Abstract
Rrp5p is the only protein so far known to be required for the processing of yeast pre-rRNA at both the early sites A0, A1 and A2 leading to 18S rRNA and at site A3, the first step specific for the pathway leading to 5.8S/25S rRNA. Previous in vivo mutational analysis of Rrp5p demonstrated that the first 8 of its 12 S1 RNA-binding motifs are involved in the formation of the ‘short’ form of 5.8S rRNA (5.8SS), which is the predominant species under normal conditions. We have constructed two strains in which the genomic RRP5 gene has been replaced by an rrp5 deletion mutant lacking either S1 motifs 3–5 (rrp5-Δ3) or 5–8 (rrp5-Δ4). The first mutant synthesizes almost exclusively 5.8SL rRNA, whereas the second one still produces a considerable amount of the 5.8SS species. Nevertheless, both mutations were found to block cleavage at site A3 completely. Instead, a novel processing event occurs at a site in a conserved stem–loop structure located between sites A2 and A3, which we have named A4. A synthetic lethality screen using the rrp5-Δ3 and rrp-Δ4 mutations identified the REX4 gene, which encodes a non-essential protein belonging to a class of related yeast proteins that includes several known 3′→5′ exonucleases. Inactivation of the REX4 gene in rrp5-Δ3 or rrp-Δ4 cells abolished cleavage at A4, restored cleavage at A3 and returned the 5.8SS:5.8SL ratio to the wild-type value. The sl phenotype of the rrp5Δ/rex4– double mutants appears to be due to a severe disturbance in ribosomal subunit assembly, rather than pre-rRNA processing. The data provide direct evidence for a crucial role of the multiple S1 motifs of Rrp5p in ensuring the correct assembly and action of the processing complex responsible for cleavage at site A3. Furthermore, they clearly implicate Rex4p in both pre-rRNA processing and ribosome assembly, even though this protein is not essential for yeast.
INTRODUCTION
Ribosome production is one of the major metabolic activities in all types of cells and this process has been highly conserved during evolution. The mature rRNA species (with the exception of eukaryotic 5S rRNA) are encoded by multiple, polycistronic units that each give rise to a single precursor transcript. This transcript then undergoes a complex series of modifications including methylation and pseudouridylation as well as endo- and exonucleolytic cleavages to remove the transcribed spacer regions. Concomitantly, the various precursor intermediates are assembled with the ribosomal proteins in an ordered fashion. In eukaryotes the majority of these reactions take place in the nucleolus and require the participation of numerous non-ribosomal, trans-acting factors.
The processing pathway leading to the eukaryotic, mature rRNA species is best characterized in the yeast Saccharomyces cerevisiae (1–3; Fig. 1). The genome of S.cerevisiae contains 150–200 tandemly repeated rDNA units, located on chromosome XII, each consisting of an 18S, 5.8S and 25S rRNA gene separated by two internal transcribed spacers (ITS1 and ITS2) and flanked by the 5′- and 3′-external transcribed spacers (ETS; Fig. 1A). Transcription of an rDNA repeat by RNA polymerase I results in a pre-rRNA that is co-transcriptionally processed at its 3′-end by Rnt1p to produce the 35S precursor (4,5). Cleavage of the 35S pre-rRNA at the early sites A0 within the 5′-ETS, A1, the mature 5′-end of 18S rRNA, and A2 within ITS1 then gives rise to the 20S and 27SA2 precursor species. Components essential for these processing cleavages include several snoRNP species as well as a host of protein factors, of which the precise role of many is still unclear (1,2,6). Inactivation of any of these trans-acting factors inhibits formation of mature 18S rRNA indicating a strong functional interdependence.
Figure 1.
Processing of pre-rRNA in S.cerevisiae. (A) Structure of the rDNA transcription unit. Mature rRNA sequences are indicated by the thick bars, transcribed spacer sequences by the thin lines. (B) The pre-rRNA processing pathway. The processing cleavages requiring Rrp5p are show in large type. Straight arrows indicate endonucleolytic cleavages, kinked arrows represent exonucleolytic processing events. Note that the manner in which the 27SA2 pre-rRNA is converted into 27SBL is not known.
The 20S pre-rRNA is exported to the cytoplasm where it is cleaved at site D to remove the remaining portion of ITS1 and generate the mature 18S rRNA (7,8). The majority (∼90%) of the 27SA2 precursor undergoes endonucleolytic cleavage at site A3 by RNaseMRP (9–12). The resulting 27SA3 pre-rRNA is further processed by the 5′→3′ exonucleases Rat1p and Xrn1p into the 27SBS species having the mature 5′-end of 5.8SS rRNA (13,14). The remaining 10% of the 27SA2 pre-rRNA is converted into the 27SBL precursor, which has the 5′-end of 5.8SL rRNA located 6 nt upstream from that of its 5.8SS counterpart. The precise mechanism of this alternative processing pathway is unknown. It is still unclear why eukaryotic cells contain two forms of 5.8S rRNA. However, differences in the pattern of translation have been observed between yeast cells having the normal, high 5.8SS:5.8SL ratio and those in which this ratio is reversed (10).
