Abstract
We explored transcriptional responses of the fission yeast Schizosaccharomyces pombe to various environmental stresses. DNA microarrays were used to characterize changes in expression profiles of all known and predicted genes in response to five stress conditions: oxidative stress caused by hydrogen peroxide, heavy metal stress caused by cadmium, heat shock caused by temperature increase to 39°C, osmotic stress caused by sorbitol, and DNA damage caused by the alkylating agent methylmethane sulfonate. We define a core environmental stress response (CESR) common to all, or most, stresses. There was a substantial overlap between CESR genes of fission yeast and the genes of budding yeast that are stereotypically regulated during stress. CESR genes were controlled primarily by the stress-activated mitogen-activated protein kinase Sty1p and the transcription factor Atf1p. S. pombe also activated gene expression programs more specialized for a given stress or a subset of stresses. In general, these “stress-specific” responses were less dependent on the Sty1p mitogen-activated protein kinase pathway and may involve specific regulatory factors. Promoter motifs associated with some of the groups of coregulated genes were identified. We compare and contrast global regulation of stress genes in fission and budding yeasts and discuss evolutionary implications.
INTRODUCTION
All cells sense and react to changes in their environment. Single-celled organisms, in particular, must contend with fluctuations in nutrients, pH, temperature, and external osmolarity, as well as exposure to UV irradiation and a range of potentially toxic environmental compounds. Appropriate responses to these environmental stresses must be induced for cell survival and proliferation. A comprehensive characterization of these responses, the mechanisms involved in sensing stress, the signaling pathways transmitting this information within the cell, and the resulting compensatory changes in physiology and gene expression, is essential in understanding how cells adapt and survive under nonideal conditions.
Exposure to low levels of stress often triggers an adaptive response resulting in a transient resistance to higher levels of the same stress. This adaptation to stress can also lead to increased resistance (or cross-protection) to other types of stress (Jamieson, 1992; Lee et al., 1995; Moradas-Ferreira and Costa, 2000). The adaptive response is short-lived and requires new protein synthesis, indicating that changes in gene expression are critical. The phenomenon of cross-protection suggests either that different stress conditions can activate similar defense mechanisms or, more broadly, that there is a general stress response that can confer a basic level of protection.
The advent of DNA microarrays (Shalon et al., 1996) allows comprehensive analyses of changes in gene expression that accompany stress responses. Whole genome expression profiling studies have revealed general responses to stress in the budding yeast Saccharomyces cerevisiae (Gasch et al., 2000; Causton et al., 2001). These studies have found that 10% to 14% of all genes are induced or repressed in response to a wide range of stresses. Induced genes are involved in various processes, including carbohydrate metabolism, detoxification of reactive oxygen species, protein folding and degradation, vacuolar and mitochondrial functions, autophagy, and metabolite transport. Repressed genes are generally involved in energy consuming and growth-related processes, including RNA processing, transcription and translation, and biosynthesis of ribosomes and nucleotides. These stereotypical changes known as the environmental stress response (ESR; Gasch et al., 2000) or the common environmental response (CER; Causton et al., 2001) are transient and graded to the type and intensity of stress (reviewed by Gasch, 2002).
In the fission yeast Schizosaccharomyces pombe, the Sty1/Spc1/Phh1p protein kinase pathway is involved in the regulation of numerous stress responses: the Sty1p kinase is phosphorylated and activated by different stress stimuli and inactivation of the kinase results in pleiotropic stress sensitivity (Millar et al., 1995; Shiozaki and Russell, 1995; Degols et al., 1996; Degols and Russell, 1997; Shieh et al., 1997). Thus, Sty1p is predicted to play a key role in mediating a general stress response. Components of this mitogen-activated protein kinase (MAPK) cascade are homologous to components of the HOG1 osmosensing MAPK pathway in S. cerevisiae and to the mammalian and Drosophila JNK and p38 stress-activated protein kinase cascades (Toone and Jones, 1998). Notably, the Sty1p, JNK, and p38 pathways are activated by a range of stresses, whereas the HOG1 pathway appears to have been specifically adapted to sense and respond to osmotic stress.
Sty1p regulates stress-dependent transcription, at least in part, through a b-ZIP transcription factor, Atf1p (Takeda et al., 1995; Shiozaki and Russell, 1996; Wilkinson et al., 1996; Yamada et al., 1999; Nguyen et al., 2000). This is analogous to the situation in mammalian cells where the transactivation potential of ATF2, a homolog of Atf1p, is regulated by the JNK and p38 protein kinases (Tibbles and Woodgett, 1999). After stress, Atf1p is phosphorylated by Sty1p and anchors Sty1p in the nucleus. Moreover, atf1 mutants show defects in stress- and Sty1p-dependent transcription. However, these mutants only display a subset of the stress sensitivities seen in sty1 mutants, suggesting that Sty1p controls other as yet unidentified proteins (Shiozaki and Russell, 1996; Wilkinson et al., 1996; Gaits et al., 1998).
The genome sequence of fission yeast has recently been reported (Wood et al., 2002), setting the stage for postgenomic approaches in this model organism. This paper describes a genome-wide study of transcriptional changes that accompany five commonly used stresses, both in wild-type as well as in sty1 and atf1 mutant cells of S. pombe. This analysis provides: a comprehensive overview of cellular responses to environmental stress; insight into how the cell integrates information concerning the state of its environment, through analysis of regulatory mutants and stress-responsive promoter elements and; a baseline from which we can compare and contrast responses to other stress stimuli as well as similar responses in other organisms.
MATERIALS AND METHODS
Strains
We used the wild-type strain 972 h- (Leupold, 1970) and two isogenic mutant strains sty1Δ (sty1::ura4+ ura4-D18 h-) and atf1Δ (atf1::ura4+ ura4-D18 h-). sty1Δ and atf1Δ were derived from auxotrophic strains (Millar et al., 1995; Takeda et al., 1995, respectively) by crossing out markers.
Stress Experiments, Cell Collection, and RNA Isolation
The three strains were cultured in yeast extract (YE) medium (http://www.bio.uva.nl/pombe/handbook/) at 30°C, shaken in flasks at 170 rpm until reaching OD600 = 0.2 (∼4 × 106 cells/ml). Cells were harvested immediately before as well as 15 and 60 min after stress treatment from the same culture. Stress conditions were as described below. Oxidative stress: hydrogen peroxide (H2O2; H1009; Sigma, St. Louis, MO) was added to a final concentration of 0.5 mM. Heavy metal stress: cadmium sulfate (CdSO4; C2919; Sigma) was added to a final concentration of 0.5 mM. Heat stress: cells were quickly transferred from 30°C to a large prewarmed flask in a 39°C water bath, reaching temperature equilibrium after 2 min. Osmotic stress: cells were grown to OD600 = 0.4, and an equal volume of prewarmed YE + 2 M sorbitol was added to a final concentration of 1 M sorbitol. Alkylating agent: methylmethane sulfonate (MMS; 64294; Fluka, Buchs, Switzerland) was added to a final concentration of 0.02% (w/v). Cells were collected by gentle centrifugation (2000 rpm for 2 min), and pellets were frozen immediately in liquid nitrogen. We isolated total RNA using a hot-phenol protocol (for details, see our website: http://www.sanger.ac.uk/PostGenomics/S_pombe/).
Target Labeling, Microarray Hybridization, and Data Acquisition
Twenty micrograms of total RNA was labeled by directly incorporating Cy3- and Cy5-dCTP through reverse transcription and the resulting cDNA was hybridized onto DNA microarrays containing probes for 99.3% (H2O2 and cadmium experiments) or 99.9% (heat, sorbitol, and MMS experiments) of all known and predicted fission yeast genes printed in duplicate onto glass slides (for details on protocols and microarrays, see our website). Microarrays were scanned using a GenePix 4000B laser scanner (Axon Instruments, Foster City, CA) and analyzed with GenePix Pro software. Unreliable signals were filtered out, and data were normalized using a customized Perl script (local adjustment of median of ratios to 1 within running windows of 1000 spots; G. Burns, R. Lyne, J. Mata, G. Rustici, D. Chen, D. Vetrie, and J. Bähler, manuscript submitted).
Experimental Design
The five stress time course experiments with the wild-type and sty1Δ strains were performed as two independent biological repeats (except the H2O2 and cadmium experiments in sty1Δ, which were done once), and the experiments with the atf1Δ cells were done once each. Labeled samples from each stress time point of the wild-type and mutant experiments were hybridized with a labeled reference pool, containing an equal amount of all the RNA samples from the wild-type time points of the corresponding stress. For duplicate experiments, the Cy dyes were swapped for the experimental and reference samples. After data acquisition and within-array normalization, the ratios of each gene (time point/reference pool) were divided by the corresponding ratios of untreated wild-type cells (0 h wild type/reference pool). Thus, the reported ratios represent the expression levels at each time point relative to the expression levels of the untreated wild-type cells from the same stress experiment. Because of the relative importance of the measurements for untreated wild-type cells, we performed two technical repeats of these arrays (with swapping of fluorochromes) and used the averaged data to “zero-transform” the data of all stress time points from wild-type and mutant cells. Expression ratios of biological repeat experiments (wild-type and sty1Δ strains) were averaged. In total, 67 microarrays were used in this study. The complete processed data set is available from our website, and all raw data will be available from the ArrayExpress repository: www.ebi.ac.uk/arrayexpress.
