Abstract
Protein kinase CK2 is a multifunctional enzyme which has long been described as a stable heterotetrameric complex resulting from the association of two catalytic (α or α′) and two regulatory (β) subunits. To track the spatiotemporal dynamics of CK2 in living cells, we fused its catalytic α and regulatory β subunits with green fluorescent protein (GFP). Both CK2 subunits contain nuclear localization domains that target them independently to the nucleus. Imaging of stable cell lines expressing low levels of GFP-CK2α or GFP-CK2β revealed the existence of CK2 subunit subpopulations exhibiting differential dynamics. Once in the nucleus, they diffuse randomly at different rates. Unlike CK2β, CK2α can shuttle, showing the dynamic nature of the nucleocytoplasmic trafficking of the kinase. When microinjected in the cytoplasm, the isolated CK2 subunits are rapidly translocated into the nucleus, whereas the holoenzyme complex remains in this cell compartment, suggesting an intramolecular masking of the nuclear localization sequences that suppresses nuclear accumulation. However, binding of FGF-2 to the holoenzyme triggers its nuclear translocation. Since the substrate specificity of CK2α is dramatically changed by its association with CK2β, the control of the nucleocytoplasmic distribution of each subunit may represent a unique potential regulatory mechanism for CK2 activity.
Protein kinase CK2 is a ubiquitous serine/threonine protein kinase, generally described as a stable α2β2 tetramer, where α and β are the catalytic and regulatory subunits, respectively (3). Although its signaling function has long remained obscure, the importance of CK2 is suggested by the evolutionary conservation of the enzyme and by the fact that the disruption of both Saccharomyces cerevisiae genes encoding CK2 catalytic subunits is a lethal event (29). In addition to its role in embryonic development and terminal differentiation, the enzyme is required for normal cell cycle progression (20, 30). Finally, a function of CK2 in cell survival has recently emerged (1).
Many of the identified CK2 substrates that are critical for cell proliferation and viability are localized in different cellular compartments. However, there is controversy as to the localization of CK2 and where its substrates are phosphorylated. Although the current prevailing view of CK2 is a tetrameric enzyme, accumulating evidence also indicates that free populations of both CK2 subunits can exist and exert specific functions in the cell (18, 37). At least in vitro, CK2β exerts a central role in modulating the catalytic activity of CK2 (26). Consequently, it is suspected that in vivo, the substrate specificity of the enzyme is likely to be determined both by subcellular localization and by affinity for its regulatory subunit that brings the kinase in proximity to the substrate.
In a previous study, the behavior of CK2 subunits fused to GFP was characterized in living cells (25). The expressed fusion proteins were functional and interacted with endogenous CK2. Both subunits were mostly nuclear in interphase and dispersed throughout the cytoplasm in mitotic cells. However, this raises the questions of how CK2 subunits are individually addressed in the nuclear compartment, whether they are restricted in their mobility in living cells, and whether their targeting sites are different when they come into contact with each other.
Here, we have analyzed the spatiotemporal organization of the CK2 subunits using live-cell imaging. We show that both nuclear import and export of CK2 subunits are regulated independently of each other and can result in rapid changes of their intracellular steady-state distribution. However, when associated in a stable holoenzyme complex, the two subunits are dynamically retargeted in the cytoplasm through their high-affinity interactions. In addition, our study also shows that the binding of fibroblast growth factor 2 (FGF-2) to the holoenzyme provokes its nuclear accumulation, supporting the concept of a signal-mediated localization, which may result in a sophisticated regulation of the kinase. The results presented here validate the targeting hypothesis for CK2 in live cells and further demonstrate the existence within the cell of a subtle equilibrium between different forms of CK2 subunits. Changes in this equilibrium may have profound effects on the cellular functions of this enzyme.
MATERIALS AND METHODS
Construction of plasmids encoding GFP-CK2 or blue fluorescent protein-CK2 subunits.
The cDNA encoding CK2α, CK2β, or deletion mutants were subcloned from pSG5 CK2α, or CK2β vectors (21) into pEGFPc1, pECFPc1, or pEYFPc1 vectors (Clontech). Mutations of green fluorescent protein (GFP)-CK2α of K75,77 and K74,75,76,77 to A were obtained using a Stratagene kit. Mutation of GFP-CK2β of C109 and C112 to S were generated according to the method given in reference 13. Generation of the CK2β3 mutant was described in reference 23.
Transient transfection and establishment of NIH 3T3 GFP-CK2 subunit cell lines.
NIH 3T3 cells were transiently transfected using Fugene 6 reagent (Roche Molecular Biochemicals) according to the manufacturer's instructions.
To generate GFP-CK2 subunits expressing cell lines, NIH 3T3 cells were transfected with linearized cDNA using Lipofectin reagent (Invitrogen) as recommended by the manufacturer.
Immunofluorescence.
All fixation, permeabilization, and immunostaining procedures were performed at room temperature. Cells grown on glass coverslips were washed in phosphate-buffered saline (PBS) and fixed for 20 min with 4% paraformaldehyde. Permeabilization was performed with 0.5% Triton X-100 in PBS for 10 min. Cells were subsequently washed in PBS and blocked in PBS supplemented with 3% nonfat milk for 30 min, and this was followed by incubation with primary polyclonal antibody (25 μg/ml) raised against the C-terminal domain of CK2β (βc). Three washes with PBS-0.05% Tween 20 were carried out before incubation with affinity-purified cyanin 3-conjugated goat anti-rabbit antibody (Jackson Immunoresearch Laboratories) for 45 min. Cells were washed in PBS, mounted with Cyti Fluor mounting medium, sealed, and left to dry before examination.
Immunoprecipitation.
NIH 3T3 cells transfected with different GFP-CK2β constructs were lysed in ice-cold 20 mM Tris-HCl (pH 7.5)-0.5 M NaCl-0.5% Triton X-100 and complete protease inhibitor cocktail (Roche) for 15 min. Cell lysates were centrifuged, and the supernatants were precleared with protein A-Sepharose beads and then incubated for 2 h with anti-CK2β serum (1/500) or anti-GFP (1/500) (Roche) followed by a 45-min incubation with protein A-Sepharose beads. Beads were washed three times in lysis buffer and used either for kinase assays or resolved by SDS-PAGE and immunoblotted as previously described (25).
Kinase assays.
CK2-specific kinase activity was determined in a 20-μl reaction mixture containing 20 mM Tris HCl (pH 7.4), 200 mM NaCl, 20 mM MgCl2, 15 μM ATP, 150 μM CK2-specific peptide substrate (RRREDEESDDEE), 5 μl of test sample, and 1 μCi of [γ-32P]ATP (3,000 Ci/mmol). The reaction mixture was incubated for 10 min at 25°C and stopped by adding 200 μg of casein and 60 μl of 4% trichloroacetic acid. Samples were incubated 30 min at 4°C and microcentrifuged. Aliquots (50 μl) of the supernatants were then spotted on P81 phosphocellulose paper (Whatman). The papers were washed three times with 0.5% phosphoric acid, and the radioactivity of the samples was finally counted in 5 ml of scintillation liquid.
Protein expression and microinjection.
