Abstract
Defective interfering (DI) RNAs of Tomato bushy stunt virus (TBSV), a plus-sense RNA virus, comprise four conserved noncontiguous regions (I through IV) derived from the viral genome. Region III, a 70-nucleotide-long sequence corresponding to a genomic segment located 378 nucleotides upstream of the 3′ terminus of the genome, has been found to enhance DI RNA accumulation by approximately 10-fold in an orientation-independent manner (D. Ray and K. A. White, Virology 256:162-171, 1999). In this study, a more detailed structure-function analysis of region III was conducted. RNA secondary-structure analyses indicated that region III contains stem-loop structures in both plus and minus strands. Through deletion analyses of a DI RNA, a primary determinant of region III activity was mapped to the 5′-proximal 35-nucleotide segment. Compensatory-type mutational analyses showed that a stem-loop structure in the minus strand of this subregion was required for enhanced DI RNA replication. The same stem-loop structure was also found to function in a position-independent manner in a DI RNA (albeit at reduced levels) and to be important for efficient accumulation within the context of the TBSV genome. Taken together, these observations suggest that the 5′-proximal segment of region III is a modular RNA replication element that functions primarily through the formation of an RNA hairpin structure in the minus strand.
Genome replication represents a fundamental step in virus reproduction. In plus-sense RNA viruses, this is a two-step process whereby minus strands are synthesized from the genome and are used subsequently as templates for the synthesis of progeny genomes. This process is asymmetrical, with viral plus strands accumulating to approximately 10- to 1,000-fold excess over minus strands (4).
Both cis-acting RNA elements and trans-acting factors (i.e., viral RNA-dependent RNA polymerase [RdRp] and/or host factors) are required for viral genome replication (4). Usually, cis-acting replication elements reside in 5′- and 3′-terminal untranslated regions of genomes (4, 8, 9), however, several internally positioned cis-acting elements have also been identified. These include RNA replication signals that are located in (i) coding regions of rhinovirus type 14 (25), poliovirus (12) and bacteriophage Qβ (22) and (ii) intergenic regions in Barley stripe mosaic virus (45), Cucumber mosaic virus (3), and Brome mosaic virus (11). The identification and analysis of cis-acting RNA replication signals can be complicated by their potential dual role in the translation of viral proteins. However, small virus-associated RNA molecules (satellite and defective interfering [DI] RNAs) have been instrumental in circumventing this problem, as many are noncoding and are amplified efficiently by viral RdRp supplied in trans. Thus, these subviral replicons are extremely useful model RNA templates for the identification and analysis of viral cis-acting replication signals.
Tomato bushy stunt virus (TBSV), the type member of the genus Tombusvirus (33), is a plus-sense single-stranded RNA plant virus whose ∼4.8-kb genome encodes five proteins with defined functions (Fig. 1) (18, 28, 31, 34, 35, 37). Prototypical DI RNAs associated with TBSV comprise four regions derived entirely from the genome (Fig. 1). Based on their correspondence to the genomic 5′ and 3′ termini and mutational analysis in vivo, regions I and IV likely contain cis-acting RNA elements important for initiation of RNA synthesis (5, 13, 14, 40, 42). This concept is consistent with in vitro studies that have identified core promoter elements necessary for plus and minus strand synthesis within regions I and IV, respectively (27, 29). Region II, which is derived from an internal coding segment corresponding to p92, is an essential element, but its function is yet undefined (5, 38). Region III (70 nucleotides) is unique in that it is derived from both viral coding and noncoding sequences (23). Previous studies have demonstrated that region III is multifunctional in that it comprises a portion of the TBSV 3′-cap-independent translational enhancer (41) and enhances DI RNA replication in vivo (32).
FIG. 1.
Schematic representation of the TBSV genome and a prototypical DI RNA. The wild-type genome is shown as a thick black line with coding regions depicted as white boxes with approximate molecular masses (in kilodaltons) of the encoded proteins (15). The regions corresponding to the two subgenomic (sg) mRNAs are shown as arrows above the genome. The prototypical DI RNA used in this and previous studies (32), DI-70F, is shown below the genome. Shaded boxes represent genomic segments maintained in the DI RNAs, whereas thin lines represent genomic segments that are absent. DI-70F is composed of four highly conserved noncontiguous regions (I to IV), the lengths (in nucleotides) of which are indicated below the shaded boxes. DI-70F has been engineered with XbaI (X) and PstI (P) restriction sites flanking region II as well as a PstI restriction site between regions III and IV.
Replication enhancers have been reported for several RNA viruses, including Alfalfa mosaic virus (36), Turnip crinkle virus (26), and bacteriophage Qβ (1). Similarly, region III of TBSV has been shown to exhibit enhancer-like properties typical of DNA transcriptional enhancers, including the ability to (i) increase basal levels of DI RNA accumulation significantly (by ∼10-fold); (ii) function independently of orientation; and (iii) retain activity at different positions (32). Similar properties have also been observed, both in vitro and in vivo, for the motif 1 hairpin in turnip crinkle virus subviral satC RNA, which is proposed to enhance plus strand RNA synthesis via recruitment of viral RdRp (26).
In this study, we deduced and dissected the structure of the replication-enhancing region III within the context of a prototypical DI RNA in order to identify and characterize its functional components. These analyses have allowed us to define a key subelement of region III that functions in an orientation- and position-independent manner. An RNA hairpin in the minus strand of this subelement was identified as a primary structural determinant of the replication-enhancing activity observed in vivo. These data are considered in relation to possible roles for this element in the viral reproductive cycle.
MATERIALS AND METHODS
Plasmid constructs.
Plasmid construct pT100 (15) and TBSV DI RNA constructs pDI-70F and pDI-72SXPΔIII have been described previously (32). Oligonucleotides used in this study are listed in Table 1. Mutants AF/AR, BF/BR, and CF/CR were made by annealing primer pairs PR31-PR32, PR33-PR34, and PR35-PR36, respectively, by heating 100 pmol of each oligonucleotide pair at 95°C for 2 min, cooling for 30 min at ambient temperature, and subsequently ligating 10 pmol of the annealed primers into a PstI-digested calf intestinal alkaline phosphatase-treated pDI-72SXPΔIII plasmid. Similarly, AF-2, AF-2a, AF-2cm, AF-MD1, AF-PD1, AF-GU2, and AF-CA2 were constructed by ligating isolated PstI-digested PCR products generated from a pDI-72SXPΔIII template with primer pairs PR45-P9, PR46-P9, PR41-P9, PR58-P9, PR60-P9, PR113-P9, and PR114-P9, respectively, into a PstI-digested calf intestinal alkaline phosphatase-treated pDI-72SXPΔIII plasmid.
TABLE 1.
Oligonucleotides used in the study
| Oligonucleotide | Positionsa | Restriction enzyme site | Sequenceb | Sensec |
|---|---|---|---|---|
| P50 | 4754-4776 | GGAACATTGCAGAAATGCAGCCC | + | |
| P51 | 1420-1443 | BamHI | GCGCCGGATCCAGCGGTGCGAAACTCCGTACTTAC | + |
| P73 | 4623-4646 | PstI | GCCGCCTGCAGCCAATTCATGCTGGCTGGTATTGC | − |
| P9 | 4757-4776 | SphI/SmaI | GGCGGCCCGCATCCCGGGCTGCATTTCTGCAATGTTCC | − |
| PF4 | 1285-1304 | XbaI | GCGCGTCTAGAAGAAACGGGAAGCTCGCTCG | + |
| PF6 | 4398-4417 | PstI | GCGCGCTGCAGAGCGAGTAAGACAGACTCTT | + |
| PF7 | 1-20 | SacI | GGCGGAGCTCTAATACGACTCACTATAGGAAATTCTCCAGGATTTCTC | + |
| PR21 | 4645-4668 | NsiI | CCGCGCATGCATATTCCTGTTTACGAAAGTTAGG | + |
| PR23 | 4398-4420 | PstI | CGCCGCTGCAGAGCGAGTAAGAGCGCCTCTTCAG | + |
| PR31 | 4398-4432 | GAGCGAGTAAGACAGACTCTTCAGTCTGAGTTTGTGCTGCA | + | |
| PR32 | 4398-4432 | GCACAAACTCAGACTGAAGAGTCTGTCTTACTCGCTCTGCA | − | |
| PR33 | 4413-4447 | GCTCTTCAGTCTGAGTTTGTGGAGATGAGTGTAAATCTGCA | + | |
| PR34 | 4413-4447 | GATTTACACTCATCTCCACAAACTCAGACTGAAGAGCTGCA | − | |
| PR35 | 4437-4467 | GGAGATGAGTGTAAATCTGGCATAGCATACAGGTTACTGCA | + | |
| PR36 | 4437-4467 | GTAACCTGTATGCTATGCCAGATTTACACTCATCTCCTGCA | − | |
| PR37 | 4686-4708 | GACCCAGACACGGTTGATCTCAC | + | |
| PR38 | 4727-4739 | GATCGCTGGAAGCACTACCGGAC | + | |
| PR41 | 4398-4432 | PstI | CCGGCCTGCAGAGCGAGTAAGAGCGCCTCTTCAGACGCACTTTGTGCTGCAG | + |
| PR44 | 4398-4435 | PstI | GCCGCCTGCAGAGCGAGTAAGAGCGCCTCTTCAGACGCAGTTTGTGGAG | + |
| PR45 | 4398-4432 | PstI | CCGGCCTGCAGAGCGAGTAAGAGCGCCTCTTCAGTCTGAGTTTGTGCTGCAG | + |
| PR46 | 4398-4432 | PstI | CGCGCCTGCAGAGCGAGTAAGACAGACTCTTCAGACGCACTTTGTGCTGCAG | + |
| PR47 | 4730-4749 | GTCCGGTAGTGCTCCAGCG | − | |
| PR48 | 4407-4426/4647-4668 | PstI | GGCGCCTGCAGGACAGACTCTTCAGTCTGAGATTCCTGTTTACGAAAGTTAGG | + |
| PR58 | 4398-4432/4647-4673 | PstI | CGGCGCTGCAGAGCGAGTAAGACAGACTCTTCAGTCTGAATTAGTGCTGCAGATTCCTG | + |
| PR60 | 4398-4432 | PstI | CCGGCCTGCAGAGCGAGTAAGATAGGCTCTTCAGTCTGAGTTTGTG | + |
| PR61 | 4407-4432/4647-4668 | PstI | CCGGCCTGCAGGACAGACTCTTCAGTCTGAGTTTGTGATTCCTGTTACGAAAGTTAGG | + |
| PR63 | 1426-1445 | GCGAAACTCCGTACTTACAC | + | |
| PR71 | 4386-4417 | BstBI | CGCATGCATCCTGCCTTCGAAAAAGAAAGCGAGTAAG | + |
| PR113 | 4398-4439/4647-4653 | PstI | CCGGCCTGCAGAGCGAGTAAGACAGATTCTTCAGTTTGAGTTTGTGCTGCAGATTCCTG | + |
| PR114 | 4398-4439/4647-4653 | PstI | CCGGCCTGCAGAGCGAGTAAGACAAACTCTTCAATCTGAGTTTGTGCTGCAGATTCCTG | + |
| PR95 | 4411-4432 | XbaI | GGCCGTCTAGACACAAACTCAGACTGAAGAGTC | − |
| PR96 | 4398-4422 | XbaI | GCGCCTCTAGAAGCGAGTAAGACAGACTCTTCAGTC | + |
| PR97 | 4398-4442/161-168 | XbaI | GCGCGTCTAGACACAAACTCAGACTGAAGAGGCGCTCTTACTCGCTTCTAGACATGTCGC | − |
| PR98 | 4399-4432 | XbaI | CCGGCTCTAGACACAAACTCGCGTCTGAAGAGTCTGTCTTACTCGC | − |
| PR99 | 4398-4432/161-168 | XbaI | CGCGGTCTAGACACAAACTCGCGTCTGAAGAGGCGCTCTTACTCGCTTCTAGACATGTCGC | − |
Coordinates correspond to those of the TBSV genome (15).
Viral sequences corresponding to the coordinates shown are underlined. Restriction enzyme sites are italicized, and individual mutations are in bold.
Refers to the sense of the oligonucleotide in reference to the plus-sense viral RNA.
III(26) and III(20) were made by ligating gel-purified PstI-SphI PCR products generated from a pDI-72SXPΔIII template with primer pairs PR61-P9 and PR48-P9, respectively, into a PstI- and SphI-digested calf intestinal alkaline phosphatase-treated pDI-72SXPΔIII plasmid. AFX, AFX-2, AFX-2a, and AFX-2cm were made by ligating isolated XbaI-digested PCR products generated from a pDI-72SXP template with primer pairs PR95-PR96, PR96-PR97, PR96-PR98, and PR96-PR99, respectively, into an XbaI-digested calf intestinal alkaline phosphatase-treated pDI-72SXPΔIII plasmid.
T100-2 and T100-2cm were made by sequentially digesting pT100 with EcoRI and BstBI and gel-purifying the ∼360-nucleotide fragment and then ligating it with a PCR product and an EcoRI- and SalI-digested calf intestinal alkaline phosphatase-treated pT100 construct. The PCR products were generated from a pDI-73 plasmid (39) with primer pairs PR23-P9 (for T100-2) and PR44-P9 (T100-2cm), and subsequently, these individual amplified segments were used as templates for a second PCR with primer pair PR71-P73 and then gel-purified following digestion with BstBI and SalI. All mutant constructs were verified by sequencing.
In vitro transcription.
Viral transcripts were synthesized in vitro by transcription of SmaI-linearized DNAs with an Ampliscribe T7 RNA polymerase transcription kit (Epicentre Technologies) as described previously (28).
Preparation and inoculation of cucumber protoplasts.
Cucumber protoplasts (var. Straight 8) were prepared and inoculation with polyethylene glycol-CaCl2 was done as described previously (39) with approximately 2 μg of T100 and 1 μg of DI RNA transcripts, unless otherwise noted.
Analysis of RNAs from protoplasts.
Total nucleic acid was harvested from protoplasts as described previously (39). Aliquots of the total nucleic acid preparations were separated in denaturing 8 M urea-4.5% polyacrylamide gels, and viral RNAs were detected by electrophoretic transfer to nylon membranes (Hybond-N; Amersham) followed by Northern blot analysis with complementary 32P-end-labeled oligonucleotides. Minus strand assays were performed as described previously (32) with the following modifications. Three-fifths of the total nucleic acid prepared from each infection was analyzed. In addition, Northern blot analysis was conducted with 32P-end-labeled oligonucleotide probes P50, PR21, PR37, and PR38. For time course experiments, oligonucleotides PF4, PF6, PF7, P51, PR35, and PR63 were used in addition to those mentioned previously. All Northern blots were quantified by radioanalytical scanning with an InstantImager (Packard Instruments). Values generated represent averages of at least three independent infections with standard errors.
RNA secondary-structure probing.
For in vitro analysis of RNA secondary structure, DI RNA transcripts (3 μg) were added to yeast RNA (3 μg) in modification buffer (25 mM Tris-HCl [pH 7.5], 200 mM NaCl, 5 mM MgCl2, and 1 mM EDTA), equilibrated (95°C for 2 min; 60°C for 10 min; and 37°C for 10 min), treated with various RNA structure-probing chemicals [diethyl pyrocarbonate (DEPC) and 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide (CMCT)] or enzymes (RNase T1 and RNase V1) and analyzed by primer extension as described previously (42). Primers PR47 and PR63 were used to analyze plus and minus strands, respectively.
RESULTS
Comparative sequence analysis of region III in tombusviruses.
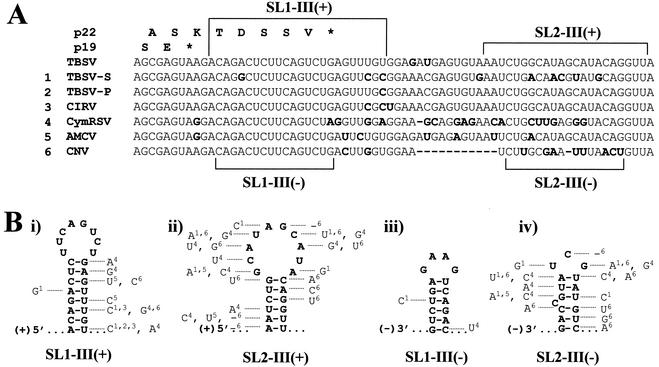
In the context of the seven members of the genus Tombusvirus for which sequence information is currently available—TBSV, TBSV-P (pepper isolate), TBSV-S (statice isolate), Carnation Italian ringspot virus, Cymbidium ringspot virus, Cucumber necrosis virus, and Artichoke mottled crinkle virus—the corresponding 5′ 28 nucleotides of region III are highly conserved, whereas the more 3′ 42 nucleotides exhibit significant variation (Fig. 2A). This high degree of sequence identity within the 5′ portion is not unexpected, as this segment corresponds to coding sequence for the last two amino acids in p19 and the last eight amino acids in p22 (23) (Fig. 2A). The variations within the 3′ half correspond primarily to substitutions, but in cucumber necrosis virus, an 11-nucleotide deletion is present.
FIG. 2.
(A) Region III RNA sequence alignment for seven members of the genus Tombusvirus. Nucleotides in lightface indicate conserved nucleotides, whereas boldface indicates variable sequences and hyphens indicate deleted nucleotides. Square brackets enclose sequences corresponding to mfold-predicted SL1-III(+), SL1-III(−), SL2-III(+), and SL2-III(−). Amino acids corresponding to the C termini of p22 and p19 for TBSV are labeled according to their one-letter abbreviations and are shown above the coding regions. CIRV, Carnation Italian ringspot virus; CymRSV, Cymbidium ringspot virus; AMCV, Artichoke mottled crinkle virus; CNV, Cucumber necrosis virus. (B) Plus and minus strand RNA secondary structures predicted by mfold to form in the 70-nucleotide region III. Variable nucleotides observed within the members of the genus Tombusvirus are shown with superscripts referring to viruses labeled in panel A. Underscores indicate deletions.
Secondary-structure analysis of full-length plus and minus strand DI RNA sequences with the computer program mfold (46) predicted two stem-loop structures in both the plus strand [SL1-III(+) and SL2-III(+)] and minus strand [SL1-III(−) and SL2-III(−)] of region III (Fig. 2B). Of the four stem-loop structures, SL1-III(−) is the most conserved between species, with single nucleotide substitutions in TBSV-S (A to G mutation at position 4413 causing a C-A mismatch) and cymbidium ringspot virus [G to A mutation at position 4424, which is predicted to maintain SL1-III(−) via a GU wobble base pair] (Fig. 2B). In contrast, comparisons of predicted secondary structures for SL2-III(−), SL1-III(+), or SL2-III(+) exhibited far less conservation (Fig. 2B).
RNA secondary-structure mapping of region III plus and minus strands.
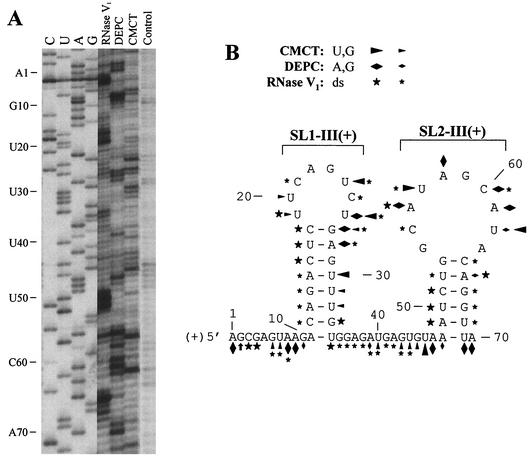
In vitro solution structure analyses of plus and minus strand sequences of region III were performed, and the results were integrated with data derived from mfold analyses (Fig. 3 and 4). Solution structures were assessed with enzymatic and chemical treatments of full-length DI-70F plus and minus strand RNA transcripts followed by primer extension analysis of the resulting modified RNA products. The enzymatic or chemical probes RNase V1 (hydrolyzes phosphodiester bonds in helical double-stranded and stacked single-stranded bases), DEPC (modifies N7 position of unpaired A and G residues), and CMCT (modifies the N3 and N2 positions of unpaired U and G residues) (24) were used in this study. Identification of individual nucleotides sensitive to chemical modification or nuclease-mediated cleavage was achieved by side-by-side comparison of corresponding primer extension cDNA products with a DNA sequencing ladder generated with identical oligonucleotide primers (Fig. 3A and 4A). It should be noted that, for these comparisons, primer extension products migrate as bands 1 nucleotide shorter than their corresponding dideoxy-terminated DNA counterparts due to differential mechanisms of termination in the reverse transcription and sequencing reactions (24).
FIG. 3.
Chemical and enzymatic probing of plussense DI-70F transcripts. (A) DI-70F RNA was treated in vitro with RNase V1, DEPC, or CMCT, and the products were subsequently analyzed by primer extension. Primer extension products generated were separated in an 8% sequencing gel. Also indicated are lanes showing untreated control transcripts extended with PR47 (control) and DI-70F plasmid DNA sequenced with PR47. (B) Optimal computer-predicted secondary-structure model of region III plus strand that includes a summary of enzymatic and chemical modifications (RNase V1, DEPC, or CMCT). Stronger and weaker relative reactivities are indicated by larger and smaller symbols, respectively.
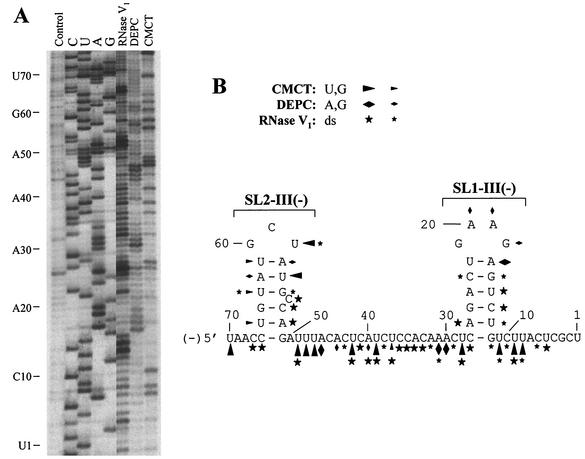
FIG. 4.
Chemical and enzymatic probing of minus-sense DI-70F. (A) Minus strand DI-70F RNA treated in vitro with RNase V1, DEPC, or CMCT and analyzed as described in the legend to Fig. 3. (B) Optimal computer-predicted secondary-structure model of region III minus strand that includes a summary of enzymatic and chemical modifications.
For the plus strand, the solution structure data were mapped onto the most stable mfold-predicted structure that included two local stem-loop structures: (i) stem-loop 1 [SL1-III(+)] (contained in the 5′ half of region III) and (ii) stem-loop 2 [SL2-III(+)] (contained in the 3′ half of region III) (Fig. 3B). The majority of the 10 nucleotides immediately 5′ proximal to SL1-III(+) (A1 to G10) were readily accessible to various single-strand-specific reagents (Fig. 3A and B), suggesting that this segment of region III is primarily unpaired. Similarly, predicted SL1-III(+) and SL2-III(+) single-stranded loops were modified preferentially by single-strand-specific probes, whereas their predicted stems were efficiently recognized by RNase V1. However, several CMCT (U30 to U32) modifications were observed within the predicted 3′ half of the stem of SL1-III(+), suggesting some flexibility of the structure under the conditions of analysis.
Previous experiments have demonstrated that region III is functional in both forward and reverse orientations, suggesting activity in both plus and minus strands (32). To identify potentially functional structures in the minus strand, similar in vitro RNA secondary-structure mapping was also carried out on full-length minus strand DI RNA transcripts. Solution structure analysis and the optimal mfold-predicted structure of region III minus strand RNA were most consistent with the presence of two stem-loop structures (Fig. 4A and 4B). SL2-III(−) is less well defined by the solution analysis but is characterized by RNase V1 reactivities within predicted double-stranded regions and a CMCT (U58) modification within the loop region. The low number of G-C base pairs and the C bulge within this stem may contribute to its instability and could explain the DEPC and CMCT reactivities observed within the predicted double-stranded region. The extended intervening segment which separates SL1-III(−) and SL2-III(−) showed numerous RNase V1 cleavages, suggesting that parts of it are double-stranded (however, the putative base-pairing partners are undefined). Other portions of this segment were accessible to DEPC and CMCT and thus single-stranded. Of the four stem-loop structures predicted by mfold in the plus and minus strands, SL1-III(−) is best supported by the solution structure data. RNase V1 is very reactive in its predicted stem, whereas loop residues were sensitive to DEPC modification (Fig. 4B).
Defining functional segments within region III.
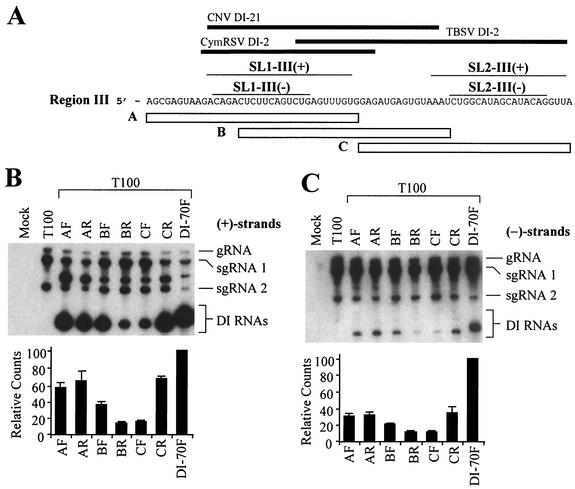
All naturally occurring tombusvirus DI RNAs maintain some portion of the 70-nucleotide-long region III present in TBSV DI-70F. For example, cucumber necrosis virus DI-21 (10) and cymbidium ringspot virus DI-2 (14) maintain 5′-proximal segments, whereas TBSV DI-2 (17) maintains a more 3′-proximal segment (Fig. 5A). The maintenance of different portions of region III within different tombusvirus DI RNAs suggests that multiple functional subelements may exist within this region.
FIG. 5.
Analysis of DI RNAs containing three different overlapping 35-nucleotide segments of region III. (A) The plus strand sequence of region III is presented. Open bars below the sequence represent segments A, B, and C. Thick black lines at the top represent corresponding region III segments present in cucumber necrosis virus (CNV) DI-21, cymbidium ringspot virus (CymRSV) DI-2, and TBSV DI-2 (10, 14, 17), and the thin lines directly below show corresponding positions of SL1-III(+), SL1-III(−), SL2-III(+), and SL2-III(−). (B) Northern blot analysis of plus strands of DI RNAs containing region III segments (outlined in Fig. 4A) introduced in either the forward (F) or reverse (R) orientation. Approximately 4.0 × 105 cucumber protoplasts were inoculated with H2O (mock) or 2 μg of T100 transcript only or coinfected with 2 μg of T100 and 1 μg of the respective DI RNAs. Total nucleic acids were isolated 24 h postinoculation, separated in 8 M urea-4.5% polyacrylamide gels, transferred to nylon membranes, and subsequently probed with a 32P-end-labeled minus-sense probe. Bands corresponding to TBSV genomic RNA (gRNA), subgenomic mRNA1 (sgRNA1), subgenomic mRNA2 (sgRNA2), and various DI RNAs are shown. The bands between sgRNA1 and sgRNA2 in coinfections represent previously described head-to-tail dimers of DI RNAs (32). Quantification of DI RNA bands (monomers only) is shown in the bar graph below the autoradiogram in panel B. (C) Northern blot analysis of corresponding minus strands for the samples in B. Minus strands were analyzed as in panel B except that membranes were hybridized with plussense probes.
To investigate this possibility, region III was divided into three overlapping 35-nucleotide-long segments (A, B, and C) (Fig. 5A). Each of these segments was used to replace the full-length region III in DI-70F (Fig. 1) in both the forward (designated F) and reverse (designated R) orientations. Note that SL1-III(+) and SL1-III(−) are present in the A segment, whereas the C segment contains SL2-III(+) and SL2-III(−) (Fig. 5A). Northern blot analysis of total nucleic acids isolated from cucumber protoplasts coinfected with transcripts of these mutant DI RNAs and the TBSV helper genome (T100) is shown in Fig. 5B. DI RNAs that contained the 3′-proximal segment C in the forward orientation (CF) accumulated to low levels, whereas accumulation of their reverse-oriented counterpart (CR) was significantly higher (Fig. 5B). In contrast, accumulation of DI RNAs containing the forward-oriented central segment B (BF) was greater relative to its oppositely oriented counterpart, BR. Interestingly, DI RNAs containing the 5′-proximal segment A accumulated to relatively high levels when present in either orientation. Thus, segment A functionally mimics the full-length 70-nucleotide region III in its ability to effectively enhance DI RNA accumulation efficiency in both the forward and reverse orientations (32). Northern blot analysis of minus strands indicated that, compared to the DI-70F control, mutant minus strand DI RNA accumulation was generally proportional to their corresponding plus strands (Fig. 5C), but for some mutants (i.e., AF, AR, BF, and CR), the relative reduction in minus strand accumulation was more pronounced than for their plus strand counterparts.
Kinetics of DI RNA accumulation.
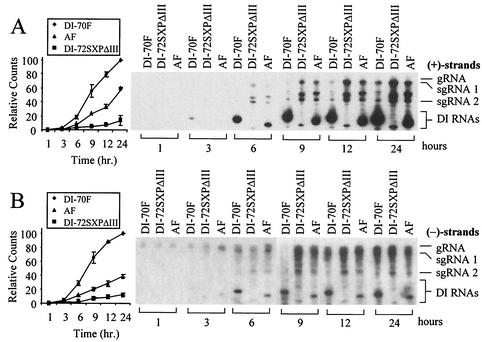
Time course experiments were conducted to determine whether there were any differences in the rates of accumulation of plus and minus strands between DI-70F, AF, and DI-72SXPΔIII, in which all of region III is deleted (32). Plus-sense DI RNAs were first detectable for both DI-70F and AF at 3 h postinoculation (Fig. 6A). Following their appearance, plus strand DI RNAs increased steadily, with AF and DI-72SXPΔIII accumulating to ∼60% and ∼10% of DI-70F levels, respectively. Minus strand DI RNAs were detectable at 3 h postinoculation for DI-70F and 6 h postinoculation for DI-72SXPΔIII and AF DI RNAs (Fig. 6B). The further accumulation of minus strand DI RNAs was also observed over the 24-h time period, with AF and DI-72SXPΔIII minus strands accumulating to ∼30% and ∼10% of DI-70F levels, respectively.
FIG. 6.
Time course analysis of accumulation of DI RNAs in protoplasts. (A) Northern blot analysis of plus strand accumulation of DI-70F, AF, and DI-72SXPΔIII in cucumber protoplasts at 1, 3, 6, 9, 12, and 24 h postinoculation as described in the legend to Fig. 5B. Graph depicts quantification of DI RNA accumulation levels by radioanalytical scanning. (B) Corresponding minus strand analysis.
Compared to DI-72SXPΔIII, the addition of segment A in AF resulted in increased accumulation of plus strands relative to minus strands. The absolute decrease in DI RNA plus strands for AF and DI-72SXPΔIII likely reflects a replication defect, as previous stability assays have demonstrated that region III does not markedly influence DI RNA plus strand degradation in vivo (32). But we cannot preclude the possibilities that region III influences DI RNA minus strand stability and that the stabilities of plus and/or minus strand DI RNAs may be altered by viral proteins present during infections, as has been demonstrated for brome mosaic virus 1a protein (20).
Minus strand stem-loop structure in segment A facilitates DI RNA accumulation.
Due to its smaller size and ability to significantly enhance replication in both orientations, AF was selected for more detailed analyses. Targeted mutations were introduced in order to disrupt and/or maintain base pairing of SL1-III(+) and/or SL1-III(−) in DI RNA AF (Fig. 7A and B). The first modification introduced into AF (mutant AF-2) was selected because it was shown previously to severely inhibit DI RNA accumulation when present in the full-length region III of DI-70F (32). We therefore wanted to determine whether the same defined modification would have a similar inhibitory effect when introduced into the smaller A segment of AF.
FIG. 7.
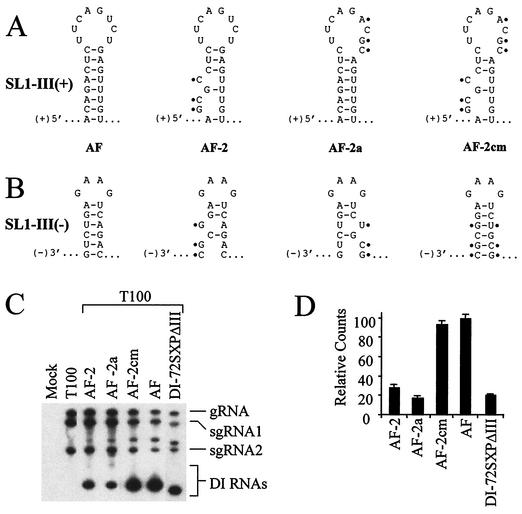
Analysis of AF mutants with modifications in SL1-III(+) and SL1-III(−). Depicted are substitutions (indicated with black dots) and their effect on SL1-III(+) (A) and SL1-III(−) (B) secondary structures, as predicted by mfold. (C) Northern blot analysis of plus strand DI RNA accumulation in protoplasts. (D) Graphic representation of the DI RNA accumulation levels in panel C.
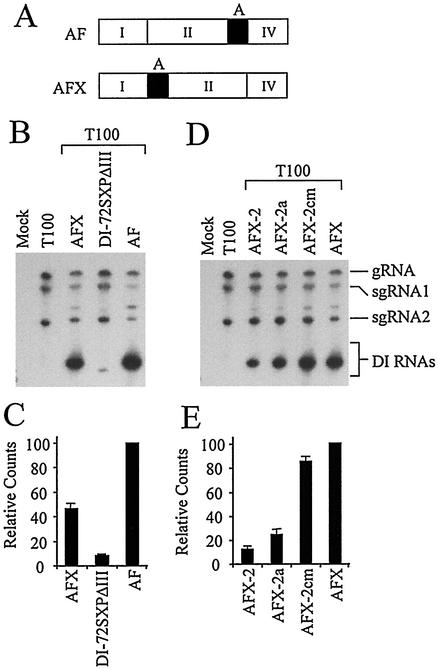
The mutant constructed, AF-2, contained three single-nucleotide substitutions predicted to disrupt the stem portions of both SL1-III(+) and SL1-III(−) (Fig. 7A and B) and exhibited accumulation levels approximately 20% that of AF (Fig. 7C and D). In AF-2a, in which the stem of SL1-III(+) but not SL1-III(−) is restored, similar low levels of accumulation were observed (Fig. 7C and D). In AF-2cm, the mutations introduced into AF-2 and AF-2a were combined to restore the stem of SL1-III(−) but not SL1-III(+). This mutant accumulated to essentially AF levels (Fig. 7C and D), suggesting that only SL1-III(−) formation is important for enhanced AF accumulation.
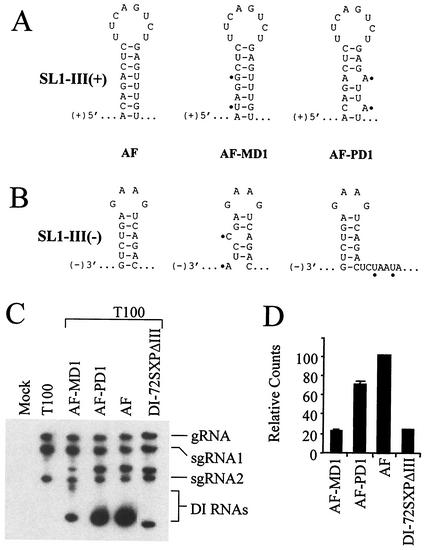
Additional sets of mutants were constructed to further test the importance of SL1-III(−) for AF accumulation. Mutations were designed to disrupt either the stem of SL1-III(+) or the stem of SL1-III(−) (Fig. 8A and B). Disrupting only SL1-III(+) in AF-PD1 reduced DI RNA accumulation modestly relative to AF, but disrupting only SL1-III(−) in AF-MD1 led to a fourfold decrease with respect to AF levels (Fig. 8C and D). These results are consistent with a role for SL1-III(−) in augmenting DI RNA accumulation.
FIG. 8.
Analysis of AF mutants with targeted disruptions in SL1-III(+) and SL1-III(−). Diagrams depict nucleotide substitutions (indicated with black dots) and their effect on SL1-III(+) (A) and/or SL1-III(−) (B) secondary structures, as predicted by mfold. (C) Northern blot analyses of DI RNA accumulation in protoplasts. (D) Graphic representation of the DI RNA accumulation levels in panel C.
The canonical nature of the base pairs in the stem of SL1-III(−) would allow the possibility of a precisely complementary structure in the plus strand, designated ASL1-III(+) (Fig. 9A). ASL1-III(+) is an alternative structure to SL1-III(+) (Fig. 8A), and the former is not strongly supported by either mfold or solution structure analysis. Nonetheless, this putative structure needed to be investigated, as the mutations targeting the SL1-III(−) (Fig. 7 and 8) would also coincidentally target this alternative plus strand structure. Therefore, to determine whether ASL1-III(+) was involved, mutants were constructed in which either SL1-III(−) or ASL1-III(+) was preferentially destabilized (Fig. 9A and B). Targeted destabilization of ASL1-III(+) in AF-CA2 did not appreciably affect DI RNA accumulation, whereas preferential disruption of SL1-III(−) in AF-GU2 resulted in a fivefold decrease in DI RNA accumulation (Fig. 9C and D). Collectively, these and the preceding data support SL1-III(−) in the minus strand as a key element for DI RNA accumulation and rule out major involvement of either of the possible stem-loop structures in the plus strand.
FIG. 9.
Analysis of AF mutants with strand-specific disruptions in SL1-III(−) and ASL1-III(+). Diagrams depict nucleotide substitutions (indicated with black dots) and their effect on (A) ASL1-III(+) or (B) SL1-III(−) secondary structures, as predicted by mfold. (C) Northern blot analyses of DI RNA accumulation in protoplasts. (D) Graphic representation of the DI RNA accumulation levels in panel C.
Defining a core element within segment A.
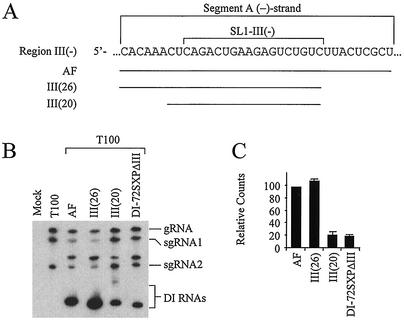
Additional derivatives of AF were constructed in order to define the boundaries of the activity of segment A (Fig. 10A). Deleting 9 nucleotides from the 3′ end of the minus strand of segment A to generate III(26) increased accumulation slightly to ∼110% of AF levels (Fig. 10B and C). This indicates that SL1-III(−)-mediated enhancement of DI RNA accumulation is not dependent on sequences 3′ to it in the minus strand sequence. Deleting an additional 6 nucleotides in mutant III(20) resulted in low levels of accumulation (∼25% of AF). This suggests that the sequence immediately 5′ to SL1-III(−) in the minus strand is important to region III function and that the maintenance of secondary structures in SL1-III(−), in addition to this upstream 6-nucleotide flanking sequence, facilitates efficient DI RNA accumulation.
FIG. 10.
Analysis of AF mutants with deletions flanking SL1-III(−). (A) Linear minus strand RNA sequence corresponding to segment A. Portions of segment A contained in mutants AF, III(26), and III(20) are indicated by black lines. (B) Northern blot analysis of DI RNA accumulation in protoplasts. (C) Graphic representation of the DI RNA accumulation levels in panel B.
Segment A enhances DI RNA accumulation from a different location in an SL1-III(−)-dependent manner.
Previously it was shown that region III enhances DI RNA accumulation when positioned either adjacent to or ∼150 nucleotides upstream of the 5′ end of region IV (32). To determine whether a more dramatic relocation could be tolerated, forward-oriented segment A was transposed with region II (Fig. 11A). Northern blot analysis indicated that this mutant (AFX) accumulated to notably higher levels than the region III-less DI-72SXPΔIII and about half that of AF (Fig. 11B and C).
FIG. 11.
Effect of segment A translocation on DI RNA accumulation. (A) Schematic diagram depicting segment A (solid black rectangle) location relative to regions I, II, and IV (white rectangles) as labeled in AF and AFX DI RNA. (B) Northern blot analysis of mutant AFX. (D) Northern blot analysis of compensatory mutants AFX-2, AF-2a, and AF-2cm (see text for details on the structure of mutants). (C and E) Graphic representation of the DI RNA accumulation levels in panels B and D, respectively.
To test whether SL1-III(−) was required for the enhanced accumulation of segment A in its new location, the stem of this structure was disrupted and then restored with a set of substitutions that were identical to those present in AF-2, AF-2a, and AF-2cm (refer to Fig. 7B). The corresponding AFX mutants are AFX-2, AFX-2a, and AFX-2cm, respectively. Both of the disrupted mutants, AFX-2 and AFX-2a, exhibited substantially decreased levels of accumulation, whereas regenerating the stem in AFX-2a restored accumulation to near AFX levels (Fig. 11D and E). These results are consistent with segment A functioning as a discrete replication enhancer element in a manner that is independent of position but dependent on SL1-III(−) formation.
Formation of SL1-III(−) is important for accumulation of TBSV genomic RNA.
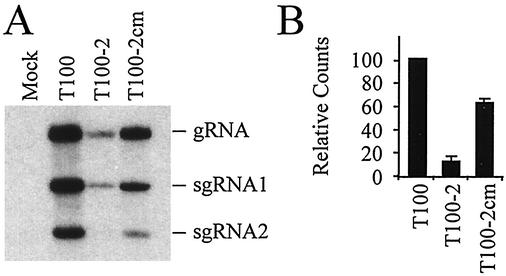
To determine whether SL1-III(−) is also required in the context of the full-length TBSV genome, compensatory mutations predicted to disrupt or maintain SL1-III(−) were introduced into the TBSV genome. Introducing a 3-nucleotide substitution to disrupt SL1-III(−) in mutant T100-2 (refer to AF-2 in Fig. 7B for RNA structures) significantly decreased genomic and subgenomic accumulation to approximately 10% of wild-type levels, whereas restoring SL1-III(−) in mutant T100-2cm (refer to AF-2cm in Fig. 7B for RNA structures) markedly recovered accumulation to 60% of wild-type levels (Fig. 12B). This finding supports the concept that SL1-III(−) is an important cis-acting element and confirms its importance within the context of the TBSV genome.
FIG. 12.
Effect of disruption and restoration of SL1-III(−) in the full-length TBSV genome. (A) Northern blot analysis of plus strand accumulation of wild-type TBSV (T100) and T100-based SL1-III(−) mutants T100-2 and T100-2cm (see text for details on the structure of mutants). (B) Graphic representation of the TBSV genomic RNA accumulation levels in panel A.
DISCUSSION
Region III is a multifunctional viral RNA element that plays at least three important roles in the TBSV life cycle, namely (i) encoding the C-terminal amino acids of p19 and p22 (15); (ii) comprising part of a translational enhancer (41); and (iii) mediating viral RNA replication (32). In this study, we analyzed the secondary structure of region III and conducted a detailed investigation of potential replication subelements within this sequence. Our results suggest that region III-mediated enhancement of TBSV DI and genomic RNA accumulation is, in part, related to the formation of a stem-loop structure, SL1-III(−), in the minus strand.
Context dependence of region III activity in tombusvirus DI RNAs.
In the genus Tombusvirus, the observation that all naturally occurring DI RNAs maintain some portion of the conserved region III sequence suggests an important role for this element in viral RNA accumulation. Previous analyses of region III function have been complicated by variability in region III size as well as contextual differences within different DI RNAs. For instance, deletion analysis has shown that region III is important for the accumulation of TBSV DI-70F (32) and cymbidium ringspot virus DI-2 (14) but not cucumber necrosis virus C2 RNA (5). In the cucumber necrosis virus DI RNA mutant, the absence of region III may be compensated for by the activity of other cis-acting RNA replication elements. Indeed, this appears to be the case for region I in TBSV DI RNAs, where some mutations within this region completely inhibit DI RNA accumulation by acting in a cis-dominant negative manner (42). But deletion of the entire region I was found to allow low-level replication that resulted from compensatory activity in region II (40).
In the present study, we have identified an important 35-nucleotide subelement (segment A) within the 5′ half of our 70-nucleotide region III of DI-70F that maintains significant activity independently of other portions of region III. Smaller naturally occurring tombusvirus DI RNAs generally maintain sections of region III that correspond to segment A and include SL1-III(−), but occasionally other sections are retained. It is possible that these different sequences are able to function efficiently in different DI RNAs due to various contextual effects or because region III sequences have redundant activities. Some type of functional redundancy was suggested by the results of scanning mutagenesis of region III in TBSV DI-70F (32) and also by studies on cymbidium ringspot virus DI RNAs, where either the 5′ or 3′ half of a region III element was sufficient for efficient DI RNA accumulation (14).
TBSV DI-2, which contains region III sequences similar to segment C, was shown previously to accumulate efficiently (17) in the absence of the segment A subelement. In contrast, DI RNA CF in this study accumulated only to low levels. Structural differences within the more extensively evolved DI-2, which is ∼190 nucleotides smaller than DI RNA CF, or the presence of additional sequences within its larger 46-nucleotide-long region III segment may account for the differences in these results.
Modular nature of segment A.
Previous analyses of region III revealed that this sequence is functional when located immediately adjacent to region IV or when it was separated from region IV by the natural intervening sequence region 3.5 (28, 32, 41). In the present study, segment A was found to maintain function when transposed with region II. This observation indicates that segment A is able to function independently of relative position and flanking sequences (Fig. 11). A major contributing factor to this modular property is the presence of a localized structure in segment A, SL1-III(−), whose formation and activity are required at both positions. Although these results suggest that segment A acts as a modular element, they do not preclude its communication with other DI RNA sequences via RNA-RNA interactions, by either the terminal loop of SL1-III(−) or the important flanking sequence immediately 5′ to this hairpin.
Previous studies have also shown that full-length region III can be inverted with little effect on DI RNA accumulation (32). We now find that segment A shares similar orientation-independent properties. The inversion of segment A would relocate SL1-III(−) to the plus strand. It is possible that in this opposite strand SL1-III(−) is still able to exert its activity. Alternatively, due to the inversion, the precise complementary structure of SL1-III(−), ASL-III(+), would be relocated to the minus strand and could functionally replace SL1-III(−). This concept is not without precedent, as functional complementary structures have been reported for the enhancer motif 1 hairpin in turnip crinkle virus (26).
Effect of segment A on viral RNA accumulation.
The kinetic analysis of DI-70F indicates that plus and minus strands accumulate coordinately throughout the 24-h incubation period (Fig. 6). This is similar to the accumulation profiles observed for turnip crinkle virus satellite RNA (26) but contrasts with those of other viruses, such as tobacco mosaic virus, in which minus strand synthesis peaks earlier in infection and subsequently decreases to low levels (19). In the absence of region III (DI-72SXPΔIII), plus and minus strands accumulate to ∼10% of DI-70F levels, indicating that region III is important but not essential for plus or minus strand synthesis (Fig. 6). Additionally, the relative ratios of plus and minus strands were similar to those for wild-type DI RNAs. However, when segment A was introduced, i.e., AF, DI RNA accumulation increased threefold for minus strands and sixfold for plus strands (Fig. 6). This enhancement of plus strand AF accumulation relative to DI-72SXPΔIII suggests that segment A may preferentially facilitate plus strand synthesis.
In a previous study, one particular scanning mutant within the 5′ half of region III (smIII-2F) was found to significantly decrease DI-70F RNA accumulation in vivo (32). In the present study, the identical modification in AF (i.e., AF-2) also dramatically reduced DI RNA accumulation (Fig. 7), which reinforces the idea that SL1-III(−) in segment A functions in a similar manner to the full-length region III. Additional substitutions that specifically restored SL1-III(−) led to recovered activity (Fig. 7). This result, in combination with those from two other sets of mutations (Fig. 8 and 9), strongly supports a functional role for SL1-III(−). Additionally, compensatory mutations in SL1-III(−) within the TBSV genome indicate that it is active in a genomic context.
It is noted that the genomic mutations also alter the C terminus of the p22 movement protein, but these modifications are not likely to influence viral replication, as p22 has been shown to be dispensable for TBSV replication in protoplasts (34). It cannot be discounted that the mutants tested also affect the translation of viral replication proteins, since region III is required for optimal translational efficiency in vivo (41). Nevertheless, the correlation between genomic accumulation levels and disruption/restoration of stem-loop-III(−) supports a minus strand activity, rather than a plus or messenger-strand activity.
Functions of segment A.
Plus strand activities for region III sequences in both protein coding and translational enhancement have been defined previously (15, 41). The results of the current study suggest an additional function for segment A that is conferred in the minus strand. Considering its minus strand location and its effect on DI RNA accumulation, it seems likely that SL1-III(−) is involved in some step of viral RNA replication. If this structure acts in cis, then it most probably functions in plus strand synthesis. Consistent with this concept, the accompanying report by Panavas and Nagy (30) demonstrates that TBSV SL1-III(−) acts as a potent enhancer of plus strand synthesis in vitro. Thus, the positive effects of SL1-III(−) observed in vivo are likely related to this defined in vitro activity. However, we cannot preclude some contribution by a plus strand activity.
For turnip crinkle virus, the motif 1 replication enhancer RNA element is believed to function in the minus strand by recruiting the viral RdRp, thereby facilitating its initiation at the plus strand promoter (26). Similarly, it is possible that TBSV SL1-III(−) functions in an analogous capacity. Its similarity in structure to the 3′-terminal tetra-loop-containing stem-loop of the TBSV minus strand promoter—that is essential for DI RNA replication in vivo (M. Fabian and K. A. White, unpublished data)—lends support to this notion. However, a recruiting role in the minus strand would require subsequent delivery of the RdRp to the plus strand promoter at the 3′ end of the template. Such long-distance interactions could be mediated by either a protein bridge or RNA-RNA interactions; the latter have been demonstrated previously for activation of subgenomic mRNA transcription in TBSV (7, 44).
Considering a genomic context, the 5′-proximal minus strand location of an enhancer of plus strand synthesis could be related to a proofreading mechanism, whereby only minus strand templates containing both the 3′-proximal plus strand promoter and the 5′-proximal enhancer are utilized efficiently by the RdRp, in effect culling out shorter, presumably defective, templates. Terminal communications between genome termini by either protein bridges (2, 16) or RNA-RNA interactions (21, 43) have been implicated in similar proofreading functions in other plus strand RNA viruses. For TBSV, the 5′-proximal minus strand location of the SL1-III(−) could also be related to a possible role in facilitating subgenomic mRNA transcription. Currently, it is believed that TBSV subgenomic mRNAs are transcribed from prematurely terminated, subgenomic mRNA-length complementary minus strands rather than the full-length minus strands of the genome (6, 7). The relative location of SL1-III(−) would allow it to also be present in these truncated minus strand templates, where it could act to stimulate subgenomic mRNA transcription. SL1-III(−) thus holds the potential to function as an enhancer of RNA synthesis in a variety of viral template contexts. This possibility and other mechanistic aspects of SL1-III(−) activity will be the focus of future investigations.
Acknowledgments
We thank Peter Nagy and members of our laboratory for reviewing the manuscript. Also, we are grateful to Peter Nagy and Tadas Panavas for sharing unpublished results.
This work was supported by grants from NSERC, PREA, and CRC to K.A.W.
REFERENCES
- 1.Barrera, I., D. Schuppli, J. M. Sogo, and H. Weber. 1993. Different mechanisms of recognition of bacteriophage Qβ plus and minus strand RNAs by Qβ replicase. J. Mol. Biol. 232:512-521. [DOI] [PubMed] [Google Scholar]
- 2.Barton, D. J., B. J. O'Donnell, and J. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boccard, F., and D. Baulcombe. 1993. Mutational analysis of cis-acting sequences and gene function in RNA3 of Cucumber mosaic virus. Virology 193:563-578. [DOI] [PubMed] [Google Scholar]
- 4.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y. C., M. Borja, H. B. Scholthof, A. O. Jackson, and T. J. Morris. 1995. Host effects and sequences essential for accumulation of defective interfering RNAs of cucumber necrosis and tomato bushy stunt tombusviruses. Virology 210:41-53. [DOI] [PubMed] [Google Scholar]
- 6.Choi, I. R., M. Ostrovsky, G. Zhang, and K. A. White. 2001. Regulatory activity of distal and core RNA elements in tombusvirus subgenomic mRNA2 transcription. J. Biol. Chem. 276:41761-41768. [DOI] [PubMed] [Google Scholar]
- 7.Choi, I. R., and K. A. White. 2002. An RNA activator of subgenomic mRNA1 transcription in tomato bushy stunt virus. J. Biol. Chem. 277:3760-3766. [DOI] [PubMed] [Google Scholar]
- 8.Dreher, T. W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151-174. [DOI] [PubMed] [Google Scholar]
- 9.Duggal, R., F. C. Lahser, and T. C. Hall. 1994. cis-acting sequences in the replication of plant viruses with plussense RNA genomes. Annu. Rev. Phytopathol. 32:287-309. [Google Scholar]
- 10.Finnen, R. L., and D. M. Rochon. 1993. Sequence and structure of defective interfering RNAs associated with cucumber necrosis virus infections. J. Gen. Virol. 74:1715-1720. [DOI] [PubMed] [Google Scholar]
- 11.French, R., and P. Ahlquist. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodfellow, I., Y. Chaudhry, A. Richardson, et al. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Havelda, Z., and J. Burgyan. 1995. 3′ terminal putative stem-loop structure required for the accumulation of cymbidium ringspot viral RNA. Virology 214:269-272. [DOI] [PubMed] [Google Scholar]
- 14.Havelda, Z., T. Dalmay, and J. Burgyan. 1995. Localization of cis-acting sequences essential for cymbidium ringspot tombusvirus defective interfering RNA replication. J. Gen. Virol. 76:2311-2316. [DOI] [PubMed] [Google Scholar]
- 15.Hearne, P. Q., D. A. Knorr, B. I. Hillman, and T. J. Morris. 1990. The complete genome structure and synthesis of infectious RNA from clones of Tomato bushy stunt virus. Virology 177:141-151. [DOI] [PubMed] [Google Scholar]
- 16.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillman, B. I., J. C. Carrington, and T. J. Morris. 1987. A defective interfering RNA that contains a mosaic of a plant virus genome. Cell 51:427-433. [DOI] [PubMed] [Google Scholar]
- 18.Hillman, B. I., P. Hearne, D. Rochon, and T. J. Morris. 1989. Organization of the Tomato bushy stunt virus genome: characterization of the coat protein gene and 3′ terminus. Virology 169:42-50. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa, M., T. Meshi, T. Ohno, and Y. Okada. 1991. Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic virus infection. J. Virol. 65:861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janda, M., and P. Ahlquist. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. USA 95:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klovins, J., and J. van Duin. 1999. A long-range pseudoknot in Qβ RNA is essential for replication. J. Mol. Biol. 294:875-884. [DOI] [PubMed] [Google Scholar]
- 23.Knorr, D. A., R. H. Mullin, P. Q. Hearne, and T. Morris. 1991. De novo generation of defective interfering RNAs of Tomato bushy stunt virus by high multiplicity passage. Virology 181:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolchanov, N. A., I. I. Titov, I. E. Vlassova, and V. V. Vlassov. 1996. Chemical and computer probing of RNA structure. Prog. Nucleic Acid Res. Mol. Biol. 53:131-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy, P. D., J. Pogany, and A. E. Simon. 1999. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus. EMBO J. 18:5653-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy, P. D., and J. Pogany. 2000. Partial purification and characterization of Cucumber necrosis virus and Tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNA-dependent RNA polymerases. Virology 276:279-288. [DOI] [PubMed] [Google Scholar]
- 28.Oster, S. K., B. Wu, and K. A. White. 1998. Uncoupled expression of p33 and p92 permits amplification of Tomato bushy stunt virus RNAs. J. Virol. 72:5845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panavas, T., J. Pogany, and P. D. Nagy. 2002. Analysis of minimal promoter sequences for plus-strand synthesis by the Cucumber necrosis virus RNA-dependent RNA polymerase. Virology 296:263-274. [DOI] [PubMed] [Google Scholar]
- 30.Panavas, T., and P. D. Nagy. 2002. The RNA replication enhancer element of tombusviruses contains two interchangeable hairpins that are functional during plus-strand synthesis. J. Virol. 77:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qu, F., and T. J. Morris. 2002. Efficient infection of Nicotiana benthamiana by Tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing. Mol. Plant-Microbe Interact. 15:193-202. [DOI] [PubMed] [Google Scholar]
- 32.Ray, D., and K. A. White. 1999. Enhancer-like properties of an RNA element that modulates tombusvirus RNA accumulation. Virology 256:162-171. [DOI] [PubMed] [Google Scholar]
- 33.Russo, M., J. Burgyan, and G. P. Martelli. 1994. Molecular biology of Tombusviridae. Adv. Virus Res. 44:381-428. [DOI] [PubMed] [Google Scholar]
- 34.Scholthof, H. B., K. B. Scholthof, M. Kikkert, and A. O. Jackson. 1995. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology 213:425-438. [DOI] [PubMed] [Google Scholar]
- 35.Scholthof, H. B., K. B. Scholthof, and A. O. Jackson. 1995. Identification of Tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell 7:1157-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Rossum, C. M. A., C. B. E. M. Reusken, F. Brederode, and J. F. Bol. 1997. The 3′ untranslated region of alfalfa mosaic virus RNA3 contains a core promoter for minus-strand RNA synthesis and an enhancer element. J. Gen. Virol. 78:3045-3049. [DOI] [PubMed] [Google Scholar]
- 37.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White, K. A., and T. J. Morris. 1994. Enhanced competitiveness of Tomato bushy stunt virus defective interfering RNAs by segment duplication or nucleotide insertion. J. Virol. 68:6092-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White, K. A., and T. J. Morris. 1994. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J. Virol. 68:14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, B., and K. A. White. 1998. Formation and amplification of a novel tombusvirus defective RNA which lacks the 5′ nontranslated region of the viral genome. J. Virol. 72:9897-9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, B., and K. A. White. 1999. A primary determinant of cap-independent translation is located in the 3′-proximal region of the Tomato bushy stunt virus genome. J. Virol. 73:8982-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, B., W. B. Vanti, and K. A. White. 2001. An RNA domain within the 5′ untranslated region of the Tomato bushy stunt virus genome modulates viral RNA replication. J. Mol. Biol. 305:741-756. [DOI] [PubMed] [Google Scholar]
- 43.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long-range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581-15591. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, G., V. Slowinski, and K. A. White. 1999. Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA 5:550-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, H., and A. O. Jackson. 1996. Analysis of cis-acting elements required for replication of barley stripe mosaic virus RNAs. Virology 219:150-160. [DOI] [PubMed] [Google Scholar]
- 46.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands.