Abstract
We describe an interaction between homeodomain-interacting protein kinase 1 (HIPK1) and Daxx, two transcriptional regulators important in transducing growth-regulatory signals. We demonstrate that HIPK1 is ubiquitously expressed in mice and humans and localizes predominantly to the nucleus. Daxx normally resides within the nucleus in promyelocytic leukemia protein (PML) oncogenic domains (PODs), where it physically interacts with PML. Under certain circumstances, Daxx is relocalized from PODs to chromatin, where it then acts as a transcriptional repressor through an association with histone deacetylase (HDAC1). We propose two novel mechanisms for regulating the activity of Daxx, both mediated by HIPK1. First, HIPK1 physically interacts with Daxx in cells and consequently relocalizes Daxx from PODs. Daxx relocalization disrupts its interaction with PML and augments its interaction with HDAC1, likely influencing Daxx activity. Although the relocalization of Daxx from PODs is phosphorylation independent, an active HIPK1 kinase domain is required, suggesting that HIPK1 autophosphorylation is important in this interaction. Second, HIPK1 phosphorylates Daxx on Ser 669, and phosphorylation of this site is important in modulating the ability of Daxx to function as a transcriptional repressor. Mutation of Daxx Ser 669 to Ala results in increased repression in three of four transcriptional reporters, suggesting that phosphorylation by HIPK1 diminishes Daxx transcriptional repression of specific promoters. Taken together, our results indicate that HIPK1 and Daxx collaborate in regulating transcription.
Homeodomain-interacting protein kinase 1 (HIPK1) is one of three closely related serine/threonine protein kinases that regulate the activity of a broad range of transcription factors (5, 10, 15, 22, 23). The HIPKs were originally identified as nuclear protein kinases that function as corepressors for various homeodomain-containing transcription factors. Subsequently, HIPK2 was shown to provide a regulatory role for a large transcriptional repressor complex, as it interacts with the repressive elements Groucho and histone deacetylase (HDAC1) (4). HIPK2 also associates with high-mobility group I (y) proteins, a family of nuclear architectural proteins that influence transcription regulation (26).
The HIPKs are related to a group of kinases that includes Yak1 (Saccharomyces cerevisiae), YakA (Dictyostelium discoideum), and Minibrain (Drosophila melanogaster, rodents, and humans) (15, 23). This group represents a family of protein kinases that regulate the transition from growth to differentiation in eukaryotic cells. The assignment of HIPKs to this group suggests a common function in growth regulation. In fact, growth-regulatory and tumor suppressor functions have been attributed to the HIPKs (5, 10, 26, 28). For example, HIPK2 transfection into cells results in a dramatic decrease in colony formation (5, 10). This outcome occurs through the cooperative activities of HIPK2 and the tumor suppressor protein p53 (5, 10). HIPK2 phosphorylates p53 on Ser 46, resulting in activation of p53-dependent transcription, cell growth regulation, and apoptosis initiation (5, 10, 31).
A potential apoptotic function was also assigned to HIPK3, as it interacts with the cell surface death receptor Fas and phosphorylates the Fas-associated death domain (FADD), a transducer of Fas-mediated apoptotic signaling (28). The association of HIPK3, a nuclear protein, with a cell surface receptor was reconciled by demonstrating diffuse cytoplasmic HIPK3 localization.
Daxx is also a nuclear protein that was shown to associate with cytoplasmic and cell surface molecules, including transforming growth factor beta and Fas (25, 32). Overexpressed Daxx was shown to bind the Fas death domain and to mediate apoptotic signaling by activating ASK-1 and subsequently the Jun-N-terminal kinase (JNK) pathway, independent of the FADD/procaspase-8 pathway (3, 32).
Within the nucleus, Daxx interacts with the promyelocytic leukemia protein (PML) and localizes to PML oncogenic domains (PODs) (17, 30, 33). Localization of Daxx to PODs correlates with Daxx's proapoptotic activity. For example, Daxx mutants that fail to localize to PODs do not facilitate Fas-induced cell death (30). Furthermore, in the absence of PML, Daxx is dispersed throughout the nucleus, and activated cell death is diminished (33). These findings indicate that Daxx and PML may cooperate in PODs in mediating apoptotic signals. This may be important in understanding the biological nature of acute promyelocytic leukemia, which is characterized by reciprocal chromosomal translocation between the pml and retinoic acid receptor α genes, resulting in the oncogenic fusion product PML-retinoic acid receptor α. In acute promyelocytic leukemia cells, Daxx does not localize to PODs. However, upon treatment with retinoic acid, which induces disease remission, Daxx relocalizes to PODs (33).
Despite evidence supporting a proapoptotic function for Daxx, other studies have demonstrated that Daxx is essential to cell survival or is antiapoptotic. Targeted disruption of Daxx in mice results in embryonic lethality accompanied by extensive apoptosis (21). Elevated apoptosis was also observed in Daxx-null embryonic stem cells (21) as well as in fibroblasts in which endogenous Daxx was depleted by RNA interference treatment (J. S. Michaelson and P. Leder, unpublished data). In myeloid precursor cells, Daxx overexpression inhibited activated cell death, indicating an antiapoptotic role for Daxx (2). Furthermore, an antiapoptotic role for Daxx in acute promyelocytic leukemia cells was proposed, as Daxx expression decreased after apoptotic induction with HDAC1 inhibitors (1). It is possible that Daxx provides bipartite functions. Under certain circumstances, Daxx may be essential for cell survival, and under other circumstances, Daxx may propagate apoptotic signals.
Similar to the HIPKs, Daxx functions as a transcriptional regulator. For example, Daxx repressed the transcriptional activities of the Pax3 and ETS1 transcription factors (11, 18). Interestingly, Daxx was not able to repress the transcriptional activities of the oncogenic fusion protein Pax3-FKHR present in an alveolar rhabdomyosarcoma. This suggests that Pax3-FKHR circumvents the transcriptional controls normally applied to Pax3 (11). Daxx also either repressed or activated Pax5-mediated transcription (6). The specific effect of Daxx on Pax5 activity varied in different B-cell lines. Activation of Pax5-mediated transcription by Daxx depended on recruitment of the histone acetyltransferase CREB binding protein.
To date, the regulation of Daxx transcriptional activity is poorly understood. It was suggested that in the absence of PML, Daxx localized to chromatin, where it recruited HDAC1 and repressed transcription (12, 17). Overexpression of PML but not the oncogenic fusion PML-retinoic acid receptor α recruits Daxx to PODs, thereby inhibiting Daxx repression (16, 17). The recruitment of Daxx from chromatin to PODs requires the secondary modification of PML by the ubiquitin-like molecule SUMO-1 (12, 16, 17). These results demonstrate that Daxx transcriptional regulatory activity is controlled in part by PML, which sequesters Daxx from condensed chromatin to PODs.
In this study, we characterized the expression and localization of HIPK1. In addition, we propose two novel mechanisms for regulating Daxx behavior, both mediated by HIPK1. HIPK1 relocates Daxx from PODs, presumably to chromatin, where Daxx participates in transcriptional regulation. In addition, HIPK1 phosphorylates Daxx on Ser 669, a site important in modulating Daxx activity.
MATERIALS AND METHODS
Northern blot hybridization.
For HIPK1 expression analysis in mice, total RNA was isolated from adult FVB mouse tissues with the Ultraspec-II RNA isolation kit (Biotecx Laboratories). RNA (10 μg) was resolved on 1.2% agarose-formaldehyde gels, transferred to Genescreen Plus nylon membranes (New England Nuclear), and cross-linked to the membranes by exposure to UV light. The membranes were prehybridized in hybridization buffer (50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 2× Denhardt's solution, 0.1% sodium dodecyl sulfate [SDS], 50 mM sodium phosphate [pH 6.5], and 20 μg of herring sperm DNA per ml) for 1 h at 42°C and hybridized with 32P-labeled cDNA probes in hybridization buffer overnight at 42°C. The membranes were washed four times in 2× SSC-0.2% SDS at room temperature for 15 min and two times in 0.1× SSC-0.2% SDS at 60°C for 30 min.
