Abstract
Porcine cells express endogenous retroviruses, some of which are infectious for human cells. To better understand the replication of these porcine endogenous retroviruses (PERVs) in cells of different types and animal species, we have performed studies of the long terminal repeat (LTR) region of known gammaretroviral isolates of PERV. Nucleotide sequence determination of the LTRs of PERV-NIH, PERV-C, PERV-A, and PERV-B revealed that the PERV-A and PERV-B LTRs are identical, whereas the PERV-NIH and PERV-C LTRs have significant sequence differences in the U3 region between each other and with the LTRs of PERV-A and PERV-B. Sequence analysis revealed a similar organization of basal promoter elements compared with other gammaretroviruses, including the presence of enhancer-like repeat elements. The sequences of the PERV-NIH and PERV-C repeat element are similar to that of the PERV-A and PERV-B element with some differences in the organization of these repeats. The sequence of the PERV enhancer-like repeat elements differs significantly from those of other known gammaretroviral enhancers. The transcriptional activities of the PERV-A, PERV-B, and PERV-C LTRs relative to each other were similar in different cell types of different animal species as determined by transient expression assays. On the other hand, the PERV-NIH LTR was considerably weaker in these cell types. The transcriptional activity of all PERV LTRs was considerably lower in porcine ST-IOWA cells than in cell lines from other species. Deletion mutant analysis of the LTR of a PERV-NIH isolate identified regions that transactivate or repress transcription depending on the cell type.
One of the major concerns regarding the use of pig cells, tissues, or organs for xenotransplantation to humans is the activation of porcine endogenous retroviruses (PERVs) and the potential risk of zoonotic infection and disease development. Gammaretroviruses from cell lines of swine origin have been characterized previously (2, 16, 29, 46) and shown to be infectious for human cells in vitro (36, 41, 47, 51). The laboratories which initially detected the human tropism of some of these porcine retroviruses have identified three classes of infectious endogenous gammaretroviruses (PERV-A, PERV-B, and PERV-C) by nucleotide sequence analysis (25) and receptor interference studies (44). In addition to these PERV classes, a more recent isolate, PERV-NIH, was rescued from National Institutes of Health (NIH) miniature pig peripheral blood mononuclear cells stimulated with mitogens and was also shown to be infectious for human cells (51). Interestingly, PERV-NIH is like PERV-A in its surface glycoprotein envelope region and like PERV-C in the C-terminal residues of the transmembrane region (52). It was also demonstrated that primary cultures of porcine endothelial cells spontaneously release PERVs capable of infecting human cells (31). In addition to gammaretroviral sequences that are present in the genome of different breeds of pig, endogenous sequences closely related to betaretroviruses have also been detected (15, 35).
Southern blot analysis suggested that there may be up to 50 copies of gammaretroviral and up to 32 copies of betaretroviral PERV sequences in the pig genome (15, 35, 36). Although it is not known exactly how many of these loci are capable of encoding infectious PERVs, recent data suggest that this number is small, at least in Large White pigs (5, 19, 32). Of even greater relevance to pig-to-human xenografts is whether the integrated PERVs are transcriptionally expressed. Studies which have examined PERV RNA expression have detected constitutive transcription in many different organs including the kidney, heart, spleen, liver, thymus, lymph nodes, and lung (1, 5, 8, 15, 35). While it has not been determined whether these organs also produce infectious virus, there is evidence for constitutive expression in NIH miniature pigs, as infectious virus was repeatedly and readily isolated from plasma (43). It will be critical to understand tissue-specific expression for the identification and breeding of source animals that do not express replication-competent PERVs.
To better understand the regulation of PERV expression in various tissues and cells, we identified regions of the long terminal repeat (LTR) that control viral transcription. Because the regulatory signals required for retroviral transcription, replication, and integration into the host cellular DNA are located within the LTR, we sequenced and performed functional analysis of the LTRs of the infectious gammaretroviral PERVs that have been identified.
MATERIALS AND METHODS
Isolation of PERV LTRs and construction of LTR-SEAP reporter gene plasmids.
Based on sequences in the GenBank database (accession no. AF038599 and Y17012), PCR primers specific for PERV-NIH, -A, -B, and -C were designed with KpnI and XhoI sites and used to amplify the LTRs of the respective viruses. PCR products were digested with KpnI and XhoI and cloned into the pCR2.1 TOPO vector (Invitrogen, Carlsbad, Calif.). Plasmid DNAs were sequenced at the Center for Molecular Medicine and Genetics DNA sequencing facility at Wayne State University. PERV LTRs were excised from the pCR2.1 TOPO clones via the KpnI and XhoI sites and inserted into the human secreted alkaline phosphatase (SEAP) basic vector (Clontech, Palo Alto, Calif.). Plasmid DNA for each LTR-SEAP clone was sequenced for verification.
Derivation of deleted LTR-SEAP constructs.
The NIH.3c LTR-SEAP reporter gene plasmid was used as a base for all of the deleted LTRs included in this study. The NIH.3c LTR was amplified from genomic DNA of PERV-NIH-3o (PERV originally isolated from NIH miniature pig peripheral blood mononuclear cells passaged through HEK 293 cells three times) and then subcloned into the LTR-SEAP reporter plasmid as described above. Deletions of the NIH.3c LTR were derived by PCR with the use of different combinations of primers and then subcloned into the pSEAP2 plasmid (Clontech). All deletions were confirmed by nucleotide sequence analysis.