Both 27SBS and 27SBL pre-rRNA are further processed in an identical manner. Cleavage at site C2 within ITS2 separates the 7S and 25.5S pre-rRNAs (15). The former is converted into 5.8S rRNA by multistep 3′→5′ exonucleolytic trimming, which involves the exosome (16–18) as well as the small exonucleases Rex1p and Rex2p (19). The portion of ITS2 still present in the 25.5S precursor is removed exonucleolytically by Rat1p and Xrn1p (15). Again, a considerable number of additional trans-acting factors have been implicated in these steps (1,2,20–27)
Although under normal conditions RNaseMRP acts upon the 27SA2 precursor, it is also able to use the 35S pre-rRNA as its substrate when cleavage at the early sites is blocked (1,2). Thus, the set of trans-acting factors required for these early cleavages on the one hand and by RNaseMRP on the other appear to be able to act independently from each other. This conclusion is supported by the fact that correct formation of 18S and 5.8S/25S rRNA occurs even if the genes are encoded in trans (28). Nevertheless, there is evidence for interdependence between formation of the small and large subunit rRNAs both from mutational analysis of ITS1 (29) and experiments involving genetic depletion of the snR30 snoRNP (13). A direct link between the processing reactions required for 18S and 5.8S rRNA formation was established with the identification of the yeast RRP5 gene (30). Genetic depletion of Rrp5p inhibits production of both 18S and 5.8SS rRNA by blocking cleavage at sites A0, A1 and A2 as well as A3. The 193 kDa Rrp5p protein has a bipartite structure: the N-terminal two-thirds contain 12 S1 RNA-binding motifs, while the remaining C-terminal region consists mainly of seven tetratricopeptide (TPR) motifs (31,32). In vivo mutational analysis has established a direct correlation between this domain structure and the function of Rrp5p in formation of the 18S and 5.8SS rRNAs, respectively. Mutations in some of the TPR motifs block the former, whereas deletions in the S1 domain strongly reduce the latter (31,32), but stimulate the production of 5.8SL rRNA, which then becomes the dominant species in the mutant cells. We have now further analyzed the effect on pre-rRNA processing of two of these mutations, rrp5-Δ3, lacking motifs 3–5, and rrp5-Δ4, lacking motifs 5–8, which change the 5.8SS:5.8SL ratio to different extents.
MATERIALS AND METHODS
Strains and plasmids
Strain YJV154 carrying a genomic, wild-type copy of the RRP5 gene under control of the GAL promoter has been described previously (31). Strains YJV306 and YJV515 were obtained by respective replacement of the wild-type RRP5 gene of strain YJV140 by either the rrp5-Δ3 or rrp-Δ4 allele constructed previously (31). Strain FVY8C (GAL-RRP5/rex–) was obtained by crossing YJV154 with YAV41 (kindly provided by Dr van Hoof), in which the REX4 gene had been inactivated by replacing it with the kanamycin gene (19), and selecting for spores that were prototrophic for uracil (the marker for the GAL-rrp5 gene) and resistant against geneticin. Strain yRP840, the parent strain of YAV41 was also obtained from Dr van Hoof. Cloning and transformation procedures were essentially as described previously (31).
Isolation of sl mutants and identification of the gene
Isolation of sl mutants was performed essentially as described previously (33). Strain YJV515 carrying the rrp5-Δ4 allele on its genome, as well as the ade2, ade3 and ura3 markers, was transformed with plasmid pHT-RRP5 containing the wild-type RRP5, as well as the ADE3 and URA3 genes. Transformants were irradiated with 254 nm UV light at a dose resulting in ∼50% survival. A total of about 900 non-sectoring (red) colonies were selected by microscopic screening of approximately 1.9 × 105 survivors. After growth on yeast extract/peptone/dextrose (YPD) plates, red colonies were restreaked on plates containing 5-fluoroorotic acid (5-FOA). Thirty-five colonies unable to grow on these plates because of their inability to lose the pHT-RRP5(URA3) plasmid were obtained. These strains were then transformed with a plasmid containing either the wild-type RRP5 gene or the rrp5-Δ4 mutant allele and 16 strains in which the former, but not the latter, gene restored sectoring were selected.
The selected strains were transformed with the pUN100 yeast genomic library. About 8 × 104 colonies were screened for restoration of sectoring resulting in 20–50 candidates for each of the sl mutants tested. Further selection on 5-FOA plates reduced the number to 20–30 candidates per strain. To exclude strains containing a library-encoded RRP5 gene the BglII restriction pattern of the recovered library plasmids was compared with that of pHT-RRP5. The plasmids were analyzed by PCR using RRP5-specific primers. About 25% of the recovered plasmids indeed contained the wild-type RRP5 gene.
Characterization of several library plasmids recovered from sl strain Y11A6 identified a 10 kb fragment originating from chromosome XV, containing five complete open reading frames (ORFs). Further subcloning and testing of the resulting plasmids for their ability to restore sectoring in Y11A6 resulted in limitation to a fragment containing the overlapping ORFs YOL079 and YOL080 present on the Watson and the Crick strand, respectively. A deletion construct removing a large portion of YOL080 without affecting YOL079 proved unable to restore sectoring, demonstrating that the former gene is the one responsible for the sl phenotype.
RNA analysis
Isolation of RNA, northern hybridization and primer extension analysis were carried out as described previously (34). The position of the primers is indicated in Figure 2A. The sequences of these primers are given in Table 1.
Figure 2.
Northern analysis of low-molecular-weight processing intermediates in rrp5-Δ3 and rrp-Δ4 cells. Cultures of strains YJV306 (rrp5-Δ3), YJV515(rrp5-Δ4) and the wild-type parental strain YJV140 were grown on YPD medium to mid-exponential phase. Samples containing identical amounts of cells were taken before addition of 0.2 M LiCl and after further incubation in the presence of LiCl for 2 and 6 h. Total RNA was isolated, separated on 8% polyacrylamide gels and blotted onto nitrocellulose filters which were then hybridized in sequence with the probes indicated in (A). (A) The region of the pre-rRNA spanning ITS1, 5.8S rRNA and part of ITS2, including the processing sites. The position of the different oligonucleotides used for northern hybridization and primer extension experiments is indicated. The sequences of these oligonucleotides are shown in Table 1. (B–H) Northern hybridization. Lanes 1–3, YJV140; lanes 4–6, YJV306; lanes 7–9, YJV515. Lanes 1, 4 and 7, samples taken before addition of LiCl; lanes 2, 5 and 8, samples taken 2 h after addition of LiCl; lanes 3, 6 and 9, samples taken 4 h after addition of LiCl. Lane 10 in (E) shows hybridization of probe 4 to RNA isolated from the wild-type strain yRP840 (19) 2 h after addition of LiCl.
Table 1. Oligonucleotides used for northern hybridization and primer extensions analysis.
| No.a | Sequence (5′→3′) |
|---|---|
| 1 | GCTCTCATGCTCTTGCCAAAAC |
| 2 | GATATGAAAACTCCACAGTG |
| 3 | TTTGGGCATTCGAGCAATCGG |
| 4 | CCAGTTACGAAAATTCTTGTTTTTGAC |
| 5 | CTGCGTTCTTGATCGATGCG |
| 6 | GAATGTTTGAGAAGGAAATGACGCTC |
| 7 | CGCCTAGACGCTCTCTTCTTA |
aThe positions of the oligonucleotides are shown in Figure 2A.
