Abstract
DNA and DNA precursors (deoxyribonucleotides) suffer damage by reactive oxygen/nitrogen species. They are important mutagens for organisms, due to their endogenous formation. Damaged DNA and nucleotides cause alterations of the genetic information by the mispairing properties of the damaged bases, such as 8-hydroxyguanine (7,8-dihydro- 8-oxoguanine) and 2-hydroxyadenine. Here, the author reviews the mutagenic potentials of damaged bases in DNA and of damaged DNA precursors formed by reactive oxygen/nitrogen species, focusing on the results obtained with synthetic oligonucleotides and 2′-deoxyribonucleoside 5′-triphosphates.
INTRODUCTION
DNA is the genetic material, which consists of base, sugar and phosphate moieties. The genetic information is maintained as the alignment of bases. Since DNA bases suffer damage by the chemical attacks, the genetic information is changed by the mispairing properties of the damaged bases thus formed. DNA is modified by many mutagens, such as reactive oxygen species (ROS), ultraviolet, X- and γ-rays and alkylating agents. ROS are recognized as very important sources of mutations and are involved in mutagenesis, carcinogenesis, aging and neurodegeneration (1–3), due to their endogenous formation in addition to their formation by many environmental mutagens and carcinogens.
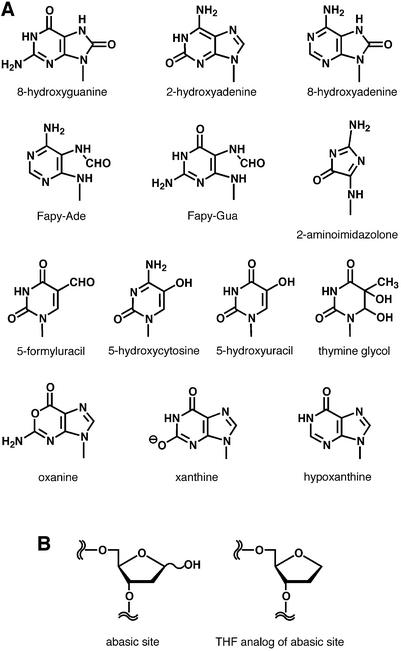
When ROS react with DNA, various kinds of DNA lesions with modified base and/or sugar moieties are produced (Fig. 1) (4,5). 8-Hydroxyguanine (8-OH-Gua, 7,8-dihydro-8-oxoguanine) (6,7) is believed to be an important lesion because of its mutagenicity in bacterial and mammalian cells, as described below. However, recent studies have suggested that other oxidative DNA lesions may be as important as 8-OH-Gua. Moreover, oxidative lesions formed in the DNA precursors are also considered to be important. The oxidative damage in DNA and in the nucleotide pool provides a basis for understanding the chemical modifications of nucleic acids that disturb the genetic information. The author now reviews the mutagenic potentials of damaged bases in DNA and of damaged DNA precursors formed by ROS. In addition, the author refers to the damage caused by reactive nitrogen species (RNS). Since the relationship between the structures of DNA damage and their mutational properties is clear, the data provided by experiments with synthetic oligonucleotides and 2′-deoxyribonucleoside 5′-triphosphates will mainly be described in this review.
Figure 1.
Structures of DNA lesions described in this paper. (A) Damaged bases produced by ROS/RNS. Only the base moiety is shown. Oxanine is the base for deoxyoxanosine 5′-triphosphate. (B) A true abasic site and its analog.
MUTAGENIC POTENTIALS OF OXIDIZED DNA LESIONS
When DNA is attacked by ROS or RNS, DNA lesions with modified base and/or sugar moieties are produced. The DNA lesions thus formed are removed by cellular enzymes, and subsequent enzymatic processes restore the original nucleotide sequences (DNA repair). However, when DNA replication occurs before the repair, the DNA polymerase(s) incorporate a 2′-deoxyribonucleoside 5′-triphosphate opposite the lesions. This translesion synthesis (TLS) is carried out by the replicative DNA polymerase (pol) in some cases, and seems to be conducted by specialized DNA polymerases (see below) in other cases, probably depending on the nature of the lesions. If DNA synthesis is completely blocked by the lesion, then it is lethal. Highly mutagenic lesions appear to block DNA synthesis incompletely, and frequently form base pairs with incorrect nucleotides. In vitro DNA synthesis experiments provide valuable information regarding these points, although they are affected by the types of DNA polymerases tested. Site-directed mutagenesis experiments using living cells provide the mutation frequency (MF), which involves the efficiency of DNA repair in addition to these points.
8-Hydroxyguanine
8-OH-Gua is now recognized as one of the most important DNA lesions, because of its prevalence in DNA and its mutagenicity in bacterial and mammalian cells, as described below. In addition, this compound is a useful marker for DNA oxidation (6,7), since it can be sensitively detected by a high-performance liquid chromatography system equipped with an electrochemical detector (HPLC-ECD). Elevation of 8-OH-Gua content in DNA has been reported in many experiments with cultured cells and animals that were exposed to ‘oxidative stress’ (6,7).
In vitro DNA synthesis experiments with synthetic templates containing 8-OH-Gua have been carried out. Shibutani et al. observed that dCMP and dAMP were incorporated opposite 8-OH-Gua with various misincorporation ratios, depending on the polymerases used [the mammalian DNA polymerases α, β and δ, and Escherichia coli pol I and the exonuclease-proficient Klenow fragment (KFexo+)] (8). We also examined the miscoding properties of 8-OH-Gua at the sites corresponding to hot spots (5′-GG*C-3′ and 5′-CG*G-3′; G* represents 8-OH-Gua) of the human c-Ha-ras protooncogene (9,10). In agreement with the results of Shibutani et al., dCMP and dAMP were incorporated. DNA pol α inserted dAMP exclusively opposite 8-OH-Gua in the 5′-GG*C-3′ sequence, and incorporated dCMP in addition to dAMP opposite 8-OH-Gua in the 5′-CG*G-3′ sequence. The influence of the neighboring sequences around 8-OH-Gua on the efficiency of nucleotide misincorporation was also observed in the case of Taq DNA pol (9,10). Efrati et al. reported that the incorporation of dAMP opposite 8-OH-Gua was slightly favored over that of dCMP, depending on the ‘downstream’ sequence context, when human DNA pol β was used (11). Interestingly, they observed a significant increase in dCMP·A and dGMP·A mispairs at the ‘upstream’ 3′-template site adjacent to the lesion. Errors at these undamaged template sites occurred in four sequence contexts with both gapped and primed single-stranded (ss) DNA templates, but not with pol α instead of pol β (11). Steady-state and pre-steady-state analyses of nucleotide insertion and extension at 8-OH-Gua by several DNA polymerases were reported by Guengerich’s group (12–14).
The mutagenic potentials of 8-OH-Gua in E.coli have been reported by several groups (15–18) using ss/double-stranded (ds) vectors (Table 1). They observed that 8-OH-Gua elicited G→T transversions with MFs of 0.7–2.2% (ss) and 0.1–0.8% (ds). The MF was increased when a mutY or mutM/mutY, but not a mutM, strain was used (17,18).
Table 1. Mutational properties of 8-OH-Gua in E.coli.
| Sequence (5′→3′)a | ds/ss | MF (%) | Mutations detectedb | Reference |
|---|---|---|---|---|
| AG*C | ss | 0.7 (G→T) | G→T (21), G→Cc | 15 |
| GG*A | dsd | 0.8 | G→T (22), G→A (1) | 16 |
| CG*T | ss | 2.2 | G→T (10) | 17 |
| CG*C | ds | 0.1 (G→T) | G→Te | 18 |
aG* represents 8-OH-Gua.
bNumber of colonies is shown in parentheses.
cIt is unclear whether this G→C mutation was induced by 8-OH-Gua, because the same mutation was observed in the case of the unmodified control DNA and the data were not represented in detail.
dApproximately 10% of the DNA did not contain 8-OH-Gua, and the complementary strand without 8-OH-Gua contained U residues.
eOther mutations were undetectable.
Several groups have reported the mutagenic potentials of 8-OH-Gua in mammalian cells (Table 2). In 1992 and 1995, we described the mutations induced by 8-OH-Gua in hot spots of the human c-Ha-ras protooncogene in mouse NIH3T3 cells (19,20). 8-OH-Gua induced various types of mutations, including the targeted G→T and G→A substitutions and the semi-targeted mutations at the 5′-flanking position in the 5′-GG*C-3′ sequence, and elicited exclusively G→T transversions in the other contexts (Table 2). These results suggest that the neighboring sequences could affect the mutational properties of 8-OH-Gua in living cells. The MFs of 8-OH-Gua were calculated as ∼1% (19,20). 8-OH-Gua in ss vectors elicited G→T transversions, with a MF of 4.0–6.8% in COS-7 cells (21–23). In addition to G→T transversions, G→A, G→C and semi-targeted mutations at the 5′-flanking positions were detected. The mutagenic potential of 8-OH-Gua in mammalian cells may depend upon the sequence context, the host cells and/or whether ss/ds vectors were used for the experiments.
Table 2. Mutational properties of 8-OH-Gua in mammalian cells.
| Sequence | ds/ss | Host | MF (%) | Mutations detected | Reference | |||
|---|---|---|---|---|---|---|---|---|
| (5′→3′)a | G→T | G→A | G→C | Others | ||||
| GG*C | ds | NIH3T3 | ∼1.4b | 8 | 3 | 1 | 2c | 19 |
| GG*C | ds | NIH3T3 | ∼1.7b | 22 | 8 | 1 | 11d | 20 |
| CG*G | ds | NIH3T3 | ∼0.6b | 14 | 1 | 0 | 0 | 20 |
| TG*G | ds | NIH3T3 | ∼1.0b | 35 | 0e | 0e | 5f | 20 |
| CG*T | ss | COS-7 | 4.2 | 15g | 0 | 1 | 2h | 21 |
| CG*G | ss | COS-7 | 4.0 | 12 | 0 | 0 | 0 | 22 |
| GG*C | ss | COS-7 | 4.0 | 12 | 0 | 0 | 1i | 22 |
| TG*G | ss | COS-7 | 5.2 | 10 | 1 | 2 | 0 | 23 |
| GG*C | ss | COS-7 | 6.8 | 22 | 3 | 0 | 3j | 23 |
aG* represents 8-OH-Gua.
bMF is calculated on the focus formation relative to that of activated c-Ha-ras genes.
cTwo mutants contained a mutation at a flanking site.
dEleven mutants contained a mutation at a flanking site.
eMutations at this position do not activate the c-Ha-ras gene. Mutants may fail to be selected due to lack of focus-formation.
fFive mutants contained a mutation at a flanking site.
gOne mutant contained a double mutation (CG*T→ATT).
hTwo mutants contained a mutation at a flanking site.
iThe mutant contained a deletion.
jThe mutants contained a mutation at a flanking site.
Synthetic oligonucleotides containing 8-OH-Gua near the 5′-end, mimicking the nucleotide incorporation step, were designed to examine their thermodynamic stabilities (24). We introduced the oxidized guanine into the sites corresponding to hot spots of the human c-Ha-ras gene. In duplexes with various bases at the 3′-end, located opposite 8-OH-Gua, remarkably high stability (a small ΔG0 value) was found in the case of the 8-OH-Gua·T pair in the 5′-GG*C-3′ sequence, while no stabilization was observed with the 8-OH-GuaT pair in the other context. These thermodynamic stabilities may be related to the mutation spectra obtained in our study using NIH3T3 cells (19,20; and see above). On the other hand, the thermodynamic stabilities did not correlate with the mutation spectra when duplexes with a central base pair involving 8-OH-Gua were examined (24). Plum et al. measured the thermodynamic stabilities of duplexes containing a base pair involving 8-OH-Gua in the center (25). In agreement with our results, they observed a lack of correlation between the thermodynamic stability of the ‘base pairs’ and the nucleotides incorporated opposite 8-OH-Gua. Thus, the stability of duplexes containing a modified base at the terminus rather than in the center could be a good parameter for nucleotide insertion.
