3 trp1 gal4Δ gal80Δ GAL2∷ADE2 lsy2∷GAL1:HIS3 met2∷GAL7:lacZ | (James et al., 1996) |
| KT1358 | MATα leu2 ura3 his3 trp1 | (Bloecher and Tatchell, 2000) |
| KT1918 | MATα leu2 his3 trp1 bni4 1∷TRP1 | |
| KT1921 | MATα leu2 ura3-52 his3 trp1 glc7∷LEU2 bni4Δ1∷TRP1 pRS316-GFP-GLC7 | This study |
| KT1922 | MATa leu2 ura3-52 his3 trp1 glc7∷LEU2 bni4Δ1∷TRP1 pRS316-GFP-GLC7 | This study |
| KT1925 | MATa leu2 ura3-52 his3 trp1 glc7∷LEU2 pRS316-GFP-GLC7 | This study |
| KT1926 | MATα leu2 ura3-52 his3 trp1 glc7∷LEU2 pRS316-GFP-GLC7 | This study |
| KT1972 | MATα leu2 his3 trp1 bni4 1∷TRP1 chs4∷TRP1 | This study |
| KT2153 | Mata leu2 his3 trp1 ura3 BNI4-YFP∷HIS3 | |
| KT2155 | Mata leu2 his3 trp1 ura3 BNI4-YFP∷HIS3 glc7-129 | |
| YAB122 | MATa leu2 his3 ura3-52 trp1 glc7∷LEU2 pAB70 | (Bloecher and Tatchell, 2000) |
| YAB608 | Mata leu2 his3 trp1 ura3∷GFP-GLC7∷URA3 | This study |
| YLK27 | MATα leu2 ura3 his3 trp1 Bni4-CFP∷kanMx6 | This study |
| YLK29 | MATα leu2 his3 ura3 trp1 glc7∷LEU2 Bni4-CFP:kanMx6 pAB70 | This study |
| YLK45 | MATα leu2 ura3 his3 trp1 Bni4-GFP∷kanMx6 | This study |
| YLK66 | MATα leu2 ura3 his3 trp1 CDC10-GFP∷kanMx6 | This study |
| YLK74 | MATα leu2 his3 trp1 bni4 1∷TRP1 ura3∷BNI4-URA3 | This study |
| YLK76 | MATα leu2 his3 trp1 bni4 1∷TRP1 ura3∷bni4 V-A/F-A A∷URA3 | This study |
| YLK78 | MATα leu2 his3 trp1 bni4 1∷TRP1 | This study |
| YLK80 | MATα leu2 his3 trp1 bni4 1∷TRP1 ura3∷BNI4-GFP∷kanMx6∷URA3 | This study |
| YLK82 | MATα leu2 his3 trp1 ura3∷BNI4-GFP∷KanMx6∷URA3 | This study |
| YLK84 | MATα leu2 his3 trp1 bni4 1∷TRP1 ura3∷bni4V-A/F-A-GFP∷kanMx6∷URA3 | This study |
| YLK86 | MATα leu2 his3 trp1 ura3∷bni4V-A/F-A-GFP∷kanMx6∷URA3 | This study |
| YLK110 | MATα leu2 his3 trp1 bni4 1∷TRP1 CDC10-YFP∷HIS3Mx6 ura3∷bni4V-A/F-A∷URA3 | This study |
| YLK112 | MATα leu2 his3 trp1 bni4 1∷TRP1 CDC10-YFP∷HIS3Mx6 ura3∷BNI4∷URA3 | This study |
| YLK160 | MATα leu2 ura3 his3 trp1 Bni4-GFP∷kanMx6 hsl1-1∷URA3 | This study |
| YLK162 | MATα leu2 his3 trp1 bni4 1∷TRP1 ura3∷bni4V-A/F-A-GFP∷KanMx6∷URA3 hsl1-1∷kanMx6 | This study |
Integrated green fluorescent protein (GFP) fusions were made according to the method described by the Yeast Resource Center at the University of Washington (http://depts.washington.edu/∼yeastrc/fm_home3.htm), which is based on the method described by Wach et al. (1997). The GFP-integration cassettes were amplified by polymerase chain reaction by using primers BGFP1 and BGFP2 for Bni4-GFP, CDC10F and CDC10R for Cdc10-GFP, CDC12F and CDC12R for Cdc12-GFP, and pLK3, pDH3, or pDH5 as templates for GFP, cyan fluorescent protein (CFP), and yellow fluorescent protein (YFP)-fusions, respectively (Primers are listed in Table 2). Cells were transformed with the amplified DNA and colonies were selected on G418-containing YEPD (GFP and CFP) or SC-His (YFP).
Table 2.
Primers used in this study
| Name | Sequence (5′–3′) |
|---|---|
| BGFP1 | ATGGAAGTACACGATGATTCGCGATGTTACACACATTTTTATGGTCGACGGATCCCCGGG |
| BGFP2 | TGTATGATTTGATTCATTTCCATTTCTCCCAGTTTTCTGCTATCGATGAATTCGAGCTCG |
| Bni4-up-2 | ATTGAGCTCCCCGGGTCGGATAGTATTTCAG |
| Bni4-D-2 | ATTAAGCTTCCCGGGCTCCCAGTTTTCTGC |
| Bni4t-2 | ATCAAGCTTCCCGGGCTAGTTTACATACACCTC |
| CDC10F | AGTCGTTCCTCAGCTCATATGTCTAGCAACGCCATTCAACGTGGTCGACGGATCCCCGGG |
| CDC10R | TTAATAACATAAGATATATAATCACCACCATTCTTATGAGATATCGATGAATTCGAGCTCG |
| CDC12F | GAGCAGGTCAAAAGCTTGCAAGTAAAAAAATCCCATTTAAAAGGTCGACGGATCCCCGGG |
| CDC12R | AGGCGTTGAAATTGACGAGACAAAGAGGAAGACATTAATTAAATCGATGAATTCGAGCTCG |
| GFP3F | GACGGATCCCCGGGTTAATTAACAGTAAAGGAGAAGAACTT |
| GFP4R | CTTATTTAGAAGTGGCGCGCCCTATTTGTATAGTTCATCCATG |
| GST1 | TCCCCCGGGGATTATGTCGGATAGTATTTCAG |
| GST2 | GCCGAAGSTTCTAATAAAAATGTGTGTAACATCGCG |
| VFA-1 | GACCAGGGTGCACGGGCTTCGTCCCAG |
| VFA-2 | CTGGGACGAAGCCCGTGCACCCTGGTC |
Plasmid Construction
Plasmids are listed in Table 3. Standard techniques were used for DNA manipulation (Maniatis et al., 1989). Restriction and modification enzymes were used as recommended by the manufacturers (Promega, Madison, WI). The Sequenase enzyme (U.S. Biochemical, Cleveland, OH) and dideoxy chain termination method (Sanger et al., 1977) were used with synthetic oligonucleotide primers (Integrated DNA Technologies, Coralville, IA) to obtain DNA sequence from double-stranded templates. Two-hybrid constructs were made in the pGAD series of vectors (James et al., 1996). pAR5 was created by cloning a PstI fragment (codons 70–892) from p366 (DeMarini et al., 1997) into pGADC1. Full-length Bni4 (pAR16), truncated Bni4 (pAR14), and Bni4V831A/F833A (pAR19) two-hybrid plasmids were obtained by PCR by using Bni4-up-2, Bni4-D-2, and Bni4t-2 primers, and p366 or pAR17 as templates. The resulting PCR products were digested with SmaI and cloned into pGADC1. To obtain pAR17, the QuikChange kit (Stratagene, La Jolla, CA) and primers VFA-1 and VFA-2 were used as recommended for site-directed mutagenesis of the VXF motif of BNI4 in p366. Mutations were screened by DNA sequence analysis. The entire sequence of the mutant allele was confirmed at Iowa State University DNA facility (Ames, IA). To construct a brighter version of GFP-Chs4, an XbaI-EcoRV fragment containing GFP-CHS4 was isolated from p326 and ligated to XbaI-SmaI-digested pUC18. A NcoI-PvuII fragment that contains mutations encoding a brighter version of GFP (Robinson et al., 1999) was used to replace the corresponding fragment of the original GFP. An XbaI-SacI fragment containing GFP-CHS4 was cloned into pRS315 and pRS313, giving pAR24 and pAR26. To obtain a GST-Bni4 fusion under control of the GAL1 promoter, restriction sites were introduced upstream (SmaI) and downstream (HindIII) of the Bni4 sequence by PCR by using primers GST1 and GST2, and either p366 for the wild type (wt) or pAR17 for the Bni4V-A/F-A mutant as templates. The resulting amplicons were ligated into SmaI-HindIII–digested pEG(KG) (Mitchell et al., 1993), creating either pLK4 (BNI4) or pLK6 (bni4V-A/F-A). Plasmid pLK3 was created by exchanging the BamHI-BssHI CFP-encoding fragment of pDH3 (Yeast Resource Center, University of Washington) for the sequence encoding GFP, which was created by PCR by using primers GFP3F and GFP4R and pTII6 (Robinson et al., 1999) as a template.
Table 3.