His6-tagged CK2α (full-length) was cloned by inserting CK2α cDNA in pET28 vector (Invitrogen), expressed in Escherichia coli BL21 and purified by sequential chromatography on nitrilotriacetic acid-agarose and heparin-Sepharose. CK2β was purified as described previously (9). Full-length His6-tagged GFP-CK2α was expressed in baculovirus-infected Sf9 cells after subcloning into pFAST Bac HTb vector (Invitrogen). The protein was purified by sequential chromatography on nitrilotriacetic acid-agarose and heparin-Sepharose. The protein was then dialysed against PBS and concentrated in a Vivaspin microconcentrator (cutoff, 30-kDa; Vivascience). Proteins (4 to 10 mg/ml) were mixed with dextran Texas Red (70 kDa; Molecular Probes) and injected into cells using a semiautomatic microinjector (Eppendorf). Time lapse imaging was performed using a Zeiss Axiovert 35 M microscope equipped with a 32× 0.4 NA LD Achrostigmat objective and a controlled-temperature stage. Time lapse sequences were recorded using a Sony Micromax 5 MHz charge-coupled device camera and IPLab Spectrum software 3.5 (Roper Scientific).
Microscopic analysis of cells expressing GFP fusion proteins.
Cells plated on glass coverslips were fixed with 4% paraformaldehyde for 30 min at 25°C, incubated for 5 min with PBS containing Hoechst 33258 (1 μg/ml), and examined using a Zeiss Axiovert 200 M microscope and 40× 1.3 or 100× 1.3 NA Plan Neofluoar objectives. In the case of confocal laser scanning microscopy, images were collected with a Leica TCS-SP2 laser scanning confocal apparatus coupled to a Leica DMIRBE microscope. The relative fluorescence intensity (RFI) was quantified as a nuclear/cytoplasmic ratio (N/C) using Leica Confocal software (version 2.0). For the other quantification experiments, 150 to 300 transfected cells were scored according to whether the fluorescence of the GFP fusion protein was higher in the nucleus than in the cytoplasm and results were expressed as percentage.
Photobleaching experiments.
Fluorescence loss in photobleaching (FLIP) experiments were performed on a Zeiss confocal microscope (LSM 410), by using the 488-nm laser line and a 63× 1.4 NA oil immersion objective. A nuclear defined region (spot) of the living cells plated in Labtek chambers (Nalgene) was repeatedly imaged and photobleached as follows: 15 s of irradiation was followed by 15 s for imaging the whole cell. This regimen was repeated for a total elapsed time of 270 s. A single z section was imaged before and at time intervals after the bleach. FLIP curves were obtained after quantification of fluorescence of the nucleus or the cytoplasm (in this case, the nucleus was excluded from the evaluated area) using NIH Image software (version 1.62). For each time, the mean RFI in the nucleus and in the cytoplasm was calculated as RFI = [Ft/F0] × 100, where F0 and Ft are the average fluorescence intensity before and after bleaching, respectively. Means for six cells at each time point were then plotted. Fluorescence change due to subsequent image acquisitions was measured on fixed cells and determined as not significant.
Human-murine heterokaryons.
Heterokaryons were prepared as previously described (38) with some modifications. Stably transfected NIH 3T3 cells were plated with HeLa cells and incubated at 37°C. The following day, cells were pretreated for 1 h with cycloheximide (100 μg/ml), fused for 2 min with 50% polyethylene glycol 3350-PBS prewarmed to 37°C, and then incubated in Dulbecco's modified Eagle medium with 10% fetal bovine serum for 2 h. Leptomycin B (LMB)-treated fusions were processed as described above, with the addition of a 1-h pretreatment with 20 nM LMB before cycloheximide treatment and the addition of LMB in the postfusion incubation. Cells were fixed with 4% paraformaldehyde for 10 min and then stained with Texas Red-phalloidin (50 μg/ml) and Hoechst.
Fluorescence correlation spectroscopy.
About 15,000 cells stably expressing either GFP, GFP-CK2α, or GFP-CK2β in Dulbecco's modified Eagle medium-10% serum were moved to eight-chamber Labtek I plates, grown for 24 h at 37°C in 5% CO2, and washed in phenol red-free medium containing 10% serum. Fluorescence correlation spectroscopy (FCS) measurements were made by using a Confocor II fluorescence correlation microscope with a 40× 1.2 NA W C-Apochromat objective (Zeiss) as described previously (40).
The illumination was provided by an ion argon laser with emission line at 488 nm, and the confocal detection volume was less than 1 fl. The fluctuation of fluorescence intensity was analyzed using an autocorrelation function [G(t)] that was fitted to simple one- or two-component three-dimensional diffusion models for GFP-expressing cells and GFP-tagged CK2 subunit-expressing cells, respectively (24). Cells were observed under a microscope and the measuring point (laser-focused point) was fixed either in nuclear or cytoplasmic space.
RESULTS
Identification of an NLS in CK2α.
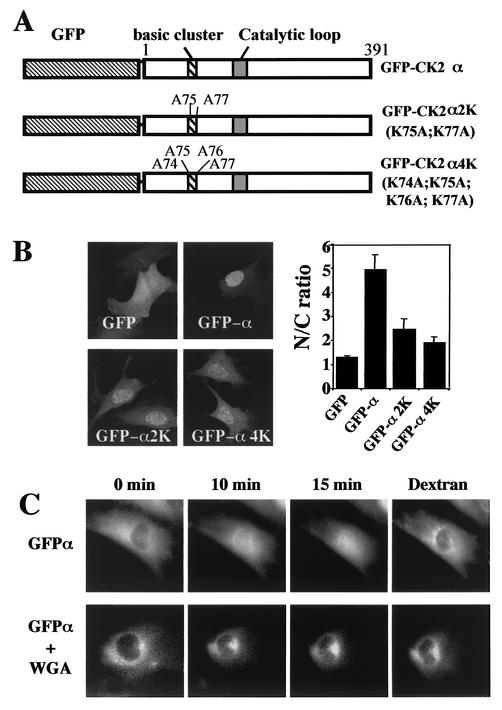
In our previous study, the N-terminal region of CK2α located between residues 74 and 77 (74KKKK77) was characterized as a functional nuclear localization signal (NLS), and it was observed that mutation of K75 and K77 severely impaired the nuclear import of the corresponding GFP-CK2α fusion constructs (25). The functionality of this NLS was further probed by mutating its basic residues to alanine in the wild-type CK2α (Fig. 1A and B). Confocal analysis showed that, in wild-type GFP-CK2α-expressing cells, staining was found mainly in the nucleus (N/C > 5). In contrast, a significant mislocalization to the cytoplasm of the expressed proteins was observed in cells transfected with different NLS mutants (N/C < 3). However, as revealed by the nuclear staining, each single mutant could still efficiently translocate to the nucleus, suggesting either that another potential NLS is present in the C-terminal domain of the protein or that CK2α may bind to NLS-bearing proteins.
FIG. 1.
Nuclear import of the GFP-CK2 α fusion protein. (A) Schematic diagram of constructs. (B) Full-length GFP-CK2α was mutated on either K75A and K77A (GFP-CK2α2K) or K74A, K75A, K76A, and K77A (GFP-CK2α4K), and the constructs were transiently transfected. Cells were observed 24 h later using a confocal microscope. Bar, 10 μm. Cells were also scored for N/C ratio as described in Materials and Methods (error bars, standard deviations). (C) Dynamics of CK2α nuclear import. His6-tagged fusion protein GFP-CK2α expressed in baculovirus-infected Sf9 cells was microinjected into the cytoplasm of NIH 3T3 cells in interphase in the absence or presence of wheat germ agglutinin (WGA). To identify the injected cells, GFP-CK2α was coinjected with dextran Texas Red. Fluorescence images were taken at the indicated time points after injection of the protein.
