Abstract
The mouse gene Fv1 encodes a saturable restriction factor that selectively blocks infection by N-tropic or B-tropic murine leukemia virus (MLV) strains. Despite the absence of an Fv1 gene, a similar activity is present in humans that blocks N-MLV infection (Ref1). Moreover, some non-human primate cell lines express a potentially related inhibitor of HIV-1 and/or SIVmac infection (Lv1). Here, we examine the spectrum of retrovirus-restricting activities expressed by human and African green monkey cell lines. Human cells restrict N-MLV and equine infectious anemia virus (EIAV), but not HIV-1, HIV-2, SIVmac or SIVagm, whilst AGM cells restrict N-MLV, EIAV, HIV-1, HIV-2 and SIVmac. Remarkably, in each example examined, restriction of infection by a given retrovirus can be abrogated at least partially by saturation with another retrovirus, provided that it is also restricted but regardless of whether it is closely related. These data suggest that restriction factors in human and non-human primate cells are able to recognize and block infection by multiple, widely divergent retroviruses and that the factors themselves may be related.
Keywords: HIV/Lv1/MLV/Ref1/restriction
Introduction
Retroviruses are dependent on a variety of host cell functions for the completion of their life cycle. As such, retroviral tropism is determined primarily by whether components of a given retrovirus are able to exploit each of the host cell factors required for viral replication. However, retroviral tropism can also be restricted by the presence of dominant-acting inhibitory activities, expressed by target cells, that prevent the efficient progression of particular steps in the viral life cycle. Presumably, these cellular inhibitors have arisen as an evolutionary consequence of pathogenic retrovirus epidemics. An example is the recently identified cellular protein, CEM15, that acts in human immunodeficiency virus type 1 (HIV-1)-producing cells to attenuate the infectivity of progeny virions. The inhibitory effect of CEM15 is counteracted by the viral Vif protein (Sheehy et al., 2002).
A distinct class of inhibitory activities is exemplified by the Fv1-mediated restriction of murine leukemia virus (MLV) infection (reviewed in Jolicoeur, 1979; Goff, 1996). Fv1 was first described as a dominant, heritable trait in inbred mouse strains that confers resistance to infection by particular MLV strains (Lilly, 1970; Pincus et al., 1971). One of the two major alleles of Fv1, Fv1n, permits infection by N-tropic but restricts B-tropic MLV strains, while the other, Fv1b, permits infection of B-tropic infection but restricts N-tropic infection. At low levels of infection, the resistance conferred by Fv1 can be substantial, but is overcome by inoculating cells with high titers of restricted virions (Decleve et al., 1975; Tennant et al., 1979; Boone et al., 1990).
Infection is blocked in restricting cells at a post-entry step, after substantial levels of reverse transcription have occurred, but before proviral integration (Jolicoeur and Baltimore, 1976; Pryciak and Varmus, 1992). The precise mechanism of action of Fv1 is unknown, but the viral determinants that confer sensitivity to restriction have been mapped to the capsid (CA) domain of MLV Gag (DesGroseillers and Jolicoeur, 1983), and a single amino acid substitution, arginine to glutamate at position CA110, is sufficient to convert an N-tropic MLV strain to B-tropism, and vice versa (Kozak and Chakraborti, 1996). The Fv1 gene has been identified by positional cloning, and encodes a protein with ∼60% homology to the Gag proteins encoded by the HERV-L and MERV-L families of endogenous retroviruses (Best et al., 1996; Benit et al., 1997). Based in part on the specific restriction of N- and B-MLV strains by reciprocal Fv1 variants, the simplest model for restriction involves direct recognition of the capsid of incoming virions by the Fv1 protein (Goff, 1996; Stoye, 1998; Bishop et al., 2001), but demonstration of a direct interaction has proved elusive.
Non-murine mammalian cells do not encode a gene with strong sequence homology to Fv1 (Best et al., 1996). Nevertheless, cells from several mammalian species, including humans, are able specifically to restrict N-tropic MLV (Towers et al., 2000, 2002). Remarkably, the same single amino acid within CA that differentiates between N- and B-tropism in mice also determines tropism for human cells. Because of this, and other similarities to Fv1-mediated restriction (see below), non-murine cells are assumed to express a factor, termed Ref1 in humans, that restricts MLV infection. In fact, the only obvious difference in the phenotypes conferred by Fv1b and Ref1 is that Fv1 acts primarily after the completion of reverse transcription, while Ref1 prevents the accumulation of reverse transcripts (Jolicoeur and Baltimore, 1976; Pryciak and Varmus, 1992; Towers et al., 2000).
Post-entry, pre-integration restrictions to retroviral tropism are not confined to MLV. HIV-1 infection of several non-human primates, for example macaques, is blocked at an early step (Shibata et al., 1995; Himathongkham and Luciw, 1996), even though the HIV-1 envelope protein is fully able to support the replication of chimeric viruses containing SIVmac Gag-Pol proteins in macaque cells (Shibata et al., 1991; Li et al., 1992, 1995; Luciw et al., 1995; Joag et al., 1996). In addition, the innate resistance of primate cells to either HIV-1 or SIVmac infection is observed even when virions are pseudotyped with the pan-tropic vesicular stomatitis virus envelope glycoprotein (VSV-G) (Hofmann et al., 1999). We have recently presented compelling evidence that the post-entry resistance of primate cells to infection by HIV-1 and SIVmac is due to the presence of an Fv1-like inhibitor (Besnier et al., 2002; Cowan et al., 2002). The resistant phenotype is dominant in human–monkey heterokaryons, is independent of the route of virus entry and acts prior to the completion of reverse transcription. Moreover, we have also demonstrated that the differential susceptibility of HIV-1 and SIVmac to restriction in some primate cell lines is determined by sequences within the CA-p2 region of Gag (Cowan et al., 2002). The restriction factor whose existence in non-human primates is inferred by these findings is termed Lv1.
One of the shared hallmarks of Fv1-, Ref1- and Lv1-mediated resistance to retroviral infection is that the block is most evident at a low multiplicity of infection (m.o.i.), and that infection is cooperative at a high m.o.i. (Decleve et al., 1975; Duran-Troise et al., 1977; Tennant et al., 1979; Besnier et al., 2002; Cowan et al., 2002; Towers et al., 2002). Where restriction is potent (≥20-fold), pronounced non-linearity, or multi-hit kinetics, are evident in viral titration curves. In other words, infection by an otherwise restricted virus is strongly facilitated by the presence of other restricted virion particles. This ‘abrogation’ of restriction by high doses of restricted virions is consistent with the presence of a saturable restriction factor and not with the lack of an essential activity in restricting target cells. Furthermore, the abrogating virus must itself be restricted, and recognized by the restriction factor, in order to saturate it and facilitate infection. For example, treatment of target cells with high levels of N-MLV abrogates Fv1b- or Ref1-mediated restriction of N-MLV reporter virus infection, whilst treatment with unrestricted B-MLV does not (Boone et al., 1990; Towers et al., 2002).
We have previously used abrogation and other assays to demonstrate that HIV-1 and SIVmac are differentially restricted in cell lines from various primate species (Besnier et al., 2002; Cowan et al., 2002). In this study, we have found that some human and non-human primate cell lines can restrict diverse retroviruses, and that in each case examined, restriction can be abrogated by a second retrovirus, provided that it is also restricted in the same cell line. Abrogation of restriction to one retrovirus by another does not appear to depend on the sequence homology between the two viruses. These data indicate that restriction factors in primates are capable of targeting multiple, widely divergent retroviruses.
Results
Restriction of N-MLV in primate cells
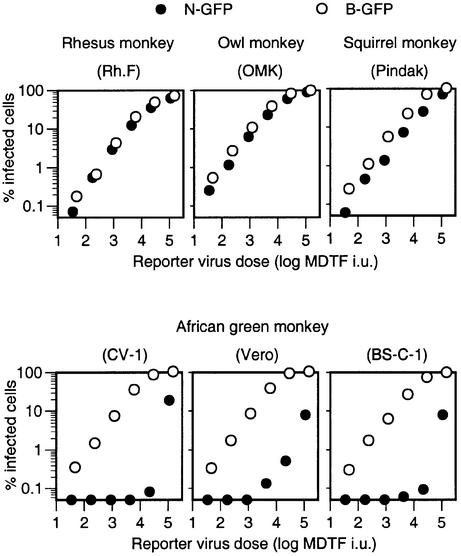
It has been shown previously that N-MLV is restricted by an Fv1-like activity in human and African green monkey (AGM) cells (Towers et al., 2000). Furthermore lentivirus infection is restricted in Old World (rhesus and African green) as well as New World (owl and squirrel) monkey cell lines (Besnier et al., 2002; Cowan et al., 2002). To determine whether the resistance to primate lentiviruses correlates with resistance to MLVs, we examined the distribution of N- or B-MLV restricting factors in primates. N-green fluorescent protein (GFP) and B-GFP vectors were titrated on a series of cell lines from primate species that we have shown previously to restrict infection by HIV-1, SIVmac or both. As is shown in Figure 1, three AGM cell lines exhibit a Ref1-like phenotype, in that they were less susceptible (>100-fold) to infection by N-GFP than by B-GFP. Moreover, at high inoculum, the titration curves of N-MLV revealed multi-hit kinetics of infection. In contrast, rhesus and owl monkey cell lines, which restrict HIV-1, did not restrict either N-GFP or B-GFP detectably. A squirrel monkey cell line that is highly resistant to SIVmac was marginally more susceptible to B-GFP than to N-GFP, but this difference was only ∼2-fold.
Fig. 1. Restriction of N-MLV in AGM cell lines but not those of other primates that restrict HIV-1 or SIVmac. Each primate cell line was infected with serially diluted N-GFP or B-GFP vectors. Each vector stock was also titrated on Fv1-null MDTF cells, and the inoculum level is given in MDTF infectious units (i.u.). The percentage of infected (GFP-positive) cells is plotted for each level of N-GFP or B-GFP inoculum.
Abrogation of restriction to MLV infection in AGM cells by primate lentiviruses
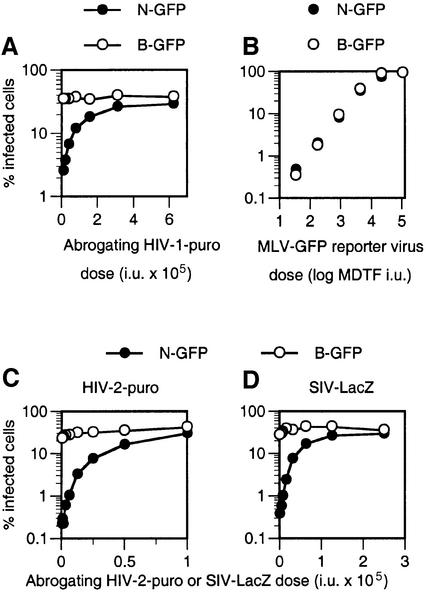
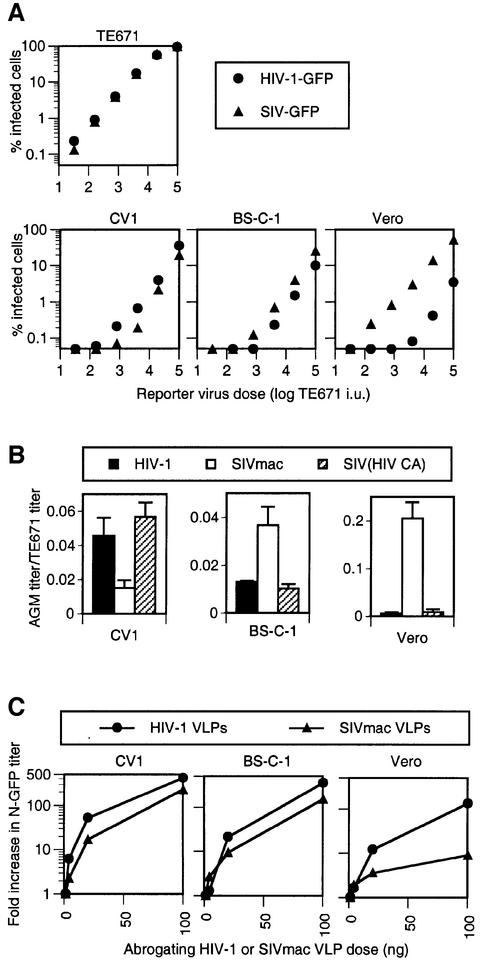
In order to investigate the relationship between factors that restrict infection by MLV and lentiviruses, we performed a series of experiments in AGM cells, which restrict both. Because restriction factors are saturable, treatment of target cells with high levels of a restricted virus can abrogate the restriction and facilitate infection by a second restricted virus. This type of experiment is shown in Figure 2A, where treatment of AGM CV-1 cells with increasing amounts of HIV-2-Puro particles can enhance infection by a fixed inoculum of a homologous HIV-2-GFP vector, in a dose-dependent manner. In contrast, HIV-2-GFP infection is not increased in non-restricting human TE671 cells by excess HIV-2 particles. This strategy can also be used to test whether factors that restrict a particular virus can be saturated by a heterologous virus, as we have shown previously for HIV-1 and SIVmac in AGM and rabbit cells (Besnier et al., 2002; Cowan et al., 2002). As is shown in Figure 2B, incubation of CV-1 target cells with increasing amounts of HIV-2 particles enhanced infection by the SIV-GFP vector by >10-fold. These data suggest that a single form of Lv1 in CV-1 cells restricts and can be saturated by HIV-1, HIV-2 and SIVmac.
Fig. 2. Saturable resistance to primate lentiviruses and N-MLV in CV-1 cells. (A) HIV-2 is restricted in CV-1 (AGM) but not in human (TE671) cells. CV-1 cells (filled symbols) or TE671 cells (open symbols) were inoculated with a fixed dose of HIV-2-GFP vector in the presence of increasing amounts of abrogating HIV-2-Puro particles. The percentage of HIV-2-GFP-infected cells as a function of abrogating virus dose is plotted. Abrogating virus dose is given in infectious units (i.u.) measured as puromycin-resistant colony formation on TE671 cells. (B) Abrogation of SIV-GFP restriction by HIV-2-Puro particles. CV-1 cells were inoculated with a fixed dose of SIV-GFP in the presence of abrogating HIV-2 particles, as in (A), and the percentage of SIV-GFP infected cells is plotted. (C) Resistance of CV-1 cell to N-GFP is saturable. CV-1 cells were inoculated with a fixed dose of N-GFP in the presence of increasing levels of abrogating N-Neo or B-Neo vector particles. Abrogating virus dose is given as MDTF i.u. as measured by G418-resistant colony formation.
We used the same abrogation of restriction strategy to confirm that the resistance of AGM cells to N-MLV infection is saturable, as has been shown previously in human cells (Towers et al., 2002). As expected, and shown in Figure 2C, treatment of CV-1 target cells with N-Neo vector dramatically facilitated infection by an N-GFP vector. This abrogation was specific, because B-Neo particles had no abrogating effect. Thus, like human cells, this AGM cell line presumably harbors a restriction factor with properties similar to those of Ref1 and Fv1b, i.e. it can be saturated by N-MLV but not B-MLV particles. Similar results were obtained in a second AGM cell line (Vero; data not shown).
Next we examined whether primate lentivirus particles could abrogate the restriction of N-MLV that is evident in AGM cells. CV-1 target cells were treated with HIV-1, HIV-2 or SIVmac particles containing a packageable vector genome encoding Puro or LacZ. Remarkably, and as shown in Figure 3A, increasing amounts of HIV-1-Puro vector particles progressively abrogated the restriction of N-GFP infection in CV-1 cells. In contrast, no effect on unrestricted B-GFP infection was observed. At saturating levels of the HIV-1-Puro vector, the restriction was completely abrogated such that N-MLV and B-MLV were equivalently infectious. Similarly, HIV-1 virus-like particles (VLPs) that lacked a genome were also capable of complete abrogation of N-MLV restriction. As is shown in Figure 3B, incubation of HIV-1 Gag-Pol VLPs with CV-1 target cells enhanced N-GFP infection such that its titration curve was linear and superimposable on that of B-GFP (compare Figure 3B with Figure 1). Moreover, both HIV-2 particles (Figure 3C) and SIVmac particles (Figure 3D) abrogated N-MLV restriction in CV-1 cells; in both cases, the inclusion of excess lentivirus particles completely restored N-GFP infection to the level obtained with B-GFP. Importantly, these results show that factors responsible for the N-MLV restriction in AGM cells can be saturated by three primate lentiviruses.
Fig. 3. Abrogation of N-MLV restriction in CV-1 cells by primate lenti virus particles. (A) Abrogation of N-GFP restriction in CV-1 cells by HIV-1 particles. CV-1 cells were inoculated with a fixed and equivalent dose of N-GFP or B-GFP reporter virus (as measured on MDTF cells) in the presence of increasing amounts of abrogating HIV-1-Puro particles. Abrogating virus dose is given as infectious units (i.u.) on TE671 cells. The percentage of N-GFP- (filled symbols) and B-GFP- (open symbols) infected cells is shown. (B) Complete abrogation of N-MLV restriction by HIV-1 particles. N-GFP (filled symbols) and B-GFP (open symbols) were titrated in CV-1 cells, precisely as in Figure 1, except that inoculation was performed in the presence of saturating levels (100 ng of p24) of genome-less HIV-1 VLPs. (C and D) Abrogation of N-MLV restriction by HIV-2 and SIVmac particles. CV-1 cells were inoculated with fixed and equivalent doses of N-GFP (filled symbols) and B-GFP (open symbols), in the presence of increasing amounts of abrogating HIV-2-Puro (C) or SIV-LacZ (D) vector particles. Abrogating virus dose is given as the titer (i.u.) as measured on TE671 cells.
Restriction factors in both human and AGM restrict a divergent non-primate lentivirus
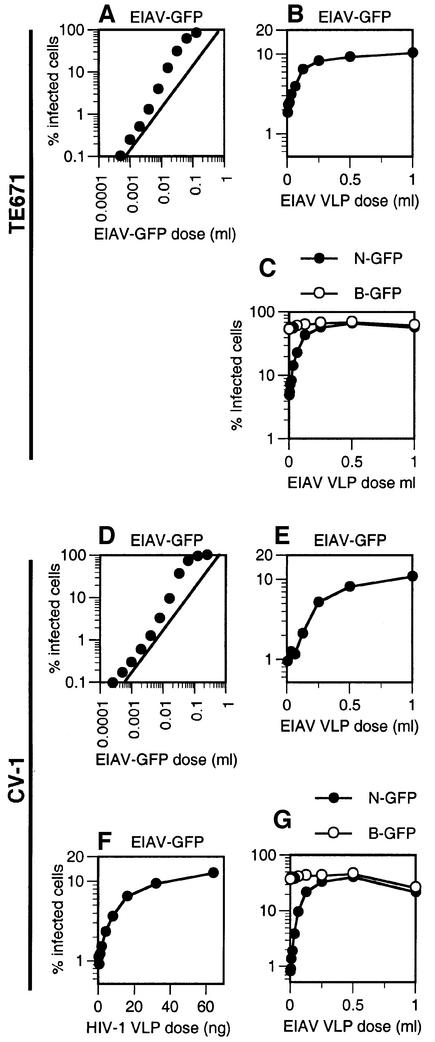
As the above data indicate, AGM cell lines have the remarkable ability to restrict infection by widely divergent primate and murine retroviruses, perhaps using a common factor, while human cells appear capable of restricting only N-MLV. We next tested a divergent non-primate lentivirus, equine infectious anemia virus (EIAV), for restriction in both human (TE671) and AGM (CV-1) cell lines. As is shown in Figure 4A, titration of an EIAV-GFP vector on TE671 cells gave rise to a curve that was non-linear at high levels of inoculum, a property that is indicative of saturable restriction. To address this more specifically, we asked whether the inclusion of additional EIAV VLPs that lacked a genome could enhance low m.o.i. infection of TE671 cells by the EIAV-GFP vector, i.e. abrogate restriction. As shown in Figure 4B, increasing amounts of EIAV VLPs indeed enhanced EIAV-GFP infection, up to 5-fold. We then showed that EIAV VLPs are also able to saturate Ref1-mediated restriction of N-MLV (Figure 4C). N-GFP infection is restored to the same level as that obtained with B-GFP, In contrast, unrestricted B-GFP infection was unaffected by EIAV VLPs.
Fig. 4. Restriction of EIAV in human and AGM cells by the same factors that restrict primate lentiviruses and N-MLV. (A) Non-linear titration curve upon infection of human (TE671) cells by EIAV. TE671 cells were inoculated with an increasing dose of EIAV-GFP. The diagonal guide line indicates a slope of 1. (B) Abrogation of EIAV restriction in TE671 cells by EIAV particles. TE671 cells were inoculated with a fixed dose of EIAV-GFP in the presence of increasing levels of EIAV VLPs. (C) Abrogation of N-MLV restriction in TE671 cells by EIAV particles. TE671 cells were inoculated with fixed and equivalent doses of N-GFP or B-GFP in the presence of increasing levels of EIAV VLPs (given in ml of transfected cell supernatant). (D) Non-linear titration curve upon infection of CV-1 cells by EIAV. CV-1 cells were inoculated with an increasing dose of EIAV-GFP, and the percentage of infected cells is plotted. (E and F) Abrogation of EIAV restriction in CV-1 cells by EIAV and HIV-1 particles. CV-1 cells were inoculated with a fixed dose of EIAV-GFP in the presence of increasing levels of (E) EIAV VLPs or (F) HIV-1-Puro particles (given in ng of p24). The percentage of EIAV-GFP infected cells as a function of abrogating particle dose is plotted. (G) Abrogation of N-MLV restriction in CV-1 cells by EIAV particles. CV-1 cells were inoculated with fixed and equivalent doses of N-GFP (filled symbols) or B-GFP (open symbols), as determined by titration on MDTF cells, in the presence of increasing levels of EIAV VLPs. The percentage of MLV-GFP-infected cells is plotted. In the absence of a convenient assay for measuring EIAV VLP concentration, a single stock of EIAV VLP-containing supernatant was used in each of the experiments presented herein, and the VLP level is given as a volume (ml) of this stock. However, each experiment was performed at least twice with independent VLP stocks, with similar results.
AGM CV-1 cells also restricted infection by EIAV-GFP. As is shown in Figure 4D, the EIAV-GFP vector titration curve exhibited noticeable non-linearity at high levels of inoculum. In addition, EIAV VLPs significantly enhanced (10-fold) low m.o.i. infection by EIAV-GFP (Figure 4E). We then asked whether saturation of Lv1 with HIV-1 particles could abrogate EIAV restriction in CV-1 cells. As is shown in Figure 4F, this indeed proved to be the case. As with EIAV particles, saturating levels of HIV-1-Puro particles enhanced EIAV infection by 10-fold. In addition, we determined whether EIAV particles could abrogate the N-GFP restriction in CV-1 cells. In fact, EIAV particles were able to abrogate N-GFP restriction completely, and restored the levels of N-GFP infection to the same as those of B-MLV (Figure 4G). Thus, it appears that factor(s) in CV-1 cells are able to restrict this equine retrovirus in addition to primate and murine retroviruses.
Primate lentivirus particles do not significantly abrogate Ref1 restriction of N-GFP in human cells
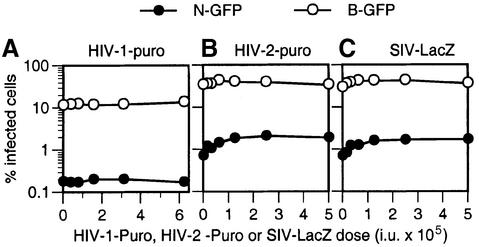
As a test of specificity, we examined whether HIV-1 and SIVmac particles could abrogate N-GFP restriction in human cells. Like AGM cells, human cells restrict N-GFP infection but, unlike AGM cells, human cells do not restrict HIV-1, HIV-2 or SIVmac (Figure 2A; Towers et al., 2000; Besnier et al., 2002; Cowan et al., 2002). Therefore, primate lentiviruses are, presumably, not recognized by Ref1 in human cells. Thus, if abrogation of N-MLV restriction by lentiviruses (Figures 3 and 4) is due to saturation of shared restriction factors, then primate lentiviruses should not exhibit this activity in human cells. Indeed, as is shown in Figure 5, N-GFP restriction was not abrogated by either HIV-1-Puro, HIV-2-Puro or SIV-LacZ vector particles in human TE671 cells. Similar results were obtained in a second N-GFP-restricting human cell line, HeLa (data not shown).
Fig. 5. Primate lentiviruses do not significantly abrogate Ref1-mediated N-MLV restriction in human cells. Human TE671 cells were inoculated with fixed and equivalent doses [in MDTF infectious units (i.u.)] of N-GFP (filled symbols) or B-GFP (open symbols) in the presence of increasing doses of abrogating HIV-1-Puro (A), HIV-2-Puro (B) or SIV-lacZ (C) particles. Higher doses of N-GFP and B-GFP were used in (B) and (C) than in (A). The percentage of MLV-GFP-infected cells is plotted as a function of abrogating virus dose given as TE671 i.u.
Partial abrogation of HIV-1 and SIVmac restriction by N-MLV particles
If, as the above data suggest, a restricted virus can abrogate the restriction of any other restricted virus, then we reasoned that N-MLV particles should reciprocally abrogate restriction of lentivirus infection. To address this, we inoculated CV-1 cells with a low, restricted dose of HIV-1-GFP or SIV-GFP reporter viruses in the presence of increasing amounts of abrogating N-Neo and B-Neo vector particles. As is shown in Figure 6A, N-Neo particles were indeed able to enhance both HIV-1 and SIVmac infection of CV-1 cells. Again this enhancement was specific, in that B-Neo particles were not able to abrogate restriction. To demonstrate further that this effect was specific to cells that restrict both primate lentiviruses and N-GFP, we simultaneously performed the same experiment using an owl monkey cell line (OMK). This cell line potently restricts HIV-1 infection but does not restrict N-GFP (Figure 1). In this case, HIV-1 infection was not enhanced by N-Neo or B-Neo particles (Figure 6A), but was enhanced >50-fold by HIV-1 VLPs (Figure 6B). Thus, abrogation of HIV-1 restriction by MLV requires that both viruses be sensitive to restriction. These data suggest that the AGM but not the owl monkey form of Lv1 can be saturated by N-MLV. It is of note that the N-Neo-mediated abrogation of HIV-1 and SIVmac restriction in CV-1 cells was incomplete. In fact, N-Neo particles enhanced HIV-1-GFP and SIV-GFP infection of CV-1 cells by only 3- to 6-fold (Figure 6A), while HIV-1 VLPs enhanced infection of each virus by up to 20- to 30-fold (Figure 6B). Increasing the amount of abrogating N-Neo particles in these assays (up to 107 infectious units/well) did not enhance restricted HIV-1-GFP or SIV-GFP infection further beyond that shown in Figure 6A.
Fig. 6. Abrogation of lentivirus restriction by N-MLV particles. (A) Abrogation of Lv1 restriction in CV-1 but not OMK cells by N-MLV particles. CV-1 and OMK cells were infected with a fixed dose of HIV-1-GFP or SIV-GFP, as indicated, in the presence of increasing doses of abrogating N-Neo (filled symbols) or B-Neo (open symbols) particles. The percentage of HIV-1-GFP- or SIV-GFP-infected cells as a function of abrogating MLV dose is plotted. Abrogating virus dose is given as infectious units (i.u.) as measured on MDTF cells. (B) Abrogation of Lv1 restriction by HIV-1 particles. CV-1 and OMK cells, as indicated, were infected with a fixed dose of HIV-1-GFP or SIV-GFP as in (A) in the presence of the indicated levels (given as ng of p24) of genome-less HIV-1 VLPs. (C) Abrogation of EIAV restriction in human cells by N-MLV particles. TE671 cells were infected with a fixed dose of EIAV-GFP in the presence of increasing amounts of N-Neo (filled symbols) or B-Neo (open symbols) particles. The percentage of EIAV-GFP-infected cells is plotted.
We also tested whether MLV particles could abrogate restriction of EIAV infection in human cells. As is shown in Figure 6C, treatment of TE671 cells with Ref1-restricted N-Neo particles enhanced infection by EIAV-GFP ∼5-fold, while treatment with unrestricted B-Neo particles had no effect. These data indicate that saturation of Ref1 in human cells can abrogate restriction of a lentivirus.