Data Evaluation, Hierarchical Clustering, and Gene Classification
We used SAM (Tusher et al., 2001) and GeneSpring (Silicon Genetics, Redwood City, CA) to discard genes that did not behave reproducibly between biological duplicate experiments. Hierarchical clustering was performed with preselected log-transformed gene sets using Cluster and TreeView software (Eisen et al., 1998), with uncentered Pearson correlations and average linkage clustering. Genes with 50% of data points missing were not used. The criteria used to select various groups of stress genes (using GeneSpring) are given below. Gene annotations were taken from GeneDB at the Sanger Institute: http://www.genedb.org/genedb/pombe/index.jsp.
Identification of CESR and SESR Genes.
Genes induced at least twofold at either 15 or 60 min were identified. Among those genes, we selected induced CESR genes as those that were up-regulated in at least four of the five stress conditions. Repressed CESR genes were selected as those being down-regulated twofold or greater in at least three of the five stress conditions. We subtracted these induced CESR genes from the genes that were up-regulated twofold or greater in at least one stress to identify induced SESR genes.
Identification of “Stress-Specific” and Super-Induced Genes.
Among the genes that were induced at least twofold at either 15 or 60 min in a given wild-type stress experiment, we selected “stress-specific” genes as those that were induced at least twice as highly in the stress of interest than in any of the other four stresses. This procedure also identified CESR genes that were more highly induced in the given stress. These “super-induced” genes were subtracted from the “stress-specific” genes and listed separately (lists available from our website).
Identification of Sty1p- and Atfp1-Dependent Genes.
We selected genes that required Sty1p and/or Atf1p for induction among the genes that were induced at least twofold in a given wild-type stress experiment and were also induced at least twice as highly in the wild-type cells than in sty1Δ or atf1Δ cells in the same stress. Among those genes, we selected genes that were Sty1p- or Atf1p-dependent in at least three of the five stress conditions. Various groups of Sty1p- and/or Atf1p-dependent genes were also selected based on cluster analysis (Figure 5A).
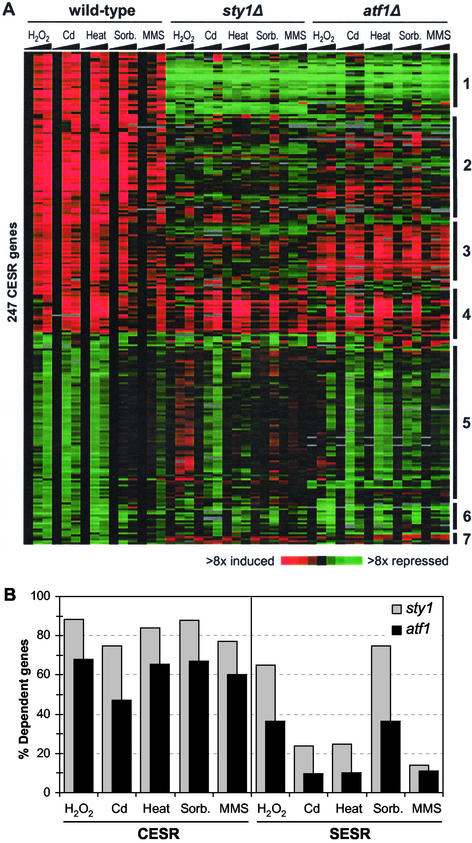
Figure 5.
Regulation of gene expression during stress. (A) The 140 induced and 109 repressed CESR repressed genes as defined in “Materials and Methods” were clustered based on their expression patterns in wild-type, sty1Δ, and atf1Δ cells in response to the five stress conditions. Details are as described in the legend of Figure 1A. Major classes of genes are indicated on the right side (see main text for details). (B) Comparison between the regulation of induced CESR and SESR genes. The histogram shows for each stress the percentages of stress-induced genes that were dependent on Sty1p (gray), as well as the percentages of Sty1p-dependent genes that were also dependent on Atf1p (black). Left side: CESR genes; right side: SESR genes.
We selected genes that required Sty1p and/or Atf1p for repression among the genes that changed <2-fold in a given wild-type stress experiment and were also induced at least twice as highly in sty1Δ or atf1Δ cells than in wild-type cells in the same stress. We then selected Sty1p- or Atf1p-repressed genes as those that were repressed in at least three of the five stress conditions. Comparison of the Sty1p- and Atf1p-repressed genes allowed us to identify genes that were repressed by both Sty1p and Atf1p and genes that were repressed by Sty1p but not by Atf1p.
Comparisons with Budding Yeast Data
Gene lists of S. cerevisiae CER/ESR genes were downloaded from the accompanying websites of Gasch et al. (2000) and Causton et al. (2001). Genes with a prospective S. pombe ortholog were determined using a table of curated orthologs created by mutual highest gene hits using FASTA aided by manual inspection of pairwise alignments and domain organization (Val Wood, personal communication; available from the Sanger Institute FTP site: http://www.sanger.ac.uk/Projects/S_pombe/ftp.shtml) and imported into GeneSpring. The total number of orthologs available at the time of analysis was 2842. CESR genes that were induced (314 genes) or repressed (424 genes) were selected based on cluster analysis (Figure 1A; lists available from our website). These genes were translated into S. cerevisiae homologs using the ortholog table and list comparisons were performed with GeneSpring.
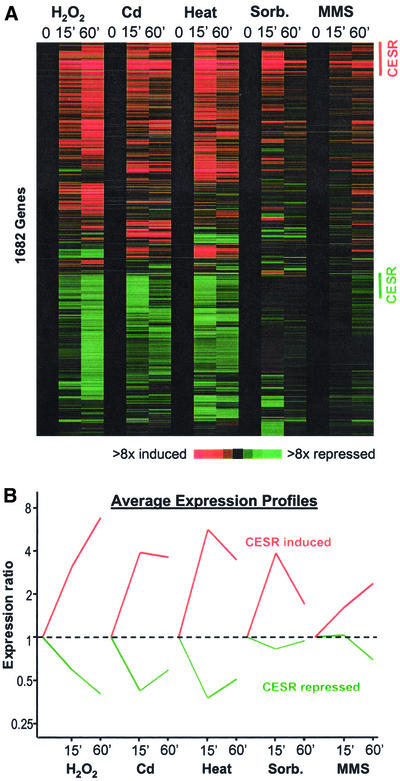
Figure 1.
Changes in gene expression in response to five environmental stresses. (A) Approximately seventeen hundred genes whose transcript levels changed significantly by at least twofold in one or more of the stress conditions were hierarchically clustered based on their expression patterns in the five time course experiments (Eisen et al., 1998; see “Materials and Methods”). Horizontal strips represent genes, and columns represent experimental time points. The fold changes in expression, relative to the untreated wild-type sample (time point 0), are color-coded as shown in the bar. The labels on the right indicate CESR induced genes (red; Table 1) and repressed genes (green) that were chosen based on conservative criteria as described in “Materials and Methods.” (B) Average expression patterns of the CESR-induced (red) and -repressed (green) genes as labeled in A in the five stress conditions.
Discovery of Statistically Significant Sequence Motifs
We searched upstream intergenic regions of limited length (up to 1000 base pairs) for sequence motifs that were statistically overrepresented for our sets of coexpressed stress-response genes. The sequences were extracted from S. pombe chromosome release 22.03.2002 on the Sanger Institute FTP site in EMBL format. The search was carried out by the SPEXS tool available online from http://ep.ebi.ac.uk (Brazma et al., 1998; Vilo, 1998). Given a set of upstream sequences, this tool searches exhaustively for all possible sequence patterns that are common to a minimum number of sequences in the set. For each of these motifs, SPEXS calculates the statistical significance of its occurrence with respect to a control set of sequences, which in our case was the total set of intergenic sequences.
Generally, we limited the query motif to substrings of arbitrary length that contained up to one “wild-card” (N), but in some cases, we used more general patterns (i.e., with one or two group character symbols). The statistical significance was calculated according to the binomial distribution (Vilo et al., 2000). To assess the significance thresholds for each set, we repeated this process on sets containing the same number of intergenic sequences selected at random, repeating the randomization three times independently. We reported only the patterns clearly above the significance threshold (with the binomial probabilities at least 10 times smaller than the lowest probabilities in any of the randomized sets) and with the selected group of upstream sequences enriched for the motif at least twofold compared with all intergenic regions.
RESULTS
Overview
We performed genome-wide expression analyses upon exposure of cells to a range of noxious conditions. Wild-type cells as well as sty1 and atf1 deletion cells were subjected to osmotic stress (1 M sorbitol), oxidative stress (0.5 mM H2O2), heavy metal stress (0.5 mM CdSO4), an alkylating agent (0.02% MMS), and heat stress (temperature shift from 30°C to 39°C). These conditions were chosen because they caused good induction of known stress genes with minimal cell death (50% viability). RNA samples were collected before and after 15 and 60 min of stress treatment and were analyzed by DNA microarray hybridization.
Figure 1A shows a cluster analysis of ∼1700 genes whose expression levels changed by twofold or greater in at least one of the stress time points. There are two main clusters: one with genes that were induced and one with genes that were repressed in response to stress. Among these stress response genes, a subset was induced or repressed by all or most stresses examined. We call these CESR genes for core environmental stress response. Other genes responded in a more stress-specific way, showing expression changes that are shared between a few, but not all, stresses (referred to as SESR genes for specific environmental stress response). A subset of the SESR genes were specifically induced in only 1 of the 5 stresses (“stress-specific” genes).
