Abstract
Type II Human DNA Topoisomerases (topos II) play an essential role in DNA replication and transcription and are important targets for cancer chemotherapeutic drugs. Topoisomerase II causes transient double-strand breaks in DNA, forming a gate through which another double helix is passed, and acts as a DNA dependent ATPase. Mutations in topoII have been linked to atypical multi-drug resistance. Both human Topoisomerase II isoforms, α and β, are targeted by amsacrine. We have used a forced molecular evolution approach to identify mutations conferring resistance to acridines. Here we report mutation βG465D, which was selected with mAMSA and DACA and is cross-resistant to etoposide, ellipticine and doxorubicin. Resistance to mAMSA appears to decrease over time indicating a previously unreported resistance mechanism. G465D lies within the B′ domain in the region that contacts the cleaved gate helix. There is a 3-fold decrease in ATP affinity and ATP hydrolysis and an altered requirement for magnesium in decatenation assays. The decatenation rate is decreased for the mutated G465D protein. And we report for the first time the use of fluorescence anisotropy with intact human topoisomerase II.
INTRODUCTION
DNA topoisomerases are a ubiquitous family of enzymes that are essential for cellular processes such as replication and transcription. This role makes them important targets for both anti-bacterial and anticancer chemotherapeutic drugs. There are two broad sub-families of topoisomerases, type I which cleave a single strand, and type II which cleave both strands of a double helix to transport a second helix through the break. The type II enzymes (Topo II) show a high degree of sequence and structural homology, with a common domain structure. There are three major proteolytic cleavage sites, termed A, B and C which break the enzyme into four domains (1,2). These three sites lie between the domains. The N-terminal domain which contains the ATP hydrolysis catalytic centre lies next to the B′ domain, and the A′ domain lies next to this. Together the B′ and A′ domains make up the cleavage core catalytic centre, and finally there is the C-terminal domain bearing a nuclear location signal (3–5) and many phosphorylation sites (6,7).
Biochemical and structural studies have elucidated a model for the mechanism of action—firstly a DNA helix is bound into a semi-circular groove with contacts in the B′ and A′ domains. This is termed the gate ‘G’ helix. ATP binding at the N-terminus leads to dimerization within the N-terminal domain which can trap a second helix, the ‘T’ helix. A conformational cascade is triggered which causes the gate helix to separate into a double-strand break through which the T-helix is transported (8,9).
ATP hydrolysis is essential for strand passage activity of topoII, although DNA cleavage may occur in its absence. The N-terminus of the enzyme has been shown to bind and hydrolyse ATP during the reaction cycle in a DNA dependent manner (10–14). Kinetic studies with yeast topoII have shown that each dimer binds two ATP molecules and hydrolyses one molecule rapidly. Pi or ADP release is the rate limiting step before hydrolysis of the second ATP (15–17). DNA transport occurs after hydrolysis of the first ATP and before hydrolysis of the second (10). It is thought that ATP binding induces dimer formation and subsequent conformational changes which close a clamp-like structure at the N-terminus to trap the T-segment of DNA, and then induce transport of the T-helix through the core of the molecule and the cleaved G-helix (17–19). Binding of just one ATP molecule has been shown to be sufficient to induce conformational change across the dimer (20,21).
Crystal structures of the N-terminal domains of GyrB and yeast topoII have been solved, and these both show two subdomains. Subdomain 1 contains an ATP-binding pocket and subdomain 2 is termed the transducer domain as it is thought to contact the core region. Residues in subdomain 2 form a loop that contacts the γ-phosphate of the ATP analogue used. This is thought to be part of a signalling mechanism within the molecule which causes a cascade of changes on ATP binding, leading ultimately to the transport of the T-helix (22,23). The N-terminal structure of the eukaryotic topoII from yeast suggests that a pore seen on dimerization isn't large enough to accommodate a double helix, and that dimerization trapping the T-helix will result in torsional strain that will probably force the T-helix through the gate helix, and into the core of the enzyme (23). This provides further evidence of the link between conformational change and progression through the reaction cycle.
Humans have two distinct topoII isoforms, α and β, which are located on different chromosomes, are encoded by different genes and show different expression patterns within the cell (24). Both of the human isoforms have been shown to be targeted by the anticancer drug mAMSA (25).
Human Topoisomerase II is a target for anticancer drugs, one important class of which is the poisons. These drugs act through stabilizing an intermediate stage of the reaction, where the enzyme is complexed with the cleaved gate helix. These complexes can be processed to double-strand DNA breaks within the cells which triggers cell death, usually by apoptosis (26).
Resistance to Topo II drugs is a clinical problem, and may arise via a variety of mechanisms. Mutations affecting the way the drug interacts with the enzyme can cause resistance (27). Alternatively reduced enzyme activity gives fewer functional molecules to target and reduced protein expression gives the same effect (28–30). The ICRF family of drugs has been found to interact with the N-terminal domain of topoII and has been shown to overlap the ATP-binding pocket and bind two halves of the dimer simultaneously as the molecule dimerizes on binding of ATP (23,31). As a result ICRF has been found to preferentially target the ATP bound form of the enzyme. Mutations in the N-terminal ATPase region have been shown to confer resistance to ICRFs specifically through inhibition of the interaction between Topo II and drug (32).
The majority of mutations conferring drug resistance have been identified in the core catalytic centre responsible for cleavage. Mutations in human topoIIα have been shown to confer resistance to mAMSA. Mutations R486K and E571K showed 100- and 25-fold resistance to mAMSA, respectively. Interestingly both these mutations showed significant cross-resistance to etoposide. ATP hydrolysis has been shown to be effected in some drug resistant mutations (33). Mutation R450Q was first identified in human leukaemic cell line CCRF-CEM having conferred resistance to teniposide, and was subsequently found to give cross-resistance to mAMSA. This mutation showed altered ATP hydrolysis activity (34,35). Resistance to a number of different drugs implies the resistance mechanism is through inhibition of the enzyme function as opposed to inhibition of specific interactions with drug.
The acridines, including mAMSA and DACA, are a family of drugs used clinically which have been shown to target both Topo IIα and Topo IIβ in vivo (25). mAMSA is established in the treatment of haematological malignancies and breast cancers (36) and DACA has entered clinical trials for the treatment of several cancers including ovarian cancer (37). The study of point mutations in topoII enzymes can help to elucidate the mechanism of resistance to topoII targeting drugs, and a random mutagenesis approach such as the one used here gives an unbiased selection of mutations that can cause drug resistance.
Here we report for the first time the characterization of a mutated human topoIIβ G465D, selected independently for resistance to both mAMSA and DACA. Interestingly resistance to mAMSA appears to reduce over time. The mutant is also cross-resistant to a number of other drugs. DNA binding of topoII to a 40 bp oligo using fluorescence anisotropy showed that although the KD was unaffected, binding was altered for βG465D, and was affected by magnesium. There is no significant difference between the relaxation activity of the βG465D mutant compared with the wild-type enzyme βWT and for decatenation when measured at a single time point; however when decatenation was measured over time the rate was significantly reduced in βG465D. ATP hydrolysis is reduced 3-fold and magnesium-binding properties measured in decatentation assays are altered.
MATERIALS AND METHODS
Chemicals and drugs
mAMSA, AMCA and mAMCA were kindly supplied by Prof. B. C. Baguley, Auckland Cancer Society Research Center, University of Auckland (Auckland, New Zealand). Etoposide, Doxorubicin and Ellipticine were purchased from Sigma. DACA was kindly supplied by Dr P. Charlton. DNA sequencing was carried out by Lark Technologies Inc. (Essex, UK). Oligonucleotides were ordered from Invitrogen (Paisley, UK). Radiochemicals were purchased from Amersham Biosciences (Little Chalfont, Buckinghamshire, UK) and enzymes from New England Biolabs (Hertfordshire, UK) or Promega (Southampton, UK). kinetoplast DNA (kDNA) was purchased from TopoGEN (Columbus, OH).
