Abstract
The finding in animal species of complexes homologous to the products of six Saccharomyces cerevisiae genes, origin of replication recognition complex (ORC), has suggested that ORC-related mechanisms have been conserved in all eukaryotes. In plants, however, the only cloned putative homologs of ORC subunits are the Arabidopsis ORC2 and the rice ORC1. Homologs of other subunits of plant origin have not been cloned and characterized. A striking observation was the absence from the Arabidopsis genome of an obvious candidate gene-homolog of ORC4. This fact raised compelling questions of whether plants, in general, and Arabidopsis, in particular, may have lost the ORC4 gene, whether ORC-homologous subunits function within a complex in plants, whether an ORC complex may form and function without an ORC4 subunit, whether a functional (but not sequence) protein homolog may have taken up the role of ORC4 in Arabidopsis, and whether lack of ORC4 is a plant feature, in general. Here, we report the first cloned and molecularly characterized five genes coding for the maize putative homologs of ORC subunits ZmORC1, ZmORC2, ZmORC3, ZmORC4 and ZmORC5. Their expression profiles in tissues with different cell-dividing activities are compatible with a role in DNA replication. Based on the potential of ORC-homologous maize proteins to bind each other in yeast, we propose a model for their possible assembly within a maize ORC. The isolation and molecular characterization of an ORC4-homologous gene from maize argues that, in its evolution, Arabidopsis may have lost the homologous ORC4 gene.
INTRODUCTION
The availability of well-defined origin of replication sequences in Saccharomyces cerevisiae [autonomous replicating sequences (ARS)] has allowed the isolation and characterization of protein factors that interact with the ARS. A six-subunit complex, origin of replication recognition complex (ORC), can recognize and specifically bind the origins of DNA replication (1). The subunits of the complex function in a genetic network, interact physically with each other, and provide a landing pad for the assembly of the pre-replicative complex (pre-RC) (reviewed in 2,3). The subsequent discovery of genes homologous to the yeast ORC subunits in higher eukaryotes was unexpected because metazoan replicators are different from the yeast ARS. Even the Schizosaccharomyces pombe origins of replication differ from the S.cerevisiae ARS and do not function as replication origins in S.cerevisiae and vice versa (4,5). The concept of a metazoan ‘origin locus’ was introduced to refer to a region permissive for initiation within which some sites were utilized more frequently than others (6). The regions serving as replication origins in higher eukaryotes also require presence of bound ORC for the assembly of the pre-RC and for the progression through the cell cycle. This has suggested that ORC has evolved as a function conserved in all eukaryotes (7–9).
In this context, it is important that no ORC of plant origin has been reported yet. In general, little is known about DNA replication and the factors involved in this process in plants, but the cloning of subunit ORC2 from Arabidopsis (10) and of ORC1 from rice (11) has made it likely that multisubunit ORC complexes might function in plants in ways similar to the other eukaryotes. Thus, it was striking that no sequence homologous to the ORC4 gene could be revealed in the Arabidopsis genome when it was searched with either the S.cerevisiae or the human ORC4 sequences as probes. In contrast, potential homologs to all other genes, including the least conserved ORC3 and ORC6, were hit by this search. The sequencing of the Arabidopsis genome is almost complete except for some gaps in the centromeric regions. Theoretically, a possibility still exists that a putative ORC4 homolog might be found in the missing sequences. However, given the fact that functional genes and unknown proteins are routinely identified through EST libraries, it appears unlikely that a highly functional gene coding for an essential protein, like ORC4, would have remained undetected. Therefore, two alternatives may be considered: either the plant ORC4 gene has diverged to such an extent that homology-search programs no longer recognize it or that, in its evolution, Arabidopsis has lost the sequence homologous to the ORC4 gene. An answer to either of these questions will have obvious implications.
The results reported here support the second alternative. Having isolated, sequenced and analyzed an ORC4-homologous gene from maize, we show that an ORC4 homolog of plant origin is well conserved in structure and immediately recognized by the available homology-search tools. This argues that should a putative Arabidopsis ORC4 sequence exist it would have been detected via its similarity to other known ORC4 genes. Putative counterparts of ORC subunits 3, 4, 5 and 6 have not been cloned and characterized for any plant species. Here, we report the first molecular characterization of five homologs of ORC subunits from maize, ZmORC1– ZmORC5, suggesting the existence of a full replication complex in plants. The expression pattern of the putative ORC subunits in various maize tissues was compatible with a possible role in DNA replication. Interaction between expressed proteins (tested in a yeast two-hybrid system) outlined a possible mode of their assembly into a putative complex. A possible model, based on their binding affinities, is proposed.
MATERIALS AND METHODS
Maize ORC cDNAs
Four maize ESTs homologous to subunits ORC1, ORC2, ORC3 and ORC4 were identified in the maize Database of Pioneer HiBred Inc., when searched with the respective human and S.cerevisiae sequences as probes. The sequences of the four EST clones were used to design primers to isolate the respective ZmORC cDNAs. No EST homologous to ORC5 was available to us. Consequently, we have used an alternative approach to clone a putative maize ORC5 cDNA, as described below.
ORC1. The partial cDNA clone (ESTP0043.cimag61) containing 1138 bp homologous to the 3′-end of known ORC1 sequences was extended upstream by PCR. A forward primer, 5′-GGG ACG ACG TGT ACG TGA AG-3′, was designed from an upstream region conserved in the ORC1 proteins of five species (see Results). The reverse primer, 5′-GTC GTT GGA TAC CCA TCC GAC TTG-3′, was complementary to a region in the available EST. Root cDNA was used as template. PCR conditions were: 95°C for 5 min, 95°C for 30 s, 50°C for 30 s, 72°C for 1 min 30 s (36 cycles), 72°C for 10 min. A PCR product of the expected size (1.5 kb) was amplified and cloned. From the newly defined 5′ sequence, a reverse primer (5′-TGG TAC GAT ACA GCT CTC TTC TCA GGT-3′) was designed for 5′ RACE using the Clontech kit according to the manufacturer’s instructions. To amplify ∼3.0 kb cDNA, the forward (5′-ACT CGT CCA CTT TTC AAA TCC ACA TTC CC-3′) and the reverse (5′-ACA TCC TTC GAT GGT TCC TTT GCA GA-3′) primers were used. The sequenced clone for ZmORC1 was submitted to GenBank (accession no. AF417481).