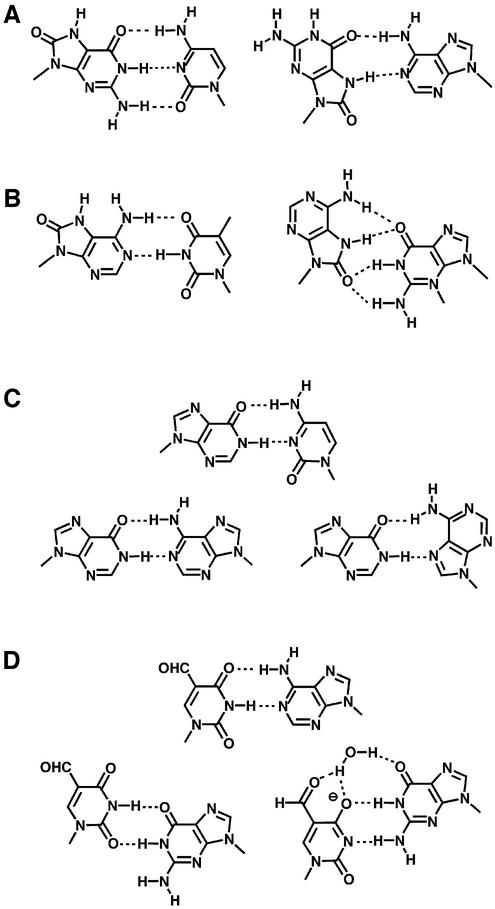
Synthetic oligonucleotides containing 8-OH-Gua were used for a structural analysis of the base pairs involving 8-OH-Gua. Figure 2A shows the hydrogen-bonding patterns of 8-OH-Gua·C and 8-OH-Gua·A pairs. It is known that 8-hydroxyguanosine takes the syn-conformation (26). An 8-OH-Gua residue in DNA adopted both the syn- and anti-orientations, depending on the opposite base (27,28). 8-OH-Gua pairs with C in the Watson–Crick geometry, and pairs with A in the Hoogsteen geometry by adopting the syn-conformation. The positions of the O6-, 7H- and O8-atoms of the syn-oriented 8-OH-Gua are similar to those of the O4-, 3H- and O2-atoms of T, which is one of the reasons for the frequent misincorporation of dAMP opposite 8-OH-Gua.
Figure 2.
Hydrogen-bonding patterns of (A) 8-OH-Gua·C and 8-OH-Gua·A pairs, (B) 8-OH-Ade·T and 8-OH-Ade·G pairs, (C) Hyp·C and Hyp·A pairs and (D) 5-CHO-Ura·A and 5-CHO-Ura·G pairs.
2-Hydroxyadenine
2-Hydroxyadenine (2-OH-Ade, 1,2-dihydro-2-oxoadenine) is another type of DNA lesion produced by ROS. The yields of 2-OH-Ade are similar to those of 8-OH-Gua in the monomeric form, although its formation in DNA is less efficient (29,30). Thus, the incorporation of 2-hydroxy-dAMP (2-OH-dAMP) from the nucleotide pool appears to be the major pathway for the formation of 2-OH-Ade in DNA. It was reported that the treatment of cultured human cells with H2O2 induced 2-OH-Ade accumulation in DNA (one-fifth of that of 8-OH-Gua) (31). 2-OH-Ade is more abundant in human cancerous tissues than in non-cancerous tissues (32). Moreover, as shown below, 2-OH-Ade possesses a mutation-inducibility similar to that of 8-OH-Gua in living cells. Thus, 2-OH-Ade seems to be another important form of DNA damage produced by ROS.
To analyze the nucleotide misincorporation induced by 2-OH-Ade in DNA, we used 12 synthetic oligonucleotides containing 2-OH-Ade with different 5′- and 3′-flanking bases as DNA templates for in vitro DNA synthesis (Table 3) (33,34). DNA polymerases inserted various nucleotides opposite 2-OH-Ade in the presence of a single dNTP, depending upon the sequence contexts and the DNA polymerases used. The two mammalian DNA polymerases incorporated dCMP most frequently among the ‘incorrect’ nucleotides, while dGMP was inserted more efficiently than dCMP and dAMP by KFexo–. Interestingly, the three DNA polymerases inserted dAMP opposite 2-OH-Ade in the 5′-TA*A-3′ (A* denotes 2-OH-Ade) sequence. This phenomenon was observed using two different templates containing the 5′-TA*A-3′ sequence. This conclusion was confirmed by a comparison of the full-length products obtained by in vitro DNA synthesis in the presence of the four nucleotides (33,34).
Table 3. Sequence dependent incorporation of nucleotides opposite 2-OH-Ade in vitro.
| Sequence of template (5′→3′)a | DNA polymerase | ||
|---|---|---|---|
| pol α | pol β | KFexo– | |
| GCGA*ATATTCCGTCAT | T>C>A∼G | T>C | T>A>G>C |
| GCCA*ATATTCCGTCAT | T>G>C>A | T>G>C | T>G>A>C |
| GCTA*ATATTCCGTCAT | T>A | A>T | T>A>G |
| GCAA*ATATTCCGTCAT | T>C∼A∼G | T>C | T>G∼A>C |
| GCGA*CTATTCCGTCAT | T>C>A | T>C | T>G>A∼C |
| GCTA*CTATTCCGTCAT | T>A∼C | T>C | T>G>A>C |
| GCGA*GTATTCCGTCAT | T>C | T>C>A∼G | T>G>A>C |
| GCTA*GTATTCCGTCAT | T>C | T>C>A | T>G>A>C |
| GCGA*TTATTCCGTCAT | T>C∼G∼A | T>C>G | T>G>A∼C |
| GCTA*TTATTCCGTCAT | T>C>G | T>C>G | T>G>C>A |
| CGTGA*CATTTCTGAT | T>C>A | C>T | T>C |
| CGTTA*AATTTCTGAT | T>A | A>T | T>A>G |
aA* represents 2-OH-Ade.
The mutations induced by 2-OH-Ade in E.coli and simian COS-7 cells were analyzed using a shuttle vector (35,36) (Table 4). The oxidized adenine was incorporated into 5′-GA*C-3′ and 5′-TA*A-3′ sequences, based on the results of the in vitro experiments. In addition, the differences in the mutagenic properties of 2-OH-Ade in the lagging and leading template strands were studied. The frequency and the spectrum of the induced mutations depended on the sequence contexts and the lagging/leading synthesis. The MFs observed in these experiments were 0.1–0.8%, which are comparable with the MFs of 8-OH-Gua in ds DNA in NIH3T3 cells (19,20). Therefore, 2-OH-Ade in DNA is as mutagenic as 8-OH-Gua. The observed –1 deletions at the 5′-GA*C-3′ and 5′-TA*A-3′ sites can be explained by the incorporation of dCMP and dAMP, respectively, opposite the 2-OH-Ade residue, and the subsequent formation of loop-out intermediates (35). These results suggest the formation of the 2-OH-Ade·C and 2-OH-Ade·A pairs in living cells.
Table 4. Mutational properties of 2-OH-Ade in E.coli and COS-7 cells.
| Sequence | ds/ss | Host | Lagging/Leading | MF (%) | Mutations detected | |||
|---|---|---|---|---|---|---|---|---|
| (5′→3′)a | A→G | A→T | A→C | ΔA | ||||
| GA*C | ss | E.coli | NAb | 0.67 | 5 | 0 | 0 | 23 |
| TA*A | ss | E.coli | NAb | 0.06 | 5 | 0 | 8 | 18 |
| GA*C | ds | E.coli | Lagging | 0.77 | 6 | 0 | 1 | 20 |
| TA*A | ds | E.coli | Lagging | 0.05 | 2 | 0 | 2 | 10 |
| GA*C | ds | E.coli | Leading | 0.20 | 14 | 0 | 0 | 5 |
| TA*A | ds | E.coli | Leading | 0.07 | 3 | 9 | 1 | 4 |
| GA*C | ds | COS-7 | Lagging | 0.60 | 3 | 0 | 0 | 21 |
| TA*A | ds | COS-7 | Lagging | 0.09 | 6 | 1 | 0 | 6 |
| GA*C | ds | COS-7 | Leading | 0.10 | 4 | 3 | 0 | 7 |
| TA*A | ds | COS-7 | Leading | 0.11 | 10 | 5 | 0 | 0 |
aA* represents 2-OH-Ade.
bNot applicable.
Synthetic oligonucleotides containing 2-OH-Ade near the 5′-end, mimicking the nucleotide incorporation step, were designed to examine their thermodynamic stabilities (37). The order of stability was T > G > C >> A in the 5′-GA*C-3′ sequence and T > A > C > G in the 5′-TA*A-3′ sequence. Since T, G and C, and T and A are the nucleotides incorporated opposite 2-OH-Ade in the 5′-GA*C-3′ and 5′-TA*A-3′ sequences, respectively, in vitro (33,34; see above), these results agree with the miscoding properties of 2-OH-Ade residues. A stable 2-OH-Ade·A pair was formed for the 5′-TA*A-3′ sequence, but not for the 5′-GA*C-3′ sequence, in molecular modeling (37).
Since no structural information concerning ‘incorrect’ base pairs involving 2-OH-Ade is available, the reasons why 2-OH-Ade is mutagenic remain to be resolved. The presence of the 2-hydroxy and 1,2-dihydro-2-oxo tautomers, and the possible presence of the syn and anti conformers, may lead to various types of base pairs involving 2-OH-Ade (38).
8-Hydroxyadenine
8-Hydroxyadenine (8-OH-Ade, 7,8-dihydro-8-oxoadenine) is an isomer of 2-OH-Ade. In vitro, 8-OH-Ade is not formed by Fenton-type oxidation, but by γ-ray irradiation (30). It was reported that 8-OH-Ade was formed in DNA by γ-irradiation of mice (39). Moreover, 8-OH-Ade is present in the neoplastic liver of fish (40) and in human cancerous tissues (32).
Shibutani et al. carried out in vitro DNA synthesis reactions catalyzed by various DNA polymerases, using synthetic templates with 8-OH-Ade, and observed the almost exclusive incorporation of dTMP opposite the base in the presence of the four nucleotides (41). In contrast, misincorporations of dGMP (pol α, pol β and KFexo+) and dAMP (KFexo+) opposite 8-OH-Ade were found in the presence of a single nucleotide (41). Guschlbauer et al. sequenced the PCR product obtained with a template containing 8-OH-Ade by the Maxam–Gilbert procedure, and found that the majority of the product contained A at the 8-OH-Ade position (42). We analyzed the nucleotides incorporated opposite 8-OH-Ade by pol α, pol β, KFexo+ and Taq DNA pol in the presence of the four nucleotides (43). In contrast to the above results, the mammalian DNA polymerases inserted incorrect nucleotides opposite 8-OH-Ade, although the prokaryotic polymerases, KFexo+ and Taq pol, exclusively inserted dTMP. Mammalian pol α misincorporated dGMP in addition to dTMP, and pol β inserted dTMP, dGMP and dAMP.
Wood et al. constructed a ss viral genome containing 8-OH-Ade (44). The mutagenic potential of 8-OH-Ade in E.coli was at least one order of magnitude less than that of 8-OH-Gua. To examine the mutagenic properties of 8-OH-Ade in mammalian cells, we introduced 8-OH-Ade into the second position of codon 61 of the human c-Ha-ras gene (43) and observed that the modified base induced targeted A→G and A→C substitutions. Since focus-formation was used as the selection pressure, only a portion of the A→C mutants might be detected (43). One clone contained a mutation at the 5′-flanking position of 8-OH-Ade. Tan et al. constructed ss phagemid DNAs containing 8-OH-Ade and transfected them into COS-7 cells (23). An 8-OH-Ade in the 5′-TA*G-3′ (A* represents 8-OH-Ade) sequence elicited an A→C transversion with an MF of 1.2%. On the other hand, 8-OH-Ade in the 5′-GA*C-3′ sequence induced an A→G transition with an MF of 0.24% (one mutant). They observed one mutant with a mutation at the 5′-flanking site.
Synthetic oligonucleotides containing 8-OH-Ade were used for structural analyses of the base pairs involving 8-OH-Ade (42,45). Figure 2B shows the hydrogen-bonding patterns of 8-OH-Ade·T and 8-OH-Ade·G pairs. It is known that 8-hydroxyadenosine takes the syn-conformation (26). An 8-OH-Ade residue in DNA adopted both the syn- and anti-orientations, depending on the opposite base. 8-OH-Ade paired with T in the Watson–Crick geometry, and paired with G in the two reverse three-center hydrogen-bonding system by adopting the syn-conformation. Since 8-OH-Ade·G pairs minimally disturb the B-DNA backbone structure (45), the formation of this base pair may cause the misincorporation of dGMP opposite 8-OH-Ade, as observed in vitro and in living cells.
Formamidopyrimidines
Formamidopyrimidine (Fapy) lesions, imidazole ring opened purines, are produced by oxidative stress. It was reported that the treatment of cultured human cells with H2O2 induced Fapy lesions in DNA (31). They are more abundant in human cancerous tissues than in non-cancerous tissues (32).
Asagoshi et al. recently prepared an oligonucleotide template containing a 5-N-methylated analog of Fapy-Gua (methyl-Fapy-Gua) and used it in in vitro DNA synthesis (46). Methyl-Fapy-Gua constituted a fairly strong, but not absolute, block to DNA synthesis catalyzed by KFexo–, and permitted TLS with limited efficiency. When methyl-Fapy-Gua was bypassed, dCMP was preferentially inserted opposite the lesion. Thus, they concluded that methyl-Fapy-Gua is potentially lethal but not pre-mutagenic.
