Abstract
Background
Chlamydiae species are of much importance from a clinical viewpoint. Their diversity both in terms of their numbers as well as clinical involvement are presently believed to be significantly underestimated. The obligate intracellular nature of chlamydiae has also limited their genetic and biochemical studies. Thus, it is of importance to develop additional means for their identification and characterization.
Results
We have carried out analyses of available chlamydiae genomes to identify sets of unique proteins that are either specific for all Chlamydiales genomes, or different Chlamydiaceae family members, or members of the Chlamydia and Chlamydophila genera, or those unique to Protochlamydia amoebophila, but which are not found in any other bacteria. In total, 59 Chlamydiales-specific proteins, 79 Chlamydiaceae-specific proteins, 20 proteins each that are specific for both Chlamydia and Chlamydophila and 445 ORFs that are Protochlamydia-specific were identified. Additionally, 33 cases of possible gene loss or lateral gene transfer were also detected.
Conclusion
The identified chlamydiae-lineage specific proteins, many of which are highly conserved, provide novel biomarkers that should prove of much value in the diagnosis of these bacteria and in exploration of their prevalence and diversity. These conserved protein sequences (CPSs) also provide novel therapeutic targets for drugs that are specific for these bacteria. Lastly, functional studies on these chlamydiae or chlamydiae subgroup-specific proteins should lead to important insights into lineage-specific adaptations with regards to development, infectivity and pathogenicity.
Background
The Chlamydiales are clinically important intracellular parasites and endosymbionts of eukaryotic hosts, and cause a wide spectrum of diseases in humans and animals [1]. Recently, the taxonomic classification of this group was revised and currently contains at least 4 distinct families (viz.Chlamydiaceae, Simkaniaceae, Parachlamydiaceae and Waddliaceae) based on >90% 16S rRNA identity and a common developmental cycle [2,3]. While chlamydial infections are best known for the genitourinary, ocular and respiratory infections they cause in humans, the chlamydiae are also quite common in many wild and domestic animals with potential for severe zoonotic disease [4]. The newly described chlamydial species Simkania and Parachlamydia have also been found associated with human respiratory infections, while Waddlia has been implicated in abortion in bovines [5-7]. These revisions reflect important changes in perceptions regarding chlamydial diversity in line with recent discoveries of novel animal isolates (Waddliae and Simkaniae) and more strikingly of chlamydia-related endosymbionts or "environmental chlamydiae" (i.e. Parachlamydiae) infecting free-living amoebae [8-10]. Free-living amoebae, important components of soil and water ecosystems, are being increasingly recognized as vectors for various bacterial human pathogens [4,7,11,12].
Species are currently recognized as belonging to the Chlamydiales according to 16S and 23S rRNA similarities and pathogenic traits [2]. As these organisms are antigenically and genetically diverse, it is important to develop additional means to identify and distinguish them unambiguously from all other bacteria. The intracellular nature of chlamydiae species has been a hindrance in understanding their genetics, physiology and development. Hence, other means that can provide some insight in these regards are needed. In recent years, the genomes of several chlamydiae species have been sequenced including five Chlamydiaceae members (viz. Chlamydia (Chl.) trachomatis, Chl. muridarum, Chlamydophila (Chlam.) pneumoniae, Chlam. caviae and Chlam. abortus [13-16] and the environmental chlamydiae species Protochlamydia amoebophila (Parachlamydiaceae) [11]. Until recently when the P. amoebophila genome (formerly Parachlamydia UWE25;[17]) sequence was released, the information for chlamydiae-like organisms was practically non-existent, and it was virtually impossible to establish a core set of genes that are common to various Chlamydiales species. The Protochlamydia genome (2.41 Mb) was found to be about twice the size of various Chlamydiaceae species (1.04–1.23 Mb) and it showed a number of important differences including the presence of a complete TCA cycle and highly modified gene order [11]. The availability of these genomes has made it possible to carry out comparative studies. Horn et al. [11] reported 711 open reading frames (ORFs) or coding sequences that were shared among all chlamydiae genomes. However, many of these proteins have homologs in bacteria outside of the Chlamydiales and the proteins which were unique to only chlamydiae were not examined. Similarly, other studies which have determined proteins common to all Chlamydiaceae genomes also included peptides which were ubiquitous among bacteria [13-16]. While the work of Kalman et al. [18] revealed a number of potentially chlamydiae-specific genes, these studies were based on only 2 genomes, Chlam. pneumoniae and Chl. trachomatis, and the study was carried out when sequence information for other bacteria was limited.
Our recent work on comparative genomics is aimed at identifying novel and distinctive molecular characteristics of different groups of bacteria, including chlamydiae, which can be used for their identification, classification as well as genetic and biochemical studies [19-23] (see also [24]). We have recently described a number of molecular signatures consisting of conserved inserts and deletions (i.e. indels) in widely distributed proteins that are distinctive characteristics of all available chlamydiae species and are not found in any other bacteria [25]. In the present study, we describe a different kind of taxonomic marker, consisting of whole proteins that are specific for different groups of chlamydiae species, which provides additional powerful means for identifying these groups of species and for understanding their evolution as well as physiological characteristics. In this work, we have carried out systematic BLAST searches on all proteins or ORFs in the genomes of a number of chlamydiae species to compile profiles of proteins that are either uniquely shared by all Chlamydiales-species or particular subgroups (family or genera) within this phylum and can likely be used as distinct molecular markers for these groups. The study of these chlamydial group specific proteins should prove instrumental in the discovery of novel physiological characteristics that are uniquely shared by this important group of pathogens.
Results
The present study was undertaken to identify unique proteins (or ORFs) which are found in the chlamydiae species at various taxonomic depths. Kalman et al [18] have previously examined the presence of chlamydiae-specific proteins in 2 sequenced genomes, Chl. trachomatis serovar D and Chlam. pneumoniae. Their study documented a number of proteins such as EUO, Gp6D, IncsB/C, LtuA/B, a number of Pmps and hypothetical proteins, which were specific for these species. However, because the number of species examined was so small, it was unclear how broadly these proteins were distributed among different chlamydiae species. At that time there was also no sequence information for the proteins from any chlamydiae-like organisms. Thus, based on this earlier study it was unclear whether the identified peptides were distinctive of different Chlamydiales, or only particular subsets of chlamydial species [18].
In order to identify chlamydiae-specific genes/proteins that are present at different phylogenetic depths, each ORF from the Chl. trachomatis and Chlam. caviae genomes were individually BLAST searched against all available databases. These species were chosen in order that sequences from both the Chlamydia and the Chlamydophila genera were represented in the searches. Results of the BLAST searches were inspected to identify proteins where either all of the observed hits were from other chlamydial species, or where all hits showing significant homology to the query protein (see Methods section) were members of the Chlamydiales. These studies have resulted in the identification of large number of signature proteins, which are uniquely found in chlamydiae species. Some of these proteins are present in all available chlamydial sequences indicating that they are likely specific for the entire Chlamydiales phylum. Other proteins were found to be specific for only the Chlamydiaceae family or the two genera, Chlamydia and the Chlamydophila, which comprise this family. Additionally, large numbers of proteins that are unique to P. amoebophila were also identified. Most of the identified chlamydiae-specific proteins or ORFs are annotated as hypothetical, hence their functions remain unknown. The genes for most of these proteins are scattered throughout the Chl. trachomatis and Chlam. caviae genomes, however a number of these hypothetical proteins were also found in clusters. Although, it is not clear whether these genes form operons, the presence of these genes in clusters suggest that they could be involved in related functions [26]. In the description of these proteins that follows, the "CT", "CC" and "PC" part of the descriptors refer to the source of the original query protein sequence from Chl. trachomatis (CT), Chlam. caviae (CCA) and P. amoebophila (PC) genomes, respectively.
Chlamydiales-specific proteins
Proteins were considered Chlamydiales-specific if they were present in all sequenced Chlamydia and Chlamydophila genomes, as well as P. amoebophila, but were absent in all other bacteria. A total of 59 Chlamydiales-specific proteins were identified (Table 1). Of these, 49 peptides were annotated hypothetical indicating that these organisms have created new strategies allowing them to pursue a parasitic lifestyle with a minimum gene complement. Upon examination of the positions of various Chlamydiales-specific proteins within the Chl. trachomatis and Chlam. caviae genomes, some of them were found to be clustered on the chromosomes (those with adjacent CT numbers). A few of these gene clusters are discussed below.
Table 1.
List of the Chlamydiales-specific proteins
| Protein Loci | Protein (Name/Size) | C. tra | C. mur | C. pne | C. cav | C. abo | Protochlam. | Next best BLAST Hit (E value, protein length) |
| CT007 (AAC67597) | hypoth, 316aa | 0 | 8e-164 | 8e-108 | 6e-108 | 3e-105 | 8e-10, 320aa | Plasmodium yoelii (0.063, 1491aa) |
| CT011 (AAC67601) | hypoth, 418aa | 0 | 0 | 7e-151 | 2e-155 | 1e-157 | 7e-47, 445aa | Desulfotalea psychrophila (1e-06, 337aa) |
| CT016 (AAC67606) | hypoth, 242aa | 2e-139 | 3e-119 | 1e-82 | 4e-89 | 3e-86 | 2e-37, 233aa | Rickettsia sibirica (0.20, 1851aa) |
| CT017 (AAC67607) | hypoth. 433aa | 0 | 0 | 1e-122 | 7e-133 | 9e-135 | 4e-52, 431aa | Thermo. tengcongensis (1e-06, 306 aa) |
| CT021 (AAC67611) | hypoth, 247aa | 2e-139 | 4e-107 | 1e-47 | 3e-56 | 4e-53 | 2e-13, 245aa | Xenopus laevis (1.7, 600aa) |
