Abstract
Zinc fingers are small protein domains in which zinc plays a structural role contributing to the stability of the domain. Zinc fingers are structurally diverse and are present among proteins that perform a broad range of functions in various cellular processes, such as replication and repair, transcription and translation, metabolism and signaling, cell proliferation and apoptosis. Zinc fingers typically function as interaction modules and bind to a wide variety of compounds, such as nucleic acids, proteins and small molecules. Here we present a comprehensive classification of zinc finger spatial structures. We find that each available zinc finger structure can be placed into one of eight fold groups that we define based on the structural properties in the vicinity of the zinc-binding site. Three of these fold groups comprise the majority of zinc fingers, namely, C2H2-like finger, treble clef finger and the zinc ribbon. Evolutionary relatedness of proteins within fold groups is not implied, but each group is divided into families of potential homologs. We compare our classification to existing groupings of zinc fingers and find that we define more encompassing fold groups, which bring together proteins whose similarities have previously remained unappreciated. We analyze functional properties of different zinc fingers and overlay them onto our classification. The classification helps in understanding the relationship between the structure, function and evolutionary history of these domains. The results are available as an online database of zinc finger structures.
INTRODUCTION
The rapid growth of structural information on proteins (1) necessitates their classification to comprehend and rationalize this variety. It is desirable to reduce all available structures to as small a number of fold groups as possible, provided that these groups present a reasonably detailed level of structural description. Such a classification would help us to understand and predict protein structure and function by assigning a protein to a fold group. Within each fold group, proteins should be classified into families based on inferred homology between them. Fold/family classification becomes exceedingly difficult for proteins that share a low level of sequence and structural similarity (2). This task is particularly challenging for small proteins, where the structure and sequence similarity statistics are marginal due to the short length of the protein chain (3). For such proteins, classification decisions cannot be made entirely automatically using structure similarity search programs like DALI (4), CE (5) or VAST (6) and a need for manual intervention exists. A combination of sequence, structural and functional information would be best to aid in the classification of these small proteins.
Several databases exist that classify proteins based on their structures, with the most widely used among them being SCOP (7), CATH (8) and FSSP (4,9,10). These databases range from being fully (FSSP) or partially (CATH) automated, to relying on manual methods (SCOP). Recently, a numerical taxonomy method based on neural networks has been proposed to identify evolutionary relationships among proteins and to classify them (11,12). Although this method is a significant advance (13) in automating the structural classification of proteins, it fails, for instance, to link the 8.3 kDa protein (gene MTH1184) of Methanobacterium thermoautotrophicum (1gh9) (14) with other members of the zinc ribbon family [see SCOP scopid = d1gh9a_ (7)], and instead recognizes it as a new fold. Thus, despite advances in the automation of protein structure classification schemes, a need for the manual classification that serves as a standard and facilitates improvement of automated methods exists.
The structures of very small protein domains are generally stabilized by the formation of disulfide bonds, or by binding to metal ions (most frequently, zinc). Metal binding increases the thermal and conformational stability of small domains but typically is not directly involved in their function. Among such domains, C2H2 zinc fingers are arguably the best studied (15–19). Initially used to define a repeated zinc-binding motif with DNA-binding properties in the Xenopus transcription factor IIIA, the term ‘zinc finger’ is now largely used to identify any compact domain stabilized by a zinc ion (18). We use it in this broader sense to describe a large group of functionally diverse and essential proteins.
To understand the structural and functional variety of all available zinc finger structures we undertook their comprehensive survey. Previous attempts have been made to classify zinc-binding sites in proteins based on ligand geometry (20–22) and the zinc fingers themselves based on the type of ligands that bind zinc (17). None of these works concentrate on the protein backbone similarity around the zinc ligands. Here we present a comprehensive classification of available zinc finger structures, i.e. small protein domains that are structured around a zinc ion, which forms part of the domain core and is sequestered from the solvent by cysteine and histidine residues. We base our classification on the structural similarity of the zinc-binding sites, namely, the spatial arrangement of secondary structural elements that contribute zinc ligands. Consequently, our fold groups (Table 1) constitute proteins that share common structural features and are frequently functionally related, but are not necessarily homologous. The structural classification of zinc finger domains should help researchers to link the structural properties of these proteins with their biological functions. Our classification is available as an online database at http://prodata.swmed.edu/zndb/. It will be updated at regular intervals as new structures become available.
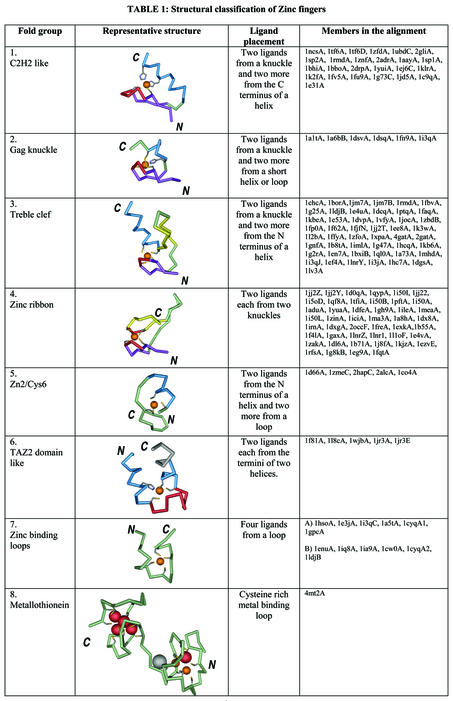
Table 1. Structural classification of zinc fingers.
Structures have been grouped into eight fold groups based on the structural properties around the zinc-binding site. The ligands that chelate zinc (colored orange) are shown as ball-and-stick. The helices are colored cyan. The zinc knuckle connecting the two strands of the β-hairpin is shown in red. The primary β-strands that neighbor the knuckle are colored in purple and other strands in yellow. Loops are shown in light green. Other parts of the structure, which do not belong to the zinc-binding region of the structure, are shown in gray. A brief description of each fold group is given. The last column lists representative members that belong to a particular fold group. No two structures in any given fold group share more than 50% identity in sequence at the zinc-binding site. The figures in this table were made using Weblab viewer Pro of Accelrys.
MATERIALS AND METHODS
Structure analysis
We searched a locally mirrored version of the PDB (1) for files that contained the string ‘ZN’ in the HETATM record. In such files the ligands of zinc were examined, and files that contained at least one zinc atom that had four ligands within a distance of 3 Å from the zinc were considered for the analysis. The sequences of individual PDB chains were extracted and clustered on the basis of sequence identity using the program BLASTCLUST (I.Dondoshansky and Y.Wolf, unpublished; ftp://ftp.ncbi.nih.gov/blast/) using an identity threshold of 50% and length coverage threshold of 90% on both sequences. The program PUU of the DaliLite suite (23) was then used to split these proteins into domains. However, not all these domains bind zinc and therefore the previously defined selection criteria were reapplied to the domains in order to filter out non-zinc-binding domains. An all against all structural alignment was initiated using the program DaliLite (23) for the selected domains. These domains were then clustered by a single-linkage clustering procedure based on their DaliLite Z-scores. All structures that aligned with a Z-score of better than 5.0 with any other structures were grouped together by single-linkage clustering. The program BESTVIEW (S.Sri Krishna and N.V.Grishin, unpublished) was then used to automatically produce stereo MOLSCRIPT (24) figures of the domains in an orientation optimal for viewing. These domains (DaliLite cluster representatives) were then visually examined and assigned into different fold groups. A total of eight fold groups were defined based on the architecture of the protein at the zinc-binding site. Some structures where the zinc-binding site does not form the core of the domain were excluded from the analysis (1ycs, 1hxq, 1fwq). Also structures where all the ligands were not either cysteine or histidine were excluded from the analysis (1dy0). The structure of DNA methylphosphotriester repair domain (1adn) of Ada (Escherichia coli) (25) was excluded from the analysis as the zinc ligands also play a catalytic role (26).
In order to locate potential zinc-binding sites in proteins where the zinc ion is not modeled in the structure, we developed a program METALBINDER that searches for two pairs of cysteine or histidine residues, whose CA atom coordinates are within a distance of 10 Å. The midpoint of the two interacting residue sets also needs to be within a distance of 10 Å. If these criteria are met, METALBINDER looks for the presence of a ZN atom within a radius of 4 Å from the center of the four CA atoms. If a ZN atom is not present, the structure is reported as a putative zinc-binding domain. Since the cutoffs used are rather relaxed, the number of false positives is high and the structures are examined manually. We have used METALBINDER to locate potential zinc-binding sites in the structures of ribosome (1fjf, 1lnr, 1jj2, 1ffk) (27–30), RNA polymerase (1i50, 1i3q) (31) and some other proteins in our analysis (1d66, 1dxg).
The structures of the zinc-binding domains were visualized and superimposed using the InsightII package (MSI) and the multiple structure-based alignments for each of the fold groups were constructed manually based on the superpositions made in InsightII. These alignments were further filtered based on the sequence identity in the aligned regions and only the structures with sequence identity of <50% to each other were retained.
Sequence analysis
Multiple sequence alignments were used to assist in constructing some structure-based alignments where the information from the structure alone was not sufficient to make a decision. These alignments were obtained using the program T-Coffee (32) from representative sequences found in PSI-BLAST (33,34) similarity searches (E-value threshold 0.01) against the non-redundant protein database (nr) maintained at the National Center for Biotechnology Information (Bethesda, MD).
RESULTS AND DISCUSSION
We detected and analyzed all potential zinc finger structures as described in Materials and Methods and classified them into eight fold groups based on the main chain conformation and secondary structure around the zinc-binding site (Table 1). All our fold groups except metallothioneins encompass more than one SCOP fold (7,35). All zinc fingers that belong to the same fold group have zinc ligands in a similar structural context. Despite this structural similarity, we do not imply that all zinc fingers within the fold group are evolutionarily related. We classify the structures within each fold group into families based on the hypothesized homology between the proteins that belong to the same family. Evolutionary relationship is inferred using the combination of sequence, structural and functional arguments. Some of our fold groups include structures that are better superposed when circular permutation is assumed. In most cases, the word ‘permutation’ is used to indicate ‘artificial’ permutation of the structures so that they are better superimposed than if they were not permuted and does not imply evolutionary events. This meaning holds for all proteins placed in different families. However, for the proteins grouped within a family and thus assumed homologous, we use ‘permutation’ in an evolutionary sense. Here we describe the eight fold groups of zinc fingers and discuss their similarities and differences with functional implications.
Fold group 1: C2H2-like finger
Domains from this group are composed of a β-hairpin followed by an α-helix that forms a left-handed ββα-unit (Table 1). Two zinc ligands are contributed by a zinc knuckle (a unique turn with the consensus sequence CPXCG) (3,36) at the end of the β-hairpin and the other two ligands come from the C-terminal end of the α-helix. The fold group consists of two families: C2H2 fingers and IAP domains.