Tudek et al. transfected phage DNA modified with dimethylsulfate and NaOH into SOS-induced E.coli, and compared the mutation spectra in the lacZα gene with and without the selective elimination of damaged bases (47). Partial elimination of the methyl-Fapy-Gua from the phage DNA by Fpg digestion resulted in a 2–3-fold decrease in G→T and G→C transversions. Selective depurination of methylated bases (37°C, 9 h, pH 7.0), which resulted in an almost complete loss of 7-methyl-Ade, caused a 9-fold decrease in A→G transitions. They concluded that methyl-Fapy-Ade induced A→G transitions, and that methyl-Fapy-Gua was at least an order of magnitude less mutagenic and elicited G→T and G→C transversions. However, the methyl substituent may affect the hydrogen-bonding, and the response of DNA polymerases to the methylated and unmethylated Fapy lesions may be very different. The recent chemical syntheses of oligonucleotides containing Fapy-Gua and Fapy-Ade (48,49) will provide clearer results.
2-Aminoimidazolone
The formation of 2-aminoimidazolone (Iz) was observed by Mn-TMPyP/KHSO5 and γ-irradiation treatments of dGuo and its derivative, respectively (50,51). In addition, photo-oxidation by riboflavin plus light of an oligodeoxyribonucleotide yielded Iz from G (52).
Kino and Sugiyama carried out an in vitro DNA synthesis, using an oligonucleotide template with Iz (53). DNA pol I and KFexo– incorporated dGMP opposite Iz. They mentioned that 98% of the template remained under the conditions used. Thus, this result suggests that Iz causes G·C→C·G transversion mutations, which are often found in ROS-treated DNA. They proposed that the base-pairing pattern of Iz·G resembles the Watson–Crick pairing of C·G (52).
Xanthine
Xanthine (Xan), the deaminated product of guanine, is formed by nitric oxide (NO) treatment of intact human cells and of aerobic solutions of DNA (54).
Eritja et al. reported that deoxyribonucleotides were incorporated at the site opposite Xan in the order of dTMP > dCMP >> dAMP and dGMP in vitro by Drosophila DNA pol α in the presence of a single nucleotide (55). We analyzed the nucleotides incorporated opposite Xan in the presence of the four substrates, and found that mouse pol α inserted dTMP and dCMP with a similar efficiency (9). In contrast, rat pol β and Taq pol incorporated dCMP almost exclusively.
A synthetic human c-Ha-ras gene with Xan at the second position of codon 12 was constructed, and this ds vector was transfected into NIH3T3 cells (56). The Xan was highly mutagenic: the MF was calculated to be 30–50% in comparison with that of an activated c-Ha-ras gene. Analyses of the genes from the transformants revealed a G→A transition almost exclusively.
Hypoxanthine
Hypoxanthine (Hyp), the deaminated product of adenine, is formed by NO treatment of intact human cells and of aerobic solutions of DNA (54).
In in vitro DNA synthesis experiments using a synthetic template containing Hyp, we observed the incorporation of dCMP opposite Hyp by Taq pol, mouse pol α and rat pol β (9). In the case of pol α, dTMP was also inserted, but much less efficiently. Ohtsuka et al. annealed a C-tailed oligonucleotide containing Hyp residues with G-tailed plasmid DNA (57). They then treated the annealed DNA with KFexo+ and transfected the DNA into E.coli after circularization. All of the Hyp positions were substituted with G, indicating the incorporation of dCMP by KFexo+.
Hill-Perkins et al. constructed a ds vector containing Hyp and found that Hyp induced A→G mutations with high frequency (MF was 11%) in E.coli (58). We replaced the second base of codon 61 of a synthetic c-Ha-ras gene with a Hyp residue in a site-specific manner (59). Transfection of this gene into NIH3T3 cells resulted in very efficient focus formation, and the MF was calculated to be 38–50%. A→G transition mutations were detected almost exclusively. Only one of the 20 clones analyzed contained an A→T mutation. These results indicate that Hyp is a highly mutagenic lesion.
The thermodynamic parameters of the self-complementary oligonucleotides with 2 bp involving Hyp in the middle were reported (60). The stability was in the order of Hyp·C > Hyp·A > Hyp·G. The duplex containing a Hyp·T pair did not show cooperative melting.
The structures of ds oligodeoxyribonucleotides containing Hyp·C and Hyp·A pairs were determined by NMR (61). Hyp can pair with C in the normal Watson–Crick manner (Fig. 2C). The formation of this base pair in the Watson–Crick geometry, together with its high stability, explains the reason for the misincorporation of dCMP opposite Hyp. On the other hand, the Hyp·A base pair structure determined by NMR, is a Watson–Crick type (61), while the Hyp·A base pair is a Hoogsteen type in crystals (Fig. 2C) (62).
5-Formyluracil
5-Formyluracil (5-CHO-Ura) is formed by the oxidation of the 5-methyl group of a thymine base. Its formation was detected in oxidation reactions (30,63–65).
Ono et al. conducted an in vitro DNA synthesis using KFexo+ (66). They observed that this polymerase incorporated dGMP in addition to dAMP. Zhang et al. reported that DNA polymerases (KFexo+, KFexo–, Tth DNA pol and Pfu DNA pol) inserted dAMP and dCMP opposite 5-CHO-Ura in vitro, in a sequence-dependent manner (67). On the other hand, Masaoka et al. found that dAMP and dGMP were incorporated opposite 5-CHO-Ura in the template DNA by KFexo– (68).
The mutational properties of 5-CHO-Ura in living cells were reported. Miyabe et al. conducted site-specific mutagenesis experiments using a ds vector and E.coli as a host (69). They observed that the MF of 5-CHO-Ura was lower than 0.05% in repair-proficient bacterial cells. 5-CHO-Ura appeared to induce substitution and deletion mutations.
We recently reported that 5-CHO-Ura residues were weakly mutagenic, and their MFs in ds plasmid vectors were 0.01–0.04% in COS-7 cells (70). These values are lower, by one order of magnitude, than the MFs of 2-OH-Ade in ds vectors in COS-7 cells and of 8-OH-Gua on a chromosome in NIH3T3 cells (19,20,36). The T→G and T→A transversions during lagging and leading strand syntheses, respectively, were the mutations found most frequently. This result suggests the formation of 5-CHO-Ura·C and 5-CHO-Ura·T base pairs.
Recently, the structures of oligonucleotides containing 5-CHO-Ura·A and 5-CHO-Ura·G base pairs were reported (71,72). The latter pairs did not adopt the Watson–Crick geometry (Fig. 2D) and this may be one reason for the infrequent misincorporation of dGMP opposite 5-CHO-Ura. Although the actual mechanisms of the induction of the T→G and T→A transversions are unknown, we hypothesized that these mutations might be explained by the participation of the formyl group in hydrogen-bonding when 5-fU adopts the syn-conformation (70).
5-Hydroxypyrimidines
5-Hydroxycytosine (5-OH-Cyt) and 5-hydroxyuracil (5-OH-Ura) are formed by the oxidation of cytosine, and the formation of the latter involves the deamination of cytosine glycol (73). Their formations were observed in vitro and in human tissues (30–32,65).
Purmal et al. prepared oligonucleotides containing 5-OH-Cyt or 5-OH-Ura in two different sequence contexts, and used them as template DNAs for in vitro DNA synthesis conducted by KFexo+ (74). In one sequence context, dGMP and dAMP were incorporated opposite 5-OH-Cyt, and dAMP was the principal nucleotide incorporated opposite 5-OH-Ura. In a second sequence context, dCMP was the predominant base incorporated opposite 5-OH-Cyt. In that same sequence context, dCMP was also the predominant nucleotide incorporated opposite 5-OH-Ura. The data suggest that 5-OH-Cyt and 5-OH-Ura have the potential to be pre-mutagenic lesions leading to C→T transitions and C→G transversions.
Kreutzer and Essigmann investigated the mutagenicity of 5-OH-Cyt and 5-OH-Ura in ss DNA in E.coli (75). The MF of 5-OH-Cyt was low (0.05%), and 5-OH-Cyt elicited C→T transitions, and to a much lesser extent, C→G transversions. In contrast, as expected, 5-OH-Ura elicited C→T mutations with high frequency (83%).
cis-Thymine glycol
cis-Thymine glycol (Tg) was detected in γ-irradiated DNA and in DNA isolated from γ-irradiated human cells (76,77). Clark and Beardsley reported that Tg in the template strand strongly blocked synthesis in in vitro DNA pol reactions (78). The bypass of Tg by KFexo+ was sequence context-dependent, and the 3′–5′ exonuclease activity of the DNA polymerase affected the bypass efficiency. McNulty et al. demonstrated that Tg blocked DNA synthesis when human pol β, viral reverse transcriptase, and Sequenase were used (79). TLS was achieved only when KFexo– was added into the reaction mixtures. In this case, the incorporations of dAMP and dGMP opposite Tg were 1700- and 240 000-fold less efficient than the insertion of dAMP opposite T. On the other hand, Purmal et al. observed that the insertion of dAMP opposite Tg by KFexo– occurred with 0.2-fold of the efficiency of that opposite T (80). The mutagenicity of Tg in E.coli was observed when Tg was present in ss DNA (MF ∼0.3%) but not in ds DNA (81). T→C transitions were exclusively generated.
An abasic site and its analogs
DNA residues that lose their bases are called abasic sites. The cleavage of the N-glycosyl bond occurs spontaneously, and is enhanced by acid treatment. It was demonstrated that ROS induced significant numbers of AP sites in calf thymus DNA (82). High steady-state levels of AP sites in rat tissues and human liver DNA were also reported (83). An increase in the number of abasic sites was also detected in human cultured cells exposed to H2O2 (82). In addition, abasic sites are formed as intermediates during the base-excision repair process conducted by DNA glycosylases, and many oxidized bases are recognized by specific glycosylases. Thus, an abasic site is one type of oxidative DNA damage.
A synthetic abasic site analog, often called a tetrahydrofuran (THF) derivative, was incorporated into synthetic oligonucleotides, which were used as templates in in vitro DNA synthesis reactions. Randall et al. used Drosophila DNA pol α in the reactions, and found that the order of insertion opposite the THF derivative was dAMP >> dGMP > dCMP and dTMP (84). Takeshita et al. used viral reverse transcriptase and calf thymus DNA pol α, and found that the former incorporated dAMP opposite the THF derivative with nearly 90% probability, while the latter inserted nucleotides in the order of dAMP (71%) >> dGMP > dTMP > dCMP (85). We carried out in vitro DNA synthesis reactions conducted with mouse pol α and Taq DNA pol, using a THF analog, and found that dAMP was almost exclusively incorporated (86). Shibutani et al. compared nucleotide incorporations opposite a THF derivative and a natural abasic site, in the presence of the four nucleotides (87). They observed that dAMP was incorporated by KFexo– and calf thymus DNA pol α opposite the two types of abasic sites, and that deletions were also induced, depending on the sequence contexts. Interestingly, when human pol β was used in in vitro DNA synthesis, the insertion of a nucleotide complementary to the 5′-adjacent base opposite the THF analog was observed (88).
Lawrence et al. constructed ss DNA containing a natural abasic site (89). Replication of the modified DNA in SOS-uninduced and -induced E.coli cells occurred with 0.3–0.6% and 5.0–6.9% efficiencies, respectively. The positions where abasic sites had been incorporated were primarily converted to T (∼50 and 80%) followed by C and A in SOS-induced cells, suggesting the predominant insertion of dAMP. The same research group prepared gapped-heteroduplexes containing a natural abasic site and transfected them into Saccharomyces cerevisiae (90). They observed that dCMP was preferentially (62–85%) inserted opposite an abasic site in yeast, in contrast to the situation in E.coli. At present, this discrepancy is interpreted to be due to the differences in the specialized DNA polymerases that bypass abasic sites: yeast Rev1 and E.coli DNA pol V (91–93).
We constructed synthetic c-Ha-ras genes with a THF analog in the first or the second position of codon 12, and transfected them into NIH3T3 cells (86). The MFs of the analog were calculated as ∼10%. The c-Ha-ras genes present in the transformants predominantly contained a point mutation to A in the modified position. In addition, point mutations in the adjacent position were also detected. These results indicate that dTMP was mainly incorporated into the sites opposite the abasic site analog (except for dCMP), and that incorrect deoxynucleotides were incorporated in the position adjacent to the abasic site analog. We also obtained similar results with a true abasic site, but the flanking mutations were detected more frequently (94). Gentil et al. constructed ds vectors containing a natural abasic site and transfected them into simian COS-7 cells (95,96). The MF was 0.6–2.6%. They observed that the mutation spectra of the abasic sites were dependent on the ‘complementary’ base of the abasic site, and that, on average, all of the nucleotides appeared to be incorporated. They also reported that the four nucleotides were inserted with a similar frequency when ss DNA containing a natural abasic site was transfected (97). The detailed mechanism of the ‘random’ mutagenesis induced by an abasic site is unknown and may be related to the involvement of different specialized DNA polymerases in mammalian cells. Table 5 summarizes the mutational properties of abasic sites and their analogs in mammalian cells.