| CT031 (AAC67621) | hypoth, 100aa | 4e-51 | 4e-42 | 2e-25 | 4e-26 | 2e-26 | 0.010, 107aa | Mus musculus (3.6, 384aa) |
| CT038 (AAC67628) | hypoth, 116aa | 1e-63 | 2e-49 | 4e-25 | 1e-25 | 2e-23 | 2e-09, 109aa | Neurospora crassa (2.8, 1223aa) |
| CT053 (AAC67644) | hypoth, 148aa | 2e-61 | 4e-57 | 2e-51 | 1e-50 | 1e-48 | 5e-29, 155aa | Xenopus tropicalis (4.7, 1049aa) |
| CT066 (AAC67657) | hypoth, 158aa | 3e-77 | 1e-75 | 8e-65 | 3e-65 | 5e-65 | 6e-41, 165aa | Vibrio parahaemolyticus (1.8, 92aa) |
| CT079 (AAC67670) | hypoth, 147aa | 4e-81 | 1e-71 | 2e-06 | 1e-29 | 1e-29 | 3e-06, 172aa | Leuconostoc mesenteroides (0.11, 263aa) |
| CT131 (AAC67722) | PTMP, 1138aa | 0 | 0 | 0 | 0 | 0 | 3e-33, 1348aa | Plasmodium yoelii (0.31, 592aa) |
| CT181 (AAC67772) | hypoth, 236aa | 7e-131 | 1e-113 | 3e-77 | 7e-81 | 4e-80 | 2e-10, 200aa | Acinetobacter sp. (0.94, 371aa) |
| CT260 (AAC67853) | hypoth, 163aa | 9e-91 | 9e-81 | 5e-62 | 3e-68 | 1e-67 | 4e-22, 159aa | Sinorhizobium meliloti (0.18, 166aa) |
| CT266 (AAC67859) | hypoth, 393aa | 0 | 5e-179 | 9e-103 | 5e-120 | 2e-119 | 4e-08, 498aa | Xanthomonas axonopodis (2.8, 189aa) |
| CT271 (AAC67864) | hypoth, 95aa | 1e-39 | 2e-36 | 7e-25 | 1e-25 | 9e-26 | 1e-10, 97aa | Gibberella zeae (3.6, 1362) |
| CT273 (AAC67866) | hypoth, 188aa | 2e-89 | 1e-77 | 2e-60 | 5e-63 | 2e-63 | 0.001, 149aa | Corynebacterium efficiens (0.34, 640aa) |
| CT274 (AAC67867) | type III secretion chaperone, 139aa | 3e-74 | 3e-70 | 1e-61 | 1e-62 | 1e-61 | 3e-31, 131aa | Synechococcus elongatus (2e-05, 363aa) |
| CT303 (AAC67896) | hypoth, 171aa | 2e-81 | 5e-66 | 2e-40 | 2e-47 | 6e-47 | 3e-04, 177aa | Yersinia pestis (1.3, 373aa) |
| CT309 (AAC67902) | hypoth, 266aa | 2e-144 | 2e-129 | 8e-86 | 1e-93 | 5e-94 | 2e-29, 263aa | Methanococcus jannaschii (2.6, 434aa) |
| CT312 (AAC67905) | pred. ferredoxin, 123aa | 6e-67 | 6e-54 | 1e-30 | 7e-36 | 1e-35 | 1e-04, 130aa | Pan troglodytes (1.2, 1524aa) |
| CT350 (AAC67945) | hypoth, 566aa | 0 | 0 | 2e-141 | 3e-157 | 1e-157 | 2e-30, 561aa | Nostoc sp. PCC 7120 (7e-06, 1547aa) |
| CT351 (AAC67946) | hypoth, 697aa | 0 | 0 | 0 | 0 | 0 | 5e-53, 720aa | Rattus norvegicus (3.4, 389aa) |
| CT355 (AAC67950) | hypoth, 353aa | 0 | 3e-161 | 3e-101 | 8e-101 | 1e-102 | 1e-21, 375aa | Chromobacterium violaceum (0.82, 556aa) |
| CT384 (AAC67980) | hypoth, 539aa | 0 | 0 | 9e-106 | 4e-113 | 1e-111 | 2e-14, 580aa | Pseudomonas fluorescens (0.29, 314aa) |
| CT387 (AAC67984) | hypoth, 691aa | 0 | 0 | 0 | 0 | 0 | 4e-160, 740aa | Rubrobacter xylanophilus (5e-10, 263aa) |
| CT421.2 (AAC68031) | hypoth, 53aa | 5e-23 | 2e-22 | 5e-19 | 7e-20 | 4e-19 | 2e-12, 52aa | no other BLAST hits |
| CT425 (AAC68022) | hypoth, 621aa | 0 | 0 | 0 | 0 | 0 | 0, 665aa | Bordetella parapertussis (4e-10, 767aa) |
| CT429 (AAC68026) | hypoth, 329aa | 4e-165 | 4e-162 | 5e-149 | 2e-151 | 7e-151 | 8e-96, 388aa | Geobacter sulfurreducens (2e-04, 146aa) |
| CT443 (AAC68042) | OmcB, 553aa | 0 | 0 | 0 | 0 | 0 | 4e-66, 558aa | Rhodopirellula baltica (3e-25, 907aa) |
| CT446 (AAC68045) | EUO protein, 183aa | 1e-78 | 8e-72 | 1e-50 | 2e-53 | 1e-52 | 4e-41, 139aa | Magnaporthe grisea (2.1, 1259aa) |
| CT474 (AAC68074) | LysM protein, 336aa | 2e-168 | 9e-116 | 5e-59 | 9e-71 | 7e-71 | 3e-04, 482aa | no other BLAST hits |
| CT504 (AAC68105) | hypoth, 288aa | 2e-127 | 1e-105 | 4e-64 | 2e-69 | 2e-68 | 1e-23, 376aa | Mus musculus (3.0, 961aa) |
| CT538 (AAC68140) | hypoth, 238aa | 8e-124 | 1e-120 | 5e-105 | 1e-108 | 1e-108 | 9e-65, 244aa | Fritschea bemisiae (3e-59, 241aa) Enterococcus faecalis (0.067, 541aa) |
| CT546 (AAC68148) | predicted OMP, 289aa | 3e-163 | 4e-157 | 6e-87 | 1e-92 | 1e-93 | 3e-23, 249aa | Legionella pneumophila (0.042, 254aa) |
| CT547 (AAC68149) | hypoth, 318aa | 2e-175 | 7e-158 | 8e-90 | 1e-92 | 2e-93 | 8e-29, 468aa | Caenorhabditis elegans (4e-04, 1168aa) |
| CT548 (AAC68150) | hypoth, 194aa | 3e-89 | 2e-67 | 1e-48 | 2e-49 | 3e-48 | 0.004, 185aa | Drosophila melanogaster (0.17, 826aa) |
| CT556 (AAC68158) | hypoth, 159aa | 1e-92 | 1e-82 | 9e-59 | 1e-60 | 1e-59 | 2e-06, 157aa | Xenopus laevis (1.8, 1636aa) |
| CT560 (AAC68162) | hypoth, 278aa | 1e-135 | 2e-125 | 1e-86 | 4e-94 | 5e-93 | 0.002, 281aa | no other BLAST hits |
| CT573 (AAC68175) | hypoth, 409aa | 0 | 1e-179 | 9e-90 | 3e-113 | 2e-106 | 1e-43, 472aa | Pseudomonas syringae (0.46, 9376aa) |
| CT583 (AAC68185) | Virulence plasmid pGP6 related protein, 263aa | 5e-134 | 6e-118 | 4e-82 | 4e-91 | 2e-92 | 1e-46, 245aa | Crocosphaera watsonii (0.036, 333aa) |
| CT590 (AAC68192) | hypoth, 954aa | 0 | 0 | 0 | 0 | 0 | 8e-96, 989aa | Porphyromonas gingivalis (0.003, 645aa) |
| CT602 (AAC68205) | hypoth, 130aa | 5e-60 | 1e-55 | 2e-35 | 3e-37 | 1e-37 | 2e-19, 131aa | Actinobacillus pleuropneumoniae (2.8, 300aa) |
| CT616 (AAC68220) | hypoth, 429aa | 0 | 0 | 2e-101 | 8e-115 | 3e-114 | 2e-09, 435aa | Giardia lamblia (0.003, 1024aa) |
| CT623 (AAC68227) | 76 kDa Homolog, 446aa | 0 | 2e-155 | 5e-152 | 1e-154 | 1e-157 | 1e-28, 470aa | Mycobacterium tuberculosis (0.18, 550aa) |
| CT631 (AAC68235) | hypoth, 84aa | 1e-32 | 2e-31 | 1e-24 | 3e-28 | 8e-28 | 1e-11, 82aa | No other BLAST hits |
| CT635 (AAC68239) | hypoth, 144aa | 2e-62 | 5e-58 | 6e-45 | 1e-50 | 2e-51 | 4e-04, 146aa | Gallus gallus (0.003, 1748aa) |
| CT642 (AAC68246) | hypoth, 271aa | 2e-134 | 2e-116 | 2e-74 | 2e-79 | 8e-81 | 2e-22, 300aa | Rattus norvegicus (1.6, 509aa) |
| CT652.1 (AAC68830) | hypoth, 59aa | 5e-26 | 3e-24 | 7e-12 | 3e-12 | 7e-13 | 3e-05, 75aa | Parachlamydia sp. (0.12, 70aa) |
| CT666 (AAC68261) | hypoth, 83aa | 5e-40 | 9e-31 | 3e-35 | 1e-36 | 9e-37 | 0.43, 92aa | Sacchromyces cerevisiae (3.7, 604aa) |
| CT667 (AAC68262) | hypoth, 149aa | 3e-78 | 1e-65 | 3e-51 | 9e-55 | 4e-55 | 8e-16, 154aa | Legionella pneumophila (0.005, 577aa) |
| CT670 (AAC68265) | hypoth, 168aa | 1e-29 | 1e-27 | 2e-23 | 4e-24 | 3e-23 | 9e-10, 165aa | No other BLAST hits |
| CT734 (AAC68329) | hypoth, 221aa | 7e-106 | 1e-95 | 2e-51 | 1e-56 | 2e-57 | 1e-08, 240aa | Caenorhabditis briggsae (1.1, 1072aa) |
| CT764 (AAC68359) | hypoth, 268aa | 2e-154 | 4e-113 | 1e-46 | 7e-59 | 4e-59 | 2e-05, 256aa | Tetraodon nigroviridis (1.5, 388aa) |
| CT788 (AAC68383) | leader (60) peptide-periplasmic, 166aa | 1e-79 | 3e-61 | 9e-25 | 4e-34 | 1e-32 | 0.003, 158aa | Caenorhabditis elegans (0.19, 2271aa) |
| CT809 (AAC68405) | hypoth, 103aa | 1e-55 | 2e-44, 146aa | 5e-17 | 2e-23 | 2e-21 | 4e-05, 158aa | Canis familiaris (2.1, 1831aa) |
| CT838 (AAC68435) | YjgP/YjgQ family protein, 366aa | 0 | 0 | e-126 | e-130 | 3e-128 | 6e-24, 360aa | Prochlorococcus marinus (0.18, 592aa) |
| CT845 (AAC68442) | hypoth, 92aa | 2e-46 | 7e-33 | 1e-19 | 2e-17 | 1e-17 | 8e-10, 85aa | Candida albicans (2.7, 557aa) |
| CT849.1 (AAC68455) | hypoth, 62aa | 2e-28 | 9e-24 | 5e-10 | 2e-09 | 1e-08 | 1e-05, 81aa | No other BLAST hits |
| CT865 (AAC68463) | hypoth, 329aa | 2e-180 | 8e-146 | 7e-76 | 1e-77 | 4e-78 | 9e-23, 352aa | Bacteroides thetaiotaomicron (0.20, 808aa) |
The Chlamydiales-specific proteins were identified by systematic BLASTP searches using Chl. trachomatis ORFs as probes, where the only significant BLAST hits obtained were for chlamydiae species (see Methods section). Accession numbers of the query sequences are shown beside the Chl. trachomatis loci (CT number) in the first column. The observed expected (E) values for various chlamydiae homologs as well as the first non-chlamydial hit in the BLAST results are given. E values of 0.0 indicate an extremely high degree of similarity between proteins sequences. The second column describes the function and the size of each query protein (protein sizes are described for other chlamydiae only if they noticeably differ from the query protein). Abbreviations are: C. tra, Chl. trachomatis; C. mur; Chl. muridarum; C. pne, Chlam. pneumoniae; C. cav, Chlam. caviae; C. abo, Chlam. abortus; hypoth, hypothetical protein; Omc; Outer membrane cytochrome; OMP, outer membrane protein; Protochlam, Protochlamydia; Pred., Predicted; PTMP, Possible transmembrane protein.
Of the proteins annotated with a predicted function, only two represent outer membrane proteins (CT131 and CT546), which is surprising in light of large expansion of genes encoding chlamydial polymorphic membrane proteins [15,27]. One of these proteins CT546 is in a cluster with two other Chlamydiales-specific proteins (CT547 and CT548), whose functions are not known, but they could be involved in related functions [26]. Another important Chlamydiales-specific protein that is involved in the formation of their cell envelope is CT443 (OmcB). OmcB is not actually intercalated into a membrane. However, this protein is 4.4% cysteine, water-soluble and crosslinked to MOMP and another protein OmcA (CT444) to form the membrane complex [28,29]. OmcA and OmcB are two of the very few chlamydiae-specific proteins that have been functionally characterized [28,29]. The OmcA (CT444) protein is 15% cysteine and so has homology only to proteins with high cysteine content. It has a classic SPII signal sequence and is a proven lipoprotein [28,29]. OmcA and OmcB are transcribed together [28,29] and the proteins that are annotated as OmcA and OmcB are present in all genome sequenced chlamydiae [13-16], including P. amoebophila [11]. However, in BLASTp searches using either the Chl. trachomatis or Chlam. caviae OmcA (CCA00184) homologs as the query proteins (with or without the low complexity filter), an OmcA homolog was not detected in P. amoebophila although a protein PC0617 (accession number YP_007016) has been annotated as its OmcA homolog. Hence, by the criteria used in this study we regard CT444 as a Chlamydiaceae-specific and not a Chlamydiales-specific protein, and we have included it in Table 2. OmcA and OmcB annotation is presently based on the high cysteine content of the predicted protein products, as the cysteine residues are the basis for OmcA and OmcB function in Chlamydiaceae. Because OmcA and OmcB annotation has changed many times in the literature over the last 15 years [28,29], these proteins are best located by BLAST search. It should be mentioned that besides the Chlamydiales a homolog of CT443 with low E value (3e-25) is also found in Rhodopirellula baltica (Table 1). The R. baltica protein (accession number CAD72259) is larger in length (907aa) in comparison to the chlamydiae homolog and the sequence similarity between these two proteins is mainly seen in the C-terminal region. Importantly, the R. baltica homolog of OmcB contains no cysteine, hence the possible significance of the presence of this homologous protein in R. baltica (Planctomycetes) is presently unclear. A close relationship of chlamydiae to the Planctomycetes, which also lack peptidoglycan in their cell wall, has been noted in earlier studies [30,31]. Although such a relationship was not supported in a later study [32], in view of the presence of this commonly shared protein, it would be of interest to reexamine the relationship between these groups based on genomic sequences.
Table 2.