ORC2. From the sequence of the partial cDNA clone (ESTP0083.cldc268) displaying homology to known ORC2 proteins, a reverse primer 5′-AAG CCA ACA AAG CAG AGC GGT TCA-3′ was designed. The forward primer (5′-GAA GCA AAA CGG ATG GCC CCG AGA-3′) was based on the sequence of a maize 5′ EST (kindly provided by S. McCormick, Berkeley). PCR was performed as above and the resulting PCR product, ZmORC2, cloned and sequenced was submitted to GenBank (accession no. AF417482).
ORC3. The partial cDNA clone (ESTP0016.ctsav84), showing dispersed homology to the human ORC3, was subjected directly to 5′ RACE. With the reverse primer (5′-CTT CGA GGG AGA GCT GCA AGA TTT CCC TTT-3′) we managed to extend the available sequence by ∼2.2 kb. Its sequence was used to design primers to amplify the ORC3 homologous cDNA (forward primer, 5′-TTC ATA CCC TCG CGA AGA CCA CTA G-3′; reverse primer, 5′-CTC AAA TGT ACC CCT TGG GCT ACA TGA-3′). The sequence of ZmORC3 is in GenBank (accession no. AF417483).
ORC4. A partial cDNA clone (ESTP0016.ctsbv18) containing a 1212 bp insert similar to known ORC4 genes was extended upstream with the reverse primer, 5′-TCC TCC TTC AGA TCA TCA AGA ACC-3′, by 5′ RACE. The recovered ∼800 bp fragment was a basis for designing a forward primer (5′-CAA GCC CCT GCT GGT TAA ACG AAT T-3′). The 5′-CAT GCC CTG CTG GTT TAA CGA ATT C-3′ sequence was used as the reverse primer in a PCR reaction amplifying the cDNA in between. The resulting 1.5 kb product (ZmORC4) was submitted to GenBank (accession no. AF417484).
ORC5. Using the human and the S.cerevisiae ORC5 sequences as probes, we were unable to identify a putative ORC5 maize EST. However, from the public databases, we successfully retrieved Arabidopsis and rice homologous sequences. These, together with the human, mouse and Xenopus laevis ORC5 sequences, were used to outline patches of conserved amino acids (see Results) and to design upstream (5′-GAC AAG CCA/C TCC GAC TTC/T GTC-3′) and downstream (5′-AAT ATA/C GCC AAG/T AGC/T CTC TCA/C AGA/T GGA AA-3′) degenerate primers. With 3′ RACE (forward primer, 5′-GCT ACT CTT GAT GCA GCT TTG TTC GAT TCG-3′) (Clontech kit) we established the 3′-end of the cDNA. The sequence (5′-GAC AAG CCA TCC GAC TTC GTC-3′) complementary to an upstream region and a sequence from the 3′-region (5′-CTA TTG ACA TAA TGC TAC CTC TGA GAC ATC TCC-3′) were used as a forward and a reverse primer, respectively, to amplify ZmORC5 cDNA (accession no. AY081944).
Gene expression
All PCR fragments were cloned with the TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, CA) and sequenced from both directions with 10–12-fold redundancy. RNA was isolated from various tissues of 13-day-old seedlings with the RNeasy Mini kit (Qiagen). cDNAs were generated by RT–PCR, using RETROscript™ (Ambio, Austin, TX) following the manufacturer’s suggestions. For semi-quantitative RT–PCR, equal amounts of RNA (1 µg) from maize roots, seedlings and mature leaves were used. For all PCR reactions, internal standards (Quantum RNATm 18S Internal Standards; Ambion) were used along with the primers designed to amplify ZmORC1, ZmORC2, ZmORC3, ZmORC4 and ZmORC5 cDNA in a multiplex PCR. This technology can modulate the amplification efficiency of a particular PCR template without affecting the performance of other targets in a multiplex PCR. PCR conditions were established such that the PCR reactions would remain in the linear range for a limited number of cycles. For the semi-quantitative RT–PCR here, the PCR conditions were optimized as follows: 95°C (1 min), 95°C (30 s), 55°C (30 s), 72°C (1 min), 30 cycles. Ready-To-Go PCR beads (Amersham) were used in all reactions to minimize variation. eIF4A was used as an amplification control under these experimental conditions.
Binding in yeast two hybrid system
The open reading frames (ORFs) of the putative maize ORC subunits were cloned in-frame in both the binding domain (BD) cloning vector pGBKT7 and in the activation domain (AD) cloning vector pGADT7 to act as ‘bait’ and ‘prey’, respectively, in the Yeast Two Hybrid Matchmaker System (Clontech). The partial ORF of ZmORC5 was also cloned in the two vectors. All possible combinations of ‘prey’ and ‘bait’ were tested in yeast strain AH109. The Matchmaker system incorporates features that reduce the incidence of false positives. It comes with both positive and negative controls. The transformants were plated on medium-stringency medium SD-his-leu-trp, and those that grew on the medium were transferred to high-stringency medium SD-ade-his-leu-trp/X-α-gal. This screen virtually eliminates false positive interactions.
Protein architecture and phylogenetic tree construction
Database searches were performed with BLASTP and PSI-BLAST programs on the NCBI non-redundant database. Determination of the modular structure of the proteins was performed using the PROFILESCAN Server from the Swiss Institute for Bioinformatics, the SMART (Simple Modular Architecture Research Tool) site and the Conserved Domain Database at NCBI. Pairwise alignments were compiled using CLUSTALX (12) and phylogenetic and molecular evolutionary analyses were conducted using MEGA version 2.1 (13). Bootstrap analysis using the neighbor joining (NJ) method (500 pseudoreplicates) and 50% majority-rule analysis were employed.