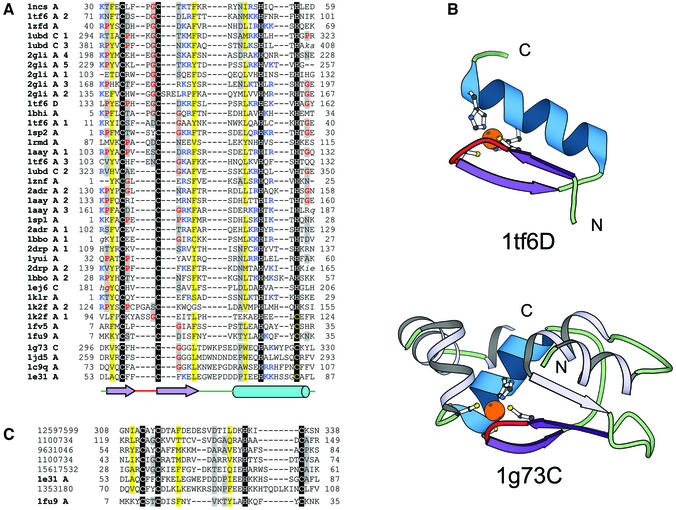
C2H2 finger family. The C2H2 zinc finger motif (classic zinc finger) was first discovered in the Xenopus laevis transcription factor IIIA, and has since been found to be present in many transcription factors and in other DNA-binding proteins (1ncs, 1zfd, 1tf6, 1ubd, 2gli, 1bhi, 1sp2, 1rmd, 2adr, 1znf, 1aay, 1sp1, 1bbo, 2drp, 1yui, 1ej6, 1klr) (16–18), which recognize specific sequences of DNA (Table 1, and Fig. 1A and B). The classical C2H2 zinc finger typically contains a repeated 28–30 amino acid sequence, including two conserved cysteines and two conserved histidine residues. However, other combinations of Cys/His as the zinc-chelating residues are possible. Nucleic acid-binding C2H2 fingers bind to the major groove of DNA through the N-terminus of the α-helix. Recognition of specific DNA sequences is achieved by the interaction of the DNA base with side chains from the surface of the α-helix (37). Although regulation of transcription seems to be the most important task performed by the C2H2 zinc fingers, recently determined structures of this class suggest their roles in mediating protein–protein interactions (1k2f, 1fv5, 1fu9) (38,39).
Figure 1.
Structure-based sequence alignment of the C2H2 finger fold group. For each sequence, the PDB entry name and chain ID (blank ID is replaced by A), fragment number (if more than one fragment is shown in the alignment), starting and ending residue numbers corresponding to the PDB numbering are given. Sequences in lower case italics are disordered in the structure. Regions in upper case italics differ in the secondary structure from the consensus secondary structure. Zinc ligands are boxed in black and non-zinc-binding residues at the same position are boxed in red. Zinc ligands in other sites are boxed in dark gray. Uncharged residues (all amino acids except D,E,K,R) in mostly hydrophobic sites are highlighted in yellow, non-hydrophobic residues (all amino acids except W,F,Y,M,L,I,V) at mostly hydrophilic sites are highlighted in light gray, small residues (G,P,A,S,C,T,V) at positions occupied by mostly small residues are shown in red letters. Charged residues K and R are marked in blue if they are present in the vicinity of a zinc-binding ligand. Long insertions are not displayed for clarity, and the number of omitted residues is specified in brackets. Regions of circular permutation are separated by a ‘|’ mark and the sequence number of the residues around the permuted region are shown in red. Sequences are further grouped into families. The same coloring and labeling scheme is used for figures of all fold groups of zinc fingers. Coloring unique to a particular figure is mentioned in its legend. A secondary structure consensus is shown below the alignment. β-strands are displayed as arrows and α-helices as cylinders. Color shading and labels of secondary structure elements correspond to those in the respective structural diagrams. Families are separated from each other by a larger spacing between the sequences. (A) Structure-based sequence alignment of domains of the C2H2 finger fold group. Here the domains are grouped into two families: the C2H2 like fingers (1ncsA, 1tf6A, 1zfdA, 1ubdC, 2gliA, 1tf6D, 1bhiA, 1sp2A, 1rmdA, 1aayA, 1znfA, 2adrA, 1sp1A, 1bboA, 2drpA, 1yuiA, 1ej6C, 1klrA, 1k2fA, 1fv5A and 1fu9A) and the IAP domains (1g73C, 1jd5A, 1c9qA and 1e31A). (B) Structure diagrams of the classical C2H2 zinc-binding motif (1tf6; chain D, residues 133–162) and BIR domain of Xiap-Bir3 domain of the human IAP (1g73 chain C, residues 296–330). The C2H2 motif found among many transcription factors is comprised of a zinc knuckle and a short β-hairpin at the N-terminus followed by a small loop and an α-helix. Two ligands each for zinc binding are contributed from the knuckle and the C-terminal part of the α-helix. The side chain of residues from the N-terminal part of the α-helix are involved in binding to the major groove of DNA in members of this class. The BIR domain, comprised of about 70 residues, has a conserved CCHC zinc-binding motif. The BIR domain is made up of a three-stranded β-sheet and four short α-helices. The colored region (non-gray) resembles a classic C2H2 motif, with two ligands each for zinc binding being contributed from a knuckle and the C-terminal part of an α-helix. The ribbon diagrams for this and all other diagrams in this manuscript were rendered by BOBSCRIPT (78,79), a modified version of MOLSCRIPT (24). (C) Alignment of representative protein sequences of IAP domains. The alignment includes apoptosis inhibitor survivin (1e31) and proteins from double-stranded DNA viruses Baculovirus (GI number 12597599, 1100734, 9631046, 1567532) and Iridovirus (GI number 1353180). The U-shaped transcription factor (1fu9) sequence is also included to better illustrate the resemblance between the IAP domains and the classical C2H2 fingers.
IAP domain family. The inhibitor of apoptosis (IAP) contains a CCHC (similar to U-shaped transcription factor from C2H2 finger family) pattern that coordinates a zinc ion (1e31, 1jd5, 1c9q, 1g73) (Fig. 1B). The IAPs have been reported to regulate programmed cell death by inhibition of caspases (40). Structures of the baculovirus inhibitor of apoptosis repeat (BIR) domains and the anti-apoptotic protein ‘survivin’ contain a conserved core made up of a central three-stranded β-sheet and four short α-helices (41). The zinc-binding region of IAP structurally resembles the classic C2H2 motif (Fig. 1B) in that the first two ligands are from a knuckle, and the other two ligands come from the C-terminal region of a broken α-helix (Fig. 1B). A PSI-BLAST alignment of the zinc-binding region of the BIR domains is shown along with the sequence of the U-shaped transcription factor (1fu9) (42) to highlight the similarities between the BIR domains and the classical C2H2 zinc fingers and to show the variability of the linker length between the last two zinc ligands in different BIR proteins (Fig. 1C). Despite the structural resemblance of the zinc-binding site in classic C2H2 domains and IAP domains, we do not have convincing evidence for homology between them and thus conservatively place them into two distinct families.
Fold group 2: Gag knuckle
The structure of this fold group is composed of two short β-strands connected by a turn (zinc knuckle) followed by a short helix or a loop (Fig. 2A and B). Two N-terminal zinc ligands are donated by the zinc knuckle and two others come from the loop or are placed at both ends of a short helix. The Gag-knuckle resembles the classical C2H2 motif with a large part of the helix and the β-hairpin truncated. Gag-knuckles are thus very short (about 20 amino acids) as compared with C2H2-like domains (about 30 residues).
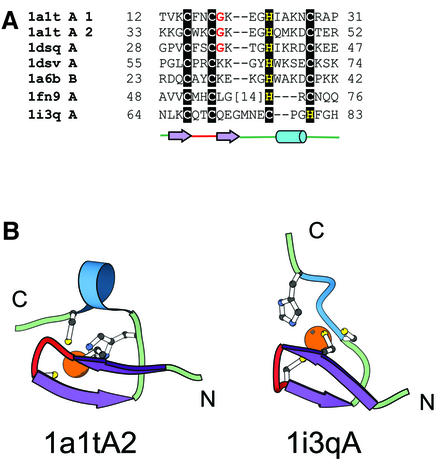
Figure 2.
‘Gag knuckle’ fold group of zinc fingers. (A) Structural alignment of members of the ‘Gag knuckle’ fold group. We define three families for this fold group. Most of the members in the alignment are from the nucleocapsid protein of retroviruses (retroviral Gag knuckle family: 1a1tA, 1dsqA, 1dsvA, 1a6bB). A zinc-binding domain from the large subunit of RNA polymerase II (polymerase Gag knuckle family: 1i3qA) and from the reovirus outer capsid protein σ3 (reovirus outer capsid protein σ3 Gag knuckle family: 1fn9A) have been included in the alignment primarily based on structural similarities to the NC proteins of retroviruses. (B) Representative Gag knuckle from HIV nucleocapsid (1a1t, chain A, residue 33–52) and RNA polymerase II large subunit (1i3q, chain A, residue 64–83). The Gag knuckle is typically characterized by a zinc knuckle, which contributes two ligands and two more ligands contributed from both ends of a small helix or a loop.
This group contains C2HC zinc fingers from the retroviral gag proteins (nucleocapsid) that are referred to in literature as zinc knuckles (16,43,44). However this term has also been used previously to describe a unique turn with the consensus sequence of CPXCG where the cysteines contribute to zinc binding (3,36). Thus for the sake of clarity we refer to the zinc finger from the retroviral gag proteins as the ‘Gag knuckle’. We consider three families.
Retroviral Gag knuckle family. In retroviral Gag knuckle, a one-turn α-helix follows the β-hairpin (Fig. 2B). The structure of this motif has been reported from the retroviral nucleocapsid (NC) protein from HIV and other related viruses (1a1t, 1a6b, 1dsq, 1dsv). The Gag knuckle binds to single-stranded RNA and is involved in recognizing specific sequences of RNA needed for viral packaging (16). Unlike the C2H2 motif, where the zinc finger is repeated many times, the Gag knuckles are mostly found as two conserved domains separated by a small linker region (44).
Polymerase Gag knuckle family. We have included the structure of a zinc finger from the A subunit of RNA polymerase II (1i3q; Fig. 2B) (31) in this fold group. The structure of the Gag knuckle from RNA polymerase II aligns with a root mean square deviation (RMSD) of 3.5–4.4 Å with different members of the retroviral NC proteins (104 atoms). The function of this zinc finger from RNA polymerase, however, remains unknown, although we can hypothesize that it may be involved in RNA-binding. In retroviral Gag-knuckles, a one-turn helix follows the hairpin. In the polymerase Gag-knuckle, this helix is substituted by a loop (Fig. 2B).
Reovirus outer capsid protein σ3 Gag knuckle family. The reoviral outer capsid protein σ3 (45) includes a zinc-binding motif that can be best described as a Gag knuckle (1fn9). The structure of the zinc-binding region resembles that from RNA polymerase and consists of a knuckle followed by a loop. Mutation of the zinc-binding residues has been shown to not affect binding of σ3 to double-stranded RNA but eliminates the ability to associate with the capsid protein µ1 (46). PSI-BLAST searches with sequences of the NC proteins of HIV, the zinc-binding region of RNA polymerase II and that from reovirus outer capsid protein σ3 fail to find links to each other, and thus we conservatively place them in different families.
Fold group 3: treble clef finger
The treble clef motif consists of a β-hairpin at the N-terminus and an α-helix at the C-terminus that contribute two ligands each for zinc binding. The first two ligands come from the zinc knuckle and the other two ligands are donated by the N-terminal turn of the helix (Fig. 3). In most treble clef fingers, a loop and a β-hairpin are present between the N-terminal β-hairpin and the C-terminal α-helix. This loop and the β-hairpin (sometimes substituted by a helix or a pair of helices) vary in length and conformation (Figs 3 and 4).