To gain insights into the dynamics of CK2α nuclear import, recombinant His6-tagged GFP-CK2α was microinjected into the cytoplasm of NIH 3T3 cells and its behavior was monitored by time lapse recordings on 12 different cells. A typical example is shown in Fig. 1C, which illustrates three time points out of 60 total images recorded over a period of 15 min. We observed that the GFP-CK2α protein was translocated into the nucleus and the process was largely completed within a period of 15 min. When GFP-CK2α was injected in the presence of the wheat germ agglutinin, the nuclear accumulation of the protein was eliminated (Fig. 1C). Since it is known that this lectin binds to the central region of the nuclear pore complex, preventing the active nuclear import of proteins, these data suggest that the nuclear uptake of CK2α is mediated by the nuclear pore complex.
The C-terminal domain of CK2β is not required for its nuclear localization.
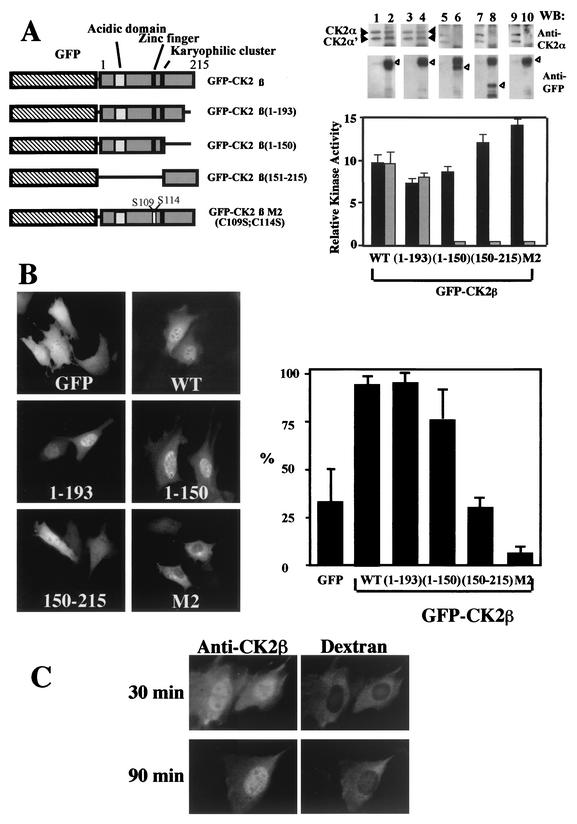
Unlike CK2α, full-length CK2β does not exhibit any recognizable consensus NLS except a karyophilic cluster (147KSSRHHH153) which resembles an extremely weak putative NLS (7). To test whether this sequence or another motif may be responsible for the nuclear localization of CK2β, we transiently expressed wild-type CK2β or several deletion mutants of CK2β in frame with GFP and analyzed their localization (Fig. 2A and B). As previously observed, wild-type GFP-CK2β or its deletion mutant with residues 1 to 193 deleted [GFP-CK2β (1-193)] accumulated in the nucleus (90% of transfected cells). Similarly, GFP-CK2β (1-150) also localized to the nucleus in 75% transfected cells, in a pattern indistinguishable from that of the GFP-CK2β (1-193) construct. In contrast, GFP-CK2β (150-215) was mislocalized, producing a diffused staining pattern throughout the cell (less than 30% of transfected cells showed a brighter fluorescence in the nucleus). As shown in the diagram (Fig. 2A) the CK2β (1-150) construct does not contain the karyophilic signal 147KSSRHHH153 but includes the zinc finger motif, which is essential for CK2β dimerization (9). Thus, cysteines 109 and 112 were replaced by serine residues to disrupt the structure of the zinc finger motif. Expression of this GFP-CK2β M2 mutant showed that the protein did not accumulate within the nucleus but remained in the cytoplasm (Fig. 2B). These results show that the region between residues 1 and 150 contains determinants for the nuclear localization of CK2β and that the integrity of the zinc finger motif is required for a correct nuclear targeting.
FIG. 2.
The C-terminal domain is not required for nuclear import of CK2β. (A) NIH 3T3 cells were transiently transfected with different GFP-CK2β constructs as illustrated in the diagram. Extracts from NIH 3T3 cells transfected with the different constructs were immunoprecipitated with anti-CK2β (black bars) or anti-GFP (grey bars) antibodies, and the corresponding immunoprecipitates were used for CK2 activity determination. Anti-CK2β immunoprecipitates (lanes 1, 3, 5, 7, and 9) or anti-GFP immunoprecipitates (lanes 2, 4, 6, 8, and 10) were also analyzed by Western blotting (WB) for the presence of CK2 subunits using anti-CK2α (solid arrowheads) or anti-GFP (open arrowheads) antibodies. WT, wild type; error bars, standard deviations. (B) Transfected cells were observed for protein localization. Bar, 10 μm. Cells were also scored according to their nuclear staining expressed as a percentage of transfected cells (error bars, standard deviations). (C) Dynamics of CK2β nuclear import. CK2β expressed in E. coli was microinjected into the cytoplasm of NIH 3T3 cells in interphase with dextran Texas Red to identify injected cells. At the indicated time points after injection, cells were fixed and CK2β was detected by indirect immunofluorescence using anti-CK2β antibodies.
We then explored the possibility that the nuclear import of CK2β could be mediated through its interaction with the CK2α subunit. To this end, immunoprecipitations were performed on extracts from NIH 3T3 cells transfected with different GFP-CK2β constructs using anti-GFP and anti-CK2β antibodies, followed by a kinase assay. CK2 activity was present in anti-CK2β immunoprecipitates from all cell extracts (Fig. 2A). In contrast, in anti-GFP immunoprecipitates, CK2 activity was detected only in extracts from cells expressing GFP-CK2β (1-193) but not GFP-CK2β (1-150), indicating that this last mutant is not competent to form a stable complex with CK2α. Immunoblot analysis confirmed that the catalytic α and α′ subunits were present in all anti-CK2β immunoprecipitates. In contrast, in anti-GFP immunoprecipitates, the catalytic subunits were detected only in extracts from cells expressing the GFP-CK2β or GFP-CK2β (1-193) fusion proteins (Fig. 2A).
Thus, CK2β is not targeted to the nucleus by virtue of a stable interaction with endogenous CK2α.
The dynamics of CK2β nuclear import was also analyzed by microinjection of the protein in the cytoplasm of NIH 3T3 cells. Because of its poor solubility at high concentrations, it was not possible to inject the GFP-CK2β fusion protein. Thus, a nonfused version of CK2β was microinjected in the cytoplasm and its localization was monitored by indirect immunofluorescence. Evidence for a nuclear accumulation of the CK2β protein appeared after 30 min and was complete at 90 min (Fig. 2C).
CK2α can shuttle between the nucleus and the cytoplasm.