Random mutagenesis, selection with drugs and mutant identification
Plasmid YEphTOP2βKLM (38), encoding recombinant human topoIIβ residues 46–1621 fused to the first five residues of yeast topoII was mutated by exposure to 0.1 M hydroxylamine at 75°C for 40 min, and selected for drug resistance using methods described previously (33,39). Mutagenized plasmid was transformed into yeast strain JN394top2-4 to select for mutations conferring resistance to mAMSA, mAMCA, AMCA and DACA. Yeast transformants were then grown in the presence of 74.7 µg/ml mAMSA or 0.5 mg/ml DACA for 96 h at 35°C. Drug resistant yeast transformants were selected by plating onto media plates lacking uracil and containing drug at the concentrations above, and grown for 3–5 days at 35°C. DNA from drug resistant colonies was isolated and the possibility of gross rearrangements or deletions eliminated by analysis of restriction digestion patterns. Selected plasmids were then retransformed into yeast to verify that resistance was plasmid borne, and mutations identified by sequencing both strands.
Owing to the large number of resistant yeast isolated it was not possible to retransform all plasmids. Plasmids not retransformed were screened for mutation G465D using restriction fragment length polymorphism (RFLP) analysis. Mutation G465D is caused by a g to a change at position 1395 and gives an additional HphI site (ggt-gat). A 1 kb region spanning the region encoding the mutation (base numbers 1200–2200) was amplified by PCR, and then digested with HphI in NEBuffer 4 (New England Biolabs) at 37°C for at least 1 h. Reaction products were then separated on a 0.8 or 1% agarose gel, stained with ethidium bromide and visualized under UV light. G465D mutation gives an additional band when compared with wild type.
In vivo characterization
To ensure resistance was due to the point mutation and not other plasmid changes, a 1674 bp fragment between sites BamHI and PmaCI containing the mutation was exchanged with the corresponding fragment of unmutagenized plasmid. DNA sequencing confirmed the presence of the mutation in the exchanged plasmid. This fragment-exchanged plasmid was used in all in vivo characterization. Sensitivity to a variety of drugs was determined by continuous exposure on agar plates containing drug. Yeast cultures of known optical density were replica plated, and grown for 3–5 days at 35°C. Fold resistance was determined from WT and G465D growth at different concentrations of drug. This method allows rapid analysis of cross-resistance to many different drugs (27). Resistance to mAMSA was further quantified by measuring the minimum lethal concentration (MLC) for βWT and βG465D. Yeast were exposed to drug in liquid culture for 6 or 24 h, and then grown on drug free plates to determine the MLC as described previously (39).
In vitro characterization
Protein purification was either as described previously (2) or modified as follows: yeast lysis was using Yeastbuster Protein Extraction Reagent (Novagen), and following elution from phosphocellulose protein was further purified on a heparin column. Expression of recombinant protein was in yeast strain Jel1Δtop1 bearing plasmid YEphTOP2βKLM or YEphTOP2βG465D.
Decatenation and relaxation assays were carried out in ‘relaxation buffer’ (50 mM Tris–HCl, pH7.5, 0.5 mM EDTA, 1 mM DTT, 100 mM KCl, 30 µg/ml BSA), plus 2 mM ATP, 10 mM MgCl2 and either 1 µg of supercoiled pBR322 plasmid DNA (for relaxation assays) or 400 ng of kDNA (for decatenation assays). The method was described previously (2,40). With time-dependent decatenation the quantified band is expressed as a percentage of total lane OD to account for background intensities.
Cleavage assays with 40 bp oligonucleotide and a 4.3 kb linearized plasmid were carried out as described previously (25).
DNA-binding measurements using fluorescence anisotropy
DNA-binding capacity was determined with purified protein and a hexachloroflourescein (HEX) labelled 40 bp double-stranded DNA oligo using fluorescence anisotropy. Measurements were carried out at 20°C using an SLM-Amnico 8100 spectrofluorometer (SLM-Aminco, Urbana, IL). The excitation wavelength was 530 nm with an excitation slit width of 8 mm and the emission wavelength was 570 nm with an emission slit width of 3 mm. A 1 ml fluorescence cuvette was used with excitation and emission paths each of 10 mm. Assays were carried out in anisotropy buffer (50 mM Tris, pH 8, 5% glycerol, 50 mM KCl, 1% Triton X-100) supplemented with 100 µg/ml acetylated BSA, and Topo II proteins were matched for buffers and salt concentration. HEX-labelled oligo (1 µM) was added to the buffer and a baseline reading taken. Protein was added as described in Results and the average anisotropy of 12 readings over 99 s measured for each titration point. MgCl2 (10 mM) was added to the buffer where described. A one-binding site hyperbola was fitted to data and the BMAX and KD calculated using GraphPad Prism 4.
ATP hydrolysis activity determination
ATP hydrolysis was determined using purified protein and a coupled assay linking ATP hydrolysis to NADH oxidation and a subsequent decrease in absorbance at 340 nm. Assays were carried out in assay buffer (50 mM Tris, pH 7.4, 66 µg/ml BSA, 1 mM DTT, 0.5 mM EDTA, 50 mM KCl and 10 mM MgCl2) using a quartz cuvette. An aliquot of 2 mM PEP, 0.1 mM NADH, 5 U of lactic dehydrogenase, 2.5 U of pyruvate kinase, 10 µg of plasmid DNA and 1 mM ATP (unless stated otherwise) were mixed and the absorbance at 340 nm monitored until stable to eliminate the effect of ADP in the sample. Protein was added at 75 nM, (unless stated otherwise), the reaction was mixed well and then absorbance at 340 nm measured for at least 5 min. The linear rate of change of Abs340 allows the rate of ATP hydrolysis to be calculated by assuming that 1 mol of NADH corresponds to 1 mol of ATP.
RESULTS
Selection of human topoisomerase IIβ mutations resistant to acridines
We have used a forced molecular evolution approach to select and identify mutations in human Topoisomerase IIβ that confer resistance to the acridines mAMSA and DACA. Mutation G465D was selected independently with both mAMSA and DACA. The plasmid encoding recombinant human Topo IIβ was randomly mutagenized by incubating with 0.1 M Hydroxylamine for 40 min at 75°C. The library of mutagenized plasmids was then transformed into the temperature sensitive yeast strain JN394top2-4: in this strain the yeast topoII is active at 25°C (the permissive temperature) but inactive at 35°C (the non-permissive temperature), so growth at the non-permissive temperature is dependent on complementation by a functional plasmid-borne topoII. The transformed yeast were selected for drug resistance by exposure to drug in liquid culture at either 74.7 µg/ml mAMSA or 0.5 mg/ml DACA, for 96 h at the non-permissive temperature. Transformants were further selected by plating onto Ura-plates containing mAMSA or DACA at the levels stated above and grown for 3–5 days at the non-permissive temperature. DNA from drug resistant colonies was isolated and subjected to restriction digest with HindIII to eliminate any plasmids that had undergone gross rearrangements or deletions. A selection of plasmids was retransformed into yeast to verify that the drug resistance was plasmid borne, and the DNA was sequenced to identify the mutation. Using this approach mutation G465D was selected and identified using this approach a total of three times, twice with mAMSA and once with DACA. A g to a change at position 1395 causes the glycine to aspartic acid change. The plasmids that had not been retransformed were screened for the presence of the G to D mutation at codon 465 using RFLP analysis as this mutation leads to a gain of an HphI site (ggtgg–ggtga), and G465D was found to have been selected once more with mAMSA.
Plasmids (500) were selected for ability to grow on plates containing mAMSA and 3 were sequenced—2 of these were found to contain a base change g–a at position 1395 which confers mutation G465D. More than 1000 plasmids were selected for ability to grow on plates containing DACA, 23 were sequenced and 1 of these was found to contain the base change g–a and so also conferred mutation G465D. Plasmids (36) selected with mAMSA, and 240 selected with DACA that were able to complement yeast topoII were screened by RFLP analysis to see if they also contained mutation G465D. One more G465D mutation from the mAMSA screen was identified giving three in total identified with mAMSA and one with DACA.
