Abstract
Human immunodeficiency virus (HIV) and simian (SIV) immunodeficiency virus entry is mediated by binding of the viral envelope glycoprotein (Env) to CD4 and chemokine receptors, CCR5 and/or CXCR4. CD4 induces extensive conformational changes that expose and/or induce formation of a chemokine receptor binding site on gp120. CD4-independent Env's of HIV type 1 (HIV-1), HIV-2, and SIV have been identified that exhibit exposed chemokine receptor binding sites and can bind directly to CCR5 or CXCR4 in the absence of CD4. While many studies have examined determinants for gp120-CCR5 binding, analysis of gp120-CXCR4 binding has been hindered by the apparently lower affinity of this interaction for X4-tropic HIV-1 isolates. We show here that gp120 proteins from two CD4-independent HIV-2 Env's, VCP and ROD/B, bind directly to CXCR4 with an apparently high affinity. By use of CXCR4 N-terminal deletion constructs, CXCR4-CXCR2 chimeras, and human-rat CXCR4 chimeras, binding determinants were shown to reside in the amino (N) terminus, extracellular loop 2 (ECL2), and ECL3. Alanine-scanning mutagenesis of charged residues, tyrosines, and phenylalanines in extracellular CXCR4 domains implicated multiple amino acids in the N terminus (E14/E15, D20, Y21, and D22), ECL2 (D187, R188, F189, Y190, and D193), and ECL3 (D262, E268, E277, and E282) in binding, although minor differences were noted between VCP and ROD/B. However, mutations in CXCR4 that markedly reduced binding did not necessarily hinder cell-cell fusion by VCP or ROD/B, especially in the presence of CD4. These gp120 proteins will be useful in dissecting determinants for CXCR4 binding and Env triggering and in evaluating pharmacologic inhibitors of the gp120-CXCR4 interaction.
Human and simian immunodeficiency viruses (HIV and SIV, respectively) enter cells through a fusion reaction triggered by the viral envelope glycoprotein (Env) and two cellular molecules: CD4 and a chemokine receptor, generally either CCR5 or CXCR4 (2, 17, 24, 29, 31, 42). The interaction of gp120 with the chemokine receptor largely accounts for differences in HIV tropism among CD4-positive cells (reviewed in references 7 and 46). In addition, chemokine receptor specificity contributes substantially to HIV pathogenesis. Viruses that use CCR5 (R5-tropic isolates) are largely responsible for HIV transmission, and individuals lacking functional CCR5 due to a 32-bp deletion in the CCR5 gene (ccr5 Δ32 allele) are highly resistant to HIV type 1 (HIV-1) infection (22, 48, 72). In approximately 50% of infected individuals, CXCR4-tropic (X4-tropic) viruses emerge later in infection, and their appearance correlates with a more rapid CD4 decline and a faster progression to AIDS (18). Dual-tropic isolates that are able to use both CCR5 and CXCR4 are also seen and may represent intermediates in the switch from CCR5 to CXCR4 tropism (29, 75). Thus, understanding the determinants for CCR5 and CXCR4 usage is critical, as it impacts both HIV transmission and progression to AIDS.
HIV Env is composed of a noncovalently associated, trimeric complex of gp120 and gp41 subunits (16, 80). CD4-gp120 binding causes extensive conformational changes in gp120 that involve movement of V1/V2 and V3 hypervariable loops and exposure and/or formation of a highly conserved domain in gp120 shown to be important for CCR5 binding (64, 70). This domain consists of residues adjacent to and within a region termed the bridging sheet, which consists of a four-stranded, antiparallel β sheet formed by the V1/V2 stem and components of the fourth conserved region (C4) of gp120 (54, 70). While the V3 loop has been shown to contribute to the specificity of CCR5 or CXCR4 utilization, conservation of the bridging-sheet region among different HIV-1, HIV-2, and SIV isolates suggests that it may represent a generic chemokine receptor binding site important for interactions with both CCR5 and CXCR4 (70).
