Abstract
Many transcription coactivators interact with nuclear receptors in a ligand- and C-terminal transactivation function (AF2)-dependent manner. These include activating signal cointegrator 2 (ASC-2), a recently isolated transcriptional coactivator molecule, which is amplified in human cancers and stimulates transactivation by nuclear receptors and numerous other transcription factors. In this report, we show that ASC-2 belongs to a steady-state complex of approximately 2 MDa (ASC-2 complex [ASCOM]) in HeLa nuclei. ASCOM contains retinoblastoma-binding protein RBQ-3, α/β-tubulins, and trithorax group proteins ALR-1, ALR-2, HALR, and ASH2. In particular, ALR-1/2 and HALR contain a highly conserved 130- to 140-amino-acid motif termed the SET domain, which was recently implicated in histone H3 lysine-specific methylation activities. Indeed, recombinant ALR-1, HALR, and immunopurified ASCOM exhibit very weak but specific H3-lysine 4 methylation activities in vitro, and transactivation by retinoic acid receptor appears to involve ligand-dependent recruitment of ASCOM and subsequent transient H3-lysine 4 methylation of the promoter region in vivo. Thus, ASCOM may represent a distinct coactivator complex of nuclear receptors. Further characterization of ASCOM will lead to a better understanding of how nuclear receptors and other transcription factors mediate transcriptional activation.
The nuclear receptor superfamily is a group of proteins that regulate, in a ligand-dependent manner, transcriptional initiation of target genes by binding to specific DNA sequences named hormone response elements (reviewed in reference 23). Functional analysis of nuclear receptors has shown that there are two major activation domains. The N-terminal domain (AF1) contains a ligand-independent activation function, whereas the ligand-binding domain (LBD) exhibits ligand-dependent transactivation function (AF2). The AF2 core region, located at the extreme C terminus of the receptor LBDs, is conserved among nuclear receptors and undergoes a major conformational change upon ligand binding (23). This region has been shown to play a critical role in mediating transactivation by serving as a ligand-dependent interaction interface with many different coactivators (reviewed in reference 9). These coactivators, including the p160 family members (i.e., SRC-1, SRC-2/GRIP1/TIF2, and SRC-3/ACTR/pCIP/AIB1/RAC3/TRAM1), CBP/p300, p/CAF, TRAP/DRIP, activating signal cointegrator 2 (ASC-2), and many others, bridge nuclear receptors and the basal transcription apparatus and/or remodel the chromatin structures (9).
Chromatin, the physiological template of all eukaryotic genetic information, undergoes a diverse array of posttranslational modifications that largely impinge on histone amino termini, thereby regulating access to the underlying DNA (reviewed in reference 12). SRC-1 and the p160 family member ACTR, along with CBP and p300, were recently shown to contain histone acetyltransferase (HAT) activities and associate with yet another HAT protein, p/CAF (9). In contrast, SMRT and N-CoR, nuclear receptor corepressors, form complexes with Sin3 and histone deacetylase proteins (9). These results are consistent with the notion that the acetylation of histones destabilizes nucleosomes and relieves transcriptional repression by allowing transcription factors to access recognition elements, whereas deacetylation of the histones stabilizes the repressed state. More recently, the histone arginine methyltransferases CARM1 and PRMT1 were newly defined as transcriptional coactivators of nuclear receptors (4, 40). NSD1 and RIZ1, two additional coregulatory proteins with the SET domain known to methylate histones (6, 16, 26, 28, 33, 35, 42, 46), were also reported (10, 50). Likewise, one can expect to identify additional coactivator molecules with other histone-modifying activities such as lysine methylation, ubiquitination, and phosphorylation. These distinct histone amino-terminal modifications can generate synergistic or antagonistic interaction affinities for chromatin-associated proteins in a combinatorial manner, which in turn dictates dynamic transitions between transcriptionally active or transcriptionally silent chromatin states (12).
A distinctive structural feature of the AF2-dependent coactivators is the presence of LXXLL signature motifs (i.e., nuclear receptor [NR] box) (9). The AF2 core region (helix 12), upon undergoing a major restructuring upon ligand binding, forms part of a charged clamp that accommodates coactivators within a hydrophobic cleft of the receptor LBD, through direct contacts with these NR boxes (9). Interestingly, the N-CoR/SMRT nuclear receptor interaction motifs exhibit a consensus sequence of I/LXXI/HI (i.e., CoRNR box, in which H indicates hydrophobic residues) (9), which interacts with specific residues in the same receptor pocket required for coactivator binding. Thus, discrimination of the subtle differences between the coactivator and corepressor interaction helices by the nuclear receptor AF2 core may provide the molecular basis for the exchange of coactivators for corepressors, with ligand-dependent formation of the charged clamp that stabilizes NR box binding and inhibits interaction with the CoRNR box helix.
ASC-2, also named AIB3, TRBP, TRAP250, NRC, and PRIP, is a novel coactivator gene amplified and overexpressed in certain human cancers (3, 8, 14, 17, 18, 19, 22, 52). Interestingly, ASC-2 contains two NR boxes. The C-terminal NR box specifically interacts with liver X receptors, and the N-terminal box binds many different nuclear receptors, including retinoic acid receptor (RAR) (19). Transgenic mice overexpressing ASC-2 fragment DN1 (ASC-2 residues 849 to 929, containing the N-terminal motif) but not DN1/m, in which the LXXLL sequences were mutated to LXXAA to disable the receptor bindings, were significantly impaired for many signaling pathways mediated by RAR and other receptors, in which DN1 competitively blocked the interaction of these receptors with the full-length endogenous ASC-2 (13a). In addition, single-cell microinjection of neutralizing antibodies against ASC-2 abolished transactivation by RAR and other nuclear receptors that interact with ASC-2 (17; our unpublished results). Taken together, ASC-2 might be a coactivator molecule crucial for the function of many different nuclear receptors in vivo.
Transcriptional coactivators often exist in steady-state complexes in vivo (9). In this study, we found that ASC-2 also belongs to a steady-state complex of approximately 2 MDa (ASC-2 complex [ASCOM]) in HeLa nuclei. Our results suggest that ASCOM may represent a new coactivator complex of nuclear receptors, which is distinct from the previously defined Swi/Snf, TRAP/DRIP, and SRC-1/CBP complexes (9, 49).
MATERIALS AND METHODS
Plasmids.
Vectors encoding glutathione S-transferase (GST) fusion proteins to histones H2A, H2B, H3, H4, and various H3 point mutants were kind gifts of Yoichi Shinkai (Kyoto University, Kyoto, Japan). PCR fragments encoding HALR residues 3712 to 4025 (i.e., HR/SET2), HALR residues 3507 to 4025 (i.e., HR/SET1), HR/SET1m1 and HR/SET1Δ (which are identical to HR/SET1 except for the deletion of four residues or the point mutation of a well-conserved glycine to serine in the SET domain), and the ASC-2 fragments DN1 and DN1/m were cloned into EcoRI and XhoI restriction sites of the GST fusion vector pGEX4T (Pharmacia) and pcDNA3 (Invitrogen), respectively. The mammalian expression vector for RAR, the transfection indicator construct pActin-β-gal, and the reporter construct β-RARE-LUC were as previously described (17).
Purification of ASCOM.