Plasmids used in this study
| Name | Description | Source |
|---|---|---|
| p326 | CEN URA3 GFP-Chs4 | (DeMarini et al., 1997) |
| p365 | pRS425 2μ LEU2 BNI4 | (DeMarini et al., 1997) |
| p366 | pRS315 CEN LEU2 BNI4 | (DeMarini et al., 1997) |
| pAR5 | pGADC1 BNI4 (aa 70-892) | This study |
| pAR14 | pGADC1 BNI4 (aa 1-779) | This study |
| pAR16 | pGADC1 BNI4 (aa 1-892) | This study |
| pAR17 | pRS315 CEN LEU2 bni4V831A/F833A | This study |
| pAR19 | pGADC1 bni4V831A/F833A (aa 70-892) | This study |
| pAR24 | pRS315 CEN LEU2 GFP-CHS4 | This study |
| pAR26 | pRS313 CEN HIS GFP-CHS4 | This study |
| pLK3 | pKanMx6 GFPS65T F64L | This study |
| pLK4 | pEG(KG) URA GAL GST-BNI4 | This study |
| pLK6 | pEG(KG) URA GAL GST-bni4V831A/F833A | This study |
| pTII6 | pRSETB GFPS65T F64L | (Robinson et al., 1999) |
| pHH149 | pAS1-GLC7 | (Wu et al., 2001) |
| pKT1703 | pAS1-glc7-129 | This study |
Immunoprecipitation and Alkaline Phosphatase Reactions
Immunoprecipitation of Bni4 and Glc7 (Figure 1B) was modified from Stuart et al. (1994). Briefly, cells were grown to mid-log phase and 50 ml was harvested. All subsequent steps were performed at 4°C. Cells were washed in breaking buffer (100 mM Tris, 200 mM NaCl, 1 mM EDTA, 5% glycerol, pH 7.0) and resuspended in 0.6 ml of breaking buffer plus 0.5% Triton X-100, 1:300 dilution protease inhibitor cocktail (1 mg of leupeptin, chymostatin, antipain, and pepstatin A in 4 ml of 50% ethanol) and 1:100 dilution of phenylmethylsulfonyl fluoride (PMSF) (saturated solution in ethanol). An equal volume of glass beads was added and cells were broken by vortexing for 10 min. The cell debris was pelleted and the supernatant transferred to a new tube. Protein G-Sepharose beads (25 μl) in radioimmunoprecipitation assay (RIPA) buffer without SDS (50 mM Tris pH 7.0, 1% Triton X-100, 0.5% sodium deoxycholate, 200 mM NaCl) was added to 90 μl of supernatant and the mixture was incubated for 1 h with rocking. The beads were pelleted, supernatant was transferred to a new tube containing primary antibody, and this solution was rocked 1 h. Protein G-Sepharose (25 μl) was added and incubated for another hour. Immunoprecipitates were pelleted and washed four times in 25% breaking buffer/75% RIPA buffer without SDS and with protease inhibitors as described above, and once in 0.5 M Tris, 0.5 M NaCl, pH 7.0. The beads were resuspended in an equal volume of 2× sample buffer (62.5 mM Tris pH 6.8, 25% glycerol, 2% SDS, 0.01% bromophenol blue, 5% β-mercaptoethanol), boiled for 3 min, and electrophoresed on 10% polyacrylamide-SDS gels. After electrophoresis, proteins were transferred to nitrocellulose for immunoblotting with subsequent detection by using the enhanced chemiluminescence system (Amersham Biosciences, Piscataway, NJ). GFP-Glc7 was immunoprecipitated with polyclonal GFP antibody (kindly provided by Dr. Nathan Davis, LSUHSC, Shreveport, LA) and detected with monoclonal anti-GFP from Molecular Probes (Eugene, OR). Bni4 was immunoprecipitated and immunoblotted with polyclonal anti-Bni4 (DeMarini et al., 1997). For the immunoprecipitation of Bni4, which preceded phosphatase treatment (Figure 8A), the following protocol was used. Cultures were grown to logarithmic phase, and cells were collected by centrifugation and resuspended in 3 volumes of ice-cold breaking buffer (100 mM Tris, 200 mM NaCl, 1 mM EDTA, 5% glycerol, pH 7.0–7.1) containing 1 mM PMSF and protease inhibitor cocktail (1 ml/20 g of wet cell pellet; Sigma-Aldrich, St. Louis, MO). Three volumes of glass beads were added and cells were shaken on a Mini-beadbeater (Biospec Products, Bartlesville, OK) four times for 30 s with 1-min breaks on ice. Cell lysates were transferred to fresh tubes. Four volumes of breaking buffer were used to wash the beads by vortexing for 5 s, after which the wash supernatant was combined with the cell lysates. Lysates were cleared by centrifugation for 20 min at 14,000 rpm (4°C) and supernatants were transferred to fresh tubes. Antibody-conjugated beads were prepared by adding 2 μl of the primary antibody (α-Bni4) to a mixture of 90 μl of 50% protein G slurry (Invitrogen, Carlsbad, CA; beads were first washed and then resuspended in RIPA buffer: 50 mM Tris pH 7.0, 1% Triton X-100, 0.5% sodium deoxycholate, 200 mM NaCl) and 0.5 ml of ice-cold phosphate-buffered saline. Mixtures were tumbled at 4°C for 1 h, and then washed twice with ice-cold RIPA buffer. Cell lysate was precleared with 50% protein G-agarose slurry at 4°C for 30 min. The beads were pelleted and the supernatant was added to the antibody-conjugated beads. Samples were incubated at 4°C for 1 h. Beads were pelleted and washed four times with a mixture of 25% RIPA and 75% breaking buffer and once with calf intestinal alkaline phosphatase (CIP) buffer (50 mM Tris-HCl, pH 8.0, 1 mM MgCl2, 0.1 mM ZnCl2) containing 1 mM PMSF and the protease inhibitor cocktail.
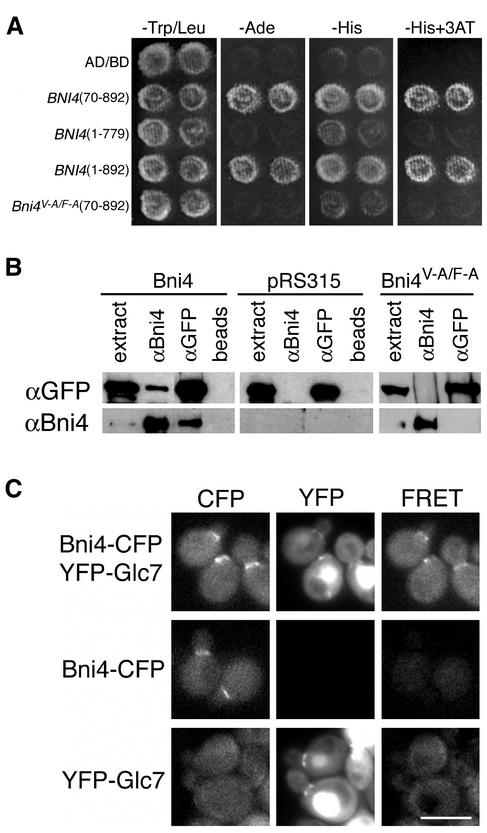
Figure 1.
Interaction between Bni4 and Glc7. (A) Two-hybrid analysis of Bni4 and Glc7. Strain PJ69-4A was cotransformed with pHH149, encoding Glc7 fused to the Gal4 DNA-binding domain, and either pAR5 (BNI4 aa 70–892), pAR14 (BNI4 aa 1–779), pAR16 (BNI4 aa 1–892), or pAR19 (bni4V-A/F-A aa 70–892), which encode the indicated Bni4 variants. Two representative transformants were plated on synthetic medium lacking tryptophane and leucine (SC-Trp/Leu) to select for the two plasmids. The two-hybrid interaction was assayed by replica plating the transformants onto SC-Ade, SC-His, or SC-His supplemented with 10 mM 3-amino-triazole (−His + 3AT). (B) Coimmunoprecipitation of Bni4 and Glc7. Strain KT1927 (GFP-GLC7, bni4::TRP1) was transformed with p366 (BNI4), the empty vector pRS316, and pAR17 (bni4V-A/F-A). Immunoprecipitates were subjected to PAGE and immunoblots were probed using anti-GFP antibody (αGFP) or anti-Bni4 antibody (αBni4). (C) Strains YLK29 (Bni4-CFP, YFP-Glc7), YLK27 (Bni4-CFP) and YAB122 (YFP-Glc7) were imaged with CFP, YFP, and FRET filter sets. Bar, 5 μm.
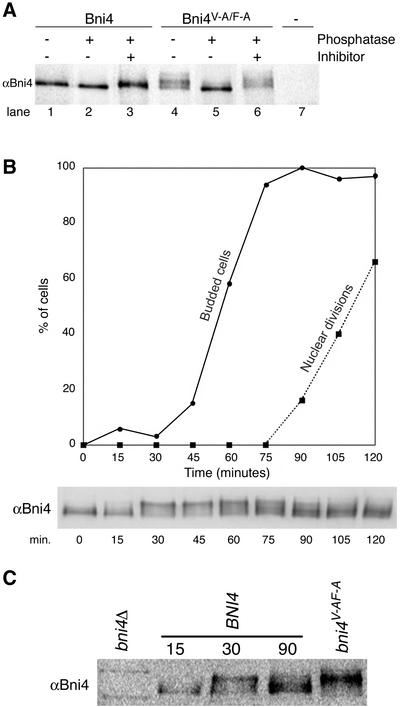
Figure 8.