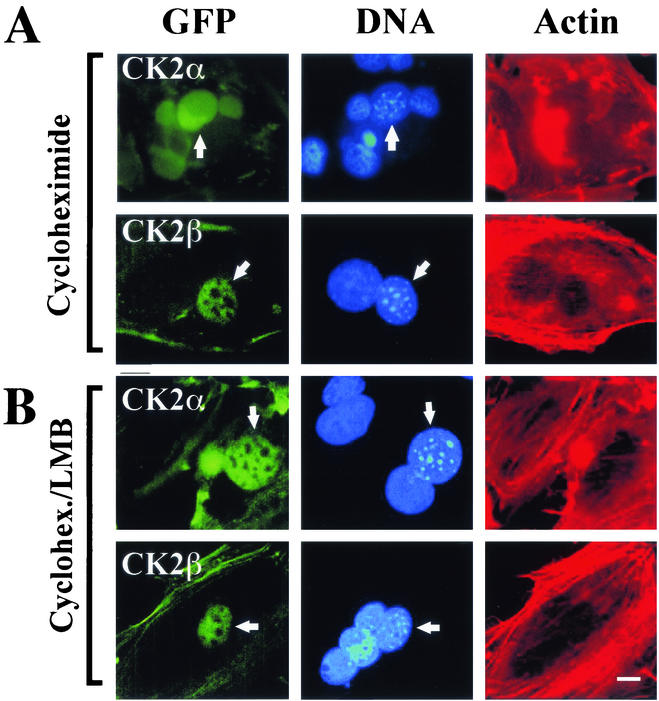
A significant fraction of CK2 subunits could be detected in the cytoplasm, raising the possibility that they can shuttle between the nuclear and the cytoplasmic compartments. To test this hypothesis, NIH 3T3 cells stably expressing the GFP-CK2 subunits were fused in the presence of polyethylene glycol to HeLa cells that did not express the fusion proteins. Heterokaryons were maintained in culture for 2 h in the presence of cycloheximide to prevent de novo synthesis of GFP-CK2 proteins. Fifty heterokaryons containing at least one transfected NIH 3T3 nucleus and one HeLa nucleus were observed. In 90% of them, GFP-CK2α was seen in both the mouse and the human nuclei, indicating that the protein was capable of traversing the nuclear membrane of the murine cell and then accumulating in human nuclei in the absence of new protein synthesis (Fig. 3A). In contrast, GFP-CK2β remained segregated in the nuclei of NIH 3T3 cells, showing that the protein was not exported under our experimental conditions. We therefore determined whether the export of CK2α from the nucleus would be ensured by a nuclear export sequence (NES)-mediated nuclear export system. To test this possibility, we used LMB, which inhibits the nuclear export of complexes consisting of CRM-1-, RanGTP-, and NES-containing proteins (14). In heterokaryons generated from LMB-treated NIH 3T3 cells, GFP-CK2α was seen only in the murine nuclei, indicating that its export requires an association with the export receptor CRM-1 (Fig. 3B). Thus, unlike CK2β, CK2α shuttled, and its apparent nuclear localization was a reflection of its nuclear import being more rapid than its export.
FIG. 3.
CK2α is a shuttling protein. Stably transfected NIH 3T3 cells expressing either GFP-CK2α or GFP-CK2β were pretreated for 1 h with cycloheximide alone (A) or LMB and cycloheximide (B) and fused to nonexpressing HeLa cells. Cells were fixed 2 h after fusion and stained with Hoechst 33258 and with Texas Red-phalloidin to reveal cytoplasmic actin filaments, facilitating detection of heterokaryons with two or more nuclei. Mouse nuclei exhibit a speckled fluorescence when stained with Hoechst 33258 (arrows), while the human nuclei stain evenly, enabling the two to be distinguished easily. GFP fluorescence in the human nucleus is indicative of export of GFP-CK2α from the mouse nucleus (arrows). Each image is representative of >50 heterokaryons analyzed on each coverslip, and two independent heterokaryon assays were performed for each experimental condition. Bar, 5 μm.
Retargeting of CK2 subunits through their high-affinity interactions.
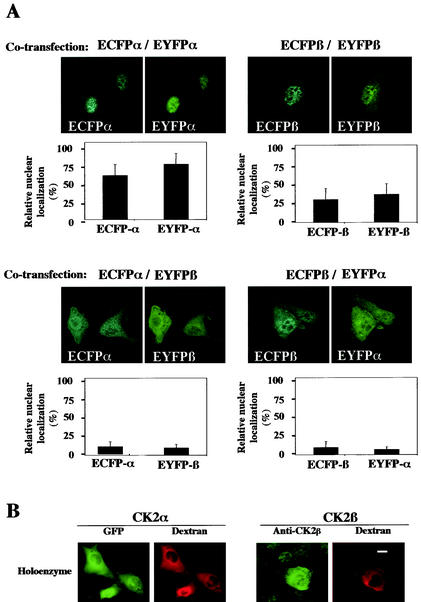
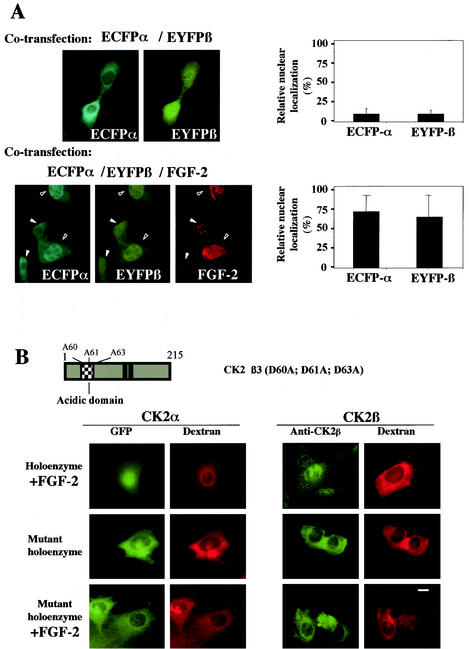
Our data support the notion that both CK2 subunits can reach the nuclear compartment independently. However, once synthesized in the cytoplasm, both proteins may also bind together with high affinity, generating the holoenzyme complex. Whether this complex can also translocate into the nucleus is unknown. To answer this question, NIH 3T3 cells were cotransfected with differently labeled CK2 subunits and dual-color imaging analysis was performed to monitor their localization after 16 h. As expected, cotransfected ECFP-CK2α/EYFP-CK2α or ECFP-CK2β/EYFP-CK2β localized to the nuclear compartment in more than 50 and 30% of cotransfected cells, respectively (Fig. 4A, upper panels). By contrast, in double-labeled live cells expressing similar levels of ECFP-CK2α/EYFP-CK2β or ECFP-CK2β/EYFP-CK2α, both subunits were mainly retained in the cytoplasm. Thus, the localization of both CK2 subunits appeared to change when they were expressed in the same living cell. Only 10% of transfected cells exhibited a brighter nuclear staining (Fig. 4A, lower panels). Further evidence for this retargeting was provided by microinjection experiments. The CK2 holoenzyme was reconstituted in vitro and microinjected in the cytoplasm of NIH 3T3 cells. A typical example of a time lapse image recording is shown in Fig. 4B, which illustrates one time point out of 60 total images recorded over a period of 15 min. Interestingly, the nuclear import of a GFP-CK2α/CK2β complex could not be observed over a period of 90 min after microinjection in the cytoplasm. These observations provide further evidence that the association of CK2 subunits in a stable holoenzyme down regulates their nuclear import.
FIG. 4.