HeLa nuclear extract (1.5 g of protein) was loaded onto a HiTrap heparin column (3 by 5 ml; Pharmacia) equilibrated with G-150 buffer (20 mM HEPES-KOH [pH 7.9], 0.5 mM EDTA, 0.05% NP-40, 10% glycerol, 1 mM dithiothreitol [DTT], protease inhibitors, 150 mM KCl). The bound proteins were eluted with 75 ml of a linear salt gradient (150 to 600 mM KCl in G buffer). The fractions immunoreactive with anti-ASC-2 were pooled (350 mM KCl) and dialyzed in Q-80 buffer (20 mM Tris-HCl [pH 7.8], 0.5 mM EDTA, 20% glycerol, 1 mM DTT, protease inhibitors, 80 mM KCl) for 2 h. The samples were loaded onto a HiTrap Q column (2 by 5 ml; Pharmacia) equilibrated with Q-100 (20 mM Tris-HCl [pH 7.8], 0.5 mM EDTA, 20% glycerol, 1 mM DTT, protease inhibitors, 100 mM KCl), and the bound proteins were eluted with 50 ml of a salt gradient from 100 to 500 mM KCl in Q buffer. The ASC-2-containing fractions were pooled (300 mM KCl, 20 mg of protein), applied to a Mono Q column (HR5/5; Pharmacia), and developed with 10 ml of a salt gradient from 120 to 800 mM KCl. Immunoreactive fractions eluting at 300 mM KCl were pooled and mixed with 60 μl of protein G-agarose conjugated with the previously defined anti-ASC-2 monoclonal antibody (A3C1) (18) at 4°C for 12 h. The beads were recovered and washed three times with 1 ml of IP-300 buffer (20 mM Tris-HCl [pH 7.8], 0.1 mM EDTA, 0.2% NP-40, 10% glycerol, 1 mM DTT, protease inhibitors, 300 mM potassium acetate) and finally washed with 500 μl of phosphate-buffered saline. Bound proteins were eluted twice with 100 μl of 100 mM glycine (pH 3.0), precipitated with 10% trichloroacetate, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis.
Antibodies.
Anti-ASC-2 monoclonal antibody (A3C1) and anti-human Med6 (anti-hMed6) polyclonal antibody from rats were as previously described (18, 20). Antibodies against CBP, SRC-2, BRG1, TRAP220, hemagglutinin (HA), α-tubulin, and β-tubulin were purchased from Santa Cruz and Boehringer Mannheim. Monoclonal antibody against SRC-1 and polyclonal antibody against methylated K4 of H3 were kind gifts from Dean Edwards and Tony Kouzarides, respectively. Anti-ASH2, anti-RBQ-3, and anti-HALR antisera were generated in rats by using, as an antigen, a recombinant ASH2 fragment (ASH2 residues 516 to 624), RBQ-3, or an HALR fragment (HALR residues 2412 to 2886) expressed in Escherichiacoli. Antisera recognizing both ALR-1 and ALR-2 were generated in rabbits against a synthetic peptide, MSPPPEESPMSP, derived from ALR-1 residues 581 to 592 and ALR-2 residues 1 to 12. Each antibody was affinity purified with recombinant proteins expressed in E. coli and rigorously tested for the specificity. Antibody coupling to protein G-agarose and immunoprecipitation were executed as previously described (20).
Transfections and immunofluorescence microscopy.
HeLa and CV-1 cells were grown in 24-well plates with medium supplemented with 10% fetal calf serum for 24 h and transfected with 100 ng of lacZ expression vector pRSV-β-gal and 100 ng of β-RARE-LUC reporter gene along with the indicated amounts of mammalian expression vectors for DN1 and DN1/m. Total amounts of the expression vectors were kept constant by adding pcDNA3. Transfections and luciferase assays were done as described previously (17), and the results were normalized to those of the lacZ expression vector. Similar results were obtained in more than two similar experiments. Immunofluorescence microscopy was done essentially as described previously (45). MCF7 cells were incubated with anti-ASC-2 monoclonal antibody (A3C1) and rabbit polyclonal antibodies against α- or β-tubulin. Cells were labeled with fluorescein isothiocyanate-conjugated goat anti-mouse and tetramethyl rhodamine isothiocyanate-conjugated goat anti-rabbit antibodies (Sigma). Coverslips were mounted on glass slides and examined with an Olympus Fluoview 300 laser scanning confocal microscope.
GST pull-down assays.
GST fusions or GST alone was expressed in E. coli, bound to glutathione-Sepharose 4B beads (Pharmacia) in binding buffer (25 mM HEPES [pH 7.8], 0.2 mM EDTA, 20% glycerol, 100 mM KCl, and 0.1% NP-40), and incubated with labeled proteins expressed by in vitro translation by using the TNT coupled transcription-translation system, with conditions as described by the manufacturer (Promega, Madison, Wis.). Specifically bound proteins were eluted from beads with 40 mM reduced glutathione in 50 mM Tris (pH 8.0) and analyzed by SDS-PAGE and autoradiography as described previously (17).
ChIP.
293T cells were grown in 10-cm-diameter dishes with medium supplemented with 10% fetal bovine serum for 24 h and transfected with the indicated amounts of mammalian expression vectors for RAR, DN1, and DN1/m. The total amounts of the expression vectors were kept constant by adding pcDNA3. Chromatin immunoprecipitation (ChIP) assays were essentially done as described previously (51). The primers used were 5′-AAGCTCTGTGAGAATCCTG-3′ and 5′-GGATCCTACCCCGACGGTG-3′, which encompass the β-RARE region and generate a 288-bp PCR fragment. The primers used for the RARE region of the p21WAF1 promoter were 5′-AGACTCTGAGCAGCCTGAG-3′ and 5′-AACCCTCATTTGCAGATGGT-3′, which generate a 258-bp PCR fragment.
Histone methyltransferase assays.
Immunoprecipitates of ASCOM-containing HiTrap Q fractions with anti-ASC-2, anti-ASH2, or anti-ALR antibodies or 10 μg of GST fusion proteins to HALR and ALR was incubated with 5 μg of the appropriate H3 peptides, purified H2A, H2B, H3, and H4, or calf thymus histones (Roche, Inc.) along with 0.55 μCi of [3H]AdoMet in methyltransferase buffer (50 mM Tris [pH 8.0], 1 mM phenylmethylsulfonyl fluoride, and 0.5 mM DTT [final concentration]) for 1 h at 30°C in a final volume of 25 μl that was spotted on Whatman P-81 and then used for scintillation counting, as described previously (33, 42). The wild-type H3 N-terminal peptide contains residues 1 to 20 of human histone H3 (ARTKQTARKSTGGKAPRKQL-C). In mutant peptides, K4, K9, K14, or K18 was changed to arginine.
RESULTS
ASC-2 exists within a steady-state complex.
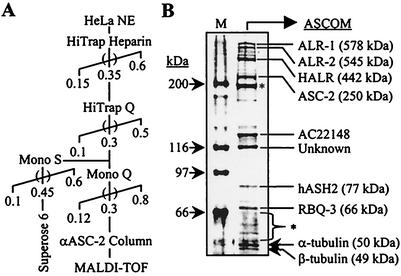
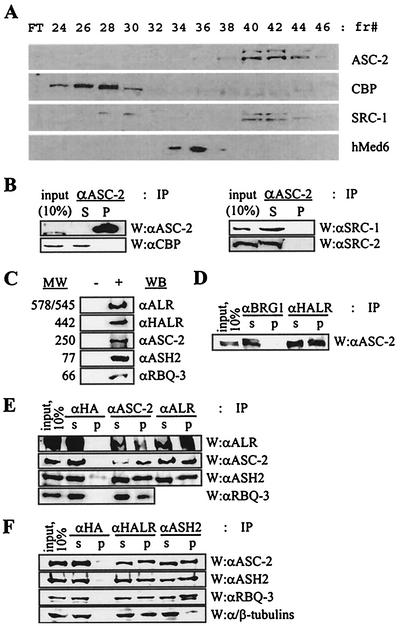
In HeLa nuclei, the 250-kDa protein ASC-2 was not observed as a free form and always existed as a significantly larger steady-state complex. To purify this complex (termed ASCOM for the ASC-2 complex), we subjected HeLa nuclear extract to a series of biochemical fractionations and immunoaffinity purification with anti-ASC-2 antibody, as shown in Fig. 1A. The final immunoaffinity purification produced many distinct protein bands (Fig. 1B). Among these, at least eight bands were reproducibly coimmunoprecipitated by anti-ASC-2 antibody throughout the purification procedures and were not present in a control immunoaffinity purification with anti-HA or anti-ASC-1 antibody (data not shown). MALDI-TOF mass spectrometry analyses identified these bands as Trx/ALL-1/MLL-related protein ALR-1 (MLL2) (GenBank accession no. 2358285, 578 kDa) and its splicing isoform ALR-2 (accession no. 2358287, 545 kDa) (32), ALR-like protein HALR (MLL3) (accession no. 10568112, 442 kDa) (43), ASC-2 (accession no. AAF13595, 250 kDa), ASH2 (accession no. 4009336, 77 kDa) (11), retinoblastoma binding protein RBQ-3 (accession no. 755750, 66 kDa) (36), α-tubulin (accession no. 4929134, 50.1 kDa), and β-tubulin (accession no. 13623683, 48.8 kDa). ALR-1 and ALR-2 differ only in their N termini (32). The composition of ASCOM was completely distinct from those of other reported coactivator complexes such as CBP/SRC-1/SRC-2, CBP/SRC-2/SRC-3/IKK (49), CBP/dTRX in Drosophilamelanogaster (31), p/CAF, and hMediator (9). Consistently, ASCOM behaved differently from complexes containing CBP, SRC-1, and hMed6 throughout the whole purification process (Fig. 2A). One obvious exception was that SRC-1 and a minor peak of CBP appeared to partly comigrate with ASC-2 in the HiTrap Q column fractionations (Fig. 2A, fractions 40 to 44). However, both SRC-1 and CBP were not coimmunoprecipitated by anti-ASC-2 antibody in these fractions (Fig. 2B). Given the fact that SRC-1 and SRC-2 can coexist in the SRC-1 complex (49), we tested whether SRC-2 is also present in these HiTrap Q fractions. As shown in Fig. 2B, SRC-2 indeed existed in these fractions but did not associate with ASC-2, like SRC-1. Taken together, we concluded that ASC-2 is tightly associated with a series of distinct proteins in vivo within a novel steady-state complex.
