Abstract
The housekeeping gene of the major adenylate kinase in Saccharomyces cerevisiae (AKY2, ADK1) is constitutively transcribed at a moderate level. The promoter has been dissected in order to define elements that effect constitutive transcription. Initiation of mRNA synthesis at the AKY2 promoter is shown to be mediated by a non-canonic core promoter, (TA)6. Nucleotide sequences 5′ of this element only marginally affect transcription suggesting that promoter activation can dispense with transactivators and essentially involves basal transcription. We show that the core promoter of AKY2 is constitutively kept free of nucleosomes. Analyses of permutated AKY2 promoter DNA revealed the presence of bent DNA. DNA structure analysis by computer and by mutation identified two kinks flanking an interstitial stretch of 65 bp of moderately bent core promoter DNA. Kinked DNA is likely incompatible with packaging into nucleosomes and responsible for positioning nucleosomes at the flanks allowing unimpeded access of the basal transcription machinery to the core promoter. The data show that in yeast, constitutive gene expression can dispense with classical transcriptional activator proteins, if two prerequisites are met: (i) the core promoter is kept free of nucleosomes; this can be due to structural properties of the DNA as an alternative to chromatin remodeling factors; and (ii) the core promoter is pre-bent to allow a high rate of basal transcription initiation.
INTRODUCTION
Transcription activation of regulated genes is generally assumed to rely on interactions of specific transcriptional activator proteins with one or more constituents of the polymerase II holoenzyme complex. The prime role of transactivators is either to effect remodeling or removal of nucleosomes repressing the promoter in its inactive state or to enhance the rate of transcription initiation by recruiting, directly or indirectly, the basal transcription machinery to the core promoter (1–7). Some factors may exert both activities. The first type of transcription factors will induce recruitment either of additional transactivators or of the basal transcription machinery to their target sequences without exerting a significant activation potential of their own (8,9). Under lining the repressive role of nucleosomes, histone deprivation and concomitant depletion of nucleosomes have been shown to lead to—at least partial—activation of inducible genes even under conditions of genetic repression. This implies that at some promoters, specific activator proteins may be dispensable, once accessibility for the transcription machinery has been established (7,10). The PHO5 promoter is one example in which positioned nucleosomes exclude transcription factors and the polymerase II holoenzyme complex from their binding sites. Transcription activation of PHO5 requires the binding of the active transactivator, Pho4p, to a low affinity motif which is permanently accessible in a gap on promoter DNA between two exactly positioned nucleosomes. Upon induction by phosphate exhaustion, binding to the low affinity site leads to removal or excessive remodeling of four accurately positioned nucleosomes which, in the repressed state, occlude the high affinity binding sites of transcription factors that are essential for gene activation (7,11,12). If, however, nucleosome depletion is achieved experimentally in vivo by switching off expression of histone H4, transcription of PHO5 is maximally induced even under repressing conditions and can dispense with additional transcription activators (7,10).
In constitutive promoters, on the other hand, a static situation is presumed to keep the promoter permanently in an activated state to allow transcription (13). These promoters likely are constitutively kept free of nucleosomes. In the case of the relatively strong promoter of the ACT1 gene encoding yeast actin it has been speculated that two binding sites for the abundant general regulatory factor Reb1p [possibly in combination or synergistically with a poly(dA·dT) element] are involved in positioning nucleosomes and in keeping the core promoter accessible (13). In the PFY1 promoter, encoding the G-actin-sequestering profilin, one binding site for the abundant regulatory factor Reb1p has been found to be necessary and sufficient to keep nucleosomes off the DNA region spanning the core promoter and the transcription initiation sites (M.Angermayr, unpublished results). However, the general regulatory factor Reb1p does not have a significant transcriptional activation potential of its own. Rather, its main role in promoters appears to rely on its property to keep a stretch of DNA in the flanks of its binding site free of nucleosomes and to position them at a distance (14,15). The lack of binding sites for classical transactivators implies that at these promoters transcription may ensue spontaneously, if the core promoter is readily accessible to polymerase II holoenzyme.
We have analyzed the promoter of the yeast major adenylate kinase (ADK1 or AKY2, called AKY2 hereafter) which is constitutively expressed at a moderate level (16,17). The protein is unusually slowly turned over, and translation rates are low (18). Adenylate kinases are ubiquitous, abundant enzymes that fulfill an essential housekeeping function. They provide the ADP required for oxidative and substrate chain phosphorylations and, because of the reversibility of the catalytic reaction, contribute to the maintenance of the homeostasis of high energy adenine nucleoside phosphate pools in the cell. We have identified the minimal promoter of the gene which mainly consists of a non-consensus TATA sequence and enables expression rates of reporter constructs that are only slightly lower than the complete HTA1–AKY2-intergenic region. Since this suggested that promoter activation of AKY2 dispenses with classical transcription factors, we have examined the possibility that structural peculiarities of the promoter DNA create a constitutively open chromatin conformation. We have found that two kinks in the DNA structure closely flanking the TATA-like element suffice to position nucleosomes at a distance and to allow moderately high levels of basal transcription without the necessity of transactivators.
MATERIALS AND METHODS
Strains and plasmids
pBluescript M13 KS(±)-based vectors (Stratagene, Heidelberg, Germany) were used as templates for promoter truncations by polymerase chain reactions (PCR), or for site-directed mutagenesis, base deletions or insertions in vitro using SOE-PCR (19). Plasmids were maintained and propagated in Escherichia coli strain XL1-Blue or SURE (both from Stratagene). The AKY2-upstream region together with the nine N-terminal coding triplets was amplified by PCR as EcoRI–BamHI fragment and ligated to the respective restriction sites of pBluescript. All constructs were then fused in the proper orientation to the bacterial lacZ gene of the yeast low copy plasmid pYLZ7. pYLZ7 was constructed from pYLZ6 (20) by inversion of the bacterial selective marker gene bla, because with very short test promoter constructs interference with the bacterial bla gene sequence has been observed. Analyses of expression from the AKY2 promoter were performed in the genetic background of yeast strain W303-1A (21) which had been transformed with the respective reporter constructs. Yeast were grown on standard media (22).