Mutation G465D confers resistance to mAMSA
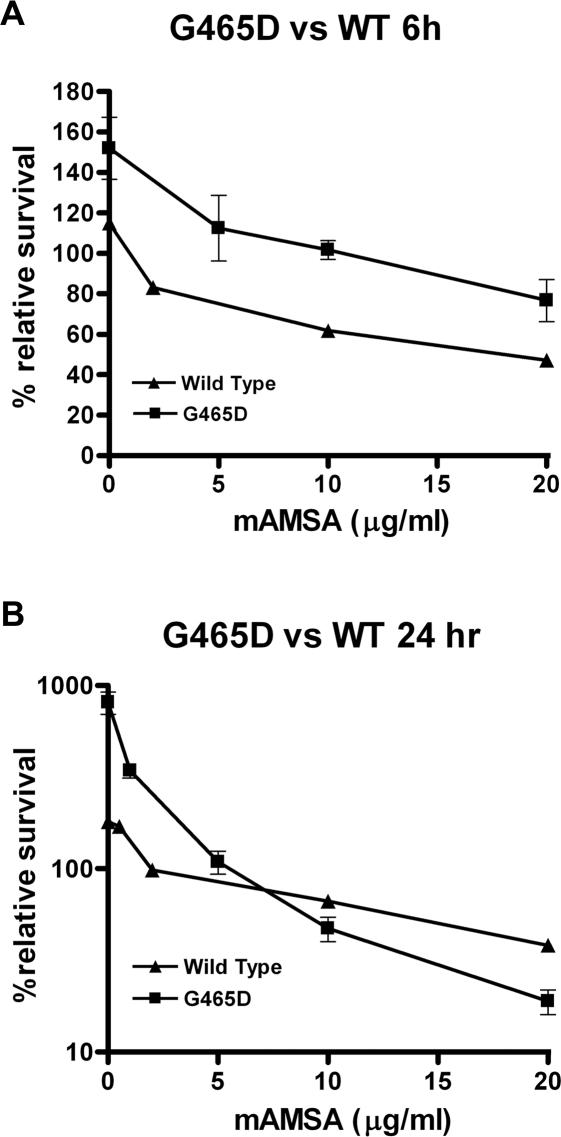
Topoisomerase mediated drug resistance can occur through decreased expression of the enzyme, perhaps through a promoter defect, giving fewer molecules to target. To verify that the point mutation identified, G465D, was responsible for the drug resistant phenotype, and not mutations elsewhere in the plasmid, fragment exchange was performed on the plasmid. A 1674 bp region containing the mutation was excised and exchanged for the corresponding segment from an unmutagenized vector. The plasmid was then transformed into JN394top2-4. The level of mAMSA drug resistance was quantified using the MLC method (39). JN394top2-4 transformed with either βWT or βG465D was grown in the presence of different concentrations of mAMSA for 6 or 24 h, then the amount of growth relative to a t0 plate quantified for both yeast strains. The βG465D transformed yeast showed ∼10-fold resistance to mAMSA at 6 h with MLC values of 1 and 10 µg/ml for βWT and βG465D, respectively (Figure 1A). This difference was statistically significant as determined by a two-tailed paired t-test (P < 0.05). Interestingly after 24 h of exposure only 2-fold resistance to mAMSA was seen with MLC values of ∼2 and 5 µg/ml seen for βWT and βG465D, respectively. This difference wasn't found to be statistically significant. (Figure 1B). Exposure of βG465D to mAMSA for 3–5 days on drug plates showed very low levels of resistance, with growth very similar to wild type (data not shown).
Figure 1.
MLC measuring survival of yeast transformed with plasmid encoded WTβ and βG465D on growing with mAMSA for 6 h (A) or 24 h (B) at 35°C. Experiments were repeated three times.
G465D confers cross-resistance to many different drugs
Mutations giving topoisomerase II drug resistance often confer cross-resistance to other drugs, so we compared the drug resistance phenotypes of yeast transformed with wild-type or mutant plasmid. The two yeast transformants were grown on YPDA plates supplemented with drugs of various concentrations, after 3–5 days the level of growth was compared. The drugs tested were from a variety of functional classes, these being acridines mAMSA, AMCA, mAMCA, DACA, the epipodophyllotoxin Etoposide, the anthracycline Doxorubicin and Ellipticine. Mutation βG465D showed resistance to all except mAMSA. The mutant also showed a 2-fold resistance to AMCA, 5-fold resistance to mAMCA and 10- to 20-fold resistance to DACA, a 4-fold resistance to etoposide, 10-fold resistance to ellipticine and 2-fold resistance to doxorubicin (data not shown).
DNA cleavage
Topo IIβ proteins were purified to 95% homogeneity as described previously (2). To try and determine the mechanism for drug resistance the DNA cleavage properties of βG465D vs βWT were assayed. The proteins were incubated with a 40 bp 32P-labelled linear DNA substrate and the reaction stopped by addition of SDS to denature the protein. Proteinase K removes the protein and allows the DNA fragments to be analysed by PAGE. Cleavage reactions were performed with either magnesium ions, the natural divalent cation for topoII, or calcium ions which are commonly used to enhance the cleavage reaction (41). Topoisomerase poisons are also known to enhance cleavage by stabilizing the cleavable complex, and cleavage assays were done in the presence and absence of mAMSA. Relative cleavage with wild-type and mutant enzyme in several experiments was calculated by quantifying the amount of end-labelled cleaved product by filmless autoradiographic analysis using a BAS-1500 (Fujifilm, Tokyo, Japan). The amount of 40 bp oligo substrate cleaved into the two products by the wild-type protein βWT in the presence of magnesium and mAMSA was quantified and this was taken as 100% and cleavage under other conditions calculated relative to this. Cleavage of wild-type protein with magnesium was stimulated 12-fold on addition of mAMSA (100%). Cleavage in the presence of calcium ions increased 5-fold (42 ± 7.7%) compared with magnesium alone (7.8 ± 2.4%), and was highest in the presence of calcium and mAMSA (376.4 ± 97.6%). βG465D cleavage with magnesium was 28.2 ± 10.3 and 82.2 ± 29% in the absence and presence of mAMSA, respectively. Calcium enhanced βG465D cleavage with values of 63.5 ± 51.9 and 250.6 ± 65% in the absence and presence of mAMSA, respectively. The average values for triplicate βG465D mAMSA cleavage experiments were lower than for the triplicate βWT experiments. However the SDs were rather large, so when a student t-test was carried out the differences were not significant.
Cleavage of the 40 bp DNA substrate can also be stabilized by etoposide. No significant difference in the ability of βWT and βG465D to cleave in the presence of etoposide was observed, with cleavage at 100 and 92 ± 8.9%, respectively.
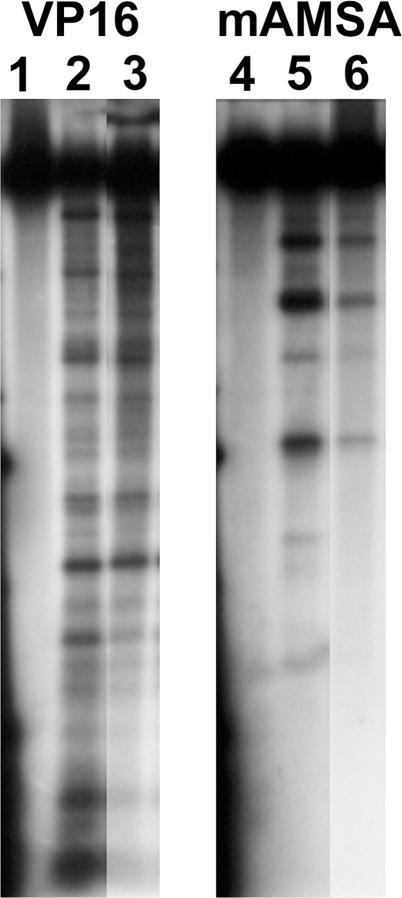
The cleavage patterns with a 32P-end-labelled 4.3 kb pBR322 fragment for βWT and βG465D enzyme were compared. The cleavage pattern with this substrate was similar to that of βWT, the same major cleavage products were seen but some bands were of lower intensity (Figure 2).
Figure 2.
Cleavage with mAMSA and a 4.3 kb linear substrate. Protein (1 pmol) was incubated with 100 µg/ml Etoposide (lanes 2–3) or 50 µg/ml mAMSA (lanes 5–6) at 37°C for 1 h before addition of 0.1% SDS and 0.5 mg/ml proteinase K. Reaction products were run overnight on a 1% agarose gel. Lanes 1 and 4 are with no protein, lanes 2 and 5 are with WTβ, and lanes 3 and 6 are βG465D.