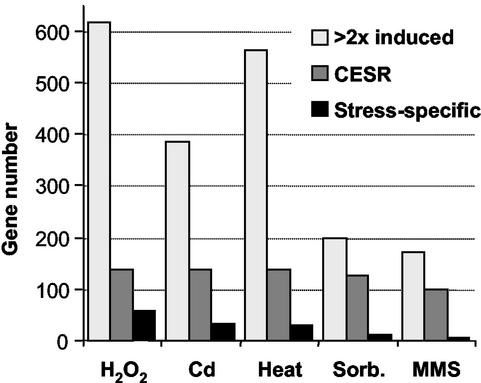
The kinetics of the transcriptional response varied for each of the stress conditions (Figure 1): whereas the responses to heat, sorbitol, and cadmium were rapid and transient, the responses to H2O2 and MMS persisted for at least 1 h. This was true for both induced and repressed genes, which behaved with similar kinetics despite opposite patterns of gene expression (Figure 1B). In addition, the magnitude of the transcriptional responses also varied for each stress under the conditions used, both in the number of genes involved and in their relative induction levels. The transcriptional responses to sorbitol and MMS were weaker than the responses to the other three stresses (Figures 1 and 2). The numbers of genes induced specifically in only one stress were relatively small and varied significantly from one stress to another (Figure 2). H2O2 and heat stress led to the induction of the largest number of genes, whereas MMS triggered expression changes in the fewest genes. The numbers and types of genes induced during stress are expected to depend also on the dose of a given stress (see “Discussion”). A list of genes that were induced threefold or greater in at least one stress, together with information on regulation in the five stresses and dependency on Sty1p and/or Atf1p, is available from our website: http://www.sanger.ac.uk/PostGenomics/S_pombe/. This site also contains a range of stress-related gene lists and a graphical gene viewer tool to check the behavior of specific genes during stress.
Figure 2.
Induction of gene expression in response to different stresses. The histogram shows the numbers of genes that are induced at least twofold, together with the number of CESR genes and the number of the “stress-specific” genes for each of the five stress experiments. CESR and “stress-specific” genes were determined using criteria described in “Materials and Methods.”
The Core Environmental Stress Response (CESR)
Induced CESR Genes.
We defined the induced CESR genes as being reproducibly induced twofold or greater in at least four of the five stresses examined. 140 induced CESR genes were identified in this way (Table 1), of which 79% have not been characterized in fission yeast. There were many more genes that were consistently induced in response to all stresses but failed to make the twofold cutoff in some of the responses (Figure 1A). Therefore, the genes in Table 1 should be regarded as a conservative representation of CESR genes. A few of these genes are in fact pseudogenes, and their stress regulation may therefore be an evolutionary remnant. The average expression profiles of the CESR genes are shown in Figure 1B.
Table 1.
Induced CESR genes
| Gene Name | Annotation |
|---|---|
| Carbohydrate metabolism | |
| ntp1 | Neutral trehalase. |
| SPACUNK4.16C | Putative alpha-trehalose-phosphate synthase. |
| tps1 | Alpha, alpha-trehalose-phosphate synthase. |
| SPAC22F8.05 | Putative alpha, alpha-trehalose-phosphate synthase. |
| SPAC4H3.03C | Conserved protein (mainly bacterial), no apparent S. cerevisiae homolog |
| zwf1 | Glucose-6-phosphate 1-dehydrogenase involved in pentose phosphate pathway |
| SPAC3C7.13C | Glucose-6-phosphate 1-dehydrogenase involved in pentose phosphate pathway |
| SPBC24C6.09C | Ortholog of human transketolase in pentose phosphate pathway |
| SPACUNK4.17 | Glucose/fructose oxidoreductase |
| SPBC12C2.04 | Similar to S. pombe CUNK4.17 glucose/fructose oxidoreductase |
| SPAPB1A11.03 | Putative FMN dependent dehydrogenase |
| exg3 | Glucan 1,3-beta-glucosidase |
| tms1 | Putative sorbitol dehydrogenase |
| gpd1 | NADH-dependent glycerol-3-phosphate dehydrogenase involved in glycerol phosphate shuttle of oxidative phosphorylation |
| gut2 | Glycerol-3-phosphate dehydrogenase involved in glycerol phosphate shuttle of oxidative phosphorylation. |
| SPBC1773.06C | Alcohol dehydrogenase |
| SPBC1289.14 | Adducin N terminal domain protein |
| SPAC513.02 | Similarity to phosphoglycerate mutases; involved in glycolytic pathway |
| SPCC306.08C | Malate dehydrogenase involved in citric acid cycle. |
| SPAC26F1.07 | Probable oxidoreductase; aldo/keto family. |
| SPBC215.11C | Putative oxidoreductase; aldo/keto family |
| SPAC19G12.09 | Putative aldose reductase; aldo/keto family; similar to S. cerevisiae Ydl124p |
| SPAC2F3.05C | Aldo/keto reductase family oxidoreductase |
| SPAC22A12.17C | Short chain dehydrogenase; possible sorbitol utilization |
| SPAC139.05 | Probable succinate semialdehyde dehydrogenase; similar to S. cerevisiae Uga2p. |
| Signaling and transcriptional regulation | |
| ptc4 | Protein phosphatase 2c isoform |
| pyp2 | Protein-tyrosine phosphatase 2; inactivates Sty1p kinase in response to stress. |
| ptc1 | Protein phosphatase 2c |
| SPBC20F10.10 | Similar to S. cerevisiae Pcl7p involved in utilization of alternative carbon source |
| SPCC1322.08 | Putative serine/threonine protein kinase, similar to S. cerevisiae Rck2p, which is a substrate for Hog1p |
| SPAC1E11.03 | Serine/threonine protein kinase; similar to S. cerevisiae Yak1p |
| SPCC1020.10 | Putative serine/threonine protein kinase; similar to S. cerevisiae Npr1p |
| cgs1 | Regulatory subunit for cAMP-dependent protein kinase A, similar to S. cerevisiae Bcy1p |
| pka1/tpk/git6 | CAMP-dependent protein kinase catalytic subunit, similar to S. cerevisiae Tpk1-3p |
| pcr1/mts2 | B zip transcription factor pcr1, similar to S. pombe Atf21p and Atf1p; roles in mating, meiosis and stress response |
| mpr1/spy1 | Stress response regulator phosphotransmitter, similar to S. cerevisiae Ypd1p, plays a role in regulation of the G(2)/M cell cycle progression. |
| atf1 | Transcription factor Atf1p |
| SPBC1105.14 | Putative Zinc-finger transcriptional activator, similar to S. cerevisiae Msn2p |
| SPAC1348.12 | Zinc finger protein |
| SPCC320.03 | Zinc-finger protein |
| SPAC57A10.09C | Nonhistone chromosomal protein 6b; similar to S. cerevisiae Nhp6bp, involved in DNA binding and bending in transcription |
| git5 | G-protein beta subunit Git5p; involved in glucose response pathway. |
| SPCP31B10.06 | C2-domain protein; synaptotagmin family (has PKC domain) |
| Lipid or fatty acid metabolism | |
| SPAC4D7.02C | Putative glycerophosphoryl diester phosphodiesterase |
| SPAPB24D3.08C | Putative NADP dependent oxidoreductase |
| SPAC4H3.08 | Putative short chain dehydrogenase; similar to S. cerevisiae Fox2p involved in peroxisomal fatty acid beta-oxidation pathway |
| SPAC521.03 | Putative short chain dehydrogenase; similar to S. cerevisiae Ymr226p |
| SPAC26F1.04C | Containing Zn-binding dehydrogenases domain; similar to S. cerevisiae Ybr026p |
| SPAC23D3.11 | Putative short chain dehydrogenase; similar to S. cerevisiae Ayr1p |
| Antioxidants | |
| trx2 | Thioredoxin II |
| zym1 | Zinc metallothionein |
| cta1 | Catalase |
| gpx1 | Glutathione peroxidase |
| grx1 | Thioltransferase |
| SPCC576.03C | Thioredoxin peroxidase |
| SPAC688.04C | Glutathione-S-transferase 3 |
| pmp20 | Peroxisomal membrane protein Pmp20p, similar to S. cerevisiae Ahp1p, an antioxidant induced by peroxide |
| DNA repair | |
| SPBC23G7.11 | DNA-3-methyladenine glycosidase; putative base excision repair-DNA repair |
| cmb1 | HMG box mismatch binding protein; likely to be involved in DNA repair |
| Transporters | |
| SPBC3H7.02 | SulP sulfate transporter |
| SPCC965.06 | Putative potassium channel subunit |
| Protein folding and protein degradation | |
| hsp16 | Heat shock protein 16 |
| hsp9 | Heat shock protein 9 |
| psi | Psi protein, heat-shock protein |
| SPAC2C4.15C | Ubiquitin regulatory domain (UBX) protein, similar to S. cerevisiae Ydr330p |
| SPCC4G3.03 | Hypothetical protein similar to S. cerevisiae YLR149C, putative apoptotic protease activating factor |
| SPCC338.12 | Sequence orphan |
| isp6 | Sexual differentiation process protein; putative subtilase-type proteinase |
| Others | |
| SPAC513.07 | Putative cinnamoyl-coa reductase, involved in vitamin/cofactor metabolism |
| plr | Pyridoxal reductase, Aldo/keto reductase family domain. Similar to S. cerevisiae Ypr127p pyridoxal reductase catalyzes the NADPH-dependent reduction of pyridoxal to form pyridoxine (vitamin B6) |
| rds1 | Stress response protein |
| SPBC2A9.02 | Putative dyhydroflavanol-4-reductase; similar to S. cerevisiae Yll056p, involved in cell stress |
| mvp1 | Putative vacuolar protein sorting protein |
| SPAC26F1.14C | Putative flavoprotein; similar to human mitochondrial apoptosis-inducing factor; involved in cell death |
| SPBC16A3.02C | Putative quinone oxidoreductase; Zn-binding oxidoreductase |
| SPBC23G7.10C | Putative NADH-dependent flavin oxidoreductase |
| SPAC2E1P3.01 | Putative dehydrogenase |
| SPBC1773.17C | Putative glycerate- and formate-dehydrogenase |
| vip1 | Protein with RNA recognition motif |
| SPAC19B12.08 | Putative protein that mediates attachment of autophagosomes to microtubules, by similarity to S. cerevisiae Aut2p |
| SPBC725.10 | Similar to peripheral-type benzodiazepine receptor |
| SPBC106.02C | ParBc-like nuclease domain; similar to S. cerevisiae YKL086W |
| SPAC22E12.03C | THIJ/PFPI family protein; putative thiamine biosynthesis enzyme |
| SPCC1183.11 | Mechano-sensitive ion channel domain |
| SPBC30D10.14 | Putative hydrolase; similar to S. cerevisiae Yal049p, which plays role in chlorocatechol degradation |
| SPAC23D3.05C | Alcohol dehydrogenase, pseudogene |
| SPAC25H1.02 | JOR domain, possibly chromatin associated |
| SPAC977.13C | Putative hydrolase, pseudogene |
Annotations for Table 1 are from GeneDB at http://www.genedb.org/genedb/pombe/index.jsp, some of them being edited by hand. Fifty-two additional proteins with no known function are not listed here (see our website for complete gene list).