A Listing of the Identified Chlamydiaceae-specific proteins
| Protein Loci | Protein (Name/Size) | C. tra | C. mur | C. pne | C. cav | C. abo | Next best BLAST hit (E value, protein length) |
| CT006 (AAC67596) | hypoth, 189aa | 3e-95 | 6e-72 | 3e-12 | 9e-10 | 2e-09 | Listeria anguillarum (0.45, 314aa) |
| CT005 (AAC67595) | hypoth, 363aa | 1e-117 | 4e-63 | 3e-18, 417aa | 8e-20 | 1e-20 | Ashbya gossypii (0.86, 2272aa) |
| CT018 (AAC 67608) | hypoth, 157aa | 3e-89 | 5e-53 | 2e-10 | 8e-09 | 3e-06 | Acetabularia acetabulum (3.9, 613aa) |
| CT046 (AAC67637) | Histone-like protein HC2, 203aa | 2e-29 | 2e-25 | 2e-05, 172aa | 8e-08, 152aa | 2e-08 | No other BLAST hits |
| CT080 (AAC67671) | LtuB protein, 97aa | 3e-50 | 4e-37 | 3e-11 | 2e-13 | 9e-16 | Clostridium tetani (0.73, 311aa) |
| CT082 (AAC67673) | hypoth, 560aa | 0 | 0 | 7e-91 | 8e-103 | 2e-102 | Saccharomycecs cerevisiae (2.0, 1537aa) |
| CT083 (AAC67674) | hypoth, 160aa | 1e-71 | 1e-57 | 7e-30 | 4e-29 | 9e-25 | Pseudomonas putida (0.17, 1162aa) |
| CT109 (AAC67700) | hypoth, 262aa | 2e-149 | 9e-133 | 8e-66 | 4e-70 | 7e-70 | Plasmodium falciparum (1.1, 1057aa) |
| CT142 (AAC67733) | hypoth, 285aa | 3e-155 | 2e-109 | 1e-39 | 9e-50 | 6e-49 | Heterobasidion annosum (5.0, 507aa) |
| CT143 (AAC67734) | hypoth, 280aa | 3e-165 | 3e-123 | 2e-36 | 9e-55 | 4e-56 | porcine (0.26, 725aa) |
| CT144 (AAC67735) | hypoth, 285aa | 2e-143 | 3e-101 | 7e-28 | 9e-34 | 6e-31 | Staphylococcus aureus (0.041, 154aa) |
| CT147 (AAC67738) | hypoth, 1449aa | 0 | 0 | 3e-71 | 4e-88 | 4e-91 | Homo sapiens (8e-10, 2442aa) |
| CT178 (AAC67769) | hypoth, 404aa | 0 | 0 | 3e-114 | 3e-120 | 4e-122 | Thiobacillus denitrificans (2e-15, 830aa) |
| CT195 (AAC67787) | hypoth, 363aa | 0 | 2e-147 | 8e-55 | 4e-71 | 1e-68 | Escherichia coli (1.5, 346aa) |
| CT232 (AAC67824) | IncB, 115aa | 7e-45 | 1e-29 | 4e-11 | 2e-08 | 7e-10 | Canis familiaris (0.59, 472aa) |
| CT233 (AAC67825) | IncC, 178aa | 9e-75 | 3e-38 | 0.52, 203aa | 4e-04 | 0.006,184aa | Tetraodon nigroviridis (0.5, 1010aa) |
| CT253 (AAC67846) | hypoth, 215aa | 5e-108 | 2e-99 | 2e-67 | 5e-72 | 8e-73 | Schizosaccharomyces pombe (0.12, 880aa) |
| CT263 (AAC67856) | hypoth, 196aa | 7e-109 | 1e-72 | 5e-23 | 2e-31 | 1e-29 | Mycobacterium bovis (2.4, 744aa) |
| CT288 (AAC67881) | hypoth, 563aa | 0 | 3e-175 | 6e-16 | 5e-23 | 1e-25 | Debaryomyces hansenii (4e-04, 2042aa) |
| CT289 (AAC67882) | hypoth, 379aa | 3e-167 | 3e-95 | 6e-27 | 6e-38 | 5e-31 | Streptococcus pyogenes (0.049, 535aa) |
| CT324 (AAC67917) | hypoth, 303aa | 1e-161 | 8e-115 | 1e-50 | 6e-55 | 1e-55 | Brucella melitensis (0.29, 263aa) |
| CT330 (AAC67925) | hypoth, 90aa | 2e-44 | 1e-32 | 3e-07 | 3e-04 | 1e-05 | Oryza sativa (2.7, 677aa) |
| CT338 (AAC67933) | hypoth, 153aa | 1e-86 | 1e-66 | 1e-23 | 2e-29 | 5e-28 | Acinetobacter sp. (1.6, 304aa) |
| CT365 (AAC67961) | hypoth, 575aa | 0 | 4e-156 | 3e-58 | 7e-63 | 1e-61 | No other BLAST hits |
| CT372 (AAC67968) | hypoth, 442aa | 0 | 0 | 1e-114 | 2e-116 | 3e-119 | Bradyrhizobium japonicum (0.016, 675aa) |
| CT377 (AAC67973) | LtuA, 46aa | 4e-20 | 8e-19 | 2e-08 | 6e-11 | 5e-10 | No other BLAST hits |
| CT383 (AAC67979) | hypoth, 243aa | 7e-140 | 1e-88 | 3e-26 | 2e-35 | 2e-28 | Burkholderia fungorum (2.9, 780aa) |
| CT392 (AAC67989) | yprS, 377aa | 0 | 2e-116 | 1e-07 | 1e-14 | 3e-07 | Saccharomyces cerevisiae (0.048, 1038aa) |
| CT412 (AAC68009) | PmpA, 975aa | 0 | 0 | 8e-141 | 3e-179 | 4e-175 | many other chlamydiae hits in between Chloroflexus aurantiacus (3e-05, 767aa) |
| CT413 (AAC68010) | PmpB, 1751aa | 0 | 0 | 0 | 0 | 0 | Chloroflexus aurantiacus, 8e-10, 767aa |
| CT421.1 (AAC68030) | hypoth, 53aa | 7e-23 | 3e-20 | 4e-15 | 6e-12 | 1e-14 | Bradyrhizobium japonicum (3.8, 803aa) |
| CT440 (AAC68039) | hypoth, 112aa | 1e-58 | 2e-45 | 6e-08 | 1e-04 | 4e-07 | Homo sapiens (0.017, 100aa) (partial) |
| CT442 (AAC68041) | 15kDa SRP, 150aa | 7e-70 | 1e-33 | 1e-04 | 1e-12 | 3e-12 | No other BLAST hits |
| CT444 (AAC68043) | OmcA, 88aa | 1e-39 | 4e-31 | 2e-12 | 6e-15 | 6e-15 | Ovis aries (0.14, 197aa) ** |
| CT466 (AAC68066) | hypoth, 109aa | 5e-48 | 7e-39 | 2e-24 | 1e-26 | 1e-27 | Geobacter metallireducens (0.55, 557aa) |
| CT471 (AAC68071) | hypoth, 200aa | 5e-112 | 1e-103 | 1e-46 | 1e-62 | 5e-63 | Azoarcus sp. (1.9, 168aa) |
| CT482 (AAC68082) | hypoth, 217aa | 1e-115 | 1e-91 | 1e-49 | 6e-60 | 4e-64 | Legionella pneumophila (1.8, 438aa) |
| CT483 (AAC68083) | hypoth, 121aa | 1e-51 | 1e-36 | 6e-06 | 2e-08 | 4e-08 | Rubrobacter xylanophilus (0.14, 293aa) |
| CT529 (AAC68131) | hypoth, 298aa | 1e-151 | 2e-91 | 4e-36 | 1e-38 | 8e-42 | Aspergillus nidulans (1.9, 274aa) |
| CT552 (AAC68154) | hypoth, 135aa | 6e-72 | 1e-41 | 5e-04 | 3e-06 | 7e-07 | Guillardia theta (0.19, 263aa) |
| CT565 (AAC68167) | hypoth, 147aa | 7e-79 | 1e-69 | 1e-41 | 3e-46 | 7e-48 | Caenorhabditis elegans (0.11, 317aa) |
| CT566 (AAC68168) | hypoth, 330aa | 8e-175 | 6e-141 | 8e-86 | 4e-89 | 1e-87 | Vibrio vulnificus (0.26, 506aa) |
| CT567 (AAC68169) | hypoth, 174aa | 1e-87 | 9e-61 | 5e-24 | 2e-17 | 2e-17 | Takifugu rubripes (4.1, 1374aa) |
| CT568 (AAC68170) | hypoth, 151aa | 2e-71 | 4e-48 | 2e-22 | 2e-18 | 5e-18 | Giardia lamblia (1.6, 6739aa) |
| CT577 (AAC68179) | hypoth, 119aa | 1e-60 | 3e-48 | 6e-16 | 3e-26 | 2e-26 | Giardia lamblia (0.050, 551aa) |
| CT578 (AAC68180) | hypoth, 487aa | 8e-175 | 3e-150 | 4e-81 | 2e-99 | 1e-103 | Candida glabrata (0.023, 592aa) |
| CT579 (AAC68181) | hypoth, 439aa | 3e-141 | 8e-115 | 2e-76 | 4e-77 | 1e-81 | No other BLAST hits |
| CT589 (AAC68191) | hypoth, 602aa | 0 | 0 | 1e-126 | 1e-145 | 3e-146 | Bacillus subtilis (0.052, 605aa) |
| CT618 (AAC68222) | hypoth, 266aa | 1e-142 | 6e-87 | 8e-10 | 4e-19, 306aa | 5e-22 | Drosophila melanogaster (0.11, 1868aa) |
| CT620 (AAC68224) | hypoth, 838aa | 0 | 0 | 0 | 1e-180 | 2e-177 | Borrelia burgdorferi (0.29, 735aa) |
| CT646 (AAC68823) | hypoth, 459aa | 0 | 0 | 3e-60 | 1e-68 | 5e-66 | Aspergillus nidulans (5.9, 629aa) |
| CT647 (AAC68824) | hypoth, 192aa | 2e-98 | 5e-65 | 2e-18 | 1e-15 | 3e-13 | Ciona intestinalis (1.8, 407aa) |
| CT651 (AAC68828) | hypoth, 608aa | 0 | 0 | 1e-97 | 7e-97 | 3e-95 | Bacteroides thetaiotaomicron (0.15, 1003aa) |
| CT654 (AAC68249) | hypoth, 162aa | 5e-81 | 9e-75 | 1e-27 | 1e-32 | 1e-33 | Mesocricetus auratus (2.5, 210aa) |
| CT668 (AAC68263) | hypoth, 223aa | 1e-114 | 9e-93 | 6e-69 | 5e-78 | 1e-76 | Bacillus halodurans (0.045, 324aa) |
| CT671 (AAC68266) | hypoth, 283aa | 2e-149 | 1e-115 | 2e-63 | 2e-76 | 1e-72 | Giberella zeae (0.053, 595aa) |
| CT681 (AAC68276) | MOMP, 393aa | 0 | 7e-176 | 2e-139 | 3e-137 | 2e-136 | No other BLAST hits except chlamydiae |
| CT695 (AAC68290) | hypoth, 398aa | 0 | 7e-147 | 6e-12 | 2e-23 | 2e-22 | Ustilago maydis (0.44, 417aa) |
| CT696 (AAC68291) | hypoth, 403aa | 0 | 3e-179 | 6e-59 | 3e-70 | 2e-73 | Mus musculus (0.031, 708aa) |
| CT712 (AAC68307) | hypoth, 390aa | 0 | 0 | 8e-101 | 1e-129 | 1e-132 | Mus musculus (0.086, 3159aa) |
| CT713 (AAC68308) | PorB, 340aa | 0 | 0 | 5e-120 | 2e-133 | 5e-132 | No other BLAST hits except chlamydiae |
| CT718 (AAC68313) | hypoth, 174aa | 3e-93 | 8e-81 | 3e-44 | 2e-46 | 6e-47 | Prochlorococcus marinus (0.82, 250aa) |
| CT744 (AAC68339) | phosphoprotein, 821aa | 0 | 0 | 2e-138 | 1e-153 | 3e-160 | Cytophaga hutchinsonii (0.38, 466aa) |
| CT753 (AAC68348) | hypoth, 74aa | 3e-34 | 4e-27 | 1e-13, 76aa | 2e-13 | 2e-13 | Cycas necrotic stunt virus (1.3, 559) |
| CT763 (AAC68358) | hypoth, 139aa | 6e-72 | 3e-52 | 2e-28 | 3e-23 | 2e-22 | Drosophila pseudoobscura (2.6, 773aa) |
| CT795 (AAC68390) | hypoth, 163aa | 1e-79 | 7e-46 | 1e-13 | 3e-13 | 2e-14 | Pan troglodytes (0.52, 924aa) |
| CT812 (AAC68408) | OmpD, 1531aa | 0 | 0 | 0 | 0 | 0 | Rhodopirelulla baltica (2e-04, 703aa) |
| CT846 (AAC68443) | hypoth, 234aa | 1e-123 | 1e-108 | 7e-67 | 3e-64 | 1e-66 | Streptomyces coelicolor (2.1, 689aa) |
| CT847 (AAC68444) | hypoth, 172aa | 2e-88 | 8e-73 | 1e-53 | 1e-53 | 2e-52 | Saccharomyces cerevisiae (0.001, 1434aa) |
| CT848 (AAC68445) | hypoth, 168aa | 1e-76 | 5e-67 | 2e-55 | 6e-54 | 8e-54 | Drosophila pseudoobscura (0.34, 1000aa) |
| CT849 (AAC68446) | hypoth, 159aa | 4e-74 | 1e-64 | 2e-57 | 1e-56 | 4e-57 | Gallus gallus (0.034, 1546aa) |
| CT860 (AAC68458) | hypoth, 493aa | 0 | 1e-132 | 2e-56 | 4e-73 | 4e-77 | Leishmania major (0.26, 1346aa) |
| CT861 (AAC68459) | hypoth, 506aa | 0 | 7e-172 | 6e-67 | 9e-74 | 5e-76 | Vibrio fischeri (0.008, 1023aa) |
| CT863 (AAC68461) | hypoth, 482aa | 0 | 2e-150 | 7e-82 | 2e-86 | 7e-90 | Mus musculus (0.006, 1043aa) |
| CT869 (AAC68467) | OmpE, 964aa | 0 | 0 | 9e-108 | 1e-119 | 8e-112 | Escherichia coli (9e-05, 1250aa) |
| CT870 (AAC68468) | OmpF, 1034aa | 0 | 0 | 1e-40, 931aa | 2e-41 | 5e-46 | long list of chlamydiae homologs ; Dictyostelium discoideum (4e-04, 485aa) |
| CT871 (AAC68469) | OmpG, 1013aa | 0 | 0 | 4e-127 | 7e-180 | 2e-175 | long list of chlamydiae homologs ; Desulfotalea psychrophila (8e-07, 2363aa) |
| CT872 (AAC68470) | OmpH, 1016aa | 0 | 0 | 6e-152, 978aa | 0 | 0 | long list of chlamydiae homologs ; Crocosphaera watsonii (0.28, 822aa) |
| CT874 (AAC68472) | OmpI, 878aa | 0 | 0 | 2e-81, 930aa | 9e-73, 942aa | 2e-77 | long list of chlamydiae homologs ; Nostoc punctiforme (0.006, 1851aa) |
These proteins are uniquely found in various Chlamydiaceae species but not detected in Protochlamydia or other bacterial groups. Other details as in Table 1. **OmcA is an annotated ORF in the P. amoebophila genome, however it could not be detected using Chl. trachomatis as probe via BLASTp and so was not considered a Chlamydiales protein; SRP, sulfur rich protein.