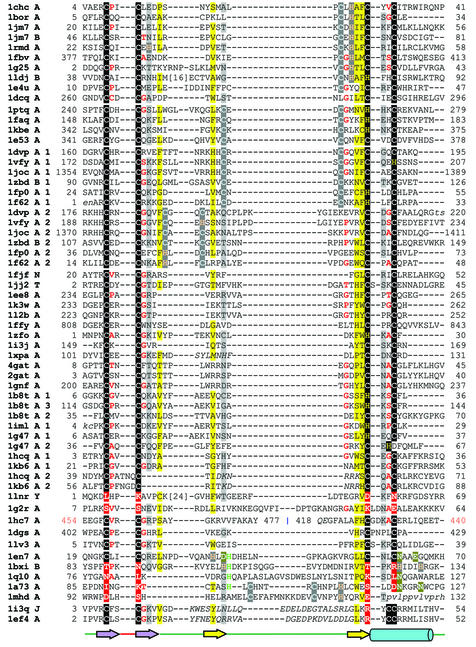
Figure 3.
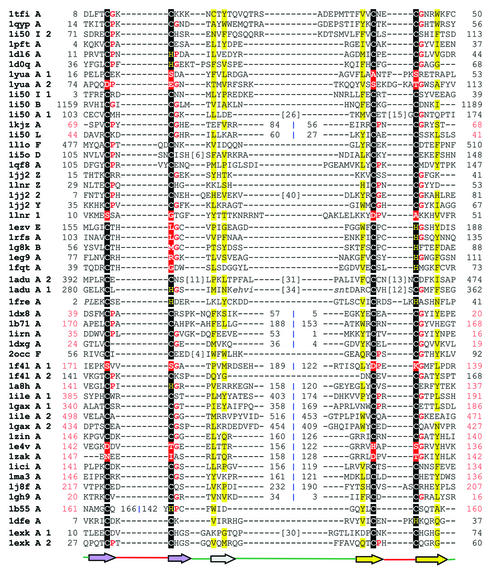
Structure-based sequence alignment of the treble clef fold group of zinc fingers. Here the structures are grouped into 10 families. The families are: RING finger-like (1chcA, 1borA, 1jm7A, 1jm7B, 1rmdA, 1fbvA, 1g25A, 1ldjB, 1e4uA, 1dcqA); protein kinase cysteine-rich domain (1ptqA, 1faqA, 1kbeA, 1e53A); phosphatidylinositol-3-phosphate-binding domain (1vfyA, 1dvpA, 1jocA, 1zbdB, 1fp0A, 1f62A); nuclear receptor-like finger (1fjfN, 1jj2T, 1ee8A, 1k3wA, 1l2bA, 1ffyA, 1zfoA, 1i3jA, 1xpaA, 4gatA, 2gatA, 1gnfA, 1b8tA, 1imlA, 1g47A, 1hcqA, 1kb6A, 1lnrY); YlxR-like hypothetical cytosolic protein (1g2rA); prolyl tRNA synthetase (1hc7A); NAD+-dependent DNA ligase treble clef domain (1dgsA); YacG-like hypothetical protein (1lv3A); His-Me endonucleases (1en7A, 1bxiB, 1ql0A, 1a73A, 1mhdA); RPB10 protein from RNA polymerase II (1i3qJ, 1ef4A). The coloring scheme follows that in Figure 1.
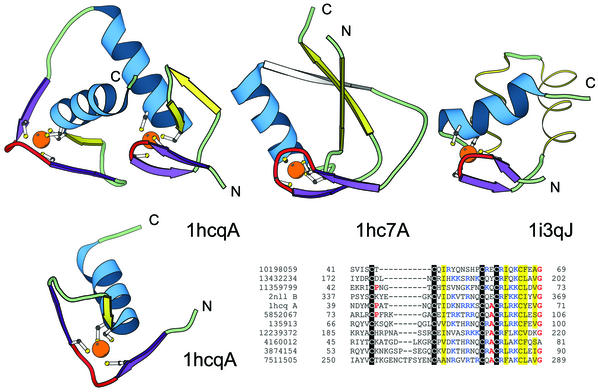
Figure 4.
Structural comparisons of representative treble clef fingers. Structural diagrams of the treble clef of the steroid hormone ‘estrogen’ receptor DNA-binding domain showing both zinc-binding sites (1hcq, chain A, residues 3–36 and 39–71), prolyl-tRNA synthetase (1hc7, chain A, residues 454–477; 418–440) and the RPB10 protein from RNA polymerase II (1i3q, chain J, residues 3–54) are shown to illustrate the wide range of variations in the structure of the treble clef finger. The N-terminal site of 1hcq is a typical treble clef motif. The C-terminal site (below), reoriented for clarity, lacks the β-hairpin shown in yellow for the N-terminal treble clef and has a distorted knuckle (red). This zinc-binding region shows some resemblance to other treble clef fingers in the placement of the ligands for zinc binding. However, the knuckle, which contributes the other two ligands in treble clef fingers, is significantly different and does not align structurally with those in other fingers. PSI-BLAST (33,34) alignment of representative nuclear receptor sequences. The sequences included in the alignment are: the nuclear factor NHR-63 (gi 10198059), peroxisome proliferator activated receptor (gi 13432234), NHR-18 (gi 11359799), retenoid X receptor (2nll), estrogen receptor (1hcq), tailless protein (TLL_DROME gi 135913), FTZ-F1 (gi 12239372), dissatisfaction (gi 4160012), NHR-67 (gi 3874154) and NHR-2 (gi 7511505). The gi accession number, start and end sequence number are given. The coloring scheme follows Figure 1.
Treble clef fingers are present in a diverse group of proteins that frequently do not share sequence and functional similarity with each other. Previous analysis (3) has revealed that proteins from seven different SCOP folds (version 1.53) (7) contain the treble clef finger as a structural core. In the present work, we detect additional members in the treble clef fold group. In some members of this fold group, the treble clef finger is the only domain present. However, in most cases, treble clef motifs are found to be incorporated in multi-domain proteins or are augmented by additional secondary structural elements. In some proteins, tandem or overlapping treble clefs are present possibly due to duplication events [LIM domain (1iml), FYVE domain (1vfy)] (3). We provisionally divide this fold group into 10 families.
RING finger-like. A number of proteins contain a conserved 40–60 residue cysteine-rich domain, which binds two zinc ions and is termed C3HC4 zinc-finger or ‘RING’ finger. Along with the classic two-zinc RING fingers (1chc, 1bor, 1jm7, 1rmd, 1fbv, 1g25, 1ldj, 1e4u) we include the Pyk2-associated protein β ARF-GAP domain (1dcq) in this family. The structure of the ARF-GAP finger is very similar to that of the RING fingers (0.94–2.13 Å; 104 atoms) and there is a residual sequence similarity. However the ARF-GAP finger lacks the second zinc-binding site that is present in the RING fingers.
Protein kinase cysteine-rich domain. This family includes the C-terminal domain of the human TfIIh P44 subunit (1e53), and cysteine-rich domains from kinases (1ptq, 1faq, 1kbe). The zinc-binding sites are at similar locations to that of RING fingers. The topological difference between the protein kinase cysteine-rich domain and RING finger structures has been explained on the basis of a circular permutation (3) and this family may be evolutionarily related to the RING finger-like proteins.
Phosphatidylinositol-3-phosphate binding domain. This family includes the zinc-binding regions of the FYVE domain (1vfy, 1dvp, 1joc) that bind phosphatidylinositol-3-phosphate with a high specificity, the effector domain of rabphilin-3a (1zbd) and the PHD zinc fingers (1f62, 1fp0). These domains bind two zinc ions and consist of an overlapping doublet of treble clef finger domains (3).
Nuclear receptor-like finger. This family mostly consists of domains that do not contain any additional secondary structural elements N- or C-terminal to the treble clef motif; however, several proteins of this family contain duplicated treble clef domains. Nuclear receptor-like fingers are mostly nucleic acid-binding proteins involved in transcription and translation and are likely to be evolutionarily related. The family contains the structures of the S14 (chain N of 1fjf) and L24E (chain T of 1jj2) ribosomal proteins, domain in MutM protein (1ee8, 1l2b), domain in endonuclease VIII (1k3w), the C-terminal domain of ile-tRNA synthetase (1ffy), the DNA repair factor XPA zinc-binding domain (1xpa), GATA-1 (4gat, 2gat, 1gnf), the nuclear receptor DNA-binding domain (1hcq, 1kb6) the LIM domain (1b8t, 1iml, 1zfo, 1g47), the I-TevI endonuclease zinc finger (1i3j) and the ribosomal protein L31 (1lnr).
A majority of these proteins have been analyzed elsewhere (3) and here we discuss three examples of the most deviant proteins that we feel belong to this family.
Nuclear receptor DNA-binding domain. The structure of the estrogen receptor DNA-binding domain (1hcq) (47) and its homologs have two zinc-binding sites (Fig. 4). Like the Gag knuckles, each steroid hormone receptor contains two repeats (domains) of a finger that binds zinc ions via four conserved cysteines. The first, N-terminal site (domain) is a typical treble clef motif where two of the ligands come from an α-helix and two more are contributed by a knuckle. The second, C-terminal site (domain) shows only partial resemblance to the treble clef finger (Fig. 4). Like in classical treble clef fingers, two of the zinc ligands are located in the N-terminal turn of an α-helix. However, the zinc knuckle, which donates the other two ligands in treble clefs, is absent in this domain and this zinc half-site appears very different structurally from the first zinc half-site of treble clef domains. Despite this structural difference, there is also some resemblance. The two cysteines in this half-site are flanked by extended regions that are conformationally similar to the short β-strands in classical treble clefs. It is likely that the second finger is a result of duplication of an ancestral treble clef domain [see SCOP scopid = d1hcqa_ (7)], after which substantial structural changes have occurred. These changes involved deterioration of the β-hairpin and the zinc-knuckle in it, which resulted in a structural reorganization of the first half-site. Our hypothesis about homology between the two zinc-binding domains is based on three lines of evidence: (i) duplications are among the most common events in molecular evolution and it may be easier for a protein to duplicate a domain than to construct one de novo; (ii) structural similarity of the second domain to the first: classical treble clef domain manifested in the second zinc sub-site and a helix complemented by the local conformational similarity in the regions of the first sub-site; (iii) variability of the second domain sequences in the region of the first sub-site, in particular, variability in the length of the linker between the first two cysteines (Fig. 4). Such variability indicates that the first sub-site may be prone to sequence, and thus structural, changes that do not affect the function of the protein, which may explain the differences from the typical treble clef arrangement of secondary structural elements.
I-TevI endonuclease zinc finger. I-TevI endonuclease belongs to the GIY-YIG family of intron-encoded endonucleases. The DNA-binding domain of intron endonuclease I-TevI (1i3j) (48) is a zinc finger that we classify as a treble clef. This domain is the shortest among treble clef fingers (Fig. 3). Its α-helix is deteriorated to a single turn. The zinc finger in I-TevI interacts with the DNA through two hydrogen bonds with the phosphate backbone at the minor groove, and is not seen to make any base-specific contacts. The general orientation of I-TevI finger on DNA is similar to the one typical for treble clef domains in which the α-helix forms most contacts with DNA. Due to the short length of the α-helix (one turn), this zinc finger also resembles the zinc ribbons and aligns with an RMSD of 1.41–5.35 Å (96 atoms) with other zinc ribbons, but lacks the third strand found in most zinc ribbons.