The induced CESR genes encode proteins that are involved in a variety of functions. Many are known or predicted to function in carbohydrate metabolism, such as the synthesis (e.g., tps1) or degradation (ntp1) of the stress-protectant sugar trehalose, the generation of NADPH reducing equivalents (zwf1 and SPAC3C7.13c, which encode glucose-6-phosphate dehydrogenases), and fructose metabolism (SPBC24C6.09c, encoding a predicted fructose-6-phosphate phosphoketolase). NADPH can also be synthesized via succinate semialdehyde dehydrogenase encoded by the CESR gene SPAC139.05. A role for this enzyme in the response to stress has been suggested in S. cerevisiae (Coleman et al., 2001). Other CESR genes encode antioxidants such as catalase (cta1), glutathione peroxidase (gpx1), thioredoxin (trx2), glutaredoxin (grx1), glutathione-S-transferase (SPAC688.04), and metallothioneine (zym1), predicted protease genes (SPCC4G3.03, SPCC338.12, and isp6) as well as a gene involved in ubiquitination (SPAC2C4.15c) and gpd1 (encoding glycerol-3-phosphate dehydrogenase). Surprisingly, we identified within the CESR only two heat shock proteins (hsp16 and hsp9) and two genes predicted to be involved in DNA repair (SPBC23G7.11 and cmb1). The CESR also included SPBC725.10 encoding a homolog of the mammalian peripheral-type benzodiazepine receptor, which is thought to control mitochondrial functions including steroid biosynthesis (Papadopoulos et al., 2001). Given the central role mitochondria play in mammalian stress/apoptotic pathways, this observation may point to a function for benzodiazepine receptors in stress resistance.
Several CESR genes encode putative or known regulatory proteins, which may either directly control the activity of stress protective proteins or may function in secondary responses facilitating adaptation to stress. These include the transcription factor genes atf1 and pcr1, which have well known roles in stress-dependent transcription, as well as signaling protein genes, including mpr1 (involved in a two-component pathway) and srk1 (encoding a kinase similar to S. cerevisiae Rck2p). Rck2p is phosphorylated by the Hog1p MAPK in response to osmotic stress, and recent studies in S. pombe have shown that Srk1p and a related protein kinase, Cmk2p, interact with, and are phosphorylated by Sty1p (Sanchez-Piris et al., 2002; Smith et al., 2002). Moreover, Sty1p is required for induction of srk1 (see below), and the Sty1p-Srk1p signaling pathway may therefore be amplified by positive feedback in response to stress. Genes encoding the protein kinase A (PKA) catalytic (pka1) and regulatory (cgs1) subunits were also induced in the CESR, as they are in S. cerevisiae. Thus, the PKA pathway may play a conserved role in regulating events after environmental stress.
Repressed CESR Genes.
Stress also triggered down-regulation of many genes (Figure 1A). Using the same criteria as for induced CESR genes, only 11 genes were identified. Gene repression was generally weaker in sorbitol and MMS stresses, although many genes are stereotypically repressed (Figure 1). Therefore, we defined repressed CESR genes as being reproducibly down-regulated twofold or greater in three of the five stresses (list available from our website). As for the induced CESR genes, the 106 repressed CESR genes are somewhat arbitrary but provide a representation of the types of genes that are consistently repressed in several stresses. These genes are mainly associated with protein synthesis (ribosome and tRNA synthesis and RNA processing, splicing, and translation initiation), transport, transcription (RNA polymerase I, II, and III subunits), cellular signaling, and cytoskeletal organization.
Comparison Between Fission and Budding Yeasts.
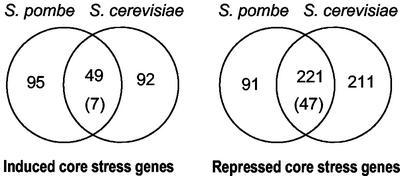
As discussed above, there are some striking similarities between the S. pombe CESR genes and the S. cerevisiae ESR/CER genes (Gasch, 2002). To see whether there is a significant overlap between the core stress genes of the two yeasts, we performed a systematic comparison. Of 364 ESR/CER genes that were induced in either or both budding yeast studies, 141 genes had fission yeast orthologs (see “Materials and Methods”). Comparing these genes with the CESR genes of Table 1 revealed an overlap of 29 genes. This is highly significant as just 3 genes are expected by chance. The number of overlapping genes can be increased by choosing the fission yeast CESR genes based on the cluster of Figure 1A. This is less conservative, and corresponds closer to the definition of the ESR/CER genes in the budding yeast studies. Of 314 such genes, 144 had budding yeast orthologs, 49 of which corresponded to ESR/CER genes (Figure 3). A corresponding analysis of repressed CESR and ESR/CER genes showed an even more significant overlap between the two yeasts (221 genes; Figure 3). This comparison shows that stress-induced changes in many genes are evolutionarily conserved in these distantly related yeasts.
Figure 3.
Comparison of core stress genes in fission and budding yeasts. Conserved genes that are part of the CESR (this study) were compared with genes that are part of the ESR and/or CER (Gasch et al., 2000; Causton et al., 2001) using an ortholog table (see “Materials and Methods”). Left side: induced genes; right side: repressed genes. The numbers of overlapping genes between the chosen gene groups are shown in the Venn diagrams (lists available from our website). Only genes with orthologs in the other yeast were included in the comparison. The numbers in brackets represent the overlap expected by chance, given the sizes of the gene sets considered and the total number of 2842 genes with orthologs. The overlaps are highly significant, both for induced genes (P ∼ 2 × 10−30) and for repressed genes (P ∼ 9 × 10−131).
The Specific Environmental Stress Response (SESR)
Having defined the CESR, we then characterized individual stresses and identified SESR genes, which are likely to play a more specific role in stress adaptation. SESR genes can be subdivided into genes that appeared to be “stress-specific” and those whose induction was shared by two or three of the stresses examined but did not meet the criteria for inclusion in the CESR. To determine the extent of overlaps between stresses, we examined the SESR genes that were induced at least twofold in more than one stress. The most extensive overlap was between H2O2 and heat stress, with 200 genes shared. There was also a substantial overlap between H2O2 and cadmium as well as between heat and cadmium stresses. Interestingly, although MMS stress led to the induction of relatively few genes, it shared most of them with H2O2 (see below). These overlaps indicate that certain stresses have similar consequences, resulting in the mobilization of the same defense genes. Below, we describe genes that were regulated in a “stress-specific” way or were super-induced in a given stress.
Hydrogen Peroxide Stress.
All cells must maintain antioxidants and enzymes to break down reactive oxygen species. Oxidative stress occurs when the cellular redox balance is upset. Transcriptional responses of S. pombe to H2O2 vary with the concentration of oxidant (Quinn et al., 2002). Here, we describe the response to an intermediate level of H2O2, which incorporates elements of low and high doses of H2O2. We identified 56 genes whose expression was altered specifically in response to oxidative stress (list available from our website). In addition, a number of genes encoding critical components of the oxidative stress response, such as catalase and peroxidases, were included within the CESR, and 12 of these were super-induced by H2O2, implying that reactive oxygen species are generated by various stresses. Of the H2O2-specific genes, trr1, SPAC869.02c, and obr1 (encoding thioredoxin reductase, a probable flavohemoprotein, and a predicted benzoquinone reductase, respectively) are also predicted to function in antioxidant pathways. H2O2 also induced genes encoding membrane transporters such as hba2 and SPBC609.04 as well as SPAC22G7.08, which encodes a predicted Ser/Thr kinase involved in regulating membrane transport and ion homeostasis. Several H2O2-specific or super-induced genes are involved in riboflavin (vitamin B6) and pyridoxine (vitamin B2) use or synthesis, suggesting an increased requirement for the B vitamin-derived coenzymes and pyridoxal phosphate. Relatively little is known about the antioxidant potential of B vitamins, although recent studies have shown that pyridoxine can quench singlet oxygen and protect against superoxide mediated damage in diabetic patients (Bilski et al., 2000; Jain and Lim, 2001).
Both H2O2 and heat stress induced the expression of Tf2 and related long-terminal-repeat containing retrotransposons. In addition, both stresses also induced two genes required for late steps in Tf2 transposition: pst1 (encoding a homolog of budding yeast Sin3p) and tlg1b (encoding a t-SNARE protein related to syntaxin). Thus, some stresses may trigger a retroviral transposition response within the cell.