Table 5. Mutational properties of abasic sites in mammalian cells.
| Sequence | Abasic site | ds/ss | Host | MF (%) | Mutations detected | Others | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|
| (5′→3′)a | X→A | X→G | X→C | X→T | ||||||
| CXT | Natural | dsb | COS-7 | 0.80 | 10 | 6 | NAc | 11 | 95 | |
| CXT | Natural | dsd | COS-7 | 0.95 | 15 | NAc | 4 | 24 | 96 | |
| CXT | Natural | dse | COS-7 | 0.61 | NAc | 4 | 3 | 4 | 96 | |
| CXT | Natural | dsf | COS-7 | 2.6 | 15 | 24 | 6 | NAc | 96 | |
| AXT | Natural | ss | COS-7 | NAc | 43 | 59 | 50 | 62 | 97 | |
| CXG | THF | dsd | NIH3T3 | ∼7.2g | 12 | NAc | 2 | 3 | 13h | 86 |
| GXC | THF | dsd | NIH3T3 | ∼13g | 16 | NAc | 0 | 4 | 5i | 86 |
| CXG | Natural | dsd | NIH3T3 | ∼7.5g | 4 | NAc | 0 | 1 | 8j | 94 |
| GXC | Natural | dsd | NIH3T3 | ∼5.8g | 5 | NAc | 0 | 2 | 9k | 94 |
aX represents an abasic site.
bComplementary base is G.
cNot applicable.
dComplementary base is C.
eComplementary base is T.
fComplementary base is A.
gMF is calculated on focus formation relative to that of activated c-Ha-ras genes.
hMutations of the 3′-flanking G position.
iMutations of the 5′-flanking G position.
jMutations of the 3′-flanking G position.
kMutations of the 5′-flanking G position.
Ide et al. measured the thermodynamic stability of model template–primer DNA duplexes containing a THF derivative at their 5′-termini (98). They observed that the duplexes containing purine bases opposite the abasic site exhibited higher stability than those with pyrimidine bases. Gelfand et al. incorporated a THF analog into the center of 13mer DNA duplexes, in which the base opposite the lesion (A, C, G or T) and the base pairs neighboring the lesion (C·G or G·C) were systematically varied (99). They concluded that the thermodynamic impact of the lesion was dominated by the identity of the neighboring base pairs, with the cross-strand partner base exerting only a secondary thermodynamic effect on the duplex properties.
Other forms of damage
Oxidation of 5-methylcytosine leads to the formation of 5-formylcytosine (5-CHO-Cyt) (100). The relative misincorporation frequency opposite 5-CHO-Cyt was similar to that opposite 5-methylcytosine during in vitro DNA synthesis conducted by KFexo– (101). In contrast, we found that 5-CHO-Cyt was mutagenic, and its MF in ds vectors was 0.03–0.28% in mammalian cells (102). The mutation spectrum of 5-CHO-Cyt was broad, and included targeted (5-CHO-Cyt→G, 5-CHO-Cyt→A and 5-CHO-Cyt→T) and untargeted mutations.
In vitro, KFexo– incorporated dAMP opposite uracil glycol, oxidized deaminated cytosine, suggesting that this modified base is highly mutagenic (80). In accordance with this result, Kreutzer and Essigmann showed that the mutagenicity of uracil glycol in ss DNA in E.coli was high (MF ∼80%) and that a C→T transition was elicited (75).
Henderson et al. constructed three ss DNAs containing a secondary oxidation product of 8-OH-Gua (oxaluric acid, oxazalone or cyanuric acid) (103). All three lesions were readily bypassed in E.coli, causing G→T transversions with frequencies of 86–97%. This result suggests that the G→T transversions observed in ROS-treated DNA may be elicited by these highly mutagenic lesions, rather than by the direct action of 8-OH-Gua.
MUTATGENIC POTENTIALS OF OXIDIZED DNA PRECURSORS
The fact that an E.coli mutator gene (mutT) encodes a hydrolyzing enzyme for 8-OH-dGTP indicates the incorporation of this oxidized nucleotide by DNA polymerases and its importance as a source of mutations (104). Tajiri et al. reported that the incorporation of 8-hydroxy-dGMP (8-OH-dGMP) from the nucleotide pool contributed almost equally to the direct oxidation of G bases in DNA and to the accumulation of 8-OH-Gua in DNA (105). In addition, the fact that the damaged nucleotides incorporated into bacterial cells elicit chromosomal gene mutations (see below) provides direct evidence that damaged DNA precursors act as mutagens. It is noteworthy that damaged DNA precursors appear to exert few biological effects if they are not incorporated into DNA by DNA polymerases. Since a damaged nucleotide ‘competes’ with the normal nucleotides and much larger amounts of normal nucleotides exist in cells, its incorporation efficiency is very important. Thus, determining the relative incorporation frequency in comparison with the normal pairing event is necessary in in vitro DNA synthesis experiments, in addition to the ratio of ‘incorrect’ to ‘correct’ insertions. In contrast, the DNA damage within DNA chains, once formed, does not ‘compete’ with the normal bases. The other important point is that a damaged precursor acts as a mutagen when the template base opposite which the damaged nucleotide is incorporated and the nucleobase incorporated opposite the damaged base in the DNA during the next round of replication are different. dUTP, formed from dCTP, does not appear to be mutagenic because dUMP is exclusively inserted opposite A, and dAMP is exclusively incorporated opposite U.
At present, the actual concentrations of oxidized DNA precursors are unknown. Shen et al. reported that the cytosolic 8-OH-Gua pool is increased by the exposure of mammalian cells to HClO and hypoxia (106). Since they used HPLC-ECD after formic acid treatment, they measured 8-OH-Gua in all monomers, including nucleosides, mono-, di- and tri-phosphate derivatives. Recently, Tassotto and Mathews tried to measure 8-OH-dGTP in E.coli by HPLC-ECD, and concluded that its intracellular concentration is below 0.34 µM (the limit of detection) (107). However, the ratio of 0.34 µM of 8-OH-dGTP to the estimated concentration of dGTP in the bacterium (100 µM) (108) is 3.4 × 10–3, and this ratio seems to be too high when the in vivo situation is considered. Since the mutagenesis by the damaged deoxyribonucleoside triphosphates certainly depends on their concentration, it is important to establish sensitive methods to determine their intracellular amounts.
8-Hydroxy-dGTP
Maki and Sekiguchi examined the incorporation of 8-OH-dGMP by the α (catalytic) subunit of E.coli DNA pol III using synthetic templates (104). 8-OH-dGMP was inserted opposite A and C with nearly equal efficiency. The kinetic parameters that they determined indicated that these incorporations were ∼30-fold less efficient than those of the normal pairing. Purmal et al. carried out a kinetic analysis of the incorporation of 8-OH-dGMP by KFexo–, and found that they were 1400–2300-fold less efficient than those of the normal pairing (109). In contrast, Einolf et al. reported that the incorporations of 8-OH-dGMP by KFexo– were 150 000–350 000-fold less efficient than those of the normal pairing (110). They also determined the kinetic parameters of 8-OH-dGMP incorporation by other DNA polymerases. The misincorporation opposite A depended upon the DNA polymerase, and the ratio of the incorporation of 8-OH-dGMP opposite A to that opposite C varied (0.05–30).
Cheng et al. added 8-OH-dGTP to the deoxyribonucleotides used for the gap-filling reaction by KFexo–, and transfected the synthesized DNA into E.coli (16). A→C transversions were almost exclusively detected. Similar results were obtained when other DNA polymerases, including the E.coli DNA pol III holoenzyme, were used (111,112). Pavlov et al. studied the effects of 8-OH-dGTP on the fidelity of replication conducted by a HeLa cell extract (112). The replicated DNA was transfected into E.coli and induced A·T→C·G mutations. The induction of A→C and A·T→C·G mutations by 8-OH-dGTP was dependent on the MutY activity: The frequency of these mutations decreased when mutY strains were used as host cells (111,112).
The mutagenicity of 8-OH-dGTP in vivo was examined by a new evaluation method using E.coli as a host (113). This method consists of the direct incorporation of a damaged nucleotide by heat treatment of E.coli cells with CaCl2, and the isolation of lacI– and lacOc mutants. We treated wild type (wt) E.coli W3110 with 8-OH-dGTP, and the frequencies of lacI– and lacOc mutations were measured. The treatment with 8-OH-dGTP increased the frequency of substitution mutations, in contrast to the treatment with either dGTP or dATP. The substitution mutation found most frequently in the 8-OH-dGTP-induced mutants was an A·T→C·G transversion. This type of mutation makes up 90% of the substitution mutations.
2-Hydroxy-dATP
We incubated synthetic oligonucleotide templates with mammalian DNA pol α in the presence of 2-hydroxy-dATP (2-OH-dATP), and found the incorporation of 2-OH-dAMP opposite T and C (29). The misincorporation of 2-OH-dAMP opposite C was observed with three different DNA templates. The kinetic parameters indicated that the incorporation of 2-OH-dAMP opposite T was disfavored by a factor of 290 over that of dAMP, and that the insertion of 2-OH-dAMP opposite C was disfavored by a factor of 1300 over that of dGMP. The incorporation of 2-OH-dAMP opposite T was favored only 4.5-fold over that opposite C. In contrast, the E.coli DNA pol III α (catalytic) subunit inserted 2-OH-dAMP opposite T and G, but not opposite C (114). This incorporation mode was observed with two different DNA templates. Based on the kinetic parameters obtained, the insertion of 2-OH-dAMP opposite T was disfavored by a factor of 70 over that of dATP. The insertion of 2-OH-dAMP opposite G was 800 times less frequent as compared with that of dCTP. Thus, the insertion of 2-OH-dAMP opposite T was calculated to be favored only 10-fold over that opposite G. Similar in vitro DNA synthesis experiments were carried out with KFexo– (114). The insertion of 2-OH-dAMP opposite T was disfavored by a factor of 90 over that of dAMP. The insertion of 2-OH-dAMP opposite G was disfavored by a factor of 20 000 over that of dCMP. The insertion of 2-OH-dAMP opposite T by KFexo– was favored 200-fold over that opposite G.
We added 2-OH-dATP to the deoxyribonucleotides used for the gap-filling reaction by the E.coli DNA pol III holoenzyme, and transfected the synthesized DNA into E.coli (111). G→T transversions were almost exclusively detected. The mutagenicity of 2-OH-dATP was higher than that of 8-OH-dGTP.
The mutagenicity of 2-OH-dATP in E.coli was examined by a new evaluation method as described above (113). When 2-OH-dATP was incorporated into wt E.coli, the MF was increased. 2-OH-dATP seemed to be more mutagenic than 8-OH-dGTP. The substitution mutation found most frequently in the 2-OH-dATP-induced mutants was a G·C→T·A transversion (84% of the total substitution mutations). This type of mutation appears to occur by the incorporation of 2-OH-dAMP opposite G residues in DNA by E.coli DNA polymerase(s), in agreement with the in vitro data. These results imply that the mutagenic potential of 2-OH-dATP is as important as that of 8-OH-dGTP, when present in the nucleotide pool.
2-Aminoimidazolone-2′-deoxyriboside 5′-triphosphate
Kino and Sugiyama examined primer extension in the presence of dGTP, dATP and dTTP, with or without 2-aminoimidazolone-2′-deoxyriboside 5′-triphosphate (53). They reported that the nucleotide was incorporated opposite G instead of dCTP, although no kinetic data were shown.
Deoxyxanthosine 5′-triphosphate and deoxyoxanosine 5′-triphosphate
Suzuki et al. examined the incorporation of deoxyxanthosine 5′-monophosphate (dXMP) and deoxyoxanosine 5′-monophosphate (dOMP) by KFexo– (115). They found that dXMP was incorporated opposite C with much less efficiency (15 000-fold) in comparison with the insertion of dGMP opposite C. The insertion of dOMP opposite C was 5000-fold less efficient than that of dGMP opposite C, and the misinsertion of dOMP opposite T occurred with a 2-fold further decreased efficiency.
5-Formyl-dUTP
Terato et al. examined the incorporation of 5-formyl-dUMP (5-CHO-dUMP) by KFexo– and observed that the damaged nucleotide was incorporated opposite A and opposite G 2- and 34 500-fold, respectively, less efficiently than the normal pairing under pH 7.4 (116). They also observed a higher misincorporation efficiency when the pH was increased. We employed 5-formyl-dUTP (5-CHO-dUTP) in an in vitro gap-filling reaction conducted by the E.coli DNA pol III holoenzyme (at pH 7.5), and transfected the synthesized DNA into E.coli (111). However, the MF was not increased significantly as compared with the control experiment, in which the gap-filling was carried out without 5-CHO-dUTP.
When we introduced 5-CHO-dUTP directly into wt E.coli, the nucleotide elicited lacI– and lacOc mutants as efficiently as 8-OH-dGTP (117). 5-CHO-dUTP induced G·C→A·T, A·T→G·C and G·C→T·A mutations. Since pol III did not seem to misincorporate 5-CHO-dUMP (111), other DNA polymerase(s) may be involved in the mutations induced by 5-CHO-dUTP in vivo.
