Abstract
Profilin is a well-characterized protein known to be important for regulating actin filament assembly. Relatively few studies have addressed how profilin interacts with other actin-binding proteins in vivo to regulate assembly of complex actin structures. To investigate the function of profilin in the context of a differentiating cell, we have studied an instructive genetic interaction between mutations in profilin (chickadee) and capping protein (cpb). Capping protein is the principal protein in cells that caps actin filament barbed ends. When its function is reduced in the Drosophila bristle, F-actin levels increase and the actin cytoskeleton becomes disorganized, causing abnormal bristle morphology. chickadee mutations suppress the abnormal bristle phenotype and associated abnormalities of the actin cytoskeleton seen in cpb mutants. Furthermore, overexpression of profilin in the bristle mimics many features of the cpb loss-of-function phenotype. The interaction between cpb and chickadee suggests that profilin promotes actin assembly in the bristle and that a balance between capping protein and profilin activities is important for the proper regulation of F-actin levels. Furthermore, this balance of activities affects the association of actin structures with the membrane, suggesting a link between actin filament dynamics and localization of actin structures within the cell.
INTRODUCTION
Regulation of actin filament dynamics is essential for many cellular processes, including cell motility, cytokinesis, cellular differentiation, and endocytosis. Many actin-binding proteins participate in regulating actin filament elongation or disassembly, and their functions have been analyzed by various in vitro assays. Yet, it is often unclear how well the biochemical properties of these proteins, as defined in vitro, correspond to their functions in vivo. Furthermore, biochemical analyses can be incomplete, not accurately reproducing the complexities of actin dynamics in the cell. Genetic studies in whole animals complement in vitro studies by providing functional information in a cellular context.
We are using the fly bristle as a model system to study actin assembly in vivo. During pupal development, the bristle cell elaborates a long process that contains prominent longitudinal bundles of actin filaments associated with the plasma membrane (Overton, 1967; Appel et al., 1993). Cuticle is deposited on the surface of this process, and then as development proceeds, the actin cytoskeleton disassembles, the cell regresses, and the remaining hollow tube of cuticle becomes the adult bristle. Mutations in actin regulatory proteins such as capping protein (cpb; Hopmann et al., 1996), profilin (chickadee; Verheyen and Cooley, 1994), ADF/cofilin (twinstar; Gunsalus et al., 1995; Chen et al., 2001), ADF/cofilin phosphatase (slingshot; Niwa et al., 2002), and twinfilin (twf; Wahlstrom et al., 2001) perturb bristle morphogenesis through effects on the actin cytoskeleton. The bristle seems to be particularly sensitive to reductions in the level of actin-binding proteins, most likely because the cells are very large and require massive amounts of actin polymerization during development.
Bristle growth is dependent on actin filament polymerization (Tilney et al., 2000a). Actin polymerization requires a free end and actin monomers competent to polymerize (Pollard et al., 2000). Capping protein and profilin are two actin-binding proteins that regulate filament elongation by modulating these two parameters. Capping protein binds to actin filament barbed ends with high affinity and prevents the addition or loss of subunits (Isenberg et al., 1980; Casella et al., 1986). Profilin is an enigmatic protein that seems to stimulate actin polymerization in some contexts and repress it in others. Profilin binds to actin monomers and was originally thought to limit actin polymerization through monomer sequestration (Carlsson et al., 1977). Subsequent data suggest profilin promotes actin polymerization at free barbed ends, but sequesters when barbed ends are capped (Pantaloni and Carlier, 1993). A few studies have shown that polymerization enhancement predominates in specific situations in vivo (Finkel et al., 1994; Rothkegel et al., 1996; Benlali et al., 2000; Boquet et al., 2000; Wolven et al., 2000; Lu and Pollard, 2001), but monomer sequestration has been invoked in others (Verheyen and Cooley, 1994). Thus, the function of profilin in a cellular context, particularly in differentiating cells of multicellular organisms, remains somewhat unclear.
In this article, we focus on an informative genetic interaction between cpb, which encodes the β subunit of capping protein, and chickadee (chic; encodes profilin). Null mutations in either gene are lethal, but partial loss-of-function alleles are viable and affect bristle development (Verheyen and Cooley, 1994; Hopmann et al., 1996). Both cpb and chic mutant flies have shortened bristles that exhibit bending, branching, and disrupted surface grooves. Previous studies have shown that in both cases, the phenotypes result from an abnormal actin cytoskeleton. Herein, we show that cpb mutations lead to dramatic increases in F-actin levels in the bristle and this is likely to be the underlying cause of the bristle phenotype. We also show that loss-of-function chic mutations suppress the cpb bristle phenotype. To our knowledge, this is the first demonstration of a genetic interaction between these two genes. This interaction is evident at the level of the actin cytoskeleton and suggests that profilin stimulates actin assembly in this context. Our results are consistent with the idea that a balance between profilin and capping protein activity contributes to the regulated assembly of actin that is required for normal bristle elongation and morphogenesis. We discuss possible mechanisms by which profilin might stimulate actin polymerization in this cell, and how the disruption of actin filament dynamics leads to disorganization of the actin bundles and abnormal bristle morphology. This work contributes to the emerging picture of how actin-binding proteins coordinate to correctly organize the actin cytoskeleton in eukaryotic cells.
MATERIALS AND METHODS
Fly Culture and Mutant Stocks
Flies were raised on standard cornmeal medium (Lewis, 1960) at 25°C. The generation of cpb6.15 and cpbF19 was described previously (Hopmann et al., 1996). The chic37 allele was provided by L. Cooley (Yale University, New Haven, CT) and the chic221 allele and UAS-profilin lines were provided by L. Jones (Yale University, New Haven, CT) and L. Cooley. The P [GAL-4] B-11 driver line was provided by J. Merriam (University of California, Los Angles, CA). The green balancer used in this study was CyO, P {GAL4-Kr.C} DC3, P {UAS-GFP.S65T} DC7, hereafter called CyO, green fluorescent protein (GFP). It is available from the Bloomington stock center (Bloomington, IN). All lines used to generate GFP marked clones were also obtained from the Bloomington stock center.
Fly Crosses and Viability Determination
Because cpb and chic reside on chromosome arm 2L, recombinant chromosomes were constructed to link chic alleles to cpbF19. Transheterozygotes were then made with the cpb6.15 allele to generate flies of the experimental genotypes as follows. cpb6.15 shv pr/CyO, GFP females were mated to males from three different lines: cpbF19 cn bw sp/CyO, GFP; cpbF19 chic37 cn/CyO, GFP; or cpbF19 chic221 cn/CyO, GFP. Seven virgin females were premated to three males in a vial for 24 h, then transferred to a bottle and incubated at 25°C. The relative viability of the different genotypes was determined by counting balancer (CyO) and nonbalancer adult progeny between days 11 and 17. If fully viable, the number of experimental (nonbalancer) progeny should be equal to one-half the number of balancer progeny. This expectation is based on the observation that balancer homozygotes never survive to adulthood, and the assumption that balancer heterozygotes are fully viable. The relative adult viability of each experimental genotype was calculated by dividing the number observed by the number expected and converting to percentage.