Bni4 is covalently modified. (A) Bni4 is phosphorylated and Bni4V-A/F-A is hyperphosphorylated. Immunoprecipitation with anti-Bni4 antibody (α-Bni4) was performed on lysates from cells expressing either Bni4 (lanes 1–3), Bni4V-A/F-A (lanes 4–6), or cells disrupted for BNI4 (lane 7) (strains YLK74, YLK76, and YLK78, respectively). Precipitates were MOCK-treated (lanes 1 and 4), treated with calf intestinal phosphatase (lanes 2 and 5), or treated with the phosphatase and the vanadate inhibitor (lanes 3 and 6). Samples were electrophoresed, blotted to membrane, and probed with α-Bni4 antibody. (B) Phosphorylation of Bni4 is cell cycle-dependent. Cells (DLY222) were arrested in G1 by incubation in medium containing sucrose. The arrest was released by exposing the cells to galactose. Samples were collected every 15 min and examined for budding (solid line), nuclear division (dotted line), and Bni4 mobility on PAGE (immunoblot), as detailed in the text. (C) Electrophoretic mobility of Bni4 in the samples collected at 30 min is similar to that of Bni4V-A/F-A from an asynchronous culture. Immunoblot with anti-Bni4 was done following SDS-PAGE separation of 15, 30, and 90 min. samples along with extract from the strain YLK76.
In vitro dephosphorylation was done as described previously (Stuart et al., 1994) with minor modifications. After immunoprecipitation, beads were resuspended in 30 μl of CIP buffer. Reaction mixtures were then incubated with or without the addition of 15 U of CIP (New England Biolabs, Beverely, MA) at 37°C for 15 min. A parallel reaction was carried out in phosphatase buffer containing CIP and 10 mM sodium orthovanadate (Sigma-Aldrich) as a phosphatase inhibitor. Immunoblots of these samples were visualized using a phosphorimager.
Microscopic Analysis
Time-lapse imaging of cells was performed as described previously (Bloecher and Tatchell, 1999) by using either a Roper MicroMAX or CoolSNAP HQ charge-coupled device camera. For imaging GFP, a 41001 filter set was used (Chroma Technology, Brattleboro, VT). For dual fluorescence imaging of CFP and YFP, a JP4 filter set (Chroma Technology) was used in which the excitation and emission filters were placed in separate computer-controlled filter wheels (Ludl Electronic Products, Hawthorne, NY). Image analysis, filter wheels, shutters, and Z axis stepping motor were controlled by IPlab Spectrum software (Scanalytics, Fairfax, VA). To measure fluorescence resonance energy transfer (FRET), images were acquired for YFP fluorescence (excitation 500 nm, emission 535 nm), CFP fluorescence (excitation 436 nm, emission 470 nm), and FRET fluorescence (excitation 436 nm, emission 535 nm). The signal in the FRET channel due to YFP and CFP alone was subtracted from the FRET signal by using IPlab software. To quantify GFP fluorescence, the average fluorescence in a small region consisting of 4 to 13 adjacent pixels was measured using the CoolSNAP HQ charge-coupled device camera. Care was taken not to photobleach the sample before image acquisition. Background fluorescence was subtracted from each measurement.
Indirect immunofluorescence was performed as described previously (Pringle et al., 1991) with the following modifications. Cells were fixed in 5% formaldehyde, washed once with PBS, and once with solution A (100 mM KPO4 pH 6.5, 0.5 mM MgCl2, 1.2 mM sorbitol). Cells were permeabilized in solution A containing 5.5% of β-glucuronidase/arylsulfatase (Roche Diagnostics, Indianapolis, IN) and 1% β-mercapto-ethanol at 37°C for 1 h. Cells were stained with anti-Bni4 (DeMarini et al., 1997). Calcofluor white staining was done as described previously (Robinson et al., 1999).
RESULTS
Bni4 Is a PP1/Glc7 Targeting Subunit
To determine whether Bni4 tethers Glc7 to the bud neck, we first confirmed the Bni4–Glc7 interaction by using two-hybrid, coimmunoprecipitation, and FRET assays (Figure 1). For the FRET assay and subsequent analysis of Bni4 in live cells, we constructed BNI4-GFP fusions by using vectors that integrated either the blue-shifted variant CFP, enhanced GFP, or the red-shifted variant YFP directly before the stop codon of the BNI4 open reading frame. The fusions were functional, as judged by normal chitin deposition and morphology in cells containing the fusions. In agreement with the location of native Bni4 detected by immunofluorescence (DeMarini et al., 1997), Bni4-GFP was often found on the cortex of unbudded cells, on the mother side of the bud neck in cells with small and medium-sized buds, and at a lower concentration on both sides of the neck in cells with large buds.
The FRET assay between a blue-shifted variant of GFP (Bni4-CFP) and the red-shifted GFP variant 10C/YFP (YFP-Glc7) (Bloecher and Tatchell, 2000) was performed by assaying the increase in YFP emission after CFP excitation. A clear FRET signal at the bud neck was only observed when both Bni4-CFP and YFP-Glc7 were expressed in the same cells (Figure 1C) and indicates that the GFP domains of the fusions are no more than 75 Å apart (Selvin, 1995). These results confirm that Bni4 interacts with Glc7 at the bud neck. Bni4 contains a putative RVXF Glc7-binding motif near its COOH terminus (G830VRF). To assess whether this sequence is necessary for Glc7 binding, we constructed missense mutations, in which the conserved valine (V831) and phenylalanine (F833) residues were changed to alanine. Valine-to-alanine and phenylalanine-to-alanine substitutions in the RVXF motif of known Glc7 binding proteins reduce or eliminate Glc7 binding (Bollen, 2001). The resulting protein, Bni4V831A/F833A, hereafter referred to as Bni4V-A/F-A, fails to associate with Glc7 in two-hybrid and IP assays (Figure 1, A and B). These results confirm that the RVXF motif in Bni4 is necessary for Glc7 binding.
To determine whether Bni4 is necessary for the accumulation of GFP-Glc7 at the incipient bud site or later in the cell cycle at the bud neck, we assayed the location of GFP-Glc7 in wild-type, bni4V-A/F-A, and bni4::TRP1 strains. GFP-Glc7 does not accumulate at the bud neck of small- and medium-budded cells in either the bni4::TRP1 (Figure 2A and attached video) or bni4V-A/F-A mutant (our unpublished data). These results show that Bni4 is required to localize Glc7 to the bud neck and place Bni4 in a growing family of PP1/Glc7 targeting subunits. However, GFP-Glc7 does accumulate at the actomyosin ring, irrespective of the BNI4 genotype, indicating that the mechanism responsible for targeting Glc7 to the neck at cytokinesis is Bni4 independent. In addition to accumulating at the bud neck, Glc7 appears at the base of mating projections (Bloecher and Tatchell, 2000) and accumulates at the prospore wall (Tachikawa et al., 2001) in a septin-dependent manner. We imaged MATa bni4::TRP1 GFP-GLC7 cells that had been arrested with alpha factor and MATa/MATα bni4::TRP1/bni4::TRP1 GFP-GLC7 diploid cells that had been induced to sporulate, but observed no difference in the location of GFP-Glc7 between bni4 null mutants and the wild type (our unpublished data). Bni4 is therefore strictly required for septin-dependent localization of Glc7 in mitotically growing cells.
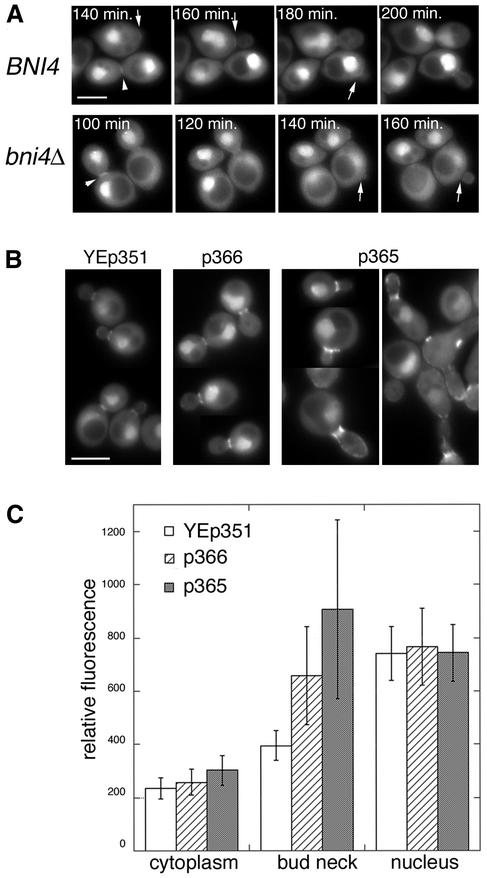
Figure 2.