PCR primers and in vitro mutagenesis
Promoter truncations were produced by PCR using upstream primers with an EcoRI restriction site and a reverse primer with a BamHI site for fusion to the lacZ reporter (restriction sites not shown). The following forward primers were used: HN, 5′-ACGGTAACATATGT-3′; H5B, 5′-CTTGAACATGATTGAGTAGC-3′; H5C, 5′-TTCACTTTGATAGTGTGACG-3′; H4, 5′-GCTCACGATTGCGCGATCC-3′; H2, 5′-CTGTCCGCAGCAGCCCGCGGC-3′; H1, 5′-ATTCGCCCATTTTTTTTTGATTTTCGAC-3′; H0, 5′-TTCACTCTGGCTAGTTTTATTAC-3′; H6, 5′-GTATATATATATACG CATAAATTTCTC-3′; H7, 5′-CGCATAAATT-TCTGAAATGG-3′. TIsh served as the reverse primer in most constructs, 5′-ATTAGGACCATTCTAATGGATTCTG-3′. Promoter mutations or internal deletions were obtained by site-directed mutagenesis using the kit and prescriptions from Stratagene. Permutation analysis was performed with DNA fragment H2/TIsh.
Tandem cloning and permutation analysis
The cloned PCR-amplified 243 bp DNA fragment H2/TIsh was consecutively restricted with BamHI (and blunted by digestion of protruding 5′ ends) and then with EcoRI (blunted by a fill-in reaction). Tandem ligation generated an EcoRI restriction site. The tandem DNA construct was digested in separate incubations using the set of restriction endonucleases indicated in Figure 2 to yield a set of DNA fragments of equal lengths permutated with respect to the fragment ends (23). After PAGE (8% gel, acrylamide:bisacrylamide = 38:2, 4°C), DNA was visualized by ethidium bromide staining.
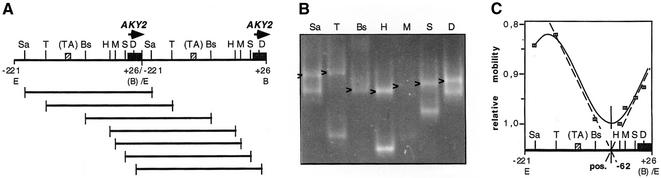
Figure 2.
Permutation analysis of AKY2 promoter DNA. (A) The DNA fragment was ligated in tandem, thereby generating an EcoRI restriction site at the junction. The construct was digested with the restriction enzymes indicated: E, EcoRI; Sa, SacII; T, TaqI; Bs, BstNI; H, HphI; M, MaeIII; S, SfaNI; D, DdeI; B, BamHI. (TA), TATA element. (B) Gel electrophoresis of the plasmid after digestion with the restriction enzymes indicated in (A). DNA was visualized by ethidium bromide staining. The set of permutated DNA fragments is marked by arrowheads. (C) Evaluation of the gel according to Thompson and Landy (39).
Analyses of chromatin structure
Crude nuclei were prepared as described (24). DNase I or micrococcal nuclease digestions were performed as described by Thoma (25). Chromatin or naked DNA was digested (5 min, 37°C) by different concentrations of DNase I (Roche, Mannheim, Germany) (chromatin at 10.0, 15.0, 20.0 or 30.0 U/ml; naked DNA at 0.01, 0.05 or 0.1 U/ml) or micrococcal nuclease (MBI Fermentas, St Leon-Rot, Germany) (chromatin at 30, 60 or 120 U/ml; naked DNA at 1.5 or 3.0 U/ml). Reactions were stopped by the addition of 0.5% SDS, 4 mM EDTA, 50 mM Tris–HCl pH 8.0 and 200 µg of Proteinase K (Merck, Darmstadt, Germany) and incubated at 37°C for 30 min. DNA was extracted twice with phenol/chloroform and precipitated by ethanol. The pellet was dissolved in TE buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA) and RNA digested with 400 µg of RNase A (Boehringer, Mannheim, Germany) at 37°C for 60 min. After extraction once with phenol/chloroform and once with chloroform, DNA was ethanol-precipitated and digested with EcoRV. Gel electrophoresis was at 100 V in 1.5% agarose gels. After Southern transfer to nylon membranes (Biodyne A, Pall, Dreieich, Germany) DNA was detected by a randomly primed, radiolabeled 527 bp HindIII–EcoRV DNA fragment which hybridized to the 3′ region of the AKY2 gene.
DNA structure analysis
DNA structure was calculated using the program BioCad which was developed in VMS-Fortran working on a DEC Microvax II GPX-station (26). The algorithm is essentially based on the wedge angles between nearest neighbor base pairs listed by Bolshoy et al. (27) and the assumption of 10.6 bp for a helical turn.
Miscellaneous procedures
The yeast expression vector pYLZ7 (above) was used for determining β-galactosidase activities of the respective promoter/lacZ fusions as described (28). Values give the mean expression activity of at least four independent clones. Generally, individual values varied in the range of 5–15%. Protein concentrations were determined according to the method described by Bradford (29). Yeast transformations were performed using the procedure described by Gietz et al. (30). Other molecular operations were performed according to standard procedures (31) or as recommended by the manufacturer.