Transcription initiation site mapping.
Total RNA was extracted from cell lines expressing PERV-A, -B, -C, and -NIH. Transcription initiation sites were mapped by using 5′ rapid amplification of cDNA ends (Life Technologies, Rockville, Md.). Amplified DNA products were agarose gel purified, cloned into the pCR2.1 TOPO vector, and subjected to DNA sequencing.
Cells used for transfections.
Human embryonic kidney cells (HEK 293 cells [ATCC CRL 1573]), human cervical epithelial cells (HeLa cells [ATCC CCL-2]), mink lung fibroblasts (ATCC CCL-64), Mus dunni tail fibroblasts (MDTF; obtained from Leonard Evans, Rocky Mountain Laboratories, Hamilton, Mont.), and swine testis cells (ST-IOWA; obtained from Richard Fister, Tufts University, Boston, Mass.) were maintained in Dulbecco's modified Eagle's medium (Gibco, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (HyClone, Logan, Utah), 2 mM glutamine, 1 mM sodium pyruvate, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. All cells were grown in 5% CO2 at 37°C.
LTR-SEAP transient expression assays.
Target cells for transfection of LTR-SEAP plasmids were seeded in 12-well dishes 1 day prior to transfections. The approximate numbers of cells per well were 8 × 104 to 10 × 104 for HEK 293, 2 × 105 for ST-IOWA, and 3 × 104 to 4 × 104 for other cell types. The next day, each well of cells was cotransfected with 0.4 μg of SEAP plasmid and 0.3 μg of cytomegalovirus-β-galactosidase plasmid (a gift of Edward Max's laboratory, Center for Biologics Evaluation and Research, Bethesda, Md.) by using the Effectene reagent kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. All constructs were transfected in duplicate or triplicate wells and assayed by at least two independent transfection experiments. Cells were incubated for an additional 72 h posttransfection, at which time the supernatant and cell lysates were collected for measurement of SEAP activity and β-galactosidase activity, respectively. P values were calculated with the Student two-tailed paired t test with the software algorithm with Microsoft Excel. P values of <0.05 were considered to represent statistically significant differences.
The SEAP activity in 90 μl of cell-free supernatant was assayed by using the reagents provided in the Great EscAPe SEAP kit (Clontech) according to the manufacturer's instructions for the chemiluminescence assay. The signal was detected with a tube luminometer (TD-20/20; Turner Design Luminometer, Sunnyvale, Calif.). The assay results for SEAP activity were normalized to the values obtained for β-galactosidase activity in order to control for differences in transfection efficiencies.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the LTR sequences of PERV-A, -B, -C, -NIH.3c, and -NIH are AF546883, AF546884, AF546885, AF546886, and AF546887, respectively.
RESULTS
Sequence comparison of PERV LTRs.
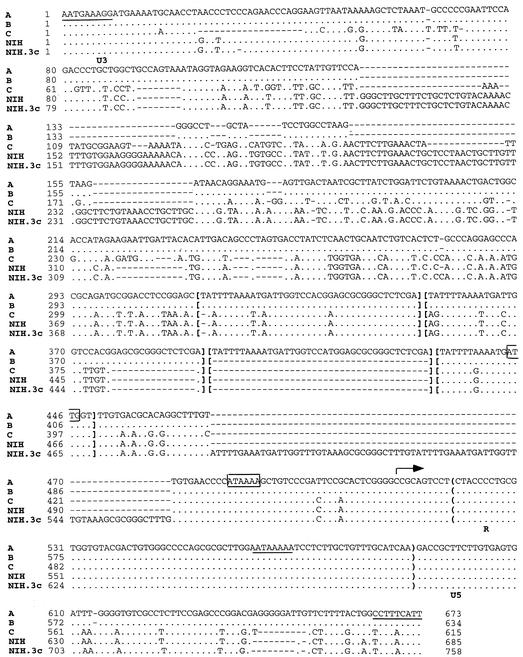
We obtained the complete nucleotide sequence of the LTRs of the known types of gammaretroviral PERVs by PCR amplification of the LTRs of either proviruses of the infected cells (PERV-NIH, -A, and -B) or a recombinant genomic clone (PERV-C) (Fig. 1). Two different PERV-NIH LTRs (PERV-NIH and PERV-NIH.3c) were obtained from virus-infected cell lines that differed in their passage number. Infectious PERV-NIH was isolated from NIH miniature pigs on HEK 293 cells (52). 293 cell lines expressing either PERV-A or PERV-B were kindly provided by Y. Takeuchi (University College London, London, United Kingdom) and C. Patience (Immerge Therapeutics, Boston, Mass.) (44). A molecular clone of PERV-C cDNA was a generous gift of J. Stoye (National Institute for Medical Research, Medical Research Council, London, United Kingdom). PERV-C has also been referred to as Shimozuma (42) and Tsukuba-1 (1).
FIG. 1.
Sequence alignment of PERV-A, -B, -C, -NIH, and -NIH.3c LTRs. U3, R, and U5 are delineated by bold letters at the start of each region. The R region is further identified with parentheses. Tandem repeated sequences are indicated by brackets. IR sequences are underlined at the 5′ and 3′ ends of the LTRs. A nonconsensus TATA sequence and reverse CAAT sequence are boxed, and a polyadenylation signal in the R region is underlined. The transcription initiation site is indicated with an arrow. The PERV-A LTR is used as the reference sequence, and nucleotides identical to PERV-A in PERV-B, -C, -NIH, and -NIH.3c are shown as dots. Nucleotide differences are identified by letters, and dashes show missing bases.