Interaction between the CK2 subunits precludes their nuclear import. (A) NIH 3T3 cells were cotransfected with different combinations of the following plasmids: enhanced cyan fluorescent protein (ECFP)-CK2α/enhanced yellow fluorescent protein (EYFP)-CK2α, ECFP-CK2β/EYFP-CK2β, ECFP-CK2α/EYFP-CK2β, or ECFP-CK2β/EYFP-CK2α. Sixteen hours after transfection a dual-color imaging analysis was performed. Cotransfected cells (150 to 300) were scored according to whether the level of fluorescence of GFP was higher in the nucleus than in the cytoplasm, and the results were expressed as a percentage of the total number of transfected cells. Error bars, standard deviations. (B) A complex between GFP-CK2α and CK2β (holoenzyme) was recombined in vitro and microinjected into the cytoplasm of NIH 3T3 cells in interphase along with 70-kDa dextran Texas Red. Localization of the GFP CK2α was followed by monitoring GFP fluorescence every 15 s. Images shown are after 10 min. Localization of CK2β was detected by indirect immunofluorescence using anti-CK2β antibodies on cells fixed 10 min after injection. Bar, 10 μm.
FGF-2 triggers the nuclear import of the CK2 holoenzyme.
Binding of FGF-2 to the acidic groove of CK2β was shown to activate the holoenzyme (6), and a functional tertiary complex between FGF-2 and the CK2 holoenzyme was demonstrated in vivo, suggesting a potential link between CK2 and the FGF-2 signaling pathway (4). The dual-color imaging analysis illustrated in Fig. 4A was repeated on NIH 3T3 cells cotransfected in the absence or presence of an FGF-2 expression plasmid. As already observed, in double-labeled live cells expressing similar levels of ECFP-CK2α/EYFP-CK2β, a significant fraction of both subunits was retained in the cytoplasm. Strikingly, in FGF-2 expressing cells, the two CK2 subunits were targeted to the nucleus (Fig. 5A). Further evidence for an FGF-2 dependent retargeting was provided by microinjection experiments. An FGF-2/GFP-CK2α/CK2β complex was microinjected into the cytoplasm of NIH 3T3 cells. It was observed that the presence of FGF-2 induced a rapid nuclear accumulation of both CK2 subunits that was observable as early as 5 to 10 min after microinjection (Fig. 5B). As a control, it was verified that binding of FGF-2 was not dissociating the holoenzyme (not shown). In contrast, a complex between FGF-2 and the holoenzyme containing CK2β3, a mutant protein which does not bind FGF-2 (4), was retained in the cytoplasm (Fig. 5B).
FIG. 5.
FGF-2 triggers the nuclear import of CK2 holoenzyme in living cells. (A) NIH 3T3 cells were cotransfected with the following plasmids: ECFP-CK2α/EYFP-CK2β in the absence or presence of an FGF-2 construct. Sixteen hours after transfection a dual-color imaging analysis was performed and the expression of FGF-2 was detected by indirect immunofluorescence using an anti-FGF-2 antibody. Note that in low-FGF-2-expressing cells (solid arrowheads), both CK2 subunits were mainly cytoplasmic, whereas in high-FGF-2-expressing cells (open arrowheads), they were nuclear. Cotransfected cells (150 to 300) were also scored according to whether the level of fluorescence of GFP was higher in the nucleus than in the cytoplasm, and the results were expressed as a percentage of the total number of transfected cells. Error bars, standard deviations. (B) A GFP-CK2α/CK2β complex was incubated with FGF-2 in vitro before injection into the cytoplasm of NIH 3T3 cells in interphase along with 70-kDa dextran Texas Red. Localization of the GFP CK2α was followed by monitoring GFP fluorescence every 15 s. Images shown are after 10 min. Localization of CK2β was detected by indirect immunofluorescence using anti-CK2β antibodies on cells fixed 10 min after injection. Microinjection of a complex between GFP CK2α and the CK2β3 mutant was performed as above in the absence or presence of FGF-2. Representative cells of at least two different experiments are shown for all panels. Bar, 10 μm. A schematic diagram of CK2β3 mutant is shown.
Altogether, these results suggest that the binding of FGF-2 to the cytoplasmic holoenzyme complex can trigger its rapid nuclear accumulation.
CK2 subunits exhibit differential mobility in the nucleus of living cells.
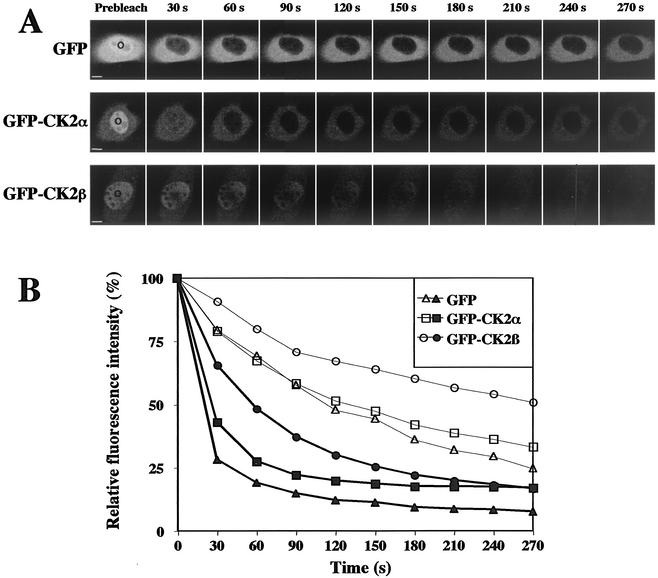
To determine whether the two CK2 subunits were mobile or participated in long-lived interactions with chromatin structures, we took advantage of the FLIP technique (42). Repeated bleaching by high-powered laser pulses in a nucleoplasmic spot of living cells expressing the GFP-tagged CK2 subunits led to a rapid loss of fluorescence in this whole compartment, indicating that all fluorescent CK2 subunits were eventually hited by a bleach pulse in the nuclear compartment (Fig. 6B). This suggests that immobile CK2 subunits are virtually absent. However, the loss of signal intensity was significantly slower than that of GFP alone, showing that both CK2 subunits were by no means as freely diffusible as unfused GFP. We noticed that the kinetics of signal loss were more rapid for GFP-CK2α than for GFP-CK2β (half lives of 30 and 60 s, respectively; n = 6 cells). This difference indicates that the two subunits may diffuse separately in the nuclear compartment. Interestingly, bleaching a nucleoplasmic spot triggered a loss of fluorescence in the cytoplasm (Fig. 6B), showing the rapid and continuous import of cytoplasmic GFP-CK2α and GFP-CK2β subunits into the nucleus.
FIG. 6.
Mobility of CK2 subunits in living cells. NIH 3T3 cells stably expressing GFP, GFP-CK2α, or GFP-CK2β were repeatedly bleached for 15 s in the same nucleoplasmic spot. (A) Images show single z sections and were obtained before (Prebleach) and after the indicated time points. The area of the bleach is indicated with a circle. Bar, 5 μm. (B) Cells were imaged between bleach pulses and the loss of fluorescence of either the whole nucleus (solid symbols) or the cytoplasm (open symbols) was expressed as relative intensity (see Materials and Methods). Curves from cells were averaged (n = 6 cells) and plotted over a 270-s time frame.
CK2 subunits exist as fast- and slow-moving subpopulations in living cells.
