Abstract
Multifunctional proteins challenge the conventional ‘one protein–one function’ paradigm. Here we note apparent multifunctional proteins with nucleic acid partners, tabulating eight examples. We then focus on eight additional cases of transcription factors that bind double-stranded DNA with sequence specificity, but that also appear to lead alternative lives as RNA-binding proteins. Exemplified by the prototypic Xenopus TFIIIA protein, and more recently by mammalian p53, this list of transcription factors includes WT-1, TRA-1, bicoid, the bacterial σ70 subunit, STAT1 and TLS/FUS. The existence of transcription factors that bind both DNA and RNA provides an interesting puzzle. Little is known concerning the biological roles of these alternative protein–nucleic acid interactions, and even less is known concerning the structural basis for dual nucleic acid specificity. We discuss how these natural examples have motivated us to identify artificial RNA sequences that competitively inhibit a DNA-binding transcription factor not known to have a natural RNA partner. The identification of such RNAs raises the possibility that RNA binding by DNA-binding proteins is more common than currently appreciated.
INTRODUCTION
Reductionist approaches to understanding the complex macromolecular partnerships in living cells involve the application of biochemical and genetic assays to specific macromolecules. These time-honored and focused strategies have been tremendously successful and are taken to provide the best evidence of molecular function, even in the face of challenges from various new parallel methods. Despite this power, one weakness of focused molecular assays is often the informal but typically implicit assumption that each macromolecule possesses one biological function to be elucidated. Once assigned, such functions allow categorization and frame an understanding of the living cell.
On the other hand, living systems are presumably not shackled by the notion that macromolecules serve single functions. Because these molecules exist in a concentrated environment with thousands of potential molecular partners, it can be argued that the selective pressure to avoid illegitimate partnerships must be at least as strong as that driving affinity for legitimate partners. It is in the context of this rich network of potential interactions that biomolecules must function. Thus, we might expect that individual molecules could participate in multiple, perhaps superficially unrelated regulatory pathways. These multiple functions may be difficult to detect by traditional reductionist approaches. Nonetheless, macromolecules with multiple functions have been identified, although such discoveries are typically unexpected.
Multifunctionality is an emerging feature of certain proteins that bind nucleic acids. Table 1 provides eight illustrative examples. This review will focus on eight additional cases of proteins that were initially categorized as DNA-binding transcription factors, but for which subsequent research has shown apparent RNA-binding activities and functions (Table 2). With the expectation that such ‘moonlighting’ by DNA-binding proteins might be more common than previously imagined, we highlight some old and new examples of this phenomenon.
Table 1. Examples of multifunctional proteins that bind nucleic acids.
| Protein | Targeta | Functionb | References |
|---|---|---|---|
| KU | dsDNA/telomerase RNA? | DNA end recognition/? | 69,70 |
| T4 gene 32 | ssDNA/mRNA | ssDNA binding/translational repression | 71–75 |
| T4 DNA polymerase | DNA/mRNA | DNA replication/translational repression | 76–78 |
| ADAR1 | Z-DNA/dsRNA | ?/adenosine deaminase | 79,80 |
| Thermus thermophilus phenylalanyl-tRNA synthetase | tRNA/dsDNA | tRNA charging/? | 81 |
| mdm2 | p53 protein/RNA | Inactivation of p53/? | 82 |
| Thymidylate synthase | –/mRNA | Thymidylate biosynthesis/translational repression | 83,84 |
| Group II intron RT/maturases | Group II intron RNA/dsDNA | RNA splicing/DNA insertion | 85–88 |
aPutative targets are indicated in the form: initially discovered target/second target.
bFunction is indicated in the form: function of complex with initially discovered target/function of complex with second target.
Table 2. Transcription factors that bind DNA and RNA.
| Protein | Targeta | Functionb | Nucleic acid binding motifc | References |
|---|---|---|---|---|
| TFIIIA | 5S rDNA/5S rRNA | Transcription factor/rRNA storage factor, autoregulation of 5S gene expression? | cys2–his2 zinc fingers 1–3/cys2–his2 zinc fingers 4–6 | 5–19 |
| WT-1 | DNA/RNA? | Tumor suppressor transcription factor/RNA splicing factor? | cys2–his2 zinc fingers/alternately spliced cys2–his2 zinc fingers with KTS insert | 20–22 |
| TRA-1 | Developmental genes/tra-2 mRNA 3′-UTR | Transcription factor/RNA nuclear export | cys2–his2 zinc fingers | 25,26 |
| Bicoid | Developmental genes/cad mRNA 3′-UTR | Transcription factor/suppressor of cad mRNA translation | Homeodomain | 27,28 |
| p53 | Cell cycle genes/p53, Cdk4 mRNA 5′-UTRs? | Transcription factor/anti-helicase?, translational repressor? | ? | 52,58,59 |
| σ70 | dsDNA/6S RNA | Bacterial promotor specificity factor/? | ? | 89 |
| STAT1 | dsDNA/TSU RNA | Transcription factor/suppressor of embryonic MHC gene expression? | ? | 1,2 |
| TLS/FUS | DNA?/RNA? | Transcription factor?/splicing regulator | Zinc finger | 4 |
aPutative targets are indicated in the form: DNA target/RNA target.
bFunction is indicated in the form: function of DNA complex/function of RNA complex.
cIndication of the protein motif employed for DNA/RNA recognition, if known.
Besides illuminating the intricacy of biological systems, the existence of transcription factors that can bind both DNA and RNA raises at least three intriguing issues. The first issue is structural. How does a single polypeptide chain make sequence-specific contacts with two nucleic acids whose secondary and tertiary structures are likely to be very different? Are distinct protein surfaces involved, or are the same amino acids used to recognize the alternate targets? Are the nucleic acids recognized competitively? Are there examples where the RNA target mimics the structure of double-stranded DNA (dsDNA)? This is not a trivial issue since DNA and RNA strands of identical sequence are expected to adopt very distinct folded structures. Even in the case of double helical polymers, A-form RNA duplexes present base functional groups in a three-dimensional context that is very different from B-form DNA. The second issue is evolutionary. When a single domain of a macromolecule participates in two discrete (perhaps mutually exclusive) regulatory interactions, which interaction represents the ‘original’ function of the molecule? The third issue is an engineering consideration that has motivated some of our own research. How difficult is it to identify specific natural or unnatural RNA ligands for DNA-binding proteins? Identifi cation of unexpected natural RNA ligands for transcription factors might be undertaken by new systematic strategies. On the other hand, the identification of unnatural RNA ligands for transcription factors might provide an interesting ‘decoy’ approach to selective RNA-based antagonism of gene function.
Many proteins that bind nucleic acids would be expected to display at least modest non-specific affinity for other nucleic acids. For the present review, we have chosen to focus on sequence-specific DNA-binding transcription factors that also appear to bind RNA with at least some sequence specificity. The eight examples selected for this review (Table 2) are not intended to be exhaustive, nor is the cited literature comprehensive. Interested readers are encouraged to consult the lead references for broader background. The authors offer their apologies in advance for oversights or cases of seminal work that have been excluded to achieve brevity. Furthermore, two of the interesting cases included in Table 2 will not be reviewed in detail. These include the potential inhibition of STAT1 transcription factor function by a non-coding RNA in embryonic suppression of MHC expression (1,2) and RNA and DNA binding by the TLS/FUS factor involved in oncogenic chromosomal translocations (3,4).