Sucrose gradient analysis
Strains YJV154 or FVY88C transformants were grown on minimal medium with galactose as the carbon source and shifted to glucose-based medium. After 24 h, cycloheximide was added at the final concentration of 50 µg/ml and the cells were cooled to 0°C. Cells were harvested by centrifugation, washed with 10 ml lysis buffer [20 mM HEPES pH 7.4, 100 mM KCl, 2 mM MgAc2, 50 µg/ml cycloheximide, 200 µg/ml heparin, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF)] and stored at –20°C. Extracts were prepared by adding 1 ml of lysis buffer to the cell pellet and vortexing with 0.5 mg glass beads 10 times for 30 s with intermittent cooling on ice. The resulting lysate was cleared by centrifuging twice in an Eppendorf centrifuge at 13 000 r.p.m. for 20 min and the OD260 was determined. About 20 OD260 units of extract were layered on 15–35% sucrose gradients in 10 mM Tris–HCl pH 7.4, 70 mM NH4Cl, 4 mM MgAc2, 1 mM DTT, and the gradients were centrifuged for 17 h at 18 000 r.p.m. in an SW28 rotor (Beckman Instruments) at 4°C. For analysis of polysome profiles we used 10–50% gradients followed by centrifugation for 17 h at 13 000 r.p.m. Gradients were analyzed by continuous scanning of the OD254.
RESULTS
The rrp5-Δ3 and rrp5-Δ4 deletions shift processing within ITS1 from site A3 to a novel, upstream site
Using a yeast strain in which the genomic RRP5 gene is conditionally expressed under control of the GAL promoter, we have shown that deletion of S1 motifs 1–2 (rrp5-Δ2), 3–5 (rrp5-Δ3) or 5–8 (rrp5-Δ4) from the N-terminal region of the Rrp5p protein lowers production of 5.8SS rRNA to different extents. At the same time formation of its 5.8SL counterpart is increased to approximately the same extent (31). The first two mutations also lowered the growth rate ∼2-fold, whereas the rrp5-Δ4 mutation did not significantly affect growth. We, therefore, decided to use the rrp5-Δ3 and rrp5-Δ4 mutations to study the role of Rrp5p in the formation of 5.8S rRNA further. As neither deletion is lethal we constructed mutant yeast strains in which the genomic copy of the RRP5 gene had been replaced by either of the mutant alleles (see Materials and Methods). The resulting YJV306 (rrp5-Δ3) and YJV515 (rrp5-Δ4) strains were found to grow 2 and 1.4 times more slowly than the parent (YJV140) wild-type strain, respectively (data not shown). In both cases this is slightly below the rate observed previously for yeast cells that have been switched from wild-type to the respective mutant Rrp5p by transfer from galactose- to glucose-based medium. This difference is probably due to residual production of wild-type Rrp5p in cells containing the GAL-rrp5 gene even when grown on glucose, whereas the integrants of course do not contain any wild-type protein.
Total RNA was isolated from exponentially growing YJV140, YJV306 and YJV515 cells and subjected to northern analysis. Because the two rrp5-Δ mutations predominantly affect synthesis of 5.8SS rRNA (31), we focused our attention on processing within ITS1, using the set of probes depicted in Figure 2A. RNA was also isolated after treating the cultures with LiCl for 2 and 6 h to facilitate detection of 5′-extended forms of 5.8S rRNA (35).
The results of the northern analysis are depicted in Figure 2B–H. In agreement with our previous results (31), YJV306 contains only a very small amount of the 5.8SS species, while the level of 5.8SL is increased ∼10-fold (Fig. 2B, lanes 4–6). The rrp5-Δ4 mutant also shows a clear predominance of 5.8SL rRNA, but still produces a considerable amount of the 5.8SS species (Fig. 2B, lanes 7–9). Again the effects of the integrated rrp5-Δ3 and rrp5-Δ4 mutant alleles are more pronounced than seen previously in the carbon-source shift experiments.
Addition of LiCl to yeast cells results in the accumulation of 5′-extended forms of 5.8S rRNA because LiCl strongly inhibits the 5′→3′ exonucleases Rat1p and Xrn1p, responsible for the 27SA3→27SBL processing step, as well as, to a lesser extent, RNase MRP which converts 27SA2 into 27SA3 pre-rRNA (35). However, cleavage of the 27SA2 and 27SA3 precursor species within ITS2 and the subsequent removal of the ITS2 spacer sequences from 7S pre-rRNA can still proceed. Figure 2C shows that upon LiCl treatment both the wild-type strain and the rrp5 deletion mutants indeed accumulate a product that hybridizes to probe 2, located between site A2 and A3, and that according to its mobility corresponds to the A2-E fragment. The same product is also recognized by probes 3 and 4 (Fig. 2D and E). Probe 6 (Fig. 2G) should not detect the A2-E fragment because it hybridizes only to RNA molecules that still contain at least a portion of ITS2. The band seen in Figure 2G, therefore, represents 7S pre-rRNA, which has almost the same size as the A2-E fragment. Probe 5 (Fig. 2F) detects both the A2-E fragment and 7S pre-rRNA. Furthermore, we can also detect the complementary D-A2 fragment of ITS1 in all three strains upon hybridization with probe 1 (Fig. 2H). Somewhat unexpectedly, we did not observe accumulation of the A3-E fragment upon hybridization with probes 4 or 5 (Fig. 2E and F). We surmise that this is due to the genetic makeup of our strains, because, as shown in Figure 2E (lane 10), the A3-E fragment could be visualized by probe 4 in another RRP5 wild-type strain, yRP840 (19), after treatment with LiCl for 2 h. Interestingly, however, probes 4 and 5 did detect a fragment in both the rrp5-Δ3 and rrp5-Δ4 strains that is absent from the wild-type cells (Fig. 2E and F, lanes 4–9). The same fragment also hybridizes with probe 3, complementary to the region of ITS1 directly upstream from site A3 (Fig. 2D, lanes 4–9), but not with either probe 2 (Fig. 2C) or probe 6 (Fig. 2G). The data indicate that in the rrp5-Δ3 and rrp5-Δ4 cells a novel processing event occurs within ITS1, either in addition to or instead of cleavage at A3. As judged from the electrophoretic mobility of the novel fragment its 5′-end, which we have called A4, is located approximately midway between A2 and A3.