The finding of only a limited number of outer membrane proteins unique to various Chlamydiales suggests that the chlamydiae species most likely favour lineage-specific mechanisms or surface receptors for interacting with their different hosts rather than sharing a homologous system. Cell surface interactions are not limited to the outer membrane proteins. The LysM domain is a widespread protein module, which the available evidence suggests is a general peptidoglycan-binding module in cell surface proteins associated with cell wall degradation [33]. A LysM domain protein (CT474) was found to be common in all members of the Chlamydiales sequenced to date. The presence of this protein is intriguing as chlamydiae are not known to contain detectable levels of peptidoglycan (PG), despite housing genes for almost a complete PG biosynthetic pathway [34-36].
In addition to interacting with their hosts via cell surface molecules, the chlamydiae are also known to utilize a type III secretion system [37]. Genes encoding the basic type III secretory apparatus have been identified in all sequenced Chlamydia, Chlamydophila and Protochlamydia genomes [11,13-15]. These surface projections are used to inject chlamydiae-derived proteins into the host cytosol from within the inclusion body [38]. Orthologs or paralogs to effector proteins commonly found in other bacteria utilizing this apparatus have not been identified in chlamydial systems although they are likely to be critical for transducing the signals required to modulate host cell function. A type III secretion chaperone (CT274) was identified in all Chlamydiales members, which may facilitate a number of processes such as adhesion, cell signaling, transport and the perpetuation of infection. Another Chlamydiales-specific protein, CT273, was found in a tight cluster with CT274. The function of CT273 is not known, but it could also play a role in type III secretion [26]. Both of these genes lie in close proximity to CT271, another Chlamydiales-specific hypothetical protein which resides in the chromosome in the opposite orientation of the above mentioned ORFs.
Due to the reduction in metabolic capabilities of the chlamydiae, it is likely that many different permeases would be necessary for acquiring sufficient levels of different substrate molecules [18,39]. Members of the YjgP/YjqE family are predicted integral membrane proteins containing 6 transmembrane regions which are predicted to function as permeases [40]. Although YjgP/YjqE proteins are distributed throughout the major domains of life, a novel protein which is indicated to be related to this family, CT838, is uniquely found in various Chlamydiales. The iron-sulfur protein ferredoxin (Fd) is an electron acceptor which participates in the redox-based metabolisms in plastids, mitochondria and bacteria [41]. A novel predicted ferredoxin is uniquely present in various Chlamydiales species (CT312) suggesting that it serves a common evolutionary adapted function in these groups of species.
Another Chlamydiales specific protein, EUO (CT446), encoded by the euo gene (early upstream open reading frame) is found just upstream of the 2 genes encoding lysine-rich proteins Hc1 and Hc2 which are highly similar to the eukaryotic H1 histone [42,43]. One study has shown that Hc1 binds to DNA, inducing nucleoid compaction observed late in the chlamydial developmental cycle when vegetative reticulate bodies differentiate into the metablocially inert infectious particles known as elementary bodies [42]. The EUO protein, which is expressed early in the chlamydial cycle, has been shown to specifically cleave the C-terminal portion of Hc1, initiating dissociation of DNA-Hc1 complexes and DNA decondensation [42-44]. Little is known about the signals that trigger the different events throughout the chlamydial life cycle. However, the unique presence of the EUO protein (CT446) in various Chlamydiales species suggests a common mechanism for chromatin remodeling.
The Chl. trachomatis locus CT583 encodes Gp6D, a genomic paralog (31% similarity) of a plasmid-born virulence factor pGp6D [45]. Three of the four sequenced Chlamydiaceae species (except Chlam. abortus) [2] have been found to contain extrachromosomal DNA elements (cryptic plasmids) believed to encode genes which might play a role in pathogenicity and/or modulating virulence [46]. P. amoebophila does not contain an extrachromosomal plasmid. The chlamydial plasmids encode 8 ORFs of significance (>100 amino acids), and although 5 have been characterized, the function of pGp6D is unknown [45]. While the plasmid-borne pGp6D is only 102aa long, the genomic Gp6D homolog found in all Chlamydiales is more than twice that size (263aa). The high degree of similarity among plasmids from both Chlamydia and Chlamydophila species suggests that an ancestral plasmid was acquired by the chlamydial lineage, perhaps before divergence of the two Chlamydiaceae genera. That plasmid-less chlamydiae are rare suggests that plasmid loss is selected against – that the plasmids do have a function specific to this family, which is apparently absent in chlamydiae-like organisms. All of the proteins in this category, which are uniquely present in various Chlamydiales, have likely evolved in the last common ancestor of all chlamydiae and were subsequently passed on to various descendent species through vertical descent.
Chlamydiaceae-specific proteins
The proteins in this grouping corresponded to those which were found strictly in species belonging to members of the Chlamydia and Chlamydophila genera, but whose homologs were not detected in P. amoebophila. Interestingly, among this category of proteins, 12 distinct gene clusters were located (described below). Of the 79 Chlamydiaceae-specific proteins identified in this work (Table 2), 60 were hypothetical. Those with predicted function are discussed below.
Chlamydiae possess an intracellular developmental cycle defined by the orderly interconversion of infectious, metabolically inactive elementary bodies (EBs) and noninfectious, dividing reticulate bodies (RBs) [1]. Only a few stage-specific genes are known, including the two late-stage genes encoding histone-like proteins. While chlamydial Hc1 (histone H1-like protein) is highly conserved (CT446), Hc2 (a second H1-like protein) is less conserved and variable in size (CT046) [42]. While both proteins are thought to participate in DNA compaction, Hc1 is found only in members of the Chlamydiales while Hc2 is found uniquely in the Chlamydiaceae, to the exclusion of Protochlamydia. Indeed, there are no other homologs of this protein outside of the Chlamydiaceae found in BLAST searches using Chl. trachomatis as the probe. LtuA and LtuB (CT377 and CT080 respectively) are two other proteins, which are uniquely shared by members of the Chlamydiaceae family. Both of these proteins are not expressed until the RBs begin to reorganize to EBs. These molecules do not bear strong homology to any other known protein and their functions are not known. The ltuB gene exhibits unusual stability properties in that the 5' end appears to break down, while the 3' end accumulates as a stable fragment of about 240 bases [47]. It has been hypothesized that the shorter ltuB RNA functions in someway in the late stage of the chlamydial developmental cycle [47]. These unique Chlamydiaceae specific proteins are likely playing distinctive stage-specific roles in the developmental cycles of the Chlamydia and Chlamydophila genera. The ltuB gene is found in close proximity to a cluster containing two other hypothetical Chlamydiaceae-specific proteins (CT082 and CT083).
A vast array of Chlamydiaceae-specific membrane proteins have been identified in our study, including Major outer membrane protein (MOMP or OmpA, CT681), polymorphic membrane protein PmpA (CT412) and PmpB (CT413) which lie in a cluster, the putative outer membrane proteins OmpD (CT812), OmpE, OmpF, OmpG, OmpH (CT869-CT872, which are clustered together on the C. trachomatis chromosome) and OmpI (CT874). Several of these predicted outer membrane proteins have recently been shown to be translated and localized to the surface of the chlamydial outer membrane [27,48]. Outer membrane proteins of microbial pathogens serve essential roles in engaging the host environment and can be important immunotherapeutic targets [49]. Besides the above outer membrane proteins, several other members of the outer membrane complex were uniquely present in all Chlamydiaceae species. Some of these e.g., porin-b(CT713) or SRP (previously annotated as CrpA) (CT442) [50], have been described as being immunogenic. PorB (CT713) is the target of neutralizing antibody responses in vitro and it lies in a cluster with another Chlamydiaceae-specific protein (CT712) of unknown function [51]. OmcA (CT444) is a well characterized chlamydial envelope-related protein (discussed in the previous section) that is found to be Chlamydiaceae-specific in our work. None of these proteins could be detected in the Protochlamydia by BLASTp searches using Chl. trachomatis proteins. Unlike the Pmps and Omps, chlamydial Inc proteins are localized to the chlamydial inclusion membrane. Different Inc proteins share minimal primary sequence identity with each other or with other proteins in the databases making the assignment of specific function difficult. Studies of individual Incs have shown that these peptides protrude from the surface of the inclusion and may mimic or bind to host cell proteins that selectively manicure the surface of the inclusion to avoid fusion with phagosomes or the host cell exocytic pathway [38,52,53]. Two Inc proteins, IncB (CT232) and IncC (CT233), were found to be unique to various Chlamydiaceae species. IncC is secreted by the type III secretion system and is expressed early during the initial stages of infection, and the gene for this protein lies adjacent to the incB loci in a cluster [37,54].
Another Chlamydiaceae specific protein, YprS (CT392), contains cysteine-rich complement-type repeats which are implicated in ligand-binding. Based on these repeat sequences, this protein bears some similarity to the insect vitellogenin and yolk protein receptors (VgR/YPR) family of proteins [55]. Many other Chlamydiaceae specific proteins of unknown functions are present in a number of different clusters including CT142-CT144, CT288/CT289, CT565-CT568, CT577-CT579, CT646/CT647, CT695/CT696 and CT846-CT849.
Chlamydia- and Chlamydophila- specific proteins
The Chlamydia and Chlamydophila constitute two of the genera within the Chlamydiaceae family [1,2]. Thus, it is likely that unique proteins will be found exclusively in the species belonging to these two groups of bacteria. The genomes of two Chlamydia (Chl. trachomatis and Chl. muridarum) and three Chlamydophila species (viz. Chlam. pneumoniae, Chlam. caviae and Chlam. abortus) have been sequenced. In our work, we have identified twenty unique proteins for each of these two genera. A brief description of these results is presented below.
Of the twenty Chlamydia-specific ORFs identified in this work (Table 3), 16 were hypothetical. Those proteins of predicted function were Incs, specifically IncD, IncE, IncF and IncG (CT115-CT118). These 4 ORFs are transcribed from a single operon in Chlamydia species and when expressed, are not uniformly distributed [56]. Instead, these peptides appear to be concentrated at discrete sites in the inclusion membrane, primarily at sites of membrane-RB contact which can be visualized by both immunofluorescence and electron microscopy [56]. Unlike IncD-G localization, chlamydial MOMP appears to be depleted at the point of contact of RBs with the inclusion membrane but is found throughout the rest of the chlamydial outer membrane [56]. The asymmetric distribution of these proteins suggests that RBs exhibit polarity. The presence of such diversity of chlamydial polypeptides in the inclusion membrane lends support to the concept that chlamydiae actively control the interactions of the inclusion with the host cell to maintain a highly specialized environment favourable to chlamydial replication, in a lineage-specific manner. The other Chlamydia-specific proteins are of unknown function and some of these are arranged in 3 additional clusters, consisting of loci CT134/CT135, CT226-CT229 and CT357/CT358.
Table 3.
Chlamydia-specific proteins
| Protein Loci | Protein Name/Size | C. tra | C. mur | Next Best Hit (E value, protein length) |
| CT115 (AAC67706) | IncD, 141aa | 4e-45 | 5e-11, 124aa | No other BLAST hits |
| CT116 (AAC67707) | IncE, 132aa | 2e-45 | 4e-14, 143aa | No other BLAST hits |
| CT117 (AAC67708) | IncF, 104aa | 3e-29 | 6e-25, 104aa | No other BLAST hits |
| CT118 (AAC67709) | IncG, 167aa | 5e-59 | 8e-11, 166aa | Mesorhizobium loti (8.2, 101aa) |
| CT135 (AAC67726) | hypoth, 360aa | 4e-179 | 4e-115, 365aa | Hepatitis C virus (0.86, 414aa) |
| CT163 (AAC67754) | hypoth, 548aa | 0 | 8e-13, 246aa | Canis familiaris (1.5, 1971aa) |
| CT174 (AAC67765) | hypoth, 151aa | 9e-81 | 4e-10, 120aa | Streptococcus pyogenes (0.066, 152aa) |
| CT192 (AAC67784) | hypoth, 257aa | 1e-120 | 2e-33, 231aa | Caenorhabditis briggsae (1.9, 1572aa) |
| CT226 (AAC67818) | hypoth, 176aa | 4e-72 | 2e-12, 166aa | Cryptosporidium parvum (0.50, 1366aa) |
| CT227 (AAC67819) | hypoth, 133aa | 4e-46 | 2e-04, 138aa | Symbiobacterium thermophilum (8.2, 143aa) |
| CT228 (AAC67820) | hypoth, 196aa | 5e-76 | 4e-28, 210aa | Campylobacter jejuni (2.5, 176aa) |
| CT229 (AAC67821) | hypoth, 215aa | 2e-86 | 4e-28, 214aa | Bos taurus (0.006, 1976aa) |
| CT249 (AAC67842) | hypoth, 116aa | 6e-41 | 3e-20, 115aa | No other BLAST hits |
| CT300 (AAC67893) | hypoth, 115aa | 2e-44 | 2e-05, 127aa | Pan troglodytes (6.3, 294aa) |
| CT326.2 (AAC67924) | hypoth, 60aa | 2e-28 | 3e-04 | No other BLAST hits |
| CT345 (AAC67940) | hypoth, 121aa | 2e-52 | 5e-24, 121aa | Haemophilus ducreyi (4.7, 221aa) |
| CT357 (AAC67953) | hypoth, 110aa | 8e-30 | 9e-07, 106aa | No other BLAST hits |
| CT358 (AAC67954) | hypoth, 178aa | 2e-79 | 8e-31, 170aa | Anabaena variabilis (0.079, 185aa) |
| CT360 (AAC67956) | hypoth, 208aa | 2e-94 | 2e-46, 212aa | Caenorhabditis Elegans (0.052, 728aa) |
| CT694 (AAC68289) | hypoth, 323aa | 2e-177 | 8e-92, 334aa | Psychrophila sp. (0.56, 335aa) |
These proteins are uniquely found in species belonging to the Chlamydia genus and are absent in the Chlamydophila and Protochlamydia.