To overexpress profilin in the developing bristle, w; P [w+ GAL4] B-11/TM3, Sb females were mated to w; l (2)/CyO; P [w+ UAS-chic]/P [w+ UAS-chic] males. The B-11 driver line is an enhancer trap that expresses GAL4 in developing bristles (J. Merriam, unpublished data). Non-Sb progeny were examined for bristle phenotypes and subjected to scanning electron microscopy.
Phenotypic Analysis of Bristles
All analyses were performed on adult flies preserved in a solution of 95% ethanol/glycerol (3:1).
To evaluate the frequency of the abnormal bristle phenotype, flies were scored for the number of bristles on the dorsal head and thorax with major defects, which were defined as sharp bends, branches, or split ends. Flies were classified as having zero, one, or two or more defective bristles.
The length of the posterior sternopleural bristle was measured under the dissecting microscope to the nearest 25 μm as described previously (Hopmann et al., 1996). Both left and right bristles were measured and treated as individual data points. Bristles that were obviously broken were not included. Average length was calculated, and error expressed as ± 1 SD.
Scanning Electron Microscopy
Preserved adult flies were processed for scanning electron microscopy as described previously (Hopmann et al., 1996). After processing, flies were mounted on stubs with carbon-adhesive tabs and carefully grounded with colloidal silver liquid (all materials available from Electron Microscopy Sciences, Fort Washington, PA). Specimens were coated and imaged as described previously (Hopmann et al., 1996).
Preparation and Staining of Pupal Pelts
Pupae of the experimental genotypes were selected after examination under an SZX-12 dissecting microscope equipped with fluorescence (Olympus, Tokyo, Japan). Balancer pupae fluoresce green because of the GFP-expressing transgene present on the CyO, GFP balancer, whereas the nonbalancer, experimental pupae are nonfluorescent. Pupae were raised at 25°C then dissected at 45–47 h after pupariation (AP). This time is near the end of bristle elongation when actin bundles are very prominent. Dissections of the dorsal epithelium were performed essentially as described by Tilney et al. (1998) with minor modifications. Pelts were fixed in 1 ml of 3.7% formaldehyde in PBSTx (1× PBS + 0.01% Triton X-100) for 30 min at room temperature, followed by 3 × 30-min washes with PBSTx.
Pelts were blocked in lectin block (1× PBSTx, 2 mM CaCl2, 5% bovine serum albumin) at least 1 h at room temperature, and then stained overnight at 4°C in 0.5 ml of fluorescein-conjugated Lycopersicon esculentum (tomato) lectin (Fluor-LE; Vector Laboratories, Burlingame, CA) at 20 μg/ml in lectin block. Pelts were washed 3 × 20 min in 1 ml of lectin block, and stained with 0.5 ml of Alexa-568-phalloidin (Molecular Probes, Eugene, OR) at 1.6 U/ml in lectin block. Pelts were washed 3 × 20 min in lectin block, and then mounted in Shandon Immumount (Shandon Southern Instruments, Sewickley, PA). Indirect flight muscles still attached to the pelt were carefully removed during the mounting procedure. Mounted pelts were examined with a laser scanning spectral confocal microscope model TCS SP2 (Leica, Heidelberg, Germany).
Generation of Marked Null Clones in Pupae
The mosaic analysis with a repressible cell marker method for induction of GFP-marked mitotic clones was used to generate cpb4.18 homozygous clones in pupae (Luo et al., 1999; Lee and Luo, 2001). cpb4.18 is a null allele and will be more fully described elsewhere (Hopmann and Miller, unpublished data). In the mosaic analysis with a repressible cell marker system, mutant clones are marked by expression of the mCD8-GFP fusion protein driven by GAL4. Because this GFP fusion is targeted to the plasma membrane, the mutant cells are nicely outlined. Expression of the GFP marker in all other cells is repressed by the presence of GAL80.
The following cross was done to generate animals of the correct genotype for clone induction: y w P [w+ UAS-mCD8-GFP]/Y; cpb4.18 shv P [ry+ FRT] 40A/+; P [w+ tubP-GAL4]/+ males X y w P [ry+ hs70-FLP] 122; P[w+ tubP-GAL80 FRT] 40A/CyO females. Eggs were collected in a food vial for 2–3 d and then adults removed. Developing embryos/larvae in the vial were heat shocked in a 38°C water bath for 2 h, and then allowed to continue development at 25°C. Pupae ∼48 h AP were directly examined under a fluorescence-equipped dissecting microscope for the presence of GFP-expressing clones. Animals with clones were dissected and fixed as described above, and stained with rhodamine-phalloidin as described previously (Hopmann et al., 1996).
RESULTS
Capping protein is αn αβ heterodimer that binds tightly to the barbed ends of actin filaments. Previous screens for mutations in the cpb gene, which encodes the β subunit, yielded two hypomorphic alleles, cpb6.15 and cpbF19 (Hopmann et al., 1996). These alleles partly complement, such that transheterozygotes survive to adulthood, although at reduced frequency. cpb6.15 and cpbF19 alleles seem to express reduced amounts of normal capping protein because 1) transheterozygous (cpb6.15/cpbF19) adults express ∼48% of normal levels of β protein with typical electrophoretic mobility (Hopmann et al., 1996), and 2) sequencing reveals no changes in the coding region of either allele (Hopmann and Miller, unpublished data).
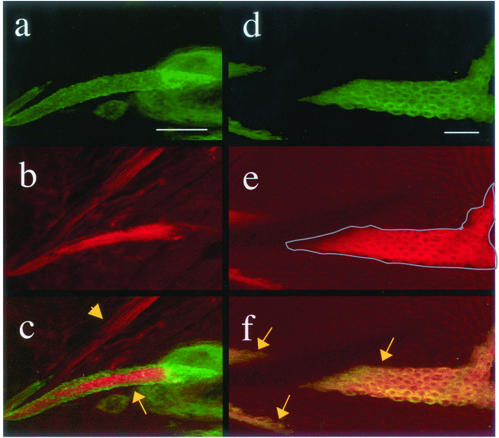
Transheterozygous adults display abnormal bristle morphology that is caused by the disorganization of actin bundles in the developing bristle (Hopmann et al., 1996; see below). Based on the in vitro biochemical properties of capping protein from other species, we predicted that the disruption of actin bundle organization in cpb6.15/cpbF19 bristles resulted from increases in the amount of F-actin caused by a reduction in barbed-end capping activity. To demonstrate that loss of capping protein function does indeed cause increased levels of F-actin, we generated mitotic clones in the pupal epidermis that were homozygous for the null allele cpb4.18. The null mutant cells were marked by the expression of mCD8-GFP, which is targeted to the plasma membrane. Pupae with clones were dissected near the end of bristle elongation, at ∼46 h AP, and then fixed and stained with rhodamine-phalloidin to label F-actin. Three clones were observed that contained elongating macrochaetae. In all three cases, the clonal macrochaetae stained much more intensely with rhodamine-phalloidin than the nonclonal macrochaetae within the same pelt. A representative example is shown in Figure 1, a–c. In this image, a clonal bristle (cpb4.18/cpb4.18), expressing mCD8-GFP (arrow in c), was located near a nonclonal bristle (cpb4.18/+; arrowhead in c) of similar size. Comparison of phalloidin fluorescence intensity between the two bristles showed more F-actin in the cpb null bristle. This difference in actin levels is also very evident in wing clones (Figure 1, d–f). In this series of images, the entire field consists of wing epithelial cells. As in the bristle clones, null cells are marked with mCD8-GFP (d). For clarity, the boundary of the largest clone is outlined in gray in e. Within in the boundary, the cells stain brightly with rhodamine-phalloidin; outside the boundary, the nonclonal cells exhibit much fainter fluorescence. This result indicates that loss of capping protein in wing epithelial cells leads to significant increases in the concentration of F-actin, as it does in bristle cells.