FIG. 1.
Purification of ASCOM. (A) The chromatography scheme for the purification of ASCOM from nuclear extract (HeLa NE) is shown. Numbers and parentheses indicate KCl molarity and pools, respectively. αASC-2, anti-ASC-2. (B) Mass spectrometric identification of polypeptides. Anti-ASC-2 antibody column eluates were run on an SDS-12% PAGE gel, silver stained, and analyzed by MALDI-TOF, with gene products and molecular masses (in parentheses) shown on the right. The masses of the marker proteins (M) are shown on the left, and asterisks indicate nonspecific bands. Antibody raised against AC22148, a protein of unknown function, revealed that it is a nonspecific contaminant. MALDI-TOF mass spectrometry analyses failed to unravel the identity of a protein of approximately 116 kDa (indicated Unknown).
FIG. 2.
Confirmation of the identity of the components of ASCOM. (A) HiTrap Q column fractions subjected to Western analysis with the indicated antibodies against ASC-2, CBP, SRC-1, and hMed6. FT, flowthrough; fr#, fraction number. (B) HiTrap Q fractions 40 to 44 were pooled and immunoprecipitated (IP) with anti-ASC-2 (αASC-2) antibody and Western (W) analyzed with the indicated antibodies. αCBP, anti-CBP; αSRC-1, anti-SRC-1; αSRC-2, anti-SRC-2. (C) Antibodies were generated against synthetic or recombinant polypeptides encoded by cDNAs isolated based on the MALDI-TOF mass spectrometry data of the purified proteins. + and −, HiTrap Q fractions containing ASCOM from HeLa nuclear extract (fractions 40 to 44) and unrelated fractions (30 to 34), respectively. Each antibody recognized a protein with the expected molecular weight (MW) (in thousands). αALR, anti-ALR; αHALR, anti-HALR; αASH2, anti-ASH2; αRBQ-3, anti-RBQ-3. (D, E, and F) HiTrap Q fractions of HeLa nuclear extract containing ASCOM (fractions 40 to 44) were immunoprecipitated with the indicated antibodies (IP), separated by SDS-4 to 6.5% PAGE, and probed with the indicated antibodies (W). αHA, anti-HA; αBRG1, anti-BRG1. S and s indicate supernatant, and P and p indicate precipitate. Ten percent of the total reaction mixture was loaded as input.
Verification of the ASCOM components.
To confirm the authenticity of the identified proteins as genuine constituents of ASCOM, we raised specific polyclonal antibodies against either synthetic or recombinant proteins derived from cognate cDNAs isolated from various cDNA libraries. Importantly, these antibodies specifically recognized each cognate band from HiTrap Q fractions 40 to 44 containing ASC-2/ASCOM but not from unrelated fractions (30 to 34) (Fig. 2C). From the ASCOM-containing HiTrap Q fractions (40 to 44), anti-HALR antibody coimmunoprecipitated ASC-2 (Fig. 2D). Similarly, antibodies against ASC-2, ALR, HALR, and ASH2 specifically coimmunoprecipitated ALR, ASC-2, ASH2, and RBQ-3 (Fig. 2E and F). In contrast, anti-HA and BRG1 antibodies did not coimmunoprecipitate any of these proteins, although these antibodies successfully depleted ectopically expressed HA-tagged proteins or BRG1 from HeLa cells (Fig. 2D, E, and F and data not shown). These results clearly demonstrate that ALR-1/2, HALR, ASH2, and RBQ-3 are genuine components of ASCOM.
Presence of α/β-tubulins in ASCOM.
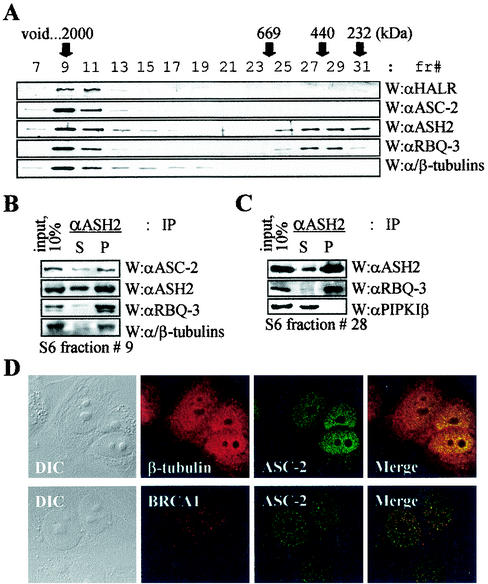
Surprisingly, α/β-tubulins were readily coimmunoprecipitated by anti-HALR antibody and less efficiently coimmunoprecipitated by ASH2 antibody from ASCOM-containing HiTrap Q fractions (Fig. 2F). Although the presence of nuclear tubulins was previously reported (2, 45), these proteins have not been shown to associate with any defined nuclear protein. Thus, we further explored this issue with more-highly purified ASCOM fractions. For this, HiTrapQ fractions containing ASCOM (fractions 40 to 44) were loaded onto a Mono S column, and anti-ASC-2-immunoreactive fractions were pooled. These fractions were then applied to a Superose 6 column and analyzed by immunoblotting, as summarized in Fig. 1A. In this Superose 6 gel filtration column, α/β-tubulins were copurified with HALR, ASC-2, ASH2, and RBQ-3, as an approximately 2-MDa complex (Fig. 3A). From Superose 6 fraction 9 containing the 2-MDa complex ASCOM, anti-ASH2 antibody coimmunoprecipitated not only ASC-2 and RBQ-3 but also α/β-tubulins (Fig. 3B). Although the amount of these nuclear α/β-tubulins associated with ASCOM is just a minor fraction of the whole-cell α/β-tubulins, antisera against α- and β-tubulins coimmunoprecipitated a detectable amount of ASC-2 from unfractionated whole-cell extracts (data not shown). Consistent with these results and a recent report (45), indirect immunofluorescence microscopy revealed the presence of tubulin in the nuclei of MCF7 cells (Fig. 3D). While α/β-tubulins were detected in both the cytoplasm and the nucleus, ASC-2 was mostly detected in the nucleus. Strongly supporting our biochemical analyses, at least some of these nuclear α/β-tubulins colocalized with ASC-2 in the nucleus, as demonstrated with confocal microscopy (Fig. 3D). In contrast, the nuclear protein BRCA1 was minimally colocalized with ASC-2. These results suggest that α/β-tubulins are genuine components of ASCOM. Importantly, the presence of α/β-tubulins raises an intriguing possibility that ASCOM may directly associate with the nuclear matrix (5). The biological significance of these findings needs to be further investigated.
FIG. 3.
α/β-Tubulins in ASCOM. (A) For the size fractionation of ASCOM, HiTrapQ fractions 40 to 44 (shown in Fig. 2A) were loaded onto a Mono S column (HR5/5; Pharmacia). Immunoreactive fractions were pooled, applied to a Superose 6 column (HR10/30; Pharmacia), and analyzed by immunoblotting as indicated. fr#, fraction number; W, Western analysis; αHALR, anti-HALR; αASC-2, anti-ASC-2; αASH2, anti-ASH2; αRBQ-3, anti-RBQ-3. (B and C) Superose 6 fraction 9 (B) or 28 (C) containing ASCOM or a smaller complex of approximately 500 kDa was immunoprecipitated with the indicated antibodies (IP), separated by SDS-4 to 6.5% PAGE, and probed with indicated antibodies (W). S and P indicate supernatant and precipitate, respectively. Ten percent of the total reaction mixture was loaded as input. αPIPKIβ, anti-PIPKIβ. (D) Cells were treated with β-tubulin, BRCA1, and ASC-2 antibodies. Fluorescein isothiocyanate (green)- and tetramethyl rhodamine isothiocyanate (red)-conjugated antibodies were used to detect ASC-2 and β-tubulin/BRCA1, respectively. Note the colocalization of ASC-2 and β-tubulin. Similar results were also obtained with α-tubulin antibody (data not shown). DIC, differential interference contrast.
Interestingly, RBQ-3 and ASH2 eluted as another distinct complex of approximately 500 kDa in the Superose 6 sizing column (Fig. 3A). Within this Superose 6 fraction (no. 28), anti-ASH2 antibody coimmunoprecipitated ASH2 and RBQ-3 but not the unrelated protein type I-β phosphatidylinositol 4-phosphate 5-kinase (Fig. 3C). We noted that this novel complex is similar in size to the previously reported Drosophila ASH2 complex (30). However, it could simply represent a submodule of ASCOM, relatively loosely attached to the putative core ASCOM module.
Notably, ALR-1, ALR-2, HALR, and ASH2 are mammalian homologues of the Drosophila trithorax group (Trx-G) proteins (reviewed in reference 7). Thus, we concluded that ASC-2 is a component of a novel nuclear steady-state complex containing RBQ-3, a subset of Trx-G proteins, and at least a subpopulation of nuclear α/β-tubulins.
H3-K4 methylase activity of ASCOM.
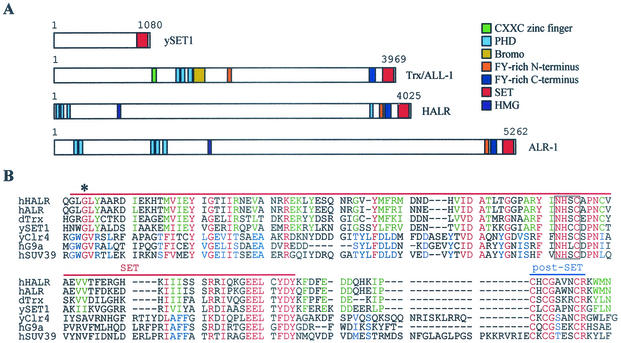
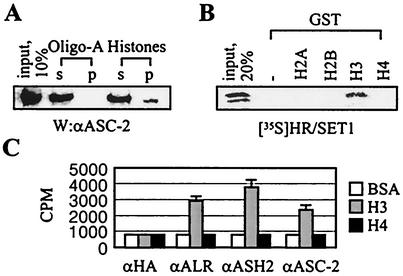
Trx-G and Polycomb group (Pc-G) proteins (7), positive and negative effectors of gene expression, are responsible for the maintenance of transcriptional regulation and provide a cellular memory mechanism throughout development. Some Trx-G and Pc-G proteins contain a 130- to 140-amino-acid motif termed the SET domain, found in a variety of chromatin-associated proteins (7). Notably, ALR-1/2 and HALR contain the SET domain at their C termini along with other known protein motifs (Fig. 4A). Interestingly, agarose beads coupled to histones but not to oligo(A) retained ASC-2 from ASCOM-containing HiTrap Q fractions 42 to 44 (Fig. 5A). These results suggest that ASC-2 or ASCOM interacts with histones. Recently, the SET domain of Drosophila Trx (dTrx) was demonstrated to specifically bind to H3 (13). Likewise, radiolabeled HR/SET1 (i.e., HALR residues 3507 to 4025) interacted with GST fusions to H3 but not to H2A, H2B, and H4 (Fig. 5B). These results imply that the interactions between ASC-2 and histones observed in the experiment for which data is shown in Fig. 5A may have resulted from direct bindings of the SET domains of HALR/ALR with H3.
FIG. 4.
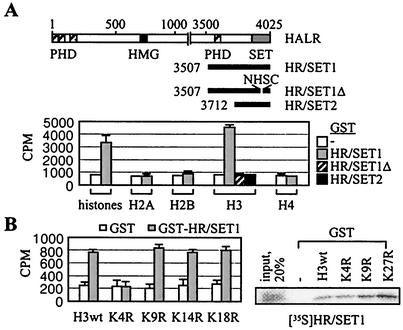
HALR/ALR SET domains. (A) Schematic representation of ySET1, hTrx/ALL-1, hHALR, and hALR-1. Various known protein motifs are color coded as indicated. (B) Amino acid alignment of SET and post-SET domains of hHALR, hALR, dTrx (accession no. AAF55041), ySET1 (accession no. AAB68867), S. pombe Clr4 (accession no. 060016), hG9a (accession no. S30385), and hSUV39 h1 (accession no. NP_003164). Amino acids conserved among all seven proteins; in hHALR, hALR, dTrx, and ySET1; and in yClr4, hG9a, and hSUV39 h1 are highlighted in red, green, and blue, respectively. Dashes indicate gaps in the alignment. The red and blue lines mark the extent of the SET and post-SET domains, respectively. The conserved block of four residues deleted in HR/SET1Δ is boxed, and the conserved glycine mutated to serine in HR/SET1m1 is marked with an asterisk.
FIG. 5.
H3 binding and methylation by ASCOM. (A) HiTrap Q column fractions containing ASCOM (fractions 40 to 44 in Fig. 2A) were incubated with either agarose beads coupled to histones or oligo(A) (Sigma), separated by SDS-7% PAGE, and probed with anti-ASC-2 (αASC-2) antibody (W). s and p indicate supernatant and precipitate, respectively. Ten percent of the total reaction mixture was loaded as input. (B) The GST pull-down experiments were done as previously described (17, 19) with GST alone or GST fusions to H2A, H2B, H3, and H4, and 20% of the total reaction mixture was loaded as input. (C) HiTrap Q column fractions containing ASCOM (fractions 40 to 44) were immunoprecipitated with the indicated antibodies and measured for methylation activities with bovine serum albumin, H3, or H4. αHA, anti-HA; αALR, anti-ALR; αASH2, anti-ASH2.