DNA binding
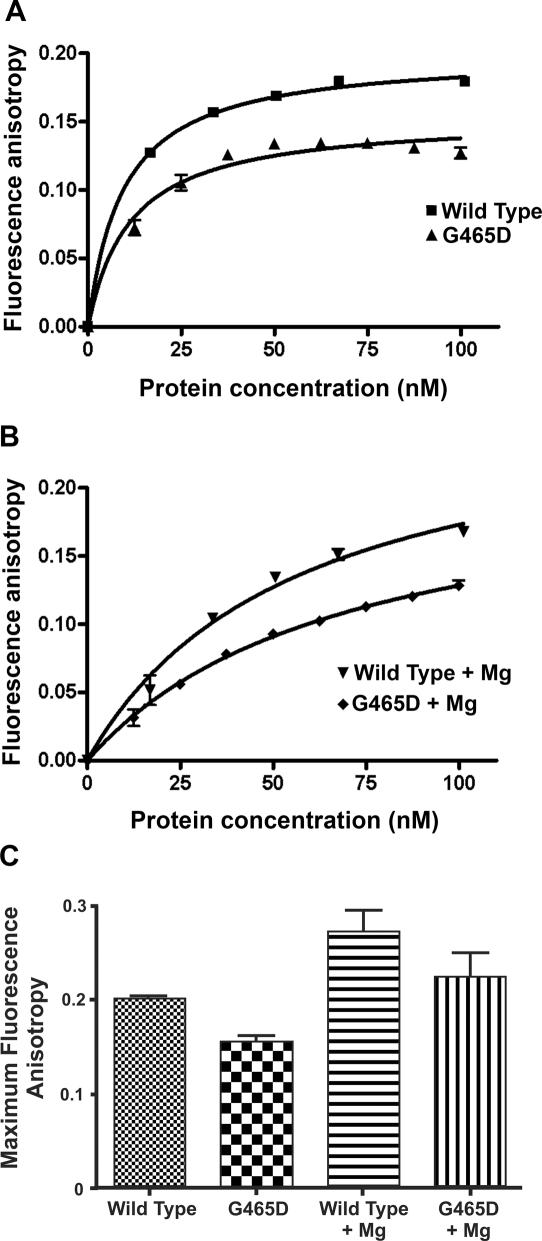
DNA binding of βWT and βG465D to a HEX-labelled 40 bp oligo was measured using fluorescence anisotropy, in the presence and absence of 10 mM MgCl2. Fluorescence anisotropy was calculated relative to the baseline reading. Without supplemented magnesium the maximum anisotropy seen for βWT and βG465D was 0.1998 ± 0.002474 and 0.1535 ± 0.004687, respectively, a significant difference as determined by a two-tailed unpaired t-test (P > 0.05; Figure 3A). When the reaction is supplemented with 10 mM MgCl2 there is an increase in βWT anisotropy to 0.2701 ± 0.02286, a significant difference (P > 0.05). βG465D also increases significantly (P < 0.0001) to 0.2180 ± 0.01203 in the presence of magnesium (Figure 3B and C). However the KD of binding is not significantly different between βWT and βG465D with values of 9.429 ± 0.6234 and 11.34 ± 1.694, respectively.
Figure 3.
Fluorescence anisotropy Bmax values for βWT and βG465D. Protein (300 nM) and HEX-labelled 40 bp oligo (1 µM) were used per experiment. MgCl2 was added to 10 mM where appropriate. Protein was added in 5 µl aliquots. For each protein concentration the average of 12 anisotropy values was taken. The average of three titrations is shown with error bars representing 1 SD from the mean. Data are presented relative to the baseline anisotropy value. (A) Anisotropy of βWT and βG465D apoproteins. (B) Anisotropy of βWT and βG465D in the presence of 10 mM MgCl2. (C) Comparison of maximum anisotropy for βWT and βG465D in the presence and absence of MgCl2.
Strand passage activity
Strand passage activities of purified proteins were assayed with relaxation and decatenation assays. A minimum of three independent experiments were carried out for each assay, for a single time point (30 min), and the wild-type activity taken as 100%. The relaxation activity of βG465D was not significantly different from βWT at 104.8 ± 8.3 and 100%, respectively. While the decatenation activity appeared slightly reduced at 60.3 ± 18.2%, this was not significantly different from βWT (100%) in a t-test.
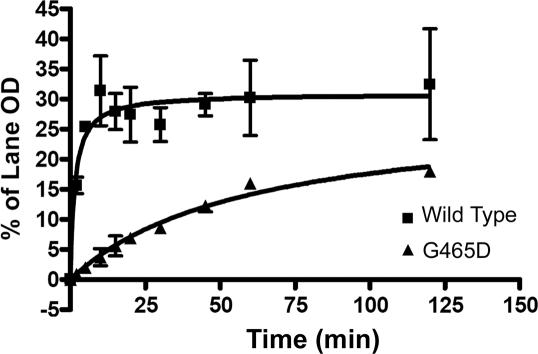
The rate of strand passage activity was also investigated by assaying the decatenation of 400 ng of kDNA by 1 U of enzyme over time. While βWT reached maximum decatenation after 20 min, βG465D begins to reach saturation after 120 min (Figure 4). The initial rate of change in intensity per minute (expressed as the change in the % lane OD per minute) for each enzyme was determined with regression analysis and found to be 4.913 ± 0.6413 and 0.4011 ± 0.1232 for βWT and βG465D, respectively, an ∼12-fold decrease in initial decatenation rate.
Figure 4.
Decatenation over time. Enzyme (1 U) was used to decatenate 400 ng of kDNA at 37°C for increasing lengths of time as shown. Bands were quantified using TINA densitometry software and then expressed as a % of the total lane OD. The average of two independent experiments is shown with error bars representing 1 SD from the mean.
An increased ATP requirement and narrower MgCl2 optima is seen with G465D
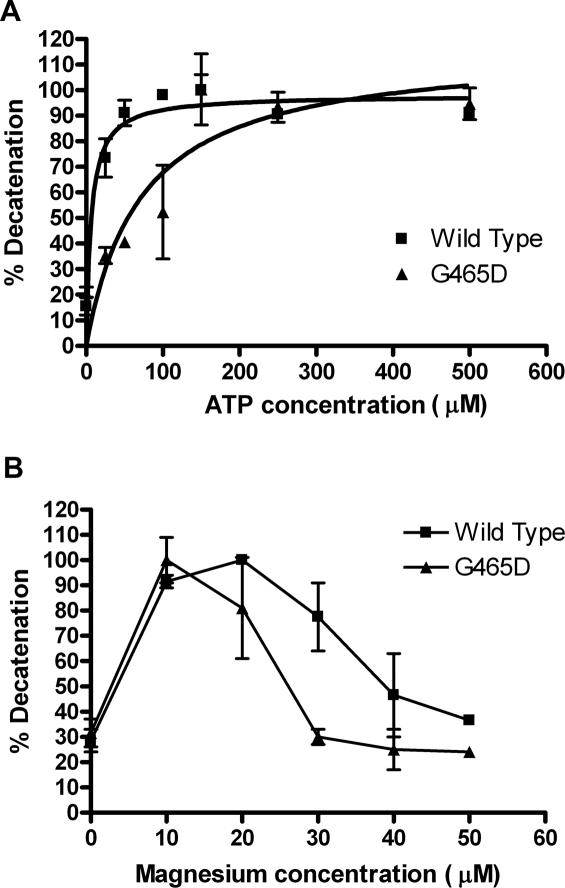
The ATP requirements of the mutant enzyme were measured using decatenation assays at a variety of ATP concentrations (Figure 5A). Interestingly the βG465D mutant showed a 3-fold increase in ATP requirement compared with the βWT. The βWT reaches maximum decatenation at ∼50 µM ATP, at which point βG465D shows only 40% decatenation. βG465D reaches 100% decatenation at 150 µM ATP, a 3-fold increase. This is confirmed with Km values; 19 µM for βWT and 62 µM for βG465D, an increase of 3.26-fold.
Figure 5.