The sequences of the LTRs of PERV-A and PERV-B are identical except for an extra 37-bp repeat element in PERV-A (described in more detail below). The sequences of these LTRs are nearly identical to those reported by Krach et al. (23), with only a 1% difference. The PERV-A and -B LTRs share 64 and 56% nucleotide sequence identity with the PERV-C and PERV-NIH LTRs, respectively. Sequence differences are located primarily in the U3 region. The nucleotide sequence of the PERV-C and PERV-NIH LTRs are 78% identical while their enhancer-like repeat sequences are completely identical. PERV-NIH.3c is identical to PERV-NIH except for two additional copies of the repeat sequence.
A common feature among the PERV LTRs is the overall length, between 600 and 800 bp (Fig. 1), which corroborates the size for the PERV-A and PERV-B LTRs reported previously (23). The organization of the PERV LTRs is similar to that of other retroviruses with well-defined U3, R, and U5 regions. A 9-bp inverted repeat (IR) sequence at the 5′ end of the LTR is identical for all four PERV LTRs. A perfect IR is present at the 3′ end of only the PERV-A and -B LTRs. In comparison, the 3′ end IR sequence of PERV-NIH and PERV-C differ by two nucleotide substitutions from the 5′ end IR. Similar to other gammaretroviruses is the presence of directly repeated sequences in the U3 region which resemble an enhancer element, a nonconsensus TATA box, and an AATAAA polyadenylation signal in the R region. The initiation site for transcription, which was mapped by using the rapid amplification of cDNA ends technique is located at nucleotides (nt) 508, 468, 361, and 530 for the PERV-A, -B, -C, and -NIH LTRs, respectively. The 5′-ATAAAA-3′ sequence, which may be a potential TATA box, was located 32 nt upstream of the identified transcription start sites. We also identified a CAAT box in the reverse orientation 5′-ATTG-3′, which is located 66 nt upstream of the start site of transcription. The U5 region for all of the PERV LTRs examined is between 73 to 77 bp long. Both PERV-NIH and PERV-C have binding sites for tRNApro, (5′-TGGGGGCTCGTCCGGGAT-3′), whereas PERV-A and -B have a tRNAgly primer binding site (5′-TGGTGCTTTGGCCGGGGA-3′) (data not shown).
PERV LTR repeated sequences.
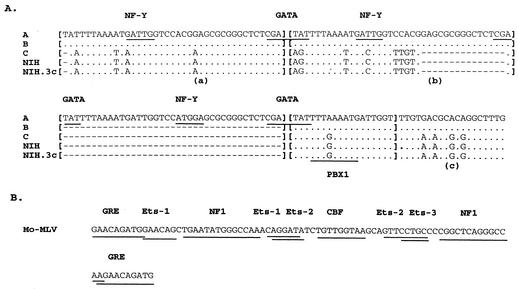
The directly repeated sequence in the PERV LTRs consists of 39 bp. This element is present 3.5 times in the PERV-A LTR and 2.5 times in the PERV-B LTR (Fig. 1 and 2A). A similar sequence with a 5-bp difference is repeated in a similar position 1.5 times in the PERV-C, PERV-NIH, and PERV-NIH.3c LTRs. However, between the 1.5 repeats in the PERV-NIH and PERV-C LTRs, there is a 24-bp sequence that resembles a truncated repeat with an 8-bp difference. Also, the PERV-NIH.3c LTR has two additional copies of a 37-bp repeat compared to a single copy of this sequence in the PERV-NIH LTR. The distance of the PERV repeated sequences from the putative promoter sequences we identified (30 nt) is much smaller than that in other gammaretroviruses (17), which have a region of substantial size (approximately 200 bp) and contain sequences important for both transcriptional regulation and the pathogenicity of some murine leukemia viruses (MLVs) (18, 37, 49, 55).
FIG. 2.
(A) Sequence alignment and identification of transcription factor-binding sites of the repeat elements of PERV-A, -B, -C, -NIH, and -NIH.3c. Each repeat element is indicated in brackets. Dashes indicate missing nucleotides. Identical nucleotides are shown as dots, and differences are indicated by letters. (a), 39-bp repeat element; (b), truncated 39-bp repeat in PERV-C, PERV-NIH, and PERV-NIH.3c; (c), 37-bp sequence repeated three times in PERV-NIH.3c. Binding sites for known transcription factors are underlined and identified by name above the sequence motif, except for PBX1 for PERV-C, -NIH, and -NIH.3c, which is identified below the sequence. Identification of the transcription factor-binding sites was made by using the TRANSFAC database. (B) Enhancer sequence and transcription factor-binding sites for Moloney MLV (Mo-MLV), which represents transcription factor-binding sites for a majority of murine leukemia retroviral enhancers. Double lines indicate overlapping binding sites.