Cadmium Stress.
The heavy metal cadmium (Cd) is a common environmental contaminant, which can react with thiol groups on proteins with myriad physiological consequences. Cells control levels of heavy metals primarily by sequestration, using either metallothioneins in mammalian cells or glutathione (GSH) and phytochelatin (PC; a polymer of GSH) in yeast and plant cells, respectively. We found that Cd elicits a complex transcriptional response, incorporating facets of heat and H2O2 stress responses in addition to more Cd-specific responses. A set of 32 genes was unique to Cd treatment and two CESR genes were super-induced in response to Cd (list available from our website). Some of these genes (e.g., SPAC869.05c, encoding a predicted sulfate transporter) presumably participate in the sulfur amino acid biosynthetic pathway. This pathway is required for the synthesis of GSH and PC, which bind Cd and are subsequently pumped into the vacuole via GSH- and PC-dependent membrane transporters such as Hmt1p. The expression levels of hmt1, the genes encoding GSH synthases (gcs1 and gsh2), and pct1 (encoding PC synthase) were not changed in our experiment. However, a gene for a GSH transporter was induced (SPAC29B12.10c; homologous to the budding yeast Hgt1p transporter). Thus, scavenging of GSH from the external milieu rather than GSH synthesis may be the initial reaction of fission yeast to Cd stress. To test this, we deleted SPAC29B12.10c and found that the mutant strain was indeed hypersensitive to Cd (W.M. Toone, unpublished results).
In S. cerevisiae, GSH is synthesized, along with the sulfur containing amino acids methionine and cysteine, through a series of reactions starting with sulfate assimilated from the environment. Although most genes in the S. cerevisiae sulfur amino acid pathway are induced by Cd (Fauchon et al., 2002), only a select group of S. pombe genes were induced. Thus, either the regulated expression of genes in the pathway differs between the two organisms, or the pathways themselves are fundamentally different.
Cd also specifically induced genes encoding potential zinc (Zn) transporters (SPBC16D10.06 and SPBP26C9.03c; homologous to budding yeast Zrt1p and Fet1p, respectively). Because Cd can displace metal ions from proteins (Stohs and Bagchi, 1995), a reason for Cd toxicity may be the depletion of Zn. Therefore we compared our Cd response with S. cerevisiae experiments monitoring the response to Zn limitation (Lyons et al., 2000). In addition to the Zn transporters above, the Cd-specific genes included two similar genes of unknown function (SPAC1348.06c and SPAC977.05c), whose S. cerevisiae homologs (YGL258w and YOR387c, respectively) were also induced during Zn limitation. Moreover, the Cd-specific gene SPAC5H10.06 had a S. cerevisiae homolog that was induced by Zn limitation (ADH4; encoding an iron-dependent, Zn-independent form of alcohol dehydrogenase). These similarities between the responses support the idea that Cd leads to Zn depletion or alters the way that this metal is utilized in the cell.
Heat Stress.
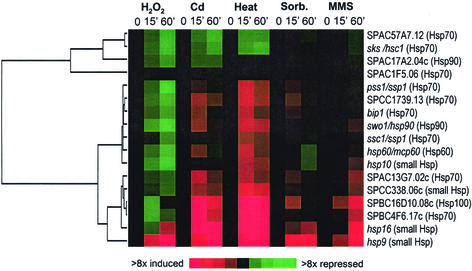
Studies in several organisms have shown that cells induce the synthesis of a set of heat shock proteins (HSPs) in response not only to heat, but also to other stresses (Cotto and Morimoto, 1999). These highly conserved proteins limit stress-induced damage by sequestering denatured proteins, thus preventing aggregation and facilitating either protein refolding or degradation. There are at least 17 members of the HSP protein families in the S. pombe genome, and their expression profiles are shown in Figure 4. Two main clusters were evident: four genes were repressed under all stresses examined, and the remaining genes were induced to varying degrees mainly by heat and, to a lesser extent, by Cd. hsp16 and hsp9 are part of the CESR (Table 1). A number of HSP-associated proteins, including those encoded by SPBC1711.08 and sti1, were among the heat-specific genes and showed similar expression profiles as the induced hsp genes. In total, 31 genes were specifically induced or super-induced by heat (list available from our website). This includes genes predicted to participate in the ubiquitin pathway, wis2 (encoding a cyclophyllin), and SPBC1711.12 (encoding a dipeptidyl peptidase), all of which are probably involved in protein folding or degradation.
Figure 4.
Expression profiles of HSPs in response to stress. Seventeen members of the conserved family of HSPs were clustered based on their expression patterns in the five time-course experiments. Details are as described in the legend of Figure 1A. A gene tree (dendrogram) based on similarities in expression across all conditions is shown on the left. Gene names are given at right, together with a classification into major Hsp families (in parentheses).
Osmotic Stress.
Exposure of cells to high osmolarity leads to dehydration, collapse of ion gradients over the plasma membrane, and decreased viability. Thirteen genes were identified as sorbitol specific, whereas eight CESR genes were super-induced by sorbitol (list available from our website). The primary response to osmotic stress is the accumulation of internal osmolytes (mainly glycerol), allowing the cell to retrieve water from the environment. Consistent with this, the CESR gene gpd1 (involved in glycerol synthesis) was most highly induced in sorbitol, whereas SPAC977.17 (encoding a predicted membrane protein regulating glycerol export) was specifically repressed in sorbitol.
Among the sorbitol-specific genes, SPAC22A12.17c and SPACUNK4.17 (encoding putative sugar oxidoreductases) may be involved in sorbitol utilization. Other genes include SPAC25B8.12c (encoding a hydrolase short chain dehydrogenase), SPCC794.04c (encoding a Major Facilitator Superfamily [MFS] transporter), as well as SPCC1183.11 and SPAC2C4.17c (encoding proteins with mechanosensitive ion channel domains). Three genes (atf21, SPCC320.03, and SPAC6B12.07c) encode predicted transcription factors that may play roles in regulating the osmotic stress response.
MMS Stress.
MMS is an SN2-type alkylating agent that predominantly methylates nitrogen atoms in purines, but will also methylate proteins. Only two genes (abc1; ABC transporter, and SPAC4F10.10c; putative role in N-glycosylation) were specifically induced by MMS, and generally few genes known to be involved in DNA repair were induced. Interestingly, there was substantial overlap between SESR genes induced by MMS treatment and those induced by exposure to H2O2. These genes include trr1 (encoding thioredoxin reductase), SPBC609.04 (encoding a MFS multidrug efflux transporter), SPBC409.13 (encoding 6,7-dimethyl-8-ribityllumazine synthase), and SPCC663.08c/SPCC663.06c (encoding short chain dehydrogenases). Similar results of overlapping responses between oxidative and MMS stress were found in S. cerevisiae (Gasch et al., 2001). These data suggest that there is either a stress component shared between MMS and H2O2 exposure, or that MMS and H2O2 can activate a common regulator, resulting in induction of a similar set of genes. A likely regulator of these genes is Pap1p in S. pombe and the homologous Yap1p in S. cerevisiae.
A prediction from this observation is that short-term exposure to H2O2 induces protection from high doses of MMS or vice versa. Therefore we tested the ability of cells pretreated with a low concentration of H2O2 (0.07 mM) for 1 h to survive treatment with a high concentration of MMS (0.2%). Similarly, we examined the ability of cells pretreated with a low dose of MMS (0.02%) to survive a potentially lethal dose of H2O2 (25 mM). Although low doses of MMS were able to protect cells from high levels of H2O2, low doses of H2O2 failed to induce protection against MMS. Interestingly, microarray analysis showed that low levels of H2O2 were able to induce oxidative stress response genes but were too low to induce CESR genes (D. Chen, unpublished data). This raises the possibility that cross protection requires induction of the CESR.
Regulation of Transcriptional Responses to Stress
The Sty1p MAPK acting on the transcription factor Atf1p plays a central role in stress responses (see “Introduction”). To characterize the roles of these proteins in regulating stress genes, we performed microarray analyses with sty1Δ and atf1Δ cells subjected to the same five environmental stresses as the wild-type cells.
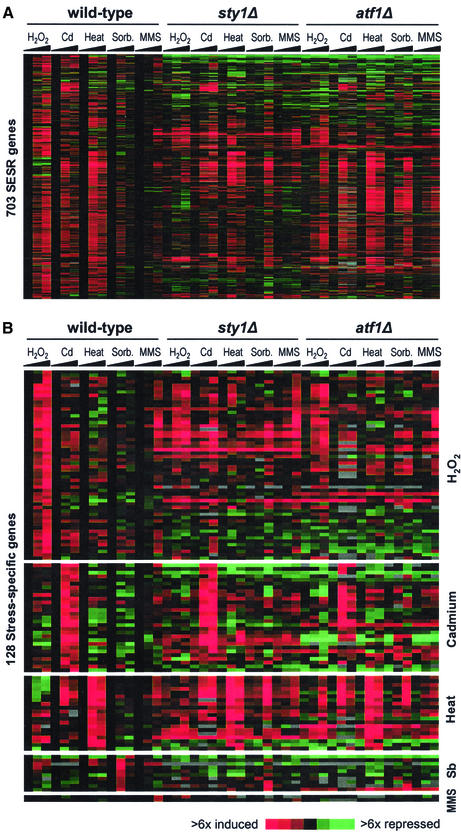
Regulation of CESR Genes.