A HIPK1 probe (open reading frame bp 2456 to 3267) was generated by PCR from mouse brain cDNA. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was a SacI-BamHI restriction product from plasmid pGEM-GAPDH encompassing exons 5 to 8 of mouse GAPDH. For HIPK expression analysis in humans, a multiple-tissue Northern blot and an RNA master blot (Clontech Laboratories) were hybridized with the HIPK1 probe described above with the manufacturer's recommended methods. The multiple-tissue Northern blot was also hybridized with a probe specific for β-actin, which was provided by the manufacturer.
Cloning of full-length HIPK1.
Full-length HIPK1 was generated from adult FVB mouse RNA (isolated as described above) by reverse transcription-PCR. Single-stranded cDNA was constructed from total RNA (5 μg) with avian myeloblastosis virus reverse transcriptase (Seikagaku America) and oligo(dT) according to the manufacturer's recommended methods. cDNA (5 μl) was used for PCR amplification of full-length HIPK1. The HIPK1 primers forward (5′-TAAGAATTCGCCACCATGGCCTCACAGCTGCAGGTG-3′) and reverse (5′-CAAGTAAGAATACTGACTGATCTTGG-3′) amplified the region from open reading frame bp 1 to 3627. HIPK1 cDNA was amplified with the Advantage-GC PCR kit (Clontech Laboratories) according to the manufacturer's recommended methods. Overhangs (3′-A) were added by incubating amplification products with 1 μl of Taq DNA polymerase (Promega) at 72°C for 15 min. The amplification product was ligated into pCR2.1 with the Original TA cloning kit (Invitrogen).
Generation of a kinase-inactive HIPK1.
A kinase-inactive version of HIPK1 (K219A) was made by changing Lys 219 to an Ala with the QuikChange site-directed mutagenesis kit (Stratagene). The following primers were used for the mutagenesis; forward, 5′-CCAAGGAAATTGTGGCCATTGCAATCTTGAAGAACCACCCCTCC-3′; and reverse, 5′-GGAGGGGTGGTTCTTCAAGATTGCAATGGCCACAATTTCCTTGG-3′.
Generation of HIPK1 antibodies.
A glutathione S-transferase (GST)-HIPK1 fusion protein was made by cloning a HIPK1 PCR amplification product (open reading frame bp 2842 to 3144) into the EcoRI and NotI sites of pGEX-6P-1 (Amersham Pharmacia). GST-HIPK1 fusions were expressed in bacteria and purified as described elsewhere (13). Covance Research Products prepared the anti-HIPK1 rabbit serum with the purified GST-HIPK1 fusion protein. HIPK1 antibodies were affinity purified from serum against the GST-HIPK1 fusion with the AminoLink Plus immobilization kit (Pierce).
Expression vectors.
Full-length HIPK1 or HIPK1 K219A containing either a Flag or Myc C-terminal epitope tag was cloned into the EcoRI site of the pβ expression vector (27). Full-length Daxx containing a Myc C-terminal epitope was cut with EcoRV and cloned into the blunted XhoI site of the pCAGGS expression vector (24). Plasmid pSG5-PML (8) was obtained from P. P. Pandolfi. The E2F1 and SP1-luciferase reporter vectors (29) were obtained from P. Farnham. The Met-luciferase reporter vector (7) was obtained from R. Maas. The CRE-luciferase reporter vector was supplied by Stratagene. The pCMVβ vector (19) was obtained from I. Skerjanc.
Immunoprecipitation and Western blot analysis.
For analysis of transfected proteins, human embryonic kidney 293 cells (2 × 104 to 2.5 × 104/cm2) were grown for 12 h in Dulbecco's modified Eagle's medium supplemented with 10% bovine calf serum (HyClone), 4 mM l-glutamine, and antibiotics (50 U of penicillin and 50 μg of streptomycin per ml). The cells were transfected with pβ, Flag epitope-tagged pβ/HIPK1, or pβ/K219A, with or without Myc epitope-tagged pCAGGS/Daxx or vectors encoding the Daxx phosphorylation mutants described below. Transfections were performed with FuGene 6 (Roche) according to the manufacturer's recommended methods. The cells were lysed 24 h after transfection in immunoprecipitation buffer (1% NP-40, 0.15 M NaCl, 0.01 M NaH2PO4, 2 mM EDTA, 50 mM NaF, 0.2 mM Na3VO4, complete protease inhibitor cocktail tablet [Roche]) and centrifuged to remove insoluble debris. Cell lysates were incubated with antibody (1 μg/300 to 500 μg of total protein) for 2 h at 4°C.
Antibodies used for immunoprecipitations included anti-HIPK1 antibody (above), anti-Flag M2 antibody (Sigma), anti-Daxx antibody (21), and anti-Myc antibody (Invitrogen). Protein A-Sepharose CL-4B (Amersham Pharmacia Biotech) or UltraLink immobilized protein A/G (Pierce) (for rabbit and mouse antibodies, respectively) (20 μl of a 1:1 solution per μg of antibody) was added and incubated for an additional 2 h at 4°C. The immobilized proteins were collected by centrifugation, washed three times with immunoprecipitation buffer, and solubilized by boiling for 5 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (0.01 M Tris-HCl [pH 6.8], 1 mM EDTA, 10% glycerol, 0.004% bromophenol blue, 4% SDS, 0.02 M dithiothreitol).
For analysis of endogenous HIPK1, nuclear lysates were prepared from confluent 293 cells with the NE-PER nuclear and cytoplasmic extraction reagents (Pierce). Immunoprecipitations were performed with anti-HIPK1 antibody or anti-Fyn antibody (Santa Cruz) with protein A-Sepharose CL-4B as described above. The proteins were resolved by SDS-PAGE and Western blotted with electrochemiluminescence (Amersham Pharmacia Biotech). Primary antibodies used for Western blotting included anti-HIPK1 antibody (1:1,000), anti-Daxx antibody (1:1,000), anti-Myc antibody (1:1,000), anti-PML (H-238) antibody (1:200; Santa Cruz), and anti-HDAC1 antibody (Upstate Biotechnology; 1:500). Secondary antibodies used for Western blotting included goat anti-rabbit immunoglobulin G (1:10,000; Southern Biotechnology) and goat anti-mouse immunoglobulin G (heavy and light chains) conjugated to horseradish peroxidase (1:10,000; Southern Biotechnology).
In vitro pulldown assay.
Daxx (open reading frame bp 1 to 2220) was cloned into the BamHI and NotI sites of pGex-6P-1. GST and Daxx-GST fusion proteins were expressed in bacteria and purified as described above. Full-length HIPK1 and MEK1 were cloned into pcDNA3.1(−)/Myc-HisB (Invitrogen), and proteins were radiolabeled from these constructs with Easy Tag Express-[35S] protein labeling mix (1,175.0 Ci/mmol) (Perkin-Elmer) with the TNT coupled reticulocyte lysate system (Promega) according to the manufacturer's recommended methods. The translation products (10 μl) were incubated with equal amounts of GST or Daxx-GST fusion proteins in GST buffer (0.5% NP-40, 20 mM Tris-Cl [pH 8], 100 mM NaCl, 1 mM EDTA, complete protease inhibitor cocktail tablet) for 3 h at 4°C. The samples were collected by centrifugation, washed three times with GST buffer, and solubilized by boiling for 5 min in SDS-PAGE sample buffer. The samples were resolved by SDS-PAGE, and the gels were dried on a Speed Gel gel drying system (Savant) and visualized by autoradiography.
In vitro kinase assay.