ZINC FINGER PROTEINS THAT BIND DNA AND RNA
Transcription factor IIIA (TFIIIA)—the original paradigm
TFIIIA in Xenopus oocytes acts both as an activator of 5S ribosomal RNA (5S rRNA) transcription (5,6) and as a storage partner for 5S rRNA as it accumulates to massive levels in the oocyte prior to ribosome assembly during later stages of oocyte maturation and early development (7,8). This remarkable discovery immediately suggested the possibility of autoregulation of 5S rRNA synthesis by product antagonism of its transcription factor (Fig. 1). Such autoregulation was initially suggested by in vitro experiments (7), but it remained unclear if a functional feedback loop exists in vivo. More recent studies in developing Xenopus embryos suggest that this proposed mode of homeostatic control of ribosomal assembly may indeed occur (9,10). Whether somatic autoregulation of 5S rRNA transcription occurs remains unknown. Interestingly, p43, a structurally similar protein from Xenopus, binds only 5S rRNA (11).
Figure 1.
Schematic depiction of 5S rRNA autoregulation that is formally possible because of the ability of Xenopus TFIIIA to bind both 5S rRNA and the dsDNA encoding it. The 5S rRNA gene is indicated as duplex DNA. In the absence of free TFIIIA (trapezoid), the gene is not transcribed (OFF). Free TFIIIA leads to DNA binding and the assembly of an active complex that recruits RNA polymerase III (ON). Accumulation of 5S rRNA has the potential to titrate levels of free TFIIIA, inhibiting transcription activation.
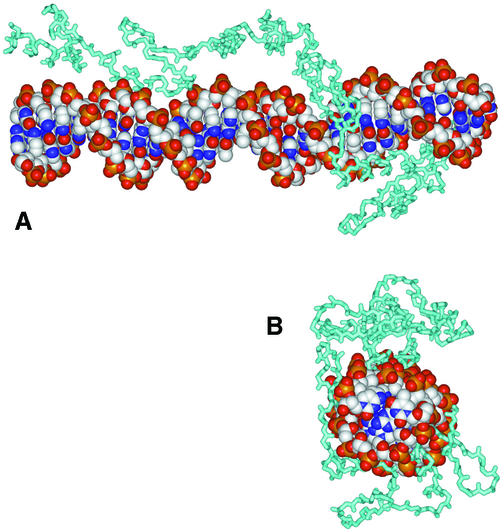
The notion that a DNA-binding transcription factor could recognize a specific RNA raised interesting structural questions. The competitive nature of DNA versus RNA binding suggested that overlapping surfaces of TFIIIA were involved in recognition of the DNA double helix and the folded 5S rRNA. Although the same nucleotide sequences are present in both targets, the folded structures of these nucleic acids are presumably very different. This conjecture was supported by the results of structural probing (12–14). TFIIIA contains nine classical cys2–his2 zinc fingers (each approximately 30 amino acids in size) arranged consecutively (15). These zinc fingers interact with ∼50 bp of dsDNA in the internal control region of the 5S gene, largely contacting one face of the DNA (16). This observation suggested that the zinc fingers did not form a spiral arrangement that tracked steadily along the major groove, but that linkers skipped across the DNA backbones at intervals. This prediction was eventually validated by an X-ray co-crystal structure of the six N-terminal zinc fingers of TFIIIA bound to duplex DNA (Fig. 2) (17).
Figure 2.
Molecular structure of a portion of TFIIIA bound to dsDNA. The six N-terminal zinc fingers of TFIIIA are shown in this rendering of the crystal structure (17). No high resolution structure of TFIIIA with 5S rRNA is available. (A) Side view of the complex. Protein (cyan backbone trace) N-terminus is at the right. DNA atoms are indicated as spheres of conventional colors (white, carbon; blue, nitrogen; red, oxygen; gold, phosphorus). (B) View of the complex from the N-terminus of the protein.
Biochemical analysis has also provided insight into the basis for zinc finger recognition of DNA versus RNA. A series of zinc finger deletions was created and analyzed in vitro by Clemens et al. (18) to study the domains within TFIIIA responsible for affinity and specificity for 5S DNA and RNA, respectively. These authors concluded that fingers 1–3 (numbered from the N-terminus of TFIIIA) contribute most of the DNA affinity, while fingers 4–6 are responsible for RNA binding. This analysis also suggested that the presence of a linker with the amino acid sequence TGEKP distinguishes the DNA-binding fingers from those specific for RNA. Complementary experiments have provided insight into the aspects of nucleic acid sequence and structure required for recognition (19). Three sequence blocks in the 5S gene reflect the linear engagement of TFIIIA along its surface. Mutations in 5S RNA suggest the roles of a key RNA bulge and base triple in TFIIIA binding. A detailed understanding of RNA recognition by TFIIIA must await a high-resolution X-ray co-crystal structure.
WT1
The notion that different zinc finger modules can preferentially recognize RNA or DNA targets is also supported by studies of the Wilms’ tumor protein. This protein is encoded by the WT1 gene and contains an N-terminal proline- and glutamine-rich regulatory domain followed by four C-terminal zinc fingers (20). Loss of this tumor suppressor protein is associated with pediatric kidney malignancies in Wilms’ tumor patients. Interestingly, the primary WT1 transcript is alternatively spliced such that the amino acids KTS are either present or absent between zinc fingers 3 and 4 (21). Protein isoforms lacking the KTS insert bind DNA sequence-specifically. In contrast, isoforms containing the KTS insert localize to regions of nuclear RNA splicing (22), and an RNA target is implied though not yet identified. In vitro RNA selection experiments with the form of the Wilms’ tumor protein lacking the KTS sequence show that even this isoform can bind both RNA and DNA in vitro (21).
TRA-1
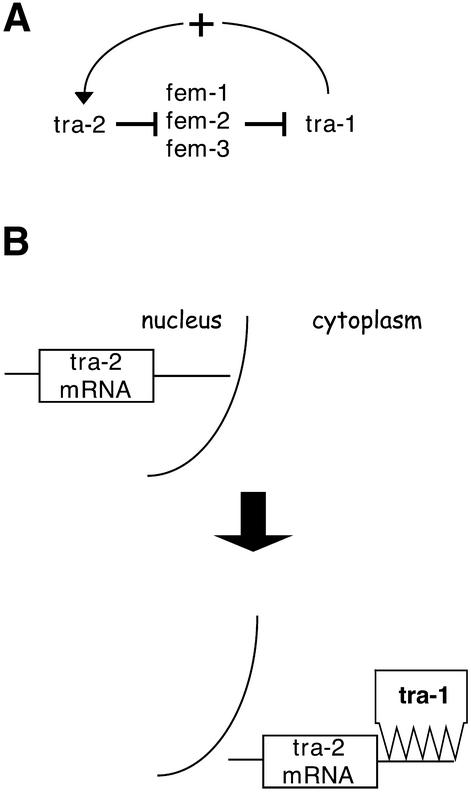
The two sexes of Caenorhabditis elegans, hermaphrodite and male, are determined by a genetic network involving the tra-1 and tra-2 genes (23,24). Expression of tra-1 promotes tra-2 activity required for proper development of the female features of the hermaphrodite. TRA-1 protein contains five zinc fingers and, like GLI family members in other organisms, binds a defined DNA sequence and regulates transcription of developmentally important genes (25,26). Genetically, tra-2 acts upstream of tra-1, but tra-1 positively feeds back on tra-2, perhaps to reinforce the female developmental switch (Fig. 3A). In elegant genetic and biochemical studies to understand the regulation of tra-2 by tra-1, Goodwin and co-workers (25) demonstrated that, unexpectedly, TRA-1 protein binds a sequence element in the 3′-untranslated region (3′-UTR) of tra-2 mRNA. This interaction induces cytoplasmic localization of the tra-2 mRNA. In their model (Fig. 3B), these authors suggest that the tra-2 mRNA is initially nuclear due to interaction with a protein factor bearing a nuclear localization signal. Binding of the tra-2 3′-UTR by TRA-1 reverses this nuclear targeting, and promotes nuclear export of both the tra-2 mRNA and bound TRA-1 protein.
