Abstract
Human 8-oxoguanine-DNA glycosylase (OGG1) is the major enzyme for repairing 8-oxoguanine (8-oxoG), a mutagenic guanine base lesion produced by reactive oxygen species (ROS). A frequently occurring OGG1 polymorphism in human populations results in the substitution of serine 326 for cysteine (S326C). The 326 C/C genotype is linked to numerous cancers, although the mechanism of carcinogenesis associated with the variant is unclear. We performed detailed enzymatic studies of polymorphic OGG1 and found functional defects in the enzyme. S326C OGG1 excised 8-oxoG from duplex DNA and cleaved abasic sites at rates 2- to 6-fold lower than the wild-type enzyme, depending upon the base opposite the lesion. Binding experiments showed that the polymorphic OGG1 binds DNA damage with significantly less affinity than the wild-type enzyme. Remarkably, gel shift, chemical cross-linking and gel filtration experiments showed that S326C both exists in solution and binds damaged DNA as a dimer. S326C OGG1 enzyme expressed in human cells was also found to have reduced activity and a dimeric conformation. The glycosylase activity of S326C OGG1 was not significantly stimulated by the presence of AP-endonuclease. The altered substrate specificity, lack of stimulation by AP-endonuclease 1 (APE1) and anomalous DNA binding conformation of S326C OGG1 may contribute to its linkage to cancer incidence.
INTRODUCTION
Reactive oxygen species (ROS) are produced as a by-product of cellular metabolism and through exposure to ultraviolet (UV) and ionizing radiation and environmental carcinogens (1–5). ROS react with DNA to produce a myriad of cytotoxic and mutagenic base lesions (5). A major base damage produced by ROS is 7,8-dihydro-8-oxoguanine (8-oxoG). Unlike normal guanine, 8-oxoG has the propensity to mispair with adenine during DNA replication, resulting in the fixation of G:C to T:A transversion mutations (6). Oxidatively modified bases, such as 8-oxoG, are repaired primarily by the base excision repair pathway (BER), the first steps of which are the recognition and excision of the damaged base by a specific DNA glycosylase. The major mammalian enzyme for removing 8-oxoG from DNA is 8-oxoguanine-DNA glycosylase (OGG1) (7–13). OGG1 is a bifunctional enzyme, having both 8-oxoG excision activity and a weak AP-lyase strand incision activity at abasic sites (7–13). Following excision of 8-oxoG by OGG1, the resultant abasic site is further processed in sequential steps by several enzymes to complete repair (14).
Studies of OGG1 knockout mice and immunodepletion experiments suggest that OGG1 is the major mammalian 8-oxoguanine repair activity in non-transcribed DNA (15–23). It is widely accepted that accumulation of oxidative DNA damage over time can lead to cancer (24). A role for OGG1 in tumor suppression is suggested by the frequent loss of the OGG1 chromosomal locus in human lung and renal cancers and by significantly lower OGG1 activity among lung cancer patients compared to controls (10,25–28). Increased late-onset lung tumors in knockout mice deficient in repair of 8-oxoG, elevated 8-oxoG levels in lung tissue of lung cancer patients and decreased repair of 8-oxoG demonstrated in several human cancer cells lines suggest that cancer and 8-oxoG repair capacity may be linked (21–23,29–33). Additional studies suggest that genomic accumulation of 8-oxoG during cellular senescence may be due in part to age-associated changes in the level and subcellular localization of OGG1 (34–36). Recently, it was reported that polymorphic S326C OGG1 expressed in human cells is excluded from nucleoli during S-phase (37). Such changes in the subcellular localization of S326C OGG1 could be related to 8-oxoguanine repair capacity in vivo. Changes in the OGG1 coding sequence that result in amino acid substitutions that affect function, abundance or intracellular location could be anticipated to impact genomic 8-oxoG levels, and thereby influence genomic stability and carcinogenesis.
Several OGG1 polymorphisms have been reported and positively correlate with a variety of cancers [reviewed in (38–41)]. A frequently occurring polymorphism results in the substitution of serine for cysteine at position 326 in the C-terminus of OGG1 (25). The allele frequency of S326C OGG1 measured in human populations ranges from 0.13 to as high as 0.62 and varies significantly with ethnicity (39). Association studies have identified that individuals homozygous for the S326C OGG1 allele have increased incidence of lung, prostate and orolaryngeal cancers (42–48). A previous study found decreased catalytic efficiency (kcat/Km) of purified polymorphic S326C OGG1 (49), while another study implicated the S326C genotype with decreased 8-oxoguanine repair capacity in vivo (50). We characterized the glycosylase and AP-lyase activities and DNA damage binding affinity of purified S326C and found novel functional defects in the polymorphic OGG1 and a distinct dimeric DNA binding conformation compared to the wild-type enzyme. Our results confirm that S326C has decreased repair activity towards 8-oxoG paired with C and further show that S326C OGG1 is particularly deficient in 8-oxoguanine excision activity when the lesion is opposite T or G. The stimulation of wild-type OGG1 by AP-endonuclease 1 (APE1) results in increased rates of 8-oxoG excision, and is believed to be an important step in the regulation and coordination of BER in vivo (51). We show that S326C OGG1 is not significantly stimulated by APE1, unlike the wild-type enzyme, thereby the coordination of BER may be perturbed during repair of 8-oxoguanine by S326C OGG1. We observed decreased repair activity and dimeric conformation of S326C OGG1 expressed in human cells, thus the altered activity and dimeric stoichiometry of the S326C OGG1 variant may be relevant in vivo.
MATERIALS AND METHODS
Cell culture
HeLa cells (CCL-2) were obtained from ATCC and maintained in EMEM medium containing 10% fetal bovine serum (Gibco) in 5% CO2 at 37°C. Cells were transfected by using Fugene 6 transfection reagent (Roche) according to the manufacturer's instructions. Nuclear extracts were prepared using NE-PER extraction reagents (Pierce).
Expression and purification of recombinant OGG1 and APE1 proteins
The human OGG1 1a coding sequence in pET-28a expression vector (Novagen) was used to express his-tagged OGG1 in BL21 Codon Plus (Stratagene), Rosetta Blue (Novagen) and BL21 Star (Invitrogen). pET-28a plasmids containing the wild-type CΔ19, S326C, S326C CΔ19 and CΔ20 OGG1 coding sequence were created with a Quik Change II XL mutagenesis kit (Stratagene) or PCR. Mammalian expression vectors encoding N-terminally FLAG-tagged OGG1s were constructed by cloning PCR amplified OGG1 genes into pCMV-2B expression vector (Stratagene). The human APE1 coding sequence in pET-15b expression vector (Novagen) was used to express his-tagged APE1 in BL21 Codon Plus. All plasmid sequences were verified by bi-directional DNA sequencing. Proteins were purified from sonicated cell-free extracts by using NTA (nickel-nitriloacetic acid) agarose (Qiagen) chromatography. His-tags were removed by incubation with biotinylated thrombin (Novagen). Thrombin was subsequently removed from cleavage reactions by incubation with streptavidin beads. Additional purification was performed by ion exchange chromatography using an AKTA FPLC purification system (Amersham). Proteins were further purified by being passed through a Q HP column (Amersham) and bound to an S HP column (Amersham) at 150 mM NaCl. Over a linear NaCl gradient of 150–600 mM, proteins were eluted at ∼350 mM NaCl. Finally, proteins were purified to homogeneity by size-exclusion chromatography using a Superdex 200 HR column (Amersham). Purified proteins were dialyzed against 20 mM Tris–HCl (pH 7.4), 300 mM NaCl, 10% glycerol and stored at −80°C.
Preparation of duplex DNA substrates
31mer oligos with the sequence 5′-GTGACTACGAGACCTXATGTGACTGAGAGAG-3′ containing 8-oxoG or uracil at position X were obtained from Midland Certified Reagent Company (Midland, TX) and Invitrogen, respectively. Damage-containing oligos were purified by high-performance liquid chromatography (HPLC) and the 8-oxoG-containing oligo was verified by mass spectrometry. Complementary HPLC-purified oligos having C, T, G or A opposite X were obtained from Invitrogen. Damage-containing oligos were annealed with a 1.2-fold excess of the complementary strand by heating to 95°C in 20 mM Tris–HCl (pH 7.4), 100 mM NaCl, 1 mM MgCl2 and slow cooling to room temperature. A duplex substrate having an abasic (AP) site opposite C (AP·C) was prepared by incubating a U·C-containing duplex with uracil DNA glycosylase (UDG) (New England Biolabs). UDG was subsequently removed by phenol–chloroform extraction and ethanol precipitation. Duplex substrates were 3′ end labeled with [α-32P]dCTP (Dupont) and Klenow exo-DNA polymerase (New England Biolabs). Unincorporated radioactivity was removed by using MicroSpin G-25 columns (Amersham).
8-oxoguanine glycosylase and AP-lyase activity assays
OGG1 proteins or nuclear extracts were incubated with DNA substrates at varying concentrations in 20 µl reactions containing 20 mM Tris–HCl (pH 7.4), 100 mM NaCl and 0.15 µg/µl BSA (New England Biolabs). To measure glycosylase activity, reactions were terminated by adding SDS and piperidine to 5% and 200 mM, respectively. Reactions were heated at 95°C for 5 min to cleave DNA at abasic sites. When measuring AP-lyase activity with an AP site substrate, reactions were terminated by adding SDS and glycerol to 5 and 10%, respectively, without heating. Reactions were electrophoresed on 20% acrylamide gels containing 7 M urea and radioactivity was quantified using a Storm phosphorimager and ImageQuant software (Molecular Dynamics). Apparent kinetic constants and were derived from Lineweaver–Burke double-reciprocal plots of kinetic data. Experiments were performed in triplicate and are shown with standard deviation.