RESULTS
Molecular characterization of the putative maize ORC homologs
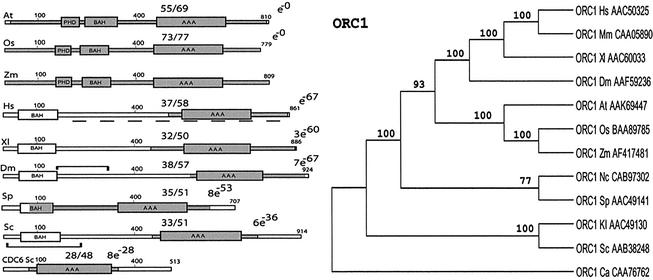
ZmORC1. The human ORC1 sequence used as a probe in a TBLASTN search recognized a maize EST containing the 3′-end of a putative maize ORC1 homolog. The 1138 bp insert was sequenced and a BLASTX search indicated significant similarity to all known ORC1 subunits. In order to obtain the entire gene sequence, we attempted a 5′ RACE with primers designed from the 5′-region of the isolated EST. This initial try was unsuccessful, probably because of the large size of the missing 5′-portion of the putative ORC1 (ZmORC1) gene. To circumvent the problem, we took advantage of a recently identified rice ORC1 gene (11). After aligning the sequences of the ORC1 homologs from rice, human, Drosophila and yeast and a putative ORC1 from the Arabidopsis database (AL049730.2), we outlined patches of amino acids conserved among all five species. Nested primers, based on a highly conserved peptide (GDDVYVK) upstream of the maize EST, were used to extend the available sequence by an additional ∼2.1 kb. From this newly sequenced region, anchor primers for a 5′ RACE were designed (see Materials and Methods). After conceptual translation of the ORF, the initiator methionine codon was defined by the start of homology with the ORC1 proteins from other species. The homology within the first amino acids of known ORC1 proteins determined the correct N-terminus of the maize protein as well. The size of ZmORC1, 809 amino acids, is comparable with the size of ORC1 from other species. It is similar to rice (73% identical, 77% similar), Arabidopsis (55% identical, 69% similar), human (37% identical, 58% similar) and S.pombe (35% identical, 51% similar) ORC1 proteins (Fig. 1, left). Maize, rice and Arabidopsis are the only species of plant origin for which data on ORC1 are available. The three proteins are similar over the entire length of their proteins. However, sequence similarity to the ORC1 of other species is restricted to the C-terminal halves of the molecules. The C-terminal region is the most highly conserved region among the ORC1 proteins of all species. This region contains a module that defines the ORC1 proteins as members of the AAA+ superfamily of ATPases (14,15).
Figure 1.
Modular architecture of ORC1 homologs and the phylogenetic tree. Proteins are drawn to scale, the figures along the bars reflect amino acid numbers. The numbers separated by a solidus show the degree of identity/similarity between the maize proteins and the other ORC subunits. Regions displaying similarity to the maize protein are shaded. The boxes correspond to the architectural motifs. The brackets in the DmORC1 and ScORC1 molecules indicate the regions binding HP1 and SIR1, respectively. The broken line under the human ORC1 corresponds to the domain interacting with HBO1. In the NJ tree branch lengths are proportional to amino acid substitutions (bar represents 10 substitutions per 100 residues). Bootstrap values (500 pseudoreplicates) are shown in association with nodes in the phylogeny. Branches with bootstrap values <50% are collapsed. Abbreviations: Sc, S.cervisiae; Sp, S.pombe; Dm, Drosophila melanogaster; At, Arabidopsis thaliana; Zm, Zea mays; Os, Oryza sativa; Xl, X.laevis; Hs, Homo sapiens; Mm, Mus musculus; Kl, K.lactis; Nc, N.crassa; Ca, Candida albicans.
It is remarkable that the N-terminal halves of the ORC1 proteins of other species also do not share sequence similarity. Thus, there is no detectable homology between the N-terminal halves of the Drosophila, the human and the yeast molecules. Nonetheless, all ORC1 homologs (including plants) contain a domain upstream of their AAA motif that was recognized as the BAH domain (Fig. 1, left) found in a variety of regulatory factors (16).
A distinctive feature of the available ORC1 proteins of plant origin is the presence of an additional motif absent from the molecules of all other species, a PHD finger, implicated in chromatin-mediated transcriptional regulation (17).
The degree of similarity between ORC1 representatives from different species is shown in the cladogram (Fig. 1, right). The ORC1 proteins from animal and from plant origin occupy two well-supported clades apparently more closely related to the ORC1 proteins from S.pombe and Neurospora crassa, than to those of Kluyveromyces lactis and S.cerevisiae.
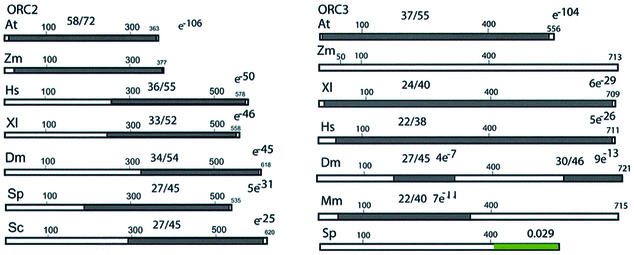
ZmORC2. In animals and in yeast, ORC2 is the second largest subunit made of about 600 amino acids. We expected that the 3′ EST at our disposal would be too short (about 243 amino acids) for a successful 5′ RACE extension. We took advantage of the sequence of a 5′ EST from a maize pollen-specific library (kindly provided by S. McCormick) to design appropriate primers and to isolate the putative maize ORC2 cDNA. Conceptual translation of the finalized cDNA sequence revealed an ORF of 377 amino acids from start to stop codons (Fig. 2).
Figure 2.
Molecular structure of ORC2 and ORC3 homologs. No architectural domains and motifs have been revealed by PROFILESCAN in any of these proteins. Annotations and abbreviations are as under Figure 1.
A BLSTP homology search found a high level of similarity (e–106) between the maize and the Arabidopsis proteins over their entire lengths (58% identical, 72% similar). The maize sequence is also similar to the human (e–50), Xenopus (e–46), Drosophila (e–45), S.pombe (5e–31) and S.cerevisiae (e–25) homologs. ZmORC2 is notably shorter than most other ORC2 homologs but is comparable to the size of the only other reported ORC2 sequence of plant origin, the Arabidopsis ORC2 (10). The entire lengths of both plant proteins correspond to the C-terminal halves of the other known ORC2 homologs and lack the N-terminal halves altogether (Fig. 2). It is remarkable that among the other species, there is a sharp drop in sequence conservation between the N- and the C-terminal halves of the ORC2 proteins as well. This drop occurs within a narrow region that maps to the N-terminal end of the plant molecules. No conserved architectural elements have been recognized in the ORC2 molecules of any species.
ZmORC3. Using the sequence of the available maize EST, we were successful in directly extending it by an additional ∼2.2 kb by 5′ RACE (see Materials and Methods). Conceptual translation of the recovered sequence, followed by BLAST searches, defined the maize protein as a 713 amino acid ORC3 homolog, ZmORC3. The similarity of ZmORC3 with reported frog and human ORC3 ORFs extended over their entire lengths, indicating that the initiator and stop codons for the maize protein were defined correctly. Regions and degrees of similarity to other species are shown in Figure 2. The initiator methionine codon was preceded by an upstream untranslated sequence (UTS) containing several stop codons in the same reading frame. The putative translated amino acids were followed by a 3′-UTS containing two stop codons. The size of the putative maize ORC3 homolog is similar to the size of animal homologs and clusters of conserved amino acids may be found at the C-terminal regions of the proteins. No homology could be revealed between ZmORC3 and the ORC3 sequences of either S.cerevisiae or S.pombe. A putative Arabidopsis protein (At5g16690) is 37% identical, 54% similar (e–104) over its entire length to the maize protein and is a probable candidate for an Arabidopsis ORC3.