Figure 3.
Schematic model for genetic and biochemical regulation of C.elegans tra-1 and tra-2. (A) Genetic regulatory relationship (26). (B) Biochemical model for post-transcriptional regulation of tra-2 mRNA localization by TRA-1 protein (25).
This model (Fig. 3B) suggests that clearance of RNA-bound TRA-1 from the nucleus could reduce its activity as a transcription factor. Such a mechanism would represent a novel RNA-based inhibition of gene expression by changing the subcellular localization of a transcription factor. It may be important to consider this regulatory model in understanding RNA binding by other transcription factors.
The detailed basis for TRA-1 recognition of both DNA and RNA remains unclear. The presence of multiple zinc fingers in TRA-1 raises the obvious analogy with both TFIIIA and WT1, suggesting that subsets of these fingers may be responsible for RNA and DNA binding. In vitro RNA binding was retained by deletion mutants of TRA-1 that contained only the first two of five zinc fingers (25).
BINDING TO DNA AND RNA BY THE HOMEODOMAIN
Bicoid
The homeodomain is a conserved sequence-specific dsDNA-binding module of approximately 60 amino acids, first identified in developmental transcription factors. Homeo domains specify helix–turn–helix protein motifs that make sequence-specific contacts in the DNA major groove. In Drosophila development, mRNA encoding the key homeodomain-containing protein bicoid (bcd) is deposited at the anterior of the egg. Upon fertilization, the translation of bcd mRNA results in a bcd anterior-to-posterior protein gradient. Local bcd concentrations determine whether bcd-responsive genes will later be activated by the protein, a key determinant in segment pattern formation. In contrast to bcd mRNA, the mRNA for a second key homeodomain transcription factor, caudal (cad), is initially evenly distributed within the early Drosophila embryo. However, in wild-type embryos, translation of cad mRNA is non-uniform, with cad mRNA translated most where bcd protein concentrations are lowest. This relationship gives rise to an inverse (posterior-to-anterior) concentration gradient of cad protein. In mutants lacking bcd protein, translation of cad mRNA leads to an abnormal, uniform distribution of cad protein across the embryo. Because this antagonistic relationship between bcd protein and cad mRNA translation is established in unfertilized eggs in the absence of transcription, a possible direct role of bcd protein in cad mRNA translation suppression was suggested.
Genetic (27) and biochemical (28) experiments were undertaken to elucidate the mechanism of cad mRNA translation suppression by bcd protein. A control element in the 3′-UTR of cad mRNA was found to be responsible for bcd suppression of cad mRNA translation (27). Further in vivo studies showed that the homeodomain region of bcd was needed for cad mRNA translational control. Remarkably, these authors were then able to show that the bcd/cad mRNA interaction was direct. Recombinant bcd protein in mammalian cell extracts or purified after expression in bacteria directly bound radiolabeled cad mRNA in vitro (27). Subsequent experiments demonstrated that binding of bcd to the cad 3′-UTR in heterologous transcripts was sufficient to block translation (27).
Similar approaches led Rivera-Pomar et al. (28) to identify proteins from Drosophila embryo nuclear extracts capable of binding to cad mRNA. These studies again led to the conclusion that the homeodomain of bcd binds directly to the 3′-UTR of cad mRNA, resulting (by an unknown mechanism) in the suppression of cad mRNA translation. This ability of a single protein to execute both transcriptional and translational control provides an efficient regulatory circuit in pattern formation. Although it seems likely that the bcd homeodomain would bind dsDNA or cad mRNA competitively, this remains unproven. As for the zinc finger examples discussed above, details of apparent homeodomain/RNA recognition remain to be elucidated.
DNA AND RNA BINDING BY THE p53 TUMOR SUPPRESSOR PROTEIN
Transcription factor biology of p53
Aptly termed the ‘guardian of the genome’, the p53 tumor suppressor protein plays multiple roles in facilitating growth arrest or apoptosis in response to DNA damage. Its vital function in growth control is evidenced by the fact that p53 mutations are among the most prevalent genetic abnormalities in human cancer (29). Perhaps because p53 is one of the most intensely studied proteins of recent years, reports continue to surface describing previously unknown functions and binding partners for this transcription factor. It is obvious that much remains to be learned of p53 functions in addition to its conventional role as a sequence-specific DNA-binding protein and transcriptional activator.
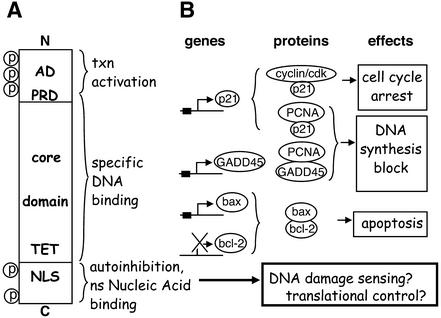
p53 is unusual among transcription factors in having two distinct binding domains for nucleic acids, each with a different specificity (Fig. 4A) (30). A sequence-specific DNA-binding domain comprises the central core of p53. The core domain is responsible for recognition of the p53 consensus sequence in promoters of target genes, and most tumor-derived missense mutations occur in this region of the protein (30). A second, non-specific, nucleic acid-binding domain lies near the C-terminus of p53. This highly basic domain appears to exhibit little, if any, sequence preference and binds with high affinity to single-stranded DNA (ssDNA) and RNA, irradiated DNA, four-way junctions, insertions, and deletions (30).
Figure 4.
Protein biochemistry and putative regulatory interactions of p53. (A) p53 protein domains. AD, activation domain; PRD, proline-rich SH3-binding domain; TET, tetramerization domain; NLS, nuclear localization sequence; P, phosphorylation sites. (B) Examples of genes regulated by p53, and their relationships to DNA damage control and cellular responses.
Binding of p53 to its cognate site in promoters activates transcription of target genes involved in growth arrest or apoptosis, thereby allowing adaptation to cellular insults. DNA damage appears to induce additional post-translational modifications of p53 that increase its promoter-binding activity. Interestingly, modification of the non-specific C-terminal nucleic acid-binding domain of p53 by phosphorylation, acetylation, antibody binding, or deletion appears to activate sequence-specific DNA binding by the core domain in vitro (31). In contrast, it has been suggested that binding of the C-terminal domain to nucleic acids sterically hinders the ability of the core domain to recognize its specific DNA-binding site (32). Thus, it appears that the C-terminal nucleic acid-binding domain of p53 plays an important regulatory role in specific DNA binding by the core domain. A more extensive review of the regulation of p53 function by its C-terminal domain is given by Ahn and Prives (30).