FCS can determine dynamics of fluorescent molecules in living cells in a small volume (24). Measuring fluctuations that are processed statistically allows the evaluation of local concentration, diffusion rates, state of aggregation, and molecular interactions of fluorescent proteins. Therefore, FCS experiments were performed on NIH 3T3 cells stably expressing GFP, GFP-CK2α, or GFP-CK2β and the collected data are illustrated in Table 1. The analysis shows that the local concentrations of both CK2 subunits in the nucleus were twofold higher than those in the cytoplasm. Data for GFP were fitted with a one-component model as previously described (11). By contrast, for both CK2 subunits present in the nuclear and the cytoplasmic compartments, two states of different spectroscopic properties could be resolved, using a two-component model. A population of fast-moving CK2 subunits was determined, displaying diffusion constants similar to that of GFP. In addition, a subpopulation of slow-moving CK2 subunits could also be visualized. Estimation of apparent molecular weight indicates that this subpopulation of CK2 subunits is obviously highly restricted in its mobility.
TABLE 1.
Summary of FCS experiments
| Cells (sample no.) | Site | Mean fluorescent-cell count/molecule ± SEM | Mean fluorescent-cell concn ± SEM (nM) | Mean diffusion constant ± SEM (cm2/s)
|
Mean % ± SEM | |
|---|---|---|---|---|---|---|
| Fast | Slow | |||||
| GFP (n = 9)b | Nucleus | 18 ± 0.7a | 472 ± 82a | (2.1 ± 0.1) × 10−7 | 100 | |
| Cytosol | 21 ± 0.4 | 327 ± 37 | (2.2 ± 0.1) × 10−7 | 100 | ||
| GFP-CK2α (n = 40)c | Nucleus | 14 ± 0.6a | 252 ± 22a | (2.7 ± 0.2) × 10−7 | 47 ± 1a | |
| (1.5 ± 0.1) × 10−8a | 53 ± 1a | |||||
| Cytosol | 16 ± 0.7 | 111 ± 12 | (2.4 ± 0.1) × 10−7 | 54 ± 2 | ||
| (1.8 ± 0.1) × 10−8 | 47 ± 2 | |||||
| GFP-CK2β (n = 40)c | Nucleus | 15 ± 0.8a | 333 ± 29a | (2.5 ± 0.2) × 10−7 | 42 ± 1 | |
| (1.6 ± 0.1) × 10−8a | 56 ± 1 | |||||
| Cytosol | 17 ± 0.9 | 185 ± 15 | (2.7 ± 0.2) × 10−7 | 43 ± 2 | ||
| (2.2 ± 0.2) × 10−8 | 57 ± 2 | |||||
Nucleus-cytoplasm significant difference (α = 0.05).
One-component fitting.
Two-component fitting.
DISCUSSION
Functional domains involved in the nucleocytoplasmic distribution of CK2 subunits.
In this study we showed that a unique cluster of predominantly basic amino acids, at position 74 to 77, closely homologous to the NLS of the simian virus 40 T antigen (22) conferred nuclear localization to CK2α and that specific lysine residues K75 and K77 are instrumental for efficient nuclear targeting. Since this basic cluster located in subdomain III of the kinase has also been clearly implicated in substrate recognition (34), one might hypothesize that this basic sequence which folds as an exposed α helix in the CK2α structure (28) represents the prototype of a bifunctional domain. Although the regulatory CK2β subunit does not contain any recognizable NLS, it is also imported to the nucleus. Disruption of the zinc finger motif generates a monomeric form of CK2β (8), and our results show that dimerization of the regulatory subunit is a prerequisite for its nuclear localization. They also show that the N-terminal half of the protein, which is unable to interact with CK2α (16; also this study), gains access to the nucleus. Thus, the region comprising positions 1 to 150 represents a minimal functional domain in CK2β: it contains both the zinc finger domain, which is pivotal in the structure of the CK2β dimer, and specific sequences that are instrumental for its nuclear import. It is possible that this region contains a nuclear import signal unrelated to the classical basic NLS domain as previously described (33). Alternatively, since in vivo, CK2β can interact with various cellular proteins (5, 10, 17), it may piggyback its way into the nucleus.
Dynamics of CK2 subunits in living cells and relevance for CK2 functions.
Real-time fluorescence microscopy was applied for the first time to analyze the behavior of CK2 subunits after microinjection in the cytoplasm of living cells. A rapid nuclear import of both CK2 subunits was observed. Although a direct comparison between the two subunits could not be performed, we suspect that the nuclear import of CK2α and CK2β is mediated by independent mechanisms. This is in keeping with the presence of a classical NLS motif in CK2α, which is absent in the regulatory subunit. We have observed that CK2α was both imported into and exported from nuclei in the absence of new protein synthesis. Since CK2α export is sensitive to LMB, it may use an exportin 1/CRM1-dependent pathway. CK2α contains a leucine-rich sequence exhibiting a strong similarity with the NES described in human immunodeficiency virus Rev (15). However, we failed to demonstrate that this highly conserved sequence is sufficient to mediate the nuclear export of a nonshuttling protein. One consequence of the CK2α shuttling would be the presence of small pools of CK2α in both the cytoplasmic and the nuclear compartments at all times. Indeed a complex between the catalytic CK2 subunit and PP2A has been observed in the cytoplasm of mitogen-starved cells (21). Thus, the shuttling of a CK2 catalytic subunit which is not associated with CK2β may allow the cell to rapidly alter the subcellular localization of the kinase to emphasize either its nuclear or cytoplasmic functions.
Our FLIP experiments show that both CK2 subunits belong to the family of highly mobile nuclear proteins, in agreement with a concept proposing rapid and continuous diffusional movement of proteins in the nuclear compartment in vivo (31). Immobile nuclear proteins such as the core histone H2B have a residence time on chromatin of several hours and show virtually no loss over FLIP period up to 60 min. In contrast, the significantly more mobile HMG proteins show loss curves similar to the ones we observed for CK2 subunits (27). Remarkably, the kinetic behavior of each CK2 subunit is different. This apparent difference in mobility suggests that, as a result of their different conformations or modes of molecular interaction, they diffuse individually at different rates. The resolution of FLIP analysis does not reach the molecular level, and our results represent the sum of movement of the CK2 subunits in all of their possible forms: bound, free, activated, inactivated, or bound to different partners. Nevertheless, new insights in this direction were provided by our FCS analysis. A subpopulation of fast-moving CK2 subunits was detected that would screen a large region of the nucleus for the presence of potential binding partners or nuclear structures. We assume that in the nucleoplasm, the two proteins can interact, at least transiently, even though our data might suggest that they are predominantly segregated in this compartment. FCS analysis also provides evidence for the existence of a subpopulation of slow-moving CK2 subunits that could represent integral components of less mobile structures, such as nuclear matrix or chromosome scaffold. Interestingly, a rapid translocation of CK2 to nuclear matrix associated with genomic activation upon induction of cell growth has been described in an in vivo physiological model (2, 19). It could be also imagined that the previously described reversible polymerization of CK2 subunits (41) might also create transient, less-mobile nuclear complexes.