Figure 1.
Loss of capping protein function leads to increased F-actin in pupal epithelial cells. Two elongating bristles from a single pupal pelt are shown (a–c). The bristle expressing the GFP marker is within a mitotic clone homozygous for cpb4.18 (arrow in c), and the unlabeled bristle is cpb4.18/+ (arrowhead in c). Note increased actin labeling in the clonal bristle, which lacks capping protein. F-actin is also elevated in a cpb4.18 pupal wing clone, similarly marked with GFP (d–f). Null clones are marked with arrows, and the largest clone is outlined in e. The rest of the field is filled with nonclonal wing epithelial cells. Clonal cells have elevated F-actin. mCD8-GFP (a and d), actin (b and e), and merge (c and f). Bars, 20 μm.
To study how profilin and capping protein work together to regulate the assembly and organization of bristle actin bundles, we looked for a genetic interaction between mutations in chickadee and cpb. chickadee (chic) is the single gene encoding profilin in flies. Several alleles of chic affect bristle morphology (Verheyen and Cooley, 1994). Some aspects of the chic bristle phenotype are reminiscent of cpb: bristles are shorter and thicker than wild type and are often bent, split, or branched. This phenotype was originally interpreted to result from increased F-actin levels, similar to cpb mutants. However, chic bristles do not display groove patterns that are as aberrant as cpb bristles. This difference corresponds to subtle differences in the actin bundle phenotypes of chic and cpb. Although chic mutant bristles have more numerous and thinner actin bundles, somewhat like cpb, the actin bundles as not as disorganized as those in cpb bristles (Verheyen and Cooley, 1994).
Given that cpb and chic mutations affect actin bundle morphology in the bristle, and the encoded proteins are known to regulate actin assembly, it seemed likely that the two genes would have a genetic interaction. We used enhancement or suppression of the cpb bristle phenotype as an assay to test the following hypotheses: if profilin's primary role in the bristle is to sequester monomer and inhibit actin polymerization, then reduction of profilin would lead to increased amounts of F-actin, and chic mutations would enhance the cpb bristle phenotype. Conversely, if profilin's main function is to stimulate actin polymerization, we would expect chic mutations to suppress the cpb bristle phenotype. Although it could be argued that this rationale is based on an oversimplified view of these proteins' functions, the results of this analysis were indeed informative.
Because null alleles of capping protein are homozygous lethal, we based our genetic interaction studies on the bristle phenotype of transheterozygous cpb6.15/cpbF19 animals. We reduced profilin levels by introducing chic alleles into this background. Two alleles of chic were used. chic37 is a hypomorphic allele that is caused by a small deletion in the 5′ untranslated region. When homozygous, chic37 reduces (but probably does not eliminate) profilin expression in the bristle and is associated with a strong bristle phenotype. chic221 is a null, homozygous lethal allele caused by a larger deletion removing part of the 5′ untranslated region as well as a large portion of the coding region (Verheyen and Cooley, 1994).
Reduction of profilin strongly suppressed multiple aspects of the cpb6.15/cpbF19 phenotype. First, heterozygosity for either chic allele improved the viability of cpb6.15/cpbF19 transheterozygotes (Table 1). cpb6.15/cpbF19 adults were observed at 43% of the expected frequency at 17 d from mating. Heterozygosity for either chic allele improved survival significantly (76% for chic37, p < 0.005; 75% for chic221, p < 0.005). Second, a developmental delay associated with loss of capping protein function was suppressed. At 13 d after mating, cpb6.15/cpbF19 adults were observed at 20% of the expected frequency, compared with 43% at day 17. Heterozygosity for chic caused the developmental delay to be less pronounced: 60% for chic37, 67% for chic221 at day 13 vs. 76 or 75% at day 17. The suppression of cpb effects by chic suggests that capping protein and profilin have opposing functions in Drosophila development.
Table 1.
Reduced profilin levels suppress cpb semilethality
| No. CyO observed | No. non-CyO expected | No. non-CyO observed | Relative adult viability, %a | |
|---|---|---|---|---|
| cpb6.15+/CyO × cpbF19+ /CyO | 208 | 104 | 45 | 43 (n = 253) |
| cpb6.15+/CyO × cpbF19 chic37/CyO | 542 | 271 | 206 | 76 (n = 748) |
| cpb6.15+/CyO × cpbF19 chic221/CyO | 604 | 302 | 228 | 75 (n = 832) |
No. observed/no. expected × 100.