(A) Dependence of decatenation activity on ATP concentration. Protein was used at a level sufficient to decatenate 80% of DNA substrate, using 400 ng of kDNA. Maximum decatenation for each protein is taken as 100% with other values expressed as a percentage of this. ATP dependent activity is shown for βWT (squares) and βG465D (triangles). Results are the average of three experiments and error bars represent 1 SD from the mean. (B) Dependence of decatenation on MgCl2. Protein was used at a level sufficient to decatenate 80% of DNA substrate, 400 ng of kDNA was used per reaction. Maximum decatenation for each protein is taken as 100% with other values expressed as a percentage of this. Magnesium dependent activity is shown for βWT (squares) and βG465D (triangles). Results are the average of two experiments and error bars represent the SD from the mean.
The response of the mutant to MgCl2, an essential cofactor, was also measured by increasing the MgCl2 concentrations in decatenation assays (Figure 5B). While the βWT and βG465D both reached maximum activity at 10 µM MgCl2, the βG465D showed a much narrower optima. The wild-type enzyme showed continuing maximum decatenation until 20 µM MgCl2, then ∼80% at 30 µM, 45–50% at 40 µM and 40% at 50 µM. This correlates with previous results (40). The βG465D mutant gave 80% decatenation at 20 µM, 30% at 30 µM and ∼20% at 40–50 µM MgCl2. The Km for both proteins is similar; however, as there is no data between 0 and 10 mM MgCl2, where activity is already maximal, this cannot be measured accurately.
Mutation G465D gives reduced ATP hydrolysis activity
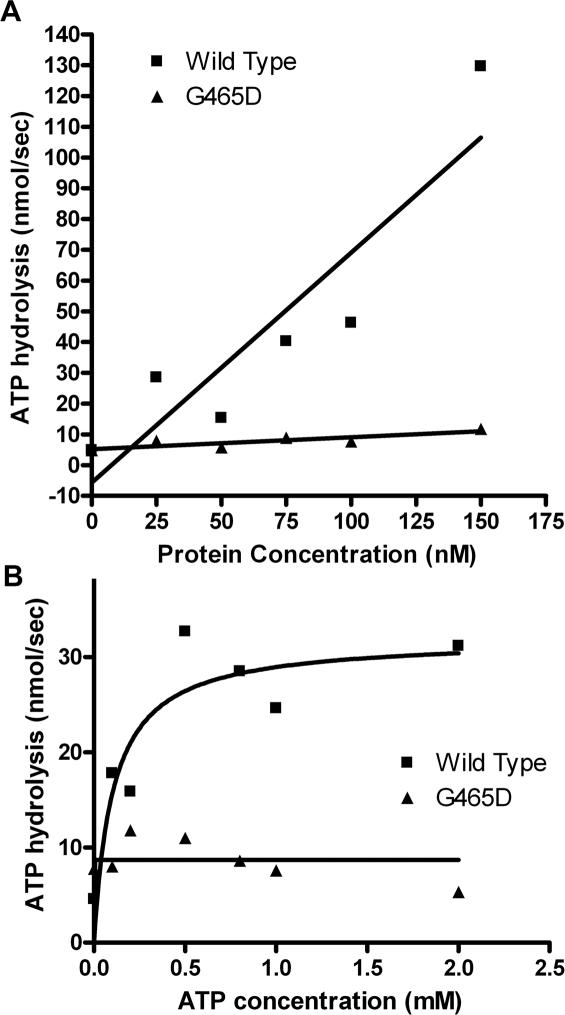
As the mutant has an increased ATP requirement for decatenation and lies towards the N-terminal ATPase domain ATP hydrolysis activity assays as described in Materials and Methods were carried out to determine if this alteration was because of impaired ATP hydrolysis activity. ATP hydrolysis assays were carried out in 10 mM MgCl2, shown to be optimal for both proteins. Assays with increasing concentrations of enzyme showed clearly that the βG465D protein had severely impaired ATP hydrolysis activity as compared with wild type. The slope determined from linear regression was 0.7477 ± 0.1744 and 0.0387 ± 0.01193 for βWT and βG465D, respectively (Figure 6A). An analysis of the Vmax of both wild-type and mutant enzymes showed that the wild-type maximum was 3.68-fold higher than that of βG465D at 31.99 nmol ATP per second and 8.702 nmol ATP per second, respectively (Figure 6B), which is in agreement with the 3-fold increase in ATP requirement to perform decatenation seen with the mutant. The ATP hydrolysis activity of human topoisomerase IIβ is unstable, and diminishes quickly after protein purification. To ensure that the ATPase activities of both proteins were optimal, the proteins were purified in the same week, flash frozen in liquid nitrogen and stored at −80°C and experiments done the following week.
Figure 6.
(A) ATP hydrolysis activity with increasing protein. Reaction carried out in reaction buffer containing 10 mM MgCl2 with 9 µg DNA and 1 mM ATP, protein was varied as shown. βG465D (triangles) and WTβ (squares) are shown. A single dataset is shown. Data were plotted in GraphPad Prism 4. (B) ATP hydrolysis with increasing ATP. Reaction was carried out in reaction buffer containing 10 mM MgCl2 with 9 µg of DNA and 75 nM protein in each case. ATP was increased as described. Maximum determined by GraphPad Prism 4. A single dataset is shown.
DISCUSSION
Using a forced molecular evolution approach, selecting with mAMSA and DACA, drug resistant mutation G465D has been identified in topoisomerase IIβ on four occasions with two selection agents. This mutation also confers resistance to several other drugs of different classes including AMCA, mAMCA, Etoposide, Ellipticine and Doxorubicin.
The resistance to DACA is very strong at 10- to 20-fold compared with wild-type enzyme. DACA is unlike the other acridines in that its exact mechanism of action is not well understood, with evidence suggesting it acts as a dual topoisomerase I and II poison, so caution must be exercised in interpreting the drug resistance data from the yeast organism. DACA may not be exclusively targeting Topo II (42,43).
An atypical drug resistance phenotype such as that given by βG465D, where resistance is conferred to a range of structurally diverse drugs, implies that the mechanism of resistance is through a reduction in enzyme function giving fewer functional molecules to target as opposed to a decrease in specific interaction with drugs. Mutations of this type have been reported previously in the B′ domain (26).
The resistance to mAMSA, the selection agent in three of the four G465D mutations identified, was measured at different time points. The MLC for strains expressing either βG465D or βWT was measured at 6 and 24 h and continuous growth on drug plates was measured after 3–5 days. A clear pattern emerged where the drug resistance decreased over time, from a 10-fold resistance after 6 h exposure, to 2-fold resistance after 24 h exposure, and then after 3–5 days βG465D showed no resistance. These data imply that, while a defect in the enzyme is likely, this defect does not permanently disable any aspect of enzyme function and merely ‘slows’ the turnover.
Indeed, the yeast carrying the mutant plasmid grows at approximately half the rate of yeast with plasmid encoding wild-type Topo IIβ (data not shown). The selection method used ensures that any mutants identified will be functional—if they weren't then the yeast would be unable to grow at the non-permissive temperature, thus the slower growth of the βG465D yeast implies that while it is functional, it is less functional than wild type. If there is less functional topoII target it would give a drug resistance phenotype.
The relaxation shows no significant difference to wild type which is unexpected as an enzyme with lower general function would be expected to have a slower reaction cycle. It is however possible that the experimental set up may be masking a slight impairment of activity. The βG465D decatenation for a single time point showed just 60.3% wild-type activity and subsequent decatenation versus time experiments showed an initial 12-fold rate reduction. Interestingly the βG465D displayed a narrower magnesium optima compared with βWT. In decatenation experiments the MgCl2 concentration was 10 µM, optimal for both βWT and βG465D proteins, and perhaps more importantly the ATP levels were saturating, enough perhaps to compensate for the enzyme's natural deficiency in ATP hydrolysis. ATP dependence assays showed maximum activity at 150 and 50 µM ATP for βG465D and βWT, respectively, and in decatenation assays ATP was at 2 mM.
The reason for this reduced decatenation rate is unclear. It is clear that the ATP requirements for βG465D differ markedly from βWT. Decatenation assays with varying concentrations of ATP showed a 3-fold decrease in ATP affinity with βG465D, and a corresponding 3-fold decrease in ATP hydrolysis activity with this mutant. This is particularly interesting as, while the mutation does lie towards the N-terminal ATPase domain, it is actually located in the B′ domain, close to where the G-helix is predicted to bind (8,44).
