Abstract
Here we report on the expression and function of RNase 7, one of the final RNase A superfamily ribonucleases identified in the human genome sequence. The human RNase 7 gene is expressed in various somatic tissues including the liver, kidney, skeletal muscle and heart. Recombinant RNase 7 is ribonucleolytically active against yeast tRNA, as expected from the presence of eight conserved cysteines and the catalytic histidine–lysine– histidine triad which are signature motifs of this superfamily. The protein is atypically cationic with an isoelectric point (pI) of 10.5. Expression of recombinant RNase 7 in Escherichia coli completely inhibits the growth of the host bacteria, similar to what has been observed for the cationic RNase, eosinophil cationic protein (ECP/RNase 3, pI 11.4). An in vitro assay demonstrates dose-dependent cytotoxicity of RNase 7 against bacteria E.coli, Pseudomonas aeruginosa and Staphylococcus aureus. While RNase 7 and ECP/RNase 3 are both cationic and share this particular aspect of functional similarity, their protein sequence identity is only 40%. Of particular interest, ECP/RNase 3’s cationicity is based on an (over)abundance of arginine residues, whereas RNase 7 includes an excess of lysine. This difference, in conjunction with the independent origins and different expression patterns, suggests that RNase 7 and ECP/RNase 3 may have been recruited to target different pathogens in vivo, if their physiological functions are indeed host defenses.
INTRODUCTION
The RNase A superfamily has been the subject of intense biochemical study for over half a century (1). The prototype, RNase A (bovine pancreatic ribonuclease) was the first enzyme to be sequenced and chemically synthesized, and the classic experiments on the denaturation and renaturation of this protein appear in virtually every biochemistry textbook. During the past two decades, dozens of novel ribonucleases of this superfamily have been identified in multiple vertebrate species (2–13). As a group, all RNase A superfamily members are secretory proteins, and include a classic hydrophobic signal peptide followed by a mature peptide with molecular mass of ∼12–16 kDa in unglycosylated state. All RNase A ribonucleases maintain three specific catalytic residues—one lysine and two histidines—that comprise a catalytic crevice, and six to eight appropriately-spaced cysteines that form three to four disulfide bonds. Except for these conserved residues, the RNases are otherwise quite divergent, with sequence identities varying from 30 to nearly 100%. Although all RNase A ribonucleases are enzymatically active, catalytic efficiency and substrate preference vary considerably.
Among the most interesting features of the RNase A superfamily is its dynamic evolutionary history. RNase 1 has provided a wealth of data for molecular systematics (14), and the great diversity in expression and function among paralogous RNase genes further makes these proteins excellent models for understanding the origin of novelty in gene family evolution (15). For example, contributions of gene duplication to organismal adaptation in changing environments were demonstrated through the study of a duplicated RNase 1 gene in a leaf-eating monkey (13). In another example, reconstruction of RNases of extinct ruminant progenitors revealed how functions of RNase 1 evolved in concert with the origin of foregut fermentation (16), and the same technology also illustrated how enhanced RNase activity was achieved in evolution by complementary advantageous amino acid substitutions (17).
Completion of the human genome sequence has permitted us to conclude that there are eight functional genes of the RNase A superfamily in the human genome (12). Despite significant understanding of the enzyme biochemistry, the physiologic role(s) of most of these RNase A ribonucleases remain unclear. Among the better characterized RNases are RNase 1, or pancreatic ribonuclease, essential for the digestion of dietary and intestinal bacterial RNAs (18) and RNase 5, or angiogenin, which can stimulate blood-vessel formation (2). Recent evidence has suggested a role for eosinophil-derived neurotoxin (EDN)/RNase 2 in host defense against respiratory viruses (19,20). Others include eosinophil cationic protein (ECP), or RNase 3, and RNases 4, 6, 7 and 8. We have previously described the unusual evolution of EDN/RNase 2 and ECP/RNase 3, which emerged as a gene pair relatively recently, sometime after the divergence of the New World and Old World monkeys (21), and have inferred that positive selection contributed to the emergence of ECP/RNase 3 as a highly cationic ribonuclease with membrane lytic and cytotoxic activities not shared by EDN/RNase 2 (15). We have noted a similar pairing of RNases 7 and 8 (12). In this work we evaluate the enzymatic and functional consequences of this cationicity, and provide some thoughts on its physiological significance.