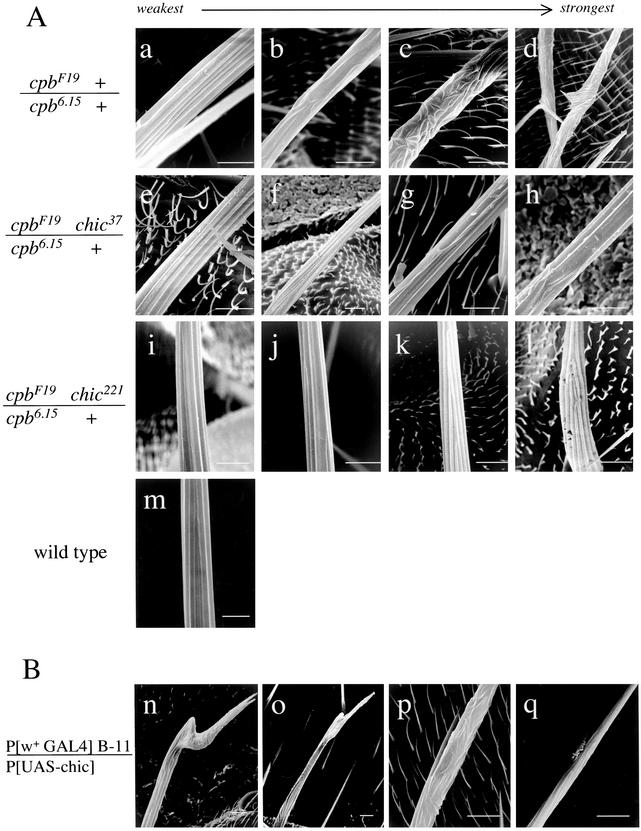
Even more striking were the effects of chic heterozygosity on the cpb bristle phenotype. cpb6.15/cpbF19 adults have a moderate bristle phenotype characterized by bending, branching, or splitting of some of the large bristles (macrochaetae). Furthermore, the cuticular surfaces of many macrochaetae seem knobby or rough. The roughening is caused by the disruption of longitudinal grooves seen in normal adult bristles (Hopmann et al., 1996) and Figure 2A). Rare cpb6.15 homozygous adult escapers have a similar bristle phenotype (Hopmann, unpublished data). In the adult, the bending, splitting and abnormal groove patterns seen in cpb6.15/cpbF19 macrochaetae were dramatically suppressed by both chic alleles. Figure 2A, a–d, shows a typical range of phenotypes of cpb6.15/cpbF19 bristles, demonstrating that although some macrochaetae were strongly affected and others mildly, all were affected to some degree. For cpb6.15 + /cpbF19 chic37, a similar phenotypic series (Figure 2, e–h) shows that weakly affected macrochaetae (Figure 2, e) were nearly indistinguishable from wild type (Figure 2, m), and that the most strongly affected macrochaetae (Figure 2, g and h) were comparable with the least affected cpb6.15/cpbF19 macrochaetae. The chic221 allele exhibited an even greater degree of phenotypic suppression (Figure 2, i–l). The majority of macrochaetae in this genotype seemed completely normal (compare with wild-type, Figure 2, m).
Figure 2.
Adult bristle phenotypes of cpb and chic mutants are consistent with opposite effects on actin polymerization. (A) cpb bristle phenotype is suppressed by chic mutations. Four scanning electron micrographs of macrochaetae from each genotype are shown to illustrate the range of phenotypic severity, from weakest (a, e, and i) to strongest (d, h, and l). Genotypes are as follows: cpbF19/cpb6.15 (a–d), cpbF19 chic37/cpb6.15 (e–h), cpbF19 chic221/cpb6.15 (i–l), and wild type (m). Bars, 10 μm. (B) Overexpression of profilin in the bristle causes a cpb-like phenotype. Four examples of affected macrochaetae from B-11 UAS-chic adults are shown (n–q), ordered from weakest to strongest phenotype with respect to the groove pattern. Bars, 10 μm.
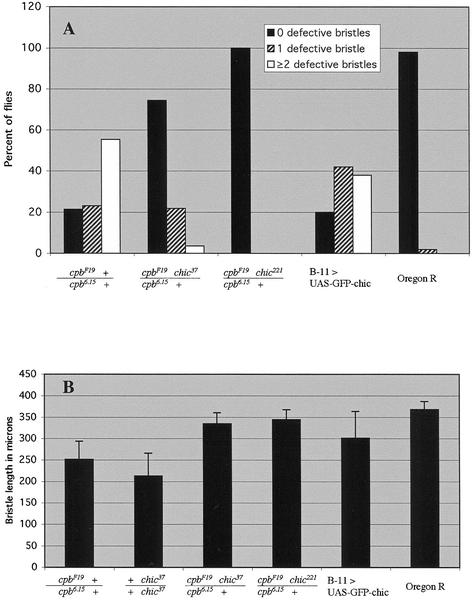
We quantified these differences in bristle phenotype in two ways. First, for each genotype we counted the number of bristles on the dorsal thorax and head that exhibited major defects (Figure 3A). Major defects were defined as sharp bends, branches, or split ends that could be easily seen under the dissecting microscope. Each fly was categorized as having zero, one, or two or more defective bristles. Although 78% of cpb6.15/cpbF19 flies had at least one major bristle defect, the presence of either chic allele greatly reduced the frequency of bristle defects. 26% of cpb6.15 + /cpbF19 chic37 flies exhibited major bristle defects, and for cpb6.15 + /cpbF19 chic221 no major defects were observed. The difference in effect of the two chic alleles correlated with the relative strength of the alleles (hypomorph vs. null) and could also be discriminated in more subtle aspects of the bristle phenotype. When viewed at low magnification, both cpb6.15/cpbF19 and cpb6.15 + /cpbF19 chic37 flies exhibited minor defects such as rough, knobby bristles, although this was less pronounced in the latter genotype. In contrast, cpb6.15 + /cpbF19 chic221 bristles seemed completely normal.
Figure 3.
cpb bristle phenotype is suppressed by chic. (A) Frequency of major bristle defects. Flies were categorized as having zero defective bristles (black bars), one defective bristle (striped bars), or two or more defective bristles (white bars). The raw data was converted to percentage of the total number of flies of each genotype. Complete genotypes are as in Figure 1. Sample sizes: cpb6.15/cpbF19 (n = 56), cpb6.15/cpbF19 chic37 (n = 55), cpb6.15/cpbF19 chic221 (n = 15), B-11 UAS-chic (n = 50), and Oregon R (wild type, n = 50). (B) Length of sternopleural bristle. Sample sizes: cpb6.15/cpbF19 (n = 30), chic37/chic37 (n = 28), cpb6.15/cpbF19 chic37 (n = 28), cpb6.15/cpbF19 chic221 (n = 28); B-11 UAS-chic (n = 43); and Oregon R (wild type, n = 22).
We also quantified the differences in the bristle phenotype by measuring the length of one particularly long bristle, the posterior sternopleural bristle, in the various genotypes. We have shown previously that reduction of capping protein concentration in cpb6.15/cpbF19 flies resulted in a significant shortening of this particular bristle compared with wild type (Hopmann et al., 1996). In the current experiment (Figure 3B), the average length of the sternopleural bristle in Oregon R flies was 368 vs. 252 μm in cpb6.15/cpbF19, an ∼30% reduction. The sternopleural bristle was even shorter in chic37 homozygotes, averaging 213 μm. However, in the background of cpb6.15/cpbF19, heterozygosity for either chic allele restored the sternopleural bristle to nearly wild-type length.
The suppression of cpb mutant phenotypes by chic suggests that profilin functions antagonistically to capping protein. Therefore, we predicted that overexpression of profilin in the bristle is likely to cause a phenotype similar to cpb loss-of-function. We tested this by combining a UAS-chic transgene to a GAL4 enhancer trap line (designated B-11) that is expressed in the elongating bristle shaft. B-11 UAS-chic pupae overexpress profilin in the bristle shaft but not in most other tissues. Adults of this genotype exhibited a strong bristle phenotype that seemed very similar to the cpb loss-of-function phenotype (Figure 2B, n–q). Macrochaetae were often bent or split at the ends, or had slender barbs branching off. This bristle phenotype was quantified in the same manner as the cpb loss of function genotypes. The frequency of major bristle defects was very similar to cpb6.15/cpbF19 (Figure 3A). Eighty percent of the B-11 UAS-chic adults had at least one abnormal bristle, compared with 78% for cpb6.15/cpbF19. Likewise, bristle length was also decreased in B-11 UAS-chic adults, although this phenotype was variable (Figure 3B). The mean length of the posterior sternopleural bristle was 301 μm, but individual bristle length ranged widely. Most strikingly, groove patterns were highly disorganized in the profilin over expressing bristles, implying the actin cytoskeleton is disorganized as it is in cpb mutants. This suggests that an increase of profilin has similar consequences as a reduction of capping protein.