Figure 5A shows a cluster analysis of the induced and repressed CESR genes under the five stress conditions in wild-type, sty1Δ, and atf1Δ strains. The majority of induced CESR genes depended on Sty1p and, to a lesser extent, on Atf1p for up-regulation. Interestingly, distinct subclusters of genes were evident within the CESR, with the major gene classes labeled in Figure 5A (lists available from our website). Class 1 genes require Sty1p and Atf1p for both basal and stress-induced gene expression; class 2 genes require Sty1p and Atf1p for full stress induction, but not for basal levels of expression; class 3 genes are dependent on Sty1p for induction, but do not generally require Atf1p; and class 4 genes are independent of both Sty1p and Atf1p for stress induction. Interestingly, these classes may correlate with gene functions. Class 1, for example, contains many genes involved in redox regulation (including cta1, grx1, and gpx1), whereas class 3 contains few genes encoding stress protective proteins but is enriched in regulatory factors (including those encoding the Srk1p kinase, Mpr1p signaling protein, Pka1p kinase, Cgs1p, the Pka1p regulatory protein, and the Ptc1p phosphatase). Thus, Sty1p appears to play multiple roles in controlling the levels of various stress response factors. The relative role of Sty1p and Atf1p in controlling the CESR is quantified in Figure 5B (left side). The majority of CESR genes were Sty1p dependent, and among these genes, the majority was also Atf1p dependent. In conclusion, Sty1p plays a critical role in regulating the CESR in all stresses, although some genes (class 4) can be induced in the absence of Sty1p. Because Sty1p controls the expression of more genes than Atf1p, it is likely that Sty1p regulates additional transcription factors.
Sty1p may also play a role in repressing CESR genes: a large proportion of them are derepressed in sty1Δ cells (class 5 genes in Figure 5A). However, the role of Sty1p in controlling these genes varied depending on the stress. For oxidative stress, the expression of 91 out of 106 repressed CESR genes were at least twofold higher in sty1Δ cells compared with wild-type cells, whereas gene repression was less dependent on Sty1p in the other stresses. Moreover, repression of CESR genes seemed to be virtually independent of Atf1p, with the exception of the few class 7 genes that were super-induced in sty1Δ and atf1Δ cells under most conditions. Class 6 genes were repressed independently of either Sty1p or Atf1p.
Regulation of SESR Genes.
Figure 6A shows a cluster analysis of the expression of induced SESR genes in the five stress conditions in wild-type, sty1Δ, and atf1Δ strains. The proportion of the Sty1p- and Atf1p-dependent genes among the SESR was smaller compared with those in the induced CESR genes (Figure 5A). This finding is quantified in Figure 5B (right side). Moreover, the contribution of Sty1p and Atf1p in regulating SESR genes varied considerably from stress to stress, e.g., Sty1p played an important role during H2O2 and sorbitol stresses but was less critical for controlling SESR genes in the other stresses.
Figure 6.
Regulation of SESR genes. (A) Approximately seven hundred SESR genes (including “stress-specific” genes) that were twofold or greater up-regulated in fewer than four stress experiments were clustered based on their expression patterns in wild-type, sty1Δ, and atf1Δ cells in response to the five stress conditions. Details are as described in the legend of Figure 1A. (B) One hundred twenty-eight “stress-specific” genes as defined in “Materials and Methods” were clustered based on their expression patterns in wild-type, sty1Δ, and atf1Δ cells in response to the five stress conditions. The stress specificities are indicated on the right side. Details are as described in the legend of Figure 1A.
We also investigated the roles of Sty1p and Atf1p in controlling “stress-specific” genes, which form a subgroup of the SESR genes (Figure 6B). As for the SESR genes as a whole, the numbers of Sty1p- and Atf1p-dependent “stress-specific” genes varied depending on the stress, but the expression of these genes was clearly less dependent on these two regulators than were the CESR genes. For example, the expression of HSP genes was independent of Sty1p or Atf1p, with transcription levels remaining unaffected or slightly induced in both deletion mutants. The only exception to this was hsp9, a CESR gene, which was Sty1p and Atf1p dependent (Figure 4). sty1Δ cells are profoundly sensitive to Cd treatment, whereas atf1Δ cells show increased resistance to Cd relative to wild-type cells (Toone et al., 1998). Sty1p was only required for induction of two out of 34 Cd-specific genes; therefore, the Cd-sensitive phenotype of sty1Δ cells may reflect their inability to induce a CESR or Sty1p may play additional roles in stress resistance that are independent of gene expression changes. Interestingly, a number of Cd-specific genes (including those encoding sulfate permeases, an F-box protein, and an MFS transporter), as well as a CESR gene encoding neutral trehalase were super-induced in atf1Δ strains. This may explain the Cd-resistant phenotype of atf1Δ cells, even in the absence of a substantial portion of the CESR.
Unlike in heat and Cd stress, H2O2- and sorbitol-specific genes were largely dependent on Sty1p. Intriguingly, sty1Δ cells showed a delayed response in sorbitol, wherein a new set of genes, not normally induced by osmotic stress, was turned on. This observation suggests that in the absence of an immediate response to osmotic stress (in sty1Δ cells), the cell compensates by activating alternative stress response pathways to deal with secondary stresses resulting from unchecked osmotic stress. This is consistent with our finding that a similar set of genes was derepressed in untreated sty1Δ cells, possibly reflecting a mechanism to compensate for the absence of basal expression levels of several CESR genes in this mutant (genes lists available from our website).
Promoter Motif Analyses.
Having defined various groups of genes that were coexpressed under different stress conditions, we performed systematic promoter analyses to detect common regulatory elements. Table 2 shows an overview of the most statistically significant patterns found. We found variations of the known and conserved ATF/CRE motif (KWCGTCA: Jones and Jones, 1989; TGACGTCA: Hai and Hartman, 2001) among the induced CESR genes (Table 2) as well as among genes that were dependent on both Sty1p and Atf1p for stress induction (K. Kivinen, unpublished results). This is consistent with the role of this motif as a binding site for the Atf1p family of transcription factors. The ATF/CRE motif was also present in a group of genes that were Sty1p-dependent in unstressed cells (Table 2), suggesting that this motif is also used for the basal level of expression of some stress genes.
Table 2.
Potential regulatory promoter motifs
| Cluster | Motif | Related motif | Frequency in cluster | Total number in genome (frequency) | Ratio | Binomial probability | Significance threshold |
|---|---|---|---|---|---|---|---|
| Induced CESR genes | TKACGT | ATF/CRE | 67/140 (47.9%) | 882 (17.8%) | 2.7 | 4.4e-16 | 1e-08 |
| Sty1p-dependent (unstressed cells) | TKAYGTCAT | ATF/CRE | 10/44 (22.7%) | 31 (0.6%) | 37.8 | 1.8e-13 | 1e-08 |
| Sty1p-, not Atf1p-dependent | TCTTNCTT | Novel | 13/20 (65%) | 658 (13.3%) | 4.9 | 1.1e-07 | 1e-06 |
| Sty1p-repressed | TTGTCTNT | Novel | 16/40 (40%) | 426 (8.5%) | 4.7 | 1.0e-07 | 1e-06 |
| Sty1p-, but not | TTTNTTTTTTAT | Novel | 9/32 (28.1%) | 116 (2.3%) | 12.1 | 3.4e-08 | 1e-06 |
| Atf1p-repressed | TNGTTAAACT | Novel | 6/32 (18.8%) | 31 (0.6%) | 30.2 | 4.5e-08 | 1e-06 |
For details on data and gene lists, see MATERIALS AND METHODS. N in the patterns means either A, C, G, or T; K means G or T; and Y means C or T.
We also discovered potential novel regulatory motifs. A pattern was found among genes that were dependent on Sty1p but not on Atf1p for stress induction (Table 2; list available from website). The significance of this motif is marginal, but all 13 occurrences were within 600 base pairs from the start codon. This motif might be required for an unidentified transcription factor that is activated by Sty1p and regulates gene expression independently of Atf1p. Interestingly, Atf1p itself contained this motif in its promoter. It is noteworthy that the ATF/CRE motif was absent within 2000 base pairs upstream of the start codon in all those genes.
Finally, we found three sequence motifs among genes that were derepressed in sty1Δ mutants. This group of genes was further divided into genes that required both Sty1p and Atf1p for repression, and those that required Sty1p but not Atf1p for repression (list available from website). One of the three motifs was found in both subgroups and can therefore be regarded as a general pattern of this gene group, whereas the two other motifs were found only among the genes that were independent of Atf1p for repression (Table 2). The two latter motifs were all found within 600 base pairs upstream of the start codons. With one exception, each motif was present only once per gene, sometimes in combination with the other motif. None of the Sty1p repressed genes showed any ATF/CRE-like motif within 2000 base pairs upstream of their start codons. In conclusion, we were able to identify both known and potentially novel motifs within some, but not all, of our groups of coexpressed genes. Further work will be required to learn more about the biological significance of these motifs.
DISCUSSION
This paper characterizes transcriptional programs of fission yeast to a range of environmental stresses. We describe a common stress response, which is regulated primarily by the Sty1p MAPK pathway. In addition to the CESR, the cell initiates gene expression programs more specific to each stress or subsets of stresses. These specialized programs may involve Sty1p and/or as yet uncharacterized stress-specific regulatory factors. It should be noted that microarrays measure differences in mRNA levels, which may reflect regulatory changes in transcription and/or in mRNA turnover (e.g., Fan et al., 2002).