We found no conserved motifs in ORC3 of any species (with PROFILESCAN) but the first 50 N-terminal amino acids region of the ZmORC3 was defined as proline-rich. None of the other species has a proline-rich N-terminus which, apparently, is a maize-specific feature.
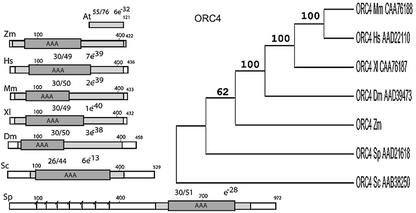
ZmORC4. A 1212 bp sequence isolated from the maize 3′ EST library (with the human ORC4 subunit as a probe) was extended upstream (∼800 bp) by 5′ RACE. The probable initiator codon was located by the beginning of homology with known ORC4 subunits within the first 10 amino acids of the compared proteins. The putative start codon is preceded by a 5′-UTS containing in-frame stop codons. Conceptually translated ORF would code for 422 amino acids (Fig. 3) similar over the entire length of the Xenopus (e–40), human (7e–39), Drosophila (3e–38) and S.cerevisiae (6e–13) proteins. The highly conserved regions among all species contain the AAA domain (in maize, amino acids 55–263). Peptide motifs corresponding to the two Walker motifs A and B (18) were also recognized in the maize protein.
Figure 3.
Modular architecture and a phylogenetic tree for ORC4 homologs. Labeling and annotations are as under Figure 1. The fission yeast ORC4 is notably larger than the homologs, containing a species-specific N-terminal half with seven AT-hook motifs, shown as vertical bars (39,49). Unrooted, NJ, bootstrap tree is shown to the right.
Use of human or yeast ORC4 sequences as a probe failed to identify a putative Arabidopsis homolog. We considered a possibility that the Arabidopsis ORC4 gene might have diverged from the yeast and mammalian genes beyond recognition. However, a search of the Arabidopsis database with the newly identified maize sequence also failed to identify a potential candidate for an Arabidopsis ORC4 homolog. Interestingly, an unknown Arabidopsis protein (AC006837) displayed 76% similarity to the C-terminal 120 amino acids of ORC4 (Fig. 3). This short (121 amino acid) unknown Arabidopsis protein is similar only to the extreme C-terminal region of the protein that does not include the AAA domain.
The relationship among reported ORC4 proteins from different species is shown in a cladogram (Fig. 3). The maize ORC4 is currently the only available protein of plant origin. The degree of its similarity with the known ORC4 has positioned ZmORC4 on a separate branch.
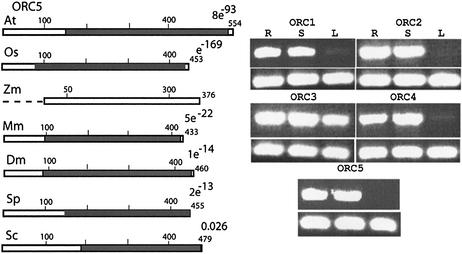
ZmORC5. We could not reveal a maize ORC5-homologous sequence in the searched databases, nor was there a relevant maize EST at our disposal. Thereby, no a priori evidence suggested that an ORC5-homologous gene existed in maize. To explore this possibility, we aligned several reported ORC5 sequences (human, mouse and Xenopus) with sequences found in the public databases that could represent Arabidopsis (At4g29910) and rice (AC084282) ORC5 homologs. Two clusters of amino acids conserved between all five proteins were outlined. An upstream domain (DKPSDFV) and a downstream conserved domain (FPLERLLAIF) were chosen to design degenerate primers and to amplify a product from maize root RNA template. After translation, the resulting ∼870 bp product amplified from a maize root cDNA template was similar to the ORC5 proteins from the other species. The 3′-end of the cDNA was established by 3′ RACE. Its analysis suggested that there were two different alleles of transcribed maize ORC5 varying in their 3′-UTS.
Attempts to recover the 5′-end of the putative maize ORC5 cDNAs were unsuccessful. Seven independent 5′ RACE experiments, using various combinations of nested anchor primers complementary to different regions of the cDNA, failed to amplify a putative 5′-end. The amplification reactions extended the available fragment in the 5′ direction but the recovered product has always remained ∼200 bp shorter when compared to the 5′-end of the putative rice ORC5 gene. Invariably, the retrieved fragments would begin with sequences corresponding to P78 of the putative rice ORC5 protein. A Walker A motif, found in ORC5 proteins, was to be located within the missing 75 N-terminal amino acids. We considered a possibility that the motif was present in the ZmORC5 molecule as well, but that for some reason we were unable to amplify it by the 5′ RACE. In an alternative approach, we designed degenerate primers from a conserved cluster of 48 amino acids present in known ORC5 proteins (the Walker A motif) and nested primers from the established maize sequences. Attempts to amplify a transcription product also failed, suggesting that ZmORC5 might not carry an ATP-binding domain.
Expression patterns of ORC-homologous maize genes
The expression levels of the five maize genes were examined in proliferating and non-proliferating tissues to test whether ORC gene activity would parallel the dividing activity of the tissue. We were interested in whether the levels of expression of all five genes, in a given tissue, would be comparable to each other. Presence of a message from each subunit in comparable amounts would be in agreement with a possibility that they might function within a stoichiometric complex.
The expression levels of the five maize genes in root tips, in 7-day-old seedlings and in mature green leaves is shown (Fig. 4, right). Reproducibly, all five genes displayed lower mRNA levels in the mature leaves, as opposed to the root tips and young seedlings. Roots and seedlings contain dividing cells, while the mature leaves are largely non-dividing. Thereby, the expression profile of the genes, paralleling cell-dividing activity, is compatible with a role in DNA replication. An interesting exception is the high expression of ZmORC3 in the leaves, implicating it in other functions, possibly even outside the complex.
Figure 4.