Of the 20 Chlamydophila-specific proteins identified in this work (Table 4), 9 are hypothetical while 11 have been minimally characterized previously. Among this group, four putative lipoproteins were identified, whose functions are not yet understood (CCA00246, CCA00575, CCA00578, CCA00738). Additionally 6 proteins have been described as putative membrane proteins in Chlam. abortus (CCA00222, CCA00261, CCA00621, CCA00360, CCA00361 and CCA00434), of which the last three are thought to be localized to the inner membrane [16]. Loci CC00360 and CC00361 are part of a cluster, which is the only cluster found among Chlamydophila-specific ORFs. This group also contained a serine-rich protein of unknown function, annotated as YwbM (CCA00062), which is absent from other chlamydiae as well as all other organisms.
Table 4.
Listing and Characteristics of Chlamydophila-specific proteins
| Protein Loci | Protein Name/Size | C. cav | C. abo | C. pne | Next Best Hit |
| CCA00062 (AAP04814) | YwbM, 463aa | 0 | 6e-113, 501aa | 1e-14, 382aa | Aspergillus nidulans (0.56, 182aa) |
| CCA00177 (AAP04928) | hypoth, 367aa | 0 | 1e-146, 370aa | 7e-35, 366aa | Tetraodon nigroviridis (2.7, 659aa) |
| CCA00189 (AAP04940) | hypoth, 316aa | 0 | 2e-111, 316aa | 3e-60, 312aa | Novosphingobium aromaticivorans (0.010, 331aa) |
| CCA00222 (AAP04973) | put. mb. protein, 123aa | 1e-55 | 6e-22, 113aa | 3e-05, 110aa | Campellolebias dorsimaculatus, (0.04, 348aa) |
| CCA00246 (AAP04997) | put. lipoprotein, 313aa | 0 | 6e-158, 314aa | 3e-96, 317aa | No other BLAST hits |
| CCA00261 (AAP05012) | put. mb. protein, 1044aa | 0 | 0, 1105aa | 3e-64, 1033aa | Capsicum annuum (0.18, 576aa) |
| CCA00296 (AAP05045) | hypoth, 448aa | 0 | 4e-74, 442aa | 2e-35, 445aa | Ralstonia eutropha (1.6, 556aa) |
| CCA00299 (AAP05048) | hypoth, 288aa | 5e-146 | 4e-96, 288aa | 3e-45, 289aa | Neurospora crassa (1.4, 541aa) |
| CCA00352 (AAP05100) | hypoth, 197aa | 3e-95 | 2e-60, 204aa | 5e-12, 212aa | Cryptophlebia leucotreta (0.016, 395aa) |
| CCA00353 (AAP05101) | hypoth, 186aa | 3e-85 | 3e-60, 205aa | 1e-11, 175aa | Mus musculus (0.12, 718aa) |
| CCA00360 (AAP05108) | put. inner mb. protein, 97aa | 4e-47 | 4e-28, 104aa | 1e-04, 101aa | Arabidopsis thaliana (0.58, 513aa) |
| CCA00361 (AAP05109) | put. inner mb. protein,, 110aa | 4e-57 | 2e-42, 110aa | 0.014, 111aa | Bacillus licheniformis (1.3, 416aa) |
| CCA00434 (AAP05180) | put. inner mb. protein, 451aa | 0 | 4e-125, 435aa | 6e-34, 424aa | other Chlamydophila hits ; Apis mellifera (0.93, 1500aa) |
| CCA00497 (AAP05241) | hypoth, 471aa | 0 | 3e-173, 471aa | 1e-55, 515aa | Trophyrema whipplei (2.9, 735aa) |
| CCA00530 (AAP05273) | hypoth, 424aa | 0 | 1e-32, 435aa | 6e-26, 435aa | Fusobacterium nucleatum (3.2, 448aa) |
| CCA00575 (AAP05317) | put. lipoprotein, 294aa | 3e-165 | 1e-97, 277aa | 3e-59, 284aa | Wolinella. succinogenes (2.5, 329aa) |
| CCA00576 (AAP05318) | hypoth 170aa | 7e-88 | 1e-43, 155aa | 2e-18, 164aa | Melanoplus sanguinipes (0.36, 254aa) |
| CCA00578 (AAP05320) | put. lipoprotein, 280aa | 1e-160 | 7e-08, 277aa | 2e-05, 284aa | Plasmodium falciparum (1.3, 294aa) |
| CCA00621 (AAP05363) | PTMP, 108aa | 3e-54 | 2e-33, 108aa | 4e-09, 161aa | Agrobacterium tumefaciens (0.34, 279aa) |
| CCA00738 (AAP05479) | lipoprotein, 95aa | 5e-35 | 3e-15, 99aa | 0.003, 99aa | Mycobacterium leprae (8.5, 574aa) |
These proteins are uniquely found in species belonging to Chlamydophila genus but they are absent in various Chlamydia and Protochlamydia species as well as all other groups of bacteria. Abbreviations: put. inner mb., putative inner membrane protein, PTMP, putative transmembrane protein.
Protochlamydia-specific proteins
Protochlamydia amoebophila provides the only completed genome of a Chlamydiales species that belongs to a family other than the Chlamydiaeceae. The comparative genomics studies by Horn et al. [11] determined which of the Protochlamydia proteins were not present in other sequenced chlamydiae genomes. Although the list compiled by these authors contained over 1000 ORFs, which were present in Protochlamydia but were absent in other Chlamydiales, their study did not examine which of these proteins were also present in other bacterial phyla. We have undertaken this analysis in order to determine Protochlamydia-specific proteins. A total of 445 Protochlamydial ORFs were identified, and all were of unknown function. The listing of these proteins is provided in the Additional file 1. Most of these peptides were large in size (>300aa). Fifty-five gene clusters were found. These clusters were located at loci PC0055/56, PC0062-64, PC0074-76, PC0080/81, PC0117/18, PC0129/30, PC0293/94, PC0296/97, PC0302-04, PC0408-11, PC0463/64, PC0529/30, PC0535/36, PC0576-80, PC0611/12, PC0698/99, PC0726/27, PC0773/74, PC0812-14, PC0829/30, PC0833/34, PC0836-43, PC0857/58, PC0869/70, PC0885/86, PC0910/11, PC0967/68, PC0982/83, PC1051/52, PC1150-55, PC1204-08, PC1223/24, PC1233/34, PC1283/84, PC1294/95, PC1380/81, PC1387/88, PC1414/15, PC1417/18, PC1517/18, PC1531/32, PC1581-83, PC1621/22, PC1642/43, PC1685-87, PC1692-94, PC1741/42, PC1773-75, PC1789/90, PC1795/96, PC1826/27, PC1870/71, PC1891-94, PC1902-04, and PC1959-61.
Cases of putative gene loss or transfer
The impact of gene acquisition and loss on bacterial genome evolution is currently a central question in microbiology which remains unresolved. It was therefore of interest to identify cases in which a gene appears to have been lost or possibly transferred from chlamydial genomes. Such cases were detected when all other species of a particular chlamydial group (e.g. Chlamydiales, Chlamydiaceae etc) contained a specific protein, with the exception of one or only a few species. In all, 33 such cases were identified among the different chlamydial groups (Table 5). Interestingly, 10 proteins which were found in the Chlamydophila and Protochlamydia were absent in the two available Chlamydia species (CCA00154, CCA00168, CCA00180, CCA00266, CCA00333, CCA00495, CCA00619, CCA00733, CCA00743, CCA00855). These genes may represent incidences of gene loss in the Chlamydia, or possible gene transfer between the Chlamydophila and Protochlamydia. Some phylogenies based on rRNA support the early separation of Chlamydia [57,58]. However, indel analysis and some protein phylogenies indicate that the Chlamydophila are ancestral to the Chlamydia [25]. Among the results, 6 proteins were found in all Chlamydiaceae species with the exception of Chl. trachomatis (CCA00231, CCA00396, CCA00425, CCA00538, CCA00827, CCA00910). Furthermore, 6 different proteins were found in all Chlamydiaceae species except for Chlam. pneumoniae (CT049, CT050, CT051, CT214, CT867, CT868). Also, 2 proteins were missing from Chlam. caviae which were found in all other available chlamydial species (CT244 and CT277), while 2 different proteins were absent in Chl. muridarum which were present in other Chlamydiaceae genomes (CCA00470, CCA00259). One protein which was unique to all Chlamydiales (CT550) and two proteins which were found in various Chlamydiaceae (CT444.1 and CT702) were also not found in the Chlam. abortus genome. One protein was found which was uniquely shared by all Chlamydia species and Chlam. pneumoniae, to the exclusion of all other Chlamydophila which may represent a lateral gene transfer (CCA00075). A single protein was absent in Chlam. pneumoniae which was present in all other Chlamydophila genomes (Chlam. caviae and Chlam. abortus) (CT785). Since Chl. trachomatis and Chlam. pneumoniae were used as probes, it is possible that other genes may have been lost from these genomes which were not identified during the searches. When the functions of these proteins are elucidated, these patterns of putative gene loss will likely shed light on their importance in regards to the differences in chlamydial biology, as well as why such gene losses may have occurred. Another well-studied example of a Chlamydiales protein that is also present in various Rickettsiales as well as various plants and algal plastids, consist of the nonmitochondrial ADP-ATP translocases (CT065 and CT495). Phylogenetic studies indicate that these genes have been transferred from a Chlamydiales ancestor to the other groups where these proteins are found [59,60].
Table 5.
Chlamydial Specific Proteins with Gene Losses in Some Species
| Loci and Accession Numbers | Loci and Accession Numbers | Loci and Accession Numbers | Loci and Accession Numbers | Loci and Accession Numbers | Loci and Accession Numbers |
| Chlamydophila and Protochlamydia proteins (absent in Chlamydia) | Chlamydiales specific proteins missing C. abortus | Chlamydiaceae proteins, missing in C. trachomatis | Chlamydiaceae proteins, missing in C. pneumoniae | Chlamydiaceae proteins, missing in C. caviae | Chlamydiaceae proteins, missing in C. muridarum |
| CCA00154 (AAP04906) | CT550 (AAC68152) | CCA00231 (AAP04982) | CT049 (AAC67640) | CT244 (AAC67837) | CCA00470 (AAP05215) |
| CCA00168 (AAP04919) | CCA00396 (AAP05142) | CT050 (AAC67641) | CT277 (AAC67870) | CCA00259 (AAP05010) | |
| CCA00180 (AAP04931) | CCA00425 (AAP05171) | CT051 (AAC67642) | |||
| CCA00266 (AAP05017) | CCA00538 (AAP05281) | CT214 (AAC67806) | |||
| CCA00333 (AAP05081) | CCA00827 (AAP05568) | CT867 (AAC68465) | |||
| CCA00495 (AAP05239) | CCA00910 (AAP05649) | CT868 (AAC68466) | |||
| CCA00619 (AAP05361) | |||||
| CCA00733 (AAP05474) | |||||
| CCA00743 (AAP05484) | |||||
| CCA00855 (AAP05596) | |||||
| Loci and Accession Numbers | Loci and Accession Numbers | Loci and Accession Numbers | Loci and Accession Number | ||
| Chlamydiaceae proteins, missing C. abortus | Chlamydophila proteins, missing C. pneumoniae | Chlamydia proteins also in C. pneumoniae | Chlamydiales, Rickettsiales and plastids | ||
| CT444.1 (AAC68048) | CCA00075 (AAP04827) | CT785 (AAC68380) | CT065 (O84068) CT495 (O84502) | ||
| CT702 (AAC68297) | CT065 (O84068) CT495 (O84502) | ||||
These chlamydiae-specific proteins are either missing in some species from particular groups or show unexpected presence in some other groups. The E values for these proteins are not listed.
Temporal expression of Chlamydiae-specific genes
The developmental cycle of chlamydiae is regulated at the transcriptional level, although this regulation is not well understood [61]. Determining the stage-specificity of different genes is important for understanding the development and function of different chlamydial species. In vivo expression studies [62], in vivo transcription studies [47], and transcriptosome studies [63] have been carried out. The different lineage-specific proteins were compared with microarray studies carried out by Nicholson et al. [63]. Of all the group-specific proteins identified in this work, 61 chlamydiae loci were determined to be regulated in this manner, and fall into one of the following 4 categories as divided by Nicholson et al. [63]): early (6-24 hours post infection (hpi)), midlate I (12-24 hpi), midlate II (18-24 hpi) or late (24-36 hpi). The results for the stage-specific expression of these proteins are summarized in Table 6. IncC, a Chlamydiaceae-specific protein, is the only identified ORF in this study to be expressed at the early time point. Of the midlate I expressed genes, all the Chlamydiales-specific proteins were hypothetical (CT053, CT066, CT273, CT504, CT635, CT666, CT667, CT670, CT734). Of the midlate I Chlamydiaceae-specific proteins, LtuA (CT377) and PmpB (CT413), Pmp F-I (CT870-2, CT874) were the only ORFs of known function, while others (CT289, CT372, CT618, CT668, CT671) remain to be understood. Interestingly, only Chlamydiaceae-specific molecules and 2 putatively lost/transferred genes were found to be expressed during the midlate II phase. These Chlamydiaceae-specific proteins consisted of the clustered loci CT412/413, as well as OmpA (CT681) and OmpB (CT713). The lost/transferred genes, CT049 and CT051, were hypothetical proteins of unknown function. Many temporally expressed chlamydiae-specific genes were found in the late phase of the developmental cycle. Of the Chlamydiales proteins, OmcA (CT444) and OmcB (CT443) were the only proteins of known function while the remaining 4 were hypothetical (CT016, CT017, CT181 and CT546). Several interesting proteins were expressed late in the Chlamydiaceae-specific category including HctB (CT046), LtuB (CT080), YprS (CT392), SRP (CT442), the clustered loci OmcA (CT444) and OmcB (CT443), PmpE (CT869) and the clustered loci CT578/579 and CT082/083. All other differentially expressed proteins during this phase were hypothetical regardless of chlamydial lineage; Chlamydiaceae (CT005, CT288, CT365, CT552, CT565, CT620, CT695, CT712, CT848), Chlamydia (CT249, CT694) and lost/transferred genes (CT050, CT214, CT702, CT868).