In order to establish whether processing at A4 indeed replaces that at A3 and to map the location of the A4 site more precisely, we carried out primer extension experiments on total RNA isolated from wild-type, rrp5-Δ3 and rrp5-Δ4 cells either before or after treatment with LiCl using probe 4. As shown in Figure 3A a stop corresponding to A3 is readily detectable in the wild-type strain upon LiCl treatment. In the two mutant strains, however, this stop has disappeared completely, indicating that cleavage at A3 is blocked completely. Instead, a new set of stops appears 34–36 nt farther upstream. Together with the results of the northern hybridization shown in Figure 2 the data firmly establish the occurrence of an alternative-processing event within ITS1 in the rrp5 deletion mutants. Furthermore, our results show that there is no absolute link between cleavage at A3 and the formation of 5.8SS rRNA. Despite the absence of any detectable processing at site A3 in the rrp5-Δ4 mutant, in these cells ∼40% of 5.8S rRNA still is the short form, which, therefore, should originate from the 27SA2 and/or 27SA4 intermediate. The existence of such a 27SA4 processing intermediate was corroborated by primer extension experiments using probes 6 and 7, located upstream and downstream of site C2 within ITS2, respectively (Fig. 2A). In both cases we observe a clear signal corresponding to site A4 in the samples isolated from LiCl-treated rrp5-Δ3 and rrp5-Δ4 mutant cells, while the signal corresponding to site A3 has disappeared. In particular the detection of site A4 with probe 7 is convincing evidence for the presence of a 27SA4 precursor.
Figure 3.

Primer extension analysis of precursor intermediates in the rrp5-Δ3 and rrp-Δ4 cells. Total RNA was isolated from exponentially growing wild-type and mutant cells immediately before and 2 h after addition of 0.2 M LiCl. Primer extension analysis was carried out using probes 4 (A), 6 (B) or 7 (C; see Fig. 2A for location of the probes). The positions of the primer extension stops corresponding to processing sites A2, A3 and A4 are indicated. In (B) and (C) only the samples from LiCl-treated cells are shown.
As shown in Figure 4 the A4 processing site maps to a region of the spacer that has been well conserved with respect to both primary and secondary structure in a wide variety of yeast species. The A4 site is located within a 5′-UCUUUG-3′ consensus sequence that is part of a stem shared by all known yeast ITS1 regions. There is no obvious structural similarity between the A4 site and either the A2 or the A3 site.
Figure 4.
Structural conservation around site A4 among various yeast species. (A) Sequence comparison of the region of ITS1 spanning site A4 that is highlighted in (B). Nucleotides in upper case are part of the stem, nucleotides in lower case form the loop of the conserved hairpin structure containing site A4. S.c., S.cerevisiae; S.t., Saccharomyces transvaalensis; T.d., Torulospora delbrückii; K.m., Kluyveromyces marxianus; H.w., Hansenula wingei; A.t., Arxioma telluris; D.h., Debaromyces hansenii; Z.b., Zygosaccharomyces bisporus. (B) Schematic representation of the structure of the S.cerevisiae ITS1 according to Yeh et al. (40). The stem–loop structure formed by the sequence shown in (A) is highlighted.
Synthetic lethality with rrp5-Δ4 identifies the REX4 gene
In order to obtain more information on the manner in which Rrp5p functions in pre-rRNA processing we carried out a synthetic lethality screen with the rrp5-Δ4 mutation using the ade2/ade3 red-white colony sectoring assay, essentially as described previously (33) (see Materials and Methods for details). Strain YJV515 carrying the rrp5-Δ4 mutation as well as the wild-type RRP5 gene on an ADE3/URA3 plasmid were mutagenized by UV irradiation and surviving colonies were screened microscopically for non-sectoring mutants, indicating the inability to lose the plasmid. The presence of a mutation that is sl with the rrp5-Δ4 deletion was then confirmed by streaking the non-sectoring mutants onto plates containing 5-fluorouracil which should inhibit growth because of the obligatory presence of the URA3 plasmid carrying the wild-type RRP5 gene. In this manner we isolated 16 mutants that regained sectoring when transformed with a wild-type RRP5 gene but not with an empty vector, demonstrating that they carry an unlinked mutation, which is sl with the rrp5-Δ4 mutation. These mutations were then characterized by transforming the strains in question with a yeast genomic library and recovering the library plasmid from resectoring colonies. Library plasmids containing the wild-type RRP5 gene were identified by PCR analysis using RRP5-specific primers. Restriction mapping and sequence analysis of the remaining plasmids recovered from one of the sl strains (Y11A6) identified a 10 kb region of chromosome XV containing two partial (IRA2 and DEC1) and four complete ORFs (YOL077, YOL078, YOL079, ATP19 and YOL080). By subcloning portions of this region the ability to restore sectoring in the Y11A6 strain was limited to a fragment that contained only ORFs YOL079 and YOL080, which are present on opposite DNA strands and overlap almost completely. To distinguish between these two ORFs, we first created a deletion that removed a large C-terminal portion of YOL080 without affecting YOL079. The resulting plasmid was unable to restore sectoring in Y11A6 cells. Secondly, mutation of the translation start codon of YOL079 did not affect the ability of the plasmid to restore sectoring. Therefore, YOL080 clearly carries the mutation that is sl with rrp5-Δ4 in these cells. Sequencing of the sl allele of YOL080 revealed a single mutation that changes the codon for Lys274 to a stop codon.
Recently, van Hoof et al. (19) showed that YOL080 encodes a member of a set of five related, non-essential, yeast proteins that includes Pan2p, the 3′→5′ exonuclease involved in initial shortening of the poly(A) tails of mRNA (36). The other four members were called Rex1p, Rex2p, Rex3p and Rex4p, the last one being the product of YOL080. The Rex proteins also contain the 3′→5′ exonuclease signature domain at their C-terminus (37,38). Rex1p–Rex3p were indeed found to be 3′→5′ exonucleases, which have distinct, but overlapping, roles in 3′-end maturation of several rRNA, snRNA and snoRNA species (19). A rex4 null mutant, however, does not display any detectable disturbance in RNA maturation (19; our unpublished data), leaving the enzymatic activity of the Rex4p protein to be established.