Unconventional nucleic acid interactions with p53
Beyond its well characterized transcription factor activity, new potential roles for p53 have recently come to light. In the late 1980s it was recognized that p53 can inhibit SV40 minichromosome replication by suppressing T-antigen DNA helicase activity (33–35). In 1993, Oberosler et al. (36) showed that p53 acts as a general anti-helicase in vitro, inhibiting several unrelated DNA and RNA helicases. Intriguingly, p53 was shown to have a much higher affinity for single-stranded rather than double-stranded nucleic acids, and RNA was found to serve as a more efficient competitor for p53 binding than ssDNA by one order of magnitude (36). The anti-helicase activity of p53 increases the rate of RNA–RNA renaturation by >200-fold, and this activity is not dependent upon base composition or length of the RNA (36).
Accumulating evidence suggests that the C-terminal domain of p53 is both necessary and sufficient for its nucleic acid annealing activity. An antibody to the p53 C-terminal, non-specific nucleic acid-binding domain (Pab421, recognizing amino acids 370–378) blocks DNA and RNA annealing activity (36), while activating sequence-specific DNA binding (31). Wu et al. (37) showed that a p53 C-terminal fragment encompassing residues 311–393 is sufficient to promote annealing of nucleic acids in vitro. Furthermore, a p53 splice variant has been detected in mice that utilizes an alternative exon at the 3′ end of the mouse p53 gene, resulting in the substitution of 17 new amino acids for the 26 C-terminal residues of the normal splice variant (38). Significantly, the p53 alternate splice variant lacks the nucleic acid annealing activity of the p53 normal splice variant (37). The fact that the two p53 splice variants appear to be expressed differentially at different times in the cell cycle (39) implies a functional relevance for the different p53 splice forms. Indeed, the p53 alternate splice form has been shown to exhibit enhanced specific DNA binding (40) but an attenuated ability to induce apoptosis (41,42).
How does p53 catalyze the annealing of complementary nucleic acid strands? Filhol et al. (43) hypothesized that p53 monomers may bind to the complementary single-stranded nucleic acids, and these are then brought in close proximity by p53 oligomerization. However, the annealing activity of p53 is separable from its ability to bind single-stranded nucleic acids, since a p53 mutant protein isolated from a human liver tumor [p53-mt276; (44)] showed reduced annealing activity although its ability to bind nucleic acids was unaffected (45). Binding of p53 does not cause major changes in RNA secondary structure (45).
Nucleic acid annealing activity by p53 might function during DNA replication, recombination, pre-mRNA splicing, RNA transport, or translation and could counteract or regulate DNA and RNA helicases (36). The fact that p53 nucleic acid annealing activity and specific DNA binding appear to be mutually exclusive suggests that p53 can exist in two very different functional states, the distribution of which may be tightly regulated. Indeed, phosphorylation of p53 at its C-terminal serine by the ubiquitously expressed serine/ threonine kinase casein kinase 2 (CK2) completely inhibits DNA annealing activity in vitro (43) while activating sequence-specific DNA binding upon UV irradiation in vivo (46).
Specific RNA binding by p53
In addition to its apparent sequence-independent RNA/DNA annealing activity, p53 has been increasingly implicated in the translational control of specific mRNA transcripts. An unexpected link to translation was first suggested by the fascinating discovery that 5.8S rRNA was found to be covalently bound to phosphoserine-389 of purified mouse p53 (47). It has since been shown that p53 can exist in a complex with Mdm2 and the large subunit-associated ribosomal protein L5 (48). Furthermore, purified Mdm2 or Mdm2/p53 binds 5S rRNA (48), presumably via the specific L5–5S rRNA interaction (49). Adding to the complexity, L5, 5S rRNA and 5.8S rRNA can form a ternary complex in solution (50). Thus, a ribonucleoprotein complex consisting of p53, Mdm2, L5 ribosomal protein, 5S rRNA and 5.8S rRNA may exist in the cell by virtue of multiple protein–protein, protein-RNA, and RNA–RNA interactions (48). This complex could conceivably function in such processes as ribosome biogenesis, ribosome transport from the nucleus, or translational regulation (48).
Fontoura et al. (51) further investigated the association of cytoplasmic p53 with ribosomes in an elegant series of cofractionation experiments. In this study it was found that mammalian p53 associates with a subset of ribosomes that comprises a small fraction of the total ribosome population (51). However, the vast majority of p53 covalently linked to 5.8S rRNA associates with ribosomes (51). Puromycin treatment blocked copurification of p53 with ribosomes, indicating that p53 is indeed associated with the translation machinery (51). Furthermore, p53 mRNA coprecipitates with ribosome-associated p53 polypeptide, indicating that p53 binds specifically to ribosomes bearing p53 mRNA and/or directly to p53 mRNA (51). Fontoura et al. (51) concluded from these studies that p53 may be involved in regulating translation of a subset of mRNAs, including its own.
Mosner et al. (52) discovered that murine p53 does in fact appear to regulate the translation of its own mRNA. Murine p53 preferentially binds the murine p53 mRNA 5′-UTR and inhibits translation of the p53 transcript in vitro (52). Stable RNA stem–loop structures are predicted to form in the 5′-UTR of p53 mRNA (52). The disruption of such secondary structures during translation is thought to require an RNA helicase, e.g. eukaryotic translation initiation factor 4A [eIF-4A; (53)]. Thus, the known, potent anti-RNA helicase activity of p53 (36) may be functionally relevant to its ability to inhibit the translation of certain mRNA transcripts. In accordance with this hypothesis, it was found that the MethA mutant p53 protein (54), which is defective in RNA–RNA annealing, binds p53 mRNA but does not inhibit translation (52). Thus, p53 may stabilize secondary structures within its own mRNA 5′-UTR, interfering with ribosome scanning. Direct autoregulation of the p53 transcript has not yet been observed in humans, although a 40-kDa protein has been shown to bind an Alu-like element in the human p53 3′-UTR to inhibit translation (55–57).
The Cdk4 mRNA provides a second example of possible translation regulation by p53. Ewen et al. (58) and Miller et al. (59) showed that wild-type p53 binds to a region of the human Cdk4 5′-UTR mapped to positions –100 to –64 upstream of the start codon. The specific binding of human p53 to this region represses human Cdk4 translation during TGF-β-induced G1 cell-cycle arrest in Mv1Lu cells (58,59). This finding provides another mechanism for p53-mediated down-regulation of Cdk4 in response to TGF-β1, in addition to the previously known transactivation of the Cdk4 inhibitor p21WAF1/CIP1 (60). p53 has a proline-rich domain (amino acid residues 61–94) that can interact with the SH3 domains of signaling proteins, possibly providing a linkage between p53 and the TGF-β receptor. Indeed, p53 amino acids 43–80 were found to be essential for translational regulation of Cdk4 (59). Cdk4 5′-UTR RNA binding is necessary but not sufficient for translation inhibition by p53, since some p53 mutants defective for translational regulation can still bind the CDK4 RNA transcript (59).