Table 6.
Stage-specific Expression of some of Chlamydiae-specific Proteins
| Stage of Expression | Chlamydiales-specific Proteins | Chlamydiaceae-specific Proteins | Chlamydia-specific Proteins | Lost or Transferred Genes |
| I-Early (6–24 hpi) | CT233 | |||
| II-Midlate (12–24 hpi) | CT053 CT635 |
CT289 CT671 |
CT360 | CT550 |
| CT066 CT666 |
CT372 CT870 |
|||
| CT273 CT667 |
CT377 CT871 |
|||
| CT504 CT670 |
CT413 CT872 |
|||
| CT590 CT734 |
CT618 CT874 |
|||
| CT668 | ||||
| III-Midlate (18–24 hpi) | CT142 | CT049 | ||
| CT143 | CT051 | |||
| CT681 | ||||
| CT713 | ||||
| IV-Late (24–36 hpi) | CT016 | CT005 CT552 |
CT249 | CT050 |
| CT017 | CT046 CT565 |
CT214 | ||
| CT181 | CT080 CT578 |
CT694 | CT702 | |
| CT443 | CT082 CT579 |
|||
| CT546 | CT083 CT620 |
CT868 | ||
| CT147 CT695 |
||||
| CT288 CT712 |
||||
| CT365 CT848 |
||||
| CT392 CT869 |
||||
| CT442 | ||||
| CT444 |
The temporal expression patterns of these proteins is based upon the microarray analysis of Nicholson et al [63]. According to the microarray profiles, significant differences were seen in the expression pattern of genes for these proteins at 4 different time points (6–24 hours post infection, 12–24 h.p.i., 18–24 h.p.i., and 24–36 h.p.i.) which correspond to different phases of the chlamydial developmental cycle where key phenotypic changes take place. It is of interest that the majority of the temporally regulated Chlamydiales and Chlamydiaceae-specific genes are expressed between 12–24 h.p.i. and 24–36 h.p.i. Such coordinate regulation suggests they play essential roles during these phases of the chlamydial cell cycle. Other chlamydiae-specific proteins which are not listed in this Table were not found to show significant differences in their temporal expression in the study by Nicholson et al. [63]. Abbreviation; h.p.i., hours post infection.
Divergence rates of Chlamydiae-specific proteins
We have also examined the sequence divergence rates for chlamydiae-specific proteins as compared to more ubiquitous proteins to determine whether the former proteins are evolving at a much faster rate. A relative measure of this was obtained by determining the degree of sequence conservation (as measured by % amino acid identity) among different chlamydial species for a number of Chl. trachomatis proteins that are either Chlamydiales-specific or which are more broadly distributed among bacteria. Results of these analysis for the two classes of proteins (i.e. Chlamydiales-specific vs broadly distributed) are presented in Tables 7A and 7B, respectively. As seen, in both cases, Chl. trachomatis proteins exhibited highest identity (~75–90%) to the Chl. muridarum homologs. This was followed by the species from the Chlamydophila genus (41–62% identity for the chlamydiae-specific proteins vs 60–80% identity for the broadly distributed group) and the P. amoebophila. The overall pattern that was observed was very similar for the two sets of proteins, although the proteins that are more broadly distributed were found to exhibit between 5–15% higher degree of conservation (i.e. slower rate of divergence) in comparison to the Chlamydiales-specific proteins.
Table 7.
Sequence Divergence of Some (A) Chlamydiales-specific vs (B) Broadly-Distributed Proteins
| A | ||||||
| Protein Loci/Identity | C. tra | C. mur | C. pneu | C. cav | C. abo | Protochlam |
| CT007 | 316/316 (100%) | 273/316 (86%) | 184/316 (58%) | 185/316 (58%) | 183/316 (57%) | 48/316 (23%) |
| CT016 | 242/242 (100%) | 205/242 (84%) | 146/246 (59%) | 153/245 (62%) | 148/245 (60%) | 83/246 (33%) |
| CT017 | 433/433 (100%) | 358/440 (81%) | 224/401 (55%) | 241/432 (55%) | 248/432 (57%) | 123/415 (29%) |
| CT021 | 247/247 (100%) | 184/247 (74%) | 102/244 (41%) | 115/244 (47%) | 108/245 (44%) | 73/254 (28%) |
| CT031 | 100/100 (100%) | 84/100 (84%) | 65/100 (65%) | 63/100 (63%) | 63/100 (63%) | 36/95 (37%) |
| CT038 | 116/116 (100%) | 95/114 (83%) | 55/112 (49%) | 52/98 (53%) | 49/98 (50%) | 34/94 (36%) |
| Average | 100% | 81.5% | 55% | 56% | 55% | 31% |
| B | ||||||
| Protein Loci/Identity | C. tra | C. mur | C. pneu | C. cav | C. abo | Protochlam |
| CT008 Ribo. HII | 300/300 (100%) | 257/301 (85%) | 186/301 (61%) | 185/300 (61%) | 185/300 (61%) | 139/301 (45%) |
| CT009 HthTR | 152/152 (100%) | 133/151 (87%) | 105/143 (69%) | 101/143 (66%) | 105/143 (69%) | 47/162 (28%) |
| CT013 Cyt. Ox. I | 446/446 (100%) | 406/445 (91%) | 317/448 (70%) | 325/446 (72%) | 328/446 (73%) | 195/465 (41%) |
| CT015 PhoH | 434/434 (100%) | 394/434 (90%) | 314/424 (72%) | 339/430 (78%) | 338/430 (77%) | 218/433 (49%) |
| CT019 IleRS | 1036/1036 (100%) | 923/1036 (89%) | 715/1043 (68%) | 729/1043 (69%) | 729/1043 (69%) | 575/1038 (55%) |
| CT022 Ribo. L31 | 108/108 (100%) | 103/108 (95%) | 90/109 (81%) | 87/208 (80%) | 86/108 (79%) | 67/117 (57%) |
| CT034 YtfF | 341/341 (100%) | 279/341 (81%) | 183/338 (53%) | 203/338 (59%) | 203/339 (59%) | 94/281 (27%) |
| Average | 100% | 88% | 67% | 69% | 69% | 43% |
The pair-wise amino acid sequence identity was determined for various chlamydial species for six C. trachomatis proteins that are specific for Chlamydiales (A) and seven other proteins that are broadly distributed in bacteria (B). The numbers in these Tables indicate the length over which the two sequences match each other as well as the % amino acid identity between the two sequences.
The abbreviations in protein names in part (B) are as follows: Ribo. HII, Ribonuclease HII; HthTR, Helix-turn-helix transcriptional regulator; Cyt. Ox. I, Cytochrome oxidase subunit I; PhoH, PhoH-ATPase; IleRS, Isoleucyl tRNA synthetase; Ribo. L31, Ribosomal L31 protein; YtfF, YytF-cationic amino acid transporter.
Conclusion
Chlamydial species are among the 'high profile' pathogens of the 21st century [1]. These species are responsible for a vast array of serious diseases in humans and animals. Our understanding of the chlamydial diversity, host range, environmental distribution, as well as clinical involvement is presently limited and significantly underestimated. In this work, we have determined a large number of proteins which are uniquely present in various known chlamydiae species at different taxonomic levels. The whole protein signatures that we have identified include 59 ORFs that are unique to various known Chlamydiales, 79 ORFs unique for the Chlamydiaceae family, 20 unique ORFs each for the Chlamydia and Chlamydophila genera, as well as 445 ORFs that are presently distinctive of the P. amoebophila (Table 8) The smaller numbers of unique ORFs that are found in the Chalmydiaceae species in comparison to the P. amoebophila may be related to their smaller genome sizes and also the fact that former species grow in a more stable niche as compared to the latter species [11]. Also, 31 cases of putatively lost genes from the different chlamydial genomes were detected. The simplest and most parsimonious explanation for these shared unique signature proteins is that they were introduced only once in a common ancestor of the indicated chlamydial taxonomic groups located at different phylogenetic depths and then vertically passed on to species that descended from that particular ancestor [64,65]. In our analysis, we have also found some examples of chlamydiae-specific proteins which show sporadic species distribution (e.g. present in only a few Chlamydia and Chlamydophila species, but not in other species from these genera). Such proteins could possibly arise from two different mechanisms. First, it is possible that such genes were introduced initially at a deeper phylogenetic level (i.e. order or family) and then selectively lost in some species from the latter branching taxa. Alternatively, their distribution can be explained if the gene in question was initially introduced in one chlamydia species and then laterally transferred to some others, as is the case for the nonmitochondrial ADP-ATP translocases [59,60]. The numbers of such proteins is relatively small in comparison to the proteins that show taxa specific distribution.
Table 8.
Summary chart of the total unique proteins found for different chlamydiae lineage
| Lineages | Number of unique ORFs (proteins) |
| Chlamydiales (C. trachomatis, C. pneumoniae, C. caviae, C. muridarum, C. abortus, P. amoebophila) | 59 |
| Chlamydiaceae (C. trachomatis, C. pneumoniae, C. caviae, C. muridarum, C. abortus) | 79 |
| Chlamydophila (C. pneumoniae, C. caviae, C. abortus) | 20 |
| Chlamydia (C. trachomatis, C. muridarum) | 20 |
| P. amoebophila | 445 |
We have recently described a number of molecular signatures consisting of conserved indels (i.e. inserts and deletions) in widely distributed proteins viz. RNA polymerase α subunit, elongation factor-Tu, elongation factor-P, DNA gyrase B and lysyl-tRNA synthetase, that are distinctive characteristics of all chlamydial species [25]. The sequence information for these proteins was also obtained from Simkania negevensis, Waddlia chondrophila, and in a number of cases for Neochlamydia hartmanellae, covering all families within the Chlamydiae phylum. The unique presence of these conserved indels in all chlamydiae species provides strong evidence that they are distinctive characteristics of the entire Chlamydiales order. In phylogenetic trees based on a combined dataset of these protein sequence as well as 16S rRNA, the traditional Chlamydiaceae species (i.e. Chlamydophila and Chlamydia) and the chlamydiae-like organisms (viz. Simkania, Waddlia and Parachlamydia) were found to form two distinct clades indicating that these two groups have diverged from each other very early in the evolution of chlamydiae. The chlamydiae-specific proteins identified in the present work provide additional powerful means for understanding and constructing a reliable phylogeny of the Chlamydiales. Based upon the shared presence of these chlamydial-specific proteins, different taxonomic clades or groups within the Chlamydiales could be identified. Figure 1 shows a tree indicating branch points marked by the different chlamydiae group-specific proteins. In addition to distinguishing P. amoebophila from other Chlamydiaceae species, these proteins also support the distinctness of the two genera (i.e. Chlamydophila and Chlamydia) within this family. The phylogenetic inferences based on these proteins are in complete agreement with the relationships observed within this phylum based on different proteins as well as 16S rRNA trees [2,8,25]. The genes encoding this set of peptides were likely acquired or evolved sometime after the divergence of the Protochlamydia and the other chlamydia-like organisms. Although sequence information for other chlamydiae (viz. Simkania and Waddlia) is lacking at present, it is likely that many of the Chlamydiales-specific proteins identified here will also be found in these species. Further, the clustering of the chlamydiae-like species in phylogenetic trees [2,25]suggests that many of the proteins which are presently indicated to be specific for P. amoebophila may also be found in the other chlamydiae-like organisms and they may provide unique molecular markers for these groups of species.
Figure 1.
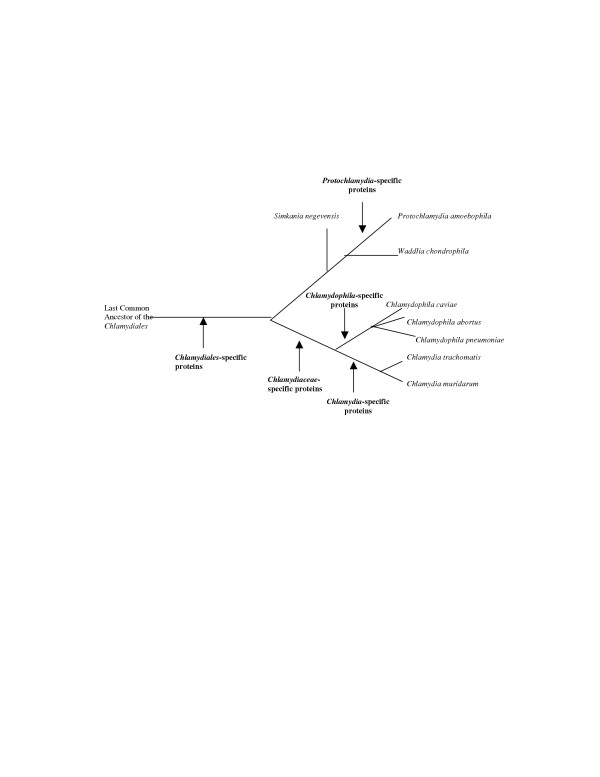
The branching pattern of the chlamydiae species based upon the distribution pattern of different chlamydiae specific proteins. The evolutionary stages where different proteins are indicated to have evolved are marked. The observed relationship is same as that seen in the phylogenetic trees based on 16S rRNA and protein sequences [1,2,25].