Ribosomal protein L31—a deteriorated treble clef finger. The structure of the large ribosomal subunit from Deinococcus radiodurans (30) reveals the L31 protein (Chain Y of 1lnr) as a treble clef finger in which the zinc-binding ligands are replaced with other residues. Despite the absence of the zinc-binding site, the structure of L31 contains all the properly oriented elements of the classical treble clef finger, such as the β-hairpin with the zinc knuckle, inserted β-hairpin and the C-terminal α-helix and thus undoubtedly belongs to this fold group (Fig. 3) along with two other ribosomal proteins L24E and S14. Another unusual feature of L31 treble clef is the presence of a long 25-residue insertion that follows the knuckle β-hairpin and is itself structured as a β-hairpin. Insertions after the knuckle β-hairpin are not uncommon among treble clefs and have been noted before to be of variable length (from 0 to 6 residues) and conformation (3), however, L31 possesses the longest insertion among them.
In this and later cases we hypothesize that complete or partial absence of the zinc ligands in zinc finger-like structures is due to a loss of ligands rather than a gain of ligands and zinc-binding property. The arguments for this hypothesis are 2-fold. First, the majority of the proteins from various phylogenetic lineages have well formed zinc sites. Typically, only a few isolated phylogenetic groups or families and maybe not even all representatives of these groups have zinc ligands absent. Second, the structures around zinc-binding sites are frequently unusual and have backbone and side-chain geometries that are probably not among the most favorable conformations in globular proteins. This geometry has probably arisen in conjunction with the zinc site formation.
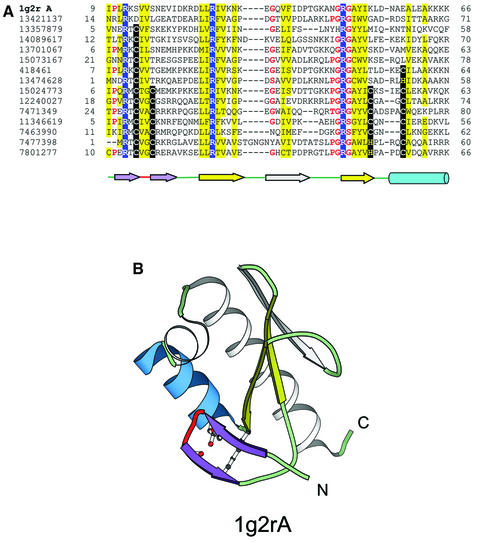
YlxR-like hypothetical cytosolic protein. The hypothetical cytosolic protein SP0554 coded by the gene from Nusa/Infb operon of Streptococcus pneumoniae [1g2r (49)] is another example of a treble clef finger with a deteriorated zinc-binding site. In contrast to the L31 protein, for which no close homologs can be found with the zinc-binding site still intact, PSI-BLAST searches reveal many such homologs for the SP0554 protein (Fig. 5A). YlxR family, to which SP0554 belongs, shows examples of partial zinc-binding site deterioration with some members retaining only one or two cysteines/histidines at the sites occupied by zinc ligands (Fig. 5A). The structure of this family is characterized by an additional β-strand inserted in the secondary β-hairpin of the treble clef (between the first knuckle and the helix) and an additional pair of α-helices at the C-terminus (Fig. 5B, shown in gray).
Figure 5.
Treble clef finger from the YlxR-like hypothetical cytosolic protein. (A) PSI-BLAST (33,34) alignment of representative homologs of the YlxR-like hypothetical cytosolic protein from the Nusa/Infb operon in S.pneumoniae. All protein sequences shown in this alignment are hypothetical proteins from bacterial sources and do not have any function assigned to them. Three conserved arginine residues are highlighted in blue. The secondary structure of 1g2r is shown below the alignment. β-Strands are displayed as arrows and α-helices are displayed as cylinders. The colors correspond to secondary structural elements shown in (B). The start and end residues of each sequence are given and the leftmost column contains the gi number of the sequence. (B) Structural diagram of the YlxR-like hypothetical cytosolic protein. This protein is a treble clef finger with an additional strand inserted between the second β-hairpin (yellow). Also the zinc-binding site of this protein is disintegrated and the structurally equivalent residues at the metal binding region are shown as ball-and-stick.
t-RNA synthetase treble clef domain. The structure of prolyl-tRNA synthetase from Thermus thermophilus (1hc7) (50) unexpectedly revealed the presence of a circularly permuted treble clef finger (Fig. 4). This is the first instance of a permuted treble clef finger seen among available protein structures. The N- and C-termini of the prolyl-tRNA synthetase treble clef are placed in the turn of the secondary β-hairpin and the α-helix is connected to the primary β-hairpin through an extended linker, part of which forms a β-strand hydrogen-bonded to the secondary β-hairpin (Fig. 4) in a manner similar to that observed in RING fingers.
NAD+-dependent DNA ligase treble clef domain. The structure of the NAD+-dependent DNA ligase from Thermus filiformis (1dgs) contains a treble clef finger inserted between the oligonucleotide binding (OB)-fold domain and helix– hairpin–helix (HhH)-containing domains (51). This zinc finger is among the shortest known treble clef domains, and shows partial similarity to zinc ribbons due to the presence of a very short helical segment at the C-terminus, which is fused with the first α-helix of an HhH hairpin, and is placed in this fold group provisionally.
YacG-like hypothetical protein. The recently determined structure of the E.coli protein YacG (52) (1lv3) is a treble clef finger since two of its zinc ligands are from a knuckle and the other two are from the first turn of a short helix. No function has been attributed to this protein. PSI-BLAST search with the YacG sequence does not find links to any other known member of the treble clef fold group and hence we place the protein in a separate family.
His-Me endonucleases. This family contains the structures of the DNase domains of colicins E7 and E9 (7cei, 1bxi), Seratia marcescens endonuclease (1ql0), intron-encoded endonuclease I-PpoI (1a73), T4 recombination endonuclease VII (1en7) and the MH1 domain of Smad (1mhd) and has been discussed in detail previously (3,53). The zinc-binding sites in all but the T4 recombination endonuclease (1en7) are deteriorated. The majority of these treble clefs are catalytic and contain an active site histidine (Fig. 3), except MH1 domain of SMAD which probably lost its catalytic activity (53).
RPB10 protein from RNA polymerase II. The RPB10 domain folds into a three-helical bundle typical of helix– turn–helix (HTH)-motif containing transcription factors (1ef4, chain J of 1i3q). However, it contains a zinc-binding site with geometry similar to the one found in treble clef fingers: two ligands come from a knuckle and two others are contributed by the N-terminus of an α-helix. In contrast to classical treble clef domains, the secondary β-hairpin in RPB10 is replaced by two α-helices. Since the secondary β-hairpin is the least conserved part of the treble clef finger and can tolerate long insertions or contain a short helix (Figs 3 and 4), we classify the RPB10 domain as a treble clef despite the replacement of the βdash;hairpin by an α-hairpin.
Fold group 4: zinc ribbon
In the zinc ribbon fold group, the ligands for zinc binding are contributed by two zinc-knuckles. The core of the structure is composed of two β-hairpins forming two structurally similar zinc-binding sub-sites (Figs 6 and 7). We call one of these hairpins a primary β-hairpin (shown in purple on Figs 6 and 7). This β-hairpin contains the N-terminal zinc sub-site in classic zinc ribbon proteins, such as the transcription initiation factor TFIIB (1pft) (54) and transcriptional elongation factor SII (TfIIS; 1tfi) (55). The other β-hairpin (secondary; shown in yellow on Figs 6 and 7) contains the C-terminal zinc sub-site in classic zinc ribbons. Typically, an additional β-strand forms hydrogen bonds with the secondary β-hairpin, thus most zinc-ribbon domains contain a three-stranded antiparallel β-sheet in their structure. The length of the β-strands in the primary β-hairpin is usually about two to four residues. The β-strands in the three-stranded sheet vary in length, but are frequently longer (4–10 residues). The distance between the two sub-sites can vary considerably and there could be additional domains inserted in between.
Figure 6.
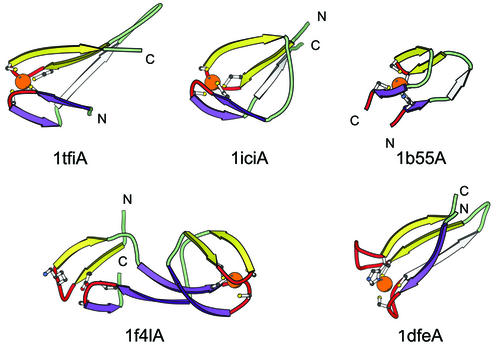
Structure-based sequence alignment of the zinc ribbon fold group. The proteins are grouped into nine families: classical zinc ribbon (1tfiA, 1qypA, 1i50I, 1pftA, 1dl6A, 1d0qA, 1yuaA, 1i50B, 1i50A, 1kjzA, 1i50L, 1l1oF, 1i5oD, 1qf8A, 1jj2Z, 1lnrZ, 1jj22, 1jj2Y, 1lnr1), the cluster binding domain of Rieske iron sulfur protein (1ezvE, 1rfsA, 1g8kB, 1eg9A, 1fqtA), the AdDBP zinc ribbons (1aduA), the B-box zinc finger (1freA), rubredoxin family (1dx8A, 1b71A, 1irnA, 1dxgA, 2occF), rubredoxin-like domains in enzymes (1f4lA, 1a8hA, 1ileA, 1gaxA, 1zinA, 1e4vA, 1zakA, 1iciA, 1ma3A, 1j8fA, 1gh9A), Btk motif (1b55A), ribosomal protein L36 (1dfeA) and the cysteine-rich domain of the chaperone protein DnaJ (1exkA). The coloring scheme follows Figure 1.
Figure 7.
Structural diagrams of the zinc ribbons. Zinc ribbons from the transcriptional factor SII, C-terminal domain (1tfi, chain A, residues 8–50), Sir2 homolog—transcriptional regulatory protein (1ici, chain A, residues 141–156; 119–134), zinc-binding domain from Bruton’s tyrosine kinase (1b55, chain A, residues 161–166; 142–160), methionyl-tRNA synthetase (1f4l, chain A, site 1: residues 171–189; 122–139, site 2: residues 141–168) are shown. Different circular permutations are seen in the ribbon. The structure diagram of the zinc ribbon from the ribosomal protein L36 from T.thermophilus (1dfe, chain A, residues 7–37) is shown to illustrate the very different orientation of the two knuckles in the structure as compared to other zinc ribbons. The coloring follows Figure 1.
The zinc ribbons are arguably the largest fold group of zinc fingers. The zinc ribbons are found in a diverse group of proteins and frequently display limited sequence similarity, which is mainly restricted to the zinc ligands and the zinc-knuckle motifs (Fig. 6). This limited sequence conservation is reflected in the structural variability of zinc ribbons. The structural analysis of zinc ribbons reveals that better superpositions are achieved for some of the structures if circular permutation is assumed. Namely, the N-terminal knuckle of one structure is superimposed with the C-terminal knuckle of another structure and the C-terminal knuckle of the first structure is superimposed with the N-terminal knuckle of the second structure (Figs 6 and 7). The superposition that assumes circular permutation allows for the inclusion of an additional β-strand (shown in gray on Figs 6 and 7). Circularly permuted versions of the zinc ribbon are found in rubredoxins. Also, circular permutation is seen among zinc ribbons that are located as insertions in larger proteins. If the zinc ribbon is present at either terminus in large proteins, then it is generally not permuted.