Although assays that evaluate the ability of Env-expressing cells to fuse with target cells expressing CD4 and CXCR4 have implicated residues on CXCR4 involved in entry and fusion (reviewed in reference 30), there is little information on the exact determinants involved in the CXCR4-gp120 binding interaction, in contrast to analyses of CCR5-gp120 binding (reviewed in reference 30). The difficulty in measuring gp120 binding to CXCR4 is the result of a markedly reduced affinity of X4-tropic gp120 proteins for CXCR4 (4, 45). By use of an optical biosensor, binding of an X4-tropic HIV-1 gp120 to CXCR4 incorporated into retrovirus particles was found to have a Kd of 500 nM (45). More recently, CXCR4-gp120 binding in the presence of soluble CD4 (sCD4) was assessed by using CXCR4 incorporated into paramagnetic proteoliposomes and found to have a Kd of 200 nM (4). In contrast, R5-tropic gp120s complexed with sCD4 bind CCR5 with dissociation constants often below 10 nM (27, 83).
Despite CD4's role in inducing conformational changes in gp120, some laboratory-adapted HIV-1 isolates as well as many primary HIV-2 and SIV strains do not require CD4 for fusion (32, 36, 38, 47, 52, 56, 68, 69). Env proteins from these CD4-independent isolates can interact directly with chemokine receptors, suggesting that their chemokine receptor binding sites are formed and exposed without the need for CD4 triggering (34, 45, 47, 52, 61). Mutations involved in the CD4-independent phenotype for a well-characterized X4-tropic HIV-1 gp120, 8x, have been shown to be located to sites flanking the bridging-sheet region, supporting the view that CD4 independence involves exposure of a chemokine receptor binding region on gp120 that is concealed prior to CD4 binding (37, 56). However, although the 8x gp120 had an exposed CXCR4 binding site, its affinity remained low (∼500 nM), indicating that these mutations did not appear to confer a high affinity for CXCR4 (45). Nonetheless, it remains possible that some CD4-independent envelopes could exhibit both enhanced exposure and increased affinity for chemokine receptors, characteristics that could promote a CD4-independent phenotype (34, 58, 61). Particularly for X4-tropic gp120s, such envelopes would be highly useful in directly evaluating determinants for gp120-CXCR4 binding.
In this study, we evaluated CXCR4 binding of gp120s from two CD4-independent HIV-2 isolates, HIV-2/vcp (38) and HIV-2/ROD/B (69). Using a cell surface gp120 binding assay, we were able to detect robust CXCR4-gp120 binding for these HIV-2 gp120s, but not for gp120s from X4-tropic HIV-1 isolates. We used these proteins to identify residues in the N terminus, the second extracellular loop (ECL2), and ECL3 of CXCR4 that are required for efficient gp120 binding. However, mutations in CXCR4 that affected binding did not necessarily abolish cell-cell fusion by VCP or ROD/B, especially in the presence of CD4. These gp120s, which exhibit an apparent increased affinity for CXCR4, will be useful in dissecting determinants for Env-CXCR4 binding both in Env and in CXCR4, in more fully characterizing the role of the bridging-sheet region in coreceptor interactions and virus entry, and in evaluating inhibitors of the gp120-CXCR4 interaction.
MATERIALS AND METHODS
Plasmids and plasmid construction.
Expression plasmids to generate secreted gp120 proteins contained stop sites at the gp120-gp41 cleavage site. HIV-1 HXBc2, 8x, HXBc2-V3BaL, and 8x-V3BaL gp120 constructs in pSP73 have been described previously (47). HXBc2 and 8x gp120 constructs were recloned with BamHI into pSP64.vE/L.T7 RNAP (58), a construct containing the synthetic vaccinia virus early/late promoter (15), replacing the T7 RNA polymerase gene with the env gene.
A VCP gp120 env clone was generated from the original pCR3.1 expression vector (38) by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) to place a stop site and an EcoRI site at the gp120-gp41 cleavage site (Trevor L. Hoffman, University of Pennsylvania, Philadelphia, Pa.). The VCP gp120 env gene was recloned into pSC59 (15) by using EcoRI, allowing regulation by the synthetic vaccinia virus early/late promoter. Because pSC59 is a derivative of pBR322 containing a low-copy-number origin of replication (15), the coding region for VCP gp120 and the synthetic vaccinia virus promoter were removed with HindIII and StuI and ligated into pSP64 (Promega, Madison, Wis.) digested with HindIII and PvuII, generating pSP64.vE/L.VCP gp120.
HIV-2/ROD/A and ROD/B gp120 constructs were generated by first removing the full-length ROD/A and ROD/B env genes with HindIII and BamHI from pCR3.1 expression vectors (58) and blunting with T4 DNA polymerase. These fragments were then ligated into pSP64.vE/L.VCP gp120 digested with EcoRI and blunted with T4 DNA polymerase, replacing the VCP gp120 env gene with the ROD/A or ROD/B env gene. Stop sites were then generated at the gp120-gp41 cleavage site with QuikChange.