The analyses of bacterial genomes have indicated that a substantial proportion of their gene repertoire is comprised of signature proteins or ORFans i.e., open reading frames which have no known homologs and consequently no known function [64-66]. The mechanisms by which such novel genes arise in different genomes are presently not clearly understood and they have been discussed in other recent publications [67-69]. However, it is possible that homologs of some of these genes/proteins are also present in other bacteria but because of their extensive sequence divergence significant similarity to the chlamydiae homologs is not observed. One example where this may be the case is that of the OmcA (CT444) protein, which is listed as a Chlamydiaceae-specific protein in this study (Table 2). An OmcA homolog is annotated in P. amoebophila genome because it is cysteine-rich and upstream of a large cysteine-rich protein, but it does not show significant similarity to the Chlamydiaceae homologs in BLASTp searches. Another protein that we have not included in any of the Tables but is of interest is CT020 (signal peptidase I). The homologs of this protein are highly conserved in all chlamydiae (E values ranging from 0 to 1e-101) and there is a large drop in E value from chlamydiae to the first non-chlamydiae hit (E value 3e-09), which is also indicated as signal peptidase I. The sequence of this protein has again greatly diverged between chlamydiae and non-chlamydiae species. Although the presence of these proteins could be explained by extensive sequence divergence, the possibility that they may have originated independently cannot be excluded. Our survey of the divergence rates of proteins that are either specific for the Chlamydiales vs those that are broadly distributed in different bacteria, indicate that Chlamydiales-specific proteins exhibit only slightly higher divergence rate (~10%) in comparison to the more broadly distributed proteins. This difference in our view is unlikely to be generally responsible for the failure to detect the orthologs of most of the chlamydial-specific proteins in other species.
Most of the chlamydiae-specific proteins that have been identified in the present work are of unknown function. Many of these genes are present in distinct clusters in genomes indicating that these clusters could comprise functional units and the corresponding proteins may be involved in related cellular functions [26,70]. The studies aimed at understanding the cellular functions of the Chlamydiales-specific proteins (i.e. their localization, expression and function) should prove highly informative for understanding chlamydial development, virulence and infectivity. The studies on the Chlamydiaceae-specific proteins should provide important insights regarding characteristics that distinguish them from other chlamydiae families. Additionally, a large number of protein coding sequences that are specific for either particular genera or different chlamydial species have been identified. The functional studies of such proteins could provide important information regarding factors, which are presumably responsible for their different properties such as tissue tropism and disease spectrum.
The identified chlamydiae-specific proteins also provide novel biomarkers that should prove of much value in the diagnosis of these bacteria and in exploration of their prevalence and diversity. The identification and screening of chlamydiae and chlamydiae-like organisms in the past has relied on a small number of biomarkers (e.g. 16S and 23S rRNA, MOMP, GroEL protein, lipopolysaccharide epitope Kdo, small cysteine-rich lipoprotein and 60-kDa cysteine-rich protein) [71-74]. In this context, our identification of numerous proteins, which are uniquely found in either various Chlamydiales, Chlamydiaceae, Chlamydophila or Chlamydia groups of species is of great interest and they provide novel and valuable means for detection of these groups of species. Many of these group-specific proteins are highly conserved and sequence alignment for one Chlamydiales-specific protein (CT429) and one Chlamydiaceae-specific protein (CT712) are presented in Figure 2. The sequence alignments of both these proteins contain many highly conserved regions that are suitable for design of PCR primers that should enable successful amplification of all species belonging to these groups. Monoclonal and polyclonal antibodies based upon different chlamydial-specific proteins provide another means for diagnostic studies. Thus, based upon different group-specific proteins that have been identified in the present work, it should be possible to develop novel diagnostics that are capable of distinguishing different groups of chlamydiae species (viz. Chlamydia, Chlamydophila, Chlamydiaceae and Chlamydiales) from each other with high degree of accuracy.
Figure 2.

Multiple sequence alignments of a representative Chlamydiales-specific protein CT429 (A), and a Chlamydiaceae-specific protein CT712 (B). Both of these proteins are of unknown function and they are uniquely found in the indicated groups of species. The sequence alignments for all available homologs for these proteins are shown. Dashes (-) in the alignments indicate identity with the amino acid on the top line (i.e. Chl. trachomatis sequence). Both these proteins contain many highly conserved regions suitable for PCR primer design that can be used for diagnostic purposes.
Methods
Identification of Chlamydiae-specific proteins
In order to identify proteins which are chlamydiae-specific, systematic BLAST searches were carried out on different proteins in the genomes of Chl. trachomatis and Chlam. caviae against all available sequences in the databases. BLASTp searches were performed using each of the proteins or ORFs in these genomes as probes to identify all other bacteria which contain related protein sequences [75]. The searches were carried out using default search parameters, as set by the protein-protein BLAST program, which included the low complexity filter. The low complexity filter removes regions of query sequence which are highly repetitive and which could lead to spurious high similarity with unrelated proteins. The results obtained were visually inspected for homologs showing specificity to the chlamydiae with no other similar homologs present in any other bacteria. This was done by analyzing expect values (E values) for all proteins. The Expect value (E value) represents the statistical significance of a hit in a BLAST search. This number equates to the number of hits that one would find by chance when searching a database of a particular size. The E values depend upon the length of the protein as well as the extent of sequence homology that is observed with any given protein. The E values which are very low (generally less than e-200) are indicated in the results obtained as '0'. In our extensive work, the E value for the top hit (i.e. to the query itself) is generally found to be '0', when the query protein is of a length longer than 325 aa. For proteins of smaller lengths, the E value for the top hit has a finite value. The E value of top hits may be slightly affected by the application of the filter which effectively shortens the query length [75]. Proteins were considered Chlamydiales-specific if all hits belonged to the chlamydiae species, or if the next best non-chlamydiae hit had an expect value which was not significant and could occur by chance. BLAST results with E values > 10-4 are in range where the observed similarity could occur by chance. All such hits were examined for protein lengths to ensure that it was similar to that of the query protein before it was considered as a related protein. Occasionally higher E-values were permitted in analysis when the length of the query protein was small as with fewer characters the E values are higher and this often produce hits with higher E-values (i.e. > 10-4) that are significant for the study. It should be mentioned that BLAST searches can sometime indicate misleading relationships, particularly when no close relatives of the query species are in the database [76]. However, in the present study where most of the reported BLAST hits are for chlamydiae species, for which there are several published genome, such a possibility is highly unlikely. For all chlamydiae-specific proteins, E-values for each chlamydiae BLAST hit, as well as the first non-chlamydial hit are reported here. Furthermore, the sizes of each protein in Chl. trachomatis (or other genomes that were used as BLAST probes) are also listed, and the sizes of other chlamydial homologs are highly similar, unless it is otherwise stated. The same criteria were used for the detection of Chlamydiaceae-specific, Chlamydia-specific and Chlamydophila-specific proteins. For clarification purposes, the chlamydiae loci and accession numbers are given as reference for the different proteins. The genome of P. amoebophila was also examined in this manner to identify proteins that are specific for Protochlamydia. Those genes which were found in all species within a given group (i.e. Chlamydiales, Chlamydiaceae etc) except for one or a few species, were considered to be due to gene loss or lateral gene transfer.
Alignments of Chlamydiae-specific proteins and percentage identity determination
The global alignments of Chlamydiales-specific protein CT429 and Chlamydiaceae-specific protein CT712 were carried out by first retrieving chlamydiae sequences from the NCBI database [77] in FASTA format. ClustalX program [78] was then used to construct both alignments using the default parameters. Pair-wise alignments of sequences for determining percentage identity between Chl. trachomatis proteins and those from other chlamydial species were carried out using the ALIGN Plus 4 program package (Scientific and Educational Software) using the BLOSUM 62 scoring matrix [79] and the default parameters of the alignment program.
Analysis of temporal expression of Chlamydiae-specific proteins
Lineage-specific chlamydial proteins were compared to the microarray data of Nicholson et al. [63]. In that work, the expression patterns of Chl. trachomatis genes were divided into different stages: early (6–24 hours post infection), midlate I (12–24 hpi), midlate II (18–24 hpi) or late (24–36 hpi). Proteins identified in that work which exhibited at least 3-fold differences in expression patterns at different stages, were then placed into one of these different groups. The information for stage-specific expression of chlamydiae-specific proteins was extracted from this study.
Authors' contributions
MSV was assigned to carry out the initial BLAST searches on Chl. trachomatis and Chlam. pneumoniae genomes to identify chlamydial specific proteins. Subsequently, EG was asked to confirm these results and she also carried out BLAST searches on Protochlamydia proteins to identify proteins that are unique to this species. EG also prepared an initial draft of the manuscript. RSG was responsible for conceiving and directing this study from the beginning to the end, for the final evaluation of all results, and for preparing the final submitted manuscript. All authors have read and approved the final manuscript.
Acknowledgments
Acknowledgements
The work was supported by a research grant from the Canadian Institute of Health Research.
Contributor Information
Emma Griffiths, Email: griffie@mcmaster.ca.
Michael S Ventresca, Email: ventrems@mcmaster.ca.
Radhey S Gupta, Email: gupta@mcmaster.ca.
References
- Kalayoglu MV, Byrne GI. Chlamydia. In: Dworkin M, editor. The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community, 3rd edition, release 3.7, http://link.springer-ny.com/link/service/books/10125/. 3rd. New York, Springer-Verlag; 2001. [Google Scholar]
- Everett KD, Bush RM, Andersen AA. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49 Pt 2:415–440. doi: 10.1099/00207713-49-2-415. [DOI] [PubMed] [Google Scholar]
- Rurangirwa FR, Dilbeck PM, Crawford TB, McGuire TC, McElwain TF. Analysis of the 16S rRNA gene of micro-organism WSU 86-1044 from an aborted bovine foetus reveals that it is a member of the order Chlamydiales: proposal of Waddliaceae fam. nov., Waddlia chondrophila gen. nov., sp. nov. Int J Syst Bacteriol. 1999;49 Pt 2:577–581. doi: 10.1099/00207713-49-2-577. [DOI] [PubMed] [Google Scholar]
- Everett KD. Chlamydia and Chlamydiales: more than meets the eye. Vet Microbiol. 2000;75:109–126. doi: 10.1016/S0378-1135(00)00213-3. [DOI] [PubMed] [Google Scholar]
- Kahane S, Greenberg D, Friedman MG, Haikin H, Dagan R. High prevalence of "Simkania Z," a novel Chlamydia-like bacterium, in infants with acute bronchiolitis. J Infect Dis. 1998;177:1425–1429. doi: 10.1086/517830. [DOI] [PubMed] [Google Scholar]
- Horn M, Wagner M, Muller KD, Schmid EN, Fritsche TR, Schleifer KH, Michel R. Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology. 2000;146 ( Pt 5):1231–1239. doi: 10.1099/00221287-146-5-1231. [DOI] [PubMed] [Google Scholar]
- Corsaro D, Venditti D. Emerging chlamydial infections. Crit Rev Microbiol. 2004;30:75–106. doi: 10.1080/10408410490435106. [DOI] [PubMed] [Google Scholar]
- Corsaro D, Valassina M, Venditti D. Increasing diversity within Chlamydiae. Crit Rev Microbiol. 2003;29:37–78. doi: 10.1080/713610404. [DOI] [PubMed] [Google Scholar]
- Horn M, Wagner M. Evidence for additional genus-level diversity of Chlamydiales in the environment. FEMS Microbiol Lett. 2001;204:71–74. doi: 10.1111/j.1574-6968.2001.tb10865.x. [DOI] [PubMed] [Google Scholar]
- Fritsche TR, Horn M, Wagner M, Herwig RP, Schleifer KH, Gautom RK. Phylogenetic diversity among geographically dispersed Chlamydiales endosymbionts recovered from clinical and environmental isolates of Acanthamoeba spp. Appl Environ Microbiol. 2000;66:2613–2619. doi: 10.1128/AEM.66.6.2613-2619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P, Nyakatura GJ, Droege M, Frishman D, Rattei T, Mewes HW, Wagner M. Illuminating the evolutionary history of chlamydiae. Science. 2004;304:728–730. doi: 10.1126/science.1096330. [DOI] [PubMed] [Google Scholar]
- Greub G, Raoult D. Parachlamydiaceae: potential emerging pathogens. Emerg Infect Dis. 2002;8:625–630. doi: 10.3201/eid0806.010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, Myers GS, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, Holtzapple E, Khouri H, Federova NB, Carty HA, Umayam LA, Haft DH, Peterson J, Beanan MJ, White O, Salzberg SL, Hsia RC, McClarty G, Rank RG, Bavoil PM, Fraser CM. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 2003;31:2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson NR, Yeats C, Bell K, Holden MT, Bentley SD, Livingstone M, Cerdeno-Tarraga AM, Harris B, Doggett J, Ormond D, Mungall K, Clarke K, Feltwell T, Hance Z, Sanders M, Quail MA, Price C, Barrell BG, Parkhill J, Longbottom D. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 2005;15:629–640. doi: 10.1101/gr.3684805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingro A, Toenshoff ER, Taylor MW, Fritsche TR, Wagner M, Horn M. 'Candidatus Protochlamydia amoebophila', an endosmbiont of Acanthamoeba spp. Int J Syst Evol Microbiol. 2005;55:1863–1866. doi: 10.1099/ijs.0.63572-0. [DOI] [PubMed] [Google Scholar]
- Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman RW, Olinger L, Grimwood J, Davis RW, Stephens RS. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- Griffiths E, Gupta RS. Protein signatures distinctive of chlamydial species: horizontal transfers of cell wall biosynthesis genes glmU from archaea to chlamydiae and murA between chlamydiae and Streptomyces. Microbiology. 2002;148:2541–2549. doi: 10.1099/00221287-148-8-2541. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS. Protein signatures distinctive of Alpha proteobacteria and its subgroups and a model for Alpha proteobacterial evolution. Crit Rev Microbiol. 2005;31:135. doi: 10.1080/10408410590922393. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Griffiths E. Critical issues in bacterial phylogeny. Theor Popul Biol. 2002;61:423–434. doi: 10.1006/tpbi.2002.1589. [DOI] [PubMed] [Google Scholar]
- Griffiths E, Gupta RS. The use of signature sequences in different proteins to determine the relative branching order of bacterial divisions: evidence that Fibrobacter diverged at a similar time to Chlamydia and the Cytophaga-Flavobacterium-Bacteroides division. Microbiology. 2001;147:2611–2622. doi: 10.1099/00221287-147-9-2611. [DOI] [PubMed] [Google Scholar]
- Gupta Lab's Bacterial Phylogeny Website:http://www.bacterialphylogeny.com. http://www bacterialphylogeny com. 2005.