The previous examples provide compelling evidence that p53 functions as an RNA-binding protein in addition to its long-recognized role as a DNA-binding protein/transcriptional activator. It is becoming increasingly evident that p53 can regulate the translation of a subset of mRNA transcripts. Possible mechanisms for this translation inhibition include stabilization of RNA secondary structures via the anti-helicase activity of p53, steric hindrance of ribosome entry, or direct interaction with the translation machinery mediated by the covalent linkage between p53 and 5.8S rRNA (61).
Although much has been learned regarding RNA binding by p53 in recent years, many unanswered questions remain. For example, how does p53 specifically recognize regions in the 5′-UTRs of certain transcripts to mediate translation inhibition? Is such binding dependent upon sequence recognition, or recognition of specific RNA secondary structures? Interestingly, positions –72 to –53 in the CDK4 5′-UTR are 84% identical among the human, mink, pig and rat sequences (59), suggesting evolutionary conservation. Can p53 inhibit the translation of other transcripts in addition to its own (mouse) and that encoding Cdk4 (mammalian)? What domain(s) of p53 are responsible for specific RNA binding and translation inhibition? The p53 C-terminus appears to be necessary but not sufficient for specific translation inhibition. Inhibition of translation of the murine p53 transcript by p53 polypeptide could be overcome by preincubation with antibody PAb421 to the p53 C-terminus (52). The isolated p53 C-terminal domain (amino acid residues 311–393) can weakly inhibit translation of the Cdk4 transcript but binds non-specifically to other RNAs as well, suggesting that additional regions of p53 may provide specificity in RNA binding (59). Finally, does RNA binding by p53 function in processes other than translation inhibition, e.g. ribosome biogenesis, pre-mRNA splicing or RNA transport? The future promises to hold further surprises with regard to the roles of this versatile and multifunctional nucleic acid-binding protein.
UNNATURAL RNA LIGANDS FOR DNA-BINDING PROTEINS: HOW DIFFICULT A QUEST?
The discovery of potential natural RNA partners for DNA-binding transcription factors has motivated our own laboratory to consider designing or selecting artificial partnerships of this kind. Of particular interest is the notion that small, folded RNAs might be encoded in designer transgenes and expressed on demand to produce molecular ‘decoys’ to competitively inhibit specific DNA-binding transcription factors. Three attractive aspects of this problem drew our attention. First, DNA-binding proteins are expected to have a general electrostatic affinity for anionic polymers such as RNA. Secondly, powerful in vitro selection methods are available for screening massive combinatorial RNA libraries (>1013 100-nt RNAs) for candidate RNA decoys (62,63). Thirdly, powerful yeast genetic selection schemes have been developed to detect and optimize novel RNA–protein interactions in eukaryotic nuclei (64,65).
Armed with these tools, we have systematically asked whether small decoy RNAs can be selected against an arbitrary mammalian transcription factor. For this purpose we have targeted the well studied p50 homodimer form of transcription factor NF-κB. This protein and related complexes bind dsDNA and activate key genes involved in inflammation and the prevention of apoptosis. No natural RNA ligand for NF-κB is known. We were therefore intrigued that in vitro selection directly yielded a 31-nt RNA motif capable of NF-κB binding in vitro with ∼1 nM affinity, indistinguishable from the affinity of the protein for its optimal DNA-binding site (Fig. 5) (66). Binding was shown to be competitive with DNA (66), and the stoichiometry of the interaction appears to be one RNA per protein dimer (67). These and other results suggest that the RNA binds in the DNA-binding cleft of the protein dimer, perhaps as a structural mimic of the normal DNA double helix target sequence. The potential imperfect A-form RNA duplex structure of the RNA aptamer may allow it to occupy a protein cavity optimized to receive a B-form DNA duplex.
Figure 5.
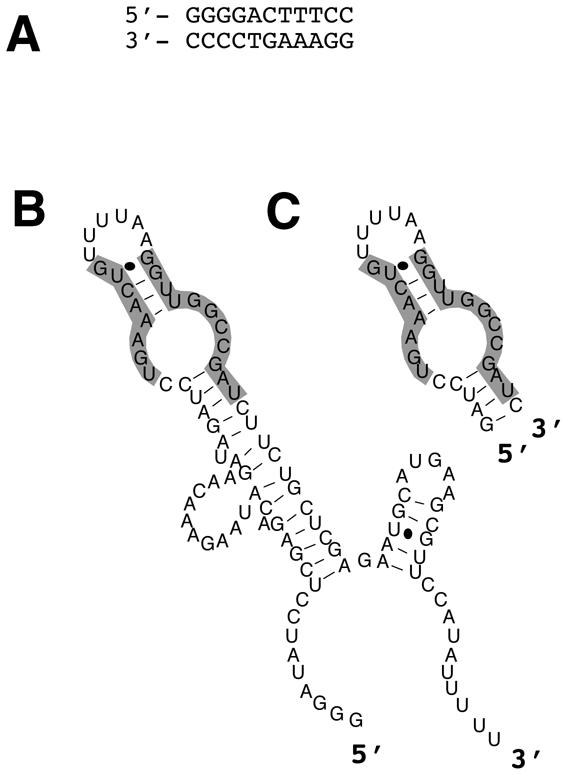
Natural and artificial nucleic acid ligands for human NF-κB. (A) Strong NF-κB recognition sequence in dsDNA, derived from the HIV-1 LTR promoter. (B) Predicted secondary structure of in vitro-selected RNA aptamer that competes with dsDNA for NF-κB binding (66). (C) Predicted secondary structure of the 31-nt core domain of the selected RNA aptamer.
Our more recent efforts have focused on the question of whether this unnatural RNA–protein interaction could be made to occur in vivo, where the in vitro-selected RNA encounters thousands of new potential protein partners and new folding and localization constraints, not to mention nucleases. Perhaps remarkably, the 31-nt RNA motif selected for NF-κB binding in vitro was found to be recognized selectively by NF-κB protein after the aptamer was engrafted into a longer hybrid RNA and expressed together with NF-κB in a sensitive yeast three-hybrid assay (68). Because the yeast three-hybrid system offers a selection for enhanced reporter expression driven by the RNA–protein interaction, this system has afforded a powerful approach to screen various aptamer variant libraries to identify RNA aptamers for NF-κB that display improved function in vivo (L.A.Cassiday and L.J.Maher III, unpublished results). Such RNA optimization may reflect improved in vivo binding affinity, or selection for other parameters such as RNA accumulation, localization, folding or avoidance of non-specific protein partners. A further avenue of research is the design of direct yeast selections for RNAs capable of disrupting DNA binding by the target transcription factor. Overall, the availability of a eukaryotic aptamer optimization strategy in the form of the yeast three-hybrid system provides a systematic method for linking the power of in vitro RNA selections to the eventual testing of RNAs as decoys in mammalian cells.
CONCLUDING QUESTIONS
This survey highlights the interesting problem of RNA binding by DNA-binding proteins and suggests at least two key questions for future investigation. The first question relates to the structural basis for RNA recognition by DNA-binding proteins. Does the folded RNA assume a structure that mimics the three-dimensional array of charges and hydrogen bond donors and acceptors presented to the protein-binding surface by duplex DNA? Insight into this problem will be gained through determination of high-resolution structures for complexes of a given transcription factor bound alternatively to RNA versus dsDNA.