293 cells were grown for 12 h as described above and transfected with pβ, Flag epitope-tagged pβ/HIPK1, Flag epitope-tagged pβ/K219A, or Myc epitope-tagged pCAGGS/Daxx. The cells were lysed 24 h after transfection in immunoprecipitation buffer. Proteins were immunoprecipitated from cell lysates as described above with anti-Flag antibody (HIPK1 and K219A) or anti-Myc antibody (Daxx). The immobilized proteins were washed three times in immunoprecipitation buffer and once in kinase buffer (20 mM Tris-Cl [pH 7.5], 10 mM MgCl2, 2 mM MnCl2, 1 mM dithiothreitol). Immobilized HIPK1 or K219A was incubated with 50 μM ATP, 5 μCi of [γ-32P]ATP, and substrate, including 1.25 μg of myelin basic protein (Sigma) or immobilized Daxx. Phosphorylation reactions were performed for 30 min at 30°C and stopped by adding 4× SDS-PAGE sample buffer containing 40 mM EDTA. Proteins were resolved by SDS-PAGE, and the gels were dried on a Speed Gel gel-drying system. Labeled proteins were visualized by autoradiography.
Immunofluorescence.
NIH 3T3 mouse fibroblasts or U2OS human fibroblasts (6.25 × 103/cm2) were grown for 12 h on two- to four-well plastic slides (Nalge Nunc) in the cell culture medium described above. The cells were transfected with Flag or Myc epitope-tagged pβ/HIPK1 or pβ/K219A, with or without pSG5-PML, Myc epitope-tagged pCAGGS/Daxx, or a vector encoding the Daxx phosphorylation mutant described below. The cells were fixed 15 to 24 h after transfection with 3.7% formaldehyde-phosphate-buffered saline (PBS), washed three times with PBS, and permeabilized with PBS containing 0.1% Triton X-100. After being washed once with PBS, the cells were stained with the indicated antibodies in PBS with 1% bovine serum albumin for 1 h at room temperature. Antibodies used included anti-HIPK1 antibody (1:200), anti-Myc antibody (1:200), anti-Flag antibody (10 μg/ml), anti-Daxx antibody (1:200), and anti-PML antibody (1:50). The cells were washed three times with PBS and stained with fluorescein isothiocyanate- or tetramethyl rhodamine isothiocyanate-conjugated goat anti-rabbit immunoglobulin antibody (1:200; Jackson Immunoresearch), fluorescein isothiocyanate- or tetramethyl rhodamine isothiocyanate-conjugated goat anti-mouse immunoglobulin antibody (1:200; Jackson Immunoresearch), and Hoechst 33258 at 10 mg/ml (1:1,000; Molecular Probes) in PBS with 1% bovine serum albumin for 30 min at room temperature. The slides were washed three times with PBS and once with distilled H2O and mounted with glass coverslips in PBS containing 15% Gelvatol (20). Fluorescent cells were examined with a Zeiss Axiophot upright fluorescence microscope, and images were captured with a SPOT-RT cooled digital camera (Diagnostic Instruments).
Identification of phosphorylated Daxx residues.
293 cells were grown for 12 h as described above on ten 10-cm cell culture dishes and transfected with Myc epitope-tagged pCAGGS/Daxx and Flag epitope-tagged pβ/HIPK1 or pβ/K219A. The Daxx protein was immunoprecipitated 24 h after transfection with an anti-Myc antibody as described above. The Daxx protein was resolved by SDS-PAGE and visualized by staining the gel with colloidal blue (Invitrogen). The various Daxx bands were excised from the gels and submitted to the Taplin Biological Mass Spectrometry Facility (Department of Cell Biology, Harvard Medical School) for microcapillary liquid chromatography-tandem mass spectrometry (LC-MS/MS). Phosphorylated Daxx residues were identified as described previously (9).
Generation of Daxx phosphorylation mutants.
Mutations in phosphorylated DAXX residues were generated with the QuikChange multisite-directed mutagenesis kit (Stratagene) with Myc epitope-tagged pCAGGS/Daxx as a template. The following primers were used for the mutagenesis; Ser 502 to Ala, 5′-GATAATGAAGGAAATGAGGCACCCACATCGCCTTCAG-3′, and Ser 669 to Ala, 5′-GTGTCCAGCCTATGCCAGCACCCCCCTTGGCCTCTG-3′. pCAGGS/Daxx-S502/669A had both Ser 502 and Ser 669 altered to alanine, while pCAGGS/Daxx-S502A and pCAGGS/Daxx-S669A had only Ser 502 or Ser 669, respectively, altered to alanine.
Transcriptional reporter assays.
HeLa human fibroblasts (2 × 104/cm2) were grown for 24 h in 35-mm cell culture dishes in the culture medium described above. The cells were transfected with pβ, pCAGGS/Daxx, or the vector encoding the Daxx phosphorylation mutant described above with the indicated luciferase reporter and pCMVβ. The cells were lysed in 1× passive lysis buffer (Promega), and the luciferase activity was measured with an Automat LB953 luminometer (Berthold) with automatic injection of the luciferase reagent (Fisher). The absolute values of luciferase activity were normalized to transfection efficiency by measuring β-galactosidase activity with the Galactostar β-galactosidase reporter kit (Tropix).
RESULTS
HIPK1 is ubiquitously expressed and localizes to nuclear speckles.
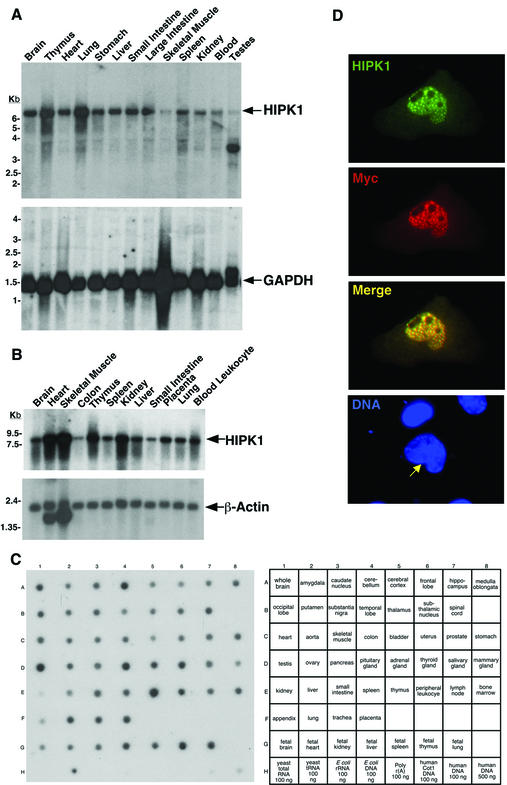
To examine the expression profile of HIPK1, mammalian multiple-tissue Northern and RNA master blots were hybridized with a HIPK1-specific probe. HIPK1 was expressed in all mouse (Fig. 1A) and human (Fig. 1B and 1C) tissues examined, although expression levels varied among tissue types. In addition, differences in relative expression levels were observed between the same mouse and human tissues. For example, HIPK1 was relatively moderately expressed in mouse skeletal muscle and heart, whereas HIPK1 was relatively strongly expressed in human skeletal muscle and heart. An aberrant transcript was detected in mouse (Fig. 1A) and human (data not shown) testes. The RNA master blot showed that HIPK1 was expressed in both adult and fetal tissues (Fig. 1C).
FIG. 1.
HIPK1 is ubiquitously expressed in mouse and human tissues and localizes to nuclear speckles. (A) Mouse multiple-tissue Northern blot probed for HIPK1 (upper panel) and GAPDH (lower panel) as a control for RNA loading. Numbers indicate the relative migration of RNA molecular size standards. (B) Human multiple-tissue Northern blot probed for HIPK1 (upper panel) and β-actin (lower panel) as a control for RNA loading. Numbers indicate the relative migration of RNA molecular size standards. (C) Human RNA master blot probed for HIPK1. The adjacent diagram indicates the RNA or DNA source for each spot. (D) Representative immunofluorescent images of a HIPK1-Myc fusion expressed in NIH 3T3 cells and stained with anti-HIPK1 antibody (green) and anti-Myc antibody (red). Overlapping localization is shown in the merged image (yellow). DNA (blue) was stained with Hoechst 33258. The yellow arrow indicates the nucleus of the HIPK1-Myc-transfected cell.