Structures that possess two knuckles in their zinc-binding sites and thus belong to this fold group fall into two distinct sub-groups defined by the geometry of zinc ligands. There exist two possible mutual orientations of the four zinc ligands placed on a tetrahedron: left-handed and right-handed. The majority of the zinc ribbon structures contain a site with left-handed geometry, namely, if the zinc ligands are numbered consecutively in the sequence and we orient the molecule with the primary hairpin above the secondary hairpin, the counter-clockwise sequence of zinc ligands is 1,4,2,3. However, a few structures, such as 1b55 (56), 1dfe (57) (Fig. 7) and both domains in 1exk (58) belong to a right-handed sub-group, for which counter-clockwise sequence of zinc ligands is 1,3,2,4. Conversion from one arrangement to the other can be rationalized through switching the places for ligands 3 and 4.
Due to the significant sequence and structural variability of zinc ribbons, our classification into families is provisional and more work is required to clarify evolutionary relationships within this fold. Evolutionary classification of this fold group is complicated by the fact that there exist zinc-binding sites similar in structure to zinc ribbons but not homologous to them. For instance, some serine proteases like the NS3 protein of hepatitis C virus (1a1r) and the guanine nucleotide exchange factor Mss4 (1fwq) have a zinc-binding site formed by two loops protruding from the structural core. However, in both structures, the zinc-binding site forms neither the core of the molecule nor the center of a separate domain and thus is not included in our analysis.
Classical zinc ribbon. This family mostly includes domains from proteins involved in the translation/transcription machinery, such as transcription factors, primases, RNA polymerases, topoisomerases and ribosomal proteins. Classical zinc ribbons are characterized by a long secondary hairpin (ribbon) and thus a longer three-stranded β-sheet as compared with members of other protein families of this fold group (Fig. 6, 1tfi, 1qyp). However, in some proteins that are probably homologous to classical zinc ribbons (e.g. zinc ribbons from ribosomal proteins), the secondary hairpin is shorter. The transcription elongation factor SII (TfIIS; 1tfi), the transcription initiation factor TFIIB (1pft, 1dl6) and the N-terminal 12 kDa fragment of DNA primase (1d0q) are typical representatives of this family. It has been argued that the C-terminal domains of prokaryotic DNA topoisomerase (1yua) are homologous to the zinc-ribbon domains in transcription factors (59). The two topoisomerase domains with available structure show statistically significant sequence similarity to the other members of the family, but do not retain the zinc ligands and probably do not bind zinc.
Several RNA polymerase subunits contain zinc ribbon domains that vary considerably in their length and sequence, but show a typical zinc ribbon structure. This group of zinc ribbons includes the polymerase proteins Rpb1, Rpb2, Rpb9 and Rpb12 (1i50 chains A, B, I, L, respectively and additionally 1qyp for Rpb9 fragment). The Rpb9 (chain I of 1i50) contains two zinc-binding domains separated by 40 residues. The structure of the C-terminal domain determined previously (1qyp) (36) was shown to form a zinc ribbon motif similar to that of the transcription factor IIS. Rpb12 (chain L of 1i50) forms a circularly permuted ribbon (Fig. 6). An unusual feature of the Rpb1 C-terminal zinc sub-site is that the two cysteine ligands are separated by an 18 residue long loop (Fig. 6).
The structure of the large γ subunit of the initiation factor e/aIF2 from Pyrococcus abyssi (1kjz) has a circularly permuted zinc ribbon. Due to its close resemblance to the transcription factors, we group it with the classical zinc ribbons.
The structure of the replication protein-A 70 kDa subunit (Rpa70) from the human single-stranded DNA (ssDNA) binding RPA trimerization core (60) contains a classic zinc ribbon inserted into the OB fold of the DNA-binding domain C. The zinc finger has been shown to modulate DNA binding (61) although the exact functional role of this zinc finger remains unclear.
Another major subfamily of the classical zinc ribbons are ribosomal proteins. Representative structures from this subfamily are the 50S ribosomal proteins L44E (chain 2 of 1jj2), L37E (chain Z of 1jj2), L37Ae (chain Y of 1jj2) from Haloarcula marismortui (29), and L32 (chain Z of 1lnr), L33 (chain 1 of 1lnr) from D.radiodurans (30). In L44E, a large insertion (∼40 residues as compared with L37Ae) exists between the two zinc-binding sub-sites. Zinc fingers are susceptible to replacements of zinc ligands and a consequent loss of zinc binding properties. This is also seen in the ribosomal protein L33 (chain 1 of 1lnr). Although the protein from D.radiodurans does not have zinc ligands, a PSI-BLAST search finds its close homologs with cysteines intact (data not shown).
Based on pronounced structural similarity and residual sequence similarity we place zinc ribbon domains of enzymes such as aspartate transcarbamoylase and casein kinase in this family (Fig. 6). The zinc ribbon of casein kinase II (1qf8) (62) aligns structurally with an RMSD of 1.13 Å (96 atoms) with TFSII (1tfi).
The cluster binding domain of Rieske iron sulfur protein. Iron sulfur proteins (ISPs) play a key role in electron transfer. The Rieske ISP is a high potential 2Fe–2S protein (63). The cluster binding domain of the Rieske ISP has a rubredoxin-like fold (1ezv, 1rfs, 1g8k, 1eg9, 1fqt), which coordinates a (63) cluster by two His and two Cys residues located in two knuckles. One of the Fe ions is coordinated by two cysteines and the other one is coordinated by two histidines. The domain is additionally stabilized by a disulfide bridge between the two knuckles, however, this disulfide link is not conserved among all structures. Also the ligand at the second position of the primary knuckle as seen in zinc ribbons and rubredoxins is not conserved among the ISP.
The adenovirus DNA-binding protein zinc ribbons. The adenovirus DNA-binding protein (AdDBP), a ssDNA-binding protein of the adenovirus E2A transcriptional unit, contains two zinc-binding motifs that are very similar to each other in structure. These zinc-binding motifs resemble the Rpb1 protein of RNA polymerase II (chain A of 1i50) in having long insertions between the two zinc sub-sites and in the region between the two cysteine ligands of the C-terminal sub-site (Fig. 6). These domains may be homologous to the classical zinc ribbons.
The B-box zinc finger. The nuclear factor Xnf7 contains a B-box (1fre) domain (64). B-box structure is composed of two loose knuckles that contribute ligands for zinc binding. The structure of Xnf7 B-box is more distant from other zinc ribbons in not having a three-stranded β-sheet. However, the structure of 1fre fails most of the PROCHECK (65) tests for the high quality structures and may not be accurate enough for detailed structural comparisons.
Rubredoxin family. Rubredoxins are low molecular weight metal-binding proteins involved in electron transfer. Representative structures of this family are the zinc- substituted rubredoxin (1dx8, 1irn), rubrerythrin (1b71), desulforedoxin (1dxg) and the polypeptide VIa of cytochrome c oxidase (chain F of 2occ). In all rubredoxins except for the cytochrome c oxidase subunit, better alignment with the classical zinc ribbons is achieved under the assumption of circular permutation, which switches the places of N- and C-terminal zinc sub-sites (Fig. 6). In such an alignment, the third β-strand in the β-sheet can be matched between rubredoxins and classical zinc ribbons.
Rubredoxin-like domains in enzymes. A wide variety of enzymes contain small zinc ribbons that appear similar and may be related to the rubredoxin domains. Similar to most rubredoxins, the majority of domains in this family are circularly permuted compared with classical zinc ribbons (Fig. 6). Known structures of these domains include aminoacyl tRNA synthetases, adenylate kinase and silent information regulator 2 (SIR2). In most of these proteins, zinc ribbons function as interaction modules, e.g. to provide a ‘lid’ for the enzyme’s active site (66).
The zinc ribbons from the structures of methionine (1f4l, 1a8h), isoleucine (1ile) and valine (1gax) aminoacyl-tRNA synthetases are shown in the alignment (Fig. 6). All these tRNA synthetases are of the class I and are characterized by an ATP binding domain with the Rossmann fold topology. Typically two zinc ribbon domains are inserted in the enzyme structure and are circularly permuted compared with classical zinc ribbons. The spacing between the two zinc sub-sites can be very large in some of these proteins, for instance in Met-tRNA synthetase, one zinc ribbon domain is inserted between the two zinc sub-sites of another zinc ribbon domain (Figs 6 and 7), with the N-terminal zinc ribbon being permuted. The N-terminal domain in E.coli structure (1f4l) has a deteriorated zinc-binding site (Figs 6 and 7). The zinc-binding site is complete in its ortholog from T.thermophilus (1a8h). Incidentally, the second zinc ribbon domain is absent in the T.thermophilus structure.
The structure of adenylate kinase from Bacillus stearothermophilus (1zin) reveals a zinc ribbon at the active site lid region. The zinc ribbon is shown to play a structural role in stabilizing the bacterial adenylate kinases (66). The structures of the enzymes from E.coli (1e4v) (67) and from maize (1zak) (68) contain the zinc ribbons, but lack the ligands for zinc binding (Fig. 6).
The SIR2 (1ici, 1ma3, 1j8f) contains a zinc ribbon domain as an insertion to the Rossmann-like fold domain (Fig. 7). Cysteines in this zinc ribbon are essential for the SIR2 function (69) and the NAD-binding pocket from the larger domain is seen to be stabilized by the presence of the zinc ribbon motif (70).
The structure of the 8.3 kDa protein (gene MTH1184) from M.thermoautotrophicum (1gh9) (14) does not contain zinc, however, the four cysteines around the potential metal-binding site and the fold of the chain argue for its classification as a zinc ribbon. MTH1184 protein does not have homologs clearly identifiable by sequence similarity searches, but is structurally more similar to proteins of this family. For instance, VAST (6) aligns 20 residues from all four β-strands of MTH1184 to isoleucyl-tRNA synthetase with RMSD of 1.0 Å.
Btk motif. In this and the next two families, the right-handed arrangement of zinc ligands is present. The Tec family of tyrosine kinases contains a zinc-binding motif (Btk motif) C-terminal from their pleckstrin-homology domain. The zinc-binding motif bears some resemblance to the zinc ribbons by having a three-stranded β-sheet and two of the zinc ligands being contributed by a β-turn in this sheet. To align all four Btk zinc ligands with zinc ribbons, we assume circular permutation of Btk motif in which the first ligand of the zinc ribbon is contributed by a C-terminal fragment of the Btk finger with the other three ligands coming from the N-terminal fragment (Fig. 7).
Ribosomal protein L36. The L36 protein (1dfe) (57) is another unusual zinc ribbon with right-handed placement of the zinc ligands. In zinc ribbons with left-handed ligand arrangement, the two knuckle hairpins are almost perpendicular to each other (Fig. 7). In L36 structure the two knuckle hairpins are parallel to each other and, as a consequence, the positions of the ligands 3 and 4 appear to be ‘flipped’ with respect to other members of the zinc ribbon fold group (Fig. 7).