(Left) Molecular structure of ORC5 and expression profiles for the maize genes. The broken line at the 5′-end of ZmORC5 indicates the putatively missing 78 amino acids. (Right) Expression profiles of ZmORC1, ZmORC2, ZmORC3, ZmORC4 and ZmORC5, in root tips, seedlings and mature leaves. The transcription of a control gene (eIF4A) is in the bottom panels.
Interaction of the ZmORCs in the yeast two-hybrid system
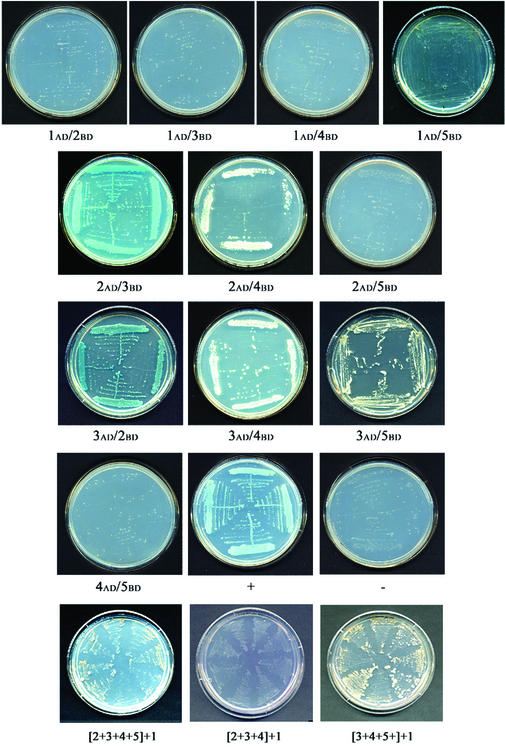
We employed the yeast two-hybrid system as an approach to study the potential of individual subunits to bind each other. The ORFs of the five genes, cloned both as ‘bait’ and as ‘prey’, were tested in all possible combinations.
ZmORC1 did not bind ZmORC2, ZmORC3 or ZmORC4, and showed only a very weak binding with ZmORC5, when tested as both AB- and BD-expressors (Fig. 5). ZmORC2 and ZmORC3 displayed the strongest affinity for each other. ZmORC2 could also bind ZmORC4, although with lesser affinity, and did not bind ZmORC5 at all. ZmORC3 displayed broader affinities, binding three subunits: ZmORC2, ZmORC4 and ZmORC5.
Figure 5.
Yeast two-hybrid binding assay. The binding of the different ZmORC subunits is shown when expressed as AD- or BD-expressors. + and – show yeast growth in a positive and a negative control, respectively. The binding of ZmORC1 to combinations of multiple co-expressed subunits is shown in the last row. The figures in the brackets correspond to the ZmORCs expressed simultaneously. In each plate, four independent interactions are shown in parallel, except in the last row where eight independent reactions are shown.
The weak binding of ZmORC1 to ZmORC5 and its inability to bind the other subunits raised a question of whether ZmORC1 would bind in a complex with the other proteins at all. Alternatively, ZmORC1 might need more than one subunit for an effective interaction within the complex. To test this possibility, we transformed yeast cells with ZmORC1 (as AD-expressor) and various combinations of ZmORC2, ZmORC3, ZmORC4 and ZmORC5 (as BD-expressors). When all five genes were co-expressed, the yeast grew under the most stringent selection conditions. Omission of ZmORC5 from the combination resulted in no colony growth, suggesting that ZmORC1 failed to bind the other subunits in the absence of ZmORC5 (Fig. 5). Co-expression of ZmORC3, ZmORC4, ZmORC5 and ZmORC1 also resulted in colony growth (Fig. 5), indicating that absence of ZmORC2 did not affect significantly the interaction of ZmORC1 with the other subunits. In contrast, none of the other combinations allowed growth in the quadruple selection media (not shown) suggesting that ZmORC3, ZmORC4 and ZmORC5 were required to effectively bind ZmORC1.
DISCUSSION
Putative maize ORC subunit homologs
ZmORC1, ZmORC4 and ZmORC5. The known ORC1, ORC4 and ORC5 are structurally related to each other and to the replication initiation protein Cdc6. They belong to the AAA+ superfamily of ATPases (14,15,19). The AAA domains containing the bipartite ATP binding and hydrolysis motifs, the Walker A and the Walker B motifs, are the most highly conserved regions between the respective proteins. The capacity to bind and/or to hydrolyze ATP varies among the different subunits. Thus, ATP binding has been demonstrated only for the yeast ORC1 and ORC5 (20–22), while only ORC1 can hydrolyze ATP (20). ATP binding by the S.cerevisiae ORC1 is required for the complex to bind the origins and ATP binding and origin binding are interdependent (20,21). ATP binding was crucial also for the function of the Drosophila ORC1, while mutations in the Walker A motif of ORC4 and ORC5 did not affect cell-free replication (23). The ORC4 of S.cerevisiae matches only weakly the Walker A motif suggesting that, probably, the motif is not involved in ATP binding in the yeast, but may be fully functional in vertebrates (24).
In maize, well-conserved Walker A and Walker B motifs were found in ZmORC1 and ZmORC4. However, the ORF of the recovered ZmORC5 contained a Walker B motif at the conserved position found in the other species but it did not carry a Walker A motif. It is interesting to point out that an ORC5 homolog without a Walker A motif was recently found in humans (24,25). Despite the absence of a Walker A motif, expressed ZmORC5 was able to bind ZmORC3 and ZmORC1 in yeast indicating that the motif might not be involved in the interaction with the other subunits. The importance of the Walker A and B motifs for the function of the maize proteins ZmORC1, ZmORC4 and ZmORC5, remains to be established.
ORC4 is highly conserved among the different species, particularly at the C-terminal half region carrying the AAA domain. The absence of an obvious ORC4 candidate in Arabidopsis raises interesting questions of how it functions without an obvious homolog of ORC4, whether this is a species-specific feature, and whether other dicots might lack an ORC4 homolog as well. An intriguing fact was the finding of a short unknown protein highly similar to the terminal 120 amino acids of ZmORC4, but lacking all other characteristic ORC4 features, including the AAA domain. It is unclear whether this putative protein could substitute for ORC4 in Arabidopsis. While these questions await their answers, one appears to be certain: failure of homology-search programs to reveal an Arabidopsis ORC4 is not due to sequence divergence of the plant counterpart.