A second question is more strategic: are there systematic and efficient approaches to identifying previously unknown RNA–protein interactions involving transcription factors? Such searches will be valuable to provide an unbiased survey of the kinds of unexpected RNA–protein interactions discussed above. Such a survey could reveal molecular partnerships that are now detected only by serendipity. Systematic screens, such as those made possible by the yeast three-hybrid system, now allow the contemplation of such studies (64).
Acknowledgments
ACKNOWLEDGEMENTS
The authors express their appreciation to Marlene Belfort, Jim Dahlberg, Bill Dynan, Betsy Goodwin, Joel Gottesfeld, Phil Hardwidge, Ralf Janknecht, Alan Lambowitz, John Peyman, Marvin Wickens, and the anonymous referees for comments and suggestions. Research in the authors’ laboratory is supported by the Mayo Foundation, and by grants from the National Institutes of Health and the American Cancer Society. L.A.C. is a predoctoral fellow of the Howard Hughes Medical Institute.
REFERENCES
- 1.Peyman J.A. (1999) Repression of major histocompatibility complex genes by a human trophoblast ribonucleic acid. Biol. Reprod., 60, 23–31. [DOI] [PubMed] [Google Scholar]
- 2.Peyman J.A. (2001) Mammalian expression cloning of two human trophoblast suppressors of major histocompatibility complex genes. Am. J. Reprod. Immunol., 45, 382–392. [DOI] [PubMed] [Google Scholar]
- 3.Perrotti D., Bonatti,S., Trotta,R., Martinez,R., Skorski,T., Salomoni,P., Grassilli,E., Iozzo,R.V., Cooper,D.R. and Calabretta,B. (1998) TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J., 17, 4442–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lerga A., Hallier,M., Delva,L., Orvain,C., Gallais,I., Marie,J. and Moreau-Gachelin,F. (2001) Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem., 276, 6807–6816. [DOI] [PubMed] [Google Scholar]
- 5.Sakonju S., Bogenhagen,D.F. and Brown,D.D. (1980) A control region in the center of the 5S RNA gene directs specific initiation of transcription: 1. The 5′ border of the region. Cell, 19, 13–25. [DOI] [PubMed] [Google Scholar]
- 6.Engelke D.R., Ng,S.-Y., Shastry,B.S. and Roeder,R.G. (1980) Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell, 19, 717–728. [DOI] [PubMed] [Google Scholar]
- 7.Pelham H.R.B. and Brown,D.D. (1980) A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc. Natl Acad. Sci. USA, 77, 4170–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honda B.M. and Roeder,R.G. (1980) Association of a 5S gene transcription factor with 5S RNA and altered levels of the factor during cell differentiation. Cell, 22, 119–126. [DOI] [PubMed] [Google Scholar]
- 9.Rollins M.B., Del Rio,S., Galey,A.L., Setzer,D.R. and Andrews,M.T. (1993) Role of TFIIIA zinc fingers in vivo: analysis of single-finger function in developing Xenopus embryos. Mol. Cell. Biol., 13, 4776–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pittman R.H., Andrews,M.T. and Setzer,D.R. (1999) A feedback loop coupling 5 S rRNA synthesis to accumulation of a ribosomal protein. J. Biol. Chem., 274, 33198–33201. [DOI] [PubMed] [Google Scholar]
- 11.Joho K.E., Darby,M.K., Crawford,E.T. and Brown,D.D. (1990) A finger protein structurally similar to TFIIIA that binds exclusively to 5S RNA in Xenopus. Cell, 61, 293–300. [DOI] [PubMed] [Google Scholar]
- 12.Andersen J., Delihas,N., Hanas,J.S. and Wu,C.-W. (1984) 5S RNA structure and interaction with transcription factor A. 1. Ribonuclease probe of the structure of 5S RNA from Xenopus laevis oocytes. Biochemistry, 23, 5752–5759. [DOI] [PubMed] [Google Scholar]
- 13.Sands M.S. and Bogenhagen,D.F. (1987) TFIIIA binds to different domains of 5S RNA and the Xenopus borealis 5S RNA gene. Mol. Cell. Biol., 7, 3985–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christiansen J., Brown,R.S., Sproat,B.S. and Garrett,R.A. (1987) Xenopus transcription factor IIIA binds primarily at junctions between double helical stems and internal loops in oocyte 5S RNA. EMBO J., 6, 453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vrana K.E., Churchill,M.E.A., Tullius,T.D. and Brown,D.D. (1988) Mapping functional regions of transcription factor TFIIIA. Mol. Cell. Biol., 8, 1684–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Churchill M.E.A., Tullius,T.D. and Klug,A. (1990) Mode of interaction of the zinc finger protein TFIIIA with a 5S RNA gene of Xenopus. Proc. Natl Acad. Sci. USA, 87, 5528–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolte R.T., Conlin,R.M., Harrison,S.C. and Brown,R.S. (1998) Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex. Proc. Natl Acad. Sci. USA, 95, 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clemens K.R., Wolf,V., McBryant,S.J., Zhang,P., Liao,X., Wright,P.E. and Gottesfeld,J.M. (1993) Molecular basis for specific recognition of both RNA and DNA by a zinc finger protein. Science, 260, 530–533. [DOI] [PubMed] [Google Scholar]
- 19.Rawlings S.L., Matt,G.D. and Huber,P.W. (1996) Analysis of the binding of Xenopus transcription factor IIIA to oocyte 5 S rRNA and to the 5 S rRNA gene. J. Biol. Chem., 271, 869–877. [DOI] [PubMed] [Google Scholar]
- 20.Rauscher F.J., Morris,J.F., Tournay,O.E., Cook,D.M. and Curran,T. (1990) Binding of Wilms’ tumor locus zinc finger protein to the EGR-1 consensus sequence. Science, 250, 1259–1262. [DOI] [PubMed] [Google Scholar]
- 21.Zhai G., Iskandar,M., Barilla,K. and Romaniuk,P.J. (2001) Characterization of RNA aptamer binding by the Wilms’ tumor suppressor protein WT1. Biochemistry, 40, 2032–2040. [DOI] [PubMed] [Google Scholar]
- 22.Larsson S.H., Charlieu,J.-P., Miyagawa,K., Engelkamp,D., Rassoulzadegan,M., Ross,A., Cuzin,F., van Heyningen,V. and Hastie,N.D. (1995) Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell, 81, 391–401. [DOI] [PubMed] [Google Scholar]
- 23.Hodgkin J. (1986) Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics, 114, 15–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgkin J.A. (1987) A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev., 1, 731–745. [DOI] [PubMed] [Google Scholar]
- 25.Graves L.E., Segal,S. and Goodwin,E.B. (1999) TRA-1 regulates the cellular distribution of the tra-2 mRNA in C. elegans. Nature, 399, 802–805. [DOI] [PubMed] [Google Scholar]
- 26.Segal S.P., Graves,L.E., Verheyden,J. and Goodwin,E.B. (2001) RNA-regulated TRA-1 nuclear export controls sexual fate. Dev. Cell, 1, 1–20. [DOI] [PubMed] [Google Scholar]
- 27.Dubnau J. and Struhl,G. (1996) RNA recognition and translational regulation by a homeodomain protein. Nature, 379, 694–699. [DOI] [PubMed] [Google Scholar]