RESULTS
Dissection of the AKY2 promoter
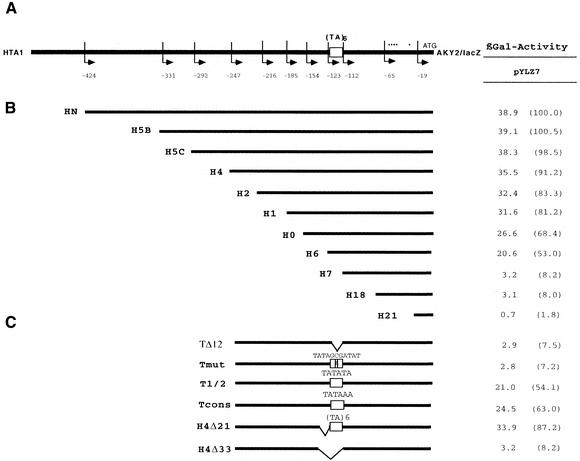
AKY2 is encoded on chromosome IV in tandem between the histone H2A-1 (HTA1) and SIR4 genes. The non-translated intergenic distance to the preceding HTA1 gene amounts to 560 bp, the non-transcribed region spans only 336 bp. The AKY2 promoter allows constitutive transcription at a moderate level (17). To define cis-acting elements required for constitutive promoter activation, we dissected the AKY2-upstream region. The 5′ flank of the gene was gradually truncated (Fig. 1). Despite progressive loss of 5′-upstream sequences (constructs H5B to H6, Fig. 1B), expression was maintained at a relatively high, only gradually declining level as long as the truncated promoter retained sequences upstream of position –154 (relative to the adenine of the translational start triplet as +1) (for activities see Fig. 1, right). The absence of any abrupt drop of the expression activity allows one to conclude that no site for a classical transcription activator has been eliminated by the deletions. In construct pH6, all sequences upstream of the sequence TATATATATATA (position –123) had been eliminated; surprisingly, this construct still displays a relatively high level of expression (>50% of the maximum), which evidently is independent of upstream elements. Expression severely drops with constructs pH7 and pH18, which have lost the (TA)6 motif, suggesting that the presumptive TATA element is functional and plays a decisive role in transcription of AKY2. Deletion of the TATA motif still allows some residual transcription, as the transcriptional initiation sites are intact, whereas in pH21, where the promoter deletion includes the transcriptional initiation sites at around position –60 (i.e. 51 bp downstream of the TATA element) promoter activity was completely abolished.
Figure 1.
Schematic drawing of the HTA1–AKY2 intergenic region. (A) Positions of 5′ ends of primers used in PCR amplification are indicated by arrows and numbers (adenine residue of the AUG translational start triplet as +1). (TA)6, TATA element. Asterisks denote transcriptional initiation sites. (B) Promoter truncations. (C) TATA mutations made in pH4. TΔ12, complete deletion of the TATA element; Tmut, the central part of the element has been interrupted by site-directed mutagenesis; Tcons, exchange of the (TA)6 element for a canonic TATAAA box. In H4Δ21, 21 bp have been deleted 5′ adjacent to the TATA-like element; in H4Δ33, 33 bp including the (TA)6 element and the 5′ kink have been deleted. Right, β-galactosidase activities (nmol/mg·min) of the respective mutant promoter/lacZ fusion constructs ligated to the low copy expression vector, pYLZ7. Numbers in brackets denote percentage of activity obtained with construct HN.
The AKY2 core promoter consists of six TA repetitions. A (TA)3 motif has been shown (32,33) to exert core promoter activity. To examine whether the (TA)6 motif is essential for transcription, we constructed a set of mutants all derived from pH4 (Fig. 1C). In pTΔ12, the 12 bp TATA-like motif has been deleted, and in pTmut the motif has been interrupted, TATAGCGATAT. In both TATA mutant promoters, transcription is severely impaired. The residual expression is considered basal transcription, a rate comparable to pH7 or pH18, which demonstrates that the (TA)6 motif directs transcription initiation. In pTcons it was replaced with a TATA box obeying the consensus, TATAAA (32,33). The canonic TATA element (pTcons) was slightly less efficient than the longer (TA)6 motif (construct pH4). In pT1/2 the length of the presumptive promoter was reduced to (TA)3 which, in fact, displays promoter activity (which is >50% of pH4).
The DNA structure of the AKY2 promoter
The above results strongly suggest that transcription activation of AKY2 can dispense with sequences 5′ of the TATA motif demonstrating that it is independent of upstream binding transactivators. This view is supported by the failure to detect presumptive binding sites for any general regulatory factor or other known transactivator by computer analysis or electrophoretic mobility shift assays either upstream of the TATA-like motif or downstream to nucleotide position –65 (data not shown). Therefore, we considered the possibility that the AKY2 promoter is activated by cis-acting elements distinct from sites of direct protein binding to DNA, and examined whether the DNA structure per se was unusual in a way that could favor assembly of the basal transcription machinery, i.e. whether the promoter region was bent. Many transcription factors including TBP have been found to bend DNA, and in some cases, among others with TBP, it has been observed that intrinsic DNA bending facilitates protein binding (34–37). Bent DNA can be identified by its retarded migration rate in acrylamide gels relative to straight DNA containing random nucleotide sequences of the same molecular mass and base composition (23). This effect is due to a lower flexibility resulting in loss of degrees of freedom in bent DNA and, thus, in an increase of the effective Stoke’s radius of the DNA fragment. The magnitude of the relative retardation is larger the closer the ends of the fragment are to one another, i.e. the larger the bend angle and the more central the bending motif are in the fragment (23,38,39). In order to test whether the AKY2 promoter is intrinisically bent, permutation analysis was performed (see Materials and Methods). Two identical 243 bp DNA fragments, H2/TIsh, containing the promoter were ligated in a tandem orientation. After restriction digestion, the relative mobilities of the set of promoter fragments (which all had the same length; Fig. 2A) were analyzed in dependence of the position of the presumed bending center with respect to the ends of the fragments (Fig. 2A and B). The resulting bend angle was determined to be 70° and the bending center to be around position –62 (Fig. 2C). The scarcity of natural restriction sites, in particular in the immediate vicinity of the core promoter, hampered correct evaluation of bending centers and bend angles. The deviation of the experimental points from a cosine function argues that more than one bending center could be present in the promoter region of the AKY2 gene.
