Abstract
Editing of misactivated amino acids by class I tRNA synthetases is encoded by a specialized internal domain specific to class I enzymes. In contrast, little is known about editing activities of the structurally distinct class II enzymes. Here we show that the class II alanyl-tRNA synthetase (AlaRS) has a specialized internal domain that appears weakly related to an appended domain of threonyl-tRNA synthetase (ThrRS), but is unrelated to that found in class I enzymes. Editing of misactivated glycine or serine was shown to require a tRNA cofactor. Specific mutations in the aforementioned domain disrupt editing and lead to production of mischarged tRNA. This class-specific editing domain was found to be essential for cell growth, in the presence of elevated concentrations of glycine or serine. In contrast to ThrRS, where the editing domain is not found in all three kingdoms of living organisms, it was incorporated early into AlaRSs and is present throughout evolution. Thus, tRNA-dependent editing by AlaRS may have been critical for making the genetic code sufficiently accurate to generate the tree of life.
Keywords: editing domain/evolution of genetic code/genetic code/tRNA cofactor/tRNA mischarging
Introduction
Aminoacyl-tRNA synthetases (aaRSs) establish the genetic code through aminoacylation reactions that match amino acids with nucleotide triplets encoded as anticodons in tRNAs (Lapointe and Giegé, 1991; Moras, 1992; Carter, 1993; Giegé et al., 1993; Martinis and Schimmel, 1995; Ibba and Söll, 2000). The general error rate for protein synthesis is estimated to be ∼1 in 3000 (Jakubowski and Goldman, 1992). If a particular synthetase fails to discriminate its cognate amino acid from a non-cognate amino acid by a factor of <3000, an editing function is predicted. This prediction has been fulfilled by the observation that those aaRSs that have discrimination factors of <3000 also have editing activities (Eldred and Schimmel, 1972; Fersht, 1985). Conservation of editing in aaRSs spans the entire tree of life (Ribas de Pouplana and Schimmel, 2001). Thus, editing appears to be essential.
The synthetases are evenly divided into two classes of 10 enzymes each. This classification scheme is based on the active site architecture shared by all members of the same class (Webster et al., 1984; Ludmerer and Schimmel, 1987; Cusack et al., 1990; Eriani et al., 1990). The general model for editing that has developed from studies of class I synthetases, such as IleRS, LeuRS and ValRS, is that the active site for amino acid activation acts as a ‘coarse sieve’ excluding amino acids with side chains larger than that of the cognate one. A second, ‘fine sieve’, hydrolyzes misactivated amino acids as aminoacyladenylate (AA- AMP) or misaminoacylated-tRNA (AA-tRNA) (Fersht, 1985; Schimmel and Schmidt, 1995). This site serves to accommodate the smaller non-cognate amino acid while sterically excluding the natural amino acid.
Editing occurs by hydrolysis of the misactivated aminoacyladenylate (pre-transfer) (Fersht, 1977; Hale and Schimmel, 1996) or aminoacylated tRNA (post-transfer) (Eldred and Schimmel, 1972):
The active site for editing by class I enzymes is encoded by an insertion (known as CP1) that splits the catalytic site for aminoacylation (Schmidt and Schimmel, 1994, 1995; Lin and Schimmel, 1996; Nureki et al., 1998; Silvian et al., 1999). For the editing reaction to occur, the misactivated or mischarged amino acid must be translocated ∼25 Å from the active site to the center for editing in CP1 (Nureki et al., 1998; Nomanbhoy et al., 1999; Hendrickson et al., 2002).
In the case of class II enzymes, such as AlaRS, little is known about editing. Early preliminary work on AlaRS, PheRS and LysRS demonstrated editing activities for these class II enzymes (Yarus, 1972; Tsui and Fersht, 1981; Jakubowski, 1997). More recent work on the class II ProRS and ThrRS elucidated editing by these enzymes as well (Beuning and Musier-Forsyth, 2000; Dock-Bregeon et al., 2000; Wong et al., 2002). In the case of ThrRS, an activity that deacylates Ser-tRNAThr has been reported and has been localized to an N-terminal domain that is fused to the catalytic body (Dock-Bregeon et al., 2000).
A question of major interest is the role of the editing domain in the establishment and maintenance of the genetic code. In the case of the class I ValRS, genetic code ambiguity is created by mutations that disrupt the center for editing. This ambiguity can lead to cell death (Döring et al., 2001). These observations are consistent with the CP1 editing domain of class I enzymes being deeply rooted and distributed through all kingdoms in the tree of life (Ribas de Pouplana and Schimmel, 2001). Thus, the CP1-encoded editing function appears to be essential for the establishment of the genetic code. In the case of class II enzymes, far less is known. Strikingly, an appended domain of ThrRS reported to deacylate Ser-tRNAThr (Dock-Bregeon et al., 2000) is not distributed through the tree of life (see below). The appended domain of bacterial and eukaryotic ThrRSs share a significant degree of sequence identity, with several residues important for the editing by bacterial enzymes being conserved among the two large clades. In Archaea, however, the equivalent region of ThrRS and the residues thought to be involved in editing are absent. This situation contrasts sharply with the results found for the aforementioned class I enzymes, where the CP1 region is conserved across all three kingdoms.