MATERIALS AND METHODS
Northern blotting
A human multiple-tissue northern membrane was purchased from Clontech (Palo Alto, CA). The membrane was prehybridized and hybridized following the manufacturer’s instructions. The hybridization was performed with the radiolabeled human RNase 7-specific oligonucleotide probe. The membrane was then washed following manufacturer’s instructions; autoradiograms were developed after 1 day exposures at –80°C. As a control, the hybridization of the human actin gene was performed with the probe provided by Clontech.
5′ and 3′ rapid amplifications of cDNA ends (RACEs)
Using RNase 7-specifc primers, we prepared cDNAs from human kidney mRNAs (Clontech) following the manufacturer’s instruction. The cDNAs were then cloned into the pCR4TOPO cloning vector (Invitrongen, CA), and multiple colonies were sequenced in both directions. The obtained sequences were assembled to get the full-length cDNA sequence.
Recombinant RNase 7 and its enzymatic activity
The signal peptide region of RNase 7 was inferred according to the known cleavage sites in other RNases (12). The mature peptide region of the RNase 7 gene was subcloned into the bacterial expression vector pFLAG CTS (Kodak, New Haven, CT) and was verified by sequencing. The vector adds the octapeptide DYKDDDDK (FLAG) to the C-terminus of the recombinant protein, which facilitates its purification and detection with M2 anti-FLAG monoclonal antibody (Sigma) without altering ribonucleolytic activity (22). Recombinant proteins were isolated, purified, and quantified as described (22). The ribonuclease activity of the recombinant proteins against a standard yeast tRNA substrate was measured in 40 mM sodium phosphate buffer (pH 7.4) at 25°C. Purified RNase was added into 0.8 ml of the aforementioned buffer with 1.42 nmol tRNA. The reaction was stopped by 0.5 ml of 20 mM lanthanum nitrate with 3% perchloric acid, and insoluble tRNA was removed by centrifugation. The amount of solubilized tRNA was determined by ultraviolet absorbance at 260 nm. The catalytic activity of the RNase was determined as the pmol of RNA digested per second per pmol of RNase (22). In the experiments, we used 13.24 pmol RNase 7. For comparison, we also examined the activities of RNase 1 and RNase 8 using 0.156 and 3.07 pmol enzymes, respectively. The results shown are normalized for 1 pmol of each RNase. The negative control prepared from a sham-isolation has an activity <3% of that of RNase 7. The average values from three experiments are presented with one standard deviation.
Antiviral assays
The activity of recombinant RNase 7 and other RNases in reducing the infectivity of respiratory syncytial virus (RSV) on HEp-2 human epithelial cells was examined by the quantitative shell vial amplification technique following the published procedure (23). Briefly, recombinant RNases were added to Hep-2 monolayers growing on coverslips (50 000 cells/coverslip) followed by ∼2000 plaque-forming units (= infectious units) of RSV-B [American Type Culture Collection (ATCC), Manassas, VA]. The vials containing virus and target cells were centrifuged for 60 min at 500 g at room temperature and then incubated at 37°C overnight, after which the coverslips were washed, acetone fixed, and stained with FITC-anti-RSV with methylene blue counterstain (Chemicon, Temecula, CA). Infected cells were identified via fluorescence microscopy.
Bacterial growth assay
Following Rosenberg (24), we examined whether inductions of the production of recombinant proteins in Escherichia coli inhibit the growth of the host bacteria. Overnight cultures of bacteria transformed with plasmids harboring the RNase 7 gene, ECP/RNase 3 gene (positive control), EDN/RNase 2 gene (negative control), pancreatic RNase/RNase 1 gene (negative control) and pFLAG CTS vector alone (negative control) were diluted 1:40 in LB broth with 100 µg/ml ampicillin. Optical densities (600 nm) were recorded at t = 0 and hourly thereafter. When exponential phase growth was achieved, 0.1 mM IPTG was added to one-half of the culture to induce the production of recombinant protein. Optical densities were recorded hourly thereafter. Each experiment was repeated three times. We are aware of the possibility that expression of human genes in E.coli may interfere with the bacterial growth because of the depletion of rare tRNAs due to a difference in codon usage between E.coli and human genes. We thus used a recently engineered E.coli strain (BL21 Codon-Plus by Stratagene, La Jolla) which has tRNA genes accommodating the codon usage of mammalian genes.