5-Hydroxy-dCTP
Purmal et al. compared the incorporations of 5-hydroxy-dCMP (5-OH-dCMP) opposite G and opposite A by KFexo– (109). 5-OH-dCMP was inserted with 0.15-fold of the efficiency of dCMP opposite G. The incorporation of 5-OH-dCMP opposite A was 1000-fold less efficient than that opposite G (7000-fold less efficient than those of the normal pairing).
Feig et al. treated dCTP with an ROS-generating system, and separated the modification products by HPLC (118). They then used the human immunodeficiency virus reverse transcriptase to incorporate each product into a DNA that contains a target gene for scoring mutations. One of the mutagenic species isolated was identified as 5-hydroxy-dCTP (5-OH-dCTP), which causes C→T transitions in E.coli. We employed 5-OH-dCTP in the in vitro gap-filling reaction conducted by the E.coli DNA pol III holoenzyme, and transfected the synthesized DNA into E.coli (111). However, the MF was not increased significantly as compared with the control experiment, in which the gap-filling was carried out without 5-OH-dCTP, in contrast to the results from Feig et al.
When we introduced 5-OH-dCTP directly into wt E.coli, the nucleotide elicited lacI– and lacOc mutants as efficiently as 8-OH-dGTP (117). 5-OH-dCTP induced A·T→C·G, A·T→G·C, and G·C→T·A mutations. However, since pol III did not seem to misincorporate 5-OH-dCMP (111), other DNA polymerase(s) may be involved in the mutations induced by 5-OH-dCTP in vivo.
Other damaged nucleotides
8-Hydroxy-dAMP was incorporated only opposite T, with 7700-fold less efficiency than that of the normal pairing by KFexo– (109). Ide et al. found that thymidine glycol 5′-monophosphate was not incorporated when KFexo+ was used (119). In contrast, they observed that 5,6-dihydrothymidine 5′-monophosphate was incorporated in place of dTMP during a primer elongation catalyzed by KFexo– (119). The rate of incorporation of 5,6-dihydrothymidine 5′-monophosphate was ∼10–25-fold lower than that of dTMP. Deoxyuridine glycol 5′-monophosphate was incorporated opposite A by KFexo– with 100-fold less efficiency than dTMP (120).
DNA REPAIR AND NUCLEOTIDE POOL SANITIZATION
As described above, many damaged nucleic acids produced by ROS/RNS, including DNA lesions and damaged DNA precursors, have mutagenic potentials. The damage in DNA, formed either directly or by the incorporation of a damaged DNA precursor, is removed by cellular DNA repair proteins. The repair efficiency of a DNA lesion is one of the determinant factors of the MF in living cells. For example, as shown above, the MF of 8-OH-Gua in E.coli is dependent on the presence of the MutM and MutY proteins, which are major repair enzymes preventing mutagenesis by the modified base (17,18). Since many reviews of DNA repair have been published (e.g. refs 4,121–123), the author will simply summarize the data on the removal of damaged nucleotides from the DNA precursor pool. Nucleotide pool sanitization is very important for organisms to remove mutagenic DNA precursors, as described below.
MutT is the gene product of the E.coli mutator gene, mutT. Disruption of the mutT gene led to an increased MF of A·T→C·G (105,124). Maki and Sekiguchi found that MutT hydrolyzes 8-OH-dGTP to form the corresponding monophosphate (104). Their report highlighted the role of ‘nucleotide pool sanitization’ to prevent the mutagenesis induced by damaged DNA precursors. A similar enzymatic activity was found in mammalian cells, and the cloning of the gene was subsequently reported by the same group (125,126). This enzyme, MTH1 (MutT Homolog), hydrolyzes 8-OH-dGTP to form the cognate monophosphate, and the MTH1 gene could partially complement a mutT-deficiency-induced mutator phenotype (126,127). In addition to 8-OH-dGTP, the human MTH1 hydrolyzes 2-OH-dATP more efficiently than 8-OH-dGTP (128). This preferential hydrolysis of 2-OH-dATP over 8-OH-dGTP was attributed to the higher affinity for 2-OH-dATP than for 8-OH-dGTP. These results imply that 2-OH-dATP is an intrinsic substrate for the human MTH1 protein. Moreover, the MTH1 protein hydrolyzes 8-hydroxy-dATP with an efficiency similar to that for 8-OH-dGTP (128). The fact that a greater number of tumors were formed in the lungs, livers and stomachs of MTH1-deficient mice than wt mice suggests that MTH1 functions as an antimutator protein (129).
Recently, we found that 2-OH-dATP, and, less efficiently, 8-OH-dGTP, were hydrolyzed by the E.coli Orf135 protein (130), suggesting that this protein may be involved in the prevention of mutations induced by these oxidized deoxynucleotides. Kobayashi et al. found that the overproduction of the E.coli ribA gene product (GTP cyclohydrolase II) reduced the increased MF level of the mutT strain to almost the normal level (131). They also observed that the purified GTP cyclohydrolase II protein efficiently hydrolyzed 8-OH-dGTP to 8-OH-dGMP. This enzyme may also protect genetic material from the untoward effects of ROS.
An inosine triphosphatase, which hydrolyzes ITP, dITP and xanthosine 5′-triphosphate, was found (132 and references therein). Although the activity has not been demonstrated, it may act on dXTP as well as dITP. Thus, this enzyme may contribute to the elimination of damaged deoxyribonucleotides formed by NO. A similar enzyme, dUTP pyrophosphatase, which hydrolyzes the deamination product of dCTP, is ubiquitously present in organisms (133 and references therein). These proteins remove damaged nucleotides in the nucleotide pool, and are called sanitizing enzymes.
ROLE OF SPECIALIZED DNA POLYMERASES
Recently, a family of DNA polymerases involved in TLS has been identified in prokaryotes and eukaryotes, including human cells (134–136). It is the accepted idea that these specialized DNA polymerases act to minimize the cell death resulting from replication blockage. For example, E.coli has three specialized DNA polymerases (DNA polymerases II, IV and V). It was shown that the bypass of N-2-acetylaminofluorene guanine adducts located within a mutation hot spot requires pol II for –2 frameshifts, but pol V for error-free TLS (137). On the other hand, error-free and –1 frameshift TLS at a benzo[a]pyrene adduct requires both pol IV and pol V (137). In the case of an abasic site, the involvement of E.coli DNA pol V in TLS has been suggested by the in vitro results (92,93), supporting the observation that TLS is not observed in an umuDC (pol V deficient) E.coli strain (138). However, the actual TLS polymerase(s) for abasic sites in mammalian cells is unclear. To date, for most oxidized DNA lesions, there has not been a distinctive correspondence of a specialized DNA pol with a DNA lesion, and the role of these polymerases in the mutagenesis remains to be resolved. In the near future, in vitro DNA synthesis reactions may be re-investigated by using the responsible DNA pol that actually bypasses the lesion in vivo.
CONCLUSION AND PERSPECTIVE
Approaches using synthetic oligodeoxyribonucleotides containing a defined DNA lesion have been successfully employed in DNA synthesis experiments in vitro and in living cells, to elucidate the mutagenic potential of the lesion of interest. To date, only the lesions that have moderate chemical stability have been examined, due to stringent conditions during the oligonucleotide synthesis. Further efforts, such as the development of new protecting groups that can be removed under very mild conditions, are necessary. Some oligonucleotides with a DNA lesion have been prepared by the post-synthetic (chemical or enzymatic) conversion of the precursor oligonucleotides (e.g. refs 70,78,89), by the enzymatic incorporation of nucleotides (ref. 74), and their combination (e.g. ref. 46). The development of these procedures is also important to synthesize oligonucleotides with specific lesions. In addition, the mutagenic potential of chemically unstable lesions must be addressed. Although they may be difficult to detect in the DNA, due to their instability, they may be highly mutagenic. In particular, the mutagenicity of an unstable lesion in living cells is important, and should be studied in detail. The hurdles toward the achievement of this issue may be overcome by the use of an oligonucleotide with a good analog, or the establishment of a novel and complete conversion method from the precursor in vector DNA.
Another important issue is the use of ds and ss vectors. In general, the repair of a DNA lesion appears to be more efficient in ds DNA than in ss DNA, in living cells. Thus, the use of a ss vector may be better to study the misincorporation frequency in cells. However, a ds vector may be more appropriate for an accurate elucidation of the mutagenicity in cells, considering both the repair and misincorporation efficiencies. The possibility that ss DNA replication may be carried out by different machinery than that for ds DNA is noteworthy. Interestingly, some DNA lesions showed different mutational properties when located in the leading and lagging template strands of ds vectors (e.g. refs 35,36,70). This is another important point to consider when evaluating the actual mutagenicity of DNA lesions in cells, in addition to the interest in discerning the mechanism of this phenomenon.
As described in this paper, the mutagenic potentials of some DNA lesions depended on the sequence context (e.g. refs 10,20,34). It is unclear whether all lesions show sequence-dependency, because very few sequence contexts have been used. Recently, Delaney and Essigmann have developed a novel system, the restriction endonuclease and post-labeling assay, and have used this method in experiments using O6-methylguanine in 16 sequence contexts (139). This method enabled them to quantify the MF very rapidly, without an analysis of each colony or plaque. This procedure seems to be very useful to examine the effects of the sequence around a DNA lesion on the mutagenicity and the mutation spectrum. Hatahet et al. synthesized oligonucleotides containing either 8-OH-Gua, a natural abasic site, or the THF derivative surrounded by four randomized bases on both the 5′ and 3′ sides, and used them as templates for synthesis by T4 DNA polymerase holoenzyme (140,141). Successful bypass products were purified after (8-OH-Gua) or without (abasic sites) DNA repair enzyme treatment, and were subcloned to determine the sequence. They observed that biases for and against certain nucleotides were readily noticeable across the entire randomized region. This type of approach appears to be useful to determine the effect of the sequence contexts and may be extended to the in vivo experiments.
Accumulating evidence indicates the importance of damaged nucleotides in the mutation process. Interestingly, Nunoshiba et al. reported that the mutations found in an E.coli strain lacking superoxide dismutases and a repressor for iron-uptake systems were G·C→T·A and A·T→C·G transversions, and they reported that these mutations might be induced by 2-OH-dATP and 8-OH-dGTP, respectively (142,143). From this viewpoint, the nucleotide pool sanitization enzymes should be studied in detail, because they may be as important as DNA repair enzymes.
In conclusion, the mutagenic potentials of oxidized nucleic acids are determined by various cellular factors: the efficiencies of the DNA repair/nucleotide pool sanitization, the misincorporation by replicative DNA polymerase(s), and the misincorporation by specialized DNA polymerase(s). Since repair and sanitization efficiencies may differ between organisms and strains, due to variations in the type and the expression of these enzymes, the mutagenicity of a DNA lesion probably depends on the cell type. Likewise, the involvement of specialized DNA polymerases will differ in each cell. Thus, DNA repair proteins, nucleotide pool sanitization enzymes, and specialized DNA polymerases that deal with a certain DNA lesion have to be examined in detail and to be corresponded to a lesion. The experimentally observed MFs will be understood by the functions of these parameters, as revealed by in vitro experiments. Chemically synthesized oligonucleotides and nucleotides will be useful to clarify the events that occur in living cells.
Acknowledgments
ACKNOWLEDGEMENTS
The author wishes to thank the collaborators who participated in the experiments described in this paper. The author also thanks Naoko Murata-Kamiya for reading the manuscript. Work in our laboratory was supported in part by Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Ames B.N. (1983) Dietary carcinogens and anticarcinogens. Science, 221, 1256–1264. [DOI] [PubMed] [Google Scholar]
- 2.Harman D. (1981) The aging process. Proc. Natl Acad. Sci. USA, 78, 7124–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames B.N., Shigenaga,M.K. and Hagen,T.M. (1993) Oxidants, antioxidants and the degenerative diseases of aging. Proc. Natl Acad. Sci. USA, 90, 7915–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace S.S. (1998) Enzymatic processing of radiation-induced free radical damage in DNA. Radiat. Res., 150, S60–S79. [PubMed] [Google Scholar]
- 5.Halliwell B. and Aruoma,O.I. (1991) DNA damage by oxygen-derived species. Its mechanism and measurement in mammalian systems. FEBS Lett., 281, 9–19. [DOI] [PubMed] [Google Scholar]
- 6.Kasai H. and Nishimura,S. (1991) Formation of 8-hydroxydeoxyguanosine in DNA by oxygen radicals and its biological significance. In Sies,H. (ed.), Oxidative Stress: Oxidants and Antioxidants. Academic Press, Inc., New York, NY, pp. 99–116.