Mutations in this region have been reported for human topoIIα and yeast topoII. The topoIIα mutation R450Q (equivalent to βK466), lies adjacent to the mutation identified here (34,35). This mutation gives a 2.5-fold resistance to mAMSA and was first identified in the drug resistant leukaemic cell line CEM/VM-1, which was found to have a 2- to 8-fold decrease in ATP hydrolysis activity. In the case of yeast topoII residue G437S, equivalent to βG464, a loss of enzyme stability, reducing the number of functional target molecules, has been reported previously as a reason for drug resistance (45). The activity profile differed to that of βG465D and αR450Q in that G437S showed drug hypersensitivity and no alteration in ATP requirement and degradation was very rapid. Extrapolating between species can be problematic and has been described previously (33). Reduced stability is not seen with βG465D, so this is unlikely to be the mechanism of resistance in this case.
The primary sequence of topoIIβ places G465 adjacent to the transducer domain of the N-terminal ATPase domain, which is thought to function by transmitting the ‘information’ of ATP hydrolysis from the N-terminus to the cleavage core through a series of conformational changes. Experiments with WTα have shown that a two-residue insert at position 408, equivalent to βE425 (46), and a deletion stretching from 350 to 407 (47) can disrupt communication between domains. In these cases inter-domainal communication was completely abolished, and it is conceivable that a residue contacting this domain may have a communication role. Additional splice variants of human topoIIα have been identified with differently sized inserts in the transducer region of the ATPase domain, at positions K321, Q355 and A401 (48). It is possible that these alternative forms will have differences in the transduction of conformational changes which could account for differences in activity in vivo.
The proximity of the residue to the transducer domain also makes it possible that this inhibits signal transduction through conformational change. The yeast core crystal structure, on which all of the topoII models are based, lacks any data for the region linking the core and ATPase domains as this was too disordered, so it is unclear how these two interact. It is likely however that the ATP hydrolysis activity of the mutant is reduced through inhibition of signal transduction through the transducer domain to the N-terminal region.
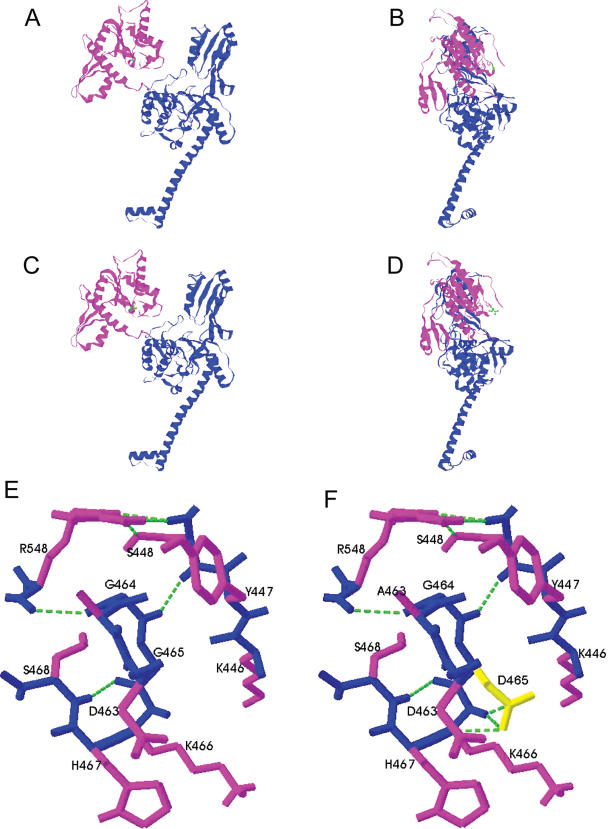
The position of G465 on the model based on the yeast structure, plus previous footprinting experiments suggest that glycine 465 lies very close to the area binding the gate helix. Lysines in yeast topoII have been identified that are protected on DNA binding, and one such residue identified was Lys 438. The equivalent residue in WTβ is K466 which is adjacent to G465 (8,44). Modelling the G465D mutation onto the yeast core structure using SwissProt shows that while the residue lies in the GyrB homology domain, and would therefore be expected to be involved in ATP hydrolysis, in eukaryotes this is actually located in the B′ domain, on the top surface of the semi-circular groove where the gate helix is thought to bind (Figure 7A–D) (8,44). The mutation of residue 465 from a glycine to an aspartic acid doesn't appear to massively change the 3D structure of the enzyme, which is expected as our selection procedure would not select a non-functional mutant. Analysis of this region shows that the mutation gives rise to some localized hydrogen bonding changes that could potentially alter the conformation of the enzyme (Figure 7E and F). The presence of the negatively charged aspartic acid creates an additional hydrogen bond to the backbone and could create more resistance to conformational change.
Figure 7.
Model of human topoIIβ monomer based on the yeast core structure [8]. The effect of the mutation was modelled using Swiss-PdbViewer 3.7, the figures were produced using RasMol. Shown are proteins WTβ in a front view (A) and a side view (B), and βG465D front view (C) and side view (D). The B′ domain of the protein is shown in magenta and the A′ domain in blue. Local hydrogen bonding changes close to residue 465 in the B′ domain are shown in WTβ (E) and βG465D (F). The backbone is shown in blue, side chains in magenta, residue 465 is shown in yellow and hydrogen bonds are shown as green dashed lines. The view in (E) and (F) is from the side, the same orientation as (B) and (D).
It is possible that the replacement of a glycine with an aspartic acid, and hence the addition of a negative charge, could alter the charge interactions involved in binding of the gate helix. Fluorescence anisotropy experiments with a 40 bp oligo showed a decreased BMAX value for βG465D, suggesting that the protein is binding in a slightly different conformation giving a consequent change in tumbling rate. However there was no significant difference in KD for the two proteins implying that binding affinity was not effected. This, along with the fact that the oligo used was not physiological, suggests that it is unlikely that impaired DNA binding is responsible for the decrease in decatenation seen with βG465D.
The narrower optimum for MgCl2 is interesting and could perhaps be explained by considering its role in the topoisomerase II mechanism. Mg2+ is thought to be utilized at both the ATP-binding domain and in the cleavage core. In the nucleotide-binding pocket Mg2+ has been shown to contact all of the phosphate groups as well as an invariant Asn residue (23,49). In the structure of the N-terminal region of human topoIIα it is also shown to hydrogen bond with a water molecule that is also forming contacts with Glu87, a residue found previously to be a general base ideally positioned to promote nucleophilic attack on the γ-phosphate (50). It is possible that the altered Mg2+ optima seen could be linked to an impairment in the nucleotide-binding pocket, whereby Glu87 is unable to promote ATP hydrolysis.
βG465 lies within the core region, where Mg2+ is thought to be involved in polarizing the active site tyrosine and stabilizing the cleavable complex intermediate, and is therefore co-ordinated near to the site of gate helix binding. There is also evidence for a structural role for Mg2+ in this region (27). In this region it is thought that Mg2+ is co-ordinated to a catalytic triad of aspartic acid residues and the active site tyrosine, allowing nucleophilic attack on the phosphate of DNA (40,51). The mutation of glycine 465 to aspartic acid introduces a residue that may also co-ordinate Mg2+. The cleavage site is likely to have a higher affinity for Mg2+ than the D465 region, which would explain why at lower Mg2+ concentrations both proteins are equally active, but at higher Mg2+ levels D465 may also co-ordinate magnesium in an inappropriate way. This additional Mg2+ associated with the protein could either interfere in the phosphoryltransfer reaction directly, or it could impair the movement of domains necessary for enzyme turnover.
The structure of human topoIIα shows that when AMPPNP (an ATP analogue) is bound the transducer domain assumes a packed conformation in which a catalytic lysine residue protrudes into the nucleotide-binding pocket and contacts the γ-phosphate. When ADP is bound however the transducer domain is rotated outward 10°, retracting the lysine from the pocket, in a conformation thought to be necessary for Pi release (49). It is thought that the Pi release event and removal of the so-called ‘switch lysine’ from the nucleotide-binding pocket may be coupled simultaneously with the separation of the G-segment in the cleavage core (52). Our data are consistent with this hypothesis, and it is probable that mutation of glycine 465 to aspartic acid impedes the communication between domains mediated by transducer region, giving a reduced ATP hydrolysis rate and slower enzyme turnover.