However, no experiments have addressed the question of whether the editing function of ThrRS is essential, as it is for ValRS. Because preliminary work reported an editing activity for AlaRS (Tsui and Fersht, 1981), and because phylogenetic relationships of AlaRS are distinct in some ways from those of other class II enzymes (Chihade et al., 2000), we set out to study in more depth the editing activity of this class II enzyme. Among other issues, we were interested in determining whether that activity could be localized to a specific domain and then disrupted. The idea was to determine whether editing was an essential function, at least under some conditions. If the editing function proved essential, we would then have a clear motivation to use phylogenetic analysis to study the distribution of the associated editing determinants for this class II enzyme through evolution. We imagined that these sorts of studies would test further and expand our understanding of the role of editing in the development and maintenance of the genetic code and the tree of life.
Results
Editing is tRNA dependent
Overall editing (pre- and post-transfer) can be monitored by following abortive ATP hydrolysis in the presence of a non-cognate amino acid. Previously, Tsui and Fersht (1981) showed that ATP hydrolysis was stimulated by AlaRS when challenged with glycine and serine, but not with alanine. We investigated aminoacyl-adenylate synthesis by AlaRS (pH 7.5, 22°C), using alanine, glycine and serine. The three amino acids were distinguished from one another mainly through their Km (0.42, 58.5 and 54.8 mM for alanine, glycine and serine, respectively), where alanine was preferred by ∼100-fold. Significantly, the kcat for activation was comparable for these amino acids (2.4, 3.0 and 1.2/s for alanine, glycine and serine, respectively). Thus, once bound, the three amino acids are treated about equivalently.
The misactivated amino acids (glycine and serine) are cleared by a hydrolytic editing activity, while alanine is not. This activity is stimulated by the addition of a transcript of tRNAAla (Figure 1A). Addition of tRNAIle or tRNALys showed no stimulation of ATPase activity (data not shown). Thus, the dependence of editing on tRNA appears to be tRNA specific. Similarly, for the class I IleRS, ValRS and LeuRS, editing activities are elicited only in the presence of the cognate tRNA (Baldwin and Berg, 1966; Fersht, 1985; Hale et al., 1997). In these instances, the tRNA appears necessary to translocate the misactivated amino acid from the active site for amino acylation to the center for editing (Nomanbhoy et al., 1999).
Fig. 1. (A) tRNA-stimulated editing by AlaRS at room temperature (22°C) (pH 7.9) in the presence of glycine and serine with and without tRNAAla. Inset: fragment 461N and wild-type AlaRS in the presence of glycine or serine and tRNAAla. (B) Sequences containing clusters of conserved amino acids in a central region in AlaRS are shown. Numbering is for E.coli AlaRS. A similar sequence of E.coli ThrRS is given below. The relative position in the sequence of the domain in AlaRS versus the analogous domain of ThrRS is depicted above.
An N-terminal 461 amino acid fragment of the 875 amino acid AlaRS contains the catalytic sites for amino acid activation and aminoacylation (Jasin et al., 1983). In contrast to wild-type AlaRS, fragment 461N showed no hydrolytic editing activity (Figure 1A, inset). Thus, we imagined that the active site for editing was encoded at least in part by a portion of the enzyme that includes amino acids on the C-terminal side of the first 461 residues.
Point mutations in AlaRS
Upon aligning 78 AlaRS sequences from all classes of organisms by ClustalW (Thompson et al., 1994), we observed that an internal sequence within A547–G675 of Escherichia coli AlaRS contains six of only 16 absolutely conserved amino acids (the remaining 10 are in fragment 461N). This sequence aligns weakly with the N-terminal domain of bacterial and eukaryotic ThrRS (Figure 1B) that is reported to be important for clearance of Ser-tRNAThr (Dock-Bregeon et al., 2000). Out of the six amino acids in this region that are conserved throughout all AlaRSs, five are also conserved in ThrRS. Four point mutations in this region of AlaRS were constructed initially: H564A, H568A, C666A and H670A. All four mutant enzymes had kcat/Km for aminoacylation within 3-fold of that of wild-type AlaRS. Consistent with the results of in vitro aminoacylation, each mutant allele complemented E.coli strain W3110 (lacIq recA Δ1 Kanr alaSΔ2) which harbors a chromosomal deletion of alaS (Jasin and Schimmel, 1984). The strain is maintained by plasmid pMJ901 (Tetr marker) which contains wild-type alaS and a temperature-sensitive replicon. This plasmid is lost at 42°C, and growth of the strain can only be provided by a second test plasmid harboring functional AlaRS.