Localization of GFP-Glc7 to the bud neck is dependent on BNI4. (A) Montage of images taken at 20-min intervals from time-lapse experiments of diploid cells homozygous for BNI4 (strain KT1925 × KT1926) or bni4::TRP1 (strain KT1921 × KT1922). Arrows designate the location of BNI4-dependent GFP-Glc7 at the bud neck and the lack of GFP-Glc7 at the neck in bin4Δ cells. Arrowheads designate the accumulation of BNI4-independent GFP-Glc7 at the bud neck during cytokinesis. (B) Images of GFP-Glc7 (strain YAB608) in cells transformed with empty vector YEp351, low-copy Bni4-expressing vector (p366) or high copy Bni4-expressing vector (p365). (C) Fluorescence levels of GFP-Glc7 were quantified in the cytoplasm, at the bud necks and in the nuclei of the cells described in B. Levels for the cytoplasm were measured in the bud. Fluorescence levels in the nucleus were measured away from the nucleolus, which contains the highest levels of Glc7. Fluorescence levels in at least 20 cells from each transformed strain were quantified. A video supplement to Figure 2A was prepared from images taken at 10-min intervals. Bars (A and B), 5 μm.
To find out whether Bni4 is a limiting determinant for Glc7 recruitment to the bud neck, we transformed a GFP-GLC7 strain with low- and high-copy plasmids expressing BNI4 and quantified the levels of GFP-Glc7 at the bud neck, in the cytoplasm, and in the nucleus. Increased gene dosage of BNI4 results in a significant increase in GFP-Glc7 at the bud neck, but not in the cytoplasm, or in the nucleus (Figure 2, B and C). Bni4 is therefore the limiting determinant for the targeting of Glc7 to the bud neck.
The morphology of cells expressing high levels of Bni4 (Figure 2B) is reminiscent of cells with defects in septin organization (DeMarini et al., 1997). To investigate the effects of Bni4 overexpression, we expressed a GST-Bni4 fusion to high levels by using a GAL1 promoter and monitored septin structure by using a Cdc10-GFP fusion. Overexpression of GST-Bni4 caused slow growth (our unpublished data) and induced an altered morphology similar to that found with strains expressing high-copy BNI4 plasmids. Growth of these cells in galactose resulted in disorder and then loss of Cdc10-GFP at the bud neck (Figure 3). Cdc10-GFP accumulated in diffuse punctate structures and longer strands at 2 h of induction. After 5 h of induction, short bars and arcs of Cdc10-GFP were common and a few small circles were observed. At longer periods of induction, Cdc10-GFP accumulated at the cell surface in small rings of fairly uniform size and intensity (Figure 3, C and D). These ring structures are unlikely to be due to the accompanying Glc7 because the same morphological perturbations and aberrant septin structures are also observed upon overexpression of GST-Bni4V-A/F-A (our unpublished data). Similar rings were observed with a Cdc12-GFP strain (our unpublished data), suggesting that the small rings may contain the normal septin complement. However, the small rings do not colocalize with Calcofluor staining (our unpublished data), and neither Bni4-GFP nor Chs4-GFP accumulates in these small rings in GST-Bni4 expressing strains, indicating that these structures do not recapitulate the septin-associated complexes that normally assemble at the mother-bud neck.
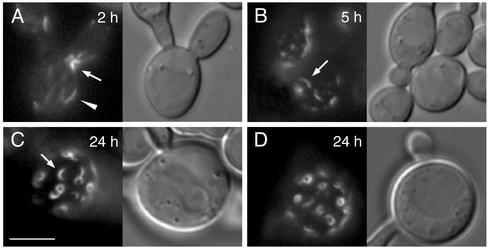
Figure 3.
Overexpression of GST-Bni4 causes elongated bud morphology and mislocalization of septins. Cells containing CDC10-GFP (YLK66) and expressing GST-Bni4 under galactose-inducible promoter (pLK4) were incubated in galactose medium at 37°C. Cells were imaged for Cdc10-GFP fluorescence at the designated times after the shift to galactose. After 2 h of incubation, Cdc10-GFP appeared in the form of patches at random locations on the cell membrane (A, arrowhead). Cdc10-GFP at the bud neck was also partially disorganized (A, arrow). At 5 h, CDC10-GFP appeared in short arced filaments dispersed in the cell membrane in addition to patches (B, arrow). Cdc10-GFP was miss-ing from bud necks. After 24 h of incubation, CDC10-GFP formed rings of ∼1 μm in diameter. Similar results were observed after incubation at 24 and 30°C. Bar, 5 μm.
Bni4 Is Required for Localization of Chs4 to Site of Bud Emergence but Not to Bud Neck at Cytokinesis
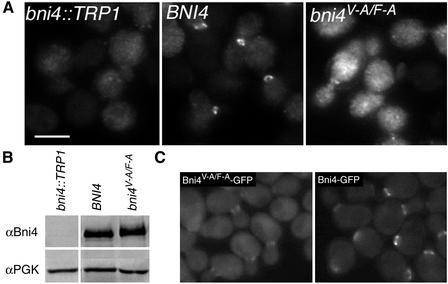
To determine whether the Bni4/Glc7 complex is required for localized chitin deposition, we examined bud scars and GFP-Chs4 accumulation in bni4V-A/F-A mutants. Calcofluor staining revealed a pattern of bud scars on bni4V-A/F-A mutant cells that closely resembled that observed on bni4::TRP1 cells (Figure 4A). These scars are enlarged, irregularly shaped, and dimmer than scars in BNI4 cells. To assess the timing of chitin deposition in bni4 mutants, we assayed chitin levels at the site of bud emergence. A septin-GFP fusion (Cdc10-GFP) was used to unequivocally identify the bud site. As shown in Figure 4B (1, 2, and 3, arrowheads), chitin is deposited at the neck in small-budded cells in the wild-type strain. In contrast, only low levels of calcofluor fluorescence were observed at the bud neck in the bni4::TRP1 (our unpublished data) and bni4V-A/F-A mutants (Figure 4B, 4–9). High levels of chitin were only observed in some bni4::TRP1 and bni4V-A/F-A cells late in the cell cycle, when the septin collar has separated into two rings (Figure 4B, 6, arrow). These results suggest that Bni4-Glc7 is required for chitin deposition early in the cell cycle.
Figure 4.
Effect of the bni4V-A/F-A mutation on chitin synthesis at the bud neck. (A) Calcofluor staining in wild-type BNI4 (YLK74), bni4V-A/F-A (YLK76), and bni4Δ1::TRP1 (YLK78) strains. Bud scars were of uniform size and shape in BNI4 strains whereas bud scars of bni4V-A/F-A and bni4Δ1::TRP1 mutants were enlarged and irregular. (B) Colocalization of the septin Cdc10-GFP and chitin in BNI4 (YLK112) and bni4V-A/F-A (YLK110) strains. In the BNI4 strain, relatively high levels of chitin were present at the bud neck throughout the cell cycle (3, arrowheads), whereas in bni4V-A/F-A cells, higher levels of chitin at the bud neck were detected only in some large-budded cells (6, arrow). Bars, 5 μm.
Figure 9.
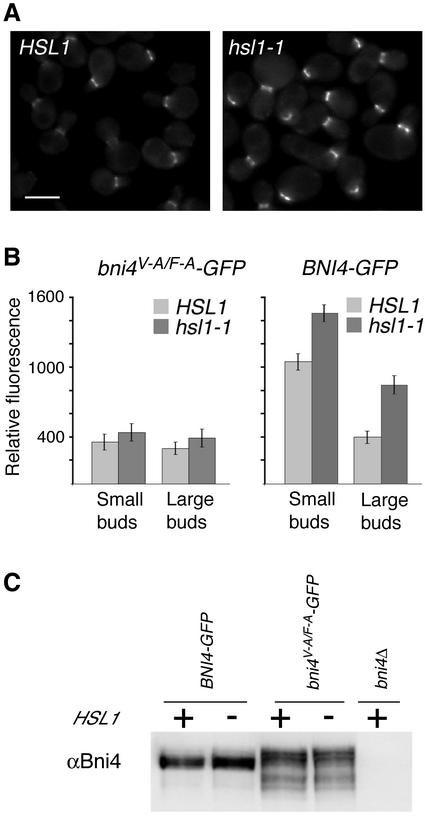
Effect of hsl1-1 deletion on the levels of Bni4-GFP and Bni4V-A/F-A-GFP at the bud neck. (A) Bni4-GFP in HSL1 (left, YLK45) and in hsl1-1 (right, YLK160) cells. Bar, 5 μm. (B) Relative fluorescence was measured for Bni4V-A/F-A-GFP (left) and Bni4-GFP (right) in HSL1 (YLK84, YLK45) and hsl1-1 (YLK162, YLK160) cells. Small-budded and large-budded cells were compared. For Bni4-GFP the difference between HSL1 and hsl1-1 in small- and large-budded cells was significant (p < 0.001). (C) Immunoblot analysis of extracts from Bni4-GFP and Bni4V-A/F-A-GFP in HSL1 and hsl1-1 strains. The immunoblots were probed with antibodies to Bni4.
Figure 6.
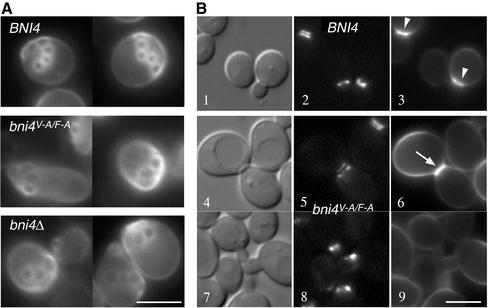
Bni4V-A/F-A fails to localize properly to the bud neck. (A) Indirect immunofluorescence of Bni4 in a bni4Δ1::TRP1 strain (YLK78), a wild-type strain (YLK74), and in a bni4V-A/F-A strain (YLK76), as described in Materials and Methods. Bni4 was visualized using anti-Bni4 antibody (DeMarini et al., 1997). Bar, 5 μm. (B) Immunoblot analysis of the strains described in A. (C) GFP fluorescence in bni4V-A/F-A:GFP (YLK84) and BNI4:GFP (YLK80) strains.