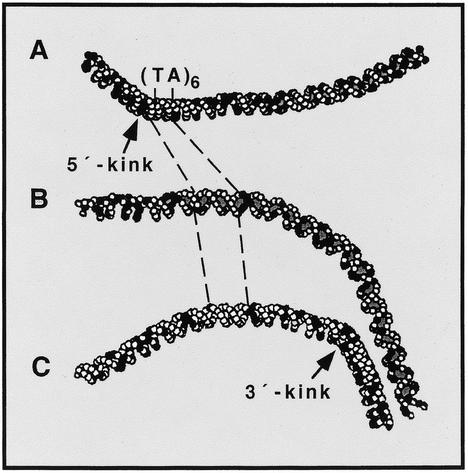
To examine this possibility and to obtain an overview of the magnitude and extension of the bend in the DNA of the AKY2 promoter, the algorithm developed for the DNA structure calculation (26) was applied to fragment H4/TIsh. The computer-derived model suggests that the complete core promoter region is slightly bent and that this structure is bordered by two kinks centered at positions –65 and –130, respectively (relative to the adenine residue of the AUG translational initiation codon as +1) (Fig. 3). The flanking kinks are not in the same plane. They form an angle of +90° between the DNA sides entering and exiting the promoter. Therefore, two different informative projections are shown, each displaying one of the kinks in the projection plane. The stronger downstream kink (Fig. 3C, ∼45°) is close to position –65 and nearly coincides with the major transcriptional initiation sites which are at positions –49 and –60. The upstream kink (Fig. 3A, ∼40°) shortly precedes the TATA element (positions –111 to –122). The third projection (Fig. 3B) reveals the intrinsic promoter bend. The computer model suggests that the promoter contains several non-straight stretches of DNA bent towards each other at different angles. Thus, the evaluation of the permutation data of Figure 2 yields the resultant overall angle.
Figure 3.
Model of the DNA structure at the core promoter of AKY2 (calculated as described in Materials and Methods). AT pairs are presented as open circles, GC pairs as filled circles. The bends are not in the same plane. (A) A projection to display the upstream kink; (B) the intrinsic curvature; and (C) the kink at the 3′ end of the core promoter. (TA)6, TATA element.
Importance of the DNA structure for transcription
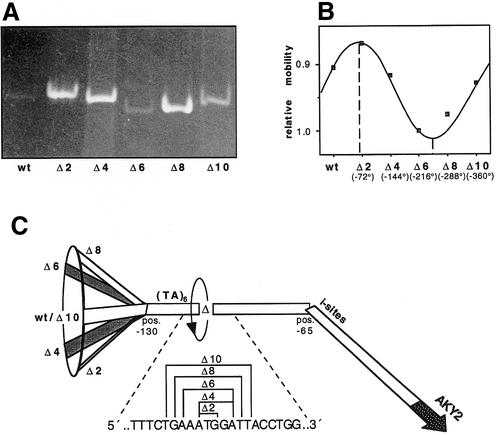
To test whether the computer-derived abnormal DNA structure has a bearing on the transcription rate, the flanking kinks and the distance between them were altered by mutation. In the first set of mutants the angle between the two kinks was altered systematically, and simultaneously the rotational orientation of the TATA motif relative to the initiation sites. The angle was varied in steps of 72° by deleting 0, 2, 4, 6, 8 or 10 bp on the 3′ side of the TATA sequence (13–23 bp downstream of the TATA element) (Fig. 4A and C). The effect of the deletions on the overall angle between the two ends of H4/TIsh was examined by gel electrophoresis (Fig. 4A and B). Simultaneously, the effect of the mutations on expression was examined by testing β-galactosidase activities expressed from the respective reporter constructs (data not shown). Figure 4A and B reveals that the electrophoretic mobility of the DNA fragments is phase-dependent: the exact phasing of the DNA (40) at the site of the deletions proves that the DNA on either side of this site is bent. The DNA fragment is retarded most, i.e. the two DNA ends lie in the same plane pointing in the same direction, when two base pairs have been deleted. This is consistent with the angle close to +90° between the ends in the wild type predicted by the computer model. Accordingly, the fragment in which 6 bp have been deleted displays the highest mobility indicating that in this mutant the two kinks are oriented in opposite directions. Although the alteration of the orientation of the TATA element relative to the downstream kink and to the initiation sites significantly changes intrinsic promoter bending, it does not affect the intrinsic bend in the core promoter and, therefore, has little influence on transcription initiation measured as β-galactosidase activity (data not shown). In line with this observation, two additional mutants, which were designed to straighten the gently curved DNA between the two kinks without altering the TATA-like motif (core promoter) or one of the flanking kinks, did not affect expression significantly (data not shown).
Figure 4.
Phasing analysis of the kinked promoter DNA. 0 (wt), 2, 4, 6, 8 or 10 bp have been deleted 3′ adjacent to the TATA element using DNA fragment H4/TIsh. (A) Ethidium bromide-stained gel. (B) Evaluation of the phasing gel showing maximum and minimum of retardation. (C) Schematic drawing of the deletion-dependent rotation of the ends. Delta denotes the point of the deletion (swivel).