It is worth noting that profilin loss of function also leads to severe bristle phenotypes that at a superficial level seem similar to the profilin overexpression phenotype (Verheyen and Cooley, 1994, Figure 7, for bristle phenotype of chic37 homozygotes). Specifically, both conditions cause bristles to be shorter and have abnormal morphology. However, analysis of scanning electron micrographs points out clear differences (compare Verheyen and Cooley, 1994, Figure 7, to Figure 2B, m–p). chic37 bristles are extremely short and highly branched, and the surface grooves are very visible and oriented longitudinally. Conversely, B-11 UAS-chic bristles are only slightly shorter than wild type, and never display the high degree in branching seen in chic37. However, the surface grooves are often shallow and very disorganized, especially in the distal half of the bristle. In all respects, the effects of profilin overexpression in the bristle are much more similar to capping protein loss of function than they are to profilin loss of function.
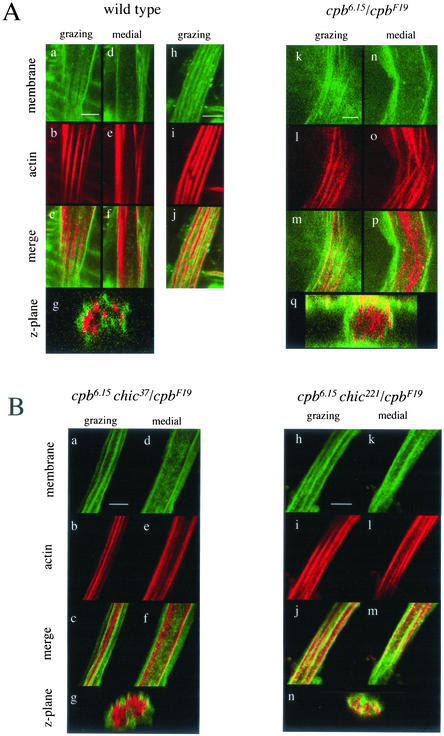
Previously, we noted that in cpb6.15/cpbF19 bristle cells the association of actin bundles with the plasma membrane seen in normal bristles is partially disrupted (Hopmann et al., 1996). To look at this more carefully we stained pupal pelts with a fluorescein-conjugated lectin from tomato (Fluor-LE) to label the plasma membrane in addition to Alexa-Fluor-568-phalloidin to label the actin bundles. In grazing longitudinal sections of wild-type bristles, actin bundles were visible as longitudinal stripes (Figure 4A, a–c). In medial longitudinal sections (Figure 4A, d–f) and z-planes extracted from a confocal series (cross sections perpendicular to longitudinal axis; Figure 4A, g), actin bundles were present at the perimeter of the cell and not in the interior. In addition, the membrane glycoproteins(s) bound by the lectin probe were excluded from the membrane domains overlying the actin bundles. In favorable images (Figure 4A, h–j), this caused the lectin labeling also to seem striped in grazing sections, and in the merged image the actin and lectin stripes were interdigitated. This alternating pattern was also evident in z-planes (Figure 4A, g) in which actin spots alternated with higher concentrations of lectin staining. In cpb6.15/cpbF19 bristle cells, the association between the actin bundles and the plasma membrane was partly disrupted such that actin bundles were visible in the interior of the bristle shaft. Although in grazing section some actin bundles were still associated with the membrane (Figure 4A, k–m), a medial section of the same bristle cell revealed many actin bundles in the center of the bristle shaft (Figure 4A, n–p). The displacement of actin bundles in the cpb mutant is also apparent in z-planes (Figure 4A, q). The disorganization of the actin cytoskeleton was reflected in the lectin staining. Although there was some striping visible in grazing sections (Figure 4A, k and m) it was not as striking as in wild type nor as well organized relative to actin. In contrast, chic37 homozygotes do not demonstrate displacement of the bristle actin bundles (our unpublished data).
Figure 4.
chic suppresses actin bundle disorganization and displacement in cpb pupal bristles. (A) cpb loss-of-function causes bristle actin bundles to become disorganized and displaced from the plasma membrane. Confocal sections of pupal bristles are from Oregon R (wild type, a–j); cpbF19 cn bw sp/cpb6.15 shv pr (k–q). Membrane was labeled with fluorescein-conjugated tomato lectin (Fluor-LE; a, d, h, k, and n) and actin was labeled with Alexa-568-phalloidin (b, e, i, l, and o). Merged images are also shown (c, f, j, m, and p). Grazing (a–c, h–j, k–m) and medial (d–f, n–p) sections are single confocal xy-planes. Z-plane cross sections (g and q) are extracted from series of xy-planes spaced 0.15 μm. For Oregon R, the z-plane is derived from a different bristle than that shown in a–f. Bars, 4 μm. (B) chic suppresses the actin bundle abnormalities of cpb pupal bristles. cpb6.15 shv pr/cpbF19 chic37 cn (a–g); cpb6.15 shv pr/cpbF19 chic221 cn (h–n). Membrane (a, d, h, and k), actin (b, e, i, and l), merge (c, f, j, and m), and z-plane merge (g and n). Bars, 4 μm.
Consistent with the adult bristle phenotypes, the disorganization of actin bundles seen in cpb6.15/cpbF19 pupal macrochaetae was suppressed by chic mutations (Figure 4B). Actin bundles in cpb6.15 + /cpbF19 chic37 (Figure 4B, a–f) and cpb6.15 + /cpbF19 chic221 (Figure 4B, h–m) bristles were more uniform in thickness and better organized than in cpb6.15/cpbF19. In fact, in most macrochaetae the actin staining was very similar to wild type. In grazing sections (Figure 4B, a–c, h–j), actin bundles were well organized and in medial sections (Figure 4B, d–f, k–m) actin bundles were mostly absent from the center of the bristle shaft and located at the cell periphery. Lectin staining was striped and alternated with actin bundles. Z-planes (Figure 4B, g and n) emphasize that both chic alleles restored association of the actin bundles with the membrane although the degree of suppression seemed greater for the chic221 allele.