Chimeras between CXCR4 and CXCR2 (60), CXCR4 constructs containing 12-, 15-, and 23-amino-acid N-terminal deletions (6), human-rat CXCR4 chimeras (10), and feline CXCR4 (82) have been described previously. CXCR4 expression constructs containing alanine substitutions were generated with QuikChange or previously described and generously provided by Anne Brelot and Marc Alizon (Institut Cochin de Génétique Moléculaire, Paris, France) (9).
HIV-2/vcp and HIV-2/ROD/B full-length Env expression plasmids in pCR3.1, used in cell-cell fusion assays, have been described elsewhere (58). CD4, CXCR4, and CCR5 in pcDNA3 (Invitrogen, Carlsbad, Calif.) and the reporter plasmid encoding luciferase under the control of a T7 promoter (T7.luciferase), used in cell-cell fusion assays, have also been described previously (35, 47, 56, 71).
Cell surface gp120 binding assay.
A cell surface gp120 binding assay (34) was modified in the following manner. Secreted gp120 was generated by infecting 293T human cells with vaccinia virus at a multiplicity of infection (MOI) of 10 for 1 h at 37°C. Since 293T cells express low levels of CXCR4, QT6 quail cells were occasionally used to generate gp120 proteins from X4-tropic HIV isolates with no effect on binding results. Wild-type vaccinia virus WR was used to generate HXBc2, 8x, VCP, ROD/A, and ROD/B gp120 proteins, since these plasmids contained vaccinia virus promoters. Vaccinia virus vTF1.1, expressing T7 RNA polymerase (1), was used to generate HXBc2-V3BaL and 8x-V3BaL gp120 proteins, since these plasmids contained T7 promoters. Vaccinia virus-infected cells were then transfected with the appropriate gp120 expression plasmids for 4 h by the standard calcium phosphate method. HIV-1 JR-FL gp120 was generated by infecting 293T cells with recombinant vaccinia virus vBD6 (27) at an MOI of 10 for 1 h at 37°C. After plasmid transfection or vBD6 infection, the medium was replaced with Dulbecco's modified Eagle medium, high glucose, with 10% fetal bovine serum (DMEM-10) containing 100 μg of rifampin/ml and cells cultured overnight at 37°C. QT6 quail target receptor cells in T-25 flasks were generated by infecting with vTF1.1, transfecting with receptor plasmids, and expressing overnight at 37°C in DMEM-10 with 100 μg of rifampin/ml as above. All receptor plasmids contained T7 promoters, thus enhancing expression levels.
After overnight expression of gp120 producer cells and receptor target cells, medium containing gp120 was clarified by low-speed centrifugation for 10 min. Unpurified proteins were used directly or incubated with phosphate-buffered saline (PBS) or 100 nM sCD4, kindly provided by Tim Hart (GlaxoSmithKline, Philadelphia, Pa.), for 1 h at room temperature. Then 1.5 ml of gp120 supernatant with or without sCD4 was overlaid on receptor cells for 1 h at 37°C. Cells were detached from flasks and washed three times with 4°C PBS, and cell pellets were lysed with ice-cold lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris [pH 8.0], and Complete protease inhibitor cocktail [Roche Molecular Biochemicals, Indianapolis, Ind.]). Alternatively, cells were also detached from flasks first, pelleted, and then bound with 200 μl of gp120-containing supernatant to reduce the binding volume with no effect on binding results. Cell lysates were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
HIV-1 gp120 proteins were detected with rabbit anti-Env sera 1169 (47), except for 8x gp120, which was detected with monoclonal antibody (MAb) J1, which recognizes constant (C) domain 2 of gp120 (63). HIV-2 gp120 proteins were detected with MAb DA6, which cross-reacts with many heterologous HIV-2 and SIV gp120 proteins (33). The appropriate peroxidase-conjugated secondary antibody (anti-rabbit or anti-mouse) was then used (Jackson ImmunoResearch Laboratories, West Grove, Pa.), and Western blots were visualized using enhanced chemiluminescence and autoradiography film.
Protein purification and binding inhibition studies.