To examine where HIPK1 resides in cells, a Myc epitope-tagged HIPK1 expression construct was expressed in NIH 3T3 cells, and the fusion protein was detected by immunofluorescence with HIPK1 and Myc antibodies (Fig. 1D). HIPK1 localized to nuclear speckles, similar to those described for HIPK2 and HIPK3 (14, 15, 28). The majority of these nuclear speckles were previously demonstrated to be distinct from PODs, although a partial overlap does exist (5, 10, 14). Localization of HIPK1 to nuclear speckles presumably occurs through the speckle retention signal and SUMO-1 modification, as described for HIPK2 (14). Although HIPK1 localized strongly to nuclear speckles, the protein was seen throughout the nucleus and was excluded only from nucleoli. In addition, smaller amounts of HIPK1 were seen diffusely in the cytoplasm, similar to HIPK3 localization (28).
HIPK1 interacts with and phosphorylates Daxx.
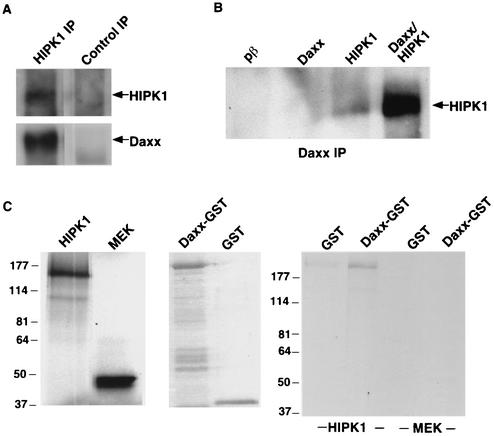
A previous study identified an interaction between HIPK3 and Daxx, although the physiological relevance of that interaction was not pursued (28). We found that HIPK1 and Daxx also interact in cells. Endogenous Daxx coimmunoprecipitated with endogenous HIPK1 (Fig. 2A). In addition, overexpressed HIPK1 coimmunoprecipitated with endogenous Daxx (HIPK1 lane) and to a much greater degree with overexpressed Daxx (Daxx/HIPK1 lane) (Fig. 2B). In this experiment, no HIPK1 was detected when an empty vector (pβ lane) or Daxx alone (Daxx lane) was expressed in cells due to the relatively low levels of endogenous HIPK1 protein.
FIG. 2.
HIPK1 interacts with Daxx in vivo and in vitro. (A) Endogenous HIPK1 was immunoprecipitated (IP) with anti-HIPK1 or a control antibody from 293 cell nuclear lysates. The immunoprecipitates were Western blotted, and the blots were probed with anti-HIPK1 antibody (upper panel) or anti-Daxx antibody (lower panel). (B) Daxx was immunoprecipitated with anti-Daxx antibody from 293 cells expressing an empty vector (pβ), pCAGGS/Daxx (Daxx), pβ/HIPK1 (HIPK1), or pCAGGS/Daxx and pβ/HIPK1 together (Daxx/HIPK1). Daxx immunoprecipitates were Western blotted, and the blots were probed with anti-HIPK1 antibody. (C) 35S-radiolabeled HIPK1 and MEK1 proteins (left panel, autoradiogram) were incubated with GST or Daxx-GST fusion proteins (center panel, colloidal blue-stained gel), and the pulled-down protein aggregates were evaluated by SDS-PAGE (right panel, autoradiogram). Numbers indicate the relative migration of protein molecular size standards (in kilodaltons).
The interaction between HIPK1 and Daxx was further verified by means of an in vitro pulldown assay (Fig. 2C). Radiolabeled HIPK1 preferentially associated with a Daxx-GST fusion protein over GST alone. The radiolabeled control, MEK1, did not bind significantly to Daxx-GST or GST alone.
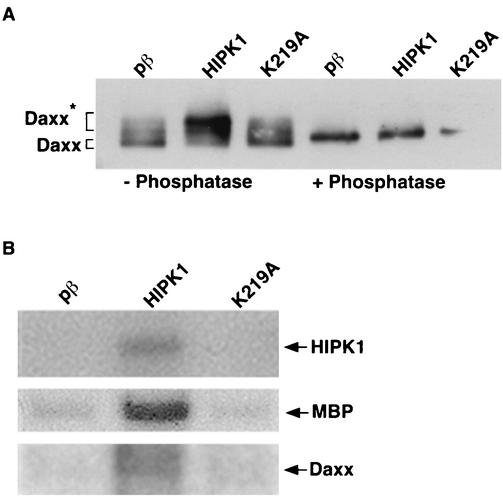
To determine whether Daxx serves as a substrate for HIPK1, Daxx was immunoprecipitated from cells cotransfected with pβ, HIPK1, or a kinase-inactive HIPK1 (K219A) (Fig. 3A). K219A was generated by mutating lysine 219 of HIPK1 to alanine, thereby disrupting the phosphotransfer reaction carried out by the kinase domain. By Western blot derived from large SDS-PAGE separations, Daxx presented as multiple bands when expressed with pβ, HIPK1, or K219A. However, when expressed with pβ or K219A, the lowest-migrating or basal phosphorylated band (Fig. 3A, Daxx) was the predominant form of Daxx. When expressed with HIPK1, the highest-migrating or hyperphosphorylated band (Fig. 3A, Daxx*) was the predominant form of Daxx. This indicated that HIPK1 shifted Daxx from a basal phosphorylated to a hyperphosphorylated state in vivo. Lambda phosphatase treatment of immunoprecipitated Daxx caused it to migrate as a single band in all samples, demonstrating that the HIPK1 modification of Daxx resulted from phosphorylation and not another form of secondary modification.
FIG. 3.
HIPK1 phosphorylates Daxx in vivo and in vitro. (A) Daxx was immunoprecipitated with anti-Myc antibody from 293 cells expressing pCAGGS/Daxx with an empty vector (pβ), pβ/HIPK1 (HIPK1), or pβ/K219A (K219A). Daxx immunoprecipitates were lambda phosphatase treated (+ Phosphatase) or mock treated (− Phosphatase) and Western blotted. The blot was probed with anti-Daxx antibody. Daxx* indicates the upper-migrating or hyperphosphorylated bands, and Daxx indicates the lower-migrating or basal phosphorylation bands in the non-phosphatase-treated samples. (B) HIPK1 was immunoprecipitated with anti-HIPK1 antibody from 293 cells expressing an empty vector (pβ), pβ/HIPK1 (HIPK1), or pβ/K219A (K219A). HIPK1 immunoprecipitates were used as the enzyme in an in vitro kinase reaction with myelin basic protein (MBP) or immunoprecipitated Daxx obtained from pCAGGS/Daxx expressed in 293 cells. HIPK1 but not pβ or K219A underwent autophosphorylation (upper panel) and phosphorylated myelin basic protein (middle panel) and Daxx (lower panel).
To determine if Daxx was a direct substrate of HIPK1, in vitro kinase assays were performed (Fig. 3B). Immunoprecipitated HIPK1 from cells transfected with pβ, HIPK1, or K219A was used as the kinase in these reactions. HIPK1 but not K219A underwent autophosphorylation and was therefore detected in this assay. Myelin basic protein, a promiscuous substrate used as a control in many in vitro kinase assays, was phosphorylated by HIPK1. Daxx was also phosphorylated by HIPK1 in vitro, demonstrating that Daxx was a direct HIPK1 substrate.
HIPK1 relocates Daxx from PODs, providing evidence for altered Daxx activity.