- 7.Kasai H. (1997) Analysis of a form of oxidative DNA damage, 8-hydroxy-2′-deoxyguanosine, as a marker of cellular oxidative stress during carcinogenesis. Mutat. Res., 387, 147–163. [DOI] [PubMed] [Google Scholar]
- 8.Shibutani S., Takeshita,M. and Grollman,A.P. (1991) DNA synthesis past the oxidation-damaged base 8-oxodG. Nature, 349, 431–434. [DOI] [PubMed] [Google Scholar]
- 9.Kamiya H., Sakaguchi,T., Murata,N., Fujimuro,M., Miura,H., Ishikawa,H., Shimizu,M., Inoue,H., Nishimura,S., Matsukage,A., Masutani,C., Hanaoka,F. and Ohtsuka,E. (1992) In vitro replication study of modified bases in ras sequences. Chem. Pharm. Bull., 40, 2792–2795. [DOI] [PubMed] [Google Scholar]
- 10.Kamiya H., Murata-Kamiya,N., Fujimuro,M., Kido,K., Inoue,H., Nishimura,S., Masutani,C., Hanaoka,F. and Ohtsuka,E. (1995) Comparison of incorporation and extension of nucleotides in vitro opposite 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) in hot spots of the c-Ha-ras gene. Jpn. J. Cancer Res., 86, 270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Efrati E., Tocco,G., Eritja,R., Wilson,S.H. and Goodman,M.F. (1999) ‘Action-at-a-distance’ mutagenesis. 8-oxo-7,8-dihydro-2′-deoxyguanosine causes base substitution errors at neighboring template sites when copied by DNA polymerase β. J. Biol. Chem., 274, 15920–15926. [DOI] [PubMed] [Google Scholar]
- 12.Lowe L.G. and Guengerich,F.P. (1996) Steady-state and pre-steady-state kinetic analysis of dNTP insertion opposite 8-oxo-7,8-dihydroguanine by Escherichia coli polymerases I exo- and II exo-. Biochemistry, 35, 9840–9849. [DOI] [PubMed] [Google Scholar]
- 13.Furge L.L. and Guengerich,F.P. (1997) Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo- and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics. Biochemistry, 36, 6475–6487. [DOI] [PubMed] [Google Scholar]
- 14.Furge L.L. and Guengerich,F.P. (1998) Pre-steady-state kinetics of nucleotide insertion following 8-oxo-7,8-dihydroguanine base pair mismatches by bacteriophage T7 DNA polymerase exo-. Biochemistry, 37, 3567–3574. [DOI] [PubMed] [Google Scholar]
- 15.Wood M.L., Dizdaroglu,M., Gajewski,E. and Essigmann,J.M. (1990) Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry, 29, 7024–7032. [DOI] [PubMed] [Google Scholar]
- 16.Cheng K.C., Cahill,D.S., Kasai,H., Nishimura,S. and Loeb,L.A. (1992) 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A→C substitutions. J. Biol. Chem., 267, 166–172. [PubMed] [Google Scholar]
- 17.Moriya M. and Grollman,A.P. (1993) Mutations in the mutY gene of Escherichia coli enhance the frequency of targeted G:C→T:A transversions induced by a single 8-oxoguanine residue in single-stranded DNA. Mol Gen. Genet., 239, 72–76. [DOI] [PubMed] [Google Scholar]
- 18.Wagner J., Kamiya,H. and Fuchs,R.P.P. (1997) Leading versus lagging strand mutagenesis induced by 7,8-dihydro-8-oxo-2′-deoxyguanosine in E. coli. J. Mol. Biol., 265, 302–309. [DOI] [PubMed] [Google Scholar]
- 19.Kamiya H., Miura,K., Ishikawa,H., Inoue,H., Nishimura,S. and Ohtsuka,E. (1992) c-Ha-ras containing 8-hydroxyguanine at codon 12 induces point mutations at the modified and adjacent positions. Cancer Res., 52, 3483–3485. [PubMed] [Google Scholar]
- 20.Kamiya H., Murata-Kamiya,N., Koizume,S., Inoue,H., Nishimura,S. and Ohtsuka,E. (1995) 8-Hydroxyguanine (7,8-dihydro-8-oxoguanine) in hot spots of the c-Ha-ras gene. Effects of sequence contexts on mutation spectra. Carcinogenesis, 16, 883–889. [DOI] [PubMed] [Google Scholar]
- 21.Moriya M. (1993) Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted GC→TA transversions in simian kidney cells. Proc. Natl Acad. Sci. USA, 90, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Page F., Margot,A., Grollman,A.P., Sarasin,A. and Gentil,A. (1995) Mutagenicity of a unique 8-oxoguanine in a human Ha-ras sequence in mammalian cells. Carcinogenesis, 16, 2779–2784. [DOI] [PubMed] [Google Scholar]
- 23.Tan X., Grollman,A.P. and Shibutani,S. (1999) Comparison of the mutagenic properties of 8-oxo-7,8-dihydro-2′-deoxyadenosine and 8-oxo-7,8-dihydro-2′-deoxyguanosine DNA lesions in mammalian cells. Carcinogenesis, 20, 2287–2292. [DOI] [PubMed] [Google Scholar]
- 24.Koizume S., Kamiya,H., Inoue,H. and Ohtsuka,E. (1994) Synthesis and thermodynamic stabilities of damaged DNA involving 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) in a ras gene fragment. Nucleosides Nucleotides, 13, 1517–1534. [Google Scholar]
- 25.Plum G.E., Grollman,A.P., Johnson,F. and Breslauer,K.J. (1995) Influence of the oxidatively damaged adduct 8-oxodeoxyguanosine on the conformation, energetics and thermodynamic stability of a DNA duplex. Biochemistry, 34, 16148–16160. [DOI] [PubMed] [Google Scholar]
- 26.Uesugi S. and Ikehara,M. (1977) Carbon-13 magnetic resonance spectra of 8-substituted purine nucleotides. Characteristic shift for the syn conformation. J. Am. Chem. Soc., 99, 3250–3253. [DOI] [PubMed] [Google Scholar]
- 27.Oda Y., Uesugi,S., Ikehara,M., Nishimura,S., Kawase,Y., Ishikawa,H., Inoue,H. and Ohtsuka,E. (1991) NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res., 19, 1407–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouchakdjian M., Bodepudi,V., Shibutani,S., Eisenberg,M., Johnson,F., Grollman,A.P. and Patel,D.J. (1991) NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn)dA(anti) alignment at lesion site. Biochemistry, 30, 1403–1412. [DOI] [PubMed] [Google Scholar]
- 29.Kamiya H. and Kasai,H. (1995) Formation of 2-hydroxydeoxyadenosine triphosphate, an oxidatively damaged nucleotide and its incorporation by DNA polymerases. J. Biol. Chem., 270, 19446–19450. [DOI] [PubMed] [Google Scholar]
- 30.Murata-Kamiya N., Kamiya,H., Muraoka,M. and Kasai,H. (1997) Comparison of oxidation products from DNA components by γ-irradiation and Fenton-type reactions. J. Radiat. Res., 38, 121–131. [DOI] [PubMed] [Google Scholar]
- 31.Jaruga P. and Dizdaroglu,M. (1996) Repair of products of oxidative DNA base damage in human cells. Nucleic Acids Res., 24, 1389–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olinski R., Zastawny,T., Budzbon,J., Skokowski,J., Zegarski,W. and Dizdaroglu,M. (1992) DNA base modifications in chromatin of human cancerous tissues. FEBS Lett., 309, 193–198. [DOI] [PubMed] [Google Scholar]
- 33.Kamiya H., Ueda,T., Ohgi,T., Matsukage,A. and Kasai,H. (1995) Misincorporation of dAMP opposite 2-hydroxyadenine, an oxidative form of adenine. Nucleic Acids Res., 23, 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamiya H. and Kasai,H. (1996) Effects of sequence contexts on misincorporation of nucleotides opposite 2-hydroxyadenine. FEBS Lett., 391, 113–116. [DOI] [PubMed] [Google Scholar]
- 35.Kamiya H. and Kasai,H. (1997) Substitution and deletion mutations induced by 2-hydroxyadenine in Escherichia coli: effects of sequence contexts in leading and lagging strands. Nucleic Acids Res., 25, 304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamiya H. and Kasai,H. (1997) Mutations induced by 2-hydroxyadenine on a shuttle vector during leading and lagging strand syntheses in mammalian cells. Biochemistry, 36, 11125–11130. [DOI] [PubMed] [Google Scholar]
- 37.Kawakami J., Kamiya,H., Yasuda,K., Fujiki,H., Kasai,H. and Sugimoto,N. (2001) Thermodynamic stability of base pairs between 2-hydroxyadenine and incoming nucleotides as a determinant of nucleotide incorporation specificity during replication. Nucleic Acids Res., 29, 3289–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamiya H. and Kasai,H. (2000) 2-Hydroxyadenine formation by reactive oxygen species and mutagenic effects. Recent Res. Dev. Biochem., 2, 41–50. [Google Scholar]
- 39.Mori T., Hori,Y. and Dizdaroglu,M. (1993) DNA base damage generated in vivo in hepatic chromatin of mice upon whole body γ-irradiation. Int. J. Radiat. Biol., 64, 645–650. [DOI] [PubMed] [Google Scholar]
- 40.Malins D.C. and Haimanot,R. (1990) 4,6-Diamino-5-formamidopyrimidine, 8-hydroxyguanine and 8-hydroxyadenine in DNA from neoplastic liver of English sole exposed to carcinogens. Biochem. Biophys. Res. Commun., 173, 614–619. [DOI] [PubMed] [Google Scholar]
- 41.Shibutani S., Bodepudi,V., Johnson,F. and Grollman,A.P. (1993) Translesional synthesis on DNA template containing 8-oxo-7,8-dihydrodeoxyadenosine. Biochemistry, 32, 4615–4621. [DOI] [PubMed] [Google Scholar]
- 42.Guschlbauer W., Duplaa,A.M., Guy,A., Teoule,R. and Fazakerley,G.V. (1991) Structure and in vitro replication of DNA templates containing 7,8-dihydro-8-oxoadenine. Nucleic Acids Res., 19, 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamiya H., Miura,H., Murata-Kamiya,N., Ishikawa,H., Sakaguchi,T., Inoue,H., Sasaki,T., Masutani,C., Hanaoka,F., Nishimura,S. and Ohtsuka,E. (1995) 8-Hydroxyadenine (7,8-dihydro-8-oxoadenine) induces misincorporation in in vitro DNA synthesis and mutations in NIH3T3 cells. Nucleic Acids Res., 23, 2893–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood M.L., Esteve,A., Morningstar,M.L., Kuziemko,G.M. and Essigmann,J.M. (1992) Genetic effects of oxidative DNA damage: comparative mutagenesis of 7,8-dihydro-8-oxoguanine and 7,8-dihydro-8-oxoadenine in Escherichia coli. Nucleic Acids Res., 20, 6023–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leonard G.A., Guy,A., Brown,T., Teoule,R. and Hunter,W.N. (1992) Conformation of guanine-8-oxoadenine base pairs in the crystal structure of d(CGCGAATT(O8A)GCG). Biochemistry, 31, 8415–8420. [DOI] [PubMed] [Google Scholar]
- 46.Asagoshi K., Terato,H., Ohyama,Y. and Ide,H. (2002) Effects of a guanine-derived formamidopyrimidine lesion on DNA replication: translesion DNA synthesis, nucleotide insertion and extension kinetics. J. Biol. Chem., 277, 14589–14597. [DOI] [PubMed] [Google Scholar]
- 47.Tudek B., Graziewicz,M., Kazanova,O., Zastawny,T.H., Obtulowicz,T. and Laval,J. (1999) Mutagenic specificity of imidazole ring-opened 7-methylpurines in M13mp18 phage DNA. Acta Biochim. Pol., 46, 785–799. [PubMed] [Google Scholar]
- 48.Haraguchi K. and Greenberg,M.M. (2001) Synthesis of oligonucleotides containing FapydG (N6-(2-deoxy-α,β-d-erythro-pentofuranosyl)-2,6-diamino-4-hydroxy-5-formamidopyrimidine). J. Am. Chem. Soc., 123, 8636–8637. [DOI] [PubMed] [Google Scholar]
- 49.Haraguchi K., Delaney,M.O., Wiederholt,C.J., Sambandam,A., Hantosi,Z. and Greenberg,M.M. (2002) Synthesis and characterization of oligodeoxynucleotides containing formamidopyrimidine lesions and nonhydrolyzable analogues. J. Am. Chem. Soc., 124, 3263–3269. [DOI] [PubMed] [Google Scholar]
- 50.Vialas C., Pratviel,G., Claparos,C. and Meunier,B. (1998) Efficient oxidation of 2′-deoxyguanosine by Mn-TMPyP/KHSO5 to imidazolone dIz without formation of 8-oxo-dG. J. Am. Chem. Soc., 120, 11548–11553. [Google Scholar]
- 51.Cadet J., Berger,M., Buchko,G.W., Joshi,S.R., Raoul,S. and Ravanat,J.L. (1994) 2,2-Diamino-4-[(3,5-di-O-acetyl-2-deoxy-β-d-erythro-pentofuranosyl)amino]-5-(2H)-oxazolone: a novel and predominant radical oxidation product of 3′,5′-di-O-acetyl-2′deoxyguanosine. J. Am. Chem. Soc., 116, 7403–7404. [Google Scholar]