Recently, several SET proteins, such as Schizosaccharomyces pombe Clr4 as well as hSUV39 h1 and mouse Suv39 h1 (mSuv39 h1), were demonstrated to methylate K9 of histone H3 (33). This resulted in a binding site for HP1 proteins, a family of heterochromatic adaptor molecules implicated in both gene silencing and the higher-order structure of chromatin (16, 27). Another SET protein, G9a, methylated H3-K9 and K27 (42). Interestingly, anti-ASH2, -ALR, and -ASC-2 antibodies precipitated weak H3 methyltransferase activities from HiTrap Q fractions 40 to 44 containing ASCOM (Fig. 5C). In contrast, these immunoprecipitates did not show any methylase activities toward bovine serum albumin or H4. These H3-specific methylase activities likely derive from the SET domains of HALR and/or ALR-1/2, as demonstrated with GST-HR/SET1 and GST-ALR/SET (Fig. 6A and data not shown). The involvement of the SET domain was confirmed, as deletion of a highly conserved 4-amino-acid motif (i.e., GST-HR/SET1Δ) abolished the methylase activity (Fig. 6A). Interestingly, GST fusion to HALR residues 3708 to 4025 (i.e., GST-HR/SET2 in Fig. 6A) had no detectable activities, suggesting that HALR residues 3507 to 3712, encompassing a PHD finger, could be important for methylase activity. Whether this region facilitates folding of the SET domain to accommodate histone tails or directly mediates the enzymatic reaction warrants further investigation.
FIG. 6.
H3-K4-specific methylation by HALR. (A) Schematic representation of HALR and three C-terminal deletion fragments of HALR with four PHD fingers, an HMG-like domain, and the C-terminal SET domain. Histone methyltransferase activity was assayed by incubation of free histones in the presence of S-adenosyl-l-[methyl-3H]methionine, and incorporated radioactivity was determined by filter binding. (B) Histone methyltransferase activity was assayed by incubation of the indicated synthetic H3 peptides in the presence of S-adenosyl-l-[methyl-3H]methionine, and incorporated radioactivity was determined by filter binding. The GST pull-down experiments were done as previously described (17, 19) with GST fusions to the N-terminal 57 residues of human H3 (wild type [wt]) and point mutants, and 20% of the total reaction mixture was loaded as input.
Interestingly, the SET domain sequences of ALR-1/2 and HALR are more homologous to those of dTrx and yeast SET1 (ySET1) than hSUV39 h1, G9a, and Clr4 (Fig. 4B). In addition, ALR-1/2, HALR, dTrx, and ySET1 do not contain the cysteine-rich pre-SET domain present in hSUV39 h1, G9a, and Clr4 (33, 42). Corroborating these differences, GST-HR/SET1-methylated H3 peptides mutated to arginine at K9, K14, and K18 but not at K4, although these mutations did not affect the H3 bindings (Fig. 6B). Thus, the H3 methylation site mediated by ASCOM in vivo is likely to be K4, although a direct demonstration in vitro (e.g., by Edman degradation) was difficult due to the very weak H3 methylation activities of HALR/ALR. Although it is currently unclear how ASCOM-mediated H3-K4 methylation may ultimately lead to local gene activation, we concluded that ALR-1/2 and HALR are enzymes capable of methylating H3-K4.
Recruitment of ASCOM to RAR in vivo.
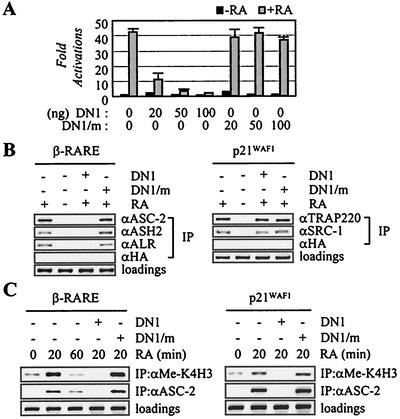
The previously described ligand-dependent protein-protein interactions between target nuclear receptors and the ASC-2 NR box (14, 19, 22) are believed to mediate the recruitment of ASCOM to receptors in vivo. Consistent with this expectation, the ASC-2 fragment containing the N-terminal LXXLL motif (i.e., DN1), but not DN1/m, in which the LXXLL motif was mutated to LXXAA to disable the receptor interactions, strongly repressed the ligand-dependent transactivation mediated by RAR in transient cotransfections, whereas the basal level of transcription was not affected (Fig. 7A). In ChIP assays, DN1, but not DN1/m, abolished the ligand-dependent recruitment of ASC-2 to the retinoid-responsive β-RARE promoter region (Fig. 7B, left panel). Importantly, DN1 specifically blocked endogenous ASC-2 to bind RAR in vivo without affecting the ligand-dependent recruitment of other LXXLL-based coactivators such as SRC-1 and TRAP220 (Fig. 7B, right panel). Thus, DN1 and its mutant version DN1/m are excellent tools to study the role of ASC-2 in nuclear receptor transactivation in vivo without the complication from other LXXLL-containing coactivators, although the basis for this specificity is not entirely clear. Nonetheless, our results demonstrate that the inhibitory effect of DN1 involves competitive, specific replacement of the endogenous full-length ASC-2 from the binding receptor. Interestingly, DN1 impaired ligand-dependent recruitment of not only ASC-2 but also other ASCOM components (ASH2 and ALR) to the retinoid-responsive β-RARE promoter region in vivo (Fig. 7B, left panel). These results are consistent with the biochemical studies which indicate that ASC-2 may only exist within a complex in vivo.
FIG. 7.
Recruitment of ASCOM in RAR transactivation. (A) The retinoid-responsive β-RARE-LUC reporter construct was cotransfected into HeLa cells, along with lacZ expression vector (100 ng) and expression vectors for DN1 (100 ng) or DN1/m (100 ng). Closed and shaded boxes indicate the absence and presence of 0.1 μM 9-cis-retinoic acid (RA), respectively. Normalized luciferase expressions from triplicate samples were calculated relative to the lacZ expressions. Similar results were obtained with CV-1 cells (data not shown). (B and C) ASCOM recruitment to β-RARE and p21WAF1 and H3-K4 methylation. 293T cells were cotransfected with expression vectors for RAR (10 ng), DN1 (100 ng), and DN1/m (100 ng) either in the absence or in the presence of 0.1 μM 9-cis-RA, as indicated. Chromatin from these cells was isolated and immunoprecipitated with the indicated antibodies. The endogenous β-RARE or p21WAF1 region present in the immunoprecipitated samples was amplified by PCR, and input PCR is shown for the loading controls.
Interestingly, retinoid-induced recruitment of ASC-2/ASCOM to β-RARE was time dependent, with a peak at 20 min after retinoid treatment, which was nicely correlated with retinoid-dependent H3-K4 methylation of the β-RARE promoter region (Fig. 7C, left panel). ASCOM could be responsible for this methylation as DN1, but not DN1/m, was inhibitory to the retinoid-induced ASCOM recruitment as well as H3-K4 methylation of the β-RARE promoter region. Similar results were also obtained with the retinoid-inducible p21WAF1 promoter (Fig. 7C, right panel). It is interesting that H3-K4 methylation, at least with these retinoid-responsive promoter regions, could be far more dynamic than was previously hypothesized (29). Thus, transactivation by nuclear receptors in vivo may require recruitment of ASCOM to target DNA response elements via the ligand-dependent interactions of ASC-2 and receptors, which may lead to subsequent, transient methylation of H3-K4 residues of the promoter region.
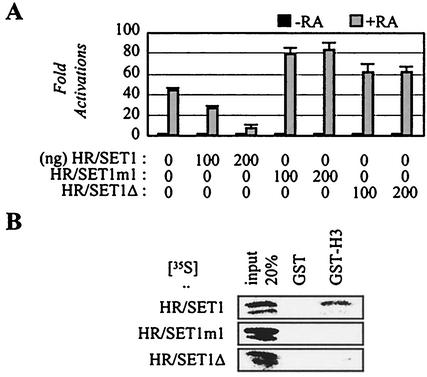
Interestingly, cotransfections of HR/SET1 inhibited transactivation by various nuclear receptors, including RAR (Fig. 8A and data not shown). Two possibilities exist. First, HR/SET1 may block the interactions of HALR/ALR-1/2 and their downstream effectors. Second, HR/SET1 may interfere with the endogenous HALR/ALR-1/2 from properly methylating their putative target substrates in vivo (H3 and/or other yet unknown substrates). Consistent with the latter possibility, mutation of a highly conserved glycine residue into serine (i.e., HR/SET1m1) or deletion of the conserved NHSC motif (i.e., HR/SET1Δ) abolished the repression (Fig. 8A). The point-mutation was based on the previously described Drosophila trxZ11 mutation that lacks H3 binding and causes homeotic transformation in the fly (13). Interestingly, the H3 binding was also impaired with HR/SET1Δ, suggesting the involvement of the NHSC motif in H3 bindings (Fig. 8B).