Enhancer elements contain binding sites for a number of nuclear transcription factors (40). It has been demonstrated that certain protein binding sites within the enhancer elements of gammaretroviruses regulate cell type-specific transcription, which in turn is a determinant of tissue tropism (45, 49, 54) and disease specificity (4, 21, 30, 33, 40). Our search of a transcription factor database (TRANSFAC) identified sites for various DNA-binding factors in the repeated sequences we identified in the PERV LTRs (Fig. 2A). Two protein binding sites common to all four PERV repeats are NF-Y and GATA. The Pbx1 binding site is present only in the PERV-C, PERV-NIH, and PERV-NIH.3c repeats. These sites are not present in the enhancers of other gammaretroviruses which show a striking conservation of other transcription factor-binding sites (17) (Fig. 2B). For comparison, Fig. 2B shows the transcription factor-binding sites for a typical mammalian gammaretrovirus enhancer, Moloney MLV. The enhancer repeats in the PERV LTRs showed no transcription factor-binding sites common to the enhancer regions of other gammaretroviruses, and they appear to bind a smaller number of factors.
Expression analysis of PERV LTRs isolated from different PERV strains.
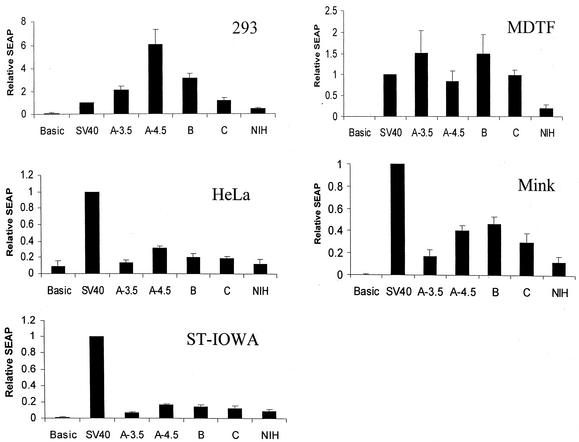
In order to functionally assess the importance of the observed nucleotide differences in the LTR sequences derived from different strains of PERV, each LTR was subcloned into an expression construct, which contained the SEAP gene, as described in Materials and Methods. We also compared an LTR containing 4.5 copies of the PERV-A repeated element (A-4.5), which was isolated from virus-infected HEK 293 cells. The relative transcriptional activity of each LTR was determined in each of five different cell lines: two human cell lines, HEK 293 and HeLa, and three nonhuman cell lines, a porcine fetal swine testis (ST-IOWA), murine MDTF, and mink epithelial (CCL-64). The results were normalized to a positive control containing the simian virus 40 (SV40) early promoter and enhancer and are shown in Fig. 3. No statistical differences were observed in transcriptional activity between any of the LTRs in the ST-IOWA, HeLa, MDTF, or mink cell lines. However, the results with HEK 293 cells were strikingly different from those with the other cell lines. The A-4.5 LTR resulted in an approximately threefold-higher level of SEAP expression than the A-3.5 LTR (P value of 0.03). Likewise, the activity observed for A-4.5 was significantly higher than those for all of the other LTRs examined in HEK 293 cells (all P values were <0.003). The PERV-NIH LTR was consistently the least active in all cell lines tested. Of note, the transcriptional activity of all LTRs relative to the SV40 promoter was significantly lower in the porcine cell line than in cells from other species. Relative SEAP values for the PERV-NIH.3c clone were consistently higher than those observed for the PERV-NIH LTR in HEK 293 cells (data not shown). In order to investigate whether the extra copies of the 37-bp sequence were responsible for the higher SEAP activity, the PERV-NIH.3c LTR was chosen for subsequent analysis.
FIG. 3.
Expression analysis of LTR-SEAP plasmids in different cell lines. Reporter constructs encoding the SEAP cDNAs downstream from the LTRs of different PERV strains were transfected into four different cell lines. A-3.5 and A-4.5 are LTRs derived from the PERV-A class and differ by the number of 39-bp repeats (A-3.5 has 3.5 repeats, and has the sequence represented in the alignment shown in Fig. 1; A-4.5 is identical to that sequence, with the exception of having 4.5 repeats). B and C are LTRs from the PERV-B and PERV-C classes, respectively. NIH is the LTR from the PERV-NIH strain. SV40 represents a positive control plasmid with the SV40 promoter-enhancer element upstream of the SEAP cDNA, and basic represents the negative control plasmid with no promoter-enhancer element upstream of the SEAP cDNA. Results shown are the means plus the standard errors of the mean for each of six separate transfection experiments performed with HEK 293 cells; three experiments with MDTF, mink, and ST-IOWA cells; and two experiments performed with HeLa cells.
Identification of LTR transcriptional regulatory sequences.
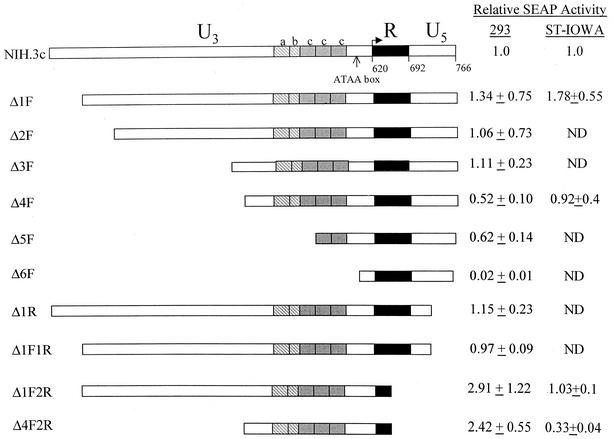
To identify PERV LTR sequences that regulate transcription, we performed a deletion analysis of the PERV-NIH.3c LTR. Deletions of different sizes were made from both the 5′ (designated F for forward) and 3′ ends (designated R for reverse) of this LTR (Fig. 4). Deleted LTRs were cloned into the SEAP expression plasmid and tested in transient expression assays as described above. Deletion of nt 1 through 297 (Δ1F through Δ3F) had no effect on the NIH.3c LTR activity in HEK 293 cells. However, when an additional 71 bp were deleted (Fig. 4, compare Δ3F and Δ4F), we detected a statistically significant twofold decrease in transcriptional activity (P value of 0.008), suggesting the presence of regulatory sequences in this region of the LTR that contribute to the activation of transcription. Removal of the a and b repeat sequences and one copy of the c repeat sequence in Δ5F did not produce a further decrease in transcriptional activity. The 5′ deletion that removed the remaining c repeat sequences and the ATAA box (Δ6F) resulted in a 50-fold decrease in activity, indicating that these sequences are essential for transcriptional activation. A 3′ deletion of 51 bp in the U5 region (Δ1R and Δ1F1R) had no effect on transcriptional activity. However, deletion of the remaining U5 sequences and 47 bp of the R region (Δ1F2R and Δ4F2R) resulted in a significant increase in NIH.3c LTR activity (P < 0.03 for Δ1F2R compared with Δ1F and P < 0.001 for Δ4F2R compared with Δ4F). This result indicated the presence of a negative regulatory element or elements in these regions of the PERV-NIH.3c LTR.
FIG. 4.
Schematic depiction of PERV-NIH LTR deletions and relative SEAP activity in HEK 293 and ST-IOWA cells. Reporter constructs carrying various portions of the NIH.3c LTR upstream of the SEAP cDNA were derived as described in Materials and Methods and are schematically depicted. The boxes with gray hatch marks or solid gray represent the repeat elements found in U3. The a box indicates the conserved 39-bp repeat, the b box indicates the 24-bp truncated repeat, and the c box indicates the 37 bp repeated three times in PERV-NIH.3c (shown Fig. 2A). The black box indicates the R region. Each reporter construct was transfected into 293 or ST-IOWA cells in at least two independent experiments containing three replicates each. Shown on the right side are the average SEAP activities for each deletion mutant ± the standard errors normalized to those observed for the NIH.3c parental LTR. ND, not done.
To test whether the 3′ deletion in the 2R clones would have the same effect in porcine cells, we tested the Δ1F, Δ1F2R, Δ4F, and Δ4F2R constructs in ST-IOWA cells. A comparison of Δ1F with Δ4F produced similar results to those seen in HEK 293 cells, which also indicated the presence of transactivating sequences in the U3 region upstream of the repeat elements that are active in ST-IOWA cells. In contrast, the effect of the 3′ deletion in ST-IOWA cells differed from that observed in HEK 293 cells; Δ1F2R had the same activity as Δ1F while Δ4F2R resulted in an approximately threefold decrease in activity relative to Δ4F. These data suggest that the presence of sequences in the R and U5 regions that upregulate rather than repress transcription. However, results from the Δ1F2R mutant suggest that additional upstream sequences in the U3 region can compensate for the loss of the Δ2R sequences. However, the presence of both regions did not have an additive effect on transcription since the SEAP activity of mutant Δ1F, which contains both the upstream and downstream sequences, was equivalent to that of either Δ4F or Δ1F2R.
DISCUSSION
By nucleotide sequence analysis of the LTRs, we have identified the basal promoter elements of transcription for the different classes of gammaretroviral PERVs that have been isolated. The PERV-C and PERV-NIH LTRs differed from those of PERV-A and PERV-B and from each other mainly in the U3 region upstream of the repeated sequences and in the U5 region (Fig. 1). In contrast, the sequence of the PERV-C LTR that we isolated differs from that reported by Scheef et al. (39) by 47%, perhaps reflecting the fact that we used a cDNA clone rather than genomic DNA as a source of the PERV-C LTR. Presumably, the sequence divergence of these PERV-C LTRs may account for the observed transcriptional activity of our isolate in contrast to the lack of detectable transcriptional activity of the previously reported one (39). Our PERV-C LTR had 84% sequence identity with that of the miniature swine lymphocyte PERV (1), with the majority of the nucleotide differences located in the U5 region. The PERV-NIH.3c LTR, which was derived from a later-passage virus than that used to isolate the PERV-NIH LTR, contains three copies of the c region of PERV-NIH. Whether these additional copies of the c region contribute to the ability of this later-passage virus to replicate at higher levels in HEK 293 cells remains to be determined. However, our deletion analysis in HEK 293 cells indicated that two copies of the c region were sufficient to confer as much transcriptional activity as all of the repeat elements together (Fig. 4, compare Δ4F with Δ5F). Additional copies of repeated sequences have been detected in the LTRs of other gammaretroviruses, which have led to an increase in replication of their viruses in target cells and an enhancement in virus-induced pathogenicity (3, 20).