Cysteine-rich domain of the chaperone protein DnaJ. The cysteine-rich domain of the chaperone DnaJ (1exk) (58) contains two zinc-binding sites, each of which is composed of two zinc knuckles. Based on this property, we place the two domains of DnaJ (one zinc-binding site in each) into the zinc ribbon fold group (Fig. 6). One of these domains is inserted between the two zinc sub-sites of the other domain. Zinc ligands have a right-handed arrangement and the β-strands characteristic of classical zinc ribbons are absent in DnaJ. Thus DnaJ domains align with other zinc ribbons with a RMSD range of 3.7–6.8 Å (96 atoms). The regions around the two zinc-binding sites in DnaJ are nearly identical to each other (RMSD of 0.88 Å; 96 atoms) and the two domains are homologous.
Fold group 5: Zn2/Cys6-like finger
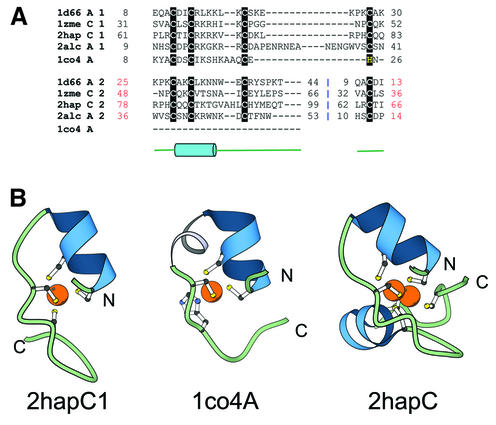
This group consists of zinc-binding domains in which two ligands are from a helix and two are from a loop (Fig. 8A and B). The first family of transcriptional regulators, the Zn2/Cys6 domain, contains two fingers. In the second family, namely, the copper responsive transcription factors (1co4) only one finger is present.
Figure 8.
Zn2/Cys6 fold group of zinc fingers. (A) Structure-based sequence alignment of members of the Zn2/Cys6 zinc finger fold group. Members of this fold group contain two zinc-binding sites with the zinc ions coordinated by six cysteine ligands. The fourth ligand to the second site is the N-terminal cysteine. The members of the first family include the transcriptional regulatory proteins Gal4 (1d66A), Hap1 (2hapC), PUT3 (1zmeC), ethanol regulon transcriptional activator (2alcA). The second family contains a zinc domain conserved in yeast copper responsive transcription factors (1co4A). (B) Structural diagrams of the N-terminal zinc-binding site of the Zn2/Cys6 fingers from the Hap1 transcription factor (2hapC1, chain C, residues 61–83) and the zinc domain conserved in yeast copper responsive transcription factors (1co4A, chain A, residues 8–26). 1co4 has only one zinc-binding site unlike the members of Zn2/Cys6 finger family (2hapC, chain C, residues 61–99) that contain two sites. The second zinc-binding site shares two ligands with the first site. Coloring scheme follows that in Figure 1.
Zn2/Cys6 finger family. The N-terminal region in several transcriptional regulators, such as Gal4 (1d66), Hap1 (2hap), PUT3 (1zme) and ethanol regulon transcriptional activator (2alc) forms a binuclear zinc cluster, in which two zinc ions are bound by six Cys residues (Fig. 8B) (16). Two of the ligands are from the N-terminus and the first turn of an α-helix and the other two are from a loop. The second zinc-binding site is best described as a circular permutation of the first, wherein the fourth ligand of the second zinc-binding site is the first ligand of the first site (Fig. 8A).
Copper responsive transcription factors. Copper response transcription factor (1co4) (71) upregulates metallothionein expression in yeast. This structure contains only the first of the two zinc-binding sites characteristic of the Zn2/Cys6 class. The region immediately after the N-terminal helix, which contributes two ligands for zinc binding, adopts a 310 helical conformation. The third and the fourth ligand for zinc binding are separated by only one residue unlike the classical Zn2/Cys6 fingers (Fig. 8A and B).
Fold group 6: TAZ2 domain-like
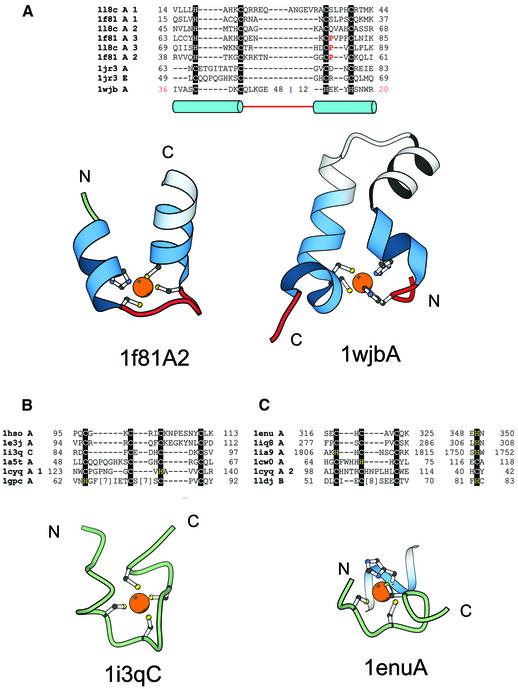
Proteins of this group are characterized by the zinc ligands that are located at the termini of α-helices. Three families are included in this fold group, namely, the TAZ1 and TAZ2 domain of the CREB-binding transcriptional adaptor protein (CBP) (1l8c, 1f81), the zinc-binding domain of DNA polymerase III γ subunit (chain A and E of 1jr3) from E.coli and the N-terminal zinc-binding domain of HIV-1 integrase (1wjb) (Fig. 9A).
Figure 9.
TAZ2 domain-like zinc finger fold group and short zinc-binding loops. (A) Structure-based sequence alignment of members of the TAZ2 domain-like zinc finger fold group. This fold group includes three families, TAZ2 domain family that consists of the structures of the three zinc-binding sites of CBP (1f81A, 1l8cA), the N-terminal zinc-binding domain of HIV-1 integrase (1wjbA) and the zinc-binding domain from DNA polymerase III γ subunit (1jr3A, 1jr3E). Structural diagrams of the second zinc-binding site of the CBP (1f81A2, chain A, residues 38–60) and the N-terminal zinc-binding domain of HIV-1 integrase (1wjbA, chain A, residues 36–48 and 12–20) are shown. Coloring scheme follows that in Figure 1. (B) Structural alignment of short zinc-binding loops present in large protein chains. The proteins in the alignment include loops from the human α alcohol dehydrogenase (1hsoA), sorbitol dehydrogenase (1e3jA), the 45 kDa polypeptide Rpb3 of the DNA-directed RNA polymerase II (1i3qC), the δ′ subunit of the clamp-loader complex of DNA polymerase III (1a5tA), intron encoded homing endonuclease I-PpoI (1cyqA) and core Gp32 ssDNA-binding protein (1gpcA). A representative figure of the RNA polymerase II protein Rpb3 (1i3qC, chain C, residue 84–97) is shown. (C) In some proteins the fourth ligand comes from a secondary structure far away in sequence from the other three ligands. This subgroup includes members from the tRNA-guanine transglycosylase (1enuA, 1iq8A), protein kinase domain of a Trp Ca-channel (1ia9A), intron encoded homing endonuclease I-PpoI (1cyqA), the Vsr endonuclease (1cw0A) and the RING finger protein Rbx1 (1ldjB). A representative figure of the zinc-binding region from tRNA-guanine transglycosylase (1enu, chain A, residue 316–325; 348–350) is shown. Three of the ligands of the zinc-binding site are from a loop and the fourth is from an α-helix.
TAZ2 domain family. The transcriptional adaptor protein CBP contains three duplicated HCCC-type zinc-binding sites that are very similar to each other. Each of the three zinc-binding sites is formed by the C-terminus of an α-helix, a short loop and the N-terminus of the next α-helix. The TAZ1 and TAZ2 domains of CBP mediate protein–protein interactions and bind to transcription factors.
Zinc-binding domain from DNA polymerase III γ subunit. The structure of DNA polymerase III γ subunit from E.coli contains a zinc-binding region which is inserted in the N-terminal ‘P loop’ type nucleotide binding fold (72). In this zinc finger, three of the four cysteines that bind zinc are from two helices and the fourth from the loop connecting the helices.
N-terminal domain of HIV-1 integrase. The N-terminal domain of HIV-1 is composed of four α-helices, two of which form an HTH motif, and contains a zinc-binding site (73). Two histidine residues from the second α-helix and two cysteines from the fourth α-helix (C-terminal) chelate a zinc ion. Compared with the other domains, the N-terminal domain of HIV-1 is circularly permuted, which is reflected in the alignment (Fig. 9A).
Fold group 7: short zinc-binding loops
This group consists of zinc-binding loops found in larger proteins. Such loops are probably stabilized by zinc and may be viewed as small but separate domains. The common structural feature of these domains is that at least three zinc ligands are very close to each other in sequence and are not incorporated into regular secondary structural elements (Fig. 9B and C). No homology is implied in the alignment shown on Figure 9B and C, just the general structural similarity. We have aligned zinc ligands in the loops from the human α alcohol dehydrogenase (1hso), sorbitol dehydrogenase (1e3j), the 45 kDa polypeptide Rpb3 of the DNA-directed RNA polymerase II (chain C of 1i3q), the δ′ subunit of the clamp-loader complex of DNA polymerase III (1a5t), intron-encoded homing endonuclease I-PpoI (1cyq), and core Gp32 ssDNA-binding protein (1gpc). In some other proteins, the fourth ligand comes from a secondary structural element far away in sequence from the rest of the three ligands (Fig. 9C). However, since the majority of zinc ligands are confined to a short loop, we feel that such three-ligand subdomains are structurally and maybe functionally similar to the four-ligand subdomains and thus include them in the same fold group. Our alignment (Fig. 9C) includes subdomains from the tRNA-guanine transglycosylase (1enu, 1iq8), protein kinase of a Trp Ca-channel (1ia9), the ‘very short patch repair’ (Vsr) endonuclease (1cw0), the intron-encoded homing endonuclease I-PpoI (1cyq) and the RING finger protein Rbx1 (1ldj). The biological roles played by these small zinc-binding domains are presently unknown and it would be of interest to investigate their functions.
Fold group 8: metallothioneins
Metallothioneins are cysteine-rich loops of about 60–70 residues that bind a variety of metals (4mt2; Table 1). No clearly defined regular secondary structural elements can be detected in metallothioneins and metal-binding sites in them do not appear similar to other proteins. Metallothioneins look like protein chains wrapped around a metal cluster with multiple cysteines liganding metals. Although the precise biological function of metallothioneins is not clear, they are known to sequester excess metal ions from the cellular environment and possibly protect from metal toxicity (74).
Functional properties of zinc-finger proteins
Proteins bind zinc as a cofactor for catalysis or as a structural stabilizer. In zinc fingers, the role of zinc is structural and zinc ions typically do not participate in the function directly. Other parts of a zinc-binding molecule bear functional importance. Small protein domains assembled around zinc ions are versatile structural templates that perform various functions. Despite their small size, zinc fingers are functionally more diverse than many larger domains and are seen to be involved in nucleic acid (DNA and RNA) binding, protein–protein interactions, binding small ligands (lipids) (3), and sometimes also possess enzymatic properties [without zinc participating in catalysis; (3,53)]. Executing these functions, zinc fingers are involved in many fundamental cellular processes, such as replication and repair, translation, programmed cell death and metal regulation.