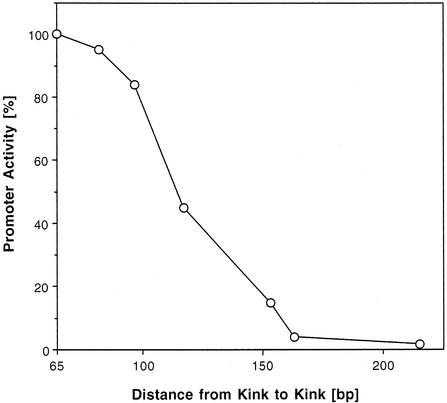
In addition, we tested the possibility that the distance between the two kinks plays a role in the transcription of AKY2 eventually, by excluding a nucleosome from the core promoter. In the wild type, the distance is shorter than required for accommodation of one nucleosome. Insertion close to the center of the DNA stretch between the two kinks (position –93) of a number of short DNA fragments varying in length from 17 to 150 bp increasingly eliminates promoter activity (Fig. 5). This manipulation enlarged the distance between the kinks from 65 bp in the wild type to a maximum of 215 bp. It appears noteworthy that expression decreased close to zero at a kink distance of ∼155 bp (insertion of 90 bp) which strikingly equals the minimum stretch of DNA occupied by a single nucleosome.
Figure 5.
Effect of DNA insertion between the kinks on β-galactosidase expression from the respective AKY2 promoter–reporter fusion constructs. Zero to 150 bp of DNA have been inserted at position –93, i.e. 19 bp downstream of the TATA element, leaving the intrinsic curvature of the TATA element (Fig. 3B) unaffected. Intervening DNA between the kinks is thereby extended from 65 to 215 bp.
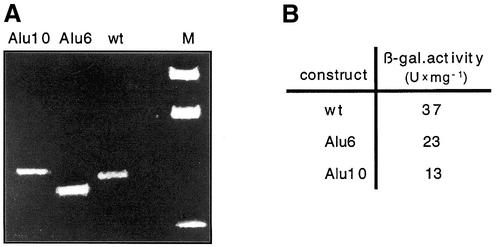
Two insertions were made in order to straighten the downstream kink at position –65. This site was chosen in order not to disturb the context of the adjacent initiation sites. Six base pairs were inserted by mutation Alu6 (sequence AATTCG) and 10 bp by Alu10 (GGGAATTCGC), thereby changing the helical phase of the DNA by close to one half turn or one full turn, respectively. The phase dependence of the effects of these mutations on the electrophoretic mobilities of the respective wild-type and mutant pH4/TIsh fragments (Fig. 6A) confirm the presence of a kink close to 3′ of position –65, the site of insertion. In these mutants, β-galactosidase reporter activities were reduced from 37 U/mg in the wild type to 23 U/mg in Alu6 and to 13 U/mg in mutant Alu10 (Fig. 6B) suggesting that the downstream kink plays a role in transcription activation.
Figure 6.
Phasing analysis of the Alu6 and Alu10 mutations. Zero, 6 or 10 bp of DNA were inserted at position –65 immediately adjacent to the calculated 3′ kink. (A) Ethidium bromide-stained phasing gel. (B) The β-galactosidase expression from the respective AKY2 promoter/lacZ fusion constructs.
The 5′ kink plays no decisive role in the activation of the AKY2 promoter as deletion of 21 bp immediately 5′ of the TATA motif (construct pH4Δ21 in Fig. 1C) had little influence on β-galactosidase activity. The activity is similar to pH4 suggesting that the core promoter activity is barely affected by the deletion and endorsing the conclusion that sequences 5′ of the core promoter have no significant importance for transcription. The bend extends into the phased AT-rich sequence in the 5′ flank of the kink so that it is, nevertheless, possible that both elements cooperate in positioning a nucleosome in the 5′ flank of the core promoter in the wild type. Simultaneous deletion of the core promoter and the upstream kink in pH4Δ33 (Fig. 1C) leads to a residual activity which compares to pH7 or pH18.
Chromatin structure at the AKY2 promoter
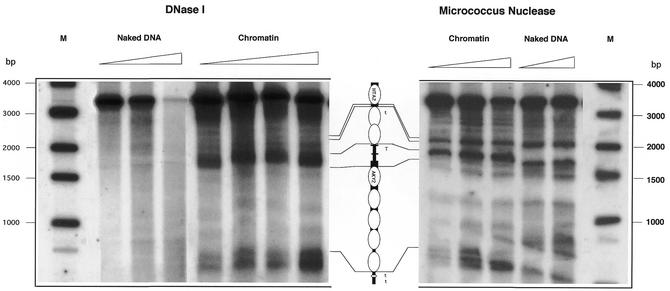
Since AKY2 is constitutively transcribed, we supposed that its promoter is permanently kept free of nucleosomes. To investigate the chromatin structure of the promoter and coding sequence of the AKY2 gene (in the genomic context), we performed in vivo footprint analyses, once by digesting chromatin with DNase I (Fig. 7, left) and once with micrococcal nuclease (Fig. 7, right). Although the two procedures yield minor differences due to the different cleavage specificities of the two nucleases, they are mutually complementary. Digestion of naked DNA served as an internal control. Consistently, we found a nucleosome-free gap in the AKY2 promoter which extends ∼100–130 bp (100 bp with micrococcus nuclease and 110–130 bp with DNase I). Calibration of the footprint signal with a DNA standard reveals that the nuclease hypersensitive site starts within the 5′ kink of the AKY2 promoter, spans the (TA)6 sequence, the downstream kink and the immediately adjacent transcriptional initiation sites. The upstream region of the AKY2 promoter is bound by two positioned nucleosomes (protected stretch 300 bp including linker) which are flanked on the 5′ side by another hypersensitive site shortly preceding the termination signal of the HTA1 gene (W.Bandlow and U.Häcker, unpublished results). On the 3′ side of the AKY2 promoter, the first and the fourth nucleosomes appear to be positioned (protected DNA 140 bp in either case), whereas the central part of the AKY2-coding region is occupied by two non-positioned nucleosomes, as expected for a moderately highly transcribed gene. In the immediate 3′ vicinity of the supposed two termination signals of AKY2 transcription (17) two strong hypersensitive regions are observed which are separated by a short protected stretch of DNA together spanning ∼90 bp, comprising the presumed (tandem) transcription terminator of the AKY2 tanscript. A schematic drawing of the averaged nucleosome positions at the AKY2 promoter is presented in the center of Figure 7.