Sodium borohydride trapping of OGG1
In a sodium borohydride trapping assay, OGG1 is reacted with an 8-oxoG·C substrate in the presence of a strong chemical reductant. After base excision, OGG1 may proceed to cleave the abasic site produced by its glycosylase activity via a β-elimination mechanism to create a single strand break (9,11,12). The reductant causes a covalent linkage of the enzyme to a reaction intermediate in the cleavage of an abasic site, thereby creating a ‘trapped’ enzyme–DNA complex (52). Chemical linkage of OGG1 to reaction intermediates was performed by incubating OGG1 enzymes (5 µM) and an 8-oxoG·C substrate (5 µM) in 20 mM Tris–HCl (pH 7.4), 100 mM NaCl, 1 mM MgCl2 with 1 mM sodium cyanoborohydride (Sigma) at 37°C for 30 min. Trapped OGG1 enzyme–DNA complexes were separated from free enzyme by SDS–PAGE electrophoresis on 4–20% acrylamide gels (Invitrogen) and quantified with Kodak 1D gel analysis software.
Chemical cross-linking of OGG1
Wild-type and polymorphic OGG1 (5 µM) were incubated with or without 8-oxoG·C substrate (10 µM) in 50 mM Tris–HCl (pH 7.4), 50 mM NaCl in the presence of 1 mM of homobifunctional sulfhydryl-reactive cross-linker BM[PEO]4 (1,11-bis-Maleimidotetraethyleneglycol) (Pierce). Reactions were incubated at 4°C for 2 h and analyzed by electrophoresis on 4–20% SDS–PAGE gels and Coomassie staining (Bio-Rad).
Determination of dissociation constants by electrophoretic mobility shift assay (EMSA)
Binding affinities were measured by incubating OGG1 proteins (5 nM) with DNA substrates (10 nM) in 20 mM Tris–HCl (pH 7.4), 50 mM NaCl, 0.15 µg/µl BSA, 15% glycerol for 5 min at 0°C. Bound complexes were separated from free substrate by electrophoresis at 4°C on native 5% acrylamide (80:1) gels containing 7 mM Tris–HCl (pH 7.4), 3 mM sodium acetate and 1 mM EDTA. No enzymatic cleavage of substrates occurred under the conditions used (data not shown). Gels were then vacuum dried and shifts quantified with a phosphorimager. Dissociation constants were calculated using the following equations.
where r =[PA] / [P] + [PA] (53); [P], [A] and [PA] represent the concentrations of enzyme, substrate and enzyme–substrate complex, respectively.
RESULTS
Functional comparison of wild-type and S326C OGG1
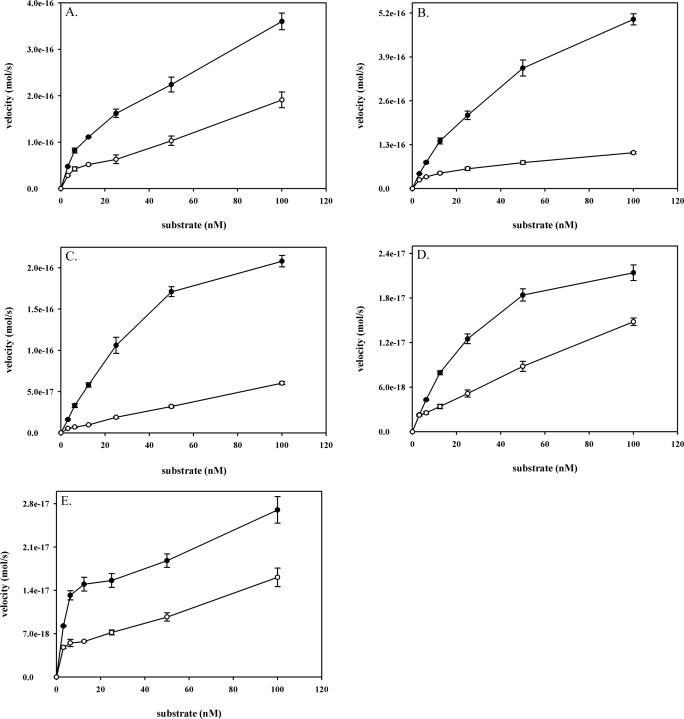
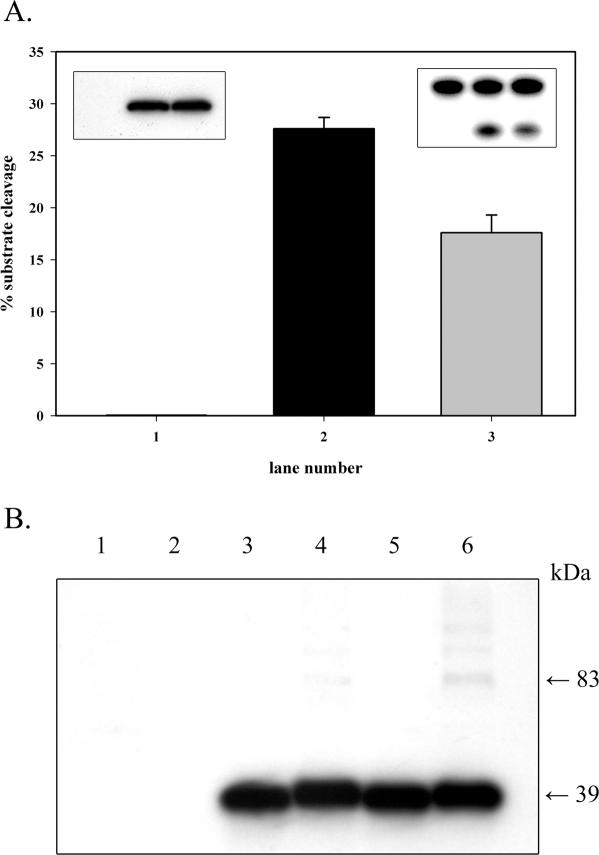
Purified wild-type and S326C OGG1 (Figure 1) were reacted with excess DNA substrates over a range of concentrations (Figure 2). Since reaction data were used to derive apparent Michaelis–Menten constants, enzyme reactions were carried out with excess substrate. A large excess of substrate over enzyme was used at each concentration point to ensure measurement of steady-state catalytic rates where the concentration of ES (enzyme–substrate complex) remains constant throughout the reaction and substrate is not limiting. With duplex substrates containing 8-oxoguanine opposite all four bases and an AP site opposite C substrate, S326C exhibited decreased glycosylase and AP-lyase activities compared to wild-type OGG1 (Figure 2A–E and Table 1). The catalytic efficiency constant (kcat/Km), a measure of how often bound molecules react to produce product, was determined for both enzymes with all substrates (Table 1). With an 8-oxoG·C substrate, S326C OGG1 had a 1.6-fold reduced catalytic efficiency, in agreement with a previous study (49). The S326C isoform also exhibited a 1.6-fold decreased efficiency in AP-lyase activity with an AP·C substrate (Table 1). With substrates containing 8-oxoguanine paired with T, G or A, efficiencies were decreased by 1.4-fold, 3-fold or were similar to the wild-type enzyme, respectively (Table 1). Catalytic constants (kcat) of S326C were at least 2-fold less than those of the wild-type enzyme (8-oxoG·C substrate) and more than 10-fold less than the wild-type (8-oxoG·G substrate). Reaction velocities (measured at a substrate concentration of 25 nM) represent actual rates of base excision for both enzymes (Figure 2 and Table 1). The S326C enzyme excised 8-oxoG opposite C and A and cleaved an AP site opposite C at rates roughly 2-fold lower than the wild-type enzyme (Table 1). Compared to the wild-type enzyme, S326C was particularly deficient in excision of 8-oxoG paired with T and G, with reaction rates decreased 3.7- and 5.6-fold, respectively (Table 1). These experiments indicate that the S326C polymorphism results in decreased rates of 8-oxoG excision, the extent of which are dependent upon the base opposite the lesion. Similar differences in the activity of wild-type and S326C OGG1 were consistently observed with multiple enzyme preparations expressed in different cell lines and with other 8-oxoguanine-containing duplex oligo substrates (data not shown). Thus, the functional defects of S326C are intrinsic to the enzyme and independent of any specific effects relating to enzyme preparation or sequence context.
Figure 1.
SDS–PAGE analysis of purified proteins. Lane 1, 0.5 µg wild-type OGG1; lane 2, 0.5 µg S326C OGG1; lane 3, 0.5 µg APE1, lane M, molecular weight marker. Proteins were purified as described in Materials and Methods and electrophoresed on a 4–20% acrylamide gel.
Figure 2.