Antibacterial assay
Pathogenic strains of bacteria Pseudomonas aeruginosa (ATCC no. 27853), E.coli (11303) and Staphylococcus aureus (27217) were purchased from ATCC. The assay procedure follows Rosenberg (24). The bacteria were grown overnight and diluted 1:1000 in 10 mM sodium phosphate buffer (pH 7.5). Two microliters [∼40 000 colony-forming units (CFUs)] of bacteria were incubated with varying concentrations of recombinant RNase 7 for 8 h at 37°C. Serial dilutions of each protein– bacteria incubation were prepared and plated, and CFUs remaining after each treatment were determined. The negative controls were identically treated, but with proteins from a sham isolation (with vector only).
RESULTS
Expression pattern of RNase 7
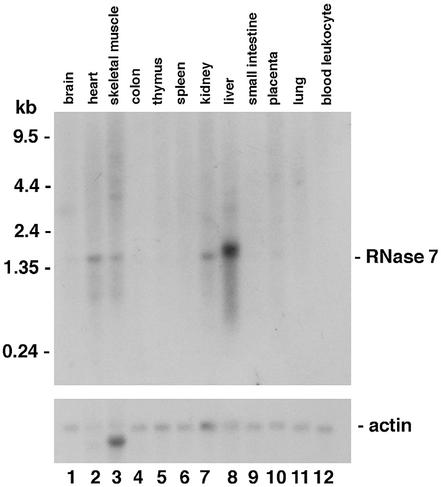
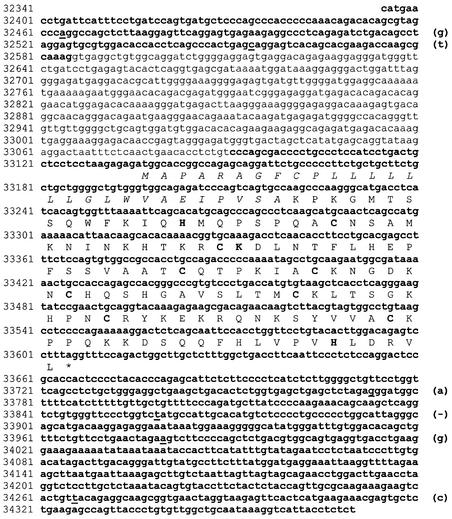
A computational search of the draft human genome sequence identified a putative ribonuclease gene named RNase 7 (12). Using the computationally predicted RNase 7 coding sequence, we designed an oligonucleotide probe, which was used to examine the expression pattern of human RNase 7 via northern analysis of mRNAs from 12 tissues (Fig. 1). RNase 7 mRNA (∼1.5 kb) was detected in liver, kidney, skeletal muscle and heart, with the strongest signal found in liver. The expression was not detected in brain, colon, thymus, spleen, small intestine, lung, leukocyte or placenta. To further confirm the expression of the RNase 7 gene and determine its exon–intron structure, we amplified and sequenced cDNAs prepared from human kidney mRNAs (Clontech) using primers designed for specific amplification of the RNase 7 gene. Both 5′ and 3′ RACEs were used to generate a full-length cDNA sequence. Figure 2 shows the genomic DNA sequence of the RNase 7 gene from the draft human genome sequence, along with the exon–intron structure inferred from our cDNA sequence. Our sequence has six nucleotide differences from the corresponding part of the draft human genome sequence, and these differences may be due to sequencing errors, mutations in cDNA synthesis and/or polymorphisms. However, none of these differences were in the protein-coding region. Human RNase 7 gene is found to have two exons, with the entire protein-coding region being encoded by exon 2. This exon–intron structure is a characteristic of RNase A gene superfamily. The single intron has 505 nt.
Figure 1.
Northern analysis of RNase 7. Total RNAs from normal human tissues indicated were probed with the 32P-radiolabeled oligonucleotide of the human RNase 7 gene. The same blot was also probed with a human actin gene fragment to control for relative loading.
Figure 2.