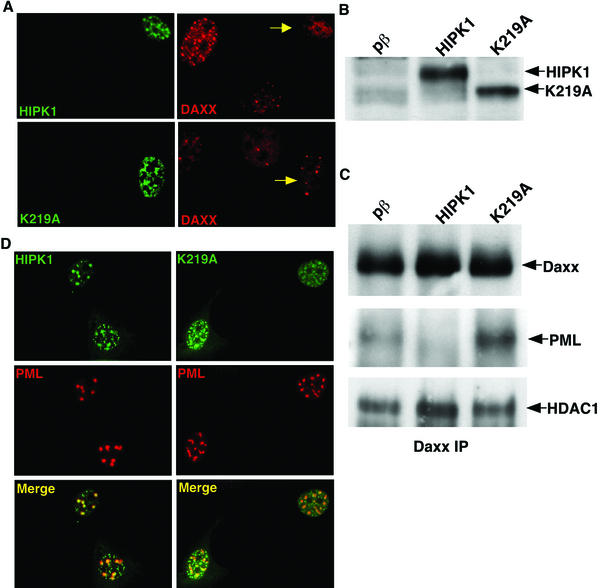
Endogenous Daxx normally localizes in part to PODs, which appear as small spots within the nucleus. Immunofluorescence revealed that increased HIPK1 expression relocalized endogenous Daxx from PODs to a diffuse localization throughout the nucleus (Fig. 4A). Increased K219A expression did not relocate endogenous Daxx from PODs (Fig. 4A). HIPK1 but not K219A relocalization of Daxx was observed in multiple human and mouse fibroblast lines (data not shown). Within PODs, Daxx is known to interact with PML (12, 16, 17). Expression of HIPK1 but not pβ or K219A disrupted this interaction, as determined by Daxx-PML coimmunoprecipitation (Fig. 4C, PML). Direct Western blot analysis of the lysates demonstrated equal transfection efficiencies for HIPK1 and K219A (Fig. 4B). In addition, equal amounts of Daxx were immunoprecipitated in each sample (Fig. 4C, Daxx). The apparent absence of phosphorylated bands in the Daxx Western blot results from a small SDS-PAGE separation. Together, the relocalization of Daxx from PODs and the disrupted Daxx-PML interaction suggest that HIPK1 sequesters Daxx from PODs to a diffuse localization throughout the nucleus, likely to chromatin.
FIG. 4.
HIPK1 relocalizes Daxx from PODs, disrupts the Daxx/PML interaction, and enhances the association between Daxx and HDAC1. (A) Representative immunofluorescent images of HIPK1-Flag and K219A-Flag fusions expressed in NIH 3T3 cells and stained with anti-Flag antibody (green) and anti-Daxx antibody (red). The yellow arrows indicate the transfected cells. (B) Western blot from 293 cells expressing pβ, pβ/HIPK1 (HIPK1), or pβ/K219A (K219A) probed with anti-HIPK1 to ensure equal HIPK1 and K219A transfection efficiencies. (C) Daxx was immunoprecipitated (IP) with anti-Daxx antibody from 293 cells expressing an empty vector (pβ), pβ/HIPK1 (HIPK1), or pβ/K219A (K219A). Daxx immunoprecipitates were Western blotted, and the blots were probed with anti-Daxx antibody (upper panel), anti-PML antibody (middle panel), or anti-HDAC1 antibody (lower panel). (D) Representative immunofluorescent images of HIPK1-Flag and K219A-Flag fusions expressed in U2OS cells with pSG5/PML and stained with anti-Flag antibody (green) and anti-PML antibody (red). Overlapping localization is shown in the merged images (yellow).
To ensure that the HIPK1 effect was specific to Daxx, POD integrity was examined by PML immunofluorescence in cells expressing increased PML with HIPK1 or K219A (Fig. 4D). PODs remained intact when HIPK1 or K219A expression was increased. The size of these PODs was augmented as a result of the increased PML expression, as PML is essential for POD formation (12). PODs also remained intact in cells with increased HIPK1 expression without PML overexpression (data not shown). HIPK1 and K219A had similar localization patterns, and both partially localized to PODs, as seen in the HIPK1-PML merged images. A partial localization of HIPK2 to PODs has also been reported (5, 10).
When not present in PODs, Daxx localizes to condensed chromatin, where it functions as a transcriptional regulator by interacting with proteins such as HDAC1 (12, 17). An association between Daxx and HDAC1 is likely required for Daxx to act as a transcriptional repressor. Therefore, to determine whether relocalization of Daxx by HIPK1 affected Daxx activity, we examined the association of Daxx with HDAC1 in the presence of pβ, HIPK1, and K219A by coimmunoprecipitation (Fig. 4C, HDAC1). Western blots containing Daxx immunoprecipitates and probed for HDAC1 showed that a Daxx-HDAC1 association was present in all samples. However, this association was increased in the presence of HIPK1 relative to the association in the presence of pβ or K219A. These findings indicate that HIPK1 may influence Daxx transcriptional repressive activity by enhancing its association with HDAC1.
Identification of phosphorylated Daxx residues.
Microcapillary liquid chromatography-tandem mass spectrometry was used to identify phosphorylated Daxx residues from samples immunoprecipitated from cells expressing increased HIPK1 or K219A (Table 1). Seven phosphorylated residues were identified by this method and were either serines or threonines. Five residues were found to be phosphorylated in the basal and hyperphosphorylated forms of Daxx and therefore are likely constitutively phosphorylated. Two residues were found phosphorylated only in the hyperphosphorylated form of Daxx, Ser 502 and Ser 669. Therefore, phosphorylation of Daxx at these sites appears to be regulated, likely by HIPK1.
TABLE 1.
Phosphorylated Daxx residues identified by LC-MS/MS
| Phosphorylated residue | Tryptic peptidea | Phosphorylation stateb |
|---|---|---|
| Ser 219 | ELDLSELDDPDSSYLQEAR | Constitutive |
| Thr 472 | EATEDEDEDLEQLQEDQGGDEEEEGGDNEGNESPTSPSDFFHR | Constitutive |
| Ser 502 | EATEDEDEDLEQLQEDQGGDEEEEGGDNEGNESPTSPSDFFHR | Regulated |
| Ser 515 | NSEQAEGLRTPEGQQK | Constitutive |
| Thr 523 | NSEPAEGLRTPEGQQK | Constitutive |
| Ser 626 | DASPPSKR | Constitutive |
| Ser 669 | EPMAQQDSGQNTSVQPMPSPPLASVASVADSSTR | Regulated |
Amino acid sequence indicates tryptic peptide from which phosphorylated Daxx residues were identified. Phosphorylated Daxx residues are indicated in bold.
Constitutive indicates that the residue was phosphorylated in the basal and hyperphosphorylated forms of Daxx. Regulated indicates that the residue was phosphorylated only in the hyperphosphorylated form of Daxx.
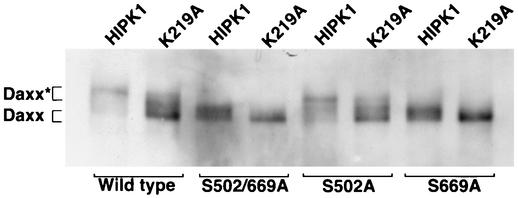
To determine if HIPK1 phosphorylates Daxx on Ser 502 or Ser 669, Daxx proteins with mutations in these residues (serine to alanine) were prepared. These mutant Daxx proteins were immunoprecipitated from cells expressing increased HIPK1 or K219A, and the phosphorylation status was evaluated by Western blot derived from large SDS-PAGE separations (Fig. 5). When expressed with HIPK1, the majority of wild-type Daxx existed in a hyperphosphorylated state (Fig. 5, Daxx*), whereas when expressed with K219A, the majority of wild-type Daxx existed in a basal phosphorylated state (Fig. 5, Daxx). Mutation of both Ser 502 and Ser 669 to alanines (S502/669A) resulted in a significant downshift in the hyperphosphorylated form of Daxx in the presence of HIPK1. Mutation of only Ser 502 to alanine (S502A) was not sufficient to cause this downshift. However, mutation of only Ser 669 to alanine (S669A) induced a significant downshift in the hyperphosphorylated form of Daxx in the presence of HIPK1. Therefore, we conclude that HIPK1 phosphorylated Daxx on Ser 669. A second band was also present in Daxx-S669A when HIPK1 levels were increased. Interestingly, in the presence of K219A, the residual levels of hyperphosphorylated Daxx disappeared in Daxx-S669A relative to wild-type Daxx (Fig. 5, compare wild type with K219A to S669A with K219A). This indicated that this site was partially phosphorylated even when HIPK1 levels were not increased, likely by endogenous HIPK1.