Figure 7.
Chromatin structure at promoter and coding regions of AKY2. Native chromatin was digested with increasing concentrations (Materials and Methods) of DNase I (left) or of micrococcal nuclease (right). Naked DNA, treated with DNase I or micrococcal nuclease, served as internal controls. The results are illustrated by the schematic drawing in the center. Thick bars, coding regions; thin bars, non-coding regions; short black boxes, the kinks flanking the TATA element (T) of AKY2. Ellipses symbolize nucleosome protected DNA, the lens denotes a short protected stretch in the terminator (t) region.
DISCUSSION
Prerequisites for constitutive expression of AKY2
To understand transcription initiation, efforts have mainly concentrated on regulated promoters. Therefore, much is known about the inducible removal of nucleosomes and the control of access of the transcription apparatus to the core promoters of regulated genes (1–4). In contrast, knowledge of the principles governing creation of a relaxed chromatin structure and/or maintenance of nucleosome-free gaps in promoters of constitutively expressed, e.g. housekeeping, genes is rather limited and—apart from the architectural protein Reb1p—little is known about transcription factors that affect the array of nucleosomes and contribute to constitutive transcription.
To investigate whether transcriptional activators are required for the transcription of AKY2 and to identify possible cis-acting sites in the upstream flank of the AKY2 promoter, the intergenic region was gradually truncated starting from close to the site where the transcripts of the upstream-encoded HTA1 gene terminated. Expression from the shortened AKY2 promoter declined only gradually as long as the deletion did not touch the TATA-like motif. A sudden loss of expression, indicative of the loss of the binding site for an upstream transcription factor or architectural protein effecting exclusion of nucleosomes as soon as the respective binding site had been deleted, was not observed. The failure to obtain a bandshift signal in an electrophoretic mobility shift assay of AKY2 promoter DNA or to detect the binding motif for any general or specific transcription factor in the databases strengthens the conclusion that trancription of AKY2 dispenses with classical transactivators or chromatin remodeling factors, such as Reb1p.
This led us to consider alternative mechanisms of constitutive gene expression. As a fundamental principle in constitutive promoter activation, a static situation is expected to set the stage for free access of the transcriptional machinery. As one scenario, the architectural protein Reb1p has been described to organize a nucleosome-free gap (3,13,14) and—in the case of the constitutive profilin or actin promoters—has been found to be essential for transcription despite the lack of a significant activation potential. However, a binding site for the abundant Reb1 protein was clearly absent.
As an alternative to a protein-mediated structural distortion of the promoter-upstream region, we examined whether a peculiar architecture of the DNA involving structural distortion can substitute for the absence of binding sites for transactivators or general transcription factors. Remarkably, Mizuno and Itoh (41) have isolated AKY2 promotor DNA in a search for intrinsically bent DNA from random yeast genomic DNA fragments, and our permutation analysis of the core promoter of AKY2 confirmed the presence of bent DNA. Moreover, we have detected that, in the AKY2 promoter, two DNA kinks placed at the edges of the core promoter can have the same effect as, e.g. Reb1p: nucleosomes are excluded from the central region between the kinks and positioned at the flanks. We also show that the particular structural deformation of the core promoter is a prerequisite to transcription initiation.
Chromatin structure at the AKY2 core promoter
In vivo footprinting of the region of the AKY2 promoter reveals a gap between positioned nucleosomes that spans ∼110 bp. It comprises the core promoter element and extends into the transcriptional initiation sites. In principle, pre-bending of DNA, e.g. by phased AT pairs, alleviates wrapping around nucleosomes and leads to nucleosome binding to preferred positions (5–7,42–44). In contrast to nucleosome-compatible bent DNA, the kinked DNA at the 3′ edge of the AKY2 promotor (45° close to position –65) likely causes inflexibility of the DNA at this site and, hence, incompatibility with wrapping around a nucleosome (43).
On the 5′ side of the core promoter, a candidate for a sequence capable of positioning the upstream-flanking nucleosome is constituted by the 5′ kink, possibly in combination with a stretch of nearly pure, slightly phased poly(dA·dT) (42) between position –133 and –176. The compatibility of poly(dA·dT) stretches with packaging into nucleosomes is controversially discussed (45–47) suggesting that it depends on the flexibility of each individual dA·dT sequence, on possible phase-dependent sequence changes, and on the proper orientation of its curvature (43,48). If these prerequisites are fulfilled, a dA·dT stretch may preferentially bind and, thereby, position (a) nucleosome(s) (42–44). Since the downstream nucleosome on the 3′ side of the core promoter appears to be positioned as well, the gap between the two nucleosomes bounding the TATA element is too short (∼110 bp) to accommodate an additional nucleosome and, thereby, excludes nucleosomes from the core promoter. Elongation of the distance to >150 bp eliminates transcription completely, in accordance with this assumption. Deletion of the upstream kink has surprisingly little influence on transcription. Presumably, this mutation leads to a random array of nucleosomes leaving access to the core promoter largely unimpaired. Similar observations, that randomly arrayed nucleosomes on a core promoter barely impede transcription, have been made with the profilin and GCY1 promoters in which nucleosome positioning elements had been removed (M.Angermayr, unpublished results). Alternatively, the (TA)6 element (possibly in combination with the rest of the T-rich upstream element) is rigid and excludes nucleosomes on its own.