Nucleotide sequence and exon–intron structure of the human RNase 7 gene. Exons are in bold type while the single intron is in normal type. The DNA sequence shown is obtained from GenBank Al157687. The differences between this sequence and the cDNA sequence we obtained from human kidney mRNAs are underlined with the cDNA sequence given in parentheses. The differences may be due to sequencing errors, RT–PCR mutations, or polymorphisms. The conceptually translated protein sequence is given below the DNA sequence, with the signal peptide italicized and the three catalytic residues and eight structural cysteines bolded.
The predicted protein sequence of human RNase 7 exhibits all characteristics of the RNase A superfamily. From homology to other RNases (12), we predict that RNase 7 includes a signal peptide of 28 amino acids and a mature peptide of 128 amino acids (Fig. 2). The mature peptide has eight cysteines at positions conserved in most RNase A ribonucleases (Fig. 2). The three catalytic residues are also present (His15, Lys38 and His123; positions refer to those of the mature peptide). Among members of the human RNase A superfamily, RNase 8 is the closest relative to RNase 7, with 78% sequence identity. RNase 6 is the next closest relative, with ∼55% sequence identity. These features suggest that RNase 7 is enzymatically active, as will be demonstrated below. The calculated isoelectric point of RNase 7 is 10.5, significantly more cationic than RNase 8 (pI 8.2) despite the aforementioned sequence identity. This is very similar to what we have observed previously in relation to the EDN/RNase 2– ECP/RNase 3 pair, which share a high amino acid sequence identity (66%) but have divergent pIs (8.9 and 11.4, respectively) (21).
Ribonuclease activity of RNase 7
We prepared recombinant proteins of the human RNase 7 gene from E.coli and examined its enzymatic activity using the standard ribonuclease assay with yeast tRNA as the substrate. As expected, RNase 7 shows the ribonuclease activity (Table 1). At the same condition, this activity is about an order lower than that of the human pancreatic RNase or RNase 1, but is about twice that of RNase 8, the closest relative to RNase 7. We were not able to determine the kinetic parameters (Kcat and Km) in the present case as the catalytic kinetics of RNase 7 against yeast tRNAs seems to be complex, and the commonly used Lineweaver–Burk plot does not apply. Nevertheless, the enzyme activities of RNase 1, 7 and 8, as shown in Table 1, were determined under the same experimental conditions with the same amount of substrates, and thus may be compared directly. In an earlier report, we presented preliminary data on the catalytic activity of RNase 7 (12), which appeared much higher than what is shown here. We are not able to replicate that early result and believe that it was likely due to an error.
Table 1. RNase activities in degrading yeast tRNAs.
| RNases | Activities ±SD |
|---|---|
| RNase 1 | 0.147 ± 0.006 |
| RNase 7 | 0.021 ± 0.002 |
| RNase 8 | 0.012 ± 0.001 |
Values are in pmol RNA digested per pmol enzyme per second. See Materials and Methods for detailed experimental conditions.
No antiviral activity
EDN/RNase 2 and, to a lesser extent, ECP/RNase 3, have been shown to reduce the infectivity of respiratory syncytial virus for target human epithelium cells in vitro (20). We tested whether RNase 7 possesses this activity. Our results were negative (data not shown). The same negative results were obtained for the closely related RNase 8 (12)
Antibacterial activity
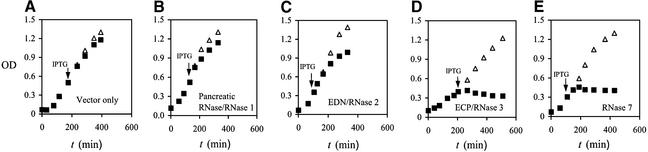
ECP/RNase 3 has been characterized as membrane lytic (25) and cytotoxic to bacteria (26). As RNase 7 is also cationic, we examined its antibacterial activity in two separate assay systems. As shown in Figure 3, induction of the expression of the ECP/RNase 3 as encoded by the pFLAG CTS plasmid of E.coli inhibits the growth of the host bacteria, whereas no inhibitory effect is seen when the plasmid contains the vector only. The inhibitory effect was also minimal in response to synthesis of RNase 1 or EDN/RNase 2, consistent with the fact that these RNases are not known to be toxic to bacteria. When the expression of the plasmid-encoding RNase 7 gene is induced, the bacterial growth is halted entirely (Fig. 3), suggesting that RNase 7 inhibits bacterial growth.
Figure 3.