FIG. 5.
HIPK1 phosphorylates Daxx on Ser 669. Daxx was immunoprecipitated with anti-Myc antibody from 293 cells expressing pCAGGS/Daxx (wild type), pCAGGS/Daxx-S502/669A (S502/669A), pCAGGS/Daxx-S502A (S502A), or pCAGGS/Daxx-S669A (S669A) with an empty vector (pβ), pβ/HIPK1 (HIPK1), or pβ/K219A (K219A). The immunoprecipitates were Western blotted, and the blot was probed with anti-Myc antibody. Daxx* indicates the upper-migrating or hyperphosphorylated bands, and Daxx indicates the lower-migrating or basally phosphorylated bands.
Interaction of HIPK1 with Daxx, not phosphorylation of Ser 669, is important in regulating Daxx localization.
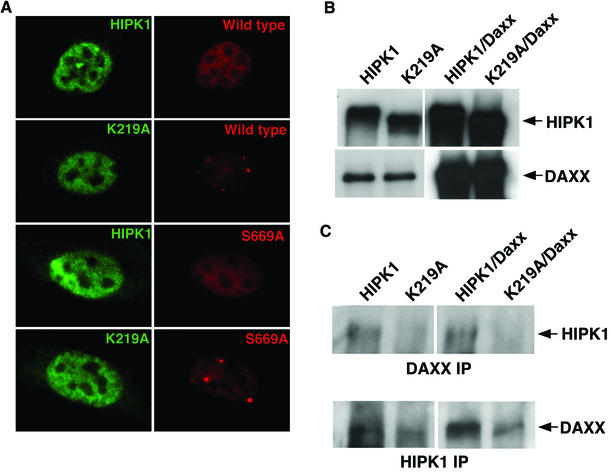
To determine if HIPK1 phosphorylation of Daxx on Ser 669 is important in relocalizing Daxx from PODs, immunofluorescence was performed on cells that expressed wild-type Daxx or Daxx-S669A with HIPK1 or K219A (Fig. 6A). Increased HIPK1 but not K219A expression relocalized Daxx-S669A from PODs to a diffuse localization throughout the nucleus, similar to wild-type Daxx. Therefore, phosphorylation of Daxx Ser 669 by HIPK1 is not necessary for Daxx relocalization.
FIG. 6.
HIPK1-Daxx interaction, not phosphorylation of Ser 669, is important in Daxx relocalization. Representative immunofluorescent images of HIPK1-Flag and K219A fusions expressed in NIH 3T3 cells with pCAGGS/Daxx (wild type) or pCAGGS/Daxx-S669A (S669A) and stained with anti-HIPK1 antibody (green) and anti-Myc antibody (red). (B) Western blots from 293 cells expressing pβ/HIPK1 (HIPK1) or pβ/K219A (K219A) alone or in combination with pCAGGS/Daxx (HIPK1/Daxx and K219A/Daxx, respectively). The blots were probed with anti-HIPK1 (upper panel) or anti-Daxx (lower panel) to ensure equal HIPK1, K219A, and Daxx transfection efficiencies. (C) Daxx and HIPK1 were immunoprecipitated (IP) with anti-Daxx and anti-HIPK1 antibodies, respectively, from 293 cells expressing pβ/HIPK1 (HIPK1) or pβ/K219A (K219A) alone or in combination with pCAGGS/Daxx (HIPK1/Daxx and K219A/Daxx, respectively). Daxx and HIPK1 immunoprecipitates were Western blotted, and the blots were probed with anti-HIPK1 (upper panel) or anti-Daxx (lower panel), respectively.
Previous work demonstrated that Daxx preferentially interacted with HIPK3 compared to a kinase-inactive HIPK3 (28). Therefore, we examined whether Daxx preferentially interacted with HIPK1 compared to K219A. HIPK1 and K219A were expressed in cells alone or in combination with Daxx. Direct Western blot analysis of lysates from these cells showed that equal amounts of HIPK1 or K219A were obtained (Fig. 6B). In addition, equal amounts of endogenous or overexpressed Daxx were also obtained. Western blots containing Daxx or HIPK1 immunoprecipitates and probed for HIPK1 or Daxx, respectively, showed that Daxx interacted with HIPK1 to a much greater degree than with K219A (Fig. 6C). This was the case for both endogenous and overexpressed Daxx. Therefore, relocalization of Daxx by HIPK1 likely occurred in large part by the direct HIPK1-Daxx interaction and independently of phosphorylation of Daxx Ser 669. This was corroborated by the colocalization of HIPK1 and Daxx (Fig. 6A).
Phosphorylation of Daxx Ser 669 is important in modulating Daxx activity.
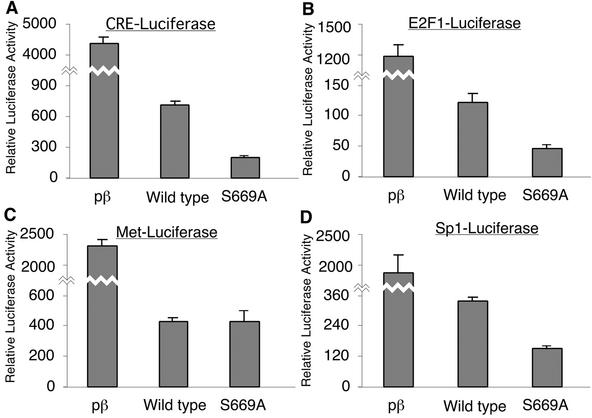
To determine the functional significance of Daxx Ser 669 phosphorylation with respect to transcriptional repression, reporter assays were performed with pβ, wild-type Daxx, and Daxx-S669A (Fig. 7). Increased expression of wild-type Daxx significantly repressed the transcriptional activities of the CRE-luciferase (Fig. 7A), E2F1-luciferase (Fig. 7B), Met-luciferase (Fig. 7C), and Sp1-luciferase (Fig. 7D) reporters relative to the levels with pβ. This is consistent with previous findings characterizing Daxx as a transcriptional repressor (6, 11, 18). Interestingly, Daxx-S669A repressed the CRE-luciferase, E2F1-luciferase, and Sp1-luciferase reporters to a greater degree than wild-type Daxx. For the Met-luciferase reporter, however, there was no significant difference in repression between wild-type Daxx and Daxx-S669A. These experiments demonstrate that phosphorylation of Daxx Ser 669 is important in modulating Daxx activity, possibly by downregulating Daxx-mediated repression.
FIG. 7.
Phosphorylation of Ser 669 modulates Daxx activity. 293 cells were transfected with the CRE-luciferase (A), E2F1-luciferase (B), Met-luciferase (C), or Sp1-Luciferase (D) reporter construct with an empty vector (pβ), pCAGGS/Daxx (wild type), or pCAGGS/Daxx-S669A (S669A) and pCMVβ. Data are presented as relative luciferase activity, determined by normalizing the absolute luciferase values for transfection efficiency by measuring the β-galactosidase activity. In each graph, the ordinate is divided into two scales.