FIG. 8.
Inhibition of RAR transactivation by HALR-SET. (A) The retinoid-responsive β-RARE-LUC reporter construct was cotransfected into HeLa cells, along with the lacZ expression vector (100 ng) and expression vectors for HR/SET1, HR/SET1m1, and HR/SET1Δ, as indicated. Closed and shaded boxes indicate the absence and presence of 0.1 μM of 9-cis-retinoic acid (RA), respectively. Normalized luciferase expressions from triplicate samples were calculated relative to that of the lacZ expressions. Similar results were obtained with CV-1 cells (data not shown). (B) GST pull-down experiments were done as previously described (17, 19) with GST alone or a GST fusion to the N-terminal 57 residues of human H3, and 20% of the total reaction mixture was loaded as input.
Overall, these results demonstrate that ASC-2 functions as a whole complex (i.e., ASCOM), and H3 binding by ASCOM and recruitment of ASCOM to target DNA response elements via the ASC-2-receptor interactions appear to be essential for transactivation by nuclear receptors in vivo.
DISCUSSION
ASC-2 is a steady-state complex homologous to the ySET1 complex.
In this report, we have described a novel nuclear steady-state complex of approximately 2 MDa from HeLa nuclei, which is associated with the previously described transcriptional coactivator molecule ASC-2 (3, 8, 14, 17, 18, 19, 22, 52). ASCOM likely represents a human orthologue of a recently reported yeast complex of approximately 440 to 1,000 kDa (25, 26, 35), which contains ySET1, yASH2, three proteins with the WD repeats (reviewed in reference 21) that are also found in RBQ-3, and a subunit called Cps40/Saf41p/Spp1, a homologue of hCGBP that recognizes unmethylated CpG islands and contains conserved cysteine-rich motifs found in hTrx/MLL and the N-terminal region of ALR-1 (44). They further demonstrated that ySET1 is an H3-K4 methylase (26, 35). However, ASC-2 homologues do not seem to exist in Saccharomyces cerevisiae, Caenorhabditiselegans, and Drosophila (data not shown), suggesting the incorporation of ASC-2 into the complex relatively later in evolution. Notably, ASH2, ALR-1, ALR-2, and HALR as well as the yeast counterparts ySET1 and yASH2 are homologues of the known members of the Drosophila Trx-G proteins (7). Thus, ASCOM and the ySET1 complex represent a distinct coactivator complex that contains a subset of defined Trx-G proteins, along with the ATPase-dependent chromatin remodeling Swi/Snf complex (7, 9) and the recently reported dTrx/dCBP complex (31). Notably, three components (i.e., ALR-1, ALR-2, and HALR) are relatively large in size (Fig. 1B), which has significantly blunted our effort to map the intrainteraction interfaces among various different components. From our recent yeast two-hybrid screening, however, ASH2 was isolated as a protein that strongly interacts with RBQ-3. Thus, ASH2-RBQ-3 may form a core submodule loosely attached to ASCOM or the putatively independent complex of approximately 500 kDa (Fig. 3A).
DN1 as a specific inhibitor of ASC-2 in vivo.
The ASC-2 fragment DN1, which interacts with RAR in an LXXLL-dependent manner, acted as a potent dominant-negative inhibitor of the RAR transactivation (Fig. 7A). Importantly, this DN1-mediated repression was recovered by overexpressed ASC-2 but not by SRC-1 or TRAP220, two well-characterized LXXLL-type coactivators (13a). In ChIP experiments with cultured cell lines, DN1 also inhibited the retinoid-dependent recruitment of ASC-2, but not of TRAP220 and SRC-1, to the retinoid-responsive p21WAF1 promoter (Fig. 7B). Furthermore, ChIP analyses of mouse embryo fibroblasts from transgenic mice overexpressing DN1, which exhibit various phenotypes derived from the compromised retinoid and other ligand signalings, indicated that the ligand-dependent recruitment of ASC-2 to the RARβ2 promoter was impaired, whereas the recruitment of TRAP220 was unaffected (13a). Similarly, microinjection of anti-ASC-2 antibody was shown to repress the RAR transactivation, which was recovered by coinjected expression vector for ASC-2, but not SRC-1 or CBP (17). However, it's important to note that DN1 can compete with recombinant TRAP220 and SRC-1 to bind receptors in vitro (data not shown). This discrepancy attests to the important fact that ASC-2, TRAP220, and SRC-1 exist within distinct steady-state complexes in vivo but not as free polypeptides, as used in our in vitro experiments. Within the context of complex, the additional ASC-2 sequences present in DN1 may serve as an interaction interface with other effectors required for the successful recruitment of ASCOM. The LXXLL motif-mediated interactions of ASC-2 with receptors may have to accompany these secondary interactions for a successful assembly of the whole ASCOM-receptor complex on the promoter region. Furthermore, it should be noted that the interactions of receptors with various cofactors are highly dynamic, not static, as recently demonstrated with estrogen and androgen receptors (37, 38). Although more work is needed to clearly understand the basis for this specificity, our results are sufficient to demonstrate that DN1 is an excellent tool to study the function of the endogenous ASC-2 complex with nuclear receptors in vivo, without any complication from other LXXLL-based coactivators such as SRC-1 and TRAP220. Thus, we concluded that the ligand-induced transient methylation of H3-K4 residues around the RARβ2 and p21WAF1 promoter regions (Fig. 7C), which was abolished by coexpressed DN1, but not by DN1/m, may have resulted from the H3-K4 methylation activities of ASCOM, although the possible involvement of or communication with other histone methylase(s) could not be excluded.
ASCOM and H3-K4 methylation.
H3-K4 methylation has been intrinsically linked to gene activation (29, 39). Recently, H3-K4 methylation mediated by a novel protein, SET9, was demonstrated to enhance histone acetylation by p300 and suppress the transcriptionally repressive SUV39 h1-mediated H3-K9 methylation as well as the binding of the NuRD repressor complex to the H3 tail (28, 46). Interestingly, the HALR-mediated H3 methylation activity in vitro was very weak relative to other histone methyltransferases (28, 33, 42, 46). In addition, the activity was not significantly affected by H3-S10 phosphorylation or H3-K14 acetylation, and almost nondetectable with mononucleosomes (data not shown). Thus, HALR/ALR may require other cofactors lost during the purification or function in a promoter-specific manner. For instance, the ySET1 complex required other components in addition to the enzymatic component itself (i.e., ySET1) for successful methylation activities (35). Thus, a series of stable cell lines that express Flag-tagged ASC-2, ASH2, and RBQ-3 are currently being developed, from which we plan to purify a functionally intact complex. In addition, it was recently demonstrated that methylation of H3-K4 by the ySET1 complex requires ubiquitination of H2B by RAD6 (6, 41). Thus, the weak in vitro methylase activity of ASCOM may also stem from the lack of ubiquitinated H2B under the reaction conditions we employed. Finally, HALR/ALR may also target nonhistone proteins, such as other coactivators, as recently documented for the H3-specific arginine methyltransferase CARM1 that methylates CBP/p300 (51). To clearly resolve these issues, we are currently constructing gene-targeted mice in which the methylase activity of HALR is specifically altered.
ASCOM and other coactivator complexes.