DeMarini et al. (1997) noted in fixed samples that the localization of GFP-Chs4 to the incipient bud was Bni4 dependent. To characterize Chs4 localization further, we imaged live cells that contained Chs4 fused to bright variant of GFP (GFPF64L S65T) (Cormack et al., 1996). This fusion complemented a chs4 null mutation when expressed from a low-copy plasmid. GFP-Chs4 was found at the highest level at the cortex in some unbudded cells, in small-budded cells, but not in medium-budded cells. Surprisingly, it was also found at high concentrations at the necks of some large-budded cells. Time-lapse imaging of this fusion (Figure 5A and attached video) revealed that GFP-Chs4 mobilizes to the site of bud emergence before bud formation (Figure 5A, 0 min, top arrow), remains at the bud neck in small-budded cells, but then rapidly disappears from the neck. At the end of the cell cycle, GFP-Chs4 again accumulates at the neck at concentrations as high or higher than those found in small-budded cells (Figure 5A, 100 min, arrowhead).
Figure 5.
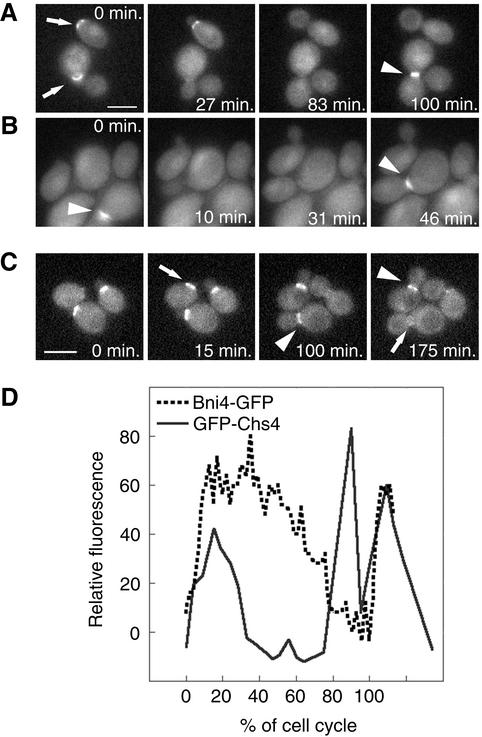
Localization of GFP-Chs4 and Bni4-GFP by time-lapse imaging. (A) In the BNI4 cells (KT1972 cotransformed with pAR26 and p366), GFP-Chs4 accumulates at the site of the presumptive bud (0 min, top arrow) and it disappears soon after bud emergence. GFP-Chs4 reappears at the neck before cytokinesis (100 arrowhead). (B) In the bni4V-A/F-A cells (KT1972 cotransformed with pAR26 and pAR17), GFP-Chs4 does not accumulate at the site of bud emergence. However, it appears at the bud neck before cytokinesis (0 and 46 min, arrowheads). (C) Localization of Bni4-GFP by time-lapse imaging (YLK80). Bni4-GFP appears at the future bud emergence site ∼15 min before budding (15 min, arrow) and stays at the bud neck at relatively high levels for 60–80% of the cell cycle (100 and 175 min, arrowheads). Close to cytokinesis, Bni4 forms a double ring that is hardly detectable (175 min, arrow). (D) Relative fluorescence of GFP-Chs4 (solid line) and Bni4-GFP (dotted line) was plotted as a function of the progress through the cell cycle (percentage of the cell cycle completed) by using the first appearance of GFP at the incipient bud site as the start point and the appearance of GFP at the incipient bud site in the mother cells in the next generation as the endpoint. Shown are representative data from three cells of each strain for which complete cell cycle data were obtained. A video supplement to Figure 5, A–C, was prepared from images taken at 5-min intervals. Bars (A and C), 5 μm.
GFP-Chs4 does not accumulate at the site of bud emergence in bni4::TRP1 (our unpublished data) and bni4V-A/F-A (Figure 5B and attached video) mutant cells but it does accumulate at the neck during cytokinesis (Figure 5B, 0 and 46 min, arrowheads). Thus, like Glc7, Chs4 requires Bni4 to accumulate at the bud neck early in the cell cycle but not at cytokinesis. We also performed time-lapse experiments on cells expressing Bni4-GFP (Figure 5C and attached video). The experiments revealed that Bni4-GFP first accumulates at the site of bud emergence before bud formation, similarly to GFP-Chs4 (15 min, arrow). In contrast to GFP-Chs4, Bni4-GFP is retained on the mother side of the bud at relatively high levels for a much longer period (100 and 175 min, arrowheads) and then mobilizes to both sides of the neck, but at a concentration of only ∼40% of that found earlier in the cell cycle (175 min, arrow). The drop of Bni4 levels at the bud neck is not simply due to a reduction in the total Bni4 concentration, because levels of Bni4 do not change significantly through the cell cycle, as judged by immunoblot analysis (see below).
To compare the dynamics of Bni4 and Chs4, we measured GFP fluorescence at the incipient bud site and at the bud neck in strains containing either Bni4-GFP or GFP-Chs4 and plotted the fluorescence values as a function of the progress through the cell cycle. As shown in Figure 5D, high levels of Bni4-GFP are retained at the neck for 60–80% of the cell cycle, whereas GFP-Chs4 is retained at the neck for roughly 30% of the cell cycle. The average time that Bni4-GFP remained at the bud neck was 145 ± 28 min (n = 14), whereas GFP-Chs4 remained at the neck for 40 ± 8 min (n = 3). GFP-Chs4 levels spike again at cytokinesis, whereas Bni4-GFP levels continue to fall at this stage.
Bni4V-A/F-A Fails to Associate Properly with Bud Neck
The null phenotype of the bni4V-A/F-A mutant (Figure 4A) suggests that Glc7 may have a critical role in recruiting chitin synthase to the bud neck. To investigate the role of Glc7 in this process, we assayed the location of wild-type Bni4 and Bni4V-A/F-A in fixed cells by indirect immunofluorescence, and directly in live cells by using GFP fusions to Bni4 and the Bni4V-A/F-A variant. Indirect immunofluorescence microscopy by using anti-Bni4 antibodies revealed that the level of Bni4V-A/F-A at the bud neck was reduced relative to that of wild-type Bni4 (Figure 6A). Only 10% (n = 150) of budded cells from a bni4V-A/F-A strain exhibited staining at the bud neck compared with 39% (n = 145) of budded cells from a wild-type strain. Immunoblot analysis of cell extracts prepared from BNI4 and bni4V-A/F-A strains showed that Bni4 and Bni4V-A/F-A were expressed at similar levels (Figure 6B). We also compared the location of Bni4-GFP and Bni4V-A/F-A-GFP fusions. The Bni4V-A/F-A-GFP fusion was only weakly localized to the bud neck (Figure 6C). We quantified the concentration of Bni4-GFP and Bni4V-A/F-A-GFP fusions at the bud neck. Before bud emergence and in small-budded cells, the levels of Bni4V-A/F-A-GFP at the site of bud emergence were only ∼40% of that observed for wild-type Bni4. However, in large-budded cells the concentrations of Bni4-GFP and Bni4V-A/F-A-GFP at the neck were similar (Figure 9B).
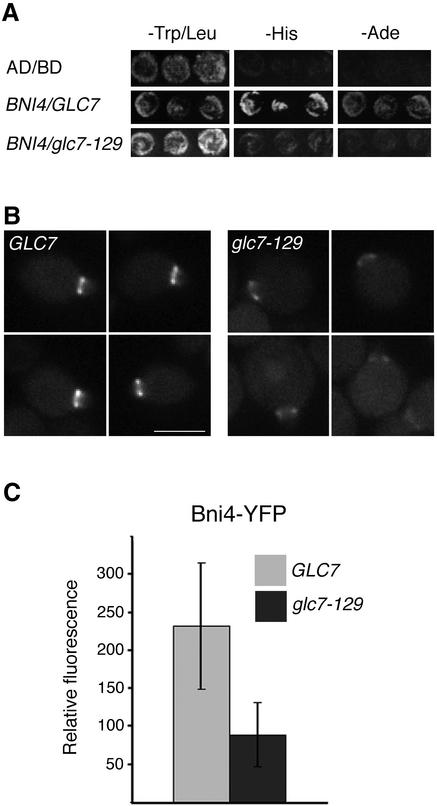
If the failure of Bni4V-A/F-A to associate properly with the neck is due to a failure to bind Glc7 then Glc7 mutants that are defective in Bni4 binding should also prevent Bni4 accumulation at the bud neck. A GFP fusion to the Glc7-129 variant fails to associate normally with the bud neck (Bloecher and Tatchell, 2000). We demonstrated by two-hybrid analysis (Figure 7A) and in coimmunoprecipitation experiments (our unpublished data) that Glc7-129 fails to interact properly with Bni4. Bni4-GFP levels were reduced in the glc7-129 mutant by at least 50% below that of the wild type (Figure 7, B and C), consistent with the hypothesis that a Bni4-Glc7 complex is required for efficient association of either of the proteins with the septin neck filaments.
Figure 7.