ZmORC1 is the largest protein among the maize subunits and has a similar size and multi-domain architecture as the ORC1 proteins from other species (22). The sequence similarity between all ORC1 homologs is confined to the C-terminal portions of their molecules containing the AAA module, while no significant sequence similarity could be revealed in their N-terminal portions. Despite this fact, all ORC1 proteins (including plants) contain a BAH domain in their N-terminal half region. The BAH domain is a widely represented conserved peptide recognized in a variety of transcriptional regulators and in DNA methyltransferases (17). The function of the domain has not been elucidated but, evidently, the linking of the BAH with the AAA domain has evolved specifically for an ORC1 function.
The yeast ORC1 BAH domain is involved in the specific binding of SIR1, resulting in a silencing complex (26). Formation of a silencing complex in Drosophila also involves ORC1, but the region specifically binding HP1 only partially overlaps the BAH domain (27,28). In the human ORC1, the region binding the histone acetylase HBO1, possibly reflecting a silencing function of mammalian ORC1, is downstream of the BAH domain (29,30). It remains to be shown whether the plant ORC1, and its BAH domain, might be involved in silencing functions.
It is interesting that the ORC1 BAH domain of the budding yeast lacks sequence similarity with the BAH domain of the fission yeast or of any other species. Likewise, the sequence similarity between the human, the Drosophila, and the three plant ORC1 proteins does not extend over their respective BAH domains. Thereby, the BAH domain is an example of conserved peptide architecture, and perhaps function, in the absence of sequence similarity. Similar examples are the conserved MAR-functions around homologous genes in different species and the conservation of the tertiary structure of the chromo domain of Archea without significant conservation of its primary sequence (31–33).
Plant ORC1 homologs are unique because they contain an additional motif, a PHD finger, absent from the molecules of the other reported species. The role of this peptide in the context of the plant ORC1 has not been established but it is clear that the PHD finger motif has been recruited for plant-specific ORC1 function.
Plant ORC2 homologs. Some ORC subunits contain potential sites for phosphorylation by cell-cycle kinases and can bind a variety of cell-cycle regulated complexes (34; and references therein). It has been suggested that phosphorylation of N-terminal regions of the ORC2 might be important for preventing re-initiation in S phase, might alter the binding of other factors, or may promote conformational changes within the ORC (2,34–37). The N-terminal halves of ORC2 subunits are highly variable among species but all of them carry conserved consensus sites for phosphorylation by Cdc2/Cdc18 (34).
In contrast, the currently available ORC2 homologs of plant origin do not carry potential Cdks sites. The putative plant homologs are unique because they lack the entire N-terminal portion altogether. This fact raises intriguing questions as to whether plant ORC2 subunits would be involved in cell-cycle control.
ZmORC3 and ZmORC6. In contrast to ORC1, ORC2, ORC4 and ORC5, which are among the most highly conserved eukaryotic proteins, ORC3 and ORC6 appear to have evolved faster (23,38,39). ORC6 is the least conserved among the six, the S.cerevisiae ORC6 being only weakly related to the S.pombe ORC6 but not to any other metazoan (39). We could not find a plausible maize ORC6 candidate in the searched databases and the lack of conserved peptides in the proteins from different species has made it impossible to isolate a putative maize ORC6 at this stage. Hence, we have no evidence of an ORC6 homolog in maize yet.
ORC3 has also considerably diverged among species (23,35,38–41). The human ORC3 does not show detectable similarity with the S.cerevisiae ORC3 and is only weakly similar to the S.pombe ORC3. Likewise, the ZmORC3 did not show detectable similarity with the yeast proteins. However, it displayed dispersed similarity with animal proteins (Fig. 2). The similarity with the DmORC3 is confined to two discrete regions of the N- and C-termini of the Drosophila protein molecule. This is consistent with the report that the N-terminal region of ORC3 is involved in ORC2 interaction, while the C-terminus is required for the ORC4 and ORC5 binding (42).
A putative Arabidopsis protein in the TAIR Database (At5g16690) is homologous to the maize protein (37% identical, 54% similar) over its entire length and might be a candidate for an Arabidopsis ORC3 subunit.
Homology relationship between the ORC subunits
Maximum parsimony cladograms ordering homologies into nested hierarchies were constructed for the five maize proteins and the respective homologs from other species. The similarity distribution patterns for ORC2, ORC3 and ORC5 (not shown) were similar to the tree shown for ORC1 (Fig. 1). In all cases, the putative plant ORC proteins were positioned in a well-supported clade, separated from the animal clade. Both animal and plant clades were well separated from the yeast. However, the cladistic analysis suggested that animal and plant proteins might be more closely related to the fission yeast than to the budding yeast proteins. An apparently similar relationship is displayed by ORC4 proteins as well but a conclusion about the members of plant origin cannot be finalized until other plant ORC4 homologs become available.
Interactions among the ZmORCs
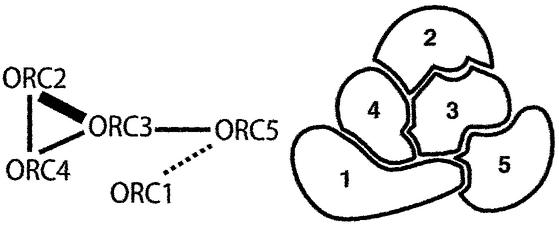
The strong affinity between ORC2 and ORC3 found in different species (25,36,40–44) was demonstrated also by the maize ZmORC2 and ZmORC3 in the yeast two-hybrid binding assays (Fig. 5). Likewise, the strong affinity of the human ORC2 for ORC4 (43) was displayed also by ZmORC2 for ZmORC4. These results are important in two aspects: they confirmed the validity of the assay and they indicated that strong ORC2–ORC3 and ORC2–ORC4 interactions are features conserved in evolution. The affinities of the ZmORCs for each other suggest that they might bind within a putative complex. Based on the binding results (Fig. 5), we proposed a model (Fig. 6) in which ZmORC3 plays a central role. It binds three subunits, ZmORC2, ZmORC4 and ZmORC5, while ZmORC1 binds the complex via ZmORC5. Presence of ZmORC3 and ZmORC4 is crucial for the binding of ZmORC5 and ZmORC1, while ZmORC2 is dispensable for the binding of ZmORC1 to the rest of the subunits. ZmORC3, ZmORC4 and ZmORC5 may act as a ‘core’ providing a required surface/architecture for the binding of ZmORC1. ZmORC2 might be outside this ZmORC1-binding core but its strong affinity for ZmORC3 and ZmORC4 implicates it in the complex assembly/function. It remains to be shown whether a putative ZmORC6 exists and whether it would bind with the other proteins.
Figure 6.