It is known that the transduction of signal through conformational change is necessary for co-ordination of core and N-terminal domains to give functional enzyme. Here we report a mutation which lies in the core domain, and yet gives a 3-fold reduction in activity at the N-terminal domain implying that transduction of signal has been affected. This mutation confers a resistance to drug that diminishes with time, suggesting that rather than being fatally impeded the enzyme is merely slowed down, a mechanism of resistance which to our knowledge is previously unreported.
Acknowledgments
We would like to acknowledge Rosalind Turnbull for preliminary experiments, and Gary Watters and Pauline Heslop for work done on the RFLP analysis, Dr Ian Cowell for help with figures, Prof. Bernard Connolly for advice regarding fluorescence anisotropy experiments and Prof. Harry Gilbert for advice with ATP hydrolysis assays. C.L. was funded by Cancer Research, UK; K.G. was funded by the Institute for Cell and Molecular Biosciences, The University of Newcastle Upon Tyne. Funding to pay the Open Access publication charges for this article was provided by Knowledge House.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindsley J.E., Wang J.C. Proteolysis patterns of epitopically labelled yeast DNA topoisomerase II suggest an allosteric transition in the enzyme induced by ATP binding. Proc. Natl Acad. Sci. USA. 1991;88:10485–10489. doi: 10.1073/pnas.88.23.10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin C.A., Marsh K.L., Wasserman R.A., Willmore E., Sayer P.J., Wang J.C., Fisher L.M. Expression, domain structure, and enzymatic properties of an active recombinant human DNA topoisomerase II beta. J. Biol. Chem. 1995;270:15739–15746. doi: 10.1074/jbc.270.26.15739. [DOI] [PubMed] [Google Scholar]
- 3.Caron P.R., Watt P., Wang J.C. The C-terminal domain of Saccharomyces cerevisiae DNA topoisomerase II. Mol. Cell. Biol. 1994;14:3197–3207. doi: 10.1128/mcb.14.5.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirski S.E., Gerlach J.H., Cummings H.J., Zirngibl R., Greer P.A., Cole S.P. Bipartite nuclear localization signals in the C terminus of human topoisomerase II alpha. Exp. Cell Res. 1997;237:452–455. doi: 10.1006/excr.1997.3805. [DOI] [PubMed] [Google Scholar]
- 5.Sengupta T., Mukherjee M., Mandal C., Das A., Majunider H.K. Functional dissection of the C-terminal domain of type II DNA topoisomerase from the kinetoplastid hemoflagellate Leishmania donovani. Nucleic Acids Res. 2003;31:5305–5316. doi: 10.1093/nar/gkg727. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Dang Q., Alghisi G.C., Gasser S.M. Phosphorylation of the C-terminal domain of yeast topoisomerase II by casein kinase II affects DNA protein interaction. J. Mol. Biol. 1994;243:10–24. doi: 10.1006/jmbi.1994.1626. [DOI] [PubMed] [Google Scholar]
- 7.Chikamori K., Grabowski D.R., Kinter M., Willard B.B., Hickson I.D., Andersen A.H., Ganapathi R., Ganapathi M.K. Phosphorylation of serine 1106 in the catalytic domain of topoisomerase IIα regulates enzymatic activity and drug sensitivity. J. Biol. Chem. 2003;278:12696–12702. doi: 10.1074/jbc.M300837200. [DOI] [PubMed] [Google Scholar]
- 8.Berger J.M., Gamblin S.J., Harrison S.C., Wang J.C. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 9.Fass D., Bogden C.E., Berger J.M. Quarternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nature Struct. Biol. 1999;6:322–326. doi: 10.1038/7556. [DOI] [PubMed] [Google Scholar]
- 10.Lindsley J.E., Wang J.C. On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. J. Biol. Chem. 1993;268:8096–8104. [PubMed] [Google Scholar]
- 11.Ali J.A., Jackson A.P., Howells A.J., Maxwell A. The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyses ATP and binds coumarin drugs. Biochemistry. 1993;32:2717–2724. doi: 10.1021/bi00061a033. [DOI] [PubMed] [Google Scholar]
- 12.Gardiner L.P., Roper D.I., Hammonds T.R., Maxwell A. The N-terminal domain of human topoisomerase II-alpha is a DNA-dependent ATPase. Biochemistry. 1998;37:16997–17004. doi: 10.1021/bi9818321. [DOI] [PubMed] [Google Scholar]
- 13.Hammonds T.R., Maxwell A. The DNA dependence of the ATPase activity of human DNA topoisomerase IIα. J. Biol. Chem. 1997;272:32696–32703. doi: 10.1074/jbc.272.51.32696. [DOI] [PubMed] [Google Scholar]
- 14.Olland S., Wang J.C. Catalysis of ATP hydrolysis by two NH(2)-terminal fragments of yeast DNA Topoisomerase II. J. Biol. Chem. 1999;274:21688–21694. doi: 10.1074/jbc.274.31.21688. [DOI] [PubMed] [Google Scholar]
- 15.Harkins T.T., Lindsley J.E. Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 1. A DNA dependent burst in ATP hydrolysis. Biochemistry. 1998;37:7292–7298. doi: 10.1021/bi9729099. [DOI] [PubMed] [Google Scholar]
- 16.Harkins T.T., Lewis T.J., Lindsley J.E. Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 2. Kinetic mechanism for the sequential hydrolysis of two ATP. Biochemistry. 1998;37:7299–7312. doi: 10.1021/bi9729108. [DOI] [PubMed] [Google Scholar]
- 17.Baird C.L., Gordon M.S., Andrenyak D.M., Marecek J.F., Lindsley J.E. The ATPase reaction cycle of yeast DNA topoisomerase II. Slow rates of ATP resynthesis and P(i) release. J. Biol. Chem. 2001;276:27893–27898. doi: 10.1074/jbc.M102544200. [DOI] [PubMed] [Google Scholar]
- 18.Roca J., Wang J.C. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 19.Williams N.L., Howells A.J., Maxwell A. Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol. 2001;306:969–984. doi: 10.1006/jmbi.2001.4468. [DOI] [PubMed] [Google Scholar]
- 20.Baird C.L., Harkins T.T., Morris S.K., Lindsley J.E. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl Acad. Sci. USA. 1999;96:13685–13690. doi: 10.1073/pnas.96.24.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skouboe C., Bjergbaek L., Oestergaard V.H., Larsen M.K., Knudsen B.R., Andersen A.H. A human topoisomerase II alpha heterodimer with only one ATP binding site can go through successive catalytic cycles. J. Biol. Chem. 2003;278:5768–5774. doi: 10.1074/jbc.M210332200. [DOI] [PubMed] [Google Scholar]
- 22.Wigley D.B., Davies G.J., Dodson E.J., Maxwell A., Dodson G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 23.Classen S., Olland S., Berger J.M. Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. Proc. Natl Acad. Sci. USA. 2003;100:10629–10634. doi: 10.1073/pnas.1832879100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen M.O., Larsen M.K., Barthelmes H.U., Hock R., Andersen C.L., Kjeldsen E., Knudsen B.R., Westergaard O., Boege F., Mielke C. Dynamics of human DNA topoisomerases IIalpha and IIbeta in living cells. J. Cell Biol. 2002;157:31–44. doi: 10.1083/jcb.200112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh K.L., Willmore E., Tinelli S., Cornarotti,M, Meczes E.L., Capranico,G, Fisher L.M., Austin C.A. Amsacrine-promoted DNA cleavage site determinants for the two human DNA topoisomerase II isoforms alpha and beta. Biochem. pharmacol. 1996;52:1675–1685. doi: 10.1016/s0006-2952(96)00516-3. [DOI] [PubMed] [Google Scholar]