VCP gp120 protein was generated as above except that serum-free DMEM was used. Supernatant containing gp120 protein was harvested 24 to 30 h later, and gp120 was purified by lectin chromatography using Galanthus nivalis-lectin coupled agarose beads (Sigma, St. Louis, Mo.) as described previously (27, 47).
Inhibition of VCP gp120-CXCR4 binding was assessed with MAbs to CXCR4 (4G10 [84], kindly provided by Chris Broder, Uniformed Services University, Bethesda, Md., 12G5 [38], and 44717.111 [R & D Systems, Minneapolis, Minn.] [6]), SDF-1α (kindly provided by Mark Marsh, University College London, London, United Kingdom), and AMD3100 (kindly provided by Anormed, Inc., Langley, British Columbia, Canada). An anti-CD4 MAb (#19) was also assessed as a control (38). The gp120 binding assay was modified as follows. CXCR4 and CD4 receptor cells were generated by vTF1.1 infection-plasmid transfection as above. After overnight expression, cells were detached, pelleted, and then resuspended in 200 μl of DMEM-10 containing the inhibitors at various concentrations and 100 μg of rifampin/ml. Cells were then incubated for 15 m at 37°C. Purified VCP gp120 was then added at a concentration of 1 μg/ml, and cells were further incubated for 1 h at 37°C. Cells were then washed, lysed, and analyzed by SDS-PAGE and Western blotting as above.
Cell-cell fusion assay.
A cell-cell fusion assay (35, 71) was modified as described previously (58). Briefly, to generate effector env-expressing cells, QT6 cells in T-25 flasks were infected with vaccinia virus strain WR at an MOI of 10 for 1 h at 37°C and transfected for 4 h by the standard calcium phosphate method with 6 μg of the desired env-expressing plasmid and 6 μg of pSP64.vE/L.T7 RNAP, a plasmid containing T7 RNA polymerase under the regulation of the synthetic vaccinia virus early and late promoter (15, 58). Cells were incubated overnight with rifampin at a concentration of 100 μg/ml. To generate target receptor cells, QT6 cells in 24-well plates were transfected with the desired receptors and the T7.luciferase reporter plasmid in a total of 1 μg by the standard calcium phosphate method for 4 h and expressed overnight. Effector cells were mixed with receptor target cells, and cell-cell fusion was assessed 7.5 h later by lysing with 0.5% NP-40-PBS. After addition of luciferase substrate (Promega), luciferase activity was quantified with a Wallac 1450 Microbeta Plus luminometer.
Analysis of surface expression of coreceptors.
Cell surface expression of coreceptors was assessed essentially as described previously (6). Briefly, QT6 quail cells were infected with vaccinia virus vTF1.1 and transfected with either CXCR4, CXCR4-CXCR2 chimeras, CXCR4 with N-terminal deletions, or CXCR4 with alanine substitutions by the standard calcium phosphate method as described above. After overnight expression, cells were detached and washed with fluorescence-activated cell sorter (FACS) buffer (PBS supplemented with 3% fetal bovine serum and 0.05% sodium azide). Cells were then incubated with anti-CXCR4 MAb 44717.111 at a final concentration of 10 μg/ml for 30 min on ice. Alternatively, anti-CXCR4 MAb 6H8 (62), kindly provided by Fernando Arenzana-Seisdedos, was used for certain CXCR4 ECL2 alanine mutants at a concentration of 25 μg/ml. Cells were then washed and incubated with phycoerythrin-conjugated anti-mouse antisera (Vector Laboratories, Inc., Burlingame, Calif.) for 30 min on ice. The cells were then washed, fixed with 1% paraformaldehyde, and analyzed with a FACScan cell analyzer (Becton Dickinson, San Jose, Calif.). Data were evaluated with CellQuest software.
RESULTS
Binding of CD4-dependent and CD4-independent HIV-1 gp120s to chemokine receptors.