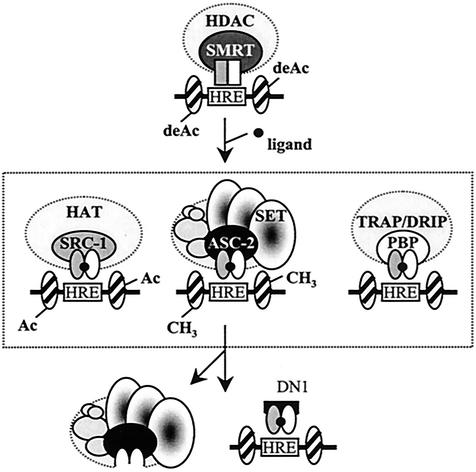
As schematically indicated in Fig. 9, unliganded nuclear receptors recruit corepressor proteins N-CoR and SMRT, which complex with various histone deacetylase enzymes that inhibit transcription (9). One primary step toward gene activation is believed to be the recruitment of proteins that disrupt chromatin formation. Coregulators equivalent to factors of the yeast Swi/Snf complex, such as BRG1, and related complexes regulate chromatin remodeling. In addition, ligand-dependent recruitment of SRC-1/CBP/p/CAF complexes brings HAT activity to nuclear receptor complexes (Fig. 9). Furthermore, biochemically isolated complexes that contain factors similar or homologous to factors in the yeast mediator complex (i.e., TRAP-SMCC-DRIP-ARC) have been isolated by coprecipitation of different transcription factors, which might be involved in directly bridging the RNA polymerase II complex with basal transcription factors (9). ASCOM is the fourth kind of nuclear receptor coactivator complex, which might be involved with methylating H3-K4 residues of the target promoter regions (Fig. 9). Nuclear receptor-mediated transactivation requires these and other coactivator complexes that can act sequentially, combinatorially, or in parallel. The selective recruitment of coactivators by individual receptors has been well established (47, 48). These selective receptor-coactivator interactions represent an efficient system through which the pleiotropic effects of nuclear receptor ligands might be mediated and are likely further determined by tissue-specific patterns of posttranslational modification of coactivators (15). In this regard, two issues will be particularly relevant for further studies of ASCOM. It will be important to examine how ASCOM is functionally interrelated to other coactivators during the transactivation processes. For instance, our preliminary results indicate that ASCOM may functionally require the intact Swi/Snf complex in vivo. Furthermore, HR/SET1, the C-terminal fragment of HALR, appears to modulate the H3 arginine methylase activities of CARM1 at least in vitro, another pivotal coactivator protein of nuclear receptors. Another equally important question will be to determine whether ASCOM can be selectively utilized under different contexts and conditions in vivo, i.e., what are the major physiological target genes of ASCOM?
FIG. 9.
Working model for ASCOM function. Upon ligand binding, nuclear receptors undergo a structural change, which signals the replacement of corepressor complexes by a series of distinct coactivator complexes. Three coactivator complexes that contain the well-defined LXXLL motif-based adaptor molecules (SRC-1, ASC-2, and PBP/TRAP220/DRIP205//TRIP2) are shown schematically. These and other coactivators can be selectively recruited to individual receptors under different promoter contexts and cellular conditions (see the text). The SET domains of HALR/ALR might, either directly or indirectly, be involved with methylating H3-K4 and/or other yet unknown substrates in vivo. DN1 competitively blocks receptors from recruiting ASCOM, but not other LXXLL-based complexes, and thus may specifically inhibit ASCOM-mediated H3-K4 methylation of the promoter region.
Nuclear tubulins in ASCOM.
This report extends the previous intriguing reports of the presence of tubulin proteins in the nucleus (24, 45). The copurification of α/β-tubulins with the other ASCOM components HALR, ASC-2, ASH2, and RBQ-3 (Fig. 3A) from the highly purified Superose 6 fractions that lack the abundant cellular free tubulins as well as the coimmunoprecipitation with ASH2 in these fractions (Fig. 3B) strongly support the authenticity of α/β-tubulins as true components of ASCOM. The indirect immunofluorescence results (Fig. 3D) also support this notion. Given the presence of α/β-tubulins in ASCOM, one interesting hypothesis to test will be whether the nuclear matrix (5) can modulate the function of ASCOM. Our preliminary results indicate that the previously defined autonomous transactivation function of ASC-2 (19, 22) is strongly induced by the tubulin-destabilizing agent vinblastin but not by the tubulin-stabilizing agent taxol (our unpublished results). Interestingly, it was previously noted that vinblastin affects biochemical processes that are seemingly unrelated to tubulin, such as DNA and RNA synthesis (2). It is possible that the effects of vinblastin on these processes could occur through an interaction with the nuclear tubulins present in ASCOM. The biological significance of the presence of α/β-tubulins in ASCOM will be an exciting future investigation.
ASCOM and human cancer.
It is noted that mutations and/or translocations into the trx/mll gene in mammals result in the development of several types of hematological malignancies (1). The ALR and HALR genes map to chromosome bands 12q12-13 and 7q36, respectively, regions associated with chromosomal aberrations encountered in human cancers (32, 43). RBQ-3 was also reported to be gene amplified in certain gliomas (36), like the ASC-2 gene in colon, breast, and lung cancers (17). Interestingly, β-tubulin was recently shown to accumulate in rapidly dividing cells, such as cancer cell lines and tissues (45). These results suggest that misregulation of different ASCOM subunits might be linked to carcinogenesis.
In conclusion, ASC-2, essential for transactivation by nuclear receptors and likely many other transcription factors, is a component of a novel human Trx-G complex capable of specifically methylating H3-K4. Further characterization of ASCOM will facilitate the unraveling of the molecular details of transcriptional control as well as cancer development.
Acknowledgments
We thank Tony Kourzarides, Yoichi Shinkai, and Dean Edwards for various reagents.
This work was supported by grant 2000-2-20900-006-1 from the Basic Research Program of the Korea Science and Engineering Foundation (to Y.C.L.) and by grants from the Postech Biotech Center (3PD02002) and GenoCheck, Inc. (to J.W.L).
Y.-H.G., Y.C.S., and D.-H.K. contributed equally to this work.