Protein–DNA interactions. Among the eight fold groups, structures of protein–DNA complexes are known for the members from the C2H2-like, treble clef and the Zn2/Cys6 fold groups. The most frequent mode of DNA binding is similar among all these DNA-binding zinc fingers, where the main interactions are formed by the side-chains of residues from an α-helix, which generally binds to DNA at the major groove. This theme of protein–DNA interactions is not restricted to zinc fingers and is seen in 28 out of 54 DNA-binding protein families (75).
The DNA-binding mode of C2H2 fingers is illustrated by the structure of protein–DNA complex (1aay), in which the α-helix of the finger interacts with the DNA major groove (Fig. 10A). All C2H2 fingers bind DNA in a similar manner. The sequence specificity and high affinity for DNA binding is achieved by the cooperative binding of the α-helices of several C2H2 zinc fingers arranged in tandem.
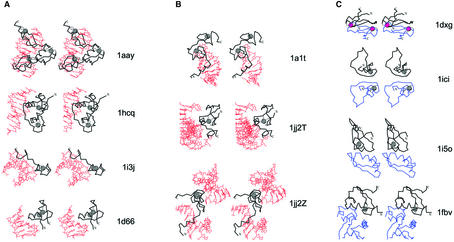
Figure 10.
Functional properties of zinc fingers. Cα traces of the zinc finger containing proteins are displayed in black with the N- and the C-terminus labeled. Zinc ions are shown as gray balls and Fe2+ ions are shown in purple. Side chains of zinc ligands or residues in corresponding sites are shown in black. Cα traces of the polypeptide chains interacting with the zinc fingers are dark blue. Nucleic acid is colored in red. (A) Protein–DNA interactions. Stereo diagrams of the Zif268 zinc finger from the C2H2-like group (1aay), the estrogen receptor (1hcq) and the intron endonuclease I-Tevi (1i3j) that belongs to the treble clef fingers and the GAL4 protein (1d66), which is a Zn2/Cys6 finger, are shown to illustrate the modes of DNA binding by zinc fingers. (B) Protein–RNA interactions. Stereo diagrams of the structure of the Gag knuckle from the HIV-1 nucleocapsid protein (1a1t) and the ribosomal proteins L24E (chain T of 1jj2; treble clef) and L37E (chain Z of 1jj2; zinc ribbon) interacting with RNA. (C) Protein–protein interactions. Stereo diagrams of desulforedoxin dimer (1dxg) and the homodimer of the zinc-binding domain of the Sir-2 homology protein (1ici). Also shown are the heterodimer of the zinc ribbon from E.coli aspartate transcarbamoylase regulatory chain (1i5o chain D; black) with the catalytic chain (1i5o chain C; blue) and the treble clef RING finger domain of the signal transduction protein Cbl in complex with ubiquitin-conjugating enzyme Ubch7 (1fbv chains A and C).
The DNA–protein interaction in the treble clef fingers is illustrated by the structures of the estrogen receptor DNA-binding domain (1hcq) (47) and the structure of the intron endonuclease I-Tevi (1i3j) (48) (Fig. 10A). The estrogen receptor belongs to the family of nuclear receptors that are involved in controlling transcription at the hormone response elements, regulated by the binding of steroid hormones. The DNA-binding regions of these nuclear receptors are comprised of two zinc-binding sites, each of which is a treble clef finger. The helices of the two fingers interact with the DNA. The N-terminal α-helix binds to the major groove of DNA and the outer β-strand of the primary β-hairpin interacts with the phosphate backbone. This mode of binding to DNA is shared by most of the treble clef fingers. However, the structure of the DNA-binding domain of the intron endonuclease I-Tevi (1i3j) is an exception to this rule in that the helix of the treble clef finger interacts with the minor rather than the major groove of the DNA and the inner β-strand of the primary β-hairpin is seen to interact with the phosphate backbone.
The Zn2/Cys6 zinc finger is comprised of two α-helices that coordinate two zinc ions via six cysteine residues. The first α-helix binds to the major groove of the DNA and recognizes specific triplets of DNA sequence (1d66, Fig. 10A). The second α-helix is involved in backbone interactions. The Zn2/Cys6 fingers generally bind to DNA as symmetrical dimers with the dimerization domain located outside the zinc-binding domain.
Protein–RNA interactions. Zinc fingers that interact with RNA were found among the structures of members from the Gag knuckle, the treble clef finger and the zinc ribbon fold groups. The structure of ribosome contains treble clef fingers and zinc ribbons forming contacts with RNA. Protein–RNA interactions are illustrated by the structure of ribosomal proteins L37E (zinc ribbon, chain Z of 1jj2) and L24E (treble clef, chain T of 1jj2) and the Gag knuckle from the HIV-1 nucleocapsid protein (1a1t) (Fig. 10B). The α-helix of the L24E treble clef finger interacts with the major groove of RNA and the mode of RNA-binding in treble clefs is similar to that of DNA-binding. The L44E, L37E, L37Ae ribosomal proteins from H.marismortui and the L32, L33 proteins from D.radiodurans and the L36 protein from T.thermophilus contain zinc ribbons. These ribbons interact mainly at the major groove of RNA with different parts of the zinc ribbon making contact with RNA.
Protein–protein interactions (homo: 1dxg, 1ici hetero: 1fbv, chain D of 1i5o). Many zinc fingers are involved in protein–protein interactions. Some of these interactions involve dimerization of zinc fingers. Such interactions are illustrated by the structures of desulforedoxin dimer (1dxg) and the zinc-binding domain of the Sir2 homology protein (1ici) (Fig. 10C). These proteins display different modes of dimerization in zinc ribbons.
Zinc fingers are known to interact with larger proteins. For instance, the structure of E.coli aspartate transcarbamoylase (76) reveals that the primary β-hairpin of zinc ribbon from the regulatory chain (D; black in Fig. 10A) interacts with the catalytic chain (C; blue in Fig. 10A). The treble clef of the RING finger domain of the signal transduction protein Cbl binds to the ubiquitin-conjugating enzyme Ubch7 (1fbv chains A and C). Residues from the α-helix and knuckle of the treble clef form the majority of contacts in this complex.
Although no structural information about the protein– protein interactions for the zinc-binding domains of the C2H2-like fingers is available, biochemical evidence points to the involvement of the C2H2 domain in mediating protein– protein interactions like in the erythroid FOG-1 and the U-shaped protein from Drosophila (38,42,77).
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Lisa Kinch and James Wrabl for critical reading of the manuscript and helpful comments.
REFERENCES
- 1.Berman H.M., Battistuz,T., Bhat,T.N., Bluhm,W.F., Bourne,P.E., Burkhardt,K., Feng,Z., Gilliland,G.L., Iype,L., Jain,S. et al. (2002) The protein data bank. Acta Crystallogr. D Biol. Crystallogr., 58, 899–907. [DOI] [PubMed] [Google Scholar]
- 2.Murzin A.G. (1998) How far divergent evolution goes in proteins. Curr. Opin. Struct. Biol., 8, 380–387. [DOI] [PubMed] [Google Scholar]
- 3.Grishin N.V. (2001) Treble clef finger—a functionally diverse zinc-binding structural motif. Nucleic Acids Res., 29, 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holm L. and Sander,C. (1997) Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res., 25, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shindyalov I.N. and Bourne,P.E. (1998) Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng., 11, 739–747. [DOI] [PubMed] [Google Scholar]
- 6.Gibrat J.F., Madej,T. and Bryant,S.H. (1996) Surprising similarities in structure comparison. Curr. Opin. Struct. Biol., 6, 377–385. [DOI] [PubMed] [Google Scholar]
- 7.LoConte L., Ailey,B., Hubbard,T.J., Brenner,S.E., Murzin,A.G. and Chothia,C. (2000) SCOP: a structural classification of proteins database. Nucleic Acids Res., 28, 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orengo C.A., Bray,J.E., Buchan,D.W., Harrison,A., Lee,D., Pearl,F.M., Sillitoe,I., Todd,A.E. and Thornton,J.M. (2002) The CATH protein family database: a resource for structural and functional annotation of genomes. Proteomics, 2, 11–21. [PubMed] [Google Scholar]
- 9.Holm L. and Sander,C. (1996) The FSSP database: fold classification based on structure-structure alignment of proteins. Nucleic Acids Res., 24, 206–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holm L. and Sander,C. (1998) Touring protein fold space with Dali/FSSP. Nucleic Acids Res., 26, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietmann S. and Holm,L. (2001) Identification of homology in protein structure classification. Nature Struct. Biol., 8, 953–957. [DOI] [PubMed] [Google Scholar]
- 12.Dietmann S., Park,J., Notredame,C., Heger,A., Lappe,M. and Holm,L. (2001) A fully automatic evolutionary classification of protein folds: Dali Domain Dictionary version 3. Nucleic Acids Res., 29, 55–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lichtarge O. (2001) Getting past appearances: the many-fold consequences of remote homology. Nature Struct. Biol., 8, 918–920. [DOI] [PubMed] [Google Scholar]
- 14.Christendat D., Yee,A., Dharamsi,A., Kluger,Y., Savchenko,A., Cort,J.R., Booth,V., Mackereth,C.D., Saridakis,V., Ekiel,I. et al. (2000) Structural proteomics of an archaeon. Nature Struct. Biol., 7, 903–909. [DOI] [PubMed] [Google Scholar]
- 15.Wolfe S.A., Grant,R.A., Elrod-Erickson,M. and Pabo,C.O. (2001) Beyond the ‘recognition code’: structures of two Cys2His2 zinc finger/TATA box complexes. Structure (Camb.), 9, 717–723. [DOI] [PubMed] [Google Scholar]
- 16.Laity J.H., Lee,B.M. and Wright,P.E. (2001) Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol., 11, 39–46. [DOI] [PubMed] [Google Scholar]
- 17.Leon O. and Roth,M. (2000) Zinc fingers: DNA binding and protein–protein interactions. Biol. Res., 33, 21–30. [DOI] [PubMed] [Google Scholar]
- 18.Klug A. and Schwabe,J.W. (1995) Protein motifs 5. Zinc fingers. FASEB J., 9, 597–604. [PubMed] [Google Scholar]
- 19.Iuchi S. (2001) Three classes of C2H2 zinc finger proteins. Cell. Mol. Life Sci., 58, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alberts I.L., Nadassy,K. and Wodak,S.J. (1998) Analysis of zinc binding sites in protein crystal structures. Protein Sci., 7, 1700–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlin S. and Zhu,Z.Y. (1997) Classification of mononuclear zinc metal sites in protein structures. Proc. Natl Acad. Sci. USA, 94, 14231–14236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlin S., Zhu,Z.Y. and Karlin,K.D. (1997) The extended environment of mononuclear metal centers in protein structures. Proc. Natl Acad. Sci. USA, 94, 14225–14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm L. and Park,J. (2000) DaliLite workbench for protein structure comparison. Bioinformatics, 16, 566–567. [DOI] [PubMed] [Google Scholar]