Growth of bacterial transformatant cultures. (A) Vector only. (B) Human pancreatic RNase/RNase 1, which is not known to have antibacterial activity in this assay. (C) Human EDN/RNase 2, which is not known to have antibacterial activity in this assay. (D) Human ECP/RNase 3, which is known to have antibacterial activity in this assay (24). (E) Human RNase 7. Optical densities (600 nm) were recorded at t = 0 and hourly thereafter. When exponential phase growth was achieved, 0.1 mM IPTG was added to one-half of the culture to induce the production of recombinant protein. Optical densities were recorded hourly thereafter. The experiments were repeated three times, and the mean values are reported. Solid squares, after IPTG induction; open triangles, no IPTG.
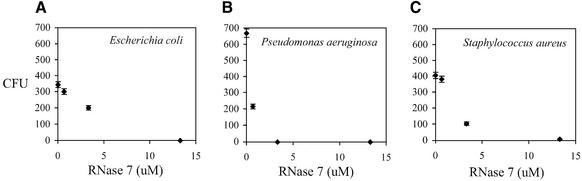
To confirm the above result, we also made recombinant proteins and tested the antibacterial function of RNase 7 in vitro. Two gram-negative (E.coli and P.aeruginosa) and one gram-positive (S.aureus) strains of pathogenic bacteria were used in this test. We found that recombinant RNase 7 protein inhibits growth of these bacteria in a dose-dependent fashion (Fig. 4). The CFU count is reduced by half in response to 1–5 µM of RNase 7. When the concentration of RNase 7 is increased to 13 µM, virtually all bacteria are killed. Our previous studies also showed that ECP/RNase 3 is antibacterial in this assay, but EDN/RNase 2 is not (22). These results indicate that the two assays we used yield consistent results on antibacterial activities of RNases.
Figure 4.
RNase 7 is toxic to bacteria (A) E.coli, (B) P.aeruginosa and (C) S.aureus. The number of CFUs after treatments of various concentrations of RNase 7 are shown with mean values and their standard deviations. ECP/RNase 3 is also known to have the antibacterial activity in this assay, but other RNases such as EDN/RNase 2 are not known to have this activity.
DISCUSSION
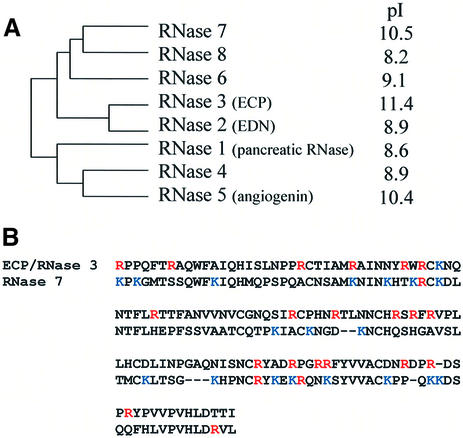
In this study, we provide a characterization of human RNase 7, a newly identified member of the RNase A superfamily. The phylogenetic relationships of the eight functional genes of the superfamily (all in chromosome 14) were studied by Zhang et al. (12) and a schematic diagram is shown here (Fig. 5A). This unrooted tree indicates that the eight genes can be separated into two major groups, with group I consisting of EDN/RNase 2, ECP/RNase 3, and RNases 6, 7 and 8, and group II consisting of RNase 1, RNase 4 and angiogenin/RNase 5. The most significant finding presented here is that RNase 7, although not closely related to RNase 3, shares atypical cationicity, and has cytotoxicity for both gram-negative and gram-positive bacteria. We previously studied the rapid evolution of ECP/RNase 3 and demonstrated positive selection as a contributory factor to the ‘cationicization’ of this protein from a more neutral ancestral sequence (15). Interestingly, while ECP/RNase 3 is rich in arginines (19 of 133 amino acids), RNase 7 has a high content of lysines (18 of 128 amino acids) (Fig. 5B). This further suggests that the cationicity was acquired independently during the evolution of ECP/RNase 3 and RNase 7, respectively. In addition to this difference, the expression patterns of the two RNases also differ substantially, as ECP/RNase 3 is exclusively found in eosinophils, while RNase 7 is expressed in multiple somatic tissues (Fig. 1). These differences suggest that although the in vitro antibacterial activities of the RNase 7 and ECP/RNase 3 appear similar, the two RNases may in fact be recruited for targeting different pathogens in vivo, if their physiological functions are indeed host defenses.