Kinetics of wild-type and S326C OGG1. Wild-type and S326C OGG1 (2.5 nM) were incubated with increasing amounts (3.25–100 nM) of duplex 8-oxoguanine substrates having C (A), T (B), G (C) or A (D) opposite 8-oxoG, or with a substrate having an abasic site opposite C (E). Glycosylase reactions with 8-oxoguanine paired with C, T and G, were incubated for 15 min at 37°C. AP-lyase reactions with the AP·C substrate and glycosylase reactions with 8-oxoG opposite A were incubated for 1 h at 37°C. Reactions were terminated and analyzed as described in Materials and Methods. Data points are means of three independent experiments with standard deviation.
Table 1.
Kinetic and dissociation constants for wild-type and polymorphic S326C OGG1
| Enzyme | Substrate | (nM) | (× 10−3) (min−1) | kcat/Km (M−1 s−1) | Δkcat/Km | Velocity (25 nM) (mol/s)(× 10−16) | ΔVelocity | (nM) | ΔKd |
|---|---|---|---|---|---|---|---|---|---|
| OGG1 | 8-oxoG·C | 16.3 ± 1.3 | 339 ± 19 | 20.7 | 1.62 | 12.7 ± 0.9 | |||
| S326C | 8-oxoG·C | 12 ± 2.1 | 150 ± 18 | 12.5 | ↓1.65 | 0.63 | ↓2.5 | 35.9 ± 2.4 | ↑2.8 |
| OGG1 | 8-oxoG·T | 40.1 ± 3.5 | 692 ± 51 | 17.3 | 2.17 | 6.9 ± 0.6 | |||
| S326C | 8-oxoG·T | 7.4 ± 0.8 | 94 ± 5 | 12.7 | ↓1.36 | 0.59 | ↓3.7 | 25.5 ± 1.8 | ↑3.7 |
| OGG1 | 8-oxoG·G | 70.7 ± 6.6 | 458 ± 39 | 6.47 | 1.06 | 15.4 ± 1.3 | |||
| S326C | 8-oxoG·G | 16.3 ± 2.3 | 35.2 ± 3 | 2.15 | ↓3 | 0.19 | ↓5.6 | 36.3 ± 2.8 | ↑2.3 |
| OGG1 | 8-oxoG·A | 44.7 ± 2.9 | 40.5 ± 2.4 | 0.90 | — | 0.12 | 6.5 ± 6.1 | ||
| S326C | 8-oxoG·A | 9.2 ± 1.2 | 9.1 ± 0.5 | 0.98 | 0.05 | ↓2.4 | 53.7 ± 3.2 | ↑8.3 | |
| OGG1 | AP·C | 5.3 ± 0.9 | 25.6 ± 0.3 | 4.8 | 0.15 | 5.9 ± 0.7 | |||
| S326C | AP·C | 3.7 ± 0.2 | 11.1 ± 0.8 | 3.0 | ↓1.6 | 0.07 | ↓2.1 | 23.4 ± 1.3 | ↑3.9 |
Actual kinetic data presented in Figure 2 were used to derive kinetic parameters from double-reciprocal Lineweaver–Burke plots. Actual electrophoretic mobility shift data from experiments presented in Figure 3 were used to calculate dissociation constants as described in Materials and Methods. All constants were calculated from experiments performed in triplicate and are shown with standard deviation.
S326C OGG1 has decreased binding affinity for 8-oxoguanine and abasic sites
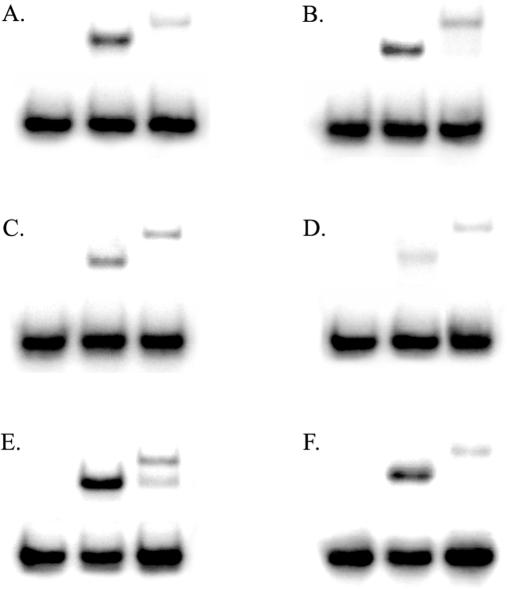
The binding affinities of wild-type and S326C OGG1 for duplex DNA substrates containing 8-oxoguanine paired with all four bases and an AP site opposite C substrate were determined by EMSA (Figure 3 and Table 1). With the exception of the 8-oxoG·A substrate, which both enzymes bound poorly, S326C exhibited markedly decreased binding affinity for 8-oxoguanine and an abasic site compared to the wild-type OGG1 (Figure 3). Dissociation constants (Kd) of S326C were significantly increased for all substrates relative to the wild-type enzyme, except with 8-oxoG·A, indicating decreased binding affinity for all major substrates. The lower DNA damage binding affinity of the polymorphic OGG1 can be seen directly in gel shifts in Figure 3 that represent actual data used to calculate dissociation constants in Table 1.
Figure 3.
EMSAs of wild-type and S326C OGG1. DNA damage binding affinities of wild-type and S326C OGG1 were measured by incubation with DNA substrates containing 8-oxoG·C (A), 8-oxoG·T (B), 8-oxoG·G (C), 8-oxoG·A (D) and AP·C (E). (F) Identical to the experiment in (A), with the addition of 1 mM DTT. Gel shifts of glycosylases were performed as described in Materials and Methods. In all panels, lane 1, no enzyme; lane 2, wild-type OGG1; lane 3, S326C OGG1.
S326C OGG1 exists as a dimer in solution and upon binding to DNA
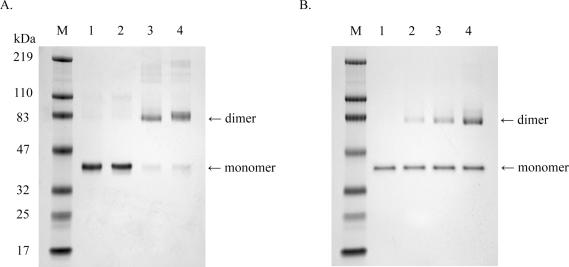
Gel shifts of wild-type and S326C OGG1 with all substrates used produced shifts of differing sizes (Figure 3A–E). Wild-type OGG1 shifts consistently produced species of a single mobility, while S326C primarily produced a higher molecular weight species consistent with an OGG1 dimer (Figure 3). To examine the possibility of dimerization of polymorphic OGG1, wild-type and S326C OGG1 were incubated with a chemical cross-linker both in the presence and absence of excess DNA substrate (Figure 4). Wild-type OGG1 was not significantly cross-linked, while S326C OGG1 cross-linked exclusively as a dimer both with and without 8-oxoguanine-containing substrate (Figure 4A). Small amounts of high molecular weight multimers could be seen in cross-linking experiments (Figure 4A, lanes 3 and 4), suggesting that, although S326C exists primarily as a dimer, multimeric complexes may form at high enzyme concentrations. Traces of wild-type OGG1 dimer could be seen after cross-linking (Figure 4A, lanes 1 and 2), so the possibility of a role for dimerization at some stage in the catalysis by wild-type OGG1 cannot be excluded. To determine if polymorphic S326C OGG1 is able to dimerize with wild-type OGG1, the wild-type enzyme was incubated either alone or with increasing amounts of S326C OGG1 in the presence of cross-linker (Figure 4B). The amount of OGG1 monomer was unchanged by the addition of excess S326C under cross-linking conditions, suggesting that the two isoforms do not form a heterodimer (Figure 4B).
Figure 4.
Dimerization of polymorphic S326C OGG1. (A) Wild-type (lanes 1 and 2) and S326C OGG1 (lanes 3 and 4) at a concentration of 5 µM were incubated with 1 mM of BM[PEO]4 cross-linker in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 10 µM duplex 8-oxoG·C substrate. Lane M, molecular weight marker. Reactions were analyzed by SDS–PAGE on 4–20% acrylamide gels. (B) Wild-type OGG1 (2.5 µM) in lanes 1–4 was incubated with 1 mM BM[PEO]4 in the presence of 0 (lane 1), 1.25 µM (lane 2), 2.5 µM (lane 3) and 5 µM (lane 4) S326C OGG1.