A model for the possible binding of the subunits within a complex. The thickness of the connecting lines reflects the relative strength of the interaction as judged from the yeast two-hybrid binding assays.
Currently, two models have been proposed for the assembly of the human ORC, based on co-immunoprecipitations and binary interactions (41,42). The two models differ slightly in the position/interaction of individual subunits. The common features between the human and the maize models are based also on the strong binary interactions between ZmORC2– ORC3, ZmORC3–ORC4 and ZmORC2–ORC4. Another prominent feature of both human and maize complexes is that binding of ORC1 apparently occurs on a core assembled by the other subunits. A difference, however, is that ZmORC1 apparently binds the complex through ZmORC5 and that ZmORC5 did not show affinity for any other subunit in the complex except ZmORC3. In the human model of Vashee et al. (41), subunits ORC2–ORC4 were suggested to form the ‘core’, while in the model proposed by Dhar et al. (42), HsORC2–HsORC5 form the core. Our results suggested that ZmORC3–ZmORC5 might form a core capable of binding ZmORC1. Whatever the minor differences and specificities of the complexes from different species might be, all reported models require ORC4, implicating it in the assembly of the ‘core’ and of the entire ORC. This fact suggests that a study of AtORC with its missing ORC4 might be revealing as to how different species assemble their ORCs.
The tissue-specific expression of the ZmORC genes was in agreement with a possible function related to DNA replication. However, an involvement of a multisubunit complex in plant DNA replication has yet to be demonstrated. In other species, roles for the complex and for individual subunits have been found beyond replication events. Thus, ORC is essential for silencing at the mating loci in S.cerevisiae (45–47), for heterochromatin association, silencing, and chromosome folding in Drosophila (26–28,48). A Drosophila ORC2 homolog is required for chorion gene amplification (44). High levels of ORC2–ORC5 were reported in non-dividing human tissues indicating involvement in roles outside the complex and DNA replication (24,25).
In maize, ZmORC3 is the only gene expressed in green leaves where the expression of the other genes is negligible. It might be expected that, in addition to its role in binding and assembling the other proteins in a complex, the maize ORC3 homolog may have pleiotropic roles beyond those requiring the whole complex.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to S. McCormick (Berkeley) for providing the sequence of the maize ORC2 5′ EST and to C. Zhang (Purdue) for carrying out the 5′ RACE on the maize ORC4 EST.
DDBJ/EMBL/GenBank accession nos+
REFERENCES
- 1.Bell S.P. and Stillman,B. (1992) ATP-dependent recognition of eukaryotic origins of replication by a multiprotein complex. Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- 2.Leatherwood J. (1998) Emerging mechanisms of eukaryotic DNA replication initiation. Curr. Opin. Genet. Dev., 10, 742–748. [DOI] [PubMed] [Google Scholar]
- 3.Takisawa H., Mimura,S. and Kubota,Y. (2000) Eukaryotic DNA replication: from pre-replication complex to initiation complex. Curr. Opin. Cell Biol., 12, 690–696. [DOI] [PubMed] [Google Scholar]
- 4.Clyne R.K. and Kelly,T.G. (1995) Genetic analysis of an ARS element from the fission yeast S.pombe. EMBO J., 14, 6348–6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubey D.D., Kim,S.M., Todorov,I.T. and Huberman,J.A. (1996) Large, complex modular structure of a fission yeast DNA replication origin. Curr. Biol., 6, 463–473. [DOI] [PubMed] [Google Scholar]
- 6.Gilbert D. (1998) Replication origins in yeast versus metazoa: separation of the haves and have nots. Curr. Opin. Genet. Dev., 8, 194–199. [DOI] [PubMed] [Google Scholar]
- 7.Cimbora D.M. and Groudine,M. (2001) The control of mammalian DNA replication: a brief history of space and timing. Cell, 104, 643–646. [PubMed] [Google Scholar]
- 8.Gilbert D. (2001) Making sense of eukaryotic DNA replication origins. Science, 294, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendez J. and Stillman,B. (2000) Chromatin association of the human origin recognition complex, CDC6 and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes during late mitosis. Mol. Cell. Biol., 20, 8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gavin K., Hidaka,M. and Stillman,B. (1995) Conserved initiator proteins in eukaryotes. Science, 270, 1667–1671. [DOI] [PubMed] [Google Scholar]
- 11.Kimura S., Ishibashi,T., Hatanaka,M., Sakakibara,Y., Hashimoto,J. and Sakaguchi,K. (2000) Molecular cloning and characterization of a plant homolog of the origin recognition complex 1. Plant Sci., 158, 33–39. [DOI] [PubMed] [Google Scholar]
- 12.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by top quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar S., Tamura,K., Jakobsen,I. and Nei,M. (2001) MEGA2: Molecular Evolutionary Genetics Analysis Software. Arizona State University, Tempe, AZ. [DOI] [PubMed]
- 14.Neuwald A.F., Aravind,L., Spouge,J.L. and Koonin,E.V. (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation and disassembly of protein complexes. Genome Res., 9, 27–43. [PubMed] [Google Scholar]
- 15.Weinreich M., Liang,C. and Stillman,B. (1999) The Cdc6 nucleotide-binding motif is required for loading Mcm proteins onto chromatin. Proc. Natl Acad. Sci. USA, 96, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aasland R., Gibson,T. and Stewart,A.F. (1995) The PHD finger: implications for chromatin mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. [DOI] [PubMed] [Google Scholar]
- 17.Callebout I., Labesse,G., Durand,P., Poupon,A., Canard,L., Chomilier,J., Henrissat,B. and Mornon,J.P. (1997) Deciphering protein sequences information through hydrophobic cluster analysis. Current status and perspectives. Cell Mol. Life Sci., 53, 621–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide-binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tugal T., Zou-Yang,H., Gavin,K., Pappin,D., Canas,B., Kobayashi,R. and Stillman,B. (1998) The Orc4p and orc5p subunits of the Xenopus and human origin recognition complex are related to ORC1 and Cdc6. J. Biol. Chem., 273, 32421–32429. [DOI] [PubMed] [Google Scholar]