- 28.Rivera-Pomar R., Niessing,D., Schmidt-Ott,U., Gehring,W.J. and Jäckle,H. (1996) RNA binding and translational suppression by bicoid. Nature, 379, 746–749. [DOI] [PubMed] [Google Scholar]
- 29.Prives C. and Hall,P.A. (1999) The p53 pathway. J. Pathol., 187, 112–126. [DOI] [PubMed] [Google Scholar]
- 30.Ahn J. and Prives,C. (2001) The C-terminus of p53: the more you learn the less you know. Nature Struct. Biol., 8, 730–732. [DOI] [PubMed] [Google Scholar]
- 31.Hupp T.R., Meek,D.W., Midgley,C.A. and Lane,D.P. (1992) Regulation of the specific DNA binding function of p53. Cell, 71, 875–886. [DOI] [PubMed] [Google Scholar]
- 32.Anderson M.E., Woelker,B., Reed,M., Wang,P. and Tegtmeyer,P. (1997) Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: implications for regulation. Mol. Cell. Biol., 17, 6255–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braithwaite A.W., Sturzbecher,H.-W., Addison,C., Palmer,C., Rudge,K. and Jenkins,J.R. (1987) Mouse p53 inhibits SV40 origin-dependent DNA replication. Nature, 329, 458–460. [DOI] [PubMed] [Google Scholar]
- 34.Sturzbecher H.-W., Brain,R., Maimets,T., Addison,C., Rudge,K. and Jenkins,J.R. (1988) Mouse p53 blocks SV40 DNA replication in vitro and downregulates T antigen DNA helicase activity. Oncogene, 3, 405–413. [PubMed] [Google Scholar]
- 35.Wang E.H., Friedman,P.N. and Prives,C. (1989) The murine p53 protein blocks replication of SV40 DNA in vitro by inhibiting the initiation functions of SV40 large T antigen. Cell, 57, 379–392. [DOI] [PubMed] [Google Scholar]
- 36.Oberosler P., Hloch,P., Ramsperger,U. and Stahl,H. (1993) p53-catalyzed annealing of complementary single-stranded nucleic acids. EMBO J., 12, 2389–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu L., Bayle,J.H., Elenbaas,B., Pavletich,N.P. and Levine,A.J. (1995) Alternatively spliced forms in the carboxy-terminal domain of the p53 protein regulate its ability to promote annealing of complementary single strands of nucleic acids. Mol. Cell. Biol., 15, 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf D., Harris,N., Goldfinger,N. and Rotter,V. (1985) Isolation of a full-length mouse cDNA clone coding for an immunologically distinct p53 molecule. Mol. Cell. Biol., 5, 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulesz-Martin M.F., Lisafeld,B., Huang,H., Kisiel,N.D. and Lee,L. (1994) Endogenous p53 protein generated from wild-type alternatively spliced p53 RNA in mouse epidermal cells. Mol. Cell. Biol., 14, 1698–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolkowicz R., Peled,A., Elkind,N.B. and Rotter,V. (1998) DNA-binding activity of wild-type p53 protein is mediated by the central part of the molecule and controlled by its C terminus. Cancer Detect. Prev., 22, 1–13. [DOI] [PubMed] [Google Scholar]
- 41.Almog N., Li,R., Peled,A., Schwartz,D., Wolkowicz,R., Goldfinger,N., Pei,H. and Rotter,V. (1997) The murine C-terminally alternatively spliced form of p53 induces attenuated apoptosis in myeloid cells. Mol. Cell. Biol., 17, 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Almog N., Goldfinger,N. and Rotter,V. (2000) p53-dependent apoptosis is regulated by a C-terminally alternatively spliced form of murine p53. Oncogene, 19, 3395–3403. [DOI] [PubMed] [Google Scholar]
- 43.Filhol O., Baudier,J., Chambaz,E.M. and Cochet,C. (1996) Casein kinase 2 inhibits the renaturation of complementary DNA strands mediated by p53 protein. Biochem. J., 316, 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volkmann M., Hofmann,W.J., Muller,M., Rath,U., Otto,G., Zentegraf,H. and Galle,P.R. (1994) p53 overexpression is frequent in European hepatocellular carcinoma and largely independent of the codon 249 hot spot mutation. Oncogene, 9, 195–204. [PubMed] [Google Scholar]
- 45.Nedbal W., Frey,M., Willemann,B., Zentgraf,H. and Sczakiel,G. (1997) Mechanistic insights into p53-promoted RNA–RNA annealing. J. Mol. Biol., 266, 677–687. [DOI] [PubMed] [Google Scholar]
- 46.Kapoor M. and Lozano,G. (1998) Functional activation of p53 via phosphorylation following DNA damage by UV but not γ radiation. Proc. Natl Acad. Sci. USA, 95, 2834–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fontoura B.M., Sorokina,E.A., David,E. and Carroll,R.B. (1992) p53 is covalently linked to 5.8S rRNA. Mol. Cell. Biol., 12, 5145–5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marechal V., Elenbaas,B., Piette,J., Nicholas,J.C. and Levine,A.J. (1994) The ribosomal L5 protein is associated with mdm-2 and mdm-2–p53 complexes. Mol. Cell. Biol., 14, 7414–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steitz J.A., Berg,C., Hendrick,J.P., La Branch-Chabot,H., Metspalu,J., Rinke,J. and Yario,T. (1988) A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J. Cell Biol., 106, 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metspalu A., Toots,I., Saarma,M. and Villems,R. (1980) The ternary complex consisting of rat liver ribosomal 5S RNA, 5.8S RNA and protein L5. FEBS Lett., 119, 81–84. [DOI] [PubMed] [Google Scholar]
- 51.Fontoura B.M., Atienza,C.A., Sorokina,E.A., Morimoto,T. and Carroll,R.B. (1997) Cytoplasmic p53 polypeptide is associated with ribosomes. Mol. Cell. Biol., 17, 3146–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mosner J., Mummenbrauer,T., Bauer,C., Sczakiel,G., Grosse,F. and Deppert,W. (1995) Negative feedback regulation of wild-type p53 biosynthesis. EMBO J., 14, 4442–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaramillo M., Dever,T.E., Merrick,W.C. and Sonenberg,N. (1991) RNA unwinding in translation: assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF-4B. Mol. Cell. Biol., 11, 5992–5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eliyahu D., Goldfinger,N., Pinhasi-Kimhi,O., Shaulsky,G., Skurnik,Y., Arai,N., Rotter,V. and Oren,M. (1988) Meth A fibrosarcoma cells express two transforming mutant p53 species. Oncogene, 3, 313–321. [PubMed] [Google Scholar]
- 55.Fu L., Minden,M.D. and Benchimol,S. (1996) Translational regulation of human p53 gene expression. EMBO J., 15, 4392–4401. [PMC free article] [PubMed] [Google Scholar]
- 56.Fu L. and Benchimol,S. (1997) Participation of the human p53 3′UTR in translational repression and activation following gamma-irradiation. EMBO J., 16, 4117–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu L., Ma,W. and Benchimol,S. (1999) A translation repressor element resides in the 3′ untranslated region of human p53 mRNA. Oncogene, 18, 6419–6424. [DOI] [PubMed] [Google Scholar]