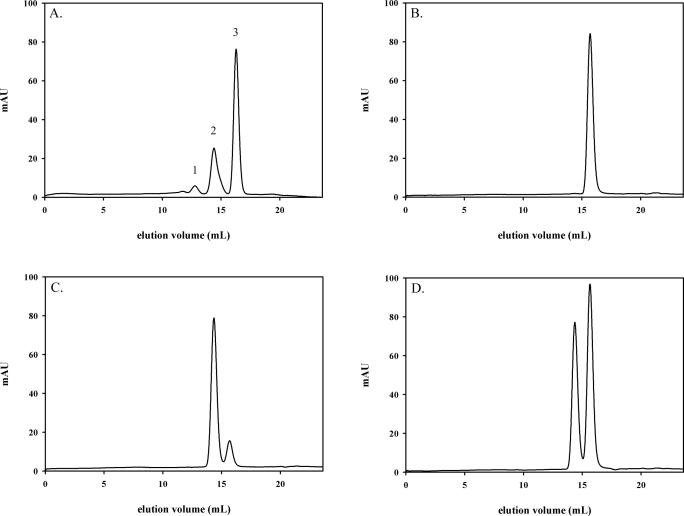
The dimeric conformation of S326C OGG1 was further investigated by size-exclusion chromatography of both OGG1 isoforms under native conditions (Figure 5). Wild-type OGG1 (Figure 5B) eluted from a Superdex 200 HR size-exclusion column (molecular weight range 10–600 kDa) later than the S326C OGG1 (Figure 5C), consistent with the larger molecular size of the S326C dimer. Approximately 90% of the S326C OGG1 exited the column as the higher molecular weight dimeric form, suggesting the polymorphic OGG1 exists primarily as a dimer in its native state (Figure 5C). Equal amounts of wild-type and S326C OGG1 were mixed and run through a size-exclusion column together (Figure 5D). The appearance of two distinct peaks identical to peaks of the individual enzymes indicates that S326C is distinct in size from and significantly larger than the wild-type enzyme, consistent with a dimeric S326C. A comparison of the elution profile of native protein markers (Figure 5A) with those of wild-type and S326C OGG1 (Figure 5B and C) indicate molecular sizes for an OGG1 monomer and dimer consistent with those observed in Figure 4A. The elution of distinct peaks also suggests that the two isoforms do not heterodimerize, consistent with the results in Figure 4B. The polymorphic enzyme could also be cross-linked as a dimer by the primary amine-reactive cross-linker BS3 [Bis (sulfosuccinimidyl) suberate] (Pierce) (data not shown). S326C bound 8-oxoguanine substrates solely as a dimer (Figure 3A–D), while an abasic site was bound both in monomeric and dimeric conformations (Figure 3E), suggesting that the type of lesion bound may influence S326C dimerization. We observed that a small fraction of S326C OGG1 produced in Escherichia coli is chemically linked as a dimer via a reducible disulfide bond (data not shown). We examined the activity and conformation of S326C OGG1 in the presence and absence of 1 mM DTT and found no significant effects on enzymatic activity or dimeric stoichiometry (Figures 3F and 6A). Thus, small amounts of disulfide linking of S326C expressed in bacteria is likely due to the close proximity of S326C OGG1 molecules in a dimer and has a negligible effect on function and substrate binding of the dimer.
Figure 5.
Size-exclusion chromatographic analysis of wild-type and S326C OGG1. (A) Non-denatured protein size markers (Sigma). Peak 1, BSA dimer (132 kDa); peak 2, BSA monomer (66 kDa) and peak 3, carbonic anydrase (29 kDa). Purified wild-type OGG1 (100 µg) was analyzed on a Superdex 200 HR column equilibrated with 20 mM Tris–HCl (pH 7.4), 300 mM NaCl at a flow rate of 0.25 ml/min (B). Identical runs were performed with 100 µg polymorphic S326C OGG1 (C) or 100 µg of both wild-type and S326C OGG1 together (D).
Figure 6.
Effect of AP-endonuclease on wild-type and S326C OGG1. (A) Glycosylase activities of wild-type (black columns) and S326C OGG1 (grey columns) (12.5 nM) were measured with an 8-oxoG·C substrate (250 nM) with or without an equimolar amount of APE1, in the presence or absence of 1 mM DTT. Inset, actual data. Lanes 1–4, wild-type OGG1; lanes 5–8, S326C; lanes 2, 4, 6 and 8, plus 1 mM DTT; lanes 3–4 and 7–8 plus 12.5 nM APE1. Inset, actual data. (B) Binding of OGG1 enzymes to an abasic site substrate after adding APE1 was measured by EMSA. Wild-type and S326C OGG1 (5 nM) were incubated with an AP site-containing duplex substrate (5 nM) in the presence of 1 mM MgCl2 at 37°C for 1 min prior to adding an equimolar amount of APE1 and incubating for an additional 1 min at 37°C. Lane 1, no enzyme; lanes 2 and 3, wild-type OGG1; lanes 4 and 5, S326C OGG1; lanes 3 and 5, plus APE1. Inset, actual data. (C) Borohydride trapping of OGG1 enzymes. Lane 1, molecular weight marker. Trapping of wild-type (lanes 2 and 3) and S326C OGG1 (lanes 4 and 5) with an 8-oxoG·C substrate in the presence (lanes 3 and 5) and absence (lanes 2 and 4) of AP-endonuclease as described in Materials and Methods. Inset, actual data.
Polymorphic S326C OGG1 is not stimulated by AP-endonuclease
It was previously shown that OGG1 binds tightly to its AP site product and cleaves it slowly, thus dissociation of OGG1 from its abasic product is rate limiting (51). APE1 can compete with OGG1 for the AP site, thereby displacing OGG1 and increasing its enzymatic turnover (51,54–56). The glycosylase activity of wild-type OGG1 was stimulated over 2-fold by the addition of an equimolar amount of purified human APE1 (Figure 6A), consistent with previous reports. In an identical reaction, S326C OGG1 was not stimulated by the addition of an equimolar amount of APE1 (Figure 6A) and was not significantly stimulated by the addition of up to a 10-fold molecular excess of APE1 (data not shown). The effect of DTT on wild-type and S326C OGG1 activity and stimulation by APE1 was measured (Figure 6A). DTT (1 mM) was slightly inhibitory of the wild-type OGG1 and had a negligible effect on the activity and APE1 stimulation of S326C OGG1 (Figure 6A). The inability of APE1 to significantly stimulate turnover of S326C OGG1 suggests that an AP site bound by S326C enzyme may be inaccessible to APE1.
S326C OGG1 is resistant to displacement from abasic sites by APE1
OGG1 bound to an AP site is readily displaced by the addition of APE1, which has high affinity and activity for the lesion (51,54–56). Displacement of wild-type and S326C OGG1 from abasic sites by APE1 was measured by EMSA and sodium borohydride trapping assay (Figure 6B and C). When wild-type and S326C OGG1 were bound to an AP site substrate and an equivalent amount of APE1 was subsequently added, wild-type OGG1 was displaced ∼30% from the abasic substrate by APE1, which competes for binding to the abasic site and rapidly cleaves it under the reaction conditions used (Figure 6B, lanes 2 and 3). In a similar reaction, S326C OGG1 was not significantly displaced by APE1 (Figure 6B, lanes 4 and 5). In a sodium borohydride trapping assay, the trapping of wild-type OGG1 to its abasic substrate was decreased roughly 3-fold by the addition of an equimolar amount of APE1 (Figure 6C, lanes 2 and 3), while trapping of S326C OGG1 was decreased relative to the wild-type OGG1 and largely resistant to competition from APE1 (Figure 6C, lanes 4 and 5). Since trapping of OGG1 requires that the enzyme be physically bound to an AP site, inhibition of trapping in the presence of APE1 indicates displacement from and cleavage of the site by APE1. Since S326C OGG1 trapping is not significantly affected by the addition of APE1, unlike the wild-type enzyme (Figure 6C), these results support the findings in Figure 6B and suggest that the polymorphic enzyme is not effectively displaced from an abasic site by APE1.
Involvement of Cysteine 326 in dimerization and reduced activity of polymorphic OGG1
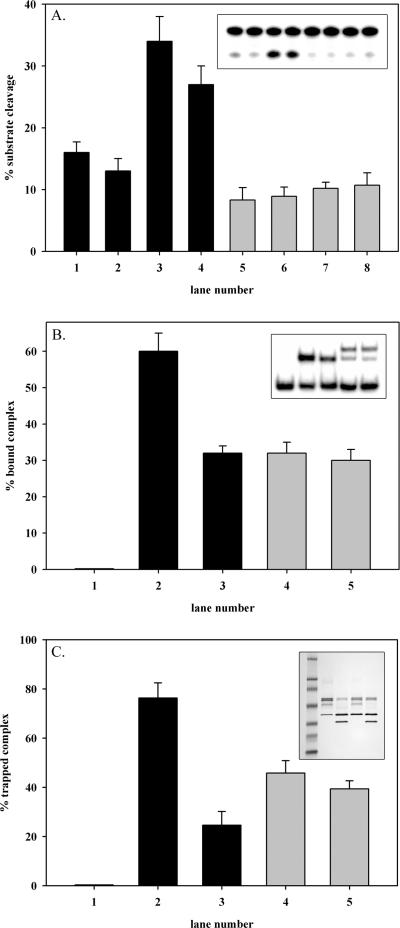
Since serine 326 is near the C-terminus of the 345 amino acid wild-type OGG1, the possibility of a role for the C-terminus in the unusual characteristics of S326C OGG1 was investigated. To determine the contribution of cysteine 326 and the OGG1 C-terminus to the observed reduction in activity and dimeric conformation of S326C OGG1, C-terminally truncated OGG1 proteins were purified as described in Materials and Methods (Figure 7A). DNA binding affinity and enzymatic activity of deletion mutants of wild-type OGG1 (wild-type CΔ19 and CΔ20) and S326C (S326C CΔ19) were examined (Figure 7B and D). The wild-type activity level and monomeric confirmation of wild-type CΔ19 and CΔ20 suggest that the C-terminus of OGG1 plays a critical role in the DNA binding affinity, but not enzymatic activity of the wild-type enzyme. Deletion of the C-terminus of S326C OGG1 up to cysteine 326 in S326C CΔ19, partially rescues the decreased activity seen in full-length S326C (Figure 7B, compare lanes 3 and 4) and reduced the fraction of the protein binding damaged DNA in a dimeric conformation (Figure 7D, compare lanes 4 and 5). These results indicate roles for the residues of S326C OGG1 C-terminal to cysteine 326 in both the dimerization and reduced activity of the polymorphic enzyme. However, the presence of cysteine 326 alone is sufficient to cause dimerization and catalytic inhibition (compare Figure 7B lanes 4 and 5; Figure 7D lanes 5 and 6). Removal of the C-terminus including serine or cysteine at position 326 (CΔ20), resulted in a protein with near wild-type activity, monomeric DNA binding, and greatly reduced DNA binding affinity.