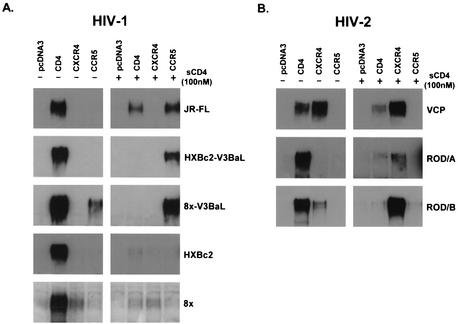
A cell surface gp120 binding assay was used to evaluate gp120-coreceptor interactions for a variety of R5- and X4-tropic isolates of HIV-1 and HIV-2. Secreted gp120 proteins were generated in 293T human cells, and binding to coreceptor-expressing target cells was determined by SDS-PAGE and Western blotting of cell lysates. As in published reports (27, 83), binding to CCR5 was detectable for the R5-tropic Env's of HIV-1 JR-FL and HXBc2 that contained the V3 loop of HIV-1/BaL (HXBc2-V3BaL), although only in the presence of sCD4 (Fig. 1A). gp120 from CD4-independent HIV-1/8x containing the HIV-1/BaL V3 loop (8x-V3BaL) could bind to CCR5 both in the presence and in the absence of sCD4 (Fig. 1A). Consistent with its reduced CXCR4 affinity, binding of X4-tropic HXBc2 gp120 to CXCR4 could not be detected with or without sCD4, and for 8x, gp120 binding to CXCR4 was barely detectable (Fig. 1A).
FIG. 1.
Binding of HIV-1 and HIV-2 gp120s to CD4, CXCR4, and CCR5. gp120 produced in 293T cells was bound to receptor-expressing QT6 cells in the presence or absence of 100 nM sCD4 as indicated. Cells were washed and lysed, and lysates were analyzed for bound gp120 by SDS-PAGE and Western blotting. HIV-1 gp120 proteins JR-FL, HXBc2-V3BaL, 8x-V3BaL, HXBc2, and 8x (A) or HIV-2 gp120 proteins VCP, ROD/A, and ROD/B (B) were evaluated on CD4, CCR5, CXCR4, or control pcDNA3-transfected cells.
In contrast, CXCR4 binding for two CD4-independent isolates, VCP and ROD/B, was readily detectable even in the absence of sCD4 (Fig. 1B). CXCR4 binding was also seen for the CD4-dependent HIV-2/ROD/A gp120, although only when sCD4 was present. Of note, binding was undetectable on CCR5, even though all three of these HIV-2 Env's were capable of using these receptors in fusion assays (58). Thus, the ability to detect gp120 binding to chemokine receptors was clearly a less sensitive indicator of coreceptor function than cell-cell fusion assays. Nonetheless, these results indicate that the VCP and ROD/B gp120s bind to CXCR4 with an affinity that is likely greater than that of HIV-1 gp120s, and that their CXCR4 binding sites are exposed and functional in the absence of CD4 triggering.
Effects of anti-CXCR4 MAbs, SDF-1α, and AMD3100 on VCP gp120 binding.
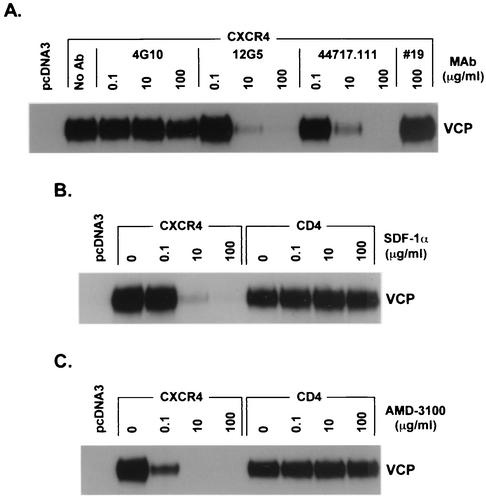
Since VCP and ROD/B gp120s could efficiently bind CXCR4, we could readily assess whether inhibitors of X4-tropic HIV viral entry also inhibited gp120 binding, and we could use domain-specific MAbs as a way to provide information on regions of CXCR4 important for Env interactions. We evaluated anti-CXCR4 MAbs (4G10, 12G5, and 44717.111), the CXCR4 natural ligand SDF-1α (8, 65), and the CXCR4 bicyclam antagonist AMD3100 (26, 73) for inhibition of VCP gp120-CXCR4 binding. Anti-CXCR4 MAbs 12G5 and 44717.111, which both bind an epitope located in ECL2 (6), inhibited VCP gp120-CXCR4 binding in a dose-dependent manner, while 4G10, which binds an epitope in the N terminus (84), did not (Fig. 2A). Anti-CD4 MAb #19, used as a control, had no effect on VCP gp120-CXCR4 binding. SDF-1α and AMD3100 also inhibited VCP gp120-CXCR4 binding while having no effect on VCP gp120-CD4 binding (Fig