Levels of Bni4-YFP at the bud neck are reduced in glc7-129 cells. (A) Two-hybrid analysis of Bni4 and Glc7. Strain PJ69-4A was cotransformed with pAR5, encoding Bni4, and either pHH149 or pKT1703, which encode Glc7 and glc7-129, respectively. (B) Bni4-YFP was visualized in either KT2153 (GLC7) or in KT2155 (glc7-129) cells. Bar, 5 μm. (C) Fluorescence at the bud neck in the small- and medium-budded cells of GLC7 or glc7-129 strains from B was measured (p < 0.001).
Bni4 Is Phosphorylated in a Cell Cycle-dependent Manner
Walsh et al. (2002) found that Bni4 migrates as two closely spaced bands on SDS-PAGE gels. We observed that the electrophoretic mobility of Bni4V-A/F-A was slower than that of wild-type Bni4 (Figure 6B) and that Bni4V-A/F-A often migrated as multiple bands. To determine whether the heterogeneity in electrophoretic mobility of Bni4 is due to phosphorylation, we treated immunoprecipitates of Bni4 with alkaline phosphatase before PAGE. Dephosphorylation caused the broad band of Bni4 cross-reacting material to migrate as a single band with a faster electrophoretic mobility (Figure 8A). Bni4V-A/F-A migrated as a single band with the same mobility as wild-type Bni4 after treatment with alkaline phosphatase (Figure 8A). These results suggest that Bni4 is phosphorylated and indicate that Bni4V-A/F-A is likely hyperphosphorylated.
As a first step in determining whether phosphorylation is responsible for the cell cycle-dependent localization of Bni4, we synchronized cells in G1 by depriving cells of G1 cyclins by using strain DLY222 (Bose et al., 2001), in which the sole G1 cyclin (Cln3) is under GAL1 control. Cells were arrested in G1 by incubation in medium containing sucrose and then transferred into medium containing galactose as the sole carbon source. Aliquots of the culture were harvested at 15-min intervals after galactose induction and assayed for cell budding, nuclear division, and Bni4 electrophoretic mobility (Figure 8B). Before galactose induction, Bni4 in the Cln3-deprived cells was in the high-mobility form, but within 15 min after Cln3 induction slower migrating forms of Bni4 were visualized. These forms predominated just before cell budding (30 min). At 60 min, forms of intermediate mobility were present and predominated at 90 min when nuclear division began. The electrophoretic mobility of Bni4 from cells at the 30- and 45-min time points was similar to that of Bni4V-A/F-A from an asynchronous culture (Figure 8, B and C). We also synchronized MATa haploid cells with the mating pheromone alpha factor and observed a similar pattern of mobility changes in Bni4 after release from alpha factor arrest (our unpublished data). Together, these results indicate that Bni4 is most likely phosphorylated at multiple sites in a cell cycle-dependent manner.
Hsl1 Kinase Regulates Association of Bni4 with Bud Neck
The cell cycle dependence of Bni4 phosphorylation and the apparent hyperphosphorylated state of Bni4V-A/F-A suggest that phosphorylation may play a key role in the localization and/or function of Bni4. If this is the case, we would expect that reduced activity of the kinase or kinases that phosphorylate Bni4 would result in an alteration in the association of Bni4 with the neck. With this rationale, we determined whether null mutations in any of several protein kinase genes raise the level of Bni4-GFP at the bud neck. We tested three kinases that are known to reside at the bud neck: Gin4, Hsl1, and Kcc4. Gin4 has a well-documented role in regulating septin morphology (Barral et al., 1999; Longtine et al., 2000). Hsl1 has a role in regulating the stability of Swe1, an inhibitory kinase of Cdc28 (Barral et al., 1999; McMillan et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000; Cid et al., 2001). Kcc4 is most similar to Hsl1 and was reported to have a partially redundant function with Gin4 and Hsl1 in some genetic backgrounds (Barral et al., 1999). We introduced the Bni4 and Bni4V-A/F-A-GFP fusions into gin4, hsl1 and kcc4 null mutants and assayed the levels of GFP fluorescence at the bud neck. By visual inspection of fluorescence images, no clear effect was found for gin4 and kcc4 null mutations, but Bni4-GFP levels at the bud neck were elevated in hsl1 null mutants (Figure 9A). Quantitation of fluorescence levels revealed a small increase in Bni4V-A/F-A-GFP and a greater increase of the wild-type Bni4-GFP levels in the hsl1 null mutant (Figure 9B). The increased fluorescence was observed both early in the cell cycle, when Bni4 levels at the bud neck are highest, and late in the cell cycle, when Bni4 levels are reduced. Immunoblot analysis revealed no obvious difference in the total level of Bni4-GFP accumulation in hsl1 null and HSL1 strains, and no differences in the phosphorylation state of Bni4-GFP, as assayed by the electrophoretic mobility of Bni4-GFP (Figure 9C).
Although loss of Hsl1 resulted in a significant increase in the level of Bni4 at the bud neck, it had a greatly reduced effect on Bni4V-A/F-A localization. This result would be expected if Hsl1 acted on Bni4-Glc7 to negatively regulate binding to the septin ring, possibly by modulating the Glc7–Bni4 interaction. Consistent with this interpretation, Bni4-GFP levels at the neck in small-budded cells were low and nearly identical in glc7-129 and glc7-129 hsl1-1 mutants (our unpublished data). These results indicate that Hsl1 affects the localization of Bni4, possibly by reducing the Bni4–Glc7 interaction.
DISCUSSION
The work presented herein and elsewhere provides a strong case that Bni4 acts as a targeting subunit for PP1c in yeast. Bni4 associates with Glc7 in two-hybrid, pull-down, and FRET assays, and contains a PP1-binding motif that is necessary for the interaction with Glc7. Glc7 is not targeted to the bud neck in bni4 null mutants, whereas enhanced expression of BNI4 results in increased Glc7 levels at the bud neck. The evolutionary importance of PP1 binding for Bni4 function is strengthened by the fact that the gene product in Candida albicans (open reading frame 6.28353) most closely related to Bni4 is most similar in the Glc7/PP1 binding domain. Although the overall sequence identity is low, the COOH terminal 63 amino acid residues of the Saccharomyces cerevisiae and C. albicans proteins are 73% identical.
Although it is clear that Bni4 targets Glc7 to the bud neck, the role of the phosphatase at this location remains obscure. Many PP1c targeting subunits bind to specific substrates to tether substrate and phosphatase directly. Given the role of Bni4 in targeting chitin synthase III to the bud neck via association with the septin ring, candidate substrates include Chs4, Chs3, or possibly septins. However, we have no evidence to date that any of these proteins are PP1 substrates. We have not observed 32P incorporation into GFP-Chs4 in vivo (Panek, unpublished observations). Although the negative evidence does not eliminate the possibility that Bni4-Glc7 may dephosphorylate Chs4, septins, or other as yet unidentified binding partners, the data presented herein suggest that Bni4 itself may be a substrate for Glc7. Bni4 is phosphorylated in vivo and Bni4V-A/F-A, which fails to associate with Glc7, is apparently hyperphosphorylated. This situation parallels that of Gac1, a glycogen targeting subunit for Glc7, which is also a phosphoprotein that is hyperphosphorylated in a strain bearing a Glc7 variant that does not bind to Gac1 (Stuart et al., 1994).
The failure of Bni4V-A/F-A to associate properly with the bud neck suggests that Glc7 facilitates association of Bni4 with septins. This could occur because Glc7 is required enzymatically, or alternatively, a Bni4–Glc7 complex might be the molecular entity that associates with the septin ring. We think it is unlikely that the two missense mutations in Bni4V-A/F-A result in a direct defect in septin binding because the Cdc10-binding domain, defined by two-hybrid analysis (DeMarini et al., 1997), does not include the COOH-terminal region required for Glc7 binding. Furthermore, levels of Bni4V-A/F-A at the bud neck are elevated in an hsl1 null background, suggesting that Bni4V-A/F-A does not have an inherent defect in septin binding.
The low level of Bni4V-A/F-A at the bud neck suggests to us that Glc7 and Bni4 are interdependent for association with the septin ring before bud emergence. Bni4 is the second Glc7-binding protein to share an interdependent role with Glc7 for septin-dependent localization. Gip1, together with Glc7, associates with septins that underlie the prospore membranes during sporulation (Tachikawa et al., 2001). In this case, there is interdependence between septins and Gip1-Glc7 for the normal development of the prospore wall. In the absence of Gip1, or in strains bearing glc7-136, a GLC7 allele whose product fails to associate with Gip1, septin filaments are not properly assembled and the prospore wall does not properly form. In contrast, loss of Bni4 does not seem to affect mitotic septin structure. However, we cannot rule out subtle effects, and the overexpression of GST-Bni4 results in formation of aberrant septin structures.