The enhancer elements of murine gammaretroviruses are usually present as directly repeated sequences of 50 to 120 nt in length (24, 26). Extensive work has demonstrated that retroviral enhancers are important determinants of not only viral replication but also disease specificity and the latency of disease induction (7, 10-13, 17, 22, 24, 27, 28, 50). It has been reported that the PERV-B repeated sequences function as an enhancer element by their ability to potentiate transcription in an orientation-independent manner (39). The direct repeats present in the PERV LTRs differ from the highly conserved sequences present in other gammaretroviral enhancers, including those for the gibbon ape leukemia virus (17), which is closely related by sequence to the PERVs (1, 25). This is the first identification of a mammalian gammaretrovirus that differs in this respect from other known gammaretroviruses. Thus, we predict that the transcription factors that may bind these PERV enhancer elements will differ from those which control the transcriptional activation of other gammaretroviruses (Fig. 2). In transient expression assays, we observed that an increase in the copy number of repeated sequences from 2.5 (PERV-B) to 4.5 (PERV-A-4.5) did not result in a concomitant increase in transcriptional activation in the cell lines we examined, except for HEK 293 cells. This result is consistent with the observation by Scheef et al. (39) that there is a threshold effect for the number of repeats that can increase transcriptional activation in only certain cell types. The statistically significant increase in activity in the LTR-driven expression of the PERV-A-4.5 LTR over the PERV-A-3.5 LTR observed only in HEK 293 cells suggests that the transcription factors necessary to interact with elements found within the repeat sequences are present in HEK 293 cells but may not be present or as active in the other cell lines tested.
We observed that the PERV-A, -B, and -C LTRs had similar transcriptional activities in cells derived from different species and representing different tissue types. Although these results suggest that PERV transcription levels may not vary in different cell types from different species once virus infection has occurred, our deletion mutant analysis, which is discussed further below, indicates that different regions of the LTR may be responsible for the regulation of transcription depending on the cell type. In comparison, the transcriptional activity of the PERV-NIH LTR was consistently lower than that of the other PERV LTRs in all of the cell lines examined. Because the direct repeats of the PERV-NIH and PERV-C are identical, these data support the idea that regulatory sequences outside of this region are responsible for the decreased transcriptional activity detectable for the PERV-NIH LTR. We predict that the region most likely to be responsible for this difference in transcription is the U3 region upstream of the direct repeats, which contains practically all of the nucleotide differences between PERV-NIH and PERV-C (Fig. 1). By a transcription factor-binding site search of the TRANSFAC database, we identified potential protein binding sites that are present in this region of the PERV-C but not in the PERV-NIH LTR, and they may be responsible for this difference. These binding sites include those for the transcription factors SOX5, Ets-1, Evi1, GATA, v-Myb, and CEBP.
It is noteworthy that the transcriptional activity of all of the PERV LTRs tested relative to the SV40 early promoter had significantly lower activity in porcine cells than in cells from other animal species. Perhaps this reflects evolutionary pressure to select for LTRs with reduced transcriptional activity once a retrovirus becomes an endogenous gene (9). Alternatively, it may be that the transcriptional activity observed in ST-IOWA cells may not be representative of primary porcine cells.
A comparison of the deletion mutants in HEK 293 and porcine ST-IOWA cells demonstrated that different regions of the LTR can either activate or repress transcription depending on the cell type. Whether this difference between the HEK 293 and ST-IOWA cells is dependent on species- or cell type-specificity or both requires further analysis. Sequences have been identified for other retroviruses in the R and U5 regions that either activate or repress transcription (6, 14, 34, 38, 48, 53).
A comparison of the Δ3F and Δ4F deletion mutants in HEK 293 cells demonstrated the presence of sequences in a 68-bp region upstream of the repeated elements that contribute to transcriptional activation. A TRANSFAC analysis of this region revealed potential protein binding sites for the REBP, AML1, AP1, Sp1, and MZF1 transcription factors, some of which may participate in the transactivation that we observed. Our data also demonstrate that the repeated sequences and ATAA nonconsensus TATA box are essential for transcriptional activation in HEK 293 cells, since deletion of these elements reduced activity by 50-fold.
Our nucleotide sequence analysis of the LTRs of different types of PERV has revealed a structural organization of basal promoter elements that is shared with other gammaretroviruses. In contrast, the repeat elements and putative transcription factor-binding sites that are conserved among the PERV LTRs appear to be unique to these LTRs among other gammaretroviruses. In addition, functional studies of LTR deletion mutants have identified regions with the ability to transactivate or repress transcription, including those that can do both depending on the cell type. This type of information will facilitate the identification of LTR regions that are essential for PERV replication in cells of different types and animal species. Further studies elucidating the cellular factors required for transcriptional activation of PERVs will be critical for the evaluation of the zoonotic risks that may be involved in the use of porcine tissues and organs in human xenotransplantation.
Acknowledgments
C.A.W. and S.L. made equal contributions to this work.
We are grateful to C. Patience, J. Stoye, and Y. Takeuchi for continuous generosity in providing PERV-infected cell lines and plasmid clones without which this study would not have been possible. We also thank Oliver Zill for technical assistance. Finally, we thank Andy Dayton and Xu Lai for critical readings of the manuscript.