REFERENCES
- 1.Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20:5695-5707. [DOI] [PubMed] [Google Scholar]
- 2.Bernstam, V. A., R. H. Gray, and I. A. Bernstein. 1980. Effects of microtubule-disrupting drugs on protein and RNA synthesis in Physarum polycephalum amoebae. Arch. Microbiol. 128:34-40. [DOI] [PubMed] [Google Scholar]
- 3.Caira, F., P. Antonson, M. Pelto-Huikko, E. Treuter, and J. A. Gustafsson. 2000. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J. Biol. Chem. 275:5308-5317. [DOI] [PubMed] [Google Scholar]
- 4.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 5.Cremer, T., G. Kreth, H. Koester, R. H. Fink, R. Heintzmann, M. Cremer, I. Solovei, D. Zink, and C. Cremer. 2000. Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Crit. Rev. Eukaryot. Gene Expr. 10:179-212. [PubMed] [Google Scholar]
- 6.Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by RAD6. J. Biol. Chem. 277:28368-28371. [DOI] [PubMed] [Google Scholar]
- 7.Francis, N. J., and R. E. Kingston. 2001. Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol. 2:409-421. [DOI] [PubMed] [Google Scholar]
- 8.Guan, X. Y., J. Xu, S. L. Anzick, H. Zhang, J. M. Trent, and P. S. Meltzer. 1996. Hybrid selection of transcribed sequences from microdissected DNA: isolation of genes within amplified region at 20q11-q13.2 in breast cancer. Cancer Res. 56:3446-3450. [PubMed] [Google Scholar]
- 9.Hermanson, O., C. K. Glass, and M. G. Rosenfeld. 2002. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol. Metab. 13:55-60. [DOI] [PubMed] [Google Scholar]
- 10.Huang, N., E. vom Baur, J. M. Garnier, T. Lerouge, J. L. Vonesch, Y. Lutz, P. Chambon, and R. Losson. 1998. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 17:3398-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikegawa, S., M. Isomura, Y. Koshizuka, and Y. Nakamura. 1999. Cloning and characterization of ASH2L and Ash2l, human and mouse homologs of the Drosophila ash2 gene. Cytogenet. Cell Genet. 84:167-172. [DOI] [PubMed] [Google Scholar]
- 12.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 13.Katsani, K. R., J. J. Arredondo, A. J. Kal, and C. P. Verrijzer. 2001. A homeotic mutation in the trithorax SET domain impedes histone binding. Genes Dev. 15:2197-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Kim, S.-W., C. Cheong, Y.-C. Sohn, Y.-H. Goo, W. Je Oh, J. H. Park, S. Y. Joe, H.-S. Kang, D.-K. Kim, C. Kee, J. W. Lee, and H.-W. Lee. 2002. Multiple developmental defects derived from impaired recruitment of ASC-2 to nuclear receptors in mice: implication for posterior lenticonus with cataract. Mol. Cell. Biol. 22:8409-8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko, L., G. R. Cardona, and W. W. Chin. 2000. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc. Natl. Acad. Sci. USA 97:6212-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko, L., G. R. Cardona, T. Iwasaki, K. S. Bramlett, T. P. Burris, and W. W. Chin. 2002. Ser-884 adjacent to the LXXLL motif of coactivator TRBP defines selectivity for ERs and TRs. Mol. Endocrinol. 16:128-140. [DOI] [PubMed] [Google Scholar]
- 16.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 17.Lee, S.-K., S. L. Anzick, J. E. Choi, L. Bubendorf, X. Y. Guan, Y. K. Jung, O. P. Kallioniemi, J. Kononen, J. M. Trent, D. Azorsa, B. H. Jhun, J. H. Cheong, Y. C. Lee, P. S. Meltzer, and J. W. Lee. 1999. A nuclear factor ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J. Biol. Chem. 274:34283-34293. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S.-K., S. Y. Na, S. Y. Jung, J. E. Choi, B. H. Jhun, J. Cheong, P. S. Meltzer, Y. C. Lee, and J. W. Lee. 2000. Activating protein-1, nuclear factor-kappaB, and serum response factor as novel target molecules of the cancer-amplified transcription coactivator ASC-2. Mol. Endocrinol. 14:915-925. [DOI] [PubMed] [Google Scholar]
- 19.Lee, S.-K., S. Y. Jung, Y. S. Kim, S. Y. Na, Y. C. Lee, and J. W. Lee. 2001. Two distinct nuclear receptor-interaction domains and CREB-binding protein-dependent transactivation function of activating signal cointegrator-2. Mol. Endocrinol. 15:241-254. [DOI] [PubMed] [Google Scholar]
- 20.Lee, Y. C., and Y.-J. Kim. 1998. Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol. Cell. Biol. 18:5364-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, D., and R. Roberts. 2001. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 58:2085-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahajan, M. A., and H. H. Samuels. 2000. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol. Cell. Biol. 20:5048-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, and R. M. Evans. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menko, A. S., and K. B. Tan. 1980. Nuclear tubulin of tissue cultured cells. Biochim. Biophys. Acta 629:359-370. [DOI] [PubMed] [Google Scholar]
- 25.Miller, T., N. J. Krogan, J. Dover, H. Erdjument-Bromage, P. Tempst, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA 98:12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy, P. L., J. Griesenbeck, R. D. Kornberg, and M. L. Cleary. 2002. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. USA 99:90-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 28.Nishioka, K., S. Chuikov, K. Sarma, H. Erdjument-Bromage, C. D. Allis, P. Tempst, and D. Reinberg. 2002. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16:479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 30.Papoulas, O., S. J. Beek, S. L. Moseley, C. M. McCallum, M. Sarte, A. Shearn, and J. W. Tamkun. 1998. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 125:3955-3966. [DOI] [PubMed] [Google Scholar]
- 31.Petruk, S., Y. Sedkov, S. Smith, S. Tillib, V. Kraevski, T. Nakamura, E. Canaani, C. M. Croce, and A. Mazo. 2001. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294:1331-1334. [DOI] [PubMed] [Google Scholar]
- 32.Prasad, R., A. B. Zhadanov, Y. Sedkov, F. Bullrich, T. Druck, R. Rallapalli, T. Yano, H. Alder, C. M. Croce, K. Huebner, A. Mazo, and E. Canaani. 1997. Structure and expression pattern of human ALR, a novel gene with strong homology to ALL-1 involved in acute leukemia and to Drosophila trithorax. Oncogene 15:549-560. [DOI] [PubMed] [Google Scholar]
- 33.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 34.Rice, J. C., and C. D. Allis. 2001. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13:263-273. [DOI] [PubMed] [Google Scholar]
- 35.Roguev, A., D. Schaft, A. Shevchenko, W. W. Pijnappel, M. Wilm, R. Aasland, and A. F. Stewart. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20:7137-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saijo, M., Y. Sakai, T. Kishino, N. Niikawa, Y. Matsuura, K. Morino, K. Tamai, and Y. Taya. 1995. Molecular cloning of a human protein that binds to the retinoblastoma protein and chromosomal mapping. Genomics 27:511-519. [DOI] [PubMed] [Google Scholar]
- 37.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 38.Shang, Y., M. Myers, and M. Brown. 2002. Formation of the androgen receptor transcription complex. Mol. Cell 9:601-610. [DOI] [PubMed] [Google Scholar]
- 39.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 96:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strahl, B. D., S. D. Briggs, C. J. Brame, J. A. Caldwell, S. S. Koh, H. Ma, R. G. Cook, J. Shabanowitz, D. F. Hunt, M. R. Stallcup, and C. D. Allis. 2001. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11:996-1000. [DOI] [PubMed] [Google Scholar]
- 41.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 42.Tachibana, M., K. Sugimoto, T. Fukushima, and Y. Shinkai. 2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276:25309-25317. [DOI] [PubMed] [Google Scholar]
- 43.Tan, Y. C., and V. T. Chow. 2001. Novel human HALR (MLL3) gene encodes a protein homologous to ALR and to ALL-1 involved in leukemia, and maps to chromosome 7q36 associated with leukemia and developmental defects. Cancer Detect. Prev. 25:454-469. [PubMed] [Google Scholar]
- 44.Voo, K. S., D. L. Carlone, B. M. Jacobsen, A. Flodin, and D. G. Skalnik. 2000. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol. Cell. Biol. 20:2108-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walss-Bass, C., K. Xu, S. David, A. Fellous, and R. F. Luduena. 2002. Occurrence of nuclear bII-tubulin in cultured cells. Cell Tissue Res. 308:215-223. [DOI] [PubMed] [Google Scholar]
- 46.Wang, H., R. Cao, L. Xia, H. Erdjument-Bromage, C. Borchers, P. Tempst, and Y. Zhang. 2001. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 8:1207-1217. [DOI] [PubMed] [Google Scholar]
- 47.Warnmark, A., T. Almlof, J. Leers, J. A. Gustafsson, and E. Treuter. 2001. Differential recruitment of the mammalian mediator subunit TRAP220 by estrogen receptors ERα and ERβ. J. Biol. Chem. 276:23397-23404. [DOI] [PubMed] [Google Scholar]
- 48.Wong, C. W., B. Komm, and B. J. Cheskis. 2001. Structure-function evaluation of ERα and β interplay with SRC family coactivators. ER selective ligands. Biochemistry 40:6756-6765. [DOI] [PubMed] [Google Scholar]
- 49.Wu, R.-C., J. Qin, Y. Hashimoto, J. Wong, J. Xu, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 2002. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) coactivator activity by IκB kinase. Mol. Cell. Biol. 22:3549-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie, M., G. Shao, I. M. Buyse, and S. Huang. 1997. Transcriptional repression mediated by the PR domain zinc finger gene RIZ. J. Biol. Chem. 272:26360-26366. [DOI] [PubMed] [Google Scholar]
- 51.Xu, W., H. Chen, K. Du, H. Asahara, M. Tini, B. M. Emerson, M. Montminy, and R. M. Evans. 2001. A transcriptional switch mediated by cofactor methylation. Science 21:2507-2511. [DOI] [PubMed] [Google Scholar]
- 52.Zhu, Y., L. Kan, C. Qi, Y. S. Kanwar, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 2000. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J. Biol. Chem. 275:13510-13516. [DOI] [PubMed] [Google Scholar]