The core promoter of the AKY2 gene is remarkable in several respects. (i) Comprising six repetitions of a TA dinucleotide, the TATA-like element is relatively long as compared with a canonic TATA box. Shortening to (TA)3 leads to decreased promoter activity. (ii) The motif is embedded into a context of intrinsically bent DNA. Both features may alleviate binding of the basal transcription machinery in the absence of transactivators. The core TATA binding protein, TBP, as well as the basal transcription factor, TFIID, have been demonstrated to bind to the major groove of the DNA and to cause its extension resulting in bending (34–36,49). Moreover, it has been observed that pre-bending of the TATA box region facilitates assembly of the basal transcription machinery with the binding of TFIID as the key event (36). Analogously, it has been found in bacterial systems that bent DNA can relieve the dependence of a promoter on a gene activator (e.g. Crp) or even make it dispensable in order to achieve transcription (50–54). The findings that deletion of 2–10 bp or point mutations that affect the orientation or the extent of curvature of the intrinsic bend between the two kinks does not affect transcription initiation rates is not in conflict with this statement, as the immediate context of the TATA-like element is not affected by these mutations and, thus, is still bent. Our experimental data allow us to conclude that <12 bp of bent DNA on the 3′ side of the TATA element suffice to enable efficient assembly of the polymerase II holoenzyme complex. Thus, the particular sequence and structure of the core promoter in combination with the structure-dependent exclusion of nucleosomes from this site appear to favor binding of the basal transcription machinery and to allow moderately high rates of transcription despite the absence of transactivators.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank W. Hörz, Institut für Physiologische Chemie, Munich, for the introduction to in vivo footprinting techniques. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to W.B. within SFB 190.
REFERENCES
- 1.Workman J.L. and Buchman,A.R. (1992) Multiple functions of nucleosomes and regulatory factors in transcription. Trends Biochem. Sci., 18, 90–95. [DOI] [PubMed] [Google Scholar]
- 2.Imbalzano A.N., Kwon,H., Green,M.R. and Kingston,R.E. (1994) Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature, 370, 481–485. [DOI] [PubMed] [Google Scholar]
- 3.Axelrod J.D., Reagan,M.S. and Majors,J. (1993) GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes Dev., 7, 857–869. [DOI] [PubMed] [Google Scholar]
- 4.Morse R.H. (1993) Nucleosome disruption by transcription factor binding in yeast. Science, 262, 1563–1566. [DOI] [PubMed] [Google Scholar]
- 5.Sivolob A.V. and Khrapunov,S.N. (1995) Translational positioning of nucleosomes on DNA: the role of sequence-dependent isotropic DNA bending stiffness J. Mol. Biol., 247, 918–931. [DOI] [PubMed] [Google Scholar]
- 6.Piña B., Barettino,D., Truss,M. and Beato,M. (1990) Structural features of a regulatory nucleosome. J. Mol. Biol., 216, 975–990. [DOI] [PubMed] [Google Scholar]
- 7.Gaudreau L., Schmid,A., Blaschke,D., Ptashne,M. and Hörz,W. (1997) RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell, 89, 55–62. [DOI] [PubMed] [Google Scholar]
- 8.Felsenfeld G. (1992) Chromatin as an essential part of the transcriptional mechanism. Nature, 355, 219–223. [DOI] [PubMed] [Google Scholar]
- 9.Beato M. and Eisfeld,K. (1998) Transcription factor access to chromatin. Nucleic Acids Res., 25, 3559–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durrin L.K., Mann,R.K. and Grunstein,M. (1992) Nucleosome loss activates CUP1 and HIS3 promoters to fully induced levels in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fascher K.-D., Schmitz,J. and Hörz,W. (1990) Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J., 9, 2523–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svaren J. and Hörz,W. (1997) Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci., 22, 93–97. [DOI] [PubMed] [Google Scholar]
- 13.McLean M., Hubberstey,A.V., Bouman,D.J., Pece,N., Mastrangelo,P. and Wildeman,A.G. (1995) Organization of the Saccharomyces cerevisiae actin gene UAS: functional significance of reiterated REB1 binding sites and AT-rich elements. Mol. Microbiol., 18, 605–614. [DOI] [PubMed] [Google Scholar]
- 14.Fedor M.J., Lue,N.F. and Kornberg,R.D. (1988) Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J. Mol. Biol., 204, 109–127. [DOI] [PubMed] [Google Scholar]
- 15.Angermayr M. and Bandlow,W. (1997) The general regulatory factor Reb1p controls basal, but not Gal4p-mediated, transcription of the GCY1 gene in yeast. Mol. Gen. Genet., 256, 682–689. [DOI] [PubMed] [Google Scholar]
- 16.Bandlow W., Strobel,G., Zoglowek,C., Oechsner,U. and Magdolen,V. (1988) Yeast adenylate kinase is active simultaneously in mitochondria and cytoplasm and is required for non-fermentative growth. Eur. J. Biochem., 178, 451–457. [DOI] [PubMed] [Google Scholar]
- 17.Oechsner U., Magdolen,V., Zoglowek,C., Häcker,U. and Bandlow,W. (1988) Yeast adenylate kinase is transcribed constitutively from a promoter in the short intergenic region to the histone H2A-1 gene. FEBS Lett., 242, 187–193. [DOI] [PubMed] [Google Scholar]
- 18.Strobel G., Zollner,A., Angermayr,M. and Bandlow,W. (2002) Competition of spontaneous protein folding and mitochondrial import causes dual subcellular location of major adenylate kinase. Mol. Biol. Cell., 13, 1439–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton R.M., Cai,Z., Ho,S.N. and Pease,L.R. (1990) Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques, 8, 528–535. [PubMed] [Google Scholar]
- 20.Hermann H., Häcker,U., Bandlow,W. and Magdolen,V. (1992) pYLZ vectors: Saccharomyces cerevisiae/Escherichia coli shuttle plasmids to analyze yeast promoters. Gene, 119, 137–141. [DOI] [PubMed] [Google Scholar]
- 21.Crivellone M.D., Wu,M. and Tzagoloff,A. (1988) Assembly of the mitochondrial membrane system. J. Biol. Chem., 263, 14323–14333. [PubMed] [Google Scholar]