- 26.Burden D.A., Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 27.Leontiou C., Lakey J.H., Austin C.A. Mutation E522K in human DNA topoisomerase IIβ confers resistance to methyl N-(4′-(9-acridinylamino)-phenyl)carbamate hydrochloride and methyl N-(4′-(9-acridinylamino)-3-methoxy-phenyl)methane sulphonamide but hypersensitivity to Etoposide. Mol. Pharm. 2004;66:430–439. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 28.Alton P.A., Harris A.L. The role of DNA topoisomerase II in drug resistance. Br. J. Haematol. 1993;85:241–245. doi: 10.1111/j.1365-2141.1993.tb03162.x. [DOI] [PubMed] [Google Scholar]
- 29.Robert J., Larsen A.K. Drug resistance to topoisomerase II inhibitors. Biochimie. 1998;80:247–254. doi: 10.1016/s0300-9084(98)80007-2. [DOI] [PubMed] [Google Scholar]
- 30.Dingemans A.M., Pinedo H.M., Giaccone G. Clinical resistance to topoisomerase-targeted drugs. Biochim. Biophys. Acta. 1998;1400:275–288. doi: 10.1016/s0167-4781(98)00141-9. [DOI] [PubMed] [Google Scholar]
- 31.Patel S., Jazrawi E., Creighton A.M., Austin C.A., Fisher L.M. Probing the interaction of the cytotoxic bisdioxopiperazine ICRF-193 with the closed enzyme clamp of human topoisomerase II alpha. Mol. Pharmacol. 2000;58:560–568. doi: 10.1124/mol.58.3.560. [DOI] [PubMed] [Google Scholar]
- 32.Renodon-Corniere A., Jensen L.H., Nitiss J.L., Jensen P.B., Sehested M. Analysis of bisdioxopiperazine dexrazoxane binding to human DNA topoisomerase II alpha: decreased binding as a mechanism of drug resistance. Biochemistry. 2003;42:9749–9754. doi: 10.1021/bi034557d. [DOI] [PubMed] [Google Scholar]
- 33.Patel S., Keller B.A., Fisher L.M. Mutations at Arg486 and Glu571 in human topoisomerase II alpha confer resistance to amsacrine: relevance for antitumor drug resistance in human cells. Mol. Pharmacol. 2000;57:784–791. doi: 10.1124/mol.57.4.784. [DOI] [PubMed] [Google Scholar]
- 34.Bugg B.A., Danks M.K., Beck W.T., Suttle D.P. Expression of a mutant DNA topoisomerase II in CCRF-CEM human leukaemic cells selected for resistance to teniposide. Proc. Natl Acad. Sci. USA. 1991;88:7654–7658. doi: 10.1073/pnas.88.17.7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsiung Y., Jannatipour M., Rose A., McMahon J., Duncan D., Nitiss J.L. Functional expression of human topoisomerase II alpha in yeast: mutations at amino acids 450 or 803 of topoisomerase II alpha result in enzymes that can confer resistance to anti-topoisomerase II agents. Cancer Res. 1996;56:91–99. [PubMed] [Google Scholar]
- 36.Bauduer F., Delmer A., Blanc M.C., Delmas-Marsalet B., Cadiou M., Rio B., Marie J.P., Zittoun R. Treatment of chronic myelogenous leukaemia in blast crisis and in accelerated phase with high or intermediate-dose cytosine arabinoside and amsacrine. Leuk. Lymphoma. 1993;10:195–200. doi: 10.3109/10428199309145883. [DOI] [PubMed] [Google Scholar]
- 37.Dittrich C., Dieras V., Kerbrat P., Punt C., Sario R., Coponigro F., Paoletti X., de Bahicourt C., Lacombe D., Fumoleau P. Phase II study of XR5000 (DACA), an inhibitor of topoisomerase I and II, administered as a 120-h infusion in patients with advanced ovarian cancer. Invest. New Drugs. 2003;21:347–352. doi: 10.1023/a:1025476813365. [DOI] [PubMed] [Google Scholar]
- 38.Meczes E.L., Marsh K.L., Fisher L.M., Rogers M.P., Austin C.A. Complementation of temperature sensitive topoisomerase II mutations in Saccharomyces cerevisiae by a human TOP2 beta construct allows the study of topoisomerase II beta inhibitors in yeast. Cancer Chemother. Pharmacol. 1997;39:367–375. doi: 10.1007/s002800050585. [DOI] [PubMed] [Google Scholar]
- 39.Nitiss J.L. Using yeast to study resistance to topoisomerase II-targeting drugs. Cancer Chemother. Pharmacol. 1994;34:S6–S13. doi: 10.1007/BF00684857. [DOI] [PubMed] [Google Scholar]
- 40.West K.L., Meczes E.L., Thorn R., Turnbull R.M., Marshall R., Austin C.A. Mutagenesis of E477 or K505 in the B′ domain of human topoisomerase II beta increases the requirement for magnesium ions during strand passage. Biochemistry. 2000;39:1223–1233. doi: 10.1021/bi991328b. [DOI] [PubMed] [Google Scholar]
- 41.Osheroff N., Zechiedrich E.L. Calcium-promoted DNA cleavage by eukaryotic topoisomerase II: trapping the covalent enzyme–DNA complex in an active form. Biochemistry. 1987;26:4303–4309. doi: 10.1021/bi00388a018. [DOI] [PubMed] [Google Scholar]
- 42.Finlay G.J., Riou J.-F., Bagulay B.C. From amsacrine to DACA (N-2-(dimethylamino)ethyl.acridines-4-carboxamide): selectivity for topoisomerases I and II among acridines derivatives. Eur. J. Cancer. 1996;32A:708–714. doi: 10.1016/0959-8049(95)00604-4. [DOI] [PubMed] [Google Scholar]
- 43.Bridewell D.J.A., Finlay G.J., Baguley B.C. Mechanism of cytotoxicity of N-2-(dimethylamino)ethyl.acridines-4-carboxamide and of its 7-chloro derivative: the roles of topoisomerases I and II. Cancer Chemother. Pharmacol. 1999;43:302–308. doi: 10.1007/s002800050899. [DOI] [PubMed] [Google Scholar]
- 44.Li W., Wang J.C. Footprinting of yeast DNA topoisomerase II lysyl side chains involved in substrate binding and interdomainal interactions. J. Biol. Chem. 1997;272:31190–31195. doi: 10.1074/jbc.272.49.31190. [DOI] [PubMed] [Google Scholar]
- 45.Sabourin M., Wilson Byl J.A., Hannah S.E., Nitiss J.L., Osheroff N. A mutant yeast topoisomerase II (top2G437S) with differential sensitivity to anticancer drugs in the presence and absence of ATP. J. Biol. Chem. 1998;273:29086–29092. doi: 10.1074/jbc.273.44.29086. [DOI] [PubMed] [Google Scholar]
- 46.Oestergaard V.H., Bjergbaek L., Skouboe C., Giangiacomo L., Knudsen B.R., Andersen A.H. The transducer domain is important for clamp operation in human topoisomerase II alpha. J. Biol. Chem. 2004;279:1684–1691. doi: 10.1074/jbc.M309624200. [DOI] [PubMed] [Google Scholar]
- 47.Bjergbaek L., Kingma P., Nielsen I.S., Wang Y., Westergaard O., Osheroff N., Andersen A.H. Communication between the ATPase and cleavage/religation domains of human topoisomerase II alpha. J. Biol. Chem. 2000;275:13041–13048. doi: 10.1074/jbc.275.17.13041. [DOI] [PubMed] [Google Scholar]
- 48.Petruti-Mot A.S., Earnshaw W.C. Two differentially spliced forms of topoisomerase IIalpha and beta mRNAs are conserved between birds and humans. Gene. 2000;258:183–192. doi: 10.1016/s0378-1119(00)00465-0. [DOI] [PubMed] [Google Scholar]
- 49.Wei H., Ruthenburg A.J., Bechis S.K., Verdine G.L. Nucleotide-dependent domain movement in the ATPase domain of a human type IIA DNA topoisomerase. J. Biol. Chem. 2005;280:37041–37047. doi: 10.1074/jbc.M506520200. [DOI] [PubMed] [Google Scholar]
- 50.Jackson A.P., Maxwell A. Identifying the catalytic residue of the ATPase reaction of DNA gyrase. Proc. Natl Acad. Sci. USA. 1993;90:11232–11236. doi: 10.1073/pnas.90.23.11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noble C.G., Maxwell A. The role of GyrB in the DNA cleavage–religation reaction of DNA gyrase: a proposed two metal-ion mechanism. J. Mol. Biol. 2002;318:361–371. doi: 10.1016/S0022-2836(02)00049-9. [DOI] [PubMed] [Google Scholar]
- 52.Schoeffler A.J., Berger J.M. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem. Soc. Trans. 2005;33:1465–1470. doi: 10.1042/BST0331465. [DOI] [PubMed] [Google Scholar]