The most significant finding in this study is the demonstration that the association of both CK2 subunits in a stable tetrameric holoenzyme eliminates their nuclear import. Binding of CK2α to CK2β may induce conformational changes allowing the anchorage of the complex in the cytoplasm. Alternatively, the CK2α NLS can be occluded in the holoenzyme through interactions with complementary segments of the CK2β subunit. Since a positively charged region (residues 69 to 80) located on the αC helix of the catalytic subunit (23) encompasses the NLS identified in this study, it is tempting to speculate that the intramolecular masking of the NLS may render the holoenzyme resistant to nuclear import or prevent the nuclear reimport of shuttling CK2α. This assumption also means that the cytoplasmic functions of CK2α may be controlled by a threshold effect in which CK2β could gradually titrate out the catalytic subunit from the nucleus. Several observations suggest that this effect can be counteracted through the binding of CK2β-specific ligands such as FGF-2: (i) cytoplasmic CK2 subunits were retargeted into the nucleus of FGF-2 expressing cells; (ii) when injected into the cytoplasm, an FGF-2-CK2 holoenzyme complex was rapidly imported into the nucleus. Together, these results support the paradigm that the interaction with CK2β influences the kinetics of CK2α shuttling. They also provide direct support for the concept that cell signaling pathways may generate allosteric binding ligands that would target the nuclear translocation of the holoenzyme. Based on the present and previous studies, we propose the model illustrated in Fig. 7 as a molecular framework in which to begin to understand the molecular details of how the subcellular localization of the CK2 subunits is regulated. In this model, the dynamic nature of nucleocytoplasmic trafficking appears to be an essential feature of CK2 subunits as regulators of important cellular functions. Evidence is accumulating now that not only does CK2 act in the form of a classical heterotetrameric holoenzyme but also that free populations of both CK2 subunits exist, either alone or acting in association with several different partners (18, 36, 37). In a similar vein, it has been observed that adenovirus infection of HeLa cells targets the catalytic and regulatory subunits to morphologically distinct structures of the cell nucleus (35).
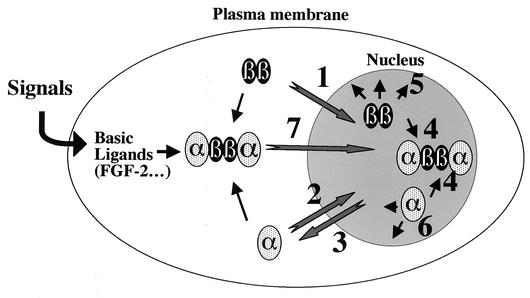
FIG. 7.
Model for intracellular dynamics of CK2 subunits. As previously suggested (12) and suggested in this study, each neo-synthesized CK2 subunit is separately imported into the nucleus through specific mechanisms (1 and 2). Unlike CK2β, nuclear CK2α can be exported back through an exportin 1/Crm1-dependent pathway (3). Once in the nucleus, CK2 subunits are freely mobile and may roam by diffusion through the nucleoplasm at a high rate, where they can either associate in a stochastic manner to generate the holoenzyme (4) or interact with appropriate binding partners or nuclear structures, leading to a subpopulation of slow-moving CK2 subunits highly restricted in their mobility (5 and 6). Likewise, cytoplasmic CK2 subunits may also bind to each other generating, the holoenzyme which would remain in the cytoplasm to phosphorylate specific protein substrates in this compartment. Cell signaling pathways may also generate allosteric binding ligands (such as FGF-2) that would target the nuclear translocation of the holoenzyme (7).
As previously observed in vitro, the molecular recognition of substrates by CK2α is dramatically changed by its association with CK2β (32). Therefore, binding of CK2β to the catalytic subunit may be important for high-affinity docking interactions, for ensuring efficient enzymatic reaction but also for regulating subcellular localization of the protein kinase that may be destined for cytoplasmic or nuclear sequestration.
Thus, our data are leading to a dynamic view of CK2 that diverges from the historic “hardwired-holoenzyme” concept in which the α and β subunits stay in a cell compartment as an indissociable complex. In this context, analysis of the biochemical mechanisms regulating the dynamic localization of CK2 subunits in space and time will be of central importance (39).
Acknowledgments
We acknowledge J. Lamarre for the critical reading of the manuscript, D. Grunwald for the confocal analysis, and T. Misteli for his helpful comments during the FLIP analysis. We thank M. Yoshida for providing us with LMB and C. Touriol for FGF-2 plasmids.
This work was supported by grants from the INSERM, the CNRS (contract 8BC06G), the Commissariat à l'Energie Atomique, the Association pour la Recherche contre le Cancer (réseau ARECA), and the Ligue Nationale contre le Cancer.
O. Filhol and A. Nueda contributed equally to this publication.
REFERENCES
- 1.Ahmed, K., D. A. Gerber, and C. Cochet. 2002. Joining the cell survival squad: an emerging role for protein kinase CK2. Trends Cell Biol. 12:226-230. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, K., S. Yenice, A. Davis, and S. A. Goueli. 1993. Association of casein kinase 2 with nuclear chromatin in relation to androgenic regulation of rat prostate. Proc. Natl. Acad. Sci. USA 90:4426-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allende, J. E., and C. C. Allende. 1995. Protein kinases. 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 9:313-323. [DOI] [PubMed] [Google Scholar]
- 4.Bailly, K., F. Soulet, D. Leroy, F. Amalric, and G. Bouche. 2000. Uncoupling of cell proliferation and differentiation activities of basic fibroblast growth factor. FASEB J. 14:333-344. [PubMed] [Google Scholar]
- 5.Boldyreff, B., and O. G. Issinger. 1997. A-Raf kinase is a new interacting partner of protein kinase CK2 beta subunit. FEBS Lett. 403:197-199. [DOI] [PubMed] [Google Scholar]
- 6.Bonnet, H., O. Filhol, I. Truchet, P. Brethenou, C. Cochet, F. Amalric, and G. Bouche. 1996. Fibroblast growth factor-2 binds to the regulatory beta subunit of CK2 and directly stimulates CK2 activity toward nucleolin. J. Biol. Chem. 271:24781-24787. [DOI] [PubMed] [Google Scholar]
- 7.Boulikas, T. 1996. Nuclear import of protein kinases and cyclins. J. Cell Biochem. 60:61-82. [DOI] [PubMed] [Google Scholar]
- 8.Canton, D. A., C. Zhang, and D. W. Litchfield. 2001. Assembly of protein kinase CK2: investigation of complex formation between catalytic and regulatory subunits using a zinc-finger-deficient mutant of CK2β. Biochem. J. 358:87-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chantalat, L., D. Leroy, O. Filhol, A. Nueda, M. J. Benitez, E. M. Chambaz, C. Cochet, and O. Dideberg. 1999. Crystal structure of the human protein kinase CK2 regulatory subunit reveals its zinc finger-mediated dimerization. EMBO J. 18:2930-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, M., and J. A. Cooper. 1997. The beta subunit of CKII negatively regulates Xenopus oocyte maturation. Proc. Natl. Acad. Sci. USA 94:9136-9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Y., J. D. Muller, Q. Ruan, and E. Gratton. 2002. Molecular brightness characterization of EGFP in vivo by fluorescence fluctuation spectroscopy. Biophys. J. 82:133-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chester, N., I. J. Yu, and D. R. Marshak. 1995. Identification and characterization of protein kinase CKII isoforms in HeLa cells. Isoform-specific differences in rates of assembly from catalytic and regulatory subunits. J. Biol. Chem. 270:7501-7514. [DOI] [PubMed] [Google Scholar]