- 24.Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- 25.Myers L.C., Verdine,G.L. and Wagner,G. (1993) Solution structure of the DNA methyl phosphotriester repair domain of Escherichia coli Ada. Biochemistry, 32, 14089–14094. [DOI] [PubMed] [Google Scholar]
- 26.Lipscomb W.N. and Strater,N. (1996) Recent advances in zinc enzymology. Chem. Rev., 96, 2375–2434. [DOI] [PubMed] [Google Scholar]
- 27.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 28.Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- 29.Klein D.J., Schmeing,T.M., Moore,P.B. and Steitz,T.A. (2001) The kink-turn: a new RNA secondary structure motif. EMBO J., 20, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harms J., Schluenzen,F., Zarivach,R., Bashan,A., Gat,S., Agmon,I., Bartels,H., Franceschi,F. and Yonath,A. (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell, 107, 679–688. [DOI] [PubMed] [Google Scholar]
- 31.Cramer P., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science, 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- 32.Notredame C., Higgins,D.G. and Heringa,J. (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol., 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 33.Altschul S.F. and Koonin,E.V. (1998) Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci., 23, 444–447. [DOI] [PubMed] [Google Scholar]
- 34.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LoConte L., Brenner,S.E., Hubbard,T.J., Chothia,C. and Murzin,A.G. (2002) SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res., 30, 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang B., Jones,D.N., Kaine,B.P. and Weiss,M.A. (1998) High-resolution structure of an archaeal zinc ribbon defines a general architectural motif in eukaryotic RNA polymerases. Structure, 6, 555–569. [DOI] [PubMed] [Google Scholar]
- 37.Wolfe S.A., Nekludova,L. and Pabo,C.O. (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys Biomol. Struct., 29, 183–212. [DOI] [PubMed] [Google Scholar]
- 38.Fox A.H., Liew,C., Holmes,M., Kowalski,K., Mackay,J. and Crossley,M. (1999) Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J., 18, 2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polekhina G., House,C.M., Traficante,N., Mackay,J.P., Relaix,F., Sassoon,D.A., Parker,M.W. and Bowtell,D.D. (2002) Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling. Nature Struct. Biol., 9, 68–75. [DOI] [PubMed] [Google Scholar]
- 40.Verhagen A.M., Coulson,E.J. and Vaux,D.L. (2001) Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol., 2, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu G., Chai,J., Suber,T.L., Wu,J.W., Du,C., Wang,X. and Shi,Y. (2000) Structural basis of IAP recognition by Smac/DIABLO. Nature, 408, 1008–1012. [DOI] [PubMed] [Google Scholar]
- 42.Liew C.K., Kowalski,K., Fox,A.H., Newton,A., Sharpe,B.K., Crossley,M. and Mackay,J.P. (2000) Solution structures of two CCHC zinc fingers from the FOG family protein U-shaped that mediate protein–protein interactions. Structure Fold. Des., 8, 1157–1166. [DOI] [PubMed] [Google Scholar]
- 43.Guo J., Wu,T., Anderson,J., Kane,B.F., Johnson,D.G., Gorelick,R.J., Henderson,L.E. and Levin,J.G. (2000) Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol., 74, 8980–8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein D.J., Johnson,P.E., Zollars,E.S., De Guzman,R.N. and Summers,M.F. (2000) The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle. Biochemistry, 39, 1604–1612. [DOI] [PubMed] [Google Scholar]
- 45.Olland A.M., Jane-Valbuena,J., Schiff,L.A., Nibert,M.L. and Harrison,S.C. (2001) Structure of the reovirus outer capsid and dsRNA-binding protein sigma3 at 1.8 Å resolution. EMBO J., 20, 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shepard D.A., Ehnstrom,J.G., Skinner,P.J. and Schiff,L.A. (1996) Mutations in the zinc-binding motif of the reovirus capsid protein delta 3 eliminate its ability to associate with capsid protein mu 1. J. Virol., 70, 2065–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwabe J.W., Chapman,L., Finch,J.T. and Rhodes,D. (1993) The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell, 75, 567–578. [DOI] [PubMed] [Google Scholar]
- 48.Van Roey P., Waddling,C.A., Fox,K.M., Belfort,M. and Derbyshire,V. (2001) Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. EMBO J., 20, 3631–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osipiuk J., Gornicki,P., Maj,L., Dementieva,I., Laskowski,R. and Joachimiak,A. (2001) Streptococcus pneumonia YlxR at 1.35 Å shows a putative new fold. Acta Crystallogr. D Biol. Crystallogr., 57, 1747–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yaremchuk A., Cusack,S. and Tukalo,M. (2000) Crystal structure of a eukaryote/archaeon-like prolyl-tRNA synthetase and its complex with tRNAPro(CGG). EMBO J., 19, 4745–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee J.Y., Chang,C., Song,H.K., Moon,J., Yang,J.K., Kim,H.K., Kwon,S.T. and Suh,S.W. (2000) Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications. EMBO J., 19, 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramelot T.A., Cort,J.R., Yee,A.A., Semesi,A., Edwards,A.M., Arrowsmith,C.H. and Kennedy,M.A. (2002) NMR structure of the Escherichia coli protein YacG: a novel sequence motif in the zinc-finger family of proteins. Proteins, 49, 289–293. [DOI] [PubMed] [Google Scholar]
- 53.Grishin N.V. (2001) Mh1 domain of Smad is a degraded homing endonuclease. J. Mol. Biol., 307, 31–37. [DOI] [PubMed] [Google Scholar]
- 54.Zhu W., Zeng,Q., Colangelo,C.M., Lewis,M., Summers,M.F. and Scott,R.A. (1996) The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nature Struct. Biol., 3, 122–124. [DOI] [PubMed] [Google Scholar]
- 55.Qian X., Gozani,S.N., Yoon,H., Jeon,C.J., Agarwal,K. and Weiss,M.A. (1993) Novel zinc finger motif in the basal transcriptional machinery: three-dimensional NMR studies of the nucleic acid binding domain of transcriptional elongation factor TFIIS. Biochemistry, 32, 9944–9959. [DOI] [PubMed] [Google Scholar]
- 56.Baraldi E., Carugo,K.D., Hyvonen,M., Surdo,P.L., Riley,A.M., Potter,B.V., O’Brien,R., Ladbury,J.E. and Saraste,M. (1999) Structure of the PH domain from Bruton’s tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate. Structure Fold. Des., 7, 449–460. [DOI] [PubMed] [Google Scholar]
- 57.Hard T., Rak,A., Allard,P., Kloo,L. and Garber,M. (2000) The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold. J. Mol. Biol., 296, 169–180. [DOI] [PubMed] [Google Scholar]
- 58.Martinez-Yamout M., Legge,G.B., Zhang,O., Wright,P.E. and Dyson,H.J. (2000) Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ. J. Mol. Biol., 300, 805–818. [DOI] [PubMed] [Google Scholar]
- 59.Grishin N.V. (2000) C-terminal domains of Escherichia coli topoisomerase I belong to the zinc-ribbon superfamily. J. Mol. Biol., 299, 1165–1177. [DOI] [PubMed] [Google Scholar]
- 60.Bochkareva E., Korolev,S., Lees-Miller,S.P. and Bochkarev,A. (2002) Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J., 21, 1855–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bochkareva E., Korolev,S. and Bochkarev,A. (2000) The role for zinc in replication protein A. J. Biol. Chem., 275, 27332–27338. [DOI] [PubMed] [Google Scholar]
- 62.Chantalat L., Leroy,D., Filhol,O., Nueda,A., Benitez,M.J., Chambaz,E.M., Cochet,C. and Dideberg,O. (1999) Crystal structure of the human protein kinase CK2 regulatory subunit reveals its zinc finger-mediated dimerization. EMBO J., 18, 2930–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iwata S., Saynovits,M., Link,T.A. and Michel,H. (1996) Structure of a water soluble fragment of the ‘Rieske’ iron-sulfur protein of the bovine heart mitochondrial cytochrome bc1 complex determined by MAD phasing at 1.5 Å resolution. Structure, 4, 567–579. [DOI] [PubMed] [Google Scholar]
- 64.Borden K.L., Lally,J.M., Martin,S.R., O’Reilly,N.J., Etkin,L.D. and Freemont,P.S. (1995) Novel topology of a zinc-binding domain from a protein involved in regulating early Xenopus development. EMBO J., 14, 5947–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- 66.Berry M.B. and Phillips,G.N.,Jr (1998) Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+. Proteins, 32, 276–288. [DOI] [PubMed] [Google Scholar]
- 67.Muller C.W. and Schulz,G.E. (1993) Crystal structures of two mutants of adenylate kinase from Escherichia coli that modify the Gly-loop. Proteins, 15, 42–49. [DOI] [PubMed] [Google Scholar]
- 68.Wild K., Grafmuller,R., Wagner,E. and Schulz,G.E. (1997) Structure, catalysis and supramolecular assembly of adenylate kinase from maize. Eur. J. Biochem., 250, 326–331. [DOI] [PubMed] [Google Scholar]
- 69.Sherman J.M., Stone,E.M., Freeman-Cook,L.L., Brachmann,C.B., Boeke,J.D. and Pillus,L. (1999) The conserved core of a human SIR2 homologue functions in yeast silencing. Mol. Biol. Cell, 10, 3045–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Min J., Landry,J., Sternglanz,R. and Xu,R.M. (2001) Crystal structure of a SIR2 homolog-NAD complex. Cell., 105, 269–279. [DOI] [PubMed] [Google Scholar]
- 71.Turner R.B., Smith,D.L., Zawrotny,M.E., Summers,M.F., Posewitz,M.C. and Winge,D.R. (1998) Solution structure of a zinc domain conserved in yeast copper-regulated transcription factors. Nature Struct. Biol., 5, 551–555. [DOI] [PubMed] [Google Scholar]
- 72.Jeruzalmi D., O’Donnell,M. and Kuriyan,J. (2001) Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell, 106, 429–441. [DOI] [PubMed] [Google Scholar]
- 73.Cai M., Zheng,R., Caffrey,M., Craigie,R., Clore,G.M. and Gronenborn,A.M. (1997) Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nature Struct. Biol., 4, 567–577. [DOI] [PubMed] [Google Scholar]
- 74.Vasak M. and Hasler,D.W. (2000) Metallothioneins: new functional and structural insights. Curr. Opin. Chem. Biol., 4, 177–183. [DOI] [PubMed] [Google Scholar]
- 75.Luscombe N.M., Austin,S.E., Berman,H.M. and Thornton,J.M. (2000) An overview of the structures of protein–DNA complexes. Genome Biol., 1, 1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Macol C.P., Tsuruta,H., Stec,B. and Kantrowitz,E.R. (2001) Direct structural evidence for a concerted allosteric transition in Escherichia coli aspartate transcarbamoylase. Nature Struct. Biol., 8, 423–426. [DOI] [PubMed] [Google Scholar]
- 77.Kowalski K., Czolij,R., King,G.F., Crossley,M. and Mackay,J.P. (1999) The solution structure of the N-terminal zinc finger of GATA-1 reveals a specific binding face for the transcriptional co-factor FOG. J. Biomol. NMR, 13, 249–262. [DOI] [PubMed] [Google Scholar]
- 78.Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model., 15, 132–134, 112–113. [DOI] [PubMed] [Google Scholar]
- 79.Esnouf R.M. (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D Biol. Crystallogr., 55, 938–940. [DOI] [PubMed] [Google Scholar]