Bni4-GFP levels at the bud neck are reduced more than twofold in glc7-129 small-budded cells, consistent with the hypothesis that Glc7 is required for Bni4 localization. However, bud scars are normal in the glc7-129 mutant (Bloecher and Tatchell, 2000) and Chs4-GFP mobilization to the incipient bud site still occurs in this mutant (our unpublished data). We don't know the reason for the lack of reciprocity in this case. It is possible that the alanine substitutions in Bni4V-A/F-A mutant protein affect some property of the protein in addition to binding Glc7. Alternatively, the results could be explained by the existence of a redundant phosphatase that can substitute for Glc7. Such a phosphatase would dephosphorylate critical residues on Bni4 in the glc7-129 strain, but would fail to interact with Bni4V-A/F-A. We have noted that two phosphatase genes whose products are closely related to Glc7, PPZ1, and PPZ2, exhibit complex genetic interactions with GLC7 (Venturi et al., 2000). Particularly relevant to this case is the observation that glc7-129 mutants are inviable in the absence of PPZ1 and PPZ2, suggesting that there is some overlap in specificity. Ppz1 is capable of associating with a number of Glc7-binding proteins, but we have not yet tested for an association between Ppz1 and Bni4. Another possibility is that Glc7-129 could exhibit reduced Bni4-binding in vivo but still bind to the extent necessary to recruit Chs4.
Based on indirect immunofluorescence studies of Bni4 (DeMarini et al., 1997) and time-lapse imaging of Bni4-GFP, Bni4 undergoes dynamic changes in its localization. It appears at the presumptive bud emergence site ∼15 min. before budding and subsequently forms a ring on the mother side of the mother-bud neck. Later in the cell cycle, Bni4 forms rings on both sides of the neck, but at a lower concentration than earlier. These dynamic changes in Bni4 localization correlate well with changes in phosphorylation, as assayed by phosphatase-dependent changes in electrophoretic mobility. Assuming that the different migrating forms of Bni4 reflect different states of phosphorylation, Bni4 undergoes complex changes in its state of phosphorylation. In G1, Bni4 migrates rapidly on SDS-PAGE, similar to the migration of dephosphorylated Bni4. Fifteen minutes before budding, Bni4 migrates more slowly on SDS-PAGE, correlating with the first appearance of Bni4 at the site of bud emergence. Later in the cell cycle, when nuclear division begins, much of the Bni4 protein migrates as a band of intermediate mobility, correlating with the time when the levels of Bni4 drop at the bud neck. These findings suggest that phosphorylation may play a role in the localization of Bni4, but we cannot at this time exclude the possibility that the changes in the phosphorylation state are a consequence of its dynamic localization. We note that Bni4V-A/F-A, which fails to localize properly to the bud neck, migrates similar to Bni4 from late G1/S, when it accumulates to high levels at the neck. This may appear as contradictory but it allows us to point out that we do not know precisely how the electrophoretic mobility of Bni4 correlates with its phosphorylation state. Nevertheless, these results indicate that Bni4 undergoes complex changes in its phosphorylation state that may be responsible for its dynamic changes in location throughout the cell cycle or for its ability to act as a scaffold for Chs4.
We found that deletion of the HSL1 gene (hsl1-1) could significantly elevate levels of wild-type Bni4 at the bud neck but it had a much smaller influence on the levels of Bni4V-A/F-A. The hsl1-1 mutation also had little influence on Bni4 levels in a glc7-129 mutant. These data suggest that Hsl1 acts by modulating the formation of the Glc7–Bni4 complex. We have no evidence that Hsl1 alters the phosphorylation state of Bni4, although our electrophoretic mobility assay for Bni4 phosphorylation could easily miss a subtle change. Hsl1 could also act directly on Glc7 at the bud neck, but there is no evidence that Glc7 is phosphorylated (Stuart et al., 1994). In contrast to Bni4, Hsl1 appears at the bud neck after bud emergence and is found on the daughter side of the neck (Barral et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000). The differences in location of the two suggest that the effect of hsl1 may be indirect. Hsl1 together with its binding partner Hsl7 are required for the inhibition of Swe1 by degradation (McMillan et al., 1999; Shulewitz et al., 1999). Whether the effect we see in hsl1 null mutants on Bni4 localization is directly due to the loss of Hsl1 or Hsl7 or due to downstream changes in Clb-Cdc28 activity will require further investigation.
Bni4 likely has roles in addition to that required for normal chitin deposition. Crh2, a glycosylphosphatidylinositol-anchored protein involved in cell wall assembly (Rodriguez-Pena et al., 2000), requires Bni4 for normal localization in large-budded cells (Rodriguez-Pena et al., 2002). This role in targeting Chr2 could explain why Bni4 is retained at the bud neck late in the cell cycle, when Chs4 has disappeared from the neck. Genetic evidence also supports multiple roles for Bni4. bni4Δ chs4Δ and bni4Δ chs3Δ double mutants grow more slowly than either chs4Δ or chs3Δ mutants. This slow-growth phenotype indicates loss of an additional function of BNI4 that further compromises the growth of strains lacking chitin synthase III. There is a precedent for functional redundancy between different cell wall components. Mutations in FKS1, encoding β-glucan synthase, and ANP1, a component of a Golgi glycosylation complex, are lethal in combination with chs3 null mutations (Osmond et al., 1999). Furthermore, an fks1 null mutant exhibits a compensatory increase in chitin synthase III activity (Garcia-Rodriguez et al., 2000). Together, these results suggest that Bni4 could target multiple cell wall biosynthetic enzymes to the bud neck.
Chs4 associates with the bud neck for a short time at bud emergence and then again late in the cell cycle by a Bni4-independent mechanism. The reappearance of Chs4 at the bud neck late in the cell cycle supports the hypothesis that chitin synthase III activity provides the auxiliary septum that forms in the absence of chitin synthase II (Schmidt et al., 2002). We do not know why Chs4 disappears from the bud neck sooner than Bni4. One possibility is that Bni4 is posttranslationally modified after bud emergence in a manner that would allow disassociation of Chs4. The cell cycle-specific changes in the phosphorylation state of Bni4 would accommodate such a model. Alternatively, Chs3 endocytosis may facilitate Chs4 removal. DeMarini et al. (1997) found that GFP-Chs4 localization to the neck was dependent upon Chs3. We don't know whether mobilization of Chs4 to the neck at cytokinesis uses a Bni4-like mechanism, but no obvious Bni4-homolog has been identified in the S. cerevisiae genome. At cytokinesis, septins form rings on both sides of the neck but judging from the time-lapse images presented in Figure 5, GFP-Chs4 accumulates as a single ring only on the mother side of the neck or directly at the actomyosin ring. Interestingly, the localization of Glc7 to the neck at this stage is also independent of Bni4. Whether Chs4 is regulated at cytokinesis by another Glc7-targeting subunit or independently of Glc7 will require further investigation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Danny Lew, Philip James, Neal Mathias, and Nathan Davis for reagents necessary to carry out this work. We also thank Lucy Robinson for reading the manuscript. This work was supported by the National Institutes of Health grant GM-47789.
Footnotes
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–06–0373. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–06–0373.