- Griffiths E, Petrich A, Gupta RS. Conserved Indels in Essential Proteins that are Distinctive Characteristics of Chlamydiales and provide novel means for their identification. Microbiology. 2005;151:2647–2657. doi: 10.1099/mic.0.28057-0. [DOI] [PubMed] [Google Scholar]
- Doerks T, von Mering C, Bork P. Functional clues for hypothetical proteins based on genomic context analysis in prokaryotes. Nucleic Acids Res. 2004;32:6321–6326. doi: 10.1093/nar/gkh973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler JE, Birkelund S, Stephens RS. Overexpression and surface localization of the Chlamydia trachomatis major outer membrane protein in Escherichia coli. Mol Microbiol. 1992;6:1087–1094. doi: 10.1111/j.1365-2958.1992.tb01545.x. [DOI] [PubMed] [Google Scholar]
- Everett KD, Hatch TP. Architecture of the cell envelope of Chlamydia psittaci 6BC. J Bacteriol. 1995;177:877–882. doi: 10.1128/jb.177.4.877-882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch TP. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J Bacteriol. 1996;178:1–5. doi: 10.1128/jb.178.1.1-5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Hatch TP, Woese CR. Eubacterial origin of chlamydiae. J Bacteriol. 1986;167:570–574. doi: 10.1128/jb.167.2.570-574.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JT, Fuerst JA, Giovannoni S, Schlesner H. The order Planctomycetales and the genera Planctomyces, Pirellula, Gemmata and Isosphaera. In: Balows A, Truper HG, Dworkin M, Harder W and Schleifer KH, editor. The Prokaryotes. 2. Vol. 1. New York, Springer-Verlag; 1992. pp. 3710–3731. [Google Scholar]
- Ward NL, Rainey FA, Hedlund BP, Staley JT, Ludwig W, Stackebrandt E. Comparative phylogenetic analyses of members of the order Planctomycetales and the division Verrucomicrobia: 23S rRNA gene sequence analysis supports the 16S rRNA gene sequence-derived phylogeny. International Journal of Systematic Bacteriology. 2000;50:1965–1972. doi: 10.1099/00207713-50-6-1965. [DOI] [PubMed] [Google Scholar]
- Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) J Mol Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- Ghuysen JM, Goffin C. Lack of cell wall peptidoglycan versus penicillin sensitivity: new insights into the chlamydial anomaly. Antimicrob Agents Chemother. 1999;43:2339–2344. doi: 10.1128/aac.43.10.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A, Rogers JC, Gilbart J, Morgan S, Davis CH, Knight S, Wyrick PB. Muramic acid is not detectable in Chlamydia psittaci or Chlamydia trachomatis by gas chromatography-mass spectrometry. Infect Immun. 1990;58:835–837. doi: 10.1128/iai.58.3.835-837.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandahl BB, Birkelund S, Christiansen G. Genome and proteome analysis of Chlamydia. Proteomics. 2004;4:2831–2842. doi: 10.1002/pmic.200400940. [DOI] [PubMed] [Google Scholar]
- Fields KA, Mead DJ, Dooley CA, Hackstadt T. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol Microbiol. 2003;48:671–683. doi: 10.1046/j.1365-2958.2003.03462.x. [DOI] [PubMed] [Google Scholar]
- Rockey DD, Lenart J, Stephens RS. Genome sequencing and our understanding of chlamydiae. Infect Immun. 2000;68:5473–5479. doi: 10.1128/IAI.68.10.5473-5479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch T. Chlamydia: old ideas crushed, new mysteries bared. Science. 1998;282:638–639. doi: 10.1126/science.282.5389.638. [DOI] [PubMed] [Google Scholar]
- NCBI Conserved Domain (YjgP_YjgQ) Website:http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=pfam03739&version=v2.03. 2006.
- Ceccarelli EA, Arakaki AK, Cortez N, Carrillo N. Functional plasticity and catalytic efficiency in plant and bacterial ferredoxin-NADP(H) reductases. Biochim Biophys Acta. 2004;1698:155–165. doi: 10.1016/j.bbapap.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Kaul R, Hoang A, Yau P, Bradbury EM, Wenman WM. The chlamydial EUO gene encodes a histone H1-specific protease. J Bacteriol. 1997;179:5928–5934. doi: 10.1128/jb.179.18.5928-5934.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Douglas AL, Hatch TP. Characterization of a Chlamydia psittaci DNA binding protein (EUO) synthesized during the early and middle phases of the developmental cycle. Infect Immun. 1998;66:1167–1173. doi: 10.1128/iai.66.3.1167-1173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CEIII, Brickman TJ, Hackstadt T. Hc1-mediated effects on DNA structure: a potential regulator of chlamydial development. Mol Microbiol. 1993;9:273–283. doi: 10.1111/j.1365-2958.1993.tb01689.x. [DOI] [PubMed] [Google Scholar]
- Thomas NS, Lusher M, Storey CC, Clarke IN. Plasmid diversity in Chlamydia. Microbiology. 1997;143 ( Pt 6):1847–1854. doi: 10.1099/00221287-143-6-1847. [DOI] [PubMed] [Google Scholar]
- Comanducci M, Ricci S, Cevenini R, Ratti G. Diversity of the Chlamydia trachomatis common plasmid in biovars with different pathogenicity. Plasmid. 1990;23:149–154. doi: 10.1016/0147-619X(90)90034-A. [DOI] [PubMed] [Google Scholar]
- Fahr MJ, Douglas AL, Xia W, Hatch TP. Characterization of late gene promoters of Chlamydia trachomatis. J Bacteriol. 1995;177:4252–4260. doi: 10.1128/jb.177.15.4252-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer RJ, Hatch TP. Characterization of outer membrane proteins in Chlamydia trachomatis LGV serovar L2. J Bacteriol. 2001;183:2686–2690. doi: 10.1128/JB.183.8.2686-2690.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenboeck B, Kousoulas KG, Storz J. Structures of and allelic diversity and relationships among the major outer membrane protein (ompA) genes of the four chlamydial species. J Bacteriol. 1993;175:487–502. doi: 10.1128/jb.175.2.487-502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starnbach MN, Loomis WP, Ovendale P, Regan D, Hess B, Alderson MR, Fling SP. An inclusion membrane protein from Chlamydia trachomatis enters the MHC class I pathway and stimulates a CD8+ T cell response. J Immunol. 2003;171:4742–4749. doi: 10.4049/jimmunol.171.9.4742. [DOI] [PubMed] [Google Scholar]
- Kawa DE, Schachter J, Stephens RS. Immune response to the Chlamydia trachomatis outer membrane protein PorB. Vaccine. 2004;22:4282–4286. doi: 10.1016/j.vaccine.2004.04.035. [DOI] [PubMed] [Google Scholar]
- Rockey DD, Scidmore MA, Bannantine JP, Brown WJ. Proteins in the chlamydial inclusion membrane. Microbes Infect. 2002;4:333–340. doi: 10.1016/S1286-4579(02)01546-0. [DOI] [PubMed] [Google Scholar]
- Hackstadt T, Fischer ER, Scidmore MA, Rockey DD, Heinzen RA. Origins and functions of the chlamydial inclusion. Trends Microbiol. 1997;5:288–293. doi: 10.1016/S0966-842X(97)01061-5. [DOI] [PubMed] [Google Scholar]
- Bannantine JP, Rockey DD, Hackstadt T. Tandem genes of Chlamydia psittaci that encode proteins localized to the inclusion membrane. Mol Microbiol. 1998;28:1017–1026. doi: 10.1046/j.1365-2958.1998.00867.x. [DOI] [PubMed] [Google Scholar]
- Sappington TW, Raikhel AS. Ligand-binding domains in vitellogenin receptors and other LDL-receptor family members share a common ancestral ordering of cysteine-rich repeats. J Mol Evol. 1998;46:476–487. doi: 10.1007/pl00006328. [DOI] [PubMed] [Google Scholar]
- Scidmore-Carlson MA, Shaw EI, Dooley CA, Fischer ER, Hackstadt T. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol Microbiol. 1999;33:753–765. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- Bush RM, Everett KD. Molecular evolution of the Chlamydiaceae. Int J Syst Evol Microbiol. 2001;51:203–220. doi: 10.1099/00207713-51-1-203. [DOI] [PubMed] [Google Scholar]
- Garner SA, Everson JS, Lambden PR, Fane BA, Clarke IN. Isolation, molecular characterisation and genome sequence of a bacteriophage (Chp3) from Chlamydophila pecorum. Virus Genes. 2004;28:207–214. doi: 10.1023/B:VIRU.0000016860.53035.f3. [DOI] [PubMed] [Google Scholar]
- Brinkman FS, Blanchard JL, Cherkasov A, Av-Gay Y, Brunham RC, Fernandez RC, Finlay BB, Otto SP, Ouellette BF, Keeling PJ, Rose AM, Hancock RE, Jones SJ, Greberg H. Evidence that plant-like genes in Chlamydia species reflect an ancestral relationship between Chlamydiaceae, cyanobacteria, and the chloroplast. Genome Res. 2002;12:1159–1167. doi: 10.1101/gr.341802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G, Raoult D. History of the ADP/ATP-translocase-encoding gene, a parasitism gene transferred from a Chlamydiales ancestor to plants 1 billion years ago. Appl Environ Microbiol. 2003;69:5530–5535. doi: 10.1128/AEM.69.9.5530-5535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Douglas AL, Hatch TP. Characterization of integration host factor (IHF) binding upstream of the cysteine-rich protein operon (omcAB) promoter of Chlamydia trachomatis LGV serovar L2. Mol Microbiol. 2001;41:451–462. doi: 10.1046/j.1365-2958.2001.02531.x. [DOI] [PubMed] [Google Scholar]
- Newhall WJ. Biosynthesis and disulfide cross-linking of outer membrane components during the growth cycle of Chlamydia trachomatis. Infect Immun. 1987;55:162–168. doi: 10.1128/iai.55.1.162-168.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson TL, Olinger L, Chong K, Schoolnik G, Stephens RS. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J Bacteriol. 2003;185:3179–3189. doi: 10.1128/JB.185.10.3179-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainth P, Gupta RS. Signature proteins that are distinctive of alpha proteobacteria. BMC Genomics. 2005;6:94. doi: 10.1186/1471-2164-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siew N, Fischer D. Twenty thousand ORFan microbial protein families for the biologist? Structure (Camb ) 2003;11:7–9. doi: 10.1016/S0969-2126(02)00938-3. [DOI] [PubMed] [Google Scholar]
- Daubin V, Lerat E, Perriere G. The source of laterally transferred genes in bacterial genomes. Genome Biol. 2003;4:R57. doi: 10.1186/gb-2003-4-9-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary Origins of Genomic Repertoires in Bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- Suyama M, Bork P. Evolution of prokaryotic gene order: genome rearrangements in closely related species. Trends Genet. 2001;17:10–13. doi: 10.1016/S0168-9525(00)02159-4. [DOI] [PubMed] [Google Scholar]
- Everett KD, Hornung LJ, Andersen AA. Rapid detection of the Chlamydiaceae and other families in the order Chlamydiales: three PCR tests. J Clin Microbiol. 1999;37:575–580. doi: 10.1128/jcm.37.3.575-580.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse K, Grossmann E, Jager C, Diller R, Hotzel H. Detection of Chlamydia suis from clinical specimens: comparison of PCR, antigen ELISA, and culture. J Microbiol Methods. 2003;54:233–238. doi: 10.1016/S0167-7012(03)00040-X. [DOI] [PubMed] [Google Scholar]
- Mardh PA. Chlamydia screening--yes, but of whom, when, by whom, and with what? Ann N Y Acad Sci. 2000;900:286–292. doi: 10.1111/j.1749-6632.2000.tb06240.x. [DOI] [PubMed] [Google Scholar]
- Watson EJ, Templeton A, Russell I, Paavonen J, Mardh PA, Stary A, Pederson BS. The accuracy and efficacy of screening tests for Chlamydia trachomatis: a systematic review. J Med Microbiol. 2002;51:1021–1031. doi: 10.1099/0022-1317-51-12-1021. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein databases search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J Mol Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information (NCBI) Website [http://www.ncbi.nlm.nih.gov/] 2005.
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]