- 20.Klemm R., Austin,R. and Bell,S. (1997) Coordinate binding of ATP and origin DNA regulates ATPase activity of the ORC. Cell, 88, 493–502. [DOI] [PubMed] [Google Scholar]
- 21.Lee D. and Bell,S. (2000) ATPase switches controlling DNA replication initiation. Curr. Opin. Genet. Dev., 12, 280–285. [DOI] [PubMed] [Google Scholar]
- 22.Bell S.P, Mitchell,J., Leber,J., Kobayashi,R. and Stillman,B. (1995) The multidomain structure of orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell, 83, 563–568. [DOI] [PubMed] [Google Scholar]
- 23.Chesnokov I., Gossen,M., Remus,D. and Botchan,M. (2000) Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes Dev., 13, 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quintana D.G., Thome,K., Hou,Z., Ligon,A.H., Morton,C. and Dutta,A. (1998) ORC5L, a new member of the human origin recognition complex, is deleted in uterine leiomyomas and malignant myeloid diseases. J. Biol. Chem., 373, 27137–27145. [DOI] [PubMed] [Google Scholar]
- 25.Thome K., Dhar,S., Quintana,D., Delmolino,L., Shahsafaei,A. and Dutta,A. (2000) Subset of human ORC subunits are expressed in non-proliferating cells and associate with non-ORC subunits. J. Biol. Chem., 276, 35233–35241. [DOI] [PubMed] [Google Scholar]
- 26.Triolo T. and Sternglantz,R. (1996) Role of interactions between the ORC and SIR1 in transcriptional silencing. Nature, 381, 251–253. [DOI] [PubMed] [Google Scholar]
- 27.Huang D., Fanti,L., Pak,D., Botchan,M., Pimpinelli,S. and Kellum,R. (1998) Distinct cytoplasmic and nuclear fractions of Drosophila HP1; their phosphorylation levels and association with ORC. J. Cell Biol., 142, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pak D., Pflumm,M., Chesnokov,I., Huang,D.W., Kellum,R., Marr,J., Romanowski,P. and Botchan,M. (1997) Association of the Origin Recognition Complex with heterochromatin and HP1 in higher eukaryotes. Cell, 91, 311–323. [DOI] [PubMed] [Google Scholar]
- 29.Iitzuka M. and Stillman,B. (1999) Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem., 274, 23027–23034. [DOI] [PubMed] [Google Scholar]
- 30.Burke T.W., Cook,J.G., Asano,M. and Nevins,J.R. (2001) Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem., 276, 15397–15408. [DOI] [PubMed] [Google Scholar]
- 31.Avramova Z., Tikhonov,A., Chen,M. and Bennetzen,J.L. (1998) Matrix-attachment regions in colinear segments of the sorghum and rice genomes. Nucleic Acids Res., 26, 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ball L.J., Murzina,N.V., Broadhurst,R.W., Raine,A.R.C., Archer,S.J., Stott,F.J., Murzin,A.G., Singh,P.B., Domaille,P.J. and Laue,E.D. (1997) Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J., 16, 2473–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tikhonov A., Bennetzen,J.L. and Avramova,Z. (2000) Structural domains and MARs along large colinear adh regions of maize and sorghum. Plant Cell, 12, 249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vas A., Mok,W. and Leatherwood,J. (2001) Control of DNA replication via Cdc2 phosphorylation sites in the origin recognition complex. Mol. Cell. Biol., 21, 5767–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carpenter P., Mueller,P. and Dunphy,W. (1996) Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature, 379, 357–360. [DOI] [PubMed] [Google Scholar]
- 36.Carpenter P. and Dunphy,W. (1998) Identification of a novel 81 kDa component of the Xenopus origin recognition complex. J. Biol. Chem., 273, 24891–24897. [DOI] [PubMed] [Google Scholar]
- 37.Leatherwood J., Lopez-Girona,A. and Russell,P. (1996) Interaction of cdc2 and cdc18 with fission yeast ORC2-like protein. Nature, 379, 360–363. [DOI] [PubMed] [Google Scholar]
- 38.Li J. and Herskowitz,I. (1992) Isolation of orc6, a component of the yeast origin recognition complex by a one-hybrid system. Science, 262, 1870–1874. [DOI] [PubMed] [Google Scholar]
- 39.Moon K.-Y., Kong,D., Lee,J.-K., Raychdhuri,S. and Hurwitz,J. (1999) Identification and reconstitution of the ORC from S.pombe. Proc. Natl Acad. Sci. USA, 96, 12367–12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto S., Quintana,D.G., Smith,P., Mihalek,R.M., Hou,Z.-H., Boynton,S., Jones,C.J., Hendricks,M., Velinzon,K., Wohlschlegel,J.A., Austin,R.J., Lane,W.S., Tully,T. and Dutta,A. (1999) latheo encodes a subunit of the origin recognition complex and disrupts neuronal proliferation and adult olfactory memory when mutant. Neuron, 23, 45–54. [DOI] [PubMed] [Google Scholar]
- 41.Vashee S., Simancek,P., Challberg,M. and Kelly,T. (2001) Assembly of the human origin recognition complex. J. Biol. Chem., 276, 26666–26673. [DOI] [PubMed] [Google Scholar]
- 42.Dhar S.K., Delmoloino,L. and Dutta,A. (2001) Architecture of the human origin replication origin. J. Biol. Chem., 276, 29067–29071. [DOI] [PubMed] [Google Scholar]
- 43.Quintana D.G., Hou,Z.-H., Thome,K., Hendricks,M., Saha,P. and Dutta,A. (1997) Identification of HsORC4, a member of the human origin of replication complex. J. Biol. Chem., 272, 28247–28251. [DOI] [PubMed] [Google Scholar]
- 44.Landis G., Kelley,R., Spradling,A. and Tower,J. (1997) The k3 gene, required for horion amplification and diploid chromososme replication encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc. Natl Acad. Sci. USA, 94, 3888–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bell S.P., Kobayashi,R. and Stillman,B. (1993) Yeast origin recognition complex functions in transcription silencing and DNA replication. Science, 262, 1844–1849. [DOI] [PubMed] [Google Scholar]
- 46.Ehrenhofer-Murray A., Gossen,M., Pak,M., Botchan,M. and Rine,J. (1995) Separation of origin recognition complex functions by cross-species complementation. Science, 70, 1671–1677. [DOI] [PubMed] [Google Scholar]
- 47.Foss M., McNally,F.J., Lauerson,P. and Rine,J. (1993) Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S.cerevisiae. Science, 262, 1838–1843. [DOI] [PubMed] [Google Scholar]
- 48.Loupart M.-L., Krause,S.A. and Heck,M.S. (2000) Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr. Biol., 10, 1547–1556. [DOI] [PubMed] [Google Scholar]
- 49.Chuang R.-Y. and Kelly,R. (1999) The fission yeast homologue of orc4 binds replication origin via multiple AT-hooks. Proc. Natl Acad. Sci. USA, 96, 2656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]