Figure 7.
Binding and catalysis by OGG1 deletion mutants. (A) Amino acid sequence of the C-termini of wild-type, wild-type CΔ19, S326C, S326C CΔ19 and CΔ20 OGG1. (B) Wild-type OGG1 (lane 1) was reacted with an 8-oxoG·C substrate as described in Figure 6. Identical reactions were carried out for wild-type CΔ19 (lane 2), S326C (lane 3), S326C CΔ19 (lane 4) and CΔ20 OGG1 (lane 5). (C) Graphical representation of actual data shown in (B). (D) Binding of OGG1 enzymes to an 8-oxoG·C substate was measured by electrophoretic shift assay as described in Figure 3. Lane 1, no enzyme; lane 2, wild-type; lane 3 wild-type CΔ19; lane 4, S326C; lane 5, S326C CΔ19; and lane 6 CΔ20 OGG1. (E) Graphical representation of actual data shown in (D). Inset, dissociation constants calculated from data shown in (D).
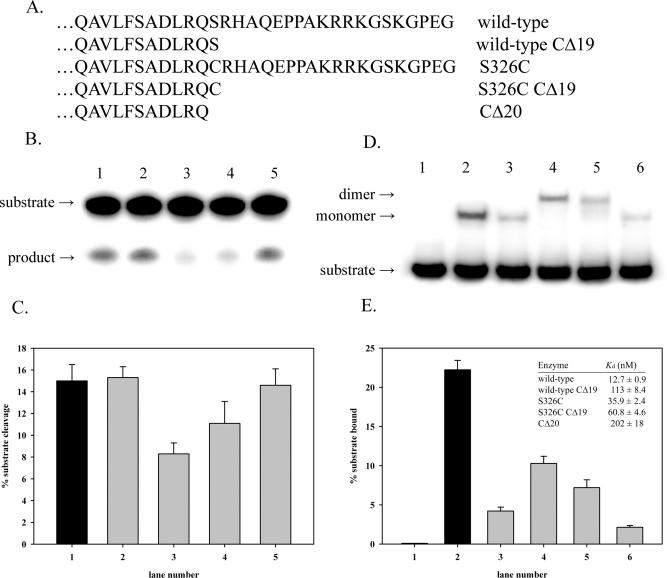
Catalytic impairment and dimerization of S326C OGG1 expressed in human cells
Nuclear extracts prepared from HeLa cells overexpressing wild-type or S326C OGG1 at identical levels were analyzed for 8-oxoG glycosylase activity (Figure 8). Cells expressing the wild-type OGG1 had significantly higher levels of 8-oxoG excision, suggesting that S326C OGG1 expressed in human cells has decreased activity relative to the wild-type enzyme (Figure 8A). These findings are consistent with previous reports suggesting polymorphic S326C OGG1 has decreased enzymatic activity in vivo (34,50), but contrasts with reports that no functional difference exists between the isoforms in human cells (57,58). Chemical cross-linking of nuclear extracts containing FLAG-tagged wild-type or S326C OGG1 is shown in Figure 8B. In the presence of cross-linker, S326C OGG1 produced primarily an ∼80 kDa species of identical size to that seen with cross-linked purified S326C protein (compare Figures 4A and 8B). As with cross-linking experiments using purified proteins, traces of a complex consistent with a dimer could be seen in cross-linked nuclear extracts of cells expressing wild-type OGG1 (Figure 8B, lane 4), suggesting that dimerization may occur to some extent with the wild-type enzyme. These results suggest that S326C OGG1 has reduced activity in vivo and may exist as a dimer when expressed in human cells.
Figure 8.
Characterization of wild-type and S326C OGG1 expressed in human cells. (A) Comparision of 8-oxoguanine glycosylase activites of HeLa nuclear extracts from cells transfected with pCMV vector (lane 1), or pCMV vector expressing N-terminally FLAG-tagged wild-type (lane 2) or S326C OGG1 (lane 3). Left inset; Anti-FLAG western blot of 2 µg of nuclear extract prepared from HeLa cells transfected with pCMV vector (lane 1), pCMV wt (lane 2) or PCMV S326C (lane 3). Right inset; actual data shown graphically in (A). (B) Anti-FLAG western blot of 4 µg of nuclear extract from cells transfected with pCMV vector (lanes 1 and 2), pCMV wt (lanes 3 and 4) or pCMV S326C (lanes 5 and 6). Nuclear extracts were incubated at 0°C overnight with (lanes 2, 4 and 6) and without (lanes 1, 3 and 5) 2 mM BM[PEO]4.
DISCUSSION
Changes in the human OGG1 coding sequence that result in amino acid substitutions and altered 8-oxoG excision activity are associated with susceptibility to various cancers (38–41). The most well documented OGG1 polymorphism (S326C) has been shown to be associated with carcinogenesis and decreased 8-oxoG repair activity in numerous studies, although reports have not been conclusive. Individuals carrying the S326C allele have been shown to be at higher risk of lung, prostate and oral cancers, but not breast cancer or basal cell carcinoma (42–48,59–61). Differences in the findings of association studies suggest that the concurrence of the S326C polymorphism with cancer incidence may be tissue specific and dependent upon specific covariate factors, including ethnicity, lifestyle practices and environmental influences. Studies of the repair function of S326C OGG1 both in vivo and in vitro suggest that the polymorphism may be associated with decreased repair activity. An initial comparison of purified glutathione S-transferase (GST)-tagged S326C and wild-type OGG1 found a 1.6-fold decrease in the efficiency of 8-oxoG excision from an 8-oxoG·C substrate by the polymorphic enzyme (49), consistent with our observations. Complementation studies in bacteria, functional analysis of nuclear extracts from 326 C/C individuals and exogenous protein expression experiments in human cancer cells suggested that the S326C isoform is active in vivo and had reduced mutation suppression ability compared to wild-type OGG1 (25,34,49,50). However, reports on the effect of the S326C amino acid substitution on directly measured OGG1 function have also not been conclusive, with some studies reporting no effect of the polymorphism on OGG1 activity (57,58). Interestingly, no effect of the S326C polymorphism on OGG1 activity was detected in both studies where OGG1 activity was measured in whole cell lysates or tissue homogenates (57,58). Previous studies have reported the presence of specific 8-oxoG binding proteins or OGG1 inhibitors in crude cell lysates, but not partially purified enzymes (15,62). We have also observed inhibition of OGG1 activity in HeLa whole cell extracts, but not nuclear extracts (data not shown). Therefore, use of crude extracts for comparative analysis of OGG1 function may potentially be influenced by cellular factors not directly related to OGG1 activity. A comparative analysis of the abilities of wild-type and S326C OGG1 to suppress mutagenesis, performed under conditions that likely reflect the in vivo situation in human cells, found a significant decrease in the ability of polymorphic S326C OGG1 to prevent mutations and suggests decreased repair function of S326C OGG1 in cells (50).
In the present study, the substrate specificity, glycosylase and AP-lyase activities and damage binding affinities of S326C OGG1 were examined and compared with those of the wild-type enzyme. Since reports of S326C enzymatic function have been controversial, we compared the activity of S326C OGG1 against the wild-type enzyme using enzymes expressed in both E.coli and in human cells. We first examined the function of purified human OGG1 enzymes expressed in bacteria (Figure 1). Our analysis of the activity of S326C OGG1 confirms and extends an earlier report that the polymorphic enzyme has slightly reduced activity towards 8-oxoG·C and AP·C substrates (49). Surprisingly, S326C OGG1 was particularly deficient in the excision of 8-oxoG paired with T and G, with the lesion being removed at rates decreased 4- and 6-fold, respectively, relative to wild-type OGG1 (Table 1). Therefore, the extent to which the S326C polymorphism negatively impacts OGG1 function is highly dependent upon the base opposite 8-oxoG. The decreased binding affinity of S326C towards all major OGG1 substrates (Table 1) is in agreement with its lower catalytic rates. However, the magnitude of decreases in the turnover rates of S326C do not correspond with decreases in directly measured S326C lesion binding affinities (Table 1). In the case of 8-oxoG·C and 8-oxoG·T, decreases in S326C substrate binding affinity correlate well with decreases in activity (Table 1). However, in the case of 8-oxoG·G, 8-oxoG·A and AP·C substrates, no direct relationship exists between changes in measured substrate binding affinities and enzymatic activity (Table 1). Therefore, the S326C polymorphism independently alters damage binding affinity and rates of base excision and strand incision by OGG1 in a manner dependent upon the lesion type and the base opposite 8-oxoguanine. Although S326C OGG1 is deficient in the repair of 8-oxoG (particularly 8-oxoG·G) compared to wild-type OGG1 (Figure 2 and Table 1) redundant repair enzymes for 8-oxoG, including NTH1 and NEIL1, are present in human cells (63,64). Thus, the S326C OGG1 polymorphism may have added importance in individuals with high levels of oxidative stress, as suggested by numerous studies which indicate that the S326C OGG1 allele may modify the effects of environmental risk factors for several types of cancer (65–69).