DISCUSSION
The HIPKs are a group of three serine/threonine protein kinases that function as transcriptional regulators. Here we report the first description of HIPK1 expression, localization, and function. In addition, two novel mechanisms are proposed for regulating the activity of Daxx, also a transcriptional regulator. In particular, we found that (i) HIPK1 is ubiquitously expressed in both mice and humans; (ii) HIPK1 localizes predominantly to nuclear speckles, with diffuse staining throughout the nucleus and to a lesser extent throughout the cytoplasm; (iii) HIPK1 physically interacts with Daxx in cells, and this interaction is dependent on an active kinase domain; (iv) HIPK1 relocalizes Daxx from PODs, largely by a direct interaction, providing evidence for altered Daxx activity; and (v) HIPK1 phosphorylates Daxx on Ser 669, and phosphorylation of this residue is important in modulating Daxx activity.
One mechanism for controlling Daxx transcriptional regulatory activity was described previously. SUMO-1-conjugated PML sequesters Daxx to PODs, thereby inhibiting transcriptional regulation by Daxx (12, 16, 17). In the absence of SUMO-1-conjugated PML or PML altogether, Daxx localizes to chromatin. When localized to chromatin, Daxx likely recruits HDAC1 and represses transcription (17). Here, we propose two novel mechanisms, mediated by HIPK1, that regulate Daxx activity. HIPK1 relocalizes Daxx from PODs, presumably to chromatin, where Daxx interacts with HDAC1. In addition, HIPK1 phosphorylates Daxx on Ser 669, a site important in modulating Daxx's transcription-repressive activity.
Transcription-repressive activities were assigned to both HIPK1 and Daxx (4, 15). HIPK2 was shown to provide a regulatory role in a large transcription-repressive complex containing the repressive elements Groucho and HDAC1 (4). Daxx localized to chromatin, where it interacted with HDAC1 and repressed transcription (12, 17). Our data indicate that HIPK1 recruits Daxx from PODs to chromatin, where these proteins likely associate with a large transcription-repressive complex containing HDAC1. Within this complex, HIPK1 provides a modulatory role, at least in part by phosphorylating Daxx.
The relocalization of Daxx from PODs by HIPK1 is dependent on an active kinase domain, as the kinase-inactive HIPK1 K219A was unable to relocate Daxx (Fig. 4A). Although K219A did not relocate Daxx from PODs, relocalization was independent of Daxx phosphorylation on Ser 669, as HIPK1 relocated a Daxx mutant with an alteration in this residue (Fig. 6A). The preferential interaction of Daxx with HIPK1 compared to K219A (Fig. 6C) indicates that Daxx relocalization is dependent on a direct HIPK1-Daxx interaction. It is possible that HIPK1 autophosphorylation is important in this interaction, justifying the necessity for an active kinase domain. Autophosphorylation may alter the HIPK1 conformation, thereby influencing its interactions.
Although increased HIPK1 expression relocalizes Daxx from PODs, POD integrity is not compromised (Fig. 4D), indicating that the HIPK1 effect is specific to Daxx. Daxx has a dynamic localization between PODs and chromatin and may oscillate between these sites in response to various cellular signals. Increased HIPK1 expression dramatically shifts Daxx localization towards chromatin. It is possible that PODs function as a Daxx warehouse, storing Daxx until its transcriptional functions are required. Alternatively, it is possible that Daxx functions both inside and outside of PODs. This is conceivable, as Daxx localization to PODs has been suggested to be necessary for it to propagate apoptotic signals (30, 33).
An opportunity exists for an overlap in HIPK1 and Daxx localization within PODs, as HIPK1 partially localized to PODs. This seems unlikely for several reasons. First, the majority of HIPK1 remains in nuclear speckles different from PODs or is diffused throughout the nucleus. Second, the relocalization of Daxx from PODs suggests that the HIPK1-Daxx interaction occurs elsewhere in the nucleus. This is supported by HIPK1 and Daxx colocalization outside of PODs (Fig. 6A).
Previous work identified an interaction between HIPK3 and Daxx (28). It was inferred that this interaction occurs at the cell periphery, as it was shown to be important in the early steps of Fas-mediated apoptosis. We found that the HIPK1-Daxx interaction occurs within the nucleus. It is possible that the HIPK3-Daxx interaction previously identified also occurred within the nucleus. It will be important to determine the extent to which the functions of the HIPKs overlap.
HIPK1 phosphorylates Daxx on Ser 669. A Daxx mutant containing an alanine in place of the serine at position 669 demonstrated the significance of phosphorylation at this site. This mutant repressed three of the four luciferase reporters examined to a greater degree than wild-type Daxx (Fig. 7). Therefore, phosphorylation at Ser 669 by HIPK1 diminishes the ability of Daxx to repress transcription. However, because the mutated Daxx influenced only three of the four luciferase reporters tested, phosphorylation of Ser 669 may provide specificity in modulating Daxx activity at specific promoters.
A role for HIPK1 in decreasing Daxx transcriptional repression by phosphorylation of Ser 669 appears contradictory to a role for HIPK1 in increasing Daxx transcriptional repression by sequestering Daxx from PODs to chromatin. However, we view these HIPK1 roles as exclusive of one another. Sequestration of Daxx to chromatin may be important in initiating or enhancing Daxx-mediated transcriptional repression. HIPK1 phosphorylation of Daxx may provide precise modifications to Daxx activity, as the Daxx phosphorylation mutant had only a modest effect on transcriptional repression relative to wild-type Daxx and only in three of the four reporters tested.
Ser 669 is phosphorylated even when HIPK1 levels are not increased, demonstrated by the decrease in the residual levels of hyperphosphorylated Daxx in the presence of K219A when this site was mutated (Fig. 5, compare wild-type Daxx with K219A to Daxx-S669 with K219A). Therefore, Daxx Ser 669 is likely phosphorylated by endogenous HIPK1. Ideally, we would like to have examined the affect of HIPK1 on Daxx more directly by including HIPK1 and K219A in the transcriptional reporter assay. Unfortunately, it was difficult to distinguish direct HIPK1 repressive effects from the indirect HIPK1 repressive effects through Daxx.
In addition to Ser 669, six other Daxx residues were found to be phosphorylated by microcapillary liquid chromatography-tandem mass spectrometry. Ser 502 was the only other regulated site of Daxx phosphorylation identified. However, our data demonstrate that Ser 502 is not phosphorylated by HIPK1. An unidentified regulated site of Daxx phosphorylation may also exist, as Daxx-Ser 669 presented as two bands when expressed with HIPK1. This is not surprising, as microcapillary liquid chromatography-tandem mass spectrometry does not normally yield complete protein sequence. The significance of phosphorylation at this site and S502A is unclear. Five Daxx residues were defined as being constitutively phosphorylated. However, lambda phosphatase treatment of Daxx did not cause a significant downshift in basal phosphorylated Daxx by Western blot (Fig. 3A). It is possible that these sites are insensitive to the phosphatase treatment. Alternatively, we have not corroborated phosphorylation at these sites by secondary methods and cannot exclude the possibility that these represent artifactual phosphorylation sites.
Our findings reveal that HIPK1 and Daxx collaborate in regulating transcription. This collaboration is likely important in cell growth and viability control. The HIPKs are related to a group of protein kinases that regulate the transition from growth to differentiation in eukaryotic cells (15, 23). More recent evidence showed that the HIPKs themselves participate in important growth-regulatory pathways (5, 10, 26, 28). In addition, Daxx is an essential gene and may under certain circumstances mediate apoptotic signals (3, 21, 25, 32). Consequently, the regulation of Daxx by HIPK1 may ultimately prove to be important in further elucidating their individual and cooperative roles in cell growth and viability.
Acknowledgments
We thank Stephen Gygi and Ross Tomaino for assistance with mass spectrometry data analysis.
J.A.E. is a Howard Hughes Medical Institute Research Fellow. J.S.M. was supported by a Breast Cancer Research Fellowship from the Department of Defense.