REFERENCES
- 1.Akiyoshi, D. E., M. Denaro, H. Zhu, J. L. Greenstein, P. Banerjee, and J. A. Fishman. 1998. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J. Virol. 72:4503-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., J. S. Porterfield, and A. T. De Madrid. 1971. C-type virus particles in pig kidney cell lines. J. Gen. Virol. 10:195-198. [DOI] [PubMed] [Google Scholar]
- 3.Belli, B., A. Patel, and H. Fan. 1995. Recombinant mink cell focus-inducing virus and long terminal repeat alterations accompany the increased leukemogenicity of the Mo+PyF101 variant of Moloney murine leukemia virus after intraperitoneal inoculation. J. Virol. 69:1037-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boral, S. L., A. Okenquist, and J. Lenz. 1989. Identification of the SL3-3 virus enhancer core as a T-lymphoma cell-specific element. J. Virol. 63:76-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch, S., C. Arnauld, and A. Jestin. 2000. Study of full-length porcine endogenous retrovirus genomes with envelope gene polymorphism in a specific-pathogen-free Large White swine herd. J. Virol. 74:8575-8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butsch, M., S. Hull, Y. Wang, T. M. Roberts, and K. Boris-Lawrie. 1999. The 5′ RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent Gag production. J. Virol. 73:4847-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatis, P. A., C. A. Holland, J. W. Hartley, W. P. Rowe, and N. Hopkins. 1983. Role for the 3′ end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc. Natl. Acad. Sci. USA 80:4408-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemenceau, B., S. Lalain, L. Martignat, and P. Sai. 1999. Porcine endogenous retroviral mRNAs in pancreas and a panel of tissues from specific pathogen-free pigs. Diabetes Metab. 25:518-525. [PubMed] [Google Scholar]
- 9.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 10.DesGroseillers, L., and P. Jolicoeur. 1984. Mapping the viral sequences conferring leukemogenicity and disease specificity in Moloney and amphotropic murine leukemia viruses. J. Virol. 52:448-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DesGroseillers, L., and P. Jolicoeur. 1984. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J. Virol. 52:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DesGroseillers, L., E. Rassart, and P. Jolicoeur. 1983. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc. Natl. Acad. Sci. USA 80:4203-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DesGroseillers, L., R. Villemur, and P. Jolicoeur. 1983. The high leukemogenic potential of Gross passage A murine leukemia virus maps in the region of the genome corresponding to the long terminal repeat and to the 3′ end of env. J. Virol. 47:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domansky, A. N., E. P. Kopantzev, E. V. Snezhkov, Y. B. Lebedev, C. Leib-Mosch, and E. D. Sverdlov. 2000. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 472:191-195. [DOI] [PubMed] [Google Scholar]
- 15.Ericsson, T., B. Oldmixon, J. Blomberg, M. Rosa, C. Patience, and G. Andersson. 2001. Identification of novel porcine endogenous betaretrovirus sequences in miniature swine. J. Virol. 75:2765-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fishman, J. A. 1998. The risk of infection in xenotransplantation. Introduction. Ann. N. Y. Acad. Sci. 862:45-51. [DOI] [PubMed] [Google Scholar]
- 17.Golemis, E. A., N. A. Speck, and N. Hopkins. 1990. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J. Virol. 64:534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanecak, R., P. K. Pattengale, and H. Fan. 1991. Deletion of a GC-rich region flanking the enhancer element within the long terminal repeat sequence alters the disease specificity of Moloney murine leukemia virus. J. Virol. 65:5357-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herring, C., G. Quinn, R. Bower, N. Parsons, N. A. Logan, A. Brawley, K. Elsome, A. Whittam, X. M. Fernandez-Suarez, D. Cunningham, D. Onions, G. Langford, and L. Scobie. 2001. Mapping full-length porcine endogenous retroviruses in a large white pig. J. Virol. 75:12252-12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland, C. A., C. Y. Thomas, S. K. Chattopadhyay, C. Koehne, and P. V. O'Donnell. 1989. Influence of enhancer sequences on thymotropism and leukemogenicity of mink cell focus-forming viruses. J. Virol. 63:1284-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollon, T., and F. K. Yoshimura. 1989. Mapping of functional regions of murine retrovirus long terminal repeat enhancers: enhancer domains interact and are not independent in their contributions to enhancer activity. J. Virol. 63:3353-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishimoto, A., M. Takimoto, A. Adachi, M. Kakuyama, S. Kato, K. Kakimi, K. Fukuoka, T. Ogiu, and M. Matsuyama. 1987. Sequences responsible for erythroid and lymphoid leukemia in the long terminal repeats of Friend-mink cell focus-forming and Moloney murine leukemia viruses. J. Virol. 61:1861-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krach, U., N. Fischer, F. Czauderna, and R. R. Tonjes. 2001. Comparison of replication-competent molecular clones of porcine endogenous retrovirus class A and class B derived from pig and human cells. J. Virol. 75:5465-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laimins, L. A., P. Gruss, R. Pozzatti, and G. Khoury. 1984. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J. Virol. 49:183-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Tissier, P., J. P. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss. 1997. Two sets of human-tropic pig retrovirus. Nature 389:681-682. [DOI] [PubMed] [Google Scholar]