Figure 5.
Independent origins of two cationic RNases. (A) Phylogeny of the human RNase A superfamily. The phylogeny is adapted from Zhang et al. (12). Isoelectric points were calculated using the Wisconsin GCG package. (B) Alignment of ECP/RNase 3 and RNase 7, showing the independent origins of the cationic residues in the two proteins. Lysines and arginines are shown in blue and red, respectively.
If RNase 7 acquired the cationicity after its divergence from RNase 8, similar to the paradigm as we understand it for EDN/RNase 2 and ECP/RNase 3, it would be very interesting to investigate the molecular bases of these two similar yet independent evolutionary innovations. The precise mechanism by which RNase 7 and ECP/RNase 3 inhibit bacterial growth is not completely clear. While earlier studies suggested that ECP/RNase 3 can create pores on bacterial cell membranes (25), the specific involvement of the arginine residues is not known. We have speculated that these positively charged arginines may be critical to the antibacterial activity because they can be used to tightly contact the negatively charged bacterial cell membrane (15), a hypothesis that has gained support from recent experimental data (E. Carreras, personal communication). However, the involvement of cationic residues in bacterial autolysin activation (27) has not yet been explored. We also noticed that angiogenin/RNase 5 has a relatively high pI (Fig. 5A), yet this RNase is not known to be antibacterial, suggesting that cationicity alone may not be sufficient for cytotoxicity. Another conundrum is the lack of dependence of ECP/RNase 3 cytotoxicity on its ribonuclease activity (24), a point that has not yet been clarified for RNase 7. It is possible that these proteins are bifunctional, i.e., they have distinct and unrelated cytotoxic and ribonuclease-dependent activities. But due to independent duplications of the eosinophil-associated RNase genes in primates and rodents, it has been difficult to evaluate this hypothesis in mouse models. It remains to be seen whether RNase 7 may be amenable for study in this context.
It is likely that all members of the human RNase A superfamily have been identified at this point (12). The functional characterization of these proteins, in the context of evolution, will help us understand how new protein function originates. For this purpose, RNase 7 and ECP/RNase 3 will serve as a good model due to their parallel acquisitions of similar but distinct forms of cationicity.
While our paper was under review, Harder and Schroder (28) published a study of human RNase 7. Their results are consistent with ours and they suggest that RNase 7 is an antimicrobial protein of healthy human skin.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Jaap Beintema and two anonymous reviewers for helpful comments. This work was supported in part by a startup fund and a Rackham grant from the University of Michigan to J.Z.
DDBJ/EMBL/GenBank accession no. AY170392
REFERENCES
- 1.D’Alessio G. and Riordan,J.F. (1997) Ribonucleases, Structures and Functions. Academic Press, San Diego, CA.
- 2.Fett J.W., Strydom,D.J., Lobb,R.R., Alderman,E.M., Bethune,J.L., Riordan,J.F. and Vallee,B.L. (1985) Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry, 24, 5480–5486. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg H.F., Tenen,D.G. and Ackerman,S.J. (1989) Molecular cloning of the human eosinophil-derived neurotoxin: a member of the ribonuclease gene family. Proc. Natl Acad. Sci. USA, 86, 4460–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg H.F., Ackerman,S.J. and Tenen,D.G. (1989) Human eosinophil cationic protein. Molecular cloning of a cytotoxin and helminthotoxin with ribonuclease activity. J. Exp. Med., 170, 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg H.F. and Dyer,K.D. (1995) Human ribonuclease 4 (RNase 4): coding sequence, chromosomal localization and identification of two distinct transcripts in human somatic tissues. Nucleic Acids Res., 23, 4290–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg H.F. and Dyer,K.D. (1996) Molecular cloning and characterization of a novel human ribonuclease (RNase k6): increasing diversity in the enlarging ribonuclease gene family. Nucleic Acids Res., 24, 3507–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larson