- 22.Sherman F., Fink,G.R. and Hicks,J. (1986) In Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Vassef A. (1992) Analysis of DNA curvature using circular permutation of 5′ end labelled fragments. Nucleic Acids Res., 20, 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almer A. and Hörz,W. (1986) Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J., 5, 2681–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thoma F. (1996) Mapping of nucleosome positions. Methods Enzymol., 274, 197–214. [DOI] [PubMed] [Google Scholar]
- 26.Schroth G.P., Siino,J.S., Cooney,C.A., Th’ng,J.P.H., Ho,P.S. and Bradbury,E.M. (1992) Intrinsically bent DNA flanks both sides of an RNA polymerase I transcription start site. Both regions display novel electrophoretic mobility. J. Biol. Chem., 267, 9958–9964. [PubMed] [Google Scholar]
- 27.Bolshoy A., McNamara,P., Harrington,R.E. and Trifonov,E.N. (1991) Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc. Natl Acad. Sci. USA, 88, 2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angermayr M. and Bandlow,W. (1997) The type of basal promoter determines the regulated or constitutive mode of transcription in the common control region of the yeast gene pair GCY1/RIO1. J. Biol. Chem., 272, 31630–31635. [DOI] [PubMed] [Google Scholar]
- 29.Bradford M.M. (1984) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 30.Gietz D., St. Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Chen W. and Struhl,K. (1988) Saturation mutagenesis of a yeast his3 ‘TATA element’: genetic evidence for a specific TATA-binding protein. Proc. Natl Acad. Sci. USA, 85, 2691–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harbury P.A. and Struhl,K. (1989) Functional distinctions between yeast TATA elements. Mol. Cell. Biol. 9, 5298–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horikoshi M., Bertuccioli,C., Takada,R., Wang,J., Yamamoto,T. and Roeder,R.G. (1992) Transcription factor TFIID induces DNA bending upon binding to the TATA element. Proc. Natl Acad. Sci. USA, 89, 1060–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starr D.B., Hoopes,B.C. and Hawley,D.K. (1995) DNA bending is an important component of site-specific recognition by the TATA binding protein. J. Mol. Biol., 250, 434–446. [DOI] [PubMed] [Google Scholar]
- 36.Parvin J.D., McCormick,R.J., Sharp,P.A. and Fisher,D.E. (1995) Pre-bending of a promoter sequence enhances affinity for the TATA-binding factor. Nature, 373, 724–727. [DOI] [PubMed] [Google Scholar]
- 37.Gaston K., Bell,A., Busby,S. and Fried,M. (1992) A comparison of the DNA bending activities of the DNA binding proteins CRP and TFIID. Nucleic Acids Res., 20, 3391–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crothers D.M., Haran,T.E. and Nadeau,J.G. (1990) Intrinsically bent DNA. J. Biol. Chem., 265, 7093–7096. [PubMed] [Google Scholar]
- 39.Thompson J.F. and Landy,A. (1988) Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res., 16, 9687–9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zinkel S.S. and Crothers,M.D. (1987) Catabolite activator protein-induced DNA bending in transcription initiation. Nature, 382, 178–181. [DOI] [PubMed] [Google Scholar]
- 41.Mizuno Y. and Itoh,K. (1988) Random cloning of bent DNA segments from Saccharomyces cerevisiae and primary characterization of their structure. Mol. Gen. Genet., 214, 249–256. [DOI] [PubMed] [Google Scholar]
- 42.Travers A.A. (1990) Why bend DNA? Cell, 60, 177–180. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka S., Zatchej,M. and Thoma,F. (1992) Artificial nucleosome positioning sequences tested in yeast minichromosomes: a strong rotational setting is not sufficient to position nucleosomes in vivo. EMBO J., 11, 1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka S., Livingstone-Zatchej,M. and Thoma,F. (1996) Chromatin structure of the yeast URA3 gene at high resolution provides insight into structure and positioning of nucleosomes in the chromosomal context. J. Mol. Biol., 257, 919–934. [DOI] [PubMed] [Google Scholar]
- 45.Struhl K. (1985) Naturally occurring poly(dA–dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc. Natl Acad. Sci. USA, 84, 8419–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iyer V. and Struhl,K. (1995) Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J., 14, 2570–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kunkel G.R. and Martinsson,H.G. (1981) Nucleosomes will not form on double-stranded RNA or over poly(dA)·poly(dT) tracts in recombinant DNA. Nucleic Acids Res., 9, 6869–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koo H.-S., Wu,H.-M. and Crothers,D.M. (1986) DNA bending at adenine·thymine tracts. Nature, 320, 501–506. [DOI] [PubMed] [Google Scholar]
- 49.Kim Y., Geiger,J.H., Hahn,S. and Sigler,P.B. (1993) Crystal structure of a yeast TBP/TATA-box complex. Nature, 365, 512–520. [DOI] [PubMed] [Google Scholar]
- 50.Mandal N., Su,W., Haber,R., Adhya,S. and Echols,H. (1990) DNA looping in cellular repression of transcription of the galactose operon. Genes Dev., 4, 410–418. [DOI] [PubMed] [Google Scholar]
- 51.Lavigne M., Herbert,M., Kolb,A. and Buc,H. (1992) Upstream curved sequences influence the initiation of transcription at the Escherichia coli galactose operon. J. Mol. Biol., 224, 293–306. [DOI] [PubMed] [Google Scholar]
- 52.Bracco L., Kotlarz,D., Kolb,A., Diekmann,S. and Buc,H. (1989) Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J., 8, 4289–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohyama T., Nagumo,M., Hirota,Y. and Sakuma,S. (1992) Alteration of the curved helical structure located in the upstream region of the β-lactamase promoter of plasmid pUC19 and its effect on transcription. Nucleic Acids Res., 20, 1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez-Martin J., Rojo,F. and de Lorenzo,V. (1994) Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev., 58, 268–290. [DOI] [PMC free article] [PubMed] [Google Scholar]