Misaminoacylation of tRNAAla
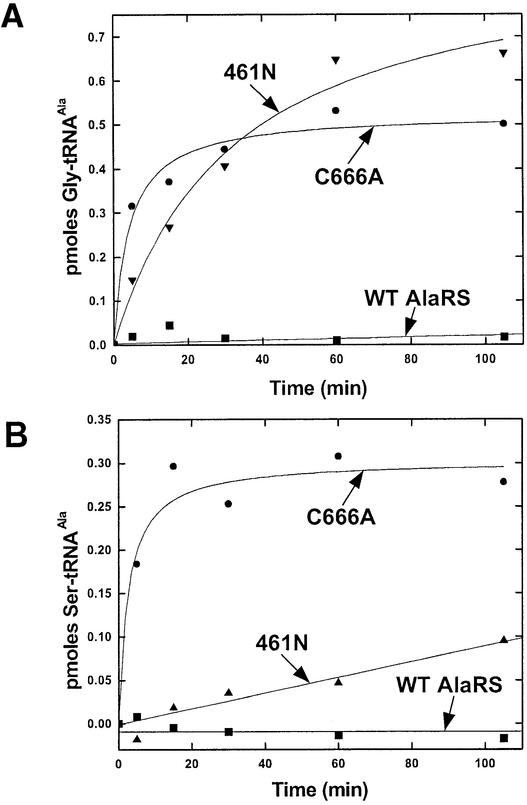
As seen in studies of class I enzymes, editing defects lead to the production of mischarged tRNA (Hendrickson et al., 2000; Döring et al., 2001; Mursinna and Martinis, 2002). With this in mind, we sought to determine whether the point mutations in AlaRS resulted in the ability to produce misaminoacylated tRNAAla with either glycine or serine. With both glycine and serine, C666A AlaRS misaminoacylated tRNAAla (Figure 2). In contrast, the wild-type enzyme did not generate significant misaminoacylated tRNAAla. Thus, the C666A substitution leads to a reduced ability to edit misaminoacylated tRNAAla. None of the three other point mutations in AlaRS generated significant quantities of misaminoacylated tRNAAla (data not shown). Therefore, we focused our experiments on C666A AlaRS. Consistent with the editing domain being contained within a region outside the catalytic machinery for aminoacylation was the demonstration that fragment 461N was also able to produce misaminoacylated tRNAAla (Figure 2).
Fig. 2. Mischarging of tRNAAla at room temperature (22°C) (pH 7.5) by C666A AlaRS and by fragment 461N with glycine (A) or serine (B).
ATP hydrolysis deficiency
Because C666A AlaRS catalyzed formation of mischarged tRNAAla, we imagined that it would be defective in the tRNAAla-dependent ATPase activity observed when glycine or serine is mixed with ATP and AlaRS (Figure 1A). Indeed, C666A AlaRS was defective in stimulating ATP hydrolysis (Figure 3A and B). This defect was seen with both glycine and serine. These data further establish that both non-cognate amino acids are cleared by the same catalytic center.
Fig. 3. Editing (ATPase) activity of wild-type and C666A AlaRS at room temperature (22°C) (pH 7.9) in the presence of glycine (A) and serine (B). tRNA-independent hydrolysis of ATP was subtracted as background. (C) Deacylation of Gly-tRNAAla or (D) Ser-tRNAAla by wild-type and C666A AlaRS and fragment 461N at room temperature (22°C) (pH 7.5).
Deacylation activity
Deacylation of misaminoacylated tRNAAla had not been investigated previously with AlaRS. To assay for this activity, a special C666A/Q584H double mutant AlaRS was used to produce misaminoacylated tRNAAla. (The Q584H mutation serendipitously was discovered to enhance further the mischarging phenotype caused by the C666A mutation.) The Q584H mutant enzyme had no defect in aminoacylation or editing. Wild-type AlaRS efficiently deacylated Gly-tRNAAla while C666A AlaRS had deacylation activity at background level (Figure 3C). C666A AlaRS also had a significantly reduced rate of deacylation towards Ser-tRNAAla as substrate (Figure 3D). In addition, fragment 461N deacylated neither Ser-tRNAAla nor Gly-tRNAAla. Even at enzyme concentrations 100 times that of wild-type AlaRS, no deacylation was observed with fragment 461N. These results are consistent with editing-requiring determinants such as C666 that are missing in fragment 461N.
Phenotypic sensitivity of AlaRS point mutants to non-cognate amino acids
Cells encoding C666A AlaRS exhibited a slow growth phenotype. To determine whether this phenotype was associated with a defect in editing, cells bearing the mutant allele were subjected to high concentrations of glycine or serine. Plates were generated with a radial gradient of glycine or serine emanating from a central well. Cells carrying a plasmid encoding C666A AlaRS were sensitive to high concentrations of glycine and serine (Figure 4A). This sensitivity is seen as a ‘halo’ of cell toxicity around the central well. In contrast, cells harboring the wild-type allele were not sensitive. This toxicity was dependent solely on the presence of non-cognate amino acid and C666A AlaRS. For example, glycine in the central well at 1.2 M was used to produce the growth defect, while the same concentration of alanine had no effect on the strain harboring C666A AlaRS. The halos of toxicity caused by non-cognate amino acid were competed out with alanine concentrations of only 10 mM (data not shown).
Fig. 4. Growth of cells containing C666A or wild-type AlaRS. (A) Growth in the presence of a radial gradient of glycine or serine. (B) A 5 µl aliquot of cells bearing either C666A or wild-type AlaRS was streaked onto plates with no amino acid, 2.5 mM serine or 80 mM glycine. All experiments were incubated for 20–24 h at 37°C.
To determine more precisely the concentrations of amino acids that result in cell death, plates with various amounts of glycine or serine were prepared. A strain bearing C666A AlaRS failed to grow on plates containing 80 mM glycine or 2.5 mM serine (Figure 4B). These effects were not due to disparate expression levels. In particular, the intracellular concentration of C666A AlaRS was comparable with that of wild-type AlaRS (as determined by SDS–PAGE). Thus, the sensitivity of C666A AlaRS to high concentrations of glycine or serine suggests strong selective pressure to maintain the editing function.
Phylogeny
We sought to determine the evolutionary origin of the editing domain of AlaRS. Because both ThrRS and AlaRS clear tRNAs that have been mischarged with serine (i.e. Ser-tRNAAla and Ser-tRNAThr), we wanted to determine whether this domain was a late addition to the AlaRS active site, recruited from a previously existing domain in ThrRS. Perhaps this domain further evolved in AlaRS to clear Gly-tRNAAla in addition to Ser-tRNAAla. From this perspective, the editing activity of AlaRS for serine could be viewed as a residual effect of having an architecture adopted from ThrRS. Contrary to this expectation, however, phylogenetic analysis showed that the evolutionary relationships of the editing domain of AlaRS are identical to those found for the AlaRS active site. This indicates that the last common ancestor of all extant AlaRSs already contained the editing domain (Figure 5A). Therefore, the domain was incorporated into AlaRS early, and essentially co-evolved with the active site-containing domain.
Fig. 5. Phylogenetic analysis of the editing domain of AlaRS. (A) Maximum parsimony tree derived from the alignment of the editing domains of AlaRS (corresponding to G535–S686 of E.coli AlaRS), rooted with AlaXp (Schimmel and Ribas De Pouplana, 2000). The numbers in branch nodes correspond to bootstrap frequencies after 1000 bootstrap cycles in parsimony and distance trees. The root of the tree was derived from the position of the AlaXp clade (shown as a red branch). The overall geometry obtained with the editing domain sequences is identical to that obtained for the active site domain alone (Chihade et al., 2000), indicating that the editing domain has evolved with the active site of the protein from the inception of modern AlaRSs. (B) Distribution of the editing domain throughout the tree of life in ThrRS (red squares) and AlaRS (blue circles).
Moreover, the editing domain in ThrRS is not detectable in archaebacterial sequences (Figure 5B). It is thus possible that the editing domain was first fixed in AlaRS and then acquired in evolution by ThrRS, or that it was present early in both enzymes and then lost in the archaeal ThrRSs.
Molecular modeling
To address the three-dimensional structural relatedness of the ThrRS and AlaRS editing domain as well as to analyze distinctions in their active sites, a model was constructed based on the AlaRS sequence containing the editing domain from G535–S686. A model was attained for E.coli AlaRS by homology modeling using the program 3D-PSSM (three-dimensional position-specific scoring matrix) (Fischer et al., 1999; Kelley et al., 2000). This model was based on the structure of E.coli ThrRS (Sankaranarayanan et al., 1999). This model was supported by a PSSM E-value of 0.000425, corresponding to a >95% confidence in the prediction.
Despite the overall reliability, an eight amino acid deletion in AlaRS resulted in removal by 3D-PSSM of the region where C666 is located. Therefore, we turned to the AlaRS sequence most similar to E.coli ThrRS. The sequence of AlaRS from Campylobacter jejuni has a smaller deletion, and the resulting model was built successfully through the region of C666 (Figure 6A). Side chains were constructed for matches with >95% confidence with SCWRL (Dunbrack, 1999). Two antiparallel β-sheets, bridged by an α-helix, surround the central α-helix to form the core of the structure. Apart from overall structural similarity, particularly of the two α-helices, there are notable differences in the predicted side chain orientations. The side chain (D180) in ThrRS whose mutation resulted in the largest editing defect (Dock-Bregeon et al., 2000) is hydrogen bonded to a tyrosine hydroxyl, while the corresponding residue in AlaRS (E664) maintains a dramatically different orientation and has no corresponding hydrogen bonding partner. The tyrosine in ThrRS is strongly conserved, while the corresponding residue in AlaRS exists mostly as phenylalanine, with the only exception being other aliphatic residues.

Fig. 6. (A) AlaRS editing domain model and known E.coli ThrRS structure (coordinates from Sankaranarayanan et al., 1999). (B) Proposed model for discrimination by the editing site of AlaRS. Top: illustration of ValRS discrimination against the hydrophobic methyl group of valine and for the hydroxyl of threonine or a side chain smaller than valine. ‘X’ denotes failure to accommodate the side chain. Bottom: AlaRS chemical discrimination by repulsion of a methyl group or accomodation through hydrogen bonding to the serine hydroxyl. The absence of a side chain allows glycine to be accomm odated.
The counterpart of the only single point mutation (C666A) of E.coli AlaRS that affected editing in all aspects (ATP hydrolysis, mischarging, deacylation and in vivo toxicity) was not detailed in the analysis of editing by ThrRS. This corresponding amino acid (C182) in E.coli ThrRS is predicted to maintain a nearly identical orientation to that of C666 in the model of AlaRS (Figure 6A). The point mutation that had the greatest effect (of those reported) on ThrRS was D180A (Dock-Bregeon et al., 2000), analogous to E664A in AlaRS. We followed up by constructing E664A AlaRS and found that the mutant enzyme failed to mischarge tRNA, deacylated mischarged tRNA with a rate commensurate with that of the wild-type enzyme, had no in vivo toxicity in the presence of glycine or serine, and had tRNA-dependent ATP hydrolysis activity only 2.5-fold lower than that of the wild-type enzyme (data not shown). Thus, although there may be a general conservation of a site able to perform hydrolysis of mischarged Ser-tRNA, it has evolved differently in the two enzymes.
Discussion
AlaRS and ThrRS are the first example in which a similar architecture is used for hydrolytic editing by class II aaRSs. No other class II enzymes encode this domain. Variations in the protein architecture of the editing domain common to ThrRS and AlaRS are required, however. AlaRS is both glycine and serine specific, while ThrRS is believed to be only serine specific because amino acid activation requires side chain hydroxyl coordination in order to orient the amino acid in the active site (Sankaranarayanan et al., 1999).
The sequence of the editing domain of AlaRS is strictly maintained in evolution through all organisms, but it is absent in ThrRS of Archaea (Figure 5B). In examining the kinetics of amino acid activation of ThrRS for serine versus threonine, serine is misactivated at a rate 1000-fold lower than threonine (Sankaranarayanan et al., 2000). This rate approaches the error rate of protein synthesis and, depending on in vivo amino acid concentrations particularly in Archaea, editing may not even be essential for ThrRS. However, the kinetics for misactivation by AlaRS are only 100- to 300-fold lower than for activation of alanine, well above the frequency of protein synthesis errors. This difference may explain why the editing domain is conserved in AlaRS throughout all classes of organisms but is not conserved in ThrRS (a growth phenotype for editing-deficient ThrRS has not been reported). Perhaps, as refinements at the level of the active site in ThrRS improved discrimination for threonine versus serine, the editing domain was lost slowly in Archaea but remains a functional remnant in bacteria and eukaryotes.
The hydrolysis of mischarged or misactivated glycine or serine but not alanine contrasts with the steric exclusion observed in class I aaRSs such as IleRS. In IleRS, the smaller misactivated valine is accommodated in the hydrolytic editing site while the larger isoluecine is sterically occluded (Nureki et al., 1998). For AlaRS, the exclusion of alanine cannot be due only to steric factors, because the hydrolytic active site must accommodate the larger serine. Thus, the hydrolytic editing site must be comprised of a pocket large enough to allow the hydroxymethyl group of serine and, therefore, to accommodate glycine. Based on size only, however, this site would also be large enough for the methyl side chain of alanine. We imagine that the pocket maintains residues equipped for hydrogen bonding to the serine hydroxyl and which at the same time serve to repel the hydrophobic methyl side chain of alanine.
The discrimination of threonine versus valine by the CP1 domain of ValRS offers this type of example. In this case, valine is excluded on the basis of the chemical nature of the side chain (Figure 6B). The threonine hydroxyl forms hydrogen bonds to conserved aspartate residues within CP1, while the methyl group of valine does not bind due to its inability to form a hydrogen bond (Fukai et al., 2000). In addition to the isosteric threonine, the CP1 domain of ValRS is able to hydrolyze two smaller misactivated amino acids, i.e. cysteine and α-aminobutyrate (Jakubowski and Fersht, 1981). This situation may be analogous to the case of AlaRS (Figure 6B) whereby alanine is chemically excluded via its side chain methyl, glycine is accommodated in the large pocket (as are cysteine and α-aminobutyrate in the CP1 of ValRS) and serine overcomes the repulsion at the β-carbon by providing a hydroxyl for hydrogen bonding into a pocket lined with hydrogen bond acceptors.
The work reported here showed that editing in class II aaRSs is essential under certain conditions (Figure 4). The essentiality of the editing function has also been shown for the class I ValRS (Döring et al., 2001). Primitive synthetases lacking editing functions would have produced statistical proteins due to high error rates in peptide synthesis. As selective pressure favored specific, defined sequences for proteins, recruitment of editing domains to aaRSs was essential. In this connection, the editing domain of AlaRS appears to be one of the most ancient, and therefore may have been critical for development of the tree of life.
Materials and methods
Construction of mutants and plasmids
Mutagenesis was applied to the alaS-encoding plasmid pQE-alaS-6H (Ribas de Pouplana and Schimmel, 1997) by using QuikChange (Stratagene, La Jolla, CA) mutagenesis. Sequences of the mutagenic primers used for this purpose are available upon request. All mutations were confirmed by DNA sequencing.
RNA preparation
A plasmid for the preparation of the in vitro transcript of E.coli tRNAAla was kindly supplied by the laboratory of Karin Musier-Forsyth (University of Minnesota, MN). The plasmid for in vitro transcription of tRNAIle was described previously (Farrow et al., 1999). tRNALys was purchased from Sigma (Saint Louis, MO). In vitro transcription was carried out as described previously (Steer and Schimmel, 1999). The transcript was refolded by heating for 3 min at 80°C, followed by slow cooling and addition of 2 mM MgCl2 at 55°C, and further cooling to room temperature. Mischarged tRNAAla was produced in vitro utilizing C666A/Q584H AlaRS. Transcript (10.8 µM) was aminoacylated in the presence of C666A/Q584H AlaRS (11.7 µM), [3H]glycine (11.8 µM) or [3H]serine (7.3 µM), and the aminoacylation buffer [50 mM HEPES pH 7.5, 20 mM KCl, 0.1 mg/ml bovine serum albumin (BSA), 20 mM β-mercaptoethanol (ME), 10 mM MgCl2]. Reactions were incubated for 60 min at room temperature, phenol extracted twice, ethanol precipitated and resuspended in H2O.
Complementation of cell growth defects
Test plasmids (Ampr marker) bearing alaS mutant alleles were transformed into strain W3110 [lacIq recA Δ1 Kanr alaSΔ2 pMJ901 (Tetr marker)] (Jasin and Schimmel, 1984). Strain W3110 bears a null allele for alaS on the chromosome. The strain is maintained by plasmid pMJ901. This plasmid bears a temperature-sensitive replicon, so that replication is aborted at 42°C. Thus, the strain has a temperature-sensitive phenotype that can be rescued by transformation with a second host plasmid that encodes an active AlaRS. After transformation of W3110 with a test plasmid, transformants were selected on LB-Kan (25 mg/l)/Amp (100 mg/l)/Tet (50 mg/l) plates at 30°C. Colonies were selected, resuspended in 15 µl of LB, an aliquot (3 µl) was plated onto LB-Kan/Amp plates and selection was carried out at 42°C, where the maintenance plasmid is lost and growth depends on the test plasmid.
Protein expression and purification
AlaRS mutant alleles that expressed proteins [from plasmid pQE-alaS-6H (lac promoter)] able to sustain cell growth were selected at 42°C in the null strain W3110. Selected colonies were grown overnight at 37°C. Overnight cultures were diluted, grown to mid-log phase and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 6–12 h. Cells were harvested by centrifugation, lysed by a French press, and batch purified by Ni-NTA chromatography. Concentrations were determined by the Bradford assay (Bradford, 1976).
Aminoacylation and total editing (ATPase) assays
ATPase assays were carried out essentially as described previously (Farrow et al., 1999) at room temperature with AlaRS (3.3 µM), tRNA transcript (15.6 µM), serine or glycine (150 mM), alanine (1.5 mM) and [γ-32P]ATP (0.3 mM). Amino acids were used at three times their Km values determined in the amino acid activation assay. For control assays, tRNAIle (21.0 µM), tRNALys (14.0 µM) or tRNAAla (21.0 µM) was added to the above assay components. Aminoacylation assays were at room temperature as previously described (Buechter and Schimmel, 1993) in standard buffer (50 mM HEPES pH 7.5, 20 mM KCl, 0.1 mg/ml BSA, 20 mM β-ME, 10 mM MgCl2). Mischarging with wild-type or mutant AlaRS (4.9 µM) or the less active fragment 461N (535 µM) was performed in the presence of tRNAAla transcript (8.1 µM) and [3H]glycine (10.7 µM) or [3H]serine (6.8 µM).
Deacylation assays
A transcript of tRNAAla (0.5 µM), mischarged with [3H]glycine or [3H]serine, was added to native AlaRS (50 nM) or fragment 461N (5.0 µM) in charging buffer (50 mM HEPES pH 7.5, 20 mM KCl, 0.1 mg/ml BSA, 20 mM β-ME, 10 mM MgCl2) at room temperature and the reaction was stopped at several time intervals by addition to a filter disc saturated in 5% trichloroacetic acid (TCA) (Farrow et al., 1999).
Analysis of toxicity of mutant alleles of alaS
Mutant alleles encoded by plasmid vector pQE-alaS-6H were transformed into strain W3110 and grown overnight in LB with 25 mg/l kanamycin and 100 mg/l ampicillin. Typically, overnight cultures were then diluted 5:1000 in minimal media. Cells were plated on to minimal media plates containing 25 mg/l kanamycin and 100 mg/l ampicillin. A radial gradient of glycine or serine was established by addition of 1.2 M glycine or 0.4 M serine (80.0 µl) to a well cut in the center of the plate. Plates were incubated for 20–24 h at 37°C and then inspected for a ‘halo of toxicity’ around the center well. For further testing the concentration tolerance of the wild-type and mutant alleles for sensitivity to serine or glycine, ampicillin/kanamycin-containing minimal media plates were poured with various amounts of amino acid. Single colonies were dissolved in minimal medium (30.0 µl) and a 5.0 µl aliquot was spotted and streaked onto the plate. Cells were grown for ∼24 h at 37°C and inspected by eye.
Sequence information and generation of the alignments
All the protein sequences were obtained from the NCBI protein database (www.ncbi.nlm.nih.gov). The complete sequences of all analyzed proteins were aligned directly using CLUSTALW (Thompson et al., 1994), and the regions corresponding to the editing domain of AlaRS identified here were edited and re-aligned with AlaXp sequences (Schimmel and Ribas De Pouplana, 2000). Initial alignments were performed with all available AlaRS sequences, but the final alignments used for comparison with previously published phylogenies of AlaRS (Chihade et al., 2000) contained a selected set of sequences. The alignments generated by CLUSTALW were edited to remove gaps, and used for the phylogenetic studies.
Phylogenetic calculations
Maximum parsimony (MP) and neighbor joining (NJ) analyses were performed with the package PHYLIP 3.57c (Felsenstein, 1988). The numbers and lengths of MP minimal trees were estimated from 1000 replicate random heuristic searches, while confidence limits of branch points were estimated from 1000 bootstrap replications. The NJ phylogeny was based on pairwise distances using the programs NEIGHBOR and PROTDIST. The Dayhoff 120 matrix was used in the PROTDIST program to estimate the expected amino acid replacements per position. The programs SEQBOOT and CONSENSE were used to estimate the confidence limits of branching points. In order to root the trees, AlaXp sequences were used as outgroup (Schimmel and Ribas De Pouplana, 2000). This widespread protein of unknown function is homologous to the editing domains of AlaRS and ThrRS, and probably represents the ancestral form of the editing domains before their incorporation to the active sites of AlaRS and ThrRS (L.Ribas de Pouplana and P.Schimmel, unpublished data).
Model building
The AlaRS sequence (C.jejuni) with greatest sequence similarity to the E.coli ThrRS sequence was submitted to fold recognition in 3D-PSSM (http://www.sbg.bio.ic.ac.uk/∼3dpssm/). SCWRL (Dunbrack, 1999) was utilized to model the side chains in 3D-PSSM for hits with >95% confidence.
Acknowledgments
Acknowledgements
This work was supported by grants GM 15539 and 23562 from the National Institutes of Health and by a fellowship from the National Foundation for Cancer Research. K.B. is an NIH postdoctoral fellow.
References
- Baldwin A.N. and Berg,P. (1966) Transfer ribonucleic acid-induced hydrolysis of valyladenylate bound to isoleucyl ribonucleic acid synthetase. J. Biol. Chem., 241, 839–845. [PubMed] [Google Scholar]
- Beuning P.J. and Musier-Forsyth,K. (2000) Hydrolytic editing by a class II aminoacyl-tRNA synthetase. Proc. Natl Acad. Sci. USA, 97, 8916–8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Buechter D.D. and Schimmel,P. (1993) Dissection of a class II tRNA synthetase: determinants for minihelix recognition are tightly associated with domain for amino acid activation. Biochemistry, 32, 5267–5272. [DOI] [PubMed] [Google Scholar]
- Carter C.W. Jr (1993) Cognition, mechanism and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem., 62, 715–748. [DOI] [PubMed] [Google Scholar]
- Chihade J.W., Brown,J.R., Schimmel,P.R. and Ribas De Pouplana,L. (2000) Origin of mitochondria in relation to evolutionary history of eukaryotic alanyl-tRNA synthetase. Proc. Natl Acad. Sci. USA, 97, 12153–12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas,C., Hartlein,M., Nassar,N. and Leberman,R. (1990) A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 Å. Nature 347, 249–255. [DOI] [PubMed] [Google Scholar]
- Dock-Bregeon A., Sankaranarayanan,R., Romby,P., Caillet,J., Springer,M., Rees,B., Francklyn,C.S., Ehresmann,C. and Moras,D. (2000) Transfer RNA-mediated editing in threonyl-tRNA synthetase. The class II solution to the double discrimination problem. Cell, 103, 877–884. [DOI] [PubMed] [Google Scholar]
- Döring V., Mootz,H.D., Nangle,L.A., Hendrickson,T.L., de Crécy-Lagard,V., Schimmel,P. and Marliere,P. (2001) Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science, 292, 501–504. [DOI] [PubMed] [Google Scholar]
- Dunbrack R.L. Jr (1999) Comparative modeling of CASP3 targets using PSI-BLAST and SCWRL. Proteins, Suppl. 3, 81–87. [DOI] [PubMed] [Google Scholar]
- Eldred E.W. and Schimmel,P.R. (1972) Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J. Biol. Chem., 247, 2961–2964. [PubMed] [Google Scholar]
- Eriani G., Delarue,M., Poch,O., Gangloff,J. and Moras,D. (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature, 347, 203–206. [DOI] [PubMed] [Google Scholar]
- Farrow M.A., Nordin,B.E. and Schimmel,P. (1999) Nucleotide determinants for tRNA-dependent amino acid discrimination by a class I tRNA synthetase. Biochemistry, 38, 16898–16903. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1988) Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet., 22, 521–565. [DOI] [PubMed] [Google Scholar]
- Fersht A.R. (1977) Editing mechanisms in protein synthesis. Rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry, 16, 1025–1030. [DOI] [PubMed] [Google Scholar]
- Fersht A. (1985) Enzyme Structure and Mechanism. W.H.Freeman and Co., New York, NY, pp. 347–368.
- Fischer D. et al. (1999) CAFASP-1: critical assessment of fully automated structure prediction methods. Proteins, Suppl. 3, 209–217. [DOI] [PubMed] [Google Scholar]
- Fukai S., Nureki,O., Sekine,S., Shimada,A., Tao,J., Vassylyev,D.G. and Yokoyama,S. (2000) Structural basis for double-sieve discrimination of l-valine from l-isoleucine and l-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase. Cell, 103, 793–803. [DOI] [PubMed] [Google Scholar]
- Giegé R., Puglisi,J.D. and Florentz,C. (1993) tRNA structure and aminoacylation efficiency. Prog. Nucleic Acid Res. Mol. Biol., 45, 129–206. [DOI] [PubMed] [Google Scholar]
- Hale S.P. and Schimmel,P. (1996) Protein synthesis editing by a DNA aptamer. Proc. Natl Acad. Sci. USA, 93, 2755–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale S.P., Auld,D.S., Schmidt,E. and Schimmel,P. (1997) Discrete determinants in transfer RNA for editing and aminoacylation. Science, 276, 1250–1252. [DOI] [PubMed] [Google Scholar]
- Hendrickson T.L., Nomanbhoy,T.K. and Schimmel,P. (2000) Errors from selective disruption of the editing center in a tRNA synthetase. Biochemistry, 39, 8180–8186. [DOI] [PubMed] [Google Scholar]
- Hendrickson T.L., Nomanbhoy,T.K., de Crécy Lagard,V., Fukai,S., Nureki,O., Yokoyama,S. and Schimmel,P. (2002) Mutational separation of two pathways for editing by a class I tRNA synthetase. Mol. Cell, 9, 353–362. [DOI] [PubMed] [Google Scholar]
- Ibba M. and Söll,D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem., 69, 617–650. [DOI] [PubMed] [Google Scholar]
- Jakubowski H. (1997) Aminoacyl thioester chemistry of class II aminoacyl-tRNA synthetases. Biochemistry, 36, 11077–11085. [DOI] [PubMed] [Google Scholar]
- Jakubowski H. and Fersht,A.R. (1981) Alternative pathways for editing non-cognate amino acids by aminoacyl-tRNA synthetases. Nucleic Acids Res., 9, 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski H. and Goldman,E. (1992) Editing of errors in selection of amino acids for protein synthesis. Microbiol. Rev., 56, 412–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M. and Schimmel,P. (1984) Deletion of an essential gene in E.coli by site-specific recombination with linear DNA fragments. J. Bacteriol., 159, 783–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M., Regan,L. and Schimmel,P. (1983) Modular arrangement of functional domains along the sequence of an aminoacyl tRNA synthetase. Nature, 306, 441–447. [DOI] [PubMed] [Google Scholar]
- Kelley L.A., MacCallum,R.M. and Sternberg,M.J. (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol., 299, 499–520. [DOI] [PubMed] [Google Scholar]
- Lapointe J. and Giegé,R. (1991) Transfer RNAs and aminoacyl-tRNA synthetases. In Trachsel,H. (ed.), Translation in Eukaryotes. CRC Press, Boca Raton, FL, pp. 35–69.
- Lin L. and Schimmel,P. (1996) Mutational analysis suggests the same design for editing activities of two tRNA synthetases. Biochemistry, 35, 5596–5601. [DOI] [PubMed] [Google Scholar]
- Ludmerer S.W. and Schimmel,P. (1987) Gene for yeast glutamine tRNA synthetase encodes a large amino terminal extension and provides strong confirmation of the signature sequence for a group of aminoacyl tRNA synthetases. J. Biol. Chem., 262, 10801–10806. [PubMed] [Google Scholar]
- Martinis S.A. and Schimmel,P. (1995) Small RNA oligonucleotide substrates for specific aminoacylations. In Söll,D. and Raj Bhandary,U.L. (eds), tRNA: Structure, Biosynthesis and Function. ASM Press, Washinton, DC, pp. 349–370.
- Moras D. (1992) Structural and functional relationships between aminoacyl-tRNA synthetases. Trends Biochem. Sci., 17, 159–164. [DOI] [PubMed] [Google Scholar]
- Mursinna R.S. and Martinis,S.A. (2002) Rational design to block amino acid editing of a tRNA synthetase. J. Am. Chem. Soc., 124, 7286–7287. [DOI] [PubMed] [Google Scholar]
- Nomanbhoy T.K., Hendrickson,T.L. and Schimmel,P. (1999) Transfer RNA-dependent translocation of misactivated amino acids to prevent errors in protein synthesis. Mol. Cell, 4, 519–528. [DOI] [PubMed] [Google Scholar]
- Nureki O. et al. (1998) Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science, 280, 578–582. [DOI] [PubMed] [Google Scholar]
- Ribas de Pouplana L. and Schimmel,P. (1997) Reconstruction of quaternary structures of class II tRNA synthetases by rational mutagenensis of a conserved domain. Biochemistry, 36, 15041–15048. [DOI] [PubMed] [Google Scholar]
- Ribas de Pouplana L. and Schimmel,P. (2001) Formation of two classes of tRNA synthetases in relation to editing functions and genetic code. Cold Spring Harbor Symp. Quant. Biol., 66, 131–166. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan R., Dock-Bregeon,A.C., Romby,P., Caillet,J., Springer,M., Rees,B., Ehresmann,C., Ehresmann,B. and Moras,D. (1999) The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell, 97, 371–381. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan R., Dock-Bregeon,A.C., Rees,B., Bovee,M., Caillet,J., Romby,P., Francklyn,C.S. and Moras,D. (2000) Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase. Nat. Struct. Biol., 7, 461–465. [DOI] [PubMed] [Google Scholar]
- Schimmel P. and Ribas De Pouplana,L. (2000) Footprints of aminoacyl-tRNA synthetases are everywhere. Trends Biochem. Sci., 25, 207–209. [DOI] [PubMed] [Google Scholar]
- Schimmel P. and Schmidt,E. (1995) Making connections: RNA-dependent amino acid recognition. Trends Biochem. Sci., 20, 1–2. [DOI] [PubMed] [Google Scholar]
- Schmidt E. and Schimmel,P. (1994) Mutational isolation of a sieve for editing in a transfer RNA synthetase. Science, 264, 265–267. [DOI] [PubMed] [Google Scholar]
- Schmidt E. and Schimmel,P. (1995) Residues in a class I tRNA synthetase which determine selectivity of amino acid recognition in the context of tRNA. Biochemistry, 34, 11204–11210. [DOI] [PubMed] [Google Scholar]
- Silvian L.F., Wang,J. and Steitz,T.A. (1999) Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science, 285, 1074–1077. [PubMed] [Google Scholar]
- Steer B.A. and Schimmel,P. (1999) Different adaptations of the same peptide motif for tRNA functional contacts by closely homologous tRNA synthetases. Biochemistry, 38, 4965–4971. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui W.C. and Fersht,A.R. (1981) Probing the principles of amino acid selection using the alanyl-tRNA synthetase from Escherichia coli. Nucleic Acids Res., 9, 4627–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster T., Tsai,H., Kula,M., Mackie,G.A. and Schimmel,P. (1984) Specific sequence homology and three-dimensional structure of an aminoacyl transfer RNA synthetase. Science, 226, 1315–1317. [DOI] [PubMed] [Google Scholar]
- Wong F.C., Beuning,P.J., Nagan,M., Shiba,K. and Musier-Forsyth,K. (2002) Functional role of the prokaryotic proline-tRNA synthetase insertion domain in amino acid editing. Biochemistry, 41, 7108–7115. [DOI] [PubMed] [Google Scholar]
- Yarus M. (1972) Phenylalanyl-tRNA synthetase and isoleucyl-tRNAPhe: a possible verification mechanism for aminoacyl-tRNA. Proc. Natl Acad. Sci. USA, 69, 1915–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]