We crossed strain YJV515 (rrp5-Δ4) with strain YAV41 in which REX4 had been inactivated by insertion of the kanamycin gene (kindly provided by Dr van Hoof). Analysis of 20 different tetrads demonstrated that viable spores producing cells that were auxotrophic for histidine (thus containing the rrp5-Δ4 allele) invariably were also sensitive to geneticine (indicating the presence of an intact REX4 gene). This further supports the existence of a genetic interaction between the RRP5 and REX4 genes. A similar cross using strain YJV306 led to the same result, showing that absence of Rex4p is also sl with the rrp5-Δ3 deletion (data not shown).
Inactivation of REX4 restores normal ITS1 processing in the rrp5-Δ strains
To study the functional relationship between Rrp5p and Rex4p we used strain YJV154 in which the genomic, wild-type RRP5 gene is under control of the inducible GAL promoter (31). This strain was crossed with the rex4 null mutant YAV41 (19) to give FVY8C (GAL-rrp5/rex4–). The latter strain was then transformed with a plasmid carrying either the rrp5-Δ3 or rrp5-Δ4 mutant allele under control of the authentic RRP5 promoter. Transformants were grown on galactose and then shifted to glucose-based medium to block production of wild-type Rrp5p. For the FVY8C transformants this causes growth to stop after ∼23 h (data not shown). Total RNA was isolated immediately before and 24 h after the shift and analyzed by northern hybridization as well as primer extension analysis; the results are shown in Figures 5 and 6, respectively.
Figure 5.
Effect of inactivation of REX4 on formation of mature rRNA in rrp5-Δ3 and rrp-Δ4. Strain FVY8C carrying a genomic wild-type RRP5 gene under control of the GAL promoter, a rex4 null allele and a plasmid-encoded rrp5-Δ3 or rrp-Δ4 gene under control of the authentic RRP5 promoter was shifted from galactose- to glucose-based medium and total RNA was isolated immediately before and 24 h after the shift. The RNA was subjected to northern analysis using probes complementary to the mature 5.8S, 18S and 25S rRNAs [(A), (C) and (D), respectively] as well as probe 3 (see Fig. 2A) located directly upstream from cleavage site A3 (B). Strain YJV154, which is isogenic with FVY8C except for the presence of a wild-type REX4 gene, was used as a control. Lanes 1, 3, 5 and 7, RNA from galactose-grown cells. Lanes 2, 4, 6 and 8, RNA from cells shifted to glucose for 24 h. The nature of the products detected is indicated.
Figure 6.

Effect of inactivation of REX4 on ITS1 processing in rrp5-Δ3 and rrp-Δ4 mutant cells. Strain FVY8C carrying a genomic wild-type RRP5 gene under control of the GAL promoter, a rex4 null allele and a plasmid-encoded rrp5-Δ3 or rrp-Δ4 gene under control of the authentic RRP5 promoter was shifted from galactose- to glucose-based medium and total RNA was isolated immediately before and 24 h after the shift. The RNA was subjected to primer extension analysis using probe 4, located between sites A3 and B1 (see Fig. 2A). The position of the stops corresponding to cleavage sites A2, A3 and A4 is indicated. Lanes 1, 3, 5 and 7, RNA from galactose-grown cells. Lanes 2, 4, 6 and 8, RNA from cells shifted to glucose for 24 h.
As is evident from Figure 5, in cells expressing wild-type Rrp5p the absence of Rex4p does not influence the formation of the mature rRNA species. When grown on galactose, the REX4 (YJV154) and rex4– (FVY8C) cells have the same, normal ratio of 5.8S:5.8SL rRNA as well as the same levels of 18S and 25S rRNA (Fig. 5A, C and D, compare lanes 5 and 7 with lanes 1 and 3, respectively). Also, northern hybridization did not detect any significant differences in the levels of the various processing intermediates between galactose-grown YJV154 and FVY8C transformants (data not shown), in agreement with previous observations (19). However, when the transformants are made dependent upon the Rrp5pΔ3 or Rrp5pΔ4 mutant protein by growing them on glucose, the lack of Rex4p does have a surprising effect: in the absence of the Rex4p protein the shift in the ratio of 5.8S:5.8SL characteristic of the respective rrp5-Δ mutation does not occur (Fig. 5B, lanes 6 and 8). Moreover, the A4-E fragment, which can be clearly seen in the rrp5-Δ3 and rrp-Δ4 cells that carry an intact REX4 gene (Fig. 5B, lanes 2 and 4), does not accumulate in their rex4– counterparts (Fig. 5B, lanes 6 and 8). Thus, inactivation of the REX4 gene corrects the abnormal processing within ITS1 caused by the rrp5-Δ3 and rrp-Δ4 mutations. There is no change in the levels of the mature, high-molecular-weight rRNA species in the rrp5-Δ3/rex4– or rrp5-Δ4/rex4– mutants (Fig. 5C and D).
Figure 6 shows the results of primer extension experiments, which are in full agreement with the conclusion drawn from the northern hybridization data. Using primer 4 (Fig. 2A) we analyzed RNA isolated from YJV154 and FVY8C cells carrying the rrp5-Δ3 or rrp-Δ4 plasmid either before or 24 h after a shift from galactose to glucose. Cells containing the intact REX4 gene show a clear reduction in the signal corresponding to the stop at A3 after the shift and the concomitant appearance of the stops corresponding to the A4 site (compare lanes 2 and 4 with lanes 1 and 3, respectively). However, in the rex4– transformants we do not see any change in the signals corresponding to the A3 and A4 cleavage sites upon repression of the wild-type RRP5 gene (compare lanes 6 and 8 with lanes 5 and 7, respectively). We conclude that the Rex4p protein is essential for redirecting ITS1 processing in both the rrp5-Δ3 and rrp5-Δ4 mutant from the normal (A3) to the alternative (A4) pathway.