- 52.Kino K., Saito,I. and Sugiyama,H. (1998) Product analysis of GG-specific photooxidation of DNA via electron transfer: 2-aminoimidazolone as a major guanine oxidation product. J. Am. Chem. Soc., 120, 7373–7374. [Google Scholar]
- 53.Kino K. and Sugiyama,H. (2001) Possible cause of GC→CG transversion mutation by guanine oxidation product, imidazolone. Chem. Biol., 8, 369–378. [DOI] [PubMed] [Google Scholar]
- 54.Nguyen T., Brunson,D., Crespi,C.L., Penman,B.W., Wishnok,J.S. and Tannenbaum,S.R. (1992) DNA damage and mutation in human cells exposed to nitric oxide in vitro. Proc. Natl Acad. Sci. USA, 89, 3030–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eritja R., Horowitz,D.M., Walker,P.A., Ziehler-Martin,J.P., Boosalis,M.S., Goodman,M.F., Itakura,K. and Kaplan,B.E. (1986) Synthesis and properties of oligonucleotides containing 2′-deoxynebularine and 2′-deoxyxanthosine. Nucleic Acids Res., 14, 8135–8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamiya H., Shimizu,M., Suzuki,M., Inoue,H. and Ohtsuka,E. (1992) Mutation induced by deoxyxanthosine in codon 12 of a synthetic c-Ha-ras gene. Nucleosides Nucleotides, 11, 247–260. [PubMed] [Google Scholar]
- 57.Ohtsuka E., Matsuki,S., Ikehara,M., Takahashi,Y. and Matsubara,K. (1985) An alternative approach to deoxyoligonucleotides as hybridization probes by insertion of deoxyinosine at ambiguous codon positions. J. Biol. Chem., 260, 2605–2608. [PubMed] [Google Scholar]
- 58.Hill-Perkins M., Jones,M.D. and Karran,P. (1986) Site-specific mutagenesis in vivo by single methylated or deaminated purine bases. Mutat. Res., 162, 153–163. [DOI] [PubMed] [Google Scholar]
- 59.Kamiya H., Miura,H., Kato,H., Nishimura,S. and Ohtsuka,E. (1992) Induction of mutation of a synthetic c-Ha-ras gene containing hypoxanthine. Cancer Res., 52, 1836–1839. [PubMed] [Google Scholar]
- 60.Kawase Y., Iwai,S., Inoue,H., Miura,K. and Ohtsuka,E. (1986) Studies on nucleic acid interactions. I. Stabilities of mini-duplexes (dG2A4XA4G2-dC2T4YT4C2) and self-complementary d(GGGAAXYTTCCC) containing deoxyinosine and other mismatched bases. Nucleic Acids Res., 14, 7727–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uesugi S., Oda,Y., Ikehara,M., Kawase,Y. and Ohtsuka,E. (1987) Identification of I:A mismatch base-pairing structure in DNA. J. Biol. Chem., 262, 6965–6968. [PubMed] [Google Scholar]
- 62.Corfield P.W., Hunter,W.N., Brown,T., Robinson,P. and Kennard,O. (1987) Inosineadenine base pairs in a B-DNA duplex. Nucleic Acids Res., 15, 7935–7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kasai H., Iida,A., Yamaizumi,Z., Nishimura,S. and Tanooka,H. (1990) 5-Formyldeoxyuridine: a new type of DNA damage induced by ionizing radiation and its mutagenicity to Salmonella strain TA102. Mutat. Res., 243, 249–253. [DOI] [PubMed] [Google Scholar]
- 64.Wakizaka A., Aiba,N., Okuhara,E. and Kawazoe,Y. (1987) Production of 5-formyluracil from thymine in an in vitro active oxygen-generating system. Biochem. Int., 14, 289–295. [PubMed] [Google Scholar]
- 65.Douki T., Delatour,T., Paganon,F. and Cadet,J. (1996) Measurement of oxidative damage at pyrimidine bases in gamma-irradiated DNA. Chem. Res. Toxicol., 9, 1145–1151. [DOI] [PubMed] [Google Scholar]
- 66.Ono A., Okamoto,T., Inada,M., Nara,H. and Matsuda,A. (1994) Nucleosides and nucleotides. 131. Synthesis and properties of oligonucleotides containing 5-formyl-2′deoxyuridine. Chem. Pharm. Bull. (Tokyo), 42, 2231–2237. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Q.M., Sugiyama,H., Miyabe,I., Matsuda,S., Saito,I. and Yonei,S. (1997) Replication of DNA templates containing 5-formyluracil, a major oxidative lesion of thymine in DNA. Nucleic Acids Res., 25, 3969–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Masaoka A., Terato,H., Kobayashi,M., Ohyama,Y. and Ide,H. (2001) Oxidation of thymine to 5-formyluracil in DNA promotes misincorporation of dGMP and subsequent elongation of a mismatched primer terminus by DNA polymerase. J. Biol. Chem., 276, 16501–16510. [DOI] [PubMed] [Google Scholar]
- 69.Miyabe I., Zhang,Q.M., Sugiyama,H., Kino,K. and Yonei,S. (2001) Mutagenic effects of 5-formyluracil on a plasmid vector during replication in Escherichia coli. Int. J. Radiat. Biol., 77, 53–58. [DOI] [PubMed] [Google Scholar]
- 70.Kamiya H., Murata-Kamiya,N., Karino,N., Ueno,Y., Matsuda,A. and Kasai,H. (2002) Induction of T→G and T→A transversions by 5-formyluracil in mammalian cells. Mutat. Res., 513, 213–222. [DOI] [PubMed] [Google Scholar]
- 71.Tsunoda M., Karino,N., Ueno,Y., Matsuda,A. and Takenaka,A. (2001) Crystalization and preliminary X-ray analysis of a DNA dodecamer containing 2′-deoxy-5-formyluridine; what is the role of magnesium cation in crystalization of Dickerson-type DNA dodecamers? Acta Cryst., D57, 245–248. [DOI] [PubMed] [Google Scholar]
- 72.Tsunoda M., Sakaue,T., Naito,S., Sunami,T., Karino,N., Ueno,Y., Matsuda,A. and Takenaka,A. (2001) X-ray analysis of DNA dodecamers containing 2′-deoxy-5-formyluridine. Nucleic Acids Res. Symp. Ser., Suppl. 1, 279–280. [DOI] [PubMed] [Google Scholar]
- 73.Dizdaroglu M., Holwitt,E., Hagan,M.P. and Blakely,W.F. (1986) Formation of cytosine glycol and 5,6-dihydroxycytosine in deoxyribonucleic acid on treatment with osmium tetroxide. Biochem J., 235, 531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Purmal A.A., Kow,Y.W. and Wallace,S.S. (1994) Major oxidative products of cytosine, 5-hydroxycytosine and 5-hydroxyuracil, exhibit sequence context-dependent mispairing in vitro. Nucleic Acids Res., 22, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kreutzer D.A. and Essigmann,J.M. (1998) Oxidized, deaminated cytosines are a source of C→T transitions in vivo. Proc. Natl Acad. Sci. USA, 95, 3578–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hegi M.E., Sagelsdorff,P. and Lutz,W.K. (1989) Detection by 32P-postlabeling of thymidine glycol in γ-irradiated DNA. Carcinogenesis, 10, 43–47. [DOI] [PubMed] [Google Scholar]
- 77.Weinfeld M., Xing,J.Z., Lee,J., Leadon,S.A., Cooper,P.K. and Le,X.C. (2001) Factors influencing the removal of thymine glycol from DNA in γ-irradiated human cells. Prog. Nucleic Acid Res. Mol. Biol., 68, 139–149. [DOI] [PubMed] [Google Scholar]
- 78.Clark J.M. and Beardsley,G.P. (1989) Template length, sequence context and 3′-5′ exonuclease activity modulate replicative bypass of thymine glycol lesions in vitro. Biochemistry, 28, 775–779. [DOI] [PubMed] [Google Scholar]
- 79.McNulty J.M., Jerkovic,B., Bolton,P.H. and Basu,A.K. (1998) Replication inhibition and miscoding properties of DNA templates containing a site-specific cis-thymine glycol or urea residue. Chem. Res. Toxicol., 11, 666–673. [DOI] [PubMed] [Google Scholar]
- 80.Purmal A.A., Lampman,G.W., Bond,J.P., Hatahet,Z. and Wallace,S.S. (1998) Enzymatic processing of uracil glycol, a major oxidative product of DNA cytosine. J. Biol. Chem., 273, 10026–10035. [DOI] [PubMed] [Google Scholar]
- 81.Basu A.K., Loechler,E.L., Leadon,S.A. and Essigmann,J.M. (1989) Genetic effects of thymine glycol: site-specific mutagenesis and molecular modeling studies. Proc. Natl Acad. Sci. USA, 86, 7677–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakamura J., La,D.K. and Swenberg,J.A. (2000) 5′-Nicked apurinic/apyrimidinic sites are resistant to β-elimination by β-polymerase and are persistent in human cultured cells after oxidative stress. J. Biol. Chem., 275, 5323–5328. [DOI] [PubMed] [Google Scholar]
- 83.Nakamura J. and Swenberg,J.A. (1999) Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res., 59, 2522–2526. [PubMed] [Google Scholar]
- 84.Randall S.K., Eritja,R., Kaplan,B.E., Petruska,J. and Goodman,M.F. (1987) Nucleotide insertion kinetics opposite abasic lesions in DNA. J. Biol. Chem., 262, 6864–6870. [PubMed] [Google Scholar]
- 85.Takeshita M., Chang,C.N., Johnson,F., Will,S. and Grollman,A.P. (1987) Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem., 262, 10171–10179. [PubMed] [Google Scholar]
- 86.Kamiya H., Suzuki,M., Komatsu,Y., Miura,H., Kikuchi,K., Sakaguchi,T., Murata,N., Masutani,C., Hanaoka,F. and Ohtsuka,E. (1992) An abasic site analogue activates a c-Ha-ras gene by a point mutation at modified and adjacent positions. Nucleic Acids Res., 20, 4409–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shibutani S., Takeshita,M. and Grollman,A.P. (1997) Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the ‘A rule’. J. Biol. Chem., 272, 13916–13922. [DOI] [PubMed] [Google Scholar]
- 88.Efrati E., Tocco,G., Eritja,R., Wilson,S.H. and Goodman,M.F. (1997) Abasic translesion synthesis by DNA polymerase beta violates the ‘A-rule’. Novel types of nucleotide incorporation by human DNA polymerase beta at an abasic lesion in different sequence contexts. J. Biol. Chem., 272, 2559–2569. [DOI] [PubMed] [Google Scholar]
- 89.Lawrence C.W., Borden,A., Banerjee,S.K. and Le Clerc,J.E. (1990) Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res., 18, 2153–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gibbs P.E. and Lawrence,C.W. (1995) Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J. Mol. Biol., 251, 229–236. [DOI] [PubMed] [Google Scholar]
- 91.Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Deoxycytidyl transferase activity of yeast REV1 protein. Nature, 382, 729–731. [DOI] [PubMed] [Google Scholar]
- 92.Tang M., Shen,X., Frank,E.G., O’Donnell,M., Woodgate,R. and Goodman,M.F. (1999) UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl Acad. Sci. USA., 96, 8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reuven N.B., Arad,G., Maor-Shoshani,A. and Livneh,Z. (1999) The mutagenesis protein UmuC is a DNA polymerase activated by UmuD’, RecA and SSB and is specialized for translesion replication. J. Biol. Chem., 274, 31763–31766. [DOI] [PubMed] [Google Scholar]
- 94.Kamiya H., Suzuki,M. and Ohtsuka,E. (1993) Mutation-spectrum of a true abasic site in codon 12 of a c-Ha-ras gene in mammalian cells. FEBS Lett., 328, 125–129. [DOI] [PubMed] [Google Scholar]
- 95.Gentil A., Renault,G., Madzak,C., Margot,A., Cabral-Neto,J.B., Vasseur,J.J., Rayner,B., Imbach,J.L. and Sarasin,A. (1990) Mutagenic properties of a unique abasic site in mammalian cells. Biochem. Biophys. Res. Commun., 173, 704–710. [DOI] [PubMed] [Google Scholar]
- 96.Gentil A., Cabral-Neto,J.B., Mariage-Samson,R., Margot,A., Imbach,J.L., Rayner,B. and Sarasin,A. (1992) Mutagenicity of a unique apurinic/apyrimidinic site in mammalian cells. J. Mol. Biol., 227, 981–984. [DOI] [PubMed] [Google Scholar]
- 97.CabralNeto J.B., Cabral,R.E., Margot,A., Le Page,F., Sarasin,A. and Gentil,A. (1994) Coding properties of a unique apurinic/apyrimidinic site replicated in mammalian cells. J. Mol. Biol., 240, 416–420. [DOI] [PubMed] [Google Scholar]