The CESR contains genes whose expressions change stereotypically with stress. We have identified ∼140 induced and ∼100 repressed genes that provide a representative sample of the general response to stress. The predicted functions of CESR genes suggest that stressed cells selectively reprogram a wide range of activities, including carbohydrate metabolism, protein synthesis, and several other metabolic functions, possibly to save energy by limiting growth-related activities and to synthesize stress-protective molecules and cofactors. CESR genes may be controlled by known and predicted regulators that form part of the CESR, such as b-ZIP and Zn-finger transcription factors, phosphatases, a Sty1p-interacting protein kinase, and components of the PKA pathway. There was significant overlap between genes of the CESR and genes of the CER/ESR, recently described in S. cerevisiae (see “Introduction”). This suggests that a general response to stress, involving similar gene sets, is evolutionarily conserved. This conservation is in contrast to genes induced during meiotic differentiation, where the overlap between S. pombe and S. cerevisiae is surprisingly small, given the large numbers of genes that are regulated (Mata et al., 2002).
The CESR and ESR/CER responses discovered through DNA microarray analysis are manifestations of the general stress response described previously (reviewed by Siderius and Mager, 1997). The general stress response was postulated to explain the phenomenon of cross-protection, wherein exposure to a nonlethal dose of one stress can protect against a potentially lethal dose of a seemingly unrelated stress. The degree of cross-protection varies depending on the stress and is not always reciprocal, indicating that stress-specific responses are required for full protection. General stress resistance is also associated with nutrient deprived cells, cells in stationary phase, and differentiated spores. Indeed, the CESR is activated during nitrogen starvation (J.M., unpublished data), and at least some of the CESR genes are also induced during sporulation (Mata et al., 2002).
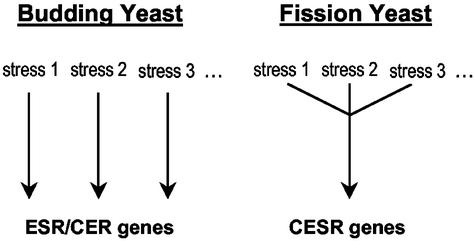
The CESR, irrespective of the type of stress, seems to be controlled predominantly by the Sty1p protein kinase and, to a lesser extent, by the Atf1p transcription factor. This is in contrast to S. cerevisiae, where the ESR/CER is not governed by one “all-purpose” regulatory system. Instead, different signaling pathways and transcription factors, acting in response to specific stress conditions, control a common set of genes (Figure 7). For example, in response to osmotic stress, the Hog1p MAPK pathway is critical for induction of ESR genes (O'Rourke et al., 2002), whereas MMS induces similar genes through a pathway that requires the Mec1p kinase (Gasch et al., 2001). Thus, S. pombe and S. cerevisiae appear to use different regulatory strategies to achieve similar outcomes.
Figure 7.
Regulation of stress genes in budding and fission yeasts. Both yeasts control a similar core group of genes in response to all or most stresses. These genes are mainly regulated by stress-specific mechanisms in budding yeast, whereas in fission yeast, the Sty1p MAPK pathway plays a central role in regulating the responses to different stresses. See main text for discussion.
The sorbitol stress response in S. pombe depended on Sty1p, both for CESR and stress-specific gene expression, possibly reflecting the evolutionary origin of the stress-activated MAPK pathway. The S. cerevisiae Hog1p pathway, which is homologous to the Sty1p pathway, is required specifically for the response to osmotic stress (O'Rourke et al., 2002). Thus, the stress-activated MAPK pathway may have evolved to control osmotic stress (as is still evident in budding yeast) and later acquired a more general role of stress regulation (as in fission yeast and Metazoa). Alternatively, the S. cerevisiae Hog1p kinase evolved away from an ancestor with a more general function.
Regulation of the CESR by Sty1p appears to be complex, with some genes requiring the kinase for both basal and stress-induced expression and others requiring it only during stress. The only known transcription factor target for Sty1p is Atf1p, which binds the conserved ATF/CRE promoter motif (Hai and Hartman, 2001). This motif was enriched in the CESR genes and other gene groups that showed Sty1p- and Atf1p-dependent transcription. The differences in expression profiles exhibited between sty1 and atf1 mutants suggest that Sty1p may interact with additional downstream regulators to control various aspects of gene expression. We identified a potential regulatory motif for an unidentified transcription factor among the genes that depended on Sty1p, but not on Atf1p, for stress induction. Interestingly, the induction of most genes directly involved in stress defense depended on Atf1p, whereas genes with regulatory functions tended to be induced by Sty1p independently of Atf1p.
Notably, a number of genes were derepressed in sty1 or atf1 mutants, both in stressed and unstressed cells. This may reflect indirect cellular responses to compensate for the problems caused by the absence of these regulators. For example, the basal gene expression levels of many CESR genes were reduced in both sty1 and atf1 mutants, which may lead to stress. However, it is possible that Sty1p and/or Atf1p also play direct roles in repressing gene expression. In fact, a previous study has suggested that Atf1p can both induce and repress genes depending on the Sty1p phosphorylation state (Degols and Russell, 1997). A role for Sty1p in down-regulating gene expression was also apparent from our finding that some CESR genes required Sty1p for repression. How Sty1p and/or Atf1p might control repression of gene expression is not known, but putative regulatory motifs were present in the promoters of Sty1p-repressed genes.
We also examined gene expression programs activated only under certain stress conditions. There were relatively few genes whose expression changes were absolutely specific to particular stresses. Analyses of SESR genes, either specific to a given stress or shared with other stresses, will aid in our understanding of the types of damage and alterations to metabolism caused by various stresses. Besides its role in the CESR, the Sty1p pathway also controls subsets of “stress-specific” genes, with the requirements varying depending on the stress. For example, heat-specific gene expression was independent of Sty1p, whereas H2O2-specific gene expression depended to a large part on Sty1p. The characterization of the cation-specific induction of cta3 in S. pombe illustrates how Sty1p may regulate stress-specific genes (Greenall et al., 2002). cta3 encodes a cation transporter, whose induction requires both activation of Sty1p and derepression of the Tup11/12p complex. In the absence of Tup11/12p, the cta3 promoter becomes responsive to other stresses regulated by Sty1p. Using similar mechanisms, Sty1p may be co-opted to regulate a range of specialized responses and thereby coordinate the CESR with the appropriate specific response.
Our results indicate that there are stress-specific regulators acting independently of Sty1p. Most of these factors, however, are as yet uncharacterized. Heat shock, for example, induced several genes functioning in protein folding or degradation in the absence of Sty1p. A subset of these genes was also induced with H2O2 and Cd. How only a portion of the heat shock response is activated by these other stresses is an interesting regulatory problem. A likely factor controlling the response to heat is Hsf1p. Intriguingly, S. pombe Hsf1p contains distinct sequences in its C-terminal domain that are responsive to different stress stimuli and are required for the activation of different subsets of heat stress proteins (Saltsman et al., 1999). This may explain how some genes of the heat shock response are also activated by other stresses.
Pap1p is a redox-sensitive transcription factor involved in regulating oxidative stress responses as well as drug and heavy metal resistance. Pap1p is particularly important for gene induction in response to low levels of H2O2, whereas Atf1p becomes the predominant transcription factor with increasing levels of H2O2, resulting in the induction of a different set of genes (Quinn et al., 2002). We have characterized this dose-dependent switch in transcription factors and target gene expression at the whole genome level (D. Chen, W.M. Toone, J. Mata, G. Burns, N. Jones, and J. Bähler, manuscript in preparation). Similarly variegated responses are likely to be found by varying the intensity of other environmental stresses. In this study, 0.02% MMS induced only two stress-specific genes and failed to induce a DNA damage response. Using a similar MMS concentration with S. cerevisiae, Gasch et al. (2001) observed induction of a surprisingly small cluster of DNA damage-specific genes. In contrast, Jelinsky et al. (1999) used a higher dose (0.1% MMS) and observed a robust DNA damage response. These studies suggest that noxious compounds may cause different types of damage and induce different responses depending on the dose used.
Stress responses varied not only in the type of genes involved but also in the kinetics of gene expression. For example, heat and sorbitol stress showed a rapid and transient response to stress, whereas gene induction during H2O2 stress continued to increase over the course of the experiment. In all cases, stress was continually applied throughout the experiment. Thus, cells appear to adapt to some situations and return to homeostasis more readily than to others, although a delayed response may also reflect the time taken for a particular stress to take effect. It would be interesting to determine if the adaptation seen in heat and osmotic stress is due to active down-regulation of Sty1p and inactivation of the response or due to a more efficient adaptation in these conditions, eliminating the need for continued Sty1p activity.
Global transcriptional responses to stress have now been examined in some detail using gene expression profiling in both S. cerevisiae and S. pombe. The transcriptional programs consist of general and stress-specific responses. Although both yeasts induce a similar set of general stress genes, there are striking differences in the regulatory pathways used for stress responses. Stress adaptation and cross protection will likely require a combination of general and specific genes, depending on the type and intensity of the stress. In addition, some stresses cause similar types of damage and therefore induce similar sets of proteins. All eukaryotic cells have mechanisms for dealing with stress. Even within the protected confines of the human body, cells can experience fluctuations in osmolarity, exposure to noxious agents, and variations in the levels of oxygen and reactive oxygen species. Some responses, such as ischemic-reperfusion injury, are closely associated with pathology. Many of the factors important for regulating stress responses, including MAPK signaling pathways and AP-1 transcription factors, are conserved from yeast to human. It is therefore likely that lessons learned from stress responses in yeast will facilitate the understanding of how cells in general respond to a changing environment.