Inactivation of REX4 in cells dependent on the Rrp5pΔ3 or Rrp5pΔ4 protein leads to severe underproduction of 80S ribosomes
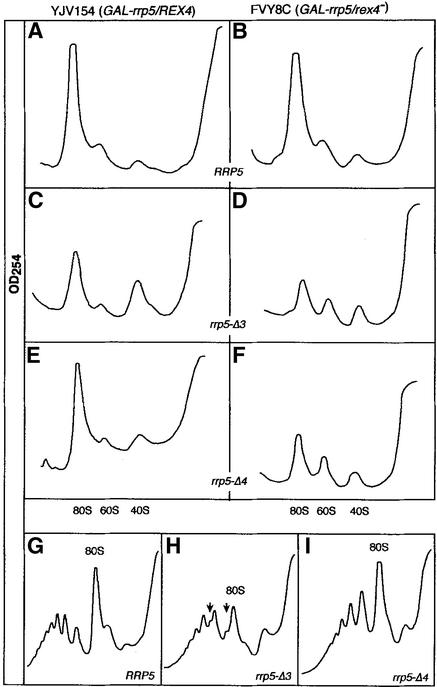
The observation that rex4– cells expressing either of the Rrp5pΔ mutant proteins possess normal levels of the pre-rRNA intermediates and mature rRNA species came as a surprise in view of the sl phenotype of such cells. In order to see whether the sl phenotype might nevertheless be due to a disturbance in ribosome biogenesis we carried out sucrose gradient analysis of extracts from YJV154 (GAL-rrp5/REX4) as well as FVY8C (GAL-rrp5/rex4–) cells carrying the wild-type RRP5 gene or either of the two rrp5 deletion alleles on a plasmid. All strains were initially grown on galactose and then shifted to glucose-based medium for 24 h prior to preparation of the extracts. As shown in Figure 7 the rrp5-Δ3 and rrp-Δ4 mutations themselves already have an effect on ribosome biogenesis. The sucrose gradient profiles of glucose-grown YJV154 transformants that carry either of the plasmid-encoded rrp5 deletion alleles reveal a significant deficit in 60S subunits apparent from the lower 60S and 80S peaks relative to the 40S subunit peak (Fig. 7, compare C and E with A). We have confirmed this by analyzing the polysome profile of these cells, which in the case of the rrp5-Δ3 mutant clearly shows the presence of ‘halfmer’ polyribosomes, mRNA molecules carrying a single 40S subunit in addition to one or more 80S ribosomes, which is characteristic of the underproduction of 60S subunits (Fig. 7H). We did not detect halfmer polyribosomes in the rrp5-Δ4 mutant (Fig. 7I) suggesting that the effect of this mutation on 60S subunit formation is less severe than that of its Δ3 counterpart. This is in agreement with the relative growth rates of cells expressing either protein (31; this paper).
Figure 7.
Effect of inactivation of the REX4 gene on ribosome biogenesis in rrp5-Δ3 and rrp-Δ4 cells. Cultures of YJV154 (GAL-rrp5/REX4) and FVY8C (GAL-rrp5/rex4–) cells carrying either the wild-type RRP5 gene or one of the rrp5-Δ deletion alleles on a plasmid were shifted from galactose- to glucose-based medium and extracts were prepared 24 h later. 20 OD260 units of extract were analyzed on 15–35% (A–F) or 10–50% sucrose gradients (G–I). Sedimentation is from right to left. (A, C and E) Extracts from strain YJV154 carrying the wild-type RRP5 (A), rrp5-Δ3 (B) or rrp-Δ4 (C) allele. (B, D and F) Extracts from strain FVY8C carrying the wild-type RRP5 (B), rrp5-Δ3 (D) or rrp5-Δ4 (F) allele. (G–I) Polysome profiles for YJV154 carrying the wild-type RRP5 (G), rrp5-Δ3 (H) or rrp5-Δ4 (I) allele. The arrows in panel H indicate the ‘halfmers’.
Introduction of the rex4 null mutation into cells producing wild-type Rrp5p has no detectable effect on the ribosome profile (Fig. 7B). A distinct change can be seen, however, in glucose-grown FVY8C (GAL-rrp5/rex4–) cells expressing the Rrp5pΔ3 or Rrp5pΔ4 mutant protein (Fig. 7D and F). Apart from the decreased level of 60S subunits discussed above, these cells show a dramatic reduction in the amount of 80S ribosomes. Despite the absence of detectable abnormalities in pre-rRNA processing in these cells, therefore, ribosome biogenesis is severely disturbed, which explains the sl phenotype of combining the rrp5-Δ3 or rrp5-Δ4 and rex4– mutations.
DISCUSSION
Genetic depletion of Rrp5p blocks synthesis of both 18S rRNA, requiring the snoRNP-dependent cleavages at A0–A2, as well as 5.8SS rRNA, which depends upon the RNaseMRP-directed cleavage at A3. This led to the hypothesis that the major role of Rrp5p is to ensure functional integration of the many trans-acting factors required for accurate and efficient pre-rRNA processing at these sites (30). Subsequent mutational analysis (31,32) demonstrated that the C-terminal region of the protein, containing seven TPR motifs, is specifically involved in 18S rRNA synthesis, whereas the N-terminal region, encompassing 12 S1 RNA-binding motifs, is crucial for production of the 5.8SS rRNA. Thus, the C-terminal domain might interact with the snoRNP complex that carries out the early processing cleavages, while the N-terminal domain assists RNase MRP. The data presented in this paper constitute direct experimental proof for the latter suggestion as they demonstrate that removal of either S1 motifs 3–5 (rrp5-Δ3) or 5–8 (rrp5-Δ4) leads to a complete block in the RNase MRP-directed cleavage at site A3 (Fig. 3).
Interestingly, our experiments reveal a second effect of the rrp5-Δ3 and rrp-Δ4 mutations, namely the occurrence of a hitherto unobserved processing event in ITS1 at a site about midway between A2 and A3 which we have designated A4 (Fig. 3). The simultaneous presence of substantial amounts of an A4-E fragment (Fig. 2D, E and F), normal amounts of the D-A2 fragment (Fig. 2H) and, in particular, the detection of the A4 site in primer extension experiments using probes 6 and 7 complementary to different regions of ITS2 clearly support the conclusion that the rrp5-Δ3 and rrp-Δ4 mutations cause the processing machinery to bypass A3 and instead to convert the 27SA2 pre-rRNA into an alternative, novel 27SA4 precursor species.
The currently available data do not allow us to decide whether formation of the 27SA4 species is an endo- or an exonucleolytic event. However, we favor the former possibility for the following reasons. First, the processing intermediate having its 5′-end at A4 accumulates in the presence of LiCl, which strongly inhibits the major 5′→3′ exonucleases Xrn1p and Rat1p known to be involved in pre-rRNA processing (Figs 2 and 3). Second, the 27SA2 pre-rRNA, the probable immediate precursor of the 27SA4 species, appears to be a poor substrate for these exonucleases, in any case, at least under normal conditions (13,14). Finally, as shown in Figure 4, the A4 site is located in a region of ITS1 that has been relatively well conserved with respect to both primary and secondary structure over a wide spectrum of yeast species, suggesting that it could be the recognition site for an endonuclease, the nature of which remains to be identified. There is no obvious structural similarity of this region with those containing other known processing sites within the pre-rRNA. It should also be noted that processing at A4 might be less precise than the standard processing events. In all cases we observe a set of three bands corresponding to consecutive nucleotides, whose relative intensity varies somewhat from one experiment to another (Figs 3 and 6).
Although the rrp5-Δ3 and rrp-Δ4 deletions cause identical changes in ITS1 processing the fate of the resulting precursors differs in the two mutants. In the rrp5-Δ3 strain subsequent processing almost exclusively follows the ‘long’ pathway leading to 5.8SL rRNA. In the rrp5-Δ4 mutant, on the other hand, substantial processing still occurs via the ‘short’ pathway, presumably by exonucleolytic degradation of the 27SA4 precursor, resulting in ∼40% 5.8SS rRNA. We propose that the presence of mutant Rrp5p protein causes a structural alteration in the processing complex that reduces either the rate of entry of the exonucleases or the rate of exonucleolytic digestion, allowing processing at site B1L to get the upper hand. Clearly, the extent of this negative effect on B1S processing is different for the two mutant proteins, being largest for the rrp5-Δ3 mutation.
A synthetic lethality screen using the rrp5-Δ4 mutation resulted in the isolation of the REX4 gene, which encodes a protein belonging to a family of non-essential 3′→5′ exonucleases. Although other members of this family were shown to be involved in pre-rRNA processing, no evidence for such a role was found in the case of the Rex4p protein (19). The data presented in Figures 5 and 6, however, clearly demonstrate that inactivation of the REX4 gene does have a rather surprising effect on pre-rRNA processing in strains expressing either of the mutant Rrp5pΔ proteins. Both the rrp5-Δ3/rex4– and rrp5-Δ4/rex4– double mutants show normal ITS1 processing as well as a wild-type 5.8SS:5.8SL ratio. We conclude that the change in ITS1 processing induced by the rrp5-Δ3 and rrp-Δ4 mutations requires intact Rex4p and that in the absence of Rex4p the mutant processing complex containing either mutant Rrp5p protein regains the ability to direct RNaseMRP to the A3 site. The molecular basis for this phenomenon remains a matter of speculation. Rex4p could participate directly in ribosome biogenesis, a conceivable hypothesis in view of the fact that its human homolog resides in the nucleolus (39). Another possibility is that Rex4p plays a role in the formation of an as yet unidentified trans-acting factor that forms part of the processing complex containing Rrp5p. In wild-type cells, however, the role of Rex4p is not critical.
The lack of any detectable abnormalities in pre-rRNA processing in the rrp5-Δ3/rex4– and rrp5-Δ4/rex4– double mutants left us without an obvious explanation for the sl phenotype of these mutants. Therefore, we considered the possibility of a defect in ribosome assembly, which for obvious reasons we studied in strains in which the wild-type RRP5 gene is conditionally expressed. Sucrose gradient analysis demonstrated that in fact the rrp5-Δ3 and rrp-Δ4 mutations by themselves already cause a significant defect in 60S subunit assembly. Extracts from cells expressing the Rrp5pΔ3 protein show a clear deficit in the large, relative to the small, subunits as well as the characteristic presence of halfmer polyribosomes (Fig. 7). This could be a consequence of the almost exclusive presence of 5.8SL rRNA in these cells which might be assembled less efficiently than its smaller counterpart, similar to 3′-extended 5.8S rRNA (17). This idea is supported by the fact that the deficit in 60S subunits, in particular as judged from the absence of detectable amounts of halfmers, is less severe in the rrp5-Δ4 mutant, which contains a higher proportion of 5.8SS rRNA (31; this paper). On the other hand, the structural abnormality of the Rrp5p protein might also directly affect 60S subunit assembly in the mutant cells.
Sucrose gradient analysis of extracts prepared from the rrp5-Δ3/rex4– and rrp5-Δ3/rex4– double mutants revealed a very low amount of 80S ribosomes (Fig. 7). The combination of mutant Rrp5p and the absence of Rex4p, therefore, appears to cause a severe defect in ribosome assembly, which would explain the sl phenotype of the double mutants. This finding further supports the hypothesis that Rex4p is involved in ribosome biogenesis, although its role only becomes manifest in the presence of the mutant Rrp5p proteins. We are presently studying the role of Rex4p in ribosome biogenesis further using both genetic and biochemical approaches.
In summary, the data presented in this paper firmly establish the role of the S1 domain of Rrp5p in assisting RNaseMRP in its cleavage at site A3. The involvement of Rrp5p in this processing step seems to be rather complex, however, because the requirement for the S1 motifs in question can be abrogated by removing Rex4p. The data also indicate a further role for Rrp5p, either directly or indirectly, in the subsequent exonucleolytic processing to site B1S and demonstrate a negative effect of the two deletion mutations on 60S subunit biogenesis. Furthermore, both genetic and biochemical evidence clearly indicates the involvement of Rex4p in the assembly of yeast ribosomes, albeit in an unusual manner.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Roy Parker and Ambro van Hoof for generously providing the strains carrying inactivated copies of the various REX genes. This work was supported in part by the Council for Chemical Sciences (CW) with financial aid from the Netherlands Foundation for Scientific research (NWO).