Evidence for polymorphism in Lv1 among African green monkeys
To examine whether the reciprocal abrogation of N-MLV and primate lentiviruses in AGM cells is unique to the CV-1 cell line, we compared the restriction properties of this cell line with two additional cell lines derived from AGMs. Each of these cell lines is derived from the same tissue (kidney) but from a separate AGM subspecies; the BS-C-1 cell line is derived from Cercopithicus aethiops aethiops (grivet), whereas Vero is derived from Cercopithicus aethiops pygerythrus (vervet). The precise origin of CV-1 cells is unclear but, based on characteristic polymorphisms in the CCR5 gene, is most probably derived from Cercopithicus aethiops tantalus (Tantalus) (Kuhmann et al., 2000). Despite their distinct origins, each of these lines potently restricts N-GFP infection (Figure 1), indicating that the presence of the restriction factor is conserved in this species.
We examined whether restrictions to HIV-1 and SIVmac infection that are evident in CV-1 cells are conserved in AGM cell lines. HIV-1-GFP and SIV-GFP stocks were normalized to give nearly identical infectivity on non-restricting human TE671 cells (Figure 7A). Thereafter, titration in AGM cells revealed both subtle and more dramatic differences in their relative susceptibility to HIV-1-GFP and SIV-GFP. In particular, both viruses appeared restricted in CV-1 and BS-C-1, with SIV-GFP slightly more so than HIV-1-GFP in CV-1 and less so in BS-C-1 (Figure 7A). In contrast, Vero cells restricted HIV-1-GFP infection to an ∼50-fold greater degree than SIV-GFP, with the latter virus appearing only modestly restricted. As is shown in Figure 7B, the differences in the degree to which HIV-1 and SIVmac were restricted in the different AGM cells was determined by the viral capsid protein. Specifically, a chimeric virus, SIV(HIV CA)-GFP that contains an HIV-1 CA protein in an otherwise SIV-GFP background, was restricted to the same degree as was HIV-1-GFP in each of the AGM cell lines. While these differences were relatively small (∼3-fold) in CV-1 and BS-C-1 cells, Vero cells restricted infection by SIV(HIV CA)-GFP ∼50-fold more potently than SIV-GFP.
Fig. 7. Phenotypic polymorphism in Lv1 among AGMs. (A) Variation in restriction of HIV-1 and SIVmac in AGM cells. HIV-1-GFP and SIV-GFP stocks were normalized to give nearly identical titration curves on TE671 cells, as shown, and titrated on CV-1, BS-C-1 and Vero cell lines. The percentage of HIV-1-GFP- and SIV-GFP-infected AGM cells for each inoculum dose, in TE671 infectious units (i.u.), is plotted. (B) The CA-p2 region of Gag determines differential restriction of HIV-1 and SIVmac on AGM cell lines. Each AGM cell line was inoculated with equivalent doses of HIV-1-GFP, SIV-GFP and SIV(HIV CA)-GFP, as indicated, normalized using TE671 target cells. The results are given as the percentage of infected AGM cells obtained, divided by the percentage of infected TE671 cells. (C) Abrogation of N-MLV restriction by HIV-1 and SIVmac VLPs on AGM cells. Each AGM cell line was inoculated with a fixed dose of N-MLV-GFP in the presence of increasing levels (given in ng of p24 or p27) of abrogating HIV-1 or SIVmac VLPs. The fold increase in the number of infected N-GFP cells as a function of abrogating VLP dose is plotted.
If the differential susceptibility of AGM cell lines to HIV-1 and SIVmac infection is due to polymorphism in a single restriction factor (Lv1) that also restricts N-MLV, then the ability of lentivirus particles to abrogate N-MLV restriction should be predicted by the degree to which the lentivirus is itself restricted. In this case, we would anticipate that HIV-1 and SIVmac particles should abrogate N-MLV restriction with similar efficiencies in CV-1 and BS-C-1 cells, but that HIV-1 particles should be significantly more potent than SIVmac particles in Vero cells.
In fact, and as shown in Figure 7C, treatment of both CV-1 and BS-C-1 cell lines with either HIV-1 or SIVmac VLPs resulted in a >100-fold increase in N-GFP titer. Conversely, when Vero cells were used as targets, HIV-1 VLPs enhanced N-GFP infection by >100-fold, but equivalent amounts of SIVmac VLPs enhanced N-GFP infection by only 9-fold. Thus, these data do suggest that Lv1 in AGM cells varies in its specificity for lentiviruses and, in some cases, can distinguish relatively closely related CA proteins (HIV-1 versus SIVmac and N-MLV versus B-MLV) while simultaneously restricting widely divergent retroviruses.
An SIV strain from AGMs is not restricted in AGM or human cells
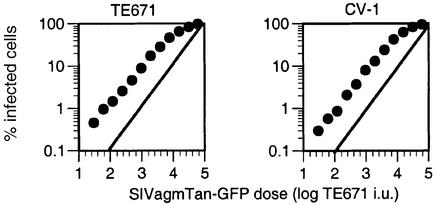
Because both AGM and human cells express restriction factors that are capable of targeting divergent retroviruses, it was of interest to determine whether a lentivirus that occurs naturally in AGMs but not in humans was recognized by restriction factors in either species. We therefore constructed a reporter virus based on an infectious clone of SIV isolated from Tantalus AGMs and tested for restriction in CV-1 (Tantalus AGM) and TE671 (human) cells. As is shown in Figure 8, the SIVagmTan-GFP titration curves were linear and nearly identical in both CV-1 and TE671 cells. Thus, it appears that neither Lv1 in AGMs nor Ref1 in humans is capable of restricting SIVagmTan.
Fig. 8. AGM and human cells do not restrict SIVagmTan infection. AGM (CV-1) and human (TE671) cell lines were infected with serially diluted SIVagmTan reporter virus. The inoculum level is given in TE671 infectious units (i.u.). The percentage of infected (GFP-positive) cells is plotted for each level of SIVagmTan-GFP inoculum.
Discussion
In this study, we have used abrogation of restriction assays to show that human and AGM cells express saturable factors that can confer resistance to infection by multiple, widely divergent retroviruses. AGM cells restrict infection by HIV-1, HIV-2, N-MLV and, at least in some cases, SIVmac and EIAV. However, human cell lines appear capable of restricting N-MLV and EIAV, but not HIV-1, HIV-2 or SIVmac. In the cell lines from other primate species that we have tested, either HIV-1 or SIVmac, but not MLV, is restricted. Thus, retrovirus restriction factors in humans, AGMs and other primates have overlapping, but non-identical specificity. Differential restriction of HIV-1 and SIVmac allows the demonstration that the target site for inhibition of primate lentiviruses is the viral capsid. Specifically, in AGM, Rhesus and owl monkey cells, a chimeric virus that is predominantly SIVmac but encodes an HIV-1 CA protein exhibits the restriction properties of HIV-1 (Figure 7; Cowan et al., 2002). Primate cells also restrict N-MLV by targeting CA. Indeed, the N-MLV and B-MLV constructs used in this study vary at only four amino acid positions, and it has been shown previously that the human (Ref1) and murine (Fv1b) N-MLV restricting factors share the same requirement for arginine at position 110 in the MLV CA protein in order to effect a block to infection (Kozak and Chakraborti, 1996; Towers et al., 2000).
Remarkably, in all examples that we have tested, incubation of target cells with restricted retrovirus particles can abrogate restriction of any other restricted retrovirus. This phenomenon is not dependent on primary sequence homology between the viruses. The sole requirement for cross-abrogation appears to be that both viruses be restricted by the target cell. Indeed, cross-abrogation is possible between unrelated viruses such as MLV and lentiviruses. These findings strongly suggest that, within a particular primate species, a single factor is able to restrict infection by very different retroviruses.
The simplest model for restriction involves a direct interaction between a restricting factor and the capsid of the incoming retrovirus (Goff, 1996; Stoye, 1998, 2002; Bishop et al., 2001). However, it would be surprising if a single restriction factor in humans and non-human primates is capable of recognizing the very divergent retroviral CA proteins present in lentiviruses and MLV. This would be even more remarkable given that the factor discriminates between the very similar N-MLV and B-MLV CA proteins and, in some cases, between the quite closely related HIV-1 and SIVmac CA proteins. Nevertheless, this scenario is possible, and a prediction of this model would be that there is a significant degree of structural conservation between the CA proteins of MLV and lentiviruses, at least in the target site for restriction, despite little sequence homology. Overall, if the CA direct-recognition model is correct, then it appears likely that restriction factors with similar specificity have arisen independently in mice and in primates, and that the primate form sometimes, most notably in AGMs, can recognize a broad array of retroviral CA proteins.
Although we favor the hypothesis that a single restriction factor in each species is responsible for its restriction characteristics, it is formally possible that the abrogating viruses used in these experiments are able to saturate multiple restriction factors present in a given cell line. Each species might then express a set of restriction factors responsible for its restriction profile. Consistent with this notion, lentivirus particles are capable of completely abrogating N-MLV restriction in both human and AGM cells, provided that they are also restricted, while restricted N-MLV particles are able to abrogate HIV-1 and SIVmac restriction in AGM cell lines only partially. However, an alternative explanation for this apparent discrepancy might lie in the different fates of the CA protein during post-entry steps of the viral life cycle. It is thought that lentivirus cores rapidly disassemble after virus entry (Farnet and Haseltine, 1991; Bukrinsky et al., 1993; Fassati and Goff, 2001; Forshey et al., 2002), which could confer partial resistance to restriction by an inhibitor that targets CA, while permitting diffusion of CA within the cell to saturate restricting factors more efficiently. Conversely, a substantial fraction of MLV CA remains associated with the viral genome for several hours after entry (Bowerman et al., 1989; Fassati and Goff, 1999), perhaps rendering the reverse transcription complex more sensitive to restriction and CA less able to act as a decoy and saturate the restriction factor.
Because the Fv1 gene appears to be derived from an endogenous retroviral gag gene (Best et al., 1996), it is reasonable to hypothesize that Lv1 and Ref1 might also have a retroviral origin. Many retroviral sequences exist in the genomes of primates, potentially providing an opportunity for the genesis of restriction factors. Thus, it is plausible that the retrovirus-restricting properties of human, AGM and other primate cells have arisen on multiple occasions, independently of each other. Alternatively, given the findings presented herein, it is also quite possible that Lv1 and Ref1 are the same factor and that the various restriction properties among primate cells simply represent divergence of a single ancestral gene. Clearly, the resolution of these issues requires the identification of the gene(s) responsible for restriction in primates.
What is clear is that cell lines from several primate species in both the New and Old Worlds express Fv1-like restricting activities, albeit with different specificities, suggesting that the occurrence of retrovirus restriction factors in primates is a widespread and probably ancient phenomenon. It seems likely that pathogenic retrovirus epidemics are responsible for the maintenance of these activities, but whether lentiviruses or other retroviruses are responsible for providing selection pressure is unclear. SIV infections are common in African primates and, among AGMs, 50% or more of adults are infected (Kanki et al., 1985; Ohta et al., 1988). Each modern AGM subspecies is segregated geographically and harbors a distinct phylogenetic subgroup of a single SIVagm lineage (Johnson et al., 1990; Allan et al., 1991; Fomsgaard et al., 1991; Hirsch et al., 1993; Jin et al., 1994). It appears likely that African primates and SIVs have co-evolved for a considerable period of time. Consistent with this notion, CCR5 alleles in AGMs, which encode the major co-receptor for SIVagm, contain an unexpectedly high frequency of non-synonymous nucleotide polymorphisms which impact their ability to mediate SIVagm infection, suggesting that SIVs have selected for change in the AGM genome (Kuhmann et al., 2000). It is perhaps not surprising that significant differences in the resistant phenotype conferred by Lv1 are apparent among closely related primates (vervet, grivet and Tantalus AGMs). Although a variety of SIV infections do not appear to be pathogenic in their natural hosts, presumably as a consequence of co-evolution, they sometimes can cause fatal disease upon transmission to a new primate species, including humans (Fultz et al., 1986; Herchenroder et al., 1989; Gao et al., 1992, 1999; Chen et al., 1996; Hahn et al., 2000). Taken together, these observations suggest that lentiviruses have imposed a selective pressure on African primates that has favored maintenance of and perhaps driven polymorphism in factors such as Lv1 that inhibit virus replication or transmission.
Many human cells express a factor, Ref1, which restricts both N-MLV and, apparently, an unrelated lentivirus, EIAV. Given that Ref1 may, in fact, be a human variant of Lv1, and that only relatively limited samples of primate lentiviruses have been examined for restriction in human cells, it is entirely possible that Ref1 has limited the transmission of these and other retroviruses to humans. However, an SIV found naturally in AGM, SIVagmTan, does not appear to be recognized by restriction factors in AGM CV1 cells or in human cells. It seems likely that there are many influences in addition to restriction factors that determine cross-species transmission of retroviruses. Moreover, while humans do not restrict HIV-1 or HIV-2 infection demonstrably, primary human cells from different individuals vary in their ability to support the replication of HIV-1 in vitro (Williams and Cloyd, 1991; Spira and Ho, 1995; Eisert et al., 2001). It is not clear at what stages of the virus life cycle this variation is manifested, but polymorphism in a restriction factor might contribute to and, more importantly, influence patterns of HIV-1 and/or HIV-2 horizontal and vertical transmission as well as disease progression. Addressing the question of whether Lv1 or Ref1 have significantly limited the prevalence of retrovirus-induced disease in primates, including humans, awaits the isolation and characterization of these important resistance factors.
Materials and methods
Cell lines
The human cell lines 293T, TE671 and HeLa were used as previously described (Besnier et al., 2002; Cowan et al., 2002), as was the mouse cell line from Mus dunni, MDTF. The rhesus monkey cell line Rh.F. was obtained from Ron Desrosiers, New England Regional Primate Research Center, and the squirrel monkey cell line was from Jonathan Scammel, University of South Alabama. The OMK cell line and African green monkey kidney cell lines CV-1, BS-C-1 and Vero were obtained from the ATCC. All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM), 10% fetal calf serum and antibiotics.
Virus and vector expression plasmids
HIV-1, HIV-2, SIVmac, MLV and EIAV reporter virus or vector stocks were generated using combinations of either two or three expression vectors. In most cases, Gag-Pol was encoded on a separate expression plasmid from the packaged viral genome; in other cases [HIV-1 and SIVmac and SIV (HIVCA)], reporter viruses were generated using plasmids that encoded the reporter vector and Gag-Pol on a single packaged genome. HIV-1 and SIVmac reporter viruses and vectors generated by two or three plasmid expression systems had essentially identical properties with respect to restriction and were used interchangeably throughout this study. Details of the reporter virus, Gag-Pol and vector expression plasmids are given in the Supplementary data available at The EMBO Journal Online. In all cases, virus and vector stocks were pseudotyped with VSV-G to enable efficient virus entry into the mammalian cell lines used in these experiments. Throughout the text, viruses are named according to the origin of the Gag-Pol (either HIV-1, HIV-2, SIVmac, SIVagmTan, EIAV, N-MLV or B-MLV) and the reporter gene encoded within the packageable genome (either GFP, Neo, Puro or LacZ). Thus, for example, an HIV-1 particle carrying a GFP reporter gene is referred to as ‘HIV-1-GFP’ and a B-tropic MLV particle carrying a Neo gene is referred to as ‘B-Neo’. In some cases, virus-like particles were generated in the absence of a packageable genome and are referred to as ‘VLPs’.
Generation of virus, vectors and virus-like particles
Virus vector and VLP stocks were made by transfecting 293T cells with Gag-Pol, packageable genome and VSV-G expression vectors as described previously (Besnier et al., 2002; Cowan et al., 2002). When required, virus stocks were concentrated 10- to 50-fold by ultracentrifugation through 20% sucrose. HIV-1, HIV-2 and SIVmac virus stocks were quantitated by titration on non-restricting human TE671 cells and/or by p24/p27 enzyme-linked immunosorbent assay (ELISA). N-MLV and B-MLV stocks were quantitated by titration on Fv1-null MDTF cells. Antibiotic-resistant colony formation was used to measure Neo and Puro vector titers, and X-gal staining was used to measure the SIV-LacZ vector. Titers of GFP vectors were measured by fluorescence-activated cell sorting (FACS; see below).
Infection and restriction of infection assays
Target cells were seeded in either 24-well (2 × 104/well) or 6-well (1 × 105/well) plates. For virus titration experiments, cells were inoculated with 2- or 5-fold serially diluted virus or vector stocks that express a GFP reporter gene (either N-GFP, B-GFP, HIV-1-GFP, HIV-2-GFP, SIV-GFP or EIAV-GFP) in the presence of 5 µg/ml polybrene. Infected target cells were enumerated 48–72 h later using a FacsCalibur instrument (Becton Dickinson) and CellQuest software.
Abrogation of restriction assay
To test whether virion particles were able to abrogate restriction of the same or a different virus, target cells were incubated with Neo, Puro or LacZ vector-containing particles, or genome-less VLPs. Restriction-abrogating particles were serially diluted 2- or 5-fold and added to target cell cultures simultaneously with a fixed dose of GFP reporter virus. The amount of GFP reporter virus used varied according to each cell line and from experiment to experiment, but was selected so that infection with a restricted virus gave low, but accurately measurable levels of infection (∼0.2–5% GFP-positive cells), and the data presented herein are representative of at least two repetitions. After 6–12 h, virus was removed and replaced with fresh medium. Thereafter, infection by the GFP reporter virus was measured as described above. In some experiments, where abrogation of N-GFP restriction was examined, B-GFP was included as a control. In these cases, equivalent titers of N-GFP and B-GFP as measured on MDTF cells were used.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Francois-Loic Cosset, Ronald Desrosiers, Heinrich Gottlinger, Andrew Lever, Kyriacos Mitrophanous, Jonathan Scammell, Adrian Thrasher and Didier Trono for gifts of reagents. The SIVagmTan clone was obtained from Beatrice Hahn through the NIH Research and Reference Reagent program. This work was supported by the Columbia Rockefeller Center for AIDS Research, the Donald A.Pels Charitable Trust, NIH Grant RO1AI50111 (to P.D.B.) and a research career development fellowship 064257 from the Wellcome Trust (to G.T.). S.P.G. is an investigator of the Howard Hughes Medical Institute.
References
- Allan J.S., Short,M., Taylor,M.E., Su,S., Hirsch,V.M., Johnson,P.R., Shaw,G.M. and Hahn,B.H. (1991) Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J. Virol., 65, 2816–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benit L., De Parseval,N., Casella,J.F., Callebaut,I., Cordonnier,A. and Heidmann,T. (1997) Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol., 71, 5652–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnier C., Takeuchi,Y. and Towers,G. (2002) Restriction of lentivirus in monkeys. Proc. Natl Acad. Sci. USA, 99, 11920–11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best S., Le Tissier,P., Towers,G. and Stoye,J.P. (1996) Positional cloning of the mouse retrovirus restriction gene Fv1. Nature, 382, 826–829. [DOI] [PubMed] [Google Scholar]
- Bishop K.N., Bock,M., Towers,G. and Stoye,J.P. (2001) Identification of the regions of Fv1 necessary for murine leukemia virus restriction. J. Virol., 75, 5182–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone L.R., Innes,C.L. and Heitman,C.K. (1990) Abrogation of Fv-1 restriction by genome-deficient virions produced by a retrovirus packaging cell line. J. Virol., 64, 3376–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B., Brown,P.O., Bishop,J.M. and Varmus,H.E. (1989) A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev., 3, 469–478. [DOI] [PubMed] [Google Scholar]
- Bukrinsky M.I., Sharova,N., McDonald,T.L., Pushkarskaya,T., Tarpley,W.G. and Stevenson,M. (1993) Association of integrase, matrix and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl Acad. Sci. USA, 90, 6125–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Telfier,P., Gettie,A., Reed,P., Zhang,L., Ho,D.D. and Marx,P.A. (1996) Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J. Virol., 70, 3617–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan S., Hatziioannou,T., Cunningham,T., Muesing,M.A., Gottlinger,H.G. and Bieniasz,P.D. (2002) Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl Acad. Sci. USA, 99, 11914–11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decleve A., Niwa,O., Gelmann,E. and Kaplan,H.S. (1975) Replication kinetics of N- and B-tropic murine leukemia viruses on permissive and nonpermissive cells in vitro. Virology, 65, 320–332. [DOI] [PubMed] [Google Scholar]
- DesGroseillers L. and Jolicoeur,P. (1983) Physical mapping of the Fv-1 tropism host range determinant of BALB/c murine leukemia viruses. J. Virol., 48, 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Troise G., Bassin,R.H., Rein,A. and Gerwin,B.I. (1977) Loss of Fv-1 restriction in Balb/3T3 cells following infection with a single N tropic murine leukemia virus particle. Cell, 10, 479–488. [DOI] [PubMed] [Google Scholar]
- Eisert V., Kreutz,M., Becker,K., Konigs,C., Alex,U., Rubsamen-Waigmann,H., Andreesen,R. and von Briesen,H. (2001) Analysis of cellular factors influencing the replication of human immuno deficiency virus type I in human macrophages derived from blood of different healthy donors. Virology, 286, 31–44. [DOI] [PubMed] [Google Scholar]
- Farnet C.M. and Haseltine,W.A. (1991) Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol., 65, 1910–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassati A. and Goff,S.P. (1999) Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol., 73, 8919–8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassati A. and Goff,S.P. (2001) Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol., 75, 3626–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomsgaard A., Hirsch,V.M., Allan,J.S. and Johnson,P.R. (1991) A highly divergent proviral DNA clone of SIV from a distinct species of African green monkey. Virology, 182, 397–402. [DOI] [PubMed] [Google Scholar]
- Forshey B.M., von Schwedler,U., Sundquist,W.I. and Aiken,C. (2002) Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol., 76, 5667–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fultz P.N., McClure,H.M., Anderson,D.C., Swenson,R.B., Anand,R. and Srinivasan,A. (1986) Isolation of a T-lymphotropic retrovirus from naturally infected sooty mangabey monkeys (Cercocebus atys). Proc. Natl Acad. Sci. USA, 83, 5286–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F. et al. (1992) Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature, 358, 495–499. [DOI] [PubMed] [Google Scholar]
- Gao F. et al. (1999) Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature, 397, 436–441. [DOI] [PubMed] [Google Scholar]
- Goff S.P. (1996) Operating under a Gag order: a block against incoming virus by the Fv1 gene. Cell, 86, 691–693. [DOI] [PubMed] [Google Scholar]
- Hahn B.H., Shaw,G.M., De Cock,K.M. and Sharp,P.M. (2000) AIDS as a zoonosis: scientific and public health implications. Science, 287, 607–614. [DOI] [PubMed] [Google Scholar]
- Herchenroder O. et al. (1989) Experimental infection of rhesus monkeys with SIV isolated from African green monkeys. Intervirology, 30, 66–72. [DOI] [PubMed] [Google Scholar]
- Himathongkham S. and Luciw,P.A. (1996) Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology, 219, 485–488. [DOI] [PubMed] [Google Scholar]
- Hirsch V.M., McGann,C., Dapolito,G., Goldstein,S., Ogen-Odoi,A., Biryawaho,B., Lakwo,T. and Johnson,P.R. (1993) Identification of a new subgroup of SIVagm in tantalus monkeys. Virology, 197, 426–430. [DOI] [PubMed] [Google Scholar]
- Hofmann W., Schubert,D., LaBonte,J., Munson,L., Gibson,S., Scammell,J., Ferrigno,P. and Sodroski,J. (1999) Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol., 73, 10020–10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M.J. et al. (1994) Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J., 13, 2935–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joag S.V., Li,Z., Foresman,L., Stephens,E.B., Zhao,L.J., Adany,I., Pinson,D.M., McClure,H.M. and Narayan,O. (1996) Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J. Virol., 70, 3189–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P.R., Fomsgaard,A., Allan,J., Gravell,M., London,W.T., Olmsted,R.A. and Hirsch,V.M. (1990) Simian immunodeficiency viruses from African green monkeys display unusual genetic diversity. J. Virol., 64, 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolicoeur P. (1979) The Fv-1 gene of the mouse and its control of murine leukemia virus replication. Curr. Top. Microbiol. Immunol., 86, 67–122. [DOI] [PubMed] [Google Scholar]
- Jolicoeur P. and Baltimore,D. (1976) Effect of Fv-1 gene product on proviral DNA formation and integration in cells infected with murine leukemia viruses. Proc. Natl Acad. Sci. USA, 73, 2236–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki P.J., Kurth,R., Becker,W., Dreesman,G., McLane,M.F. and Essex,M. (1985) Antibodies to simian T-lymphotropic retrovirus type III in African green monkeys and recognition of STLV-III viral proteins by AIDS and related sera. Lancet, 1, 1330–1332. [DOI] [PubMed] [Google Scholar]
- Kozak C.A. and Chakraborti,A. (1996) Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology, 225, 300–305. [DOI] [PubMed] [Google Scholar]
- Kuhmann S.E., Platt,E.J., Kozak,S.L. and Kabat,D. (2000) Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol., 74, 7005–7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lord,C.I., Haseltine,W., Letvin,N.L. and Sodroski,J. (1992) Infection of cynomolgus monkeys with a chimeric HIV-1/SIVmac virus that expresses the HIV-1 envelope glycoproteins. J. AIDS, 5, 639–646. [PubMed] [Google Scholar]
- Li J.T., Halloran,M., Lord,C.I., Watson,A., Ranchalis,J., Fung,M., Letvin,N.L. and Sodroski,J.G. (1995) Persistent infection of macaques with simian–human immunodeficiency viruses. J. Virol., 69, 7061–7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly F. (1970) Fv-2: identification and location of a second gene governing the spleen focus response to Friend leukemia virus in mice. J. Natl Cancer Inst., 45, 163–169. [PubMed] [Google Scholar]
- Luciw P.A., Pratt-Lowe,E., Shaw,K.E., Levy,J.A. and Cheng-Mayer,C. (1995) Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc. Natl Acad. Sci. USA, 92, 7490–7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y., Masuda,T., Tsujimoto,H., Ishikawa,K., Kodama,T., Morikawa,S., Nakai,M., Honjo,S. and Hayami,M. (1988) Isolation of simian immunodeficiency virus from African green monkeys and seroepidemiologic survey of the virus in various non-human primates. Int. J. Cancer, 41, 115–122. [DOI] [PubMed] [Google Scholar]
- Pincus T., Hartley,J.W. and Rowe,W.P. (1971) A major genetic locus affecting resistance to infection with murine leukemia viruses. I. Tissue culture studies of naturally occurring viruses. J. Exp. Med., 133, 1219–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak P.M. and Varmus,H.E. (1992) Fv-1 restriction and its effects on murine leukemia virus integration in vivo and in vitro. J. Virol., 66, 5959–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis,N.C., Choi,J.D. and Malim,M.H. (2002) Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature, 418, 646–650. [DOI] [PubMed] [Google Scholar]
- Shibata R., Kawamura,M., Sakai,H., Hayami,M., Ishimoto,A. and Adachi,A. (1991) Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J. Virol., 65, 3514–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata R., Sakai,H., Kawamura,M., Tokunaga,K. and Adachi,A. (1995) Early replication block of human immunodeficiency virus type 1 in monkey cells. J. Gen. Virol., 76, 2723–2730. [DOI] [PubMed] [Google Scholar]
- Spira A.I. and Ho,D.D. (1995) Effect of different donor cells on human immunodeficiency virus type 1 replication and selection in vitro. J. Virol., 69, 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye J.P. (1998) Fv1, the mouse retrovirus resistance gene. Rev. Sci. Technol., 17, 269–277. [DOI] [PubMed] [Google Scholar]
- Stoye J.P. (2002) An intracellular block to primate lentivirus replication. Proc. Natl Acad. Sci. USA, 99, 11549–11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant R.W., Otten,J.A., Brown,A., Yang,W.K. and Kennel,S.J. (1979) Characterization of Fv-1 host range strains of murine retroviruses by titration and p30 protein characteristics. Virology, 99, 349–357. [DOI] [PubMed] [Google Scholar]
- Towers G., Bock,M., Martin,S., Takeuchi,Y., Stoye,J.P. and Danos,O. (2000) A conserved mechanism of retrovirus restriction in mammals. Proc. Natl Acad. Sci. USA, 97, 12295–12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers G., Collins,M. and Takeuchi,Y. (2002) Abrogation of Ref1 retrovirus restriction in human cells. J. Virol., 76, 2548–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L.M. and Cloyd,M.W. (1991) Polymorphic human gene(s) determines differential susceptibility of CD4 lymphocytes to infection by certain HIV-1 isolates. Virology, 184, 723–728. [DOI] [PubMed] [Google Scholar]