- 98.Ide H., Murayama,H., Sakamoto,S., Makino,K., Honda,K., Nakamuta,H., Sasaki,M. and Sugimoto,N. (1995) On the mechanism of preferential incorporation of dAMP at abasic sites in translesional DNA synthesis. Role of proofreading activity of DNA polymerase and thermodynamic characterization of model template-primers containing an abasic site. Nucleic Acids Res., 23, 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gelfand C.A., Plum,G.E., Grollman,A.P., Johnson,F. and Breslauer,K.J. (1998) Thermodynamic consequences of an abasic lesion in duplex DNA are strongly dependent on base sequence. Biochemistry, 37, 7321–7327. [DOI] [PubMed] [Google Scholar]
- 100.Murata-Kamiya N., Kamiya,H., Karino,N., Ueno,Y., Matsuda,A. and Kasai,H. (1999) Formation of 5-formyl-2′-deoxycytidine from 5-methyl-2′-deoxycytidine in duplex DNA by Fenton-type reactions and γ-irradiation. Nucleic Acids Res., 27, 4385–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Karino N., Ueno,Y. and Matsuda,A. (2001) Synthesis and properties of oligonucleotides containing 5-formyl-2′-deoxycytidine: in vitro DNA polymerase reactions on DNA templates containing 5-formyl-2′-deoxycytidine. Nucleic Acids Res. 29, 2456–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kamiya H., Tsuchiya,H., Karino,N., Ueno,Y., Matsuda,A. and Harashima,H. (2002) Mutagenicity of 5-formylcytosine, an oxidation product of 5-methylcytosine, in DNA in mammalian cells. J. Biochem., 132, 551–555. [DOI] [PubMed] [Google Scholar]
- 103.Henderson P.T., Delaney,J.C., Gu,F., Tannenbaum,S.R. and Essigmann,J.M. (2002) Oxidation of 7,8-dihydro-8-oxoguanine affords lesions that are potent sources of replication errors in vivo. Biochemistry, 41, 914–921. [DOI] [PubMed] [Google Scholar]
- 104.Maki H. and Sekiguchi,M. (1992) MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature, 355, 273–275. [DOI] [PubMed] [Google Scholar]
- 105.Tajiri T., Maki,H. and Sekiguchi,M. (1995) Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res., 336, 257–267. [DOI] [PubMed] [Google Scholar]
- 106.Shen Z., Wu,W. and Hazen,S.L. (2000) Activated leukocytes oxidatively damage DNA, RNA and the nucleotide pool through halide-dependent formation of hydroxyl radical. Biochemistry, 39, 5474–5482. [DOI] [PubMed] [Google Scholar]
- 107.Tassotto M.L. and Mathews,C.K. (2002) Assessing the metabolic function of the MutT 8-oxodeoxyguanosine triphosphatase in Escherichia coli by nucleotide pool analysis. J. Biol. Chem., 277, 15807–15812. [DOI] [PubMed] [Google Scholar]
- 108.Mathews C.K. (1972) Biochemistry of deoxyribonucleic acid-defective amber mutants of bacteriophage T4. 3. Nucleotide pools. J. Biol. Chem., 247, 7430–7438. [PubMed] [Google Scholar]
- 109.Purmal A.A., Kow,Y.W. and Wallace,S.S. (1994) 5-Hydroxypymidine deoxynucleoside triphosphates are more efficiently incorporated into DNA by exonuclease-free Klenow fragment than 8-oxopurine deoxynucleoside triphosphates. Nucleic Acids Res., 22, 3930–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Einolf H.J., Schnetz-Boutaud,N. and Guengerich,F.P. (1998) Steady-state and pre-steady-state kinetic analysis of 8-oxo-7,8-dihydroguanosine triphosphate incorporation and extension by replicative and repair DNA polymerases. Biochemistry, 37, 13300–13312. [DOI] [PubMed] [Google Scholar]
- 111.Kamiya H. and Kasai,H. (2000) 2-Hydroxy-dATP is incorporated opposite G by Escherichia coli DNA polymerase III resulting in high mutagenicity. Nucleic Acids Res., 28, 1640–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pavlov Y.I., Minnick,D.T., Izuta,S. and Kunkel,T.A. (1994) DNA replication fidelity with 8-oxodeoxyguanosine triphosphate. Biochemistry, 33, 4695–4701. [DOI] [PubMed] [Google Scholar]
- 113.Inoue M., Kamiya,H., Fujikawa,K., Ootsuyama,Y., Murata-Kamiya,N., Osaki,T., Yasumoto,K. and Kasai,H. (1998) Induction of chromosomal gene mutations in Escherichia coli by direct incorporation of oxidatively damaged nucleotides. J. Biol. Chem., 273, 11069–11074. [DOI] [PubMed] [Google Scholar]
- 114.Kamiya H., Maki,H. and Kasai,H. (2000) Two DNA polymerases of Escherichia coli display distinct misinsertion specificities for 2-hydroxy-dATP during DNA synthesis. Biochemistry, 39, 9508–9513. [DOI] [PubMed] [Google Scholar]
- 115.Suzuki T., Yoshida,M., Yamada,M., Ide,H., Kobayashi,M., Kanaori,K., Tajima,K. and Makino,K. (1998) Misincorporation of 2′-deoxyoxanosine 5′-triphosphate by DNA polymerases and its implication for mutagenesis. Biochemistry, 37, 11592–11598. [DOI] [PubMed] [Google Scholar]
- 116.Terato H., Masaoka,A., Kobayashi,M., Fukushima,S., Ohyama,Y., Yoshida,M. and Ide,H. (1999) Enzymatic repair of 5-formyluracil. II. Mismatch formation between 5-formyluracil and guanine during DNA replication and its recognition by two proteins involved in base excision repair (AlkA) and mismatch repair (MutS). J. Biol. Chem., 274, 25144–25150. [DOI] [PubMed] [Google Scholar]
- 117.Fujikawa K., Kamiya,H. and Kasai,H. (1998) The mutations induced by oxidatively damaged nucleotides, 5-formyl-dUTP and 5-hydroxy-dCTP, in Escherichia coli. Nucleic Acids Res., 26, 4582–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Feig D.I., Sowers,L.C. and Loeb,L.A. (1994) Reverse chemical mutagenesis: identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc. Natl Acad. Sci. USA, 91, 6609–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ide H., Melamede,R.J. and Wallace,S.S. (1987) Synthesis of dihydrothymidine and thymidine glycol 5′-triphosphates and their ability to serve as substrates for Escherichia coli DNA polymerase I. Biochemistry, 26, 964–969. [DOI] [PubMed] [Google Scholar]
- 120.Purmal A.A., Bond,J.P., Lyons,B.A., Kow,Y.W. and Wallace,S.S. (1998) Uracil glycol deoxynucleoside triphosphate is a better substrate for DNA polymerase I Klenow fragment than thymine glycol deoxynucleoside triphosphate. Biochemistry, 37, 330–338. [DOI] [PubMed] [Google Scholar]
- 121.Demple B. and Harrison,L. (1994) Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem., 63, 915–948. [DOI] [PubMed] [Google Scholar]
- 122.Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- 123.Scharer O.D. and Jiricny,J. (2001) Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays, 23, 270–281. [DOI] [PubMed] [Google Scholar]
- 124.Yanofsky C., Cox,E.C. and Horn,V. (1966) The unusual mutagenic specificity of an E. coli mutator gene. Proc. Natl Acad. Sci. USA, 55, 274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mo J.Y., Maki,H. and Sekiguchi,M., (1992) Hydrolytic elimination of a mutagenic nucleotide, 8-oxodGTP, by human 18-kilodalton protein: sanitization of nucleotide pool. Proc. Natl Acad. Sci. USA, 89, 11021–11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sakumi K., Furuichi,M., Tsuzuki,T., Kakuma,T., Kawabata,S., Maki,H. and Sekiguchi,M. (1993) Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem., 268, 23524–23530. [PubMed] [Google Scholar]
- 127.Furuichi M., Yoshida,M.C., Oda,H., Tajiri,T., Nakabeppu,Y., Tsuzuki,T. and Sekiguchi,M. (1994) Genomic structure and chromosome location of the human mutT homologue gene MTH1 encoding 8-oxo-dGTPase for prevention of A:T to C:G transversion. Genomics, 24, 485–490. [DOI] [PubMed] [Google Scholar]
- 128.Fujikawa K., Kamiya,H., Yakushiji,H., Fujii,Y., Nakabeppu,Y. and Kasai,H. (1999) The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J. Biol. Chem., 274, 18201–18205. [DOI] [PubMed] [Google Scholar]
- 129.Tsuzuki T., Egashira,A., Igarashi,H., Iwakuma,T., Nakatsuru,Y., Tominaga,Y., Kawate,H., Nakao,K., Nakamura,K., Ide,F., Kura,S., Nakabeppu,Y., Katsuki,M.T., Ishikawa,M. and Sekiguchi,M. (2001) Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase. Proc. Natl Acad. Sci. USA, 98, 11456–11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kamiya H., Murata-Kamiya,N., Iida,E. and Harashima,H. (2001) Hydrolysis of oxidized Nucleotides by the Escherichia coli Orf135 protein. Biochem. Biophys. Res. Commun., 288, 499–502. [DOI] [PubMed] [Google Scholar]
- 131.Kobayashi M., Ohara-Nemoto,Y., Kaneko,M., Hayakawa,H., Sekiguchi,M. and Yamamoto,K. (1998) Potential of Escherichia coli GTP cyclohydrolase II for hydrolyzing 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem., 273, 26394–26399. [DOI] [PubMed] [Google Scholar]
- 132.Lin S., McLennan,A.G., Ying,K., Wang,Z., Gu,S., Jin,H., Wu,C., Liu,W., Yuan,Y., Tang,R., Xie,Y. and Mao,Y. (2001) Cloning, expression and characterization of a human inosine triphosphate pyrophosphatase encoded by the itpa gene. J. Biol. Chem., 276, 18695–18701. [DOI] [PubMed] [Google Scholar]
- 133.McIntosh E.M., Ager,D.D., Gadsden,M.H. and Haynes,R.H. (1992) Human dUTP pyrophophatase: cDNA sequence and potential biological importance of the enzyme. Proc. Natl Acad. Sci. USA, 89, 8020–8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang Z. (2001) Translesion synthesis by the UmuC family of DNA polymerases. Mutat. Res., 486, 59–70. [DOI] [PubMed] [Google Scholar]
- 135.Livneh Z. (2001) DNA damage control by novel DNA polymerases: translesion replication and mutagenesis. J. Biol. Chem., 276, 25639–25642. [DOI] [PubMed] [Google Scholar]
- 136.Friedberg E.C., Wagner,R. and Radman,M. (2002) Specialized DNA polymerases, cellular survival and the genesis of mutations. Science, 296, 1627–1630. [DOI] [PubMed] [Google Scholar]
- 137.Napolitano R., Janel-Bintz,R., Wagner,J. and Fuchs,R.P. (2000) All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J., 19, 6259–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lawrence C.W., Borden,A. and Woodgate,R. (1996) Analysis of the mutagenic properties of the UmuDC, MucAB and RumAB proteins, using a site-specific abasic lesion. Mol. Gen. Genet., 251, 493–498. [DOI] [PubMed] [Google Scholar]
- 139.Delaney J.C. and Essigmann,J.M. (2001) Effect of sequence context on O6-methylguanine repair and replication in vivo. Biochemistry, 40, 14968–14975. [DOI] [PubMed] [Google Scholar]
- 140.Hatahet Z., Zhou,M., Reha-Krantz,L.J., Morrical,S.W. and Wallace,S.S. (1998) In search of a mutational hotspot. Proc. Natl Acad. Sci. USA, 95, 8556–8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hatahet Z., Zhou,M., Reha-Krantz,L.J., Ide,H., Morrical,S.W. and Wallace,S.S. (1998) In vitro selection of sequence contexts which enhance bypass of abasic sites and tetrahydrofuran by T4 DNA polymerase holoenzyme. J. Mol. Biol., 286, 1045–1057. [DOI] [PubMed] [Google Scholar]
- 142.Nunoshiba T., Obata,F., Boss,A.C., Oikawa,S., Mori,T., Kawanishi,S. and Yamamoto,K. (1999) Role of iron and superoxide for generation of hydroxyl radical, oxidative DNA lesions and mutagenesis in Escherichia coli. J. Biol. Chem., 274, 34832–34837. [DOI] [PubMed] [Google Scholar]
- 143.Nunoshiba T., Watanabe,T., Nakabeppu,Y. and Yamamoto,K. (2002) Mutagenic target for hydroxyl radicals generated in Escherichia coli mutant deficient in Mn- and Fe-superoxide dismutases and Fur, a repressor for iron-uptake systems. DNA Repair, 1, 411–418. [DOI] [PubMed] [Google Scholar]