REFERENCES
- Alms GR, Sanz P, Carlson M, Haystead TA. Reg1p targets protein phosphatase 1 to dephosphorylate hexokinase II in Saccharomyces cerevisiae: characterizing the effects of a phosphatase subunit on the yeast proteome. EMBO J. 1999;18:4157–4168. doi: 10.1093/emboj/18.15.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen PR, Lacroix FB, Villa-Moruzzi E, Margolis RL. Differential subcellular localization of protein phosphatase-1 alpha, gamma1, and delta isoforms during both interphase and mitosis in mammalian cells. J Cell Biol. 1998;141:1207–1215. doi: 10.1083/jcb.141.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beullens M, Van Eynde A, Vulsteke V, Connor J, Shenolikar S, Stalmans W, Bollen M. Molecular determinants of nuclear protein phosphatase-1 regulation by NIPP-1. J Biol Chem. 1999;274:14053–14061. doi: 10.1074/jbc.274.20.14053. [DOI] [PubMed] [Google Scholar]
- Bloecher A, Tatchell K. Defects in Saccharomyces cerevisiae protein phosphatase type I activate the spindle/kinetochore checkpoint. Genes Dev. 1999;13:517–522. doi: 10.1101/gad.13.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloecher A, Tatchell K. Dynamic localization of protein phosphatase type 1 in the mitotic cell cycle of S. cerevisiae. J Cell Biol. 2000;149:125–140. doi: 10.1083/jcb.149.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen M. Combinatorial control of protein phosphatase-1. Trends Biochem Sci. 2001;26:426–431. doi: 10.1016/s0968-0004(01)01836-9. [DOI] [PubMed] [Google Scholar]
- Bollen M, Stalmans W. The structure, role, and regulation of type 1 protein phosphatases. Crit Rev Biochem Mol Biol. 1992;27:227–281. doi: 10.3109/10409239209082564. [DOI] [PubMed] [Google Scholar]
- Bose I, Irazoqui JE, Moskow JJ, Bardes ES, Zyla TR, Lew DJ. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J Biol Chem. 2001;276:7176–7186. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- Bulawa C. CSD2, CSD3, and CSD4, genes required for chitin synthesis in Saccharomyces cerevisiae: the CSD2 gene product is related to chitin synthases and to developmentally regulated proteins in Rhizobium species and Xenopus laevis. Mol Cell Biol. 1992;12:1764–1776. doi: 10.1128/mcb.12.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa CE. Genetics and molecular biology of chitin synthesis in fungi. Annu Rev Microbiol. 1993;47:505–534. doi: 10.1146/annurev.mi.47.100193.002445. [DOI] [PubMed] [Google Scholar]
- Cabib E, Sburlati A, Bowers B, Silverman SJ. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J Cell Biol. 1989;108:1665–1672. doi: 10.1083/jcb.108.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WJ, Santos B, Duran A, Cabib E. Are yeast chitin synthases regulated at the transcriptional or the posttranslational level? Mol Cell Biol. 1994a;14:7685–7694. doi: 10.1128/mcb.14.12.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WJ, Sburlati A, Cabib E. Chitin synthase 3 from yeast has zymogenic properties that depend on both the CAL1 and the CAL3 genes. Proc Natl Acad Sci USA. 1994b;91:4727–4730. doi: 10.1073/pnas.91.11.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JS, Schekman RW. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p [published erratum appears in J. Cell Biol. (1996) 135, 1925] J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid VJ, Shulewitz MJ, McDonald KL, Thorner J. Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle. Mol Biol Cell. 2001;12:1645–1669. doi: 10.1091/mbc.12.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACs-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Cos T, Ford RA, Trilla JA, Duran A, Cabib E, Roncero C. Molecular analysis of Chs3p participation in chitin synthase III activity. Eur J Biochem. 1998;256:419–426. doi: 10.1046/j.1432-1327.1998.2560419.x. [DOI] [PubMed] [Google Scholar]
- Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- DeMarini DJ, Adams AE, Fares H, De Virgilio C, Valle G, Chuang JS, Pringle JR. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombek KM, Voronkova V, Raney A, Young ET. Functional analysis of the yeast GLC7-binding protein Reg1 identifies a PP1-binding motif as essential for repression of ADH2 expression. Mol Cell Biol. 1999;19:6029–6040. doi: 10.1128/mcb.19.9.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997;16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rodriguez LJ, Trilla JA, Castro C, Valdivieso MH, Duran A, Roncero C. Characterization of the chitin biosynthesis process as a compensatory mechanism in the fks1 mutant of Saccharomyces cerevisiae. FEBS Lett. 2000;478:84–88. doi: 10.1016/s0014-5793(00)01835-4. [DOI] [PubMed] [Google Scholar]
- He B, Gross M, Roizman B. The gamma134.5 protein of herpes simplex virus 1 has the structural and functional attributes of a protein phosphatase 1 regulatory subunit and is present in a high molecular weight complex with the enzyme in infected cells. J Biol Chem. 1998;273:20737–20743. doi: 10.1074/jbc.273.33.20737. [DOI] [PubMed] [Google Scholar]
- Hsieh-Wilson LC, Allen PB, Watanabe T, Nairn AC, Greengard P. Characterization of the neuronal targeting protein spinophilin and its interactions with protein phosphatase-1. Biochemistry. 1999;38:4365–4373. doi: 10.1021/bi982900m. [DOI] [PubMed] [Google Scholar]
- Huang H-b, Horiuchi A, Watanabe T, Shih S-R, Tsay H-J, Li H-C, Greengard P, Nairn AC. Characterization of the inhibition of protein phosphatase-1 by DARPP-32 and inhibitor-2. J Biol Chem. 1999;274:7870–7878. doi: 10.1074/jbc.274.12.7870. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1994. [Google Scholar]
- Kwon YG, Huang HB, Desdouits F, Girault JA, Greengard P, Nairn AC. Characterization of the interaction between DARPP-32 and protein phosphatase 1 (PP-1): DARPP-32 peptides antagonize the interaction of PP-1 with binding proteins. Proc Natl Acad Sci USA. 1997;94:3536–3541. doi: 10.1073/pnas.94.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Theesfeld CL, McMillan JN, Weaver E, Pringle JR, Lew DJ. Septin-dependent assembly of a cell cycle-regulatory module in saccharomyces cerevisiae. Mol Cell Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, Sambrook TJ, Fritsch EF. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- McMillan JN, Longtine MS, Sia RA, Theesfeld CL, Bardes ES, Pringle JR, Lew DJ. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol Cell Biol. 1999;19:6929–6939. doi: 10.1128/mcb.19.10.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Marshall TK, Deschenes RJ. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- Ono N, Yabe T, Sudoh M, Nakajima T, Yamada-Okabe T, Arisawa M, Yamada-Okabe H. The yeast Chs4 protein stimulates the trypsin-sensitive activity of chitin synthase 3 through an apparent protein-protein interaction. Microbiology. 2000;146:385–391. doi: 10.1099/00221287-146-2-385. [DOI] [PubMed] [Google Scholar]
- Osmond BC, Specht CA, Robbins PW. Chitin synthase III: synthetic lethal mutants and “stress related” chitin synthesis that bypasses the CSD3/CHS6 localization pathway. Proc Natl Acad Sci USA. 1999;96:11206–11210. doi: 10.1073/pnas.96.20.11206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pammer M, Briza P, Ellinger A, Schuster T, Stucka R, Feldmann H, Breitenbach M. DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast. 1992;8:1089–1099. doi: 10.1002/yea.320081211. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Robinson LC, Bradley C, Bryan JD, Jerome A, Kweon Y, Panek HR. The Yck2 yeast casein kinase 1 isoform shows cell cycle-specific localization to sites of polarized growth and is required for proper septin organization. Mol Biol Cell. 1999;10:1077–1092. doi: 10.1091/mbc.10.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Pena JM, Cid VJ, Arroyo J, Nombela C. A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol Cell Biol. 2000;20:3245–3255. doi: 10.1128/mcb.20.9.3245-3255.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Pena JM, Rodriguez C, Alvarez A, Nombela C, Arroyo J. Mechanisms for targeting of the Saccharomyces cerevisiae GPI-anchored cell wall protein Crh2p to polarised growth sites. J Cell Sci. 2002;115:2549–2558. doi: 10.1242/jcs.115.12.2549. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sburlati A, Cabib E. Chitin synthetase 2, a presumptive participant in septum formation in Saccharomyces cerevisiae. J Biol Chem. 1986;261:15147–15152. [PubMed] [Google Scholar]
- Schmidt M, Bowers B, Varma A, Roh DH, Cabib E. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J Cell Sci. 2002;115:293–302. doi: 10.1242/jcs.115.2.293. [DOI] [PubMed] [Google Scholar]
- Selvin PR. Fluorescence resonance energy transfer. Methods Enzymol. 1995;246:300–334. doi: 10.1016/0076-6879(95)46015-2. [DOI] [PubMed] [Google Scholar]
- Shenolikar S. Protein serine/threonine phosphatases - new avenues for cell regulation. Annu Rev Cell Biol. 1994;10:55–86. doi: 10.1146/annurev.cb.10.110194.000415. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Shulewitz MJ, Inouye CJ, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S, Sburlati A, Slater M, Cabib E. Chitin synthase 2 is essential for septum formation and cell division in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1988;85:4735–4739. doi: 10.1073/pnas.85.13.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits GJ, van den Ende H, Klis FM. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology. 2001;147:781–794. doi: 10.1099/00221287-147-4-781. [DOI] [PubMed] [Google Scholar]
- Stark MJR. Yeast protein serine/threonine phosphatases: multiple roles and diverse regulation. Yeast. 1996;12:1647–1675. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1647::AID-YEA71%3E3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Stuart JS, Frederick DL, Varner CM, Tatchell K. The mutant type 1 protein phosphatase encoded by glc7-1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol Cell Biol. 1994;14:896–905. doi: 10.1128/mcb.14.2.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachikawa H, Bloecher A, Tatchell K, Neiman AM. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J Cell Biol. 2001;155:797–808. doi: 10.1083/jcb.200107008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trilla JA, Cos T, Duran A, Roncero C. Characterization of CHS4 (CAL2), a gene of Saccharomyces cerevisiae involved in chitin biosynthesis and allelic to SKT5 and CSD4. Yeast. 1997;13:795–807. doi: 10.1002/(SICI)1097-0061(199707)13:9<795::AID-YEA139>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Trilla JA, Duran A, Roncero C. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae [published erratum appears in J. Cell Biol. (1999) 146, following 264] J Cell Biol. 1999;145:1153–1163. doi: 10.1083/jcb.145.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J, Song W, Carlson M. Protein phosphatase type 1 interacts with proteins required for meiosis and other cellular processes in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4199–4206. doi: 10.1128/mcb.16.8.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, et al. A. comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae [see comments] Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Valdivia RH, Baggott D, Chuang JS, Schekman RW. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Venturi GM, Bloecher A, Williams-Hart T, Tatchell K. Genetic interactions between GLC7, PPZ1 and PPZ2 in Saccharomyces cerevisiae. Genetics. 2000;155:69–83. doi: 10.1093/genetics/155.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules fro PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Walsh EP, Lamont DJ, Beattie KA, Stark MJ. Novel interactions of Saccharomyces cerevisiae type 1 protein phosphatase identified by single-step affinity purification and mass spectrometry. Biochemistry. 2002;41:2409–2420. doi: 10.1021/bi015815e. [DOI] [PubMed] [Google Scholar]
- Wu X, Hart H, Cheng C, Roach PJ, Tatchell K. Characterization of Gac1p, a regulatory subunit of protein phosphatase type I involved in glycogen accumulation in Saccharomyces cerevisiae. Mol Genet Genomics. 2001;265:622–635. doi: 10.1007/s004380100455. [DOI] [PubMed] [Google Scholar]
- Ziman M, Chuang JS, Schekman RW. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol Biol Cell. 1996;7:1909–1919. doi: 10.1091/mbc.7.12.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Chuang JS, Tsung M, Hamamoto S, Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.