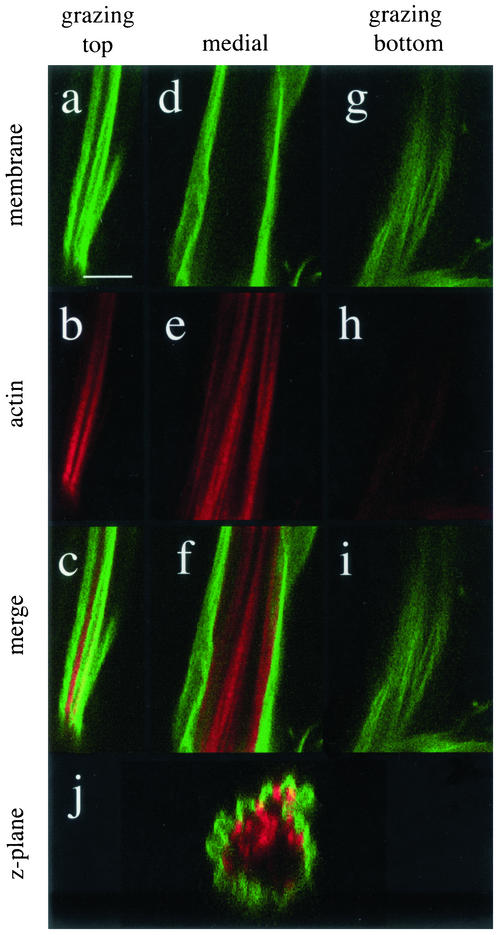
If the adult phenotype of bristles overexpressing profilin truly reflects a similar mechanism as the cpb loss of function, one would expect the phenotype of actin bundles in the pupal bristle to resemble cpb. To test this, P[GAL4] B-11/P [UAS-chic] pupae were stained for actin and membrane as described above (Figure 5). The actin bundle phenotype seen in these pupae was strikingly similar to the cpb transheterozygote. Actin bundles were more numerous and heterogeneous and were partially displaced from the plasma membrane. Although the top grazing section (a–c) showed bundles still opposed to the membrane and striped lectin labeling, the medial section (d–f) revealed bundles in the center and the bottom grazing section (g–i) showed less organized membrane labeling and no actin bundles. The z-plane (j) further illustrates how bundles from one side of the bristle seemed to be mislocalized in the center of the bristle shaft. This displacement of actin bundles from the membrane is very similar to that seen in cpb loss of function mutants, and very different from defects seen in chic loss of function mutants.
Figure 5.
Profilin overexpression mimics capping protein loss of function effects on bristle actin bundles. Confocal sections from a P[GAL4] B-11/P [UAS-chic] pupa. Membrane was labeled with Fluor-LE (a, d, and g) and actin with Alex-568-phalloidin (b, e, and h), merge (c, f, and i), and z-plane (j). Bars, 4 μm.
DISCUSSION
Capping protein loss of function leads to dramatic increases in F-actin in the fly bristle, resulting in aberrant organization of the actin cytoskeleton. Reduction of profilin suppresses the disorganized actin phenotype caused by reduction of capping protein function, suggesting that profilin promotes actin assembly in the elongating bristle. These results emphasize the idea that the balance of activities of actin-binding proteins is critical for assembling actin structures that are organized and positioned properly.
Numerous studies have demonstrated the importance of the actin cytoskeleton for the normal elongation and morphogenesis of the fly bristle. Tilney et al. (2000a) showed that inhibitors of actin polymerization significantly decreased the elongation rates of bristles whereas inhibitors of microtubule polymerization had little effect. The morphology of bristle actin bundles is affected by changes in the amount of cross-linking proteins (Tilney et al., 2000b) as well as mutations in genes that encode regulators of actin dynamics, including ADF/cofilin (twinstar; Gunsalus et al., 1995; Chen et al., 2001), twinfilin (Wahlstrom et al., 2001), and ADF/cofilin phosphatase (slingshot; Niwa et al., 2002). Yet many of these alterations do not cause severely displaced and disoriented actin bundles. In contrast, mutations in capping protein strongly affect not only the amount of F-actin but also the position and orientation of actin structures. In this regard, the phenotype of twinfilin (twf) mutants is particularly noteworthy. Twinfilin is a monomer-sequestering protein that is structurally related to ADF/cofilin (Palmgren et al., 2002). In twf mutant bristles, F-actin levels are increased and the actin bundles are very disorganized, like they are in cpb mutants. Furthermore, the actin bundles show the same dramatic displacement from the membrane in twf as they do in cpb. This contrasts with the phenotype of chic bristles, which do not show displacement of bundles, and underscores the fact that although twinfilin and profilin both have sequestering activity in vitro, they clearly have different roles in vivo.
What the analysis of individual mutant phenotypes does not tell us is how the different actin regulatory proteins work together to generate normal actin bundles. Our analysis of cpb chic double mutants demonstrates this clearly. Because the original phenotypic characterization of cpb and chic single mutants suggested that they both led to increased levels of F-actin, our original expectation was that chic loss of function would enhance cpb loss of function. Instead, we observed the opposite effect. This approach has yielded valuable insights regarding the importance of the balance of capping protein and profilin activities in normal cells. In other cases, mutant combinations do exhibit predictable phenotypes. For example, double heterozygous combinations of twf and tsr, which encodes ADF/cofilin, exhibit a moderate bristle phenotype even though the single mutant heterozygotes show little or no bristle phenotype (Wahlstrom et al., 2001). This is consistent with the proposed function of both proteins: reduction of twinfilin leads to increases in F-actin assembly due to reduced sequestering activity, and reduction of ADF/cofilin leads to a decreased rate of actin depolymerization. Thus, it is expected that the two mutations behave synergistically and cause an increase in F-actin. We anticipate that additional mutant combinations will be equally informative about the complex interplay of activities required to construct normal actin bundles, at present, formulating a model that incorporates the many different actin regulators is difficult because there is limited data of this type available.
Our results support the idea that profilin has polymerization-promoting activity, as demonstrated by previous work. Expression of vertebrate or plant profilins in mammalian tissue culture cells led to increases in F-actin (Finkel et al., 1994; Rothkegel et al., 1996) and profilin null clones in the developing Drosophila eye exhibited greatly reduced levels of F-actin (Benlali et al., 2000).
However, the observation that profilin acts in an opposite manner to capping protein, seeming to stimulate actin polymerization in the fly bristle seems at first difficult to reconcile with the original characterization of the chic bristle phenotype. In chic mutants, the elongating bristle seemed to have an increased number of actin bundles that were thinner than wild-type bundles (Verheyen and Cooley, 1994). This phenotype was thought to reflect an overall increase in the amount of F-actin, which is consistent with a monomer-sequestering role for profilin. We suggest two possible explanations for this seeming paradox. First, biochemical data on profilin activity have shown that its activity is dependent on the state of the barbed ends. Profilin-actin can add to free barbed ends but not to capped ones (Pollard and Cooper, 1984; Pantaloni and Carlier, 1993; Kang et al., 1999). Thus, in wild-type bristles, barbed ends may be maximally capped (except at the growing tip) and profilin's primary function would be to sequester monomer. In a chic mutant bristle, reduction in profilin-mediated sequestering activity might lead to the observed increase in F-actin. We would then predict that when capping protein is reduced, barbed ends are not maximally capped and thus, profilin's polymerization-promoting activity would predominate, which is consistent with our observations.