K.A., Olson,E.V., Madden,B.J., Gleich,G.J., Lee,N.A. and Lee,J.J. (1996) Two highly homologous ribonuclease genes expressed in mouse eosinophils identify a larger subgroup of the mammalian ribonuclease superfamily. Proc. Natl Acad. Sci. USA, 93, 12370–12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beintema J.J. and Kleineidam,R.G. (1998) The ribonuclease A superfamily: general discussion. Cell. Mol. Life Sci., 54, 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J., Dyer,K.D. and Rosenberg,H.F (2000) Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc. Natl Acad. Sci. USA, 97, 4701–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao Y.D., Huang,H.C., Leu,Y.J., Wei,C.W., Tang,P.C. and Wang,S.C. (2000) Purification and cloning of cytotoxic ribonucleases from Rana catesbeiana (bullfrog). Nucleic Acids Res., 28, 4097–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg H.F., Zhang,J., Liao,Y.D. and Dyer,K.D. (2001) Rapid diversification of RNase A superfamily ribonucleases from the bullfrog, Rana catesbeiana. J. Mol. Evol., 53, 31–38. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J., Dyer,K.D. and Rosenberg,H.F. (2002) RNase 8, a novel RNase A superfamily ribonuclease expressed uniquely in placenta. Nucleic Acids Res., 30, 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Zhang,Y.P. and Rosenberg,H.F. (2002) Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nature Genet., 30, 411–415. [DOI] [PubMed] [Google Scholar]
- 14.Beintema J.J., Fitch,W.M. and Carsana,A. (1986) Molecular evolution of pancreatic-type ribonucleases. Mol. Biol. Evol., 3, 262–275. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J., Rosenberg,H.F. and Nei,M. (1998) Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl Acad. Sci. USA, 95, 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jermann T.M., Opitz,J.G., Stackhouse,J. and Benner,S.A. (1995) Reconstructing the evolutionary history of the artiodactyl ribonuclease superfamily. Nature, 374, 57–59. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J. and Rosenberg,H.F. (2002) Complementary advantageous substitutions in the evolution of an antiviral RNase of higher primates. Proc. Natl Acad. Sci. USA, 99, 5486–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barnard E.A. (1969) Biological function of pancreatic ribonuclease. Nature, 221, 340–344. [DOI] [PubMed] [Google Scholar]
- 19.Domachowske J.B., Dyer,K.D., Bonville,C.A. and Rosenberg,H.F. (1998) Recombinant human eosinophil-derived neurotoxin/RNase 2 functions as an effective antiviral agent against respiratory syncytial virus. J. Infect. Dis., 177, 1458–1464. [DOI] [PubMed] [Google Scholar]
- 20.Rosenberg H.F. and Domachowske,J.B. (2001) Eosinophils, eosinophil ribonucleases and their role in host defense against respiratory virus pathogens. J. Leukoc. Biol., 70, 691–698. [PubMed] [Google Scholar]
- 21.Rosenberg H.F., Dyer,K.D., Tiffany,H.L. and Gonzalez,M. (1995) Rapid evolution of a unique family of primate ribonuclease genes. Nature Genet., 10, 219–223. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg H.F. and Dyer,K.D. (1995) Eosinophil cationic protein and eosinophil-derived neurotoxin. Evolution of novel function in a primate ribonuclease gene family. J. Biol. Chem., 270, 21539–21544. [DOI] [PubMed] [Google Scholar]
- 23.Domachowske J.B. and Bonville,C.A. (1998) Overnight titration of human respiratory syncytial virus using quantitative shell vial amplification. Biotechniques, 25, 644–647. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg H.F. (1995) Recombinant human eosinophil cationic protein. Ribonuclease activity is not essential for cytotoxicity. J. Biol. Chem., 270, 7876–7881. [DOI] [PubMed] [Google Scholar]
- 25.Young J.D., Peterson,C.G.B., Venge,P. and Cohn,G.J. (1986) Mechanism of membrane damage mediated by human eosinophil cationic protein. Nature, 321, 613–616. [DOI] [PubMed] [Google Scholar]
- 26.Lehrer R.I., Szklarek,D., Barton,A., Ganz,T., Hamann,K.J. and Gleich,G.J. (1989) Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J. Immunol., 142, 4428–4434. [PubMed] [Google Scholar]
- 27.Lewis K. (2000) Programmed death in bacteria. Microbiol. Mol. Biol. Rev., 64, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harder J. and Schroder J.M. (2002) RNase 7: a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem., 277, 46779–46784. [DOI] [PubMed] [Google Scholar]