- 58.Ewen M.E., Oliver,C.J., Sluss,H.K., Miller,S.J. and Peeper,D.S. (1995) p53-dependent repression of cdk4 translation in TGF-beta-induced G(1) cell-cycle arrest. Genes Dev., 9, 204–217. [DOI] [PubMed] [Google Scholar]
- 59.Miller S.J., Suthiphongchai,T., Zambetti,G.P. and Ewen,M.E. (2000) p53 binds selectively to the 5′ untranslated region of cdk4, an RNA element necessary and sufficient for transforming growth factor β- and p53-mediated translational inhibition of cdk4. Mol. Cell. Biol., 20, 8420–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harper J.W., Adami,G.R., Wei,N., Keyomarsi,D. and Elledge,S.J. (1993) The p21 cdk-interaction protein cip-1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell, 75, 805–816. [DOI] [PubMed] [Google Scholar]
- 61.Ewen M.E. and Miller,S.J. (1996) p53 and translational control. Biochim. Biophys. Acta, 1242, 181–184. [DOI] [PubMed] [Google Scholar]
- 62.Tuerk C. and Gold,L. (1990) Systemic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 63.Ellington A.D. and Szostak,J.W. (1990) In vitro selection of RNA molecules that bind specific ligands. Nature, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 64.SenGupta D.J., Zhang,B.L., Kraemer,B., Pochart,P., Fields,S. and Wickens,M. (1996) A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl Acad. Sci. USA, 93, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sengupta D., Wickens,M. and Fields,S. (1999) Identification of RNAs that bind a specific protein using the yeast three-hybrid system. RNA, 5, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lebruska L.L. and Maher,L.J.,III (1999) Selection and characterization of an RNA decoy for transcription factor NF-κB. Biochemistry, 38, 3168–3174. [DOI] [PubMed] [Google Scholar]
- 67.Cassiday L.A., Lebruska,L.L., Benson,L.M., Naylor,S., Owen,W.G. and Maher,L.J.,III. (2002) Binding stoichiometry of an RNA aptamer and its transcription factor target. Anal. Biochem., 306, 290–297. [DOI] [PubMed] [Google Scholar]
- 68.Cassiday L.A. and Maher,L.J.,III (2001) In vivo recognition of an RNA aptamer by its transcription factor target. Biochemistry, 40, 2433–2438. [DOI] [PubMed] [Google Scholar]
- 69.Yoo S. and Dynan,W.S. (1998) Characterization of the RNA binding properties of Ku protein. Biochemistry, 37, 1336–1343. [DOI] [PubMed] [Google Scholar]
- 70.Peterson S.E., Stellwagen,A.E., Diede,S.J., Singer,M.S., Haimberger,Z.W., Johnson,C.O., Tzoneva,M. and Gottschling,D.E. (2001) The function of a stem–loop in telomerase RNA is linked to the DNA repair protein Ku. Nature Genet., 27, 64–67. [DOI] [PubMed] [Google Scholar]
- 71.Russel M., Gold,L., Morrissett,H. and O’Farrell,P.Z. (1976) Translational, autogenous regulation of gene 32 expression during bacteriophage T4 infection. J. Biol. Chem., 251, 7263–7270. [PubMed] [Google Scholar]
- 72.Krisch H.M. and Allet,B. (1982) Nucleotide sequences involved in bacteriophage T4 gene 32 translational self-regulation. Proc. Natl Acad. Sci. USA, 79, 4937–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McPheeters D.S., Stormo,G.D. and Gold,L. (1988) Autogenous regulatory site on the bacteriophage T4 gene 32 messenger RNA. J. Mol. Biol., 201, 517–535. [DOI] [PubMed] [Google Scholar]
- 74.Shamoo Y., Webster,K.R., Williams,K.R. and Konigsberg,W.H. (1991) A retrovirus-like zinc domain is essential for translational repression of bacteriophage T4 gene 32. J. Biol. Chem., 266, 7967–7970. [PubMed] [Google Scholar]
- 75.Shamoo Y., Tam,A., Konigsberg,W.H. and Williams,K.R. (1993) Translational repression by the bacteriophage T4 gene 32 protein involves specific recognition of an RNA pseudoknot structure. J. Mol. Biol., 232, 89–104. [DOI] [PubMed] [Google Scholar]
- 76.Andrake M., Guild,N., Hsu,T., Gold,L., Tuerk,C. and Karam,J. (1988) DNA polymerase of bacteriophage T4 is an autogenous translational repressor. Proc. Natl Acad. Sci. USA, 85, 7942–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tuerk C., Eddy,S., Parma,D. and Gold,L. (1990) Autogenous translational operator recognized by bacteriophage T4 DNA polymerase. J. Mol. Biol., 213, 749–761. [DOI] [PubMed] [Google Scholar]
- 78.Pavlov A.R. and Karam,J.D. (2000) Nucleotide-sequence-specific and non-specific interactions of T4 DNA polymerase with its own mRNA. Nucleic Acids Res., 28, 4657–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berger I., Winston,W., Manoharan,R., Schwartz,T., Alfken,J., Kim,Y.G., Lowenhaupt,K., Herbert,A. and Rich,A. (1998) Spectroscopic characterization of a DNA-binding domain, Zα, from the editing enzyme, dsRNA adenosine deaminase: evidence for left-handed Z-DNA in the Zα-DNA complex. Biochemistry, 37, 13313–13321. [DOI] [PubMed] [Google Scholar]
- 80.Schwartz T., Rould,M.A., Lowenhaupt,K., Herbert,A. and Rich,A. (1999) Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science, 284, 1841–1845. [DOI] [PubMed] [Google Scholar]
- 81.Lechler A. and Kreutzer,R. (1998) The phenylalanyl-tRNA synthetase specifically binds DNA. J. Mol. Biol., 278, 897–901. [DOI] [PubMed] [Google Scholar]
- 82.Burch L.R., Midgley,C.A., Currie,R.A., Lane,D.P. and Hupp,T.R. (2000) Mdm2 binding to a conformationally sensitive domain on p53 can be modulated by RNA. FEBS Lett., 472, 93–98. [DOI] [PubMed] [Google Scholar]
- 83.Chu E. and Allegra,C.J. (1996) The role of thymidylate synthase as an RNA binding protein. Bioessays, 18, 191–198. [DOI] [PubMed] [Google Scholar]
- 84.Lin X., Mizunuma,N., Chen,T., Copur,S.M., Maley,G.F., Liu,J., Maley,F. and Chu,E. (2000) In vitro selection of an RNA sequence that interacts with high affinity with thymidylate synthase. Nucleic Acids Res., 28, 4266–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ho Y., Kim,S.-J. and Waring,R.B. (1997) A protein encoded by a group I intron in Aspergillus nidulans directly assists RNA splicing and is a DNA endonuclease. Proc. Natl Acad. Sci. USA, 94, 8994–8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wank H., SanFilippo,J., Singh,R.N., Matsuura,M. and Lambowitz,A.M. (1999) A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol. Cell, 4, 239–250. [DOI] [PubMed] [Google Scholar]
- 87.Saldanha R., Chen,B., Wank,H., Matsuura,M., Edwards,J. and Lambowitz,A.M. (1999) RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry, 38, 9069–9083. [DOI] [PubMed] [Google Scholar]
- 88.Singh N.N. and Lambowitz,A.M. (2001) Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J. Mol. Biol., 309, 361–386. [DOI] [PubMed] [Google Scholar]
- 89.Wassarman K.M. and Storz,G. (2000) 6S RNA regulates E. coli RNA polymerase activity. Cell, 101, 613–623. [DOI] [PubMed] [Google Scholar]