Since the highly purified S326C OGG1 used to determine lesion binding affinities was a single species on an SDS–PAGE gel (Figure 1), the presence of two bands in S326C gel shifts (Figure 3) suggested that the polymorphic enzyme might exist as a dimer. We investigated this possibility by chemical cross-linking experiments which indicate that S326C OGG1, but not the wild-type enzyme can be cross-linked both with and without a damaged DNA substrate (Figure 4A). Native size-exclusion experiments further support the distinct dimeric conformation of S326C OGG1 (Figure 5), with native wild-type and S326C OGG1 having elution profiles consistent with expected molecular sizes for an OGG1 monomer and dimer, respectively. Mixing of S326C and wild-type OGG1 enzymes in both cross-linking and native chromatography experiments suggest that the two isoforms do not heterodimerize (Figures 4B and 5D), although the possibility of some level of interaction between the isoforms in vivo cannot be excluded. Interestingly, S326C bound an AP·C substrate as both a monomer and dimer (Figure 3E). Since the polymorphic enzyme is essentially completely dimerized in the absence of substrate, it therefore seems likely that binding of S326C OGG1 to an abasic site substrate may induce conformational changes in the dimeric enzyme that lead to its dissociation into monomers.
The unexpected constitutive dimerization of the polymorphic OGG1 has implicit functional effects on enzymatic activity. Presumably, only one molecule of OGG1 can engage a single 8-oxoguanine at once, so the binding of two molecules at one lesion would by definition result in an unproductive binding by one member of the S326C dimer. This in part explains why measured reaction velocities of S326C OGG1 are at least 2-fold less than those of the wild-type enzyme (Table 1). However, larger differences in the rate of base excision by S326C, i.e. 6-fold lower than the wild-type with an 8-oxoG·G substrate, cannot be explained solely by an unproductive binding due to dimerization and suggest a possible allosteric effect caused by dimerization that is dependent upon the base opposite 8-oxoG and results in significantly decreased glycosylase activity. While the precise catalytic mechanism and subunit interface of the S326C dimer remain to be determined, dimerization of the protein in the absence of DNA and the minimal 2-fold reduction in reaction rate (Table 1) suggest that one molecule of a preformed dimer may bind DNA, rather than both members of the dimer directly binding to damage sites. The decrease in both the turnover rate and damage binding affinity of S326C suggest that the subunit interface of the S326C dimer may alter both binding and catalysis by the member of the dimer engaging DNA damage. Additionally, partial physical occlusion of the active site of the binding member of a S326C dimer by its subunit partner and structural perturbation of S326C independent of dimerization cannot be ruled out.
Binding and functional studies of C-terminally truncated OGG1 enzymes (Figure 7) indicate roles for both cysteine 326 and the downstream C-terminus in the reduced enzymatic function and dimerization of the polymorphic enzyme. Removal of the C-terminal 19 amino acids from S326C OGG1 reduced damage binding affinity and dimerization and restored some enzymatic activity to the polymorphic enzyme. Based on these observations, we hypothesize that the dimer interface of S326C is comprised of the C-terminus of the enzyme, including cysteine 326. Interestingly, our results suggest that the C-terminal 20 amino acids of OGG1 have a role primarily in DNA binding, but not catalysis, in the wild-type enzyme, since wild-type CΔ19 and CΔ20 have near wild-type activity, but significantly lower damage binding affinity than the wild-type enzyme (Figure 7). Surprisingly, tight substrate binding by OGG1 does not appear to be an absolute requirement for activity, since terminally truncated enzymes with reduced substrate binding have wild-type activity. The slightly decreased binding affinity of CΔ20 compared to wild-type CΔ19 suggests that the amino acid at position 326 may have an minor individual effect on DNA binding. In contrast, the C-terminal 19 amino acids of S326C OGG1 have an effect on both DNA binding and catalysis, presumably due to their involvement in the homodimer interface. Published co-crystal structures of OGG1 with 8-oxoG and abasic site analog [tetrahydrofuran; (THF)] substrates were obtained with N- and C-terminally (NΔ12-CΔ18) truncated OGG1 proteins (70,71). Limited proteolytic digestion of full-length OGG1 produced the NΔ12-CΔ18 OGG1 core domain, thus the C-terminal 18 amino acids of OGG1 were proposed to form an unstructured peptide (70). Interestingly, serine 326 is located at the boundary of the enzyme core domain (70), thus its substitution for another amino acid could potentially influence both the core structure and conformation of the downstream C-terminus. Based on our binding measurements, the binding of full-length wild-type OGG1 to DNA is significantly tighter than that of wild-type CΔ19 or CΔ20 and may involve additional or distinct protein–DNA contacts mediated via the C-terminal 20 amino acids (Figure 7D and E). The decreased damage binding affinity of S326C OGG1 may in part be explained by the involvement of the C-terminal 20 amino acids in dimerization, rather than DNA binding.
It was previously shown that OGG1 turnover is significantly stimulated by AP-endonuclease in vitro (51,54,55). OGG1 binds tightly to the abasic site produced by its glycosylase activity and cleaves the site slowly, thus the enzyme is product inhibited and product release is rate limiting in the excision of 8-oxoG by OGG1. APE1 has high affinity for and activity towards the abasic site, thereby competing for the AP site with OGG1 and quickly cleaving it, thereby enhancing OGG1 turnover (51,54,55). The high affinity and low activity of S326C OGG1 for abasic sites (Table 1) suggest that the polymorphic enzyme is also product inhibited. The failure of APE1 to stimulate S326C turnover (Figure 6A) suggests that the dimeric binding mode of the polymorphic enzyme may prevent access to the AP site by APE1. We directly measured physical displacement of S326C and wild-type OGG1 from an AP site substrate by APE1 using both electrophoretic mobility shift and a trapping assay (Figure 6B and C). In both cases, addition of APE1 failed to significantly displace S326C OGG1 and suggests that AP sites bound by S326C OGG1 may be inaccessible by APE1.
A comparison of the enzymatic activity of wild-type and S326C OGG1 expressed at identical levels in human cells showed a decrease in 8-oxoG excision with the polymorphic S326C enzyme consistent with the decrease in activity observed with purified S326C (Figure 8A). Cross-linking of nuclear extracts from cells expressing wild-type or S326C OGG1 produced results similar to those observed with purified enzymes and suggest that S326C OGG1 may exist as a dimer inside the nucleus of human cells (Figure 8B). We speculate that the dimeric conformation of polymorphic S326C could have multiple consequences in vivo. The decreased enzymatic activity of dimeric S326C OGG1 may be further compounded in cells by the apparent resistance of the isoform to stimulation by an enzyme acting subsequent to OGG1 in the BER pathway (Figure 6). Additionally, it seems probable that the dimerization of S326C could influence intracellular localization and reported interactions of the wild-type OGG1 with other DNA repair proteins, such as XRCC1 (72), that may play crucial roles in the regulation of 8-oxoG repair in vivo. To our knowledge, no report exists regarding differences in the spectrum of mutations occurring in individuals carrying either wild-type or S326C OGG1 alleles. In previous studies, it has been shown that wild-type OGG1 expressed in human cells suppresses G:C to T:A transversion mutations and that S326C OGG1 has a lower ability to prevent such mutations in vivo (50,73). An examination of the differences between the in vivo mutation suppression abilities of wild-type and S326C OGG1 with defined lesions containing 8-oxoG paired not only with cytosine, but also thymine and guanine, may reveal additional consequences of the S326C polymorphism that could affect genomic stability. While further studies are required to elucidate the effects of the S326C polymorphism on the efficacy and regulation of 8-oxoguanine repair in vivo, the catalytic and stoichiometric changes in OGG1 and resistance to displacement from AP sites resulting from the S326C substitution reported here may be involved in the pathological association of this prevalent polymorphism.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging. The authors thank Drs Sebastian Fugmann and Michael Pazin (NIA) for critical reading of this manuscript. Funding to pay the Open Access publication charges for this article was provided by the NIA Intramural Research Program.
Conflict of interest statement. None declared.