REFERENCES
- 1.Amin, H. M., S. Saeed, and S. Alkan. 2001. Histone deacetylase inhibitors induce caspase-dependent apoptosis and downregulation of daxx in acute promyelocytic leukaemia with t(15;17). Br. J. Haematol. 115:287-297. [DOI] [PubMed] [Google Scholar]
- 2.Cermak, L., A. Imova, A. Pintzas, V. Horeji, and L. Andera. 2001. Molecular mechanisms involved in CD43-mediated apoptosis of TF-1 cells: roles of transcription, Daxx expression and adhesion molecules. J. Biol. Chem. 31:31.. [DOI] [PubMed] [Google Scholar]
- 3.Chang, H. Y., H. Nishitoh, X. Yang, H. Ichijo, and D. Baltimore. 1998. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science 281:1860-1863. [DOI] [PubMed] [Google Scholar]
- 4.Choi, C. Y., Y. H. Kim, H. J. Kwon, and Y. Kim. 1999. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J. Biol. Chem. 274:33194-33197. [DOI] [PubMed] [Google Scholar]
- 5.D'Orazi, G., B. Cecchinelli, T. Bruno, I. Manni, Y. Higashimoto, S. Saito, M. Gostissa, S. Coen, A. Marchetti, G. Del Sal, G. Piaggio, M. Fanciulli, E. Appella, and S. Soddu. 2002. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 4:11-19. [DOI] [PubMed] [Google Scholar]
- 6.Emelyanov, A. V., C. R. Kovac, M. A. Sepulveda, and B. K. Birshtein. 2002. The interaction of Pax5 (BSAP) with Daxx can result in transcriptional activation in B cells. J. Biol. Chem. 17:17.. [DOI] [PubMed] [Google Scholar]
- 7.Epstein, J. A., D. N. Shapiro, J. Cheng, P. Y. Lam, and R. L. Maas. 1996. Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc. Natl. Acad. Sci. USA 93:4213-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fagioli, M., M. Alcalay, P. P. Pandolfi, L. Venturini, A. Mencarelli, A. Simeone, D. Acampora, F. Grignani, and P. G. Pelicci. 1992. Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene 7:1083-1091. [PubMed] [Google Scholar]
- 9.Gygi, S. P., D. K. Han, A. C. Gingras, N. Sonenberg, and R. Aebersold. 1999. Protein analysis by mass spectrometry and sequence database searching: tools for cancer research in the post-genomic era. Electrophoresis 20:310-319. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann, T. G., A. Moller, H. Sirma, H. Zentgraf, Y. Taya, W. Droge, H. Will, and M. L. Schmitz. 2001. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 4:1-10. [DOI] [PubMed] [Google Scholar]
- 11.Hollenbach, A. D., J. E. Sublett, C. J. McPherson, and G. Grosveld. 1999. The Pax3-FKHR oncoprotein is unresponsive to the Pax3-associated repressor hDaxx. EMBO J. 18:3702-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss 3rd, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaelin, W. G., Jr., W. Krek, W. R. Sellers, J. A. DeCaprio, F. Ajchenbaum, C. S. Fuchs, T. Chittenden, Y. Li, P. J. Farnham, M. A. Blanar, et al. 1992. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell 70:351-364. [DOI] [PubMed] [Google Scholar]
- 14.Kim, Y. H., C. Y. Choi, and Y. Kim. 1999. Covalent modification of the homeodomain-interacting protein kinase 2 (HIPK2) by the ubiquitin-like protein SUMO-1. Proc. Natl. Acad. Sci. USA 96:12350-12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, Y. H., C. Y. Choi, S. J. Lee, M. A. Conti, and Y. Kim. 1998. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J. Biol. Chem. 273:25875-25879. [DOI] [PubMed] [Google Scholar]
- 16.Lehembre, F., S. Muller, P. P. Pandolfi, and A. Dejean. 2001. Regulation of Pax3 transcriptional activity by SUMO-1-modified PML. Oncogene 20:1-9. [DOI] [PubMed] [Google Scholar]
- 17.Li, H., C. Leo, J. Zhu, X. Wu, J. O'Neil, E. J. Park, and J. D. Chen. 2000. Sequestration and inhibition of Daxx-mediated transcriptional repression by promyelocytic leukemia protein. Mol. Cell. Biol. 20:1784-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, R., H. Pei, D. K. Watson, and T. S. Papas. 2000. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene 19:745-753. [DOI] [PubMed] [Google Scholar]
- 19.MacGregor, G. R., and C. T. Caskey. 1989. Construction of plasmids that express E. coli beta-galactosidase in mammalian cells. Nucleic Acids Res. 17:2365.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin, S. S., and P. Leder. 2001. Human MCF10A mammary epithelial cells undergo apoptosis following actin depolymerization that is independent of attachment and rescued by Bcl-2. Mol. Cell. Biol. 21:6529-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michaelson, J. S., D. Bader, F. Kuo, C. Kozak, and P. Leder. 1999. Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev. 13:1918-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Missero, C., M. T. Pirro, S. Simeone, M. Pischetola, and R. Di Lauro. 2001. The DNA glycosylase T:G mismatch-specific thymine DNA glycosylase represses thyroid transcription factor-1-activated transcription. J. Biol. Chem. 276:33569-33575. [DOI] [PubMed] [Google Scholar]
- 23.Moilanen, A. M., U. Karvonen, H. Poukka, O. A. Janne, and J. J. Palvimo. 1998. Activation of androgen receptor function by a novel nuclear protein kinase. Mol. Biol. Cell 9:2527-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 25.Perlman, R., W. P. Schiemann, M. W. Brooks, H. F. Lodish, and R. A. Weinberg. 2001. transforming growth factor beta-induced apoptosis is mediated by the adapter protein Daxx that facilitates JNK activation. Nat. Cell Biol. 3:708-714. [DOI] [PubMed] [Google Scholar]
- 26.Pierantoni, G. M., M. Fedele, F. Pentimalli, G. Benvenuto, R. Pero, G. Viglietto, M. Santoro, L. Chiariotti, and A. Fusco. 2001. High mobility group I (Y) proteins bind HIPK2, a serine-threonine kinase protein which inhibits cell growth. Oncogene 20:6132-6141. [DOI] [PubMed] [Google Scholar]
- 27.Pinkas, J., and P. Leder. 2002. MEK1 signaling mediates transformation and metastasis of EpH4 mammary epithelial cells independent of epithelial to mesenchymal transition. Cancer Res. 65:4781-4790. [PubMed]
- 28.Rochat-Steiner, V., K. Becker, O. Micheau, P. Schneider, K. Burns, and J. Tschopp. 2000. FIST/HIPK3: a Fas/FADD-interacting serine/threonine kinase that induces FADD phosphorylation and inhibits Fas-mediated Jun NH2-terminal kinase activation. J. Exp. Med. 192:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slansky, J. E., Y. Li, W. G. Kaelin, and P. J. Farnham. 1993. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol. Cell. Biol. 13:1610-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torii, S., D. A. Egan, R. A. Evans, and J. C. Reed. 1999. Human Daxx regulates Fas-induced apoptosis from nuclear PML oncogenic domains (PODs). EMBO J. 18:6037-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, Y., K. M. Debatin, and H. Hug. 2001. HIPK2 overexpression leads to stabilization of p53 protein and increased p53 transcriptional activity by decreasing Mdm2 protein levels. BMC Mol. Biol. 2:8. [DOI] [PMC free article] [PubMed]
- 32.Yang, X., R. Khosravi-Far, H. Y. Chang, and D. Baltimore. 1997. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 89:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong, S., P. Salomoni, S. Ronchetti, A. Guo, D. Ruggero, and P. P. Pandolfi. 2000. Promyelocytic leukemia protein (PML) and Daxx participate in a novel nuclear pathway for apoptosis. J. Exp. Med. 191:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]