REFERENCES
- 1.Kressler D., Linder,P. and De La Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 216–311. [DOI] [PubMed] [Google Scholar]
- 3.Lafontaine D.L. and Tollervey,D. (2001) The function and synthesis of ribosomes. Nature Rev. Mol. Cell. Biol., 2, 514–520. [DOI] [PubMed] [Google Scholar]
- 4.Abou-Elela S., Igel,H. and Ares,M.,Jr (1996) RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell, 85, 115–124. [DOI] [PubMed] [Google Scholar]
- 5.Kufel J., Dichtl,B. and Tollervey,D. (1999) Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA, 5, 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dragon F., Gallagher,J.E.G., Compagnone-Post,P.A., Mitchell,B.A., Porwancher,K.A., Wehner,K.A., Wormsley,S., Settlage,R.E., Shabanowitz,J., Osheim,Y., Beyer,A.L., Hunt,D.F. and Baserga,S.J. (2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature, 417, 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moy T.I. and Silver,P.A. (1999) Nuclear export of the small ribosomal subunit requires the Ran-GTPase cycle and certain nucleoporins. Genes Dev., 13, 2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanrobays E., Gleizes,P.-E., Bousquet-Antonelli,C., Noaillac-Depeyre,J., Caizergues-Ferrer,M. and Gélugne,J.-P. (2001) Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J., 20, 4204–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu S., Archer,R.H., Zengel,J.M. and Lindahl,L. (1994) The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc. Natl Acad. Sci. USA, 91, 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitt M.E. and Clayton,D.A. (1993) Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 7935–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dichtl B. and Tollervey,D. (1997) Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J., 16, 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lygerou Z., Allmang,C., Tollervey,D. and Séraphin,B. (1996) Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science, 272, 268–270. [DOI] [PubMed] [Google Scholar]
- 13.Henry Y., Wood,H., Morrissey,J.P., Petfalski,E., Kearsey,S. and Tollervey,D. (1994) The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J., 13, 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petfalski E., Dandekar,T., Henry,Y. and Tollervey,D. (1998) Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol., 18, 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geerlings T., Vos,J.C. and Raué,H.A. (2000) The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′→3′ exonucleases. RNA, 6, 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell P., Petfalski,E. and Tollervey,D. (1996) The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev., 10, 502–513. [DOI] [PubMed] [Google Scholar]
- 17.Briggs M.W., Burkard,K.T.D. and Butler,J.C. (1998) Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem., 273, 13255–13263. [DOI] [PubMed] [Google Scholar]
- 18.Allmang C., Kufel,J., Chanfreau,G., Mitchell,P., Petfalski,E. and Tollervey,D. (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Hoof A., Lennertz,P. and Parker,R. (2000) Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J., 19, 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burger F., Daugeron,M.-C. and Linder,P. (2000) Dbp10p, a putative RNA helicase from Saccharomyces cerevisiae, is required for ribosome biogenesis. Nucleic Acids Res., 28, 2315–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunbar D.A., Dragon,F., Lee,S.J. and Baserga,S.J. (2000) A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl Acad. Sci. USA, 97, 13027–13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strezoska Z., Pestov,D.G. and Lau,L.F. (2000) Bop1 Is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol., 20, 5516–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue Y., Bai,X., Lee,I., Kallstrom,G., Ho,J., Brown,J., Stevens,A. and Johnson,A.W. (2000) Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol., 20, 4006–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louk T.L., Aitchison,J.D., Maguire,S. and Wozniak,R.W. (2001) Rrb1p, a yeast nuclear WD-repeat protein involved in the regulation of ribosome biosynthesis. Mol. Cell. Biol., 21, 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestov D.G., Stockelman,M.G., Sttrezoska,Z. and Lau,L.F. (2001) ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res., 29, 3621–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saveanu C., Bienvenu,D., Namane,A., Gleizes,P.-E., Gas,N., Jacquier,A. and Fromont-Racine,M. (2001) Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J., 20, 6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fatica A., Cronshaw,A.D., Dlakic,M. and Tollervey,D. (2002) Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell, 9, 341–351. [DOI] [PubMed] [Google Scholar]
- 28.Liang W.-Q. and Fournier,M.J. (1997) Synthesis of functional eukaryotic ribosomal RNAs in trans: development of a novel in vitro rDNA system for dissecting ribosome biogenesis. Proc. Natl Acad. Sci. USA, 94, 2864–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allmang C., Henry,Y., Morrissey,J.P., Wood,H., Petfalski,E. and Tollervey,D. (1996) Processing of the yeast pre-rRNA at sites A2 and A3 is linked. RNA, 2, 60–73. [PMC free article] [PubMed] [Google Scholar]
- 30.Venema J. and Tollervey,D. (1996) RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J., 15, 5701–5714. [PMC free article] [PubMed] [Google Scholar]
- 31.Eppens E.A., Rensen,S., Granneman,S., Raué,H.A. and Venema,J. (1999) The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA, 5, 779–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torchet C., Jacq,C. and Hermann-le Denmat,S. (1998) Two mutant forms of the S1/TPR-containing protein Rrp5p affect the 18S rRNA synthesis in Saccharomyces cerevisiae. RNA, 4, 1636–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venema J., Bousquet-Antonelli,C., Gelugne,J.-P., Caizergues-Ferrer,M. and Tollervey,D. (1997) Rok1p is a putative RNA helicase required for rRNA processing. Mol. Cell. Biol., 17, 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venema J., Vos,H., Faber,A.W., van Venrooij,W.J. and Raué,H.A. (2000) Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for the early pre-rRNA processing cleavages and requires box C for its association. RNA, 6, 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dichtl B., Stevens,A. and Tollervey,D. (1997) Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J., 16, 7184–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown C.E. and Sachs,A.B. (1998) Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol., 18, 6548–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moser M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res., 25, 5110–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuo Y. and Deutscher,M.P. (2001) Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res., 29, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersen J.S., Lyon,C.E., Fox,A.H., Leung,A.K.L., Lam,Y.W., Steen,H., Mann,M. and Lamond,A.I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol., 12, 1–11. [DOI] [PubMed] [Google Scholar]
- 40.Yeh L.-C., Thweatt,R. and Lee,J.C. (1990) Internal transcribed spacer 1 of the yeast precursor ribosomal RNA. Higher order structure and common structural motifs. Biochemistry, 29, 5911–5918. [DOI] [PubMed] [Google Scholar]