- 13.Deng, W. P., and J. A. Nickoloff. 1992. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem. 200:81-88. [DOI] [PubMed] [Google Scholar]
- 14.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 15.Gerace, L. 1995. Nuclear export signals and the fast track to the cytoplasm. Cell 82:341-344. [DOI] [PubMed] [Google Scholar]
- 16.Graham, K. C., and D. W. Litchfield. 2000. The regulatory beta subunit of protein kinase CK2 mediates formation of tetrameric CK2 complexes. J. Biol. Chem. 275:5003-5010. [DOI] [PubMed] [Google Scholar]
- 17.Grein, S., K. Raymond, C. Cochet, W. Pyerin, E. M. Chambaz, and O. Filhol. 1999. Searching interaction partners of protein kinase CK2beta subunit by two-hybrid screening. Mol. Cell. Biochem. 191:105-109. [PubMed] [Google Scholar]
- 18.Guerra, B., S. Siemer, B. Boldyreff, and O. G. Issinger. 1999. Protein kinase CK2: evidence for a protein kinase CK2β subunit fraction, devoid of the catalytic CK2α subunit, in mouse brain and testicles. FEBS Lett. 462:353-357. [DOI] [PubMed] [Google Scholar]
- 19.Guo, C., S. Yu, A. T. Davis, and K. Ahmed. 1999. Nuclear matrix targeting of the protein kinase CK2 signal as a common downstream response to androgen or growth factor stimulation of prostate cancer cells. Cancer Res. 59:1146-1151. [PubMed] [Google Scholar]
- 20.Hanna, D. E., A. Rethinaswamy, and C. V. Glover. 1995. Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae. J. Biol. Chem. 270:25905-25914. [DOI] [PubMed] [Google Scholar]
- 21.Heriche, J. K., F. Lebrin, T. Rabilloud, D. Leroy, E. M. Chambaz, and Y. Goldberg. 1997. Regulation of protein phosphatase 2A by direct interaction with casein kinase 2α. Science 276:952-955. [DOI] [PubMed] [Google Scholar]
- 22.Kalderon, D., B. L. Roberts, W. D. Richardson, and A. E. Smith. 1984. A short amino acid sequence able to specify nuclear location. Cell 39:499-509. [DOI] [PubMed] [Google Scholar]
- 23.Leroy, D., J. K. Heriche, O. Filhol, E. M. Chambaz, and C. Cochet. 1997. Binding of polyamines to an autonomous domain of the regulatory subunit of protein kinase CK2 induces a conformational change in the holoenzyme. A proposed role for the kinase stimulation. J. Biol. Chem. 272:20820-20827. [DOI] [PubMed] [Google Scholar]
- 24.Maiti, S., U. Haupts, and W. W. Webb. 1997. Fluorescence correlation spectroscopy: diagnostics for sparse molecules. Proc. Natl. Acad. Sci. USA 94:11753-11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martel, V., O. Filhol, A. Nueda, D. Gerber, M. J. Benitez, and C. Cochet. 2001. Visualization and molecular analysis of nuclear import of protein kinase CK2 subunits in living cells. Mol. Cell. Biochem. 227:81-90. [PubMed] [Google Scholar]
- 26.Meggio, F., B. Boldyreff, O. Marin, L. A. Pinna, and O. G. Issinger. 1992. Role of the beta subunit of casein kinase-2 on the stability and specificity of the recombinant reconstituted holoenzyme. Eur. J. Biochem. 204:293-297. [DOI] [PubMed] [Google Scholar]
- 27.Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D. T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877-881. [DOI] [PubMed] [Google Scholar]
- 28.Niefind, K., B. Guerra, L. A. Pinna, O. G. Issinger, and D. Schomburg. 1998. Crystal structure of the catalytic subunit of protein kinase CK2 from Zea mays at 2.1 A resolution. EMBO J. 17:2451-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padmanabha, R., J. L. Chen-Wu, D. E. Hanna, and C. V. Glover. 1990. Isolation, sequencing, and disruption of the yeast CKA2 gene: casein kinase II is essential for viability in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:4089-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pepperkok, R., P. Lorenz, W. Ansorge, and W. Pyerin. 1994. Casein kinase II is required for transition of G0/G1, early G1, and G1/S phases of the cell cycle. J. Biol. Chem. 269:6986-6991. [PubMed] [Google Scholar]
- 31.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 32.Pinna, L. A., and F. Meggio. 1997. Protein kinase CK2 (“casein kinase-2”) and its implication in cell division and proliferation. Prog. Cell Cycle Res. 3:77-97. [DOI] [PubMed] [Google Scholar]
- 33.Pollard, V. W., W. M. Michael, S. Nakielny, M. C. Siomi, F. Wang, and G. Dreyfuss. 1996. A novel receptor-mediated nuclear protein import pathway. Cell 86:985-994. [DOI] [PubMed] [Google Scholar]
- 34.Sarno, S., B. Boldyreff, O. Marin, B. Guerra, F. Meggio, O. G. Issinger, and L. A. Pinna. 1995. Mapping the residues of protein kinase CK2 implicated in substrate recognition: mutagenesis of conserved basic residues in the alpha-subunit. Biochem. Biophys. Res. Commun. 206:171-179. [DOI] [PubMed] [Google Scholar]
- 35.Souquere-Besse, S., E. Pichard, O. Filhol, V. Legrand, M. Rosa-Calatrava, A. G. Hovanessian, C. Cochet, and F. Puvion-Dutilleul. 2002. Adenovirus infection targets the cellular protein kinase CK2 and RNA-activated protein kinase (PKR) into viral inclusions of the cell nucleus. Microsc. Res. Technol. 56:465-478. [DOI] [PubMed] [Google Scholar]
- 36.Stalter, G., S. Siemer, E. Becht, M. Ziegler, K. Remberger, and O. G. Issinger. 1994. Asymmetric expression of protein kinase CK2 subunits in human kidney tumors. Biochem. Biophys. Res. Commun. 202:141-147. [DOI] [PubMed] [Google Scholar]
- 37.Stigare, J., N. Buddelmeijer, A. Pigon, and E. Egyhazi. 1993. A majority of casein kinase II alpha subunit is tightly bound to intranuclear components but not to the beta subunit. Mol. Cell. Biochem. 129:77-85. [DOI] [PubMed] [Google Scholar]
- 38.Stommel, J. M., N. D. Marchenko, G. S. Jimenez, U. M. Moll, T. J. Hope, and G. M. Wahl. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18:1660-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teruel, M. N., and T. Meyer. 2000. Translocation and reversible localization of signaling proteins: a dynamic future for signal transduction. Cell 103:181-184. [DOI] [PubMed] [Google Scholar]
- 40.Tilo, J., and Jankan, R. 2001. Confocor 2, the second generation of fluorescence correlation microscopes, p. 331-345. In R. Rigler and E. S. Elson (ed.), Fluorescence correlation spectroscopy: theory and applications. Springer-Verlag, Berlin, Germany.
- 41.Valero, E., S. De Bonis, O. Filhol, R. H. Wade, J. Langowski, E. M. Chambaz, and C. Cochet. 1995. Quaternary structure of casein kinase 2. Characterization of multiple oligomeric states and relation with its catalytic activity. J. Biol. Chem. 270:8345-8352. [DOI] [PubMed] [Google Scholar]
- 42.White, J., and E. Stelzer. 1999. Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 9:61-65. [DOI] [PubMed] [Google Scholar]