REFERENCES
- 1.Doetsch P.W., Zasatawny T.H., Martin A.M., Dizdaroglu M. Monomeric base damage products from adenine, guanine, and thymine induced by exposure of DNA to ultraviolet radiation. Biochemistry. 1995;34:737–742. doi: 10.1021/bi00003a005. [DOI] [PubMed] [Google Scholar]
- 2.Dizdaroglu M. Formation of an 8-hydroxyguanine moiety in deoxyribonucleic acid on gamma-irradiation in aqueous solution. Biochemistry. 1985;24:4476–4481. doi: 10.1021/bi00337a032. [DOI] [PubMed] [Google Scholar]
- 3.Kasai H., Nishimura S. Formation of 8-hydroxyguanosine in DNA by oxygen radicals and its biological significance. In: Seis H., editor. Oxidative Stress: Oxidants and Antioxidants. London: Academic Press; 1991. pp. 99–116. [Google Scholar]
- 4.Cadet J., Berger M., Douki T., Ravanat J.L. Oxidative damage to DNA: formation, measurement, and biological significance. Rev. Physiol. Biochem. Pharmacol. 1997;131:1–87. doi: 10.1007/3-540-61992-5_5. [DOI] [PubMed] [Google Scholar]
- 5.Barnes D.E., Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 6.Shibutani S., Takeshita M., Grollman A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 7.Aburatani H., Hippo Y., Ishida T., Takashima R., Matsuba C., Kodama T., Takao M., Yasui A., Yamamoto K., Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 8.Arai K., Morishita K., Shinmura K., Kohno T., Kim S.R., Nohmi T., Taniwaki M., Ohwada S., Yokota J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 9.Bjoras M., Luna L., Johnsen B., Hoff E., Haug T., Rognes T., Seeberg E. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 1997;16:6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu R., Nash H.M., Verdine G.L. A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Curr. Biol. 1997;7:397–407. doi: 10.1016/s0960-9822(06)00187-4. [DOI] [PubMed] [Google Scholar]
- 11.Radicella J.P., Dherin C., Desmaze C., Fox M.S., Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roldan-Arjona T., Wei Y.F., Carter K.C., Klungland A., Anselmino C., Wang R.P., Augustus M., Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl Acad. Sci. USA. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenquist T.A., Zharkov D.O., Grollman A.P. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl Acad. Sci. USA. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ide H., Kotera M. Human DNA glycosylases involved in the repair of oxidatively damaged DNA. Biol. Pharm. Bull. 2004;27:480–485. doi: 10.1248/bpb.27.480. [DOI] [PubMed] [Google Scholar]
- 15.Monden Y., Arai T., Asano M., Ohtsuka E., Aburatani H., Nishimura S. Human MMH (OGG1) type 1a protein is a major enzyme for repair of 8-hydroxyguanine lesions in human cells. Biochem. Biophys. Res. Commun. 1999;258:605–610. doi: 10.1006/bbrc.1999.0649. [DOI] [PubMed] [Google Scholar]
- 16.Klungland A., Rosewell I., Hollenbach S., Larsen E., Daly G., Epe B., Seeberg E., Lindahl T., Barnes D.E. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl Acad. Sci. USA. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minowa O., Arai T., Hirano M., Monden Y., Nakai S., Fukuda M., Itoh M., Takano H., Hippou Y., Aburatani H., et al. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl Acad. Sci. USA. 2000;97:4156–4161. doi: 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Page F., Klungland A., Barnes D.E., Sarasin A., Boiteux S. Transcription coupled repair of 8-oxoguanine in murine cells: the ogg1 protein is required for repair in nontranscribed sequences but not in transcribed sequences. Proc. Natl Acad. Sci. USA. 2000;97:8397–8402. doi: 10.1073/pnas.140137297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Souza-Pinto N.C., Eide L., Hogue B.A., Thybo T., Stevnsner T., Seeberg E., Klungland A., Bohr V.A. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 20.Arai K., Morishita K., Shinmura K., Kohno T., Kim S.R., Nohmi T., Taniwaki M., Ohwada S., Yokota J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 21.Sakumi K., Tominaga Y., Furuichi M., Xu P., Tsuzuki T., Sekiguchi M., Nakabeppu Y. Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res. 2003;63:902–905. [PubMed] [Google Scholar]
- 22.Xie Y., Yang H., Cunanan C., Okamoto K., Shibata D., Pan J., Barnes D.E., Lindahl T., McIlhatton M., Fishel R., et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- 23.Russo M.T., De Luca G., Degan P., Parlanti E., Dogliotti E., Barnes D.E., Lindahl T., Yang H., Miller J.H., Bignami M. Accumulation of the oxidative base lesion 8-hydroxyguanine in DNA of tumor-prone mice defective in both the Myh and Ogg1 DNA glycosylases. Cancer Res. 2004;64:4411–4414. doi: 10.1158/0008-5472.CAN-04-0355. [DOI] [PubMed] [Google Scholar]
- 24.Ames B.N., Shigenaga M.K., Hagen T.M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl Acad. Sci. USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohno T., Shinmura K., Tosaka M., Tani M., Kim S.R., Sugimura H., Nohmi T., Kasai H., Yokota J. Genetic polymorphisms and alternative splicing of the hOGG1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene. 1998;16:3219–3225. doi: 10.1038/sj.onc.1201872. [DOI] [PubMed] [Google Scholar]
- 26.Audebert M., Chevillard S., Levalois C., Gyapay G., Vieillefond A., Klijanienko J., Vielh P., El Naggar A.K., Oudard S., Boiteux S., et al. Alterations of the DNA repair gene OGG1 in human clear cell carcinomas of the kidney. Cancer Res. 2000;60:4740–4744. [PubMed] [Google Scholar]
- 27.Wikman H., Risch A., Klimek F., Schmezer P., Spiegelhalder B., Dienemann H., Kayser K., Schulz V., Drings P., Bartsch H. hOGG1 polymorphism and loss of heterozygosity (LOH): significance for lung cancer susceptibility in a caucasian population. Int. J. Cancer. 2000;88:932–937. doi: 10.1002/1097-0215(20001215)88:6<932::aid-ijc15>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 28.Paz-Elizur T., Krupsky M., Blumenstein S., Elinger D., Schechtman E., Livneh Z. DNA repair activity for oxidative damage and risk of lung cancer. J. Natl Cancer Inst. 2003;95:1312–1319. doi: 10.1093/jnci/djg033. [DOI] [PubMed] [Google Scholar]
- 29.Inoue M., Osaki T., Noguchi M., Hirohashi S., Yasumoto K., Kasai H. Lung cancer patients have increased 8-hydroxydeoxyguanosine levels in peripheral lung tissue DNA. Jpn. J. Cancer Res. 1998;89:691–695. doi: 10.1111/j.1349-7006.1998.tb03272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mambo E., Nyaga S.G., Bohr V.A., Evans M.K. Defective repair of 8-hydroxyguanine in mitochondria of MCF-7 and MDA-MB-468 human breast cancer cell lines. Cancer Res. 2002;62:1349–1355. [PubMed] [Google Scholar]
- 31.Parker A.R., O'Meally R.N., Oliver D.H., Hua L., Nelson W.G., DeWeese T.L., Eshleman J.R. 8-Hydroxyguanosine repair is defective in some microsatellite stable colorectal cancer cells. Cancer Res. 2002;62:7230–7233. [PubMed] [Google Scholar]
- 32.Trzeciak A.R., Nyaga S.G., Jaruga P., Lohani A., Dizdaroglu M., Evans M.K. Cellular repair of oxidatively induced DNA base lesions is defective in prostate cancer cell lines, PC-3 and DU-145. Carcinogenesis. 2004;25:1359–1370. doi: 10.1093/carcin/bgh144. [DOI] [PubMed] [Google Scholar]
- 33.Mambo E., Chatterjee A., de Souza-Pinto N.C., Mayard S., Hogue B.A., Hoque M.O., Dizdaroglu M., Bohr V.A., Sidransky D. Oxidized guanine lesions and hOgg1 activity in lung cancer. Oncogene. 2005;24:4496–4508. doi: 10.1038/sj.onc.1208669. [DOI] [PubMed] [Google Scholar]
- 34.Chen S.K., Hsieh W.A., Tsai M.H., Chen C.C., Hong A.I., Wei Y.H., Chang W.P. Age-associated decrease of oxidative repair enzymes, human 8-oxoguanine DNA glycosylases (hOgg1), in human aging. J. Radiat. Res. 2003;44:31–35. doi: 10.1269/jrr.44.31. (Tokyo) [DOI] [PubMed] [Google Scholar]
- 35.Szczesny B., Hazra T.K., Papaconstantinou J., Mitra S., Boldogh I. Age-dependent deficiency in import of mitochondrial DNA glycosylases required for repair of oxidatively damaged bases. Proc. Natl Acad. Sci. USA. 2003;100:10670–10675. doi: 10.1073/pnas.1932854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang M.K., Kim R.H., Shin K.H., Zhong W., Faull K.F., Park N.H. Senescence-associated decline in the intranuclear accumulation of hOGG1-alpha and impaired 8-oxo-dG repair activity in senescing normal human oral keratinocytes in vivo. Exp. Cell Res. 2005;310:186–195. doi: 10.1016/j.yexcr.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 37.Luna L., Rolseth V., Hildrestrand G.A., Otterlei M., Dantzer F., Bjoras M., Seeberg E. Dynamic relocalization of hOGG1 during the cell cycle is disrupted in cells harbouring the hOGG1-Cys326 polymorphic variant. Nucleic Acids Res. 2005;33:1813–1824. doi: 10.1093/nar/gki325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goode E.L., Ulrich C.M., Potter J.D. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol. Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 39.Weiss J.M., Goode E.L., Ladiges W.C., Ulrich C.M. Polymorphic variation in hOGG1 and risk of cancer: a review of the functional and epidemiologic literature. Mol. Carcinog. 2005;42:127–141. doi: 10.1002/mc.20067. [DOI] [PubMed] [Google Scholar]
- 40.Hung R.J., Hall J., Brennan P., Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am. J. Epidemiol. 2005;162:925–942. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 41.Nohmi T., Kim S.R., Yamada M. Modulation of oxidative mutagenesis and carcinogenesis by polymorphic forms of human DNA repair enzymes. Mutat. Res. 2005;591:60–73. doi: 10.1016/j.mrfmmm.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 42.Sugimura H., Kohno T., Wakai K., Nagura K., Genka K., Igarashi H., Morris B.J., Baba S., Ohno Y., Gao C., et al. hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Cancer Epidemiol. Biomarkers Prev. 1999;8:669–674. [PubMed] [Google Scholar]
- 43.Le Marchand L., Donlon T., Lum-Jones A., Seifried A., Wilkens L.R. Association of the hOGG1 Ser326Cys polymorphism with lung cancer risk. Cancer Epidemiol. Biomarkers Prev. 2002;11:409–412. [PubMed] [Google Scholar]
- 44.Xu J., Zheng S.L., Turner A., Isaacs S.D., Wiley K.E., Hawkins G.A., Chang B.L., Bleecker E.R., Walsh P.C., Meyers D.A., et al. Associations between hOGG1 sequence variants and prostate cancer susceptibility. Cancer Res. 2002;62:2253–2257. [PubMed] [Google Scholar]
- 45.Elahi A., Zheng Z., Park J., Eyring K., McCaffrey T., Lazarus P. The human OGG1 DNA repair enzyme and its association with orolaryngeal cancer risk. Carcinogenesis. 2002;23:1229–1234. doi: 10.1093/carcin/23.7.1229. [DOI] [PubMed] [Google Scholar]
- 46.Chen L., Elahi A., Pow-Sang J., Lazarus P., Park J. Association between polymorphism of human oxoguanine glycosylase 1 and risk of prostate cancer. J. Urol. 2003;170:2471–2474. doi: 10.1097/01.ju.0000087498.23008.bb. [DOI] [PubMed] [Google Scholar]
- 47.Park J., Chen L., Tockman M.S., Elahi A., Lazarus P. The human 8-oxoguanine DNA N-glycosylase 1 (hOGG1) DNA repair enzyme and its association with lung cancer risk. Pharmacogenetics. 2004;14:103–109. doi: 10.1097/00008571-200402000-00004. [DOI] [PubMed] [Google Scholar]
- 48.Zienolddiny S., Campa D., Lind H., Ryberg D., Skaug V., Stangeland L., Phillips D.H., Canzian F., Haugen A. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27:560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 49.Dherin C., Radicella J.P., Dizdaroglu M., Boiteux S. Excision of oxidatively damaged DNA bases by the human alpha-hOgg1 protein and the polymorphic alpha-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999;27:4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamane A., Kohno T., Ito K., Sunaga N., Aoki K., Yoshimura K., Murakami H., Nojima Y., Yokota J. Differential ability of polymorphic OGG1 proteins to suppress mutagenesis induced by 8-hydroxyguanine in human cell in vivo. Carcinogenesis. 2004;25:1689–1694. doi: 10.1093/carcin/bgh166. [DOI] [PubMed] [Google Scholar]
- 51.Hill J.W., Hazra T.K., Izumi T., Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dodson M.L., Michaels M.L., Lloyd R.S. Unified catalytic mechanism for DNA glycosylases. J. Biol. Chem. 1994;269:32709–32712. [PubMed] [Google Scholar]
- 53.Price N.C., Dwek R.A. Principles and Problems in Physical Chemistry for Biochemists. 2nd edn. Oxford: Oxford University Press; 1982. [Google Scholar]
- 54.Vidal A.E., Hickson I.D., Boiteux S., Radicella J.P. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res. 2001;29:1285–1292. doi: 10.1093/nar/29.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saitoh T., Shinmura K., Yamaguchi S., Tani M., Seki S., Murakami H., Nojima Y., Yokota J. Enhancement of OGG1 protein AP lyase activity by increase of APEX protein. Mutat. Res. 2001;486:31–40. doi: 10.1016/s0921-8777(01)00078-7. [DOI] [PubMed] [Google Scholar]
- 56.Hazra T.K., Hill J.W., Izumi T., Mitra S. Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions. In: Moldave K., editor. Progress in Nucleic Acid Research and Molecular Biology. Vol. 68. San Diego: Academic Press; 2001. pp. 193–205. [DOI] [PubMed] [Google Scholar]
- 57.Janssen K., Schlink K., Gotte W., Hippler B., Kaina B., Oesch F. DNA repair activity of 8-oxoguanine DNA glycosylase 1 (OGG1) in human lymphocytes is not dependent on genetic polymorphism Ser326/Cys326. Mutat. Res. 2001;486:207–216. doi: 10.1016/s0921-8777(01)00096-9. [DOI] [PubMed] [Google Scholar]
- 58.Park Y.J., Choi E.Y., Choi J.Y., Park J.G., You H.J., Chung M.H. Genetic changes of hOGG1 and the activity of oh8Gua glycosylase in colon cancer. Eur. J. Cancer. 2001;37:340–346. doi: 10.1016/s0959-8049(00)00388-9. [DOI] [PubMed] [Google Scholar]
- 59.Vogel U., Nexo B.A., Olsen A., Thomsen B., Jacobsen N.R., Wallin H., Overvad K., Tjonneland A. No association between OGG1 Ser326Cys polymorphism and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2003;12:170–171. [PubMed] [Google Scholar]
- 60.Choi J.Y., Hamajima N., Tajima K., Yoo K.Y., Yoon K.S., Park S.K., Kim S.U., Lee K.M., Noh D.Y., Ahn S.H., et al. hOGG1 Ser326Cys polymorphism and breast cancer risk among Asian women. Breast Cancer Res. Treat. 2003;79:59–62. doi: 10.1023/a:1023305826726. [DOI] [PubMed] [Google Scholar]
- 61.Vogel U., Olsen A., Wallin H., Overvad K., Tjonneland A., Nexo B.A. No association between OGG1 Ser326Cys and risk of basal cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 2004;13:1680–1681. [PubMed] [Google Scholar]
- 62.Hazra T.K., Izumi T., Maidt L., Floyd R.A., Mitra S. The presence of two distinct 8-oxoguanine repair enzymes in human cells: their potential complementary roles in preventing mutation. Nucleic Acids Res. 1998;26:5116–5122. doi: 10.1093/nar/26.22.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsumoto Y., Zhang Q.M., Takao M., Yasui A., Yonei S. Escherichia coli Nth and human hNTH1 DNA glycosylases are involved in removal of 8-oxoguanine from 8-oxoguanine/guanine mispairs in DNA. Nucleic Acids Res. 2001;29:1975–1981. doi: 10.1093/nar/29.9.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hazra T.K., Izumi T., Boldogh I., Imhoff B., Kow Y.W., Jaruga P., Dizdaroglu M., Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl Acad. Sci. USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takezaki T., Gao C.M., Wu J.Z., Li Z.Y., Wang J.D., Ding J.H., Liu Y.T., Hu X., Xu T.L., Tajima K., et al. hOGG1 Ser(326)Cys polymorphism and modification by environmental factors of stomach cancer risk in Chinese. Int. J. Cancer. 2002;99:624–627. doi: 10.1002/ijc.10400. [DOI] [PubMed] [Google Scholar]
- 66.Kim J.I., Park Y.J., Kim K.H., Song B.J., Lee M.S., Kim C.N., Chang S.H. hOGG1 Ser326Cys polymorphism modifies the significance of the environmental risk factor for colon cancer. World J. Gastroenterol. 2003;9:956–960. doi: 10.3748/wjg.v9.i5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kau H.C., Tsai C.C., Hsu W.M., Liu J.H., Wei Y.H. Genetic polymorphism of hOGG1 and risk of pterygium in Chinese. Eye. 2004;18:635–639. doi: 10.1038/sj.eye.6700738. [DOI] [PubMed] [Google Scholar]
- 68.Aka P., Mateuca R., Buchet J.P., Thierens H., Kirsch-Volders M. Are genetic polymorphisms in OGG1, XRCC1 and XRCC3 genes predictive for the DNA strand break repair phenotype and genotoxicity in workers exposed to low dose ionising radiations? Mutat. Res. 2004;556:169–181. doi: 10.1016/j.mrfmmm.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 69.Lan Q., Mumford J.L., Shen M., Demarini D.M., Bonner M.R., He X., Yeager M., Welch R., Chanock S., Tian L., et al. Oxidative damage-related genes AKR1C3 and OGG1 modulate risks for lung cancer due to exposure to PAH-rich coal combustion emissions. Carcinogenesis. 2004;25:2177–2181. doi: 10.1093/carcin/bgh240. [DOI] [PubMed] [Google Scholar]
- 70.Bruner S.D., Norman D.P., Verdine G.L. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 71.Norman D.P., Bruner S.D., Verdine G.L. Coupling of substrate recognition and catalysis by a human base-excision DNA repair protein. J. Am. Chem. Soc. 2001;123:359–360. doi: 10.1021/ja003144m. [DOI] [PubMed] [Google Scholar]
- 72.Marsin S., Vidal A.E., Sossou M., Menissier-de Murcia J., Le Page F., Boiteux S., de Murcia G., Radicella J.P. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J. Biol. Chem. 2003;278:44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 73.Sunaga N., Kohno T., Shinmura K., Saitoh T., Matsuda T., Saito R., Yokota J. OGG1 protein suppresses G:C→T:A mutation in a shuttle vector containing 8-hydroxyguanine in human cells. Carcinogenesis. 2001;22:1355–1362. doi: 10.1093/carcin/22.9.1355. [DOI] [PubMed] [Google Scholar]