ACKNOWLEDGMENTS
We thank Val Wood for support with gene annotations and information on orphans and orthologs, Dave Vetrie for microarray printing, Jaak Vilo for help with SPEXS software, Audrey Gasch and Helen Causton for critical reading of the manuscript, Helen Parkinson and Rob Andrews for help with preparing data for ArrayExpress, Jonathan Millar for a sty1Δ strain, Roger Pettet for development of the gene expression views, and the developers of GeneDB for providing this useful database. We apologize to colleagues in the field for not citing all relevant papers due to space limitations. D.C. and W.M.T. were supported by the EMF Biological Research Trust. This research was funded by Cancer Research UK (Jones and Bähler laboratories), and A.B. was partly funded by the TEMBLOR grant from the European Commission.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0499. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–08–0499.
REFERENCES
- Bilski P, Li MY, Ehrenshaft M, Daub ME, Chignell CF. Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem Photobiol. 2000;71:129–134. doi: 10.1562/0031-8655(2000)071<0129:sipvbp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Brazma A, Jonassen I, Vilo J, Ukkonen E. Predicting gene regulatory elements in silico on a genomic scale. Genome Res. 1998;8:1202–1215. doi: 10.1101/gr.8.11.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ST, Fang TK, Rovinsky SA, Turano FJ, Moye-Rowley WS. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J Biol Chem. 2001;276:244–250. doi: 10.1074/jbc.M007103200. [DOI] [PubMed] [Google Scholar]
- Cotto JJ, Morimoto RI. Stress-induced activation of the heat-shock response: cell and molecular biology of heat-shock factors. Biochem Soc Symp. 1999;64:105–118. [PubMed] [Google Scholar]
- Degols G, Russell P. Discrete roles of the Spc1 kinase and the Atf1 transcription factor in the UV response of Schizosaccharomyces pombe. Mol Cell Biol. 1997;17:3356–3363. doi: 10.1128/mcb.17.6.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degols G, Shiozaki K, Russell P. Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol Cell Biol. 1996;16:2870–2877. doi: 10.1128/mcb.16.6.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Yang X, Wang W, Wood III WH, Becker KG, Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc Natl Acad Sci USA. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauchon M, Lagniel G, Aude JC, Lombardia L, Soularue P, Petat C, Marguerie G, Sentenac A, Werner M, Labarre J. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol Cell. 2002;9:713–723. doi: 10.1016/s1097-2765(02)00500-2. [DOI] [PubMed] [Google Scholar]
- Gaits F, Degols G, Shiozaki K, Russell P. Phosphorylation and association with the transcription factor Atf1 regulate localization of Spc1/StyI stress-activated kinase in fission yeast. Genes Dev. 1998;12:1464–1473. doi: 10.1101/gad.12.10.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A.P. (2003). The environmental stress response: a common yeast response to environmental stresses. In: Yeast Stress Responses, ed S. Hohmann and P. Mager, Springer Verlag: (in press).
- Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenall A, Hadcroft AP, Malakasi P, Jones N, Morgan BA, Hoffman CS, Whitehall SK. Role of the fission yeast Tup1-like repressors and the Prr1 transcription factor in the response to salt stress. Mol Biol Cell, 2002;13:2977–2989. doi: 10.1091/mbc.01-12-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- Jain SK, Lim G. Pyridoxine and pyridoxamine inhibits superoxide radicals and prevents lipid peroxidation, protein glycosylation, and (Na+ + K+)-ATPase activity reduction in high glucose-treated human erythrocytes. Free Radic Biol Med. 2001;30:232–237. doi: 10.1016/s0891-5849(00)00462-7. [DOI] [PubMed] [Google Scholar]
- Jamieson DJ. Saccharomyces cerevisiae has distinct adaptive responses to both hydrogen peroxide and menadione. J Bacteriol. 1992;174:6678–6681. doi: 10.1128/jb.174.20.6678-6681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RH, Jones NC. Mammalian cAMP-responsive element can activate transcription in yeast and binds a yeast factor(s) that resembles the mammalian transcription factor ANF. Proc Natl Acad Sci USA. 1989;86:2176–2180. doi: 10.1073/pnas.86.7.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Dawes IW, Roe JH. Adaptive response of Schizosaccharomyces pombe to hydrogen peroxide and menadione. Microbiology. 1995;141:3127–3132. doi: 10.1099/13500872-141-12-3127. [DOI] [PubMed] [Google Scholar]
- Leupold U. Genetical methods for Schizosaccharomyces pombe. Methods Cell Physiol. 1970;4:169–177. [Google Scholar]
- Lyons TJ, Gasch AP, Gaither LA, Botstein D, Brown PO, Eide DJ. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad Sci USA. 2000;97:7957–7962. doi: 10.1073/pnas.97.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bähler J. The transcriptional program of sexual differentiation in fission yeast. Nature Genet, 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- Millar JB, Buck V, Wilkinson MG. Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev. 1995;9:2117–2130. doi: 10.1101/gad.9.17.2117. [DOI] [PubMed] [Google Scholar]
- Moradas-Ferreira P, Costa V. Adaptive response of the yeast Saccharomyces cerevisiae to reactive oxygen species: defenses, damage and death. Redox Rep. 2000;5:277–285. doi: 10.1179/135100000101535816. [DOI] [PubMed] [Google Scholar]
- Nguyen AN, Lee A, Place W, Shiozaki K. Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol Biol Cell. 2000;11:1169–1181. doi: 10.1091/mbc.11.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Herskowitz I, O'Shea EK. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 2002;18:405–412. doi: 10.1016/s0168-9525(02)02723-3. [DOI] [PubMed] [Google Scholar]
- Papadopoulos V, Amri H, Li H, Yao Z, Brown RC, Vidic B, Culty M. Structure, function and regulation of the mitochondrial peripheral-type benzodiazepine receptor. Therapie. 2001;56:549–556. [PubMed] [Google Scholar]
- Quinn J, Findlay VJ, Dawson K, Millar JB, Jones N, Morgan BA, Toone WM. Distinct regulatory proteins control the graded transcriptional response to increasing H2O2 levels in the fission yeast Schizosaccharomyces pombe. Mol Biol Cell. 2002;13:805–816. doi: 10.1091/mbc.01-06-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltsman KA, Prentice HL, Kingston RE. Mutations in the Schizosaccharomyces pombe heat shock factor that differentially affect responses to heat and cadmium stress. Mol Gen Genet. 1999;261:161–169. doi: 10.1007/s004380050953. [DOI] [PubMed] [Google Scholar]
- Sanchez-Piris M, Posas F, Alemany V, Winge I, Hidalgo E, Bachs O, Aligue R. The serine/threonine kinase Cmk2 is required for oxidative stress response in fission yeast. J Biol Chem. 2002;277:17722–17727. doi: 10.1074/jbc.M200104200. [DOI] [PubMed] [Google Scholar]
- Shalon D, Smith SJ, Brown PO. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- Shieh JC, Wilkinson MG, Buck V, Morgan BA, Makino K, Millar JB. The Mcs4 response regulator coordinately controls the stress-activated Wak1-Wis1-StyI MAP kinase pathway and fission yeast cell cycle. Genes Dev. 1997;11:1008–1022. doi: 10.1101/gad.11.8.1008. [DOI] [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Counteractive roles of protein phosphatase 2C (PP2C) and a MAP kinase kinase homolog in the osmoregulation of fission yeast. EMBO J. 1995;14:492–502. doi: 10.1002/j.1460-2075.1995.tb07025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- Siderius M, Mager WH. General stress response. In: Hohmann S, Mager W, editors. Search of a Common Denominator in Yeast Stress Responses. New York: Springer-Verlag; 1997. [Google Scholar]
- Smith DA, Toone WM, Chen Murchie D, Bähler J, Jones N, Morgan BA, Quinn J. The Srk1 protein kinase is a target for the StyI stress-activated MAPK in fission yeast. J Biol Chem. 2002;277:33411–33421. doi: 10.1074/jbc.M204593200. [DOI] [PubMed] [Google Scholar]
- Stohs SJ, Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free Radic Biol Med. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- Takeda T, Toda T, Kominami K, Kohnosu A, Yanagida M, Jones N. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 1995;14:6193–6208. doi: 10.1002/j.1460-2075.1995.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbles LA, Woodgett JR. The stress-activated protein kinase pathways. Cell Mol Life Sci. 1999;55:1230–1254. doi: 10.1007/s000180050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone WM, Jones N. Stress-activated signaling pathways in yeast. Genes Cells. 1998;3:485–498. doi: 10.1046/j.1365-2443.1998.00211.x. [DOI] [PubMed] [Google Scholar]
- Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, Jones N. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase. StyI/Spc1. Genes Dev. 1998;12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilo J. Discovering Frequent Patterns from Strings. Technical Report C-1998–9. Finland: Department of Computer Science, University of Helsinki; 1998. [Google Scholar]
- Vilo J, Brazma A, Jonassen I, Robinson A, Ukkonen E. Mining for putative regulatory elements in the yeast genome using gene expression data. Proc Int Conf Intell Syst Mol Biol. 2000;8:384–394. [PubMed] [Google Scholar]
- Wilkinson MG, Samuels M, Takeda T, Toone WM, Shieh JC, Toda T, Millar JB, Jones N. The Atf1 transcription factor is a target for the StyI stress-activated MAP kinase pathway in fission yeast. Genes Dev. 1996;10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]
- Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Yamada K, Nakagawa CW, Mutoh N. Schizosaccharomyces pombe homologue of glutathione peroxidase, which does not contain selenocysteine, is induced by several stresses and works as an antioxidant. Yeast. 1999;15:1125–1132. doi: 10.1002/(SICI)1097-0061(199908)15:11<1125::AID-YEA442>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]