Another interpretation of the chic bristle phenotype is suggested by the results of inhibitor studies performed on cultured Drosophila pupae (Tilney et al., 2000a; Guild et al., 2002). Exposure of cultured pupae to cytochalasin D, an inhibitor of actin polymerization, causes the actin bundles in elongating bristles to fall apart by splitting into thinner subbundles, reminiscent of chic mutant bristles that exhibit an increased number of thinner bundles. The similarity of these two phenotypes suggests that continued actin polymerization is required to maintain the integrity of actin bundles, and reductions in actin polymerization cause the actin bundles to “unravel.” Although it is clear that profilin can promote actin polymerization, the mechanism by which it does this is less well understood. Studies in yeast have demonstrated that profilin's nucleotide exchange activity is required for its function (Wolven et al., 2000; Lu and Pollard, 2001). Because ATP-actin is more readily incorporated onto barbed ends of filaments (Pollard, 1986), this activity can explain profilin's effects on actin assembly (Blanchoin and Pollard, 1998; Didry et al., 1998). However, there is reason to believe Drosophila profilin may not work this way. Plant profilins do not catalyze nucleotide exchange (Perelroizen et al., 1996; Eads et al., 1998), and some even seem to repress it (Kovar et al., 2001). A comprehensive mutational analysis of profilin in fission yeast (Schizosaccharomyces pombe) by Lu and Pollard (2001) has identified tyrosine79 as critical to its ability to stimulate nucleotide exchange. When tyrosine79 is mutated to arginine, S. pombe profilin loses its exchange activity. Notably, the majority of plant profilins naturally contain arginine at the comparable position, whereas all characterized vertebrate profilins, which tend to have very high exchange activity, contain aspartate. Thus, there is a correlation between arginine at position 79 and low activity, tyrosine and moderate activity, and aspartate and high activity. Interestingly, Drosophila has arginine: it is the only nonplant profilin, besides that of shrimp, known to have arginine at this position (Lu and Pollard, 2001). The exchange activity of Drosophila profilin is unknown, but it seems reasonable to predict that Drosophila profilin has low activity.
Although plant profilins do not enhance nucleotide exchange by actin monomers, some stimulate actin polymerization in vitro in thymosin-β4/actin solutions (Perelroizen et al., 1996). Thymosin-β4 is a true monomer sequestering protein in that T-β4-actin cannot add to a growing filament, whereas profilin-actin adds readily to the barbed ends of actin filaments. Profilin is thought to shuttle monomer out of the T-β4 pool (Pantaloni and Carlier, 1993), and this may be the relevant mechanism in other cell types. Studies in Drosophila may prove useful in elucidating the details as well as the physiological relevance of alternate mechanisms of profilin activity.
In this article, as well as previous work we have demonstrated that a reduction of capping protein function leads to increased F-actin and abnormal actin organization. It is likely that the aberrant actin cytoskeleton underlies all of the defects observed in the adult bristle such as decreased length, bending, branching, and abnormal groove patterns. Although some of the correlations between the actin abnormalities and adult phenotypes are fairly obvious, it may seem counterintuitive that increases in F-actin levels would lead to shorter bristles. One might expect increased F-actin polymerization to give rise to longer bristles. Indeed, Tilney et al. (2000a,b) have shown that treatment of cultured pupae with jasplakinolide, a drug that stabilizes F-actin, increases the growth rate of the bristle shaft. However, their experiments were done for 6–7 h, whereas bristle elongation takes ∼16 h at 25°C. Perhaps the increased growth rate would not be maintained were it possible to expose the growing bristle to drug for the entire elongation period. We hypothesize that in cpb mutants, actin is overpolymerized at the beginning of bristle elongation. Some component required for actin bundle assembly may be limiting in the bristle (Tilney et al., 2000b); therefore, in a cpb mutant bristle, the limiting component would be prematurely depleted due to the increase in F-actin. Comparing the growth rates of wild-type and mutant bristles can test this idea.
Although our data demonstrate that reduction of capping protein function leads to increases in F-actin, we have not quantified these changes. It would have been desirable to measure the concentrations of F-actin in the various mutant genotypes directly, but technical limitations prevented us from doing so in a controlled manner. Phalloidin staining often varies greatly between experiments, so the subtle differences we might expect to see in different genotypes could be obscured. We are currently developing more quantitative methods for measuring actin in situ.
One of the most puzzling features of the cpb mutant phenotype is the displacement of actin bundles from the membrane. An increase in the amount of F-actin in the bristle does not, by itself, seem to explain this phenotype. In bristles where the cross-linking protein fascin is overexpressed, F-actin amounts are increased and bundles are considerably larger, but they do not show significant displacement from the membrane (Tilney et al., 2000b). A structural function of capping protein in physically linking the bundles to the plasma membrane would explain this phenotype. Previous studies in chicken myoblasts have uncovered a structural requirement for capping protein in organizing actin filaments within the sarcomere (Schafer et al., 1995). However, a structural role seems unlikely given that the displacement of bundles is suppressed when profilin dosage is reduced. Instead, the proper regulation of actin assembly may be important for the positioning of actin bundles. As noted above, twf mutant bristles also exhibit this displacement phenotype. Because capping protein and twinfilin are known to associate in yeast (Palmgren et al., 2001), this raises the interesting possibility that these two proteins work together in regulating actin assembly such that the association of bundles with the membrane is established and/or maintained. Intriguingly, treatment of cultured pupae with okadaic acid, an inhibitor of protein phosphatases, causes a similar displacement of actin bundles (Tilney et al., 2000b), suggesting the phosphorylation status of one or more proteins may be relevant.
In this article, we have shown that the balanced activities of capping protein and profilin are essential in the regulation of actin dynamics and organization in the elongating Drosophila bristle. Our data are consistent with the emerging idea that the activity of profilin is context dependent, and that in many cells, profilin promotes actin assembly. Our data also suggest that perturbations of actin dynamics in the bristle lead to a striking displacement of actin bundles from the membrane. In the future, we hope to clarify the role of capping protein in the bristle and better understand how it is integrated with the many other actin regulators functioning in the bristle such that actin bundles are correctly assembled and positioned.
ACKNOWLEDGMENTS
We are grateful to Lynn Cooley and Lynn Jones for the UAS-chic transgene and chic221 allele, and John Merriam for the B-11 driver, as well as the Bloomington stock center for numerous stocks. We thank John Cooper, Melissa Kramer, Magdalena Benzanilla, Phil Harries, Debbie Frank, Tatsuhiko Noguchi, and Aaron Rogat for comments on the manuscript, and Ian Duncan for helpful discussions. We also thank Linda Drury, Lorie Weishaar, and Julie Morrison for technical assistance, and Mike Veith for help with microscopy. Finally, we thank the two reviewers of this manuscript for their exceptionally pertinent and constructive criticisms. This work was supported in its early stages by a grant from the National Science Foundation, and subsequently by a grant from the American Heart Association.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0300. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0300.
REFERENCES
- Appel LF, Prout M, Abu-Shumays R, Hammonds A, Garbe JC, Fristrom D, Fristrom J. The Drosophila Stubble-stubbloid gene encodes an apparent transmembrane serine protease required for epithelial morphogenesis. Proc Natl Acad Sci USA. 1993;90:4937–4941. doi: 10.1073/pnas.90.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlali A, Draskovic I, Hazelett DJ, Treisman JE. Act up controls actin polymerization to alter cell shape and restrict Hedgehog signaling in the Drosophila eye disc. Cell. 2000;101:271–281. doi: 10.1016/s0092-8674(00)80837-5. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J Biol Chem. 1998;273:25106–25111. doi: 10.1074/jbc.273.39.25106. [DOI] [PubMed] [Google Scholar]
- Boquet I, Boujemaaa R, Carlier M F, Preat T. Ciboulot regulates actin assembly during Drosophila brain metamorphosis. Cell. 2000;102:797–808. doi: 10.1016/s0092-8674(00)00068-4. [DOI] [PubMed] [Google Scholar]
- Carlsson L, Nystrom LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- Casella JF, Maack DJ, Lin S. Purification and initial characterization of a protein from skeletal muscle that caps the barbed ends of actin filaments. J Biol Chem. 1986;261:10915–10921. [PubMed] [Google Scholar]
- Chen J, Godt D, Gunsalus K, Kiss I, Goldberg M, Laski FA. Cofilin/ADF is required for cell motility during Drosophila ovary development and oogenesis. Nat Cell Biol. 2001;3:204–209. doi: 10.1038/35055120. [DOI] [PubMed] [Google Scholar]
- Didry D, Carlier M-F, Pantaloni D. Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J Biol Chem. 1998;273:25602–25611. doi: 10.1074/jbc.273.40.25602. [DOI] [PubMed] [Google Scholar]
- Eads JC, Mahoney NM, Vorobiev S, Bresnick AR, Wen KK, Rubenstein PA, Haarer BK, Almo SC. Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry. 1998;37:11171–11181. doi: 10.1021/bi9720033. [DOI] [PubMed] [Google Scholar]
- Finkel T, Theriot J A, Dise K R, Tomaselli G F, Goldschmidt-Clermont P J. Dynamic actin structures stabilized by profilin. Proc Natl Acad Sci USA. 1994;91:1510–1514. doi: 10.1073/pnas.91.4.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guild GM, Connelly PS, Vranich KA, Shaw MK, Tilney LG. Actin filament turnover removes bundles from Drosophila bristle cells. J Cell Sci. 2002;115:641–653. doi: 10.1242/jcs.115.3.641. [DOI] [PubMed] [Google Scholar]
- Gunsalus KC, Bonaccorsi S, Williams E, Verni F, Gatti M, Goldberg ML. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann R, Cooper JA, Miller KG. Actin organization, bristle morphology, and viability are affected by actin capping protein mutations in Drosophila. J Cell Biol. 1996;133:1293–1305. doi: 10.1083/jcb.133.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg G, Aebi U, Pollard TD. An actin-binding protein from Acanthamoeba regulates actin filament polymerization and interactions. Nature. 1980;288:455–459. doi: 10.1038/288455a0. [DOI] [PubMed] [Google Scholar]
- Kang F, Purich DL, Southwick FS. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J Biol Chem. 1999;274:36963–36972. doi: 10.1074/jbc.274.52.36963. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Yang P, Sale WS, Drobak BK, Staiger CJ. Chlamydomonas reinhardtii produces a profilin with unusual biochemical properties. J Cell Sci. 2001;114:4293–4305. doi: 10.1242/jcs.114.23.4293. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A new standard food medium. Drosoph Inf Serv. 1960;34:117–118. [Google Scholar]
- Lu J, Pollard TD. Profilin binding to poly-l-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Lee T, Nardine T, Null B, Reuter J. Using the MARCM system to positively mark mosaic clones in Drosophila. Drosoph Inf Serv. 1999;82:102–105. [Google Scholar]
- Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- Overton J. The fine structure of developing bristles in wild type and mutant Drosophila melanogaster. J Morphol. 1967;122:367–379. doi: 10.1002/jmor.1051220406. [DOI] [PubMed] [Google Scholar]
- Palmgren S, Ojala PJ, Wear MA, Cooper JA, Lappalainen P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J Cell Biol. 2001;155:251–260. doi: 10.1083/jcb.200106157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren S, Vartiainen M, Lappalainen P. Twinfilin, a molecular mailman for actin monomers. J Cell Sci. 2002;115:881–886. doi: 10.1242/jcs.115.5.881. [DOI] [PubMed] [Google Scholar]
- Pantaloni D, Carlier MF. How profilin promotes actin filament assembly in the presence of thymosin beta 4. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- Perelroizen I, Didry D, Christensen H, Chua NH, Carlier MF. Role of nucleotide exchange and hydrolysis in the function of profilin in action assembly. J Biol Chem. 1996;271:12302–12309. doi: 10.1074/jbc.271.21.12302. [DOI] [PubMed] [Google Scholar]
- Pollard T. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 1984;23:6631–6641. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Rothkegel M, Mayboroda O, Rohde M, Wucherpfennig C, Valenta R, Jockusch BM. Plant and animal profilins are functionally equivalent and stabilize microfilaments in living animal cells. J Cell Sci. 1996;109:83–90. doi: 10.1242/jcs.109.1.83. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Hug C, Cooper JA. Inhibition of CapZ during myofibrillogenesis alters assembly of actin filaments. J Cell Biol. 1995;128:61–70. doi: 10.1083/jcb.128.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Connelly PS, Vranich KA, Shaw MK, Guild GM. Why are two different cross-linkers necessary for actin bundle formation in vivo and what does each cross-link contribute? J Cell Biol. 1998;143:121–133. doi: 10.1083/jcb.143.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L, Connelly P, Vranich K, Shaw M, Guild G. Actin filaments and microtubules play different roles during bristle elongation in Drosophila. J Cell Sci. 2000a;113:1255–1265. doi: 10.1242/jcs.113.7.1255. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Connelly PS, Vranich KA, Shaw MK, Guild GM. Regulation of actin filament cross-linking and bundle shape in Drosophila bristles. J Cell Biol. 2000b;148:87–100. doi: 10.1083/jcb.148.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development. 1994;120:717–728. doi: 10.1242/dev.120.4.717. [DOI] [PubMed] [Google Scholar]
- Wahlstrom G, Vartiainen M, Yamamoto L, Mattila PK, Lappalainen P, Heino TI. Twinfilin is required for actin-dependent developmental processes in Drosophila. J Cell Biol. 2001;155:787–796. doi: 10.1083/jcb.200108022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolven AK, Belmont LD, Mahoney NM, Almo SC, Drubin DG. In vivo importance of actin nucleotide exchange catalyzed by profilin. J Cell Biol. 2000;150:895–904. doi: 10.1083/jcb.150.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]