- 26.Levinson, B., G. Khoury, G. V. Woude, and P. Gruss. 1982. Activation of SV40 genome by 72-base pair tandem repeats of Moloney sarcoma virus. Nature 295:568-572. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, A. F., T. Stacy, W. R. Green, L. Taddesse-Heath, J. W. Hartley, and N. A. Speck. 1999. Core-binding factor influences the disease specificity of Moloney murine leukemia virus. J. Virol. 73:5535-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y., E. Golemis, J. W. Hartley, and N. Hopkins. 1987. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J. Virol. 61:693-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieber, M. M., C. J. Sherr, G. J. Todaro, R. E. Benveniste, R. Callahan, and H. G. Coon. 1975. Isolation from the Asian mouse Mus caroli of an endogenous type C virus related to infectious primate type C viruses. Proc. Natl. Acad. Sci. USA 72:2315-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LoSardo, J., A. L. Boral, and J. Lenz. 1990. Relative importance of elements within the SL3-3 virus enhancer for T-cell specificity. J. Virol. 64:1756-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, U., V. Kiessig, J. H. Blusch, A. Haverich, K. von der Helm, T. Herden, and G. Steinhoff. 1998. Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet 352:692-694. [DOI] [PubMed] [Google Scholar]
- 32.Niebert, M., C. Rogel-Gaillard, P. Chardon, and R. R. Tonjes. 2002. Characterization of chromosomally assigned replication-competent gamma porcine endogenous retroviruses derived from a large white pig and expression in human cells. J. Virol. 76:2714-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsson, P., B. Hallberg, A. Thornell, and T. Grundstrom. 1989. Mutant analysis of protein interactions with a nuclear factor I binding site in the SL3-3 virus enhancer. Nucleic Acids Res. 17:4061-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okumura, K., G. Sakaguchi, S. Takagi, K. Naito, T. Mimori, and H. Igarashi. 1996. Sp1 family proteins recognize the U5 repressive element of the long terminal repeat of human T cell leukemia virus type I through binding to the CACCC core motif. J. Biol. Chem. 271:12944-12950. [DOI] [PubMed] [Google Scholar]
- 35.Patience, C., W. M. Switzer, Y. Takeuchi, D. J. Griffiths, M. E. Goward, W. Heneine, J. P. Stoye, and R. A. Weiss. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 75:2771-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nature Med. 3:282-286. [DOI] [PubMed] [Google Scholar]
- 37.Reuss, F. U., B. Berdel, M. Ploss, and R. Heber. 2001. Replication of enhancer-deficient amphotropic murine leukemia virus in human cells. Proc. Natl. Acad. Sci. USA 98:10898-10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell, R. A., Y. Zeng, O. Erlwein, B. R. Cullen, and M. O. McClure. 2001. The R region found in the human foamy virus long terminal repeat is critical for both Gag and Pol protein expression. J. Virol. 75:6817-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheef, G., N. Fischer, U. Krach, and R. R. Tonjes. 2001. The number of a U3 repeat box acting as an enhancer in long terminal repeats of polytropic replication-competent porcine endogenous retroviruses dynamically fluctuates during serial virus passages in human cells. J. Virol. 75:6933-6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speck, N. A., and D. Baltimore. 1987. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol. Cell. Biol. 7:1101-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Specke, V., S. Rubant, and J. Denner. 2001. Productive infection of human primary cells and cell lines with porcine endogenous retroviruses. Virology 285:177-180. [DOI] [PubMed] [Google Scholar]
- 42.Suzuka, I., N. Shimizu, K. Sekiguchi, H. Hoshino, M. Kodama, and K. Shimotohno. 1986. Molecular cloning of unintegrated closed circular DNA of porcine retrovirus. FEBS Lett. 198:339-343. [DOI] [PubMed] [Google Scholar]
- 43.Takefman, D. M., S. Wong, T. Maudru, K. Peden, and C. A. Wilson. 2001. Detection and characterization of porcine endogenous retrovirus in porcine plasma and porcine factor VIII. J. Virol. 75:4551-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. Le Tissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thornell, A., B. Hallberg, and T. Grundstrom. 1988. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol. Cell. Biol. 8:1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Todaro, G. J., R. E. Benveniste, M. M. Lieber, and C. J. Sherr. 1974. Characterization of a type C virus released from the porcine cell line PK(15). Virology 58:65-74. [DOI] [PubMed] [Google Scholar]
- 47.Tonjes, R. R., F. Czauderna, N. Fischer, U. Krach, K. Boller, P. Chardon, C. Rogel-Gaillard, M. Niebert, G. Scheef, A. Werner, and R. Kurth. 2000. Molecularly cloned porcine endogenous retroviruses replicate on human cells. Transplant. Proc. 32:1158-1161. [DOI] [PubMed] [Google Scholar]
- 48.Trubetskoy, A. M., S. A. Okenquist, and J. Lenz. 1999. R region sequences in the long terminal repeat of a murine retrovirus specifically increase expression of unspliced RNAs. J. Virol. 73:3477-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tupper, J. C., H. Chen, E. F. Hays, G. C. Bristol, and F. K. Yoshimura. 1992. Contributions to transcriptional activity and to viral leukemogenicity made by sequences within and downstream of the MCF13 murine leukemia virus enhancer. J. Virol. 66:7080-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogt, M., C. Haggblom, S. Swift, and M. Haas. 1985. Envelope gene and long terminal repeat determine the different biological properties of Rauscher, Friend, and Moloney mink cell focus-inducing viruses. J. Virol. 55:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, C. A., S. Wong, J. Muller, C. E. Davidson, T. M. Rose, and P. Burd. 1998. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J. Virol. 72:3082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, C. A., S. Wong, M. VanBrocklin, and M. J. Federspiel. 2000. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J. Virol. 74:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, P., M. Zemba, M. Aboud, R. M. Flugel, and M. Lochelt. 1997. Deletion analysis of both the long terminal repeat and the internal promoters of the human foamy virus. Virus Genes 15:17-23. [DOI] [PubMed] [Google Scholar]
- 54.Yoshimura, F. K., J. Tupper, and K. Diem. 1989. Differential DNA binding of nuclear proteins to a long terminal repeat region of the MCF13 and AKV murine leukemia viruses. J. Virol. 63:4945-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshimura, F. K., T. Wang, and M. Cankovic. 1999. Sequences between the enhancer and promoter in the long terminal repeat affect murine leukemia virus pathogenicity and replication in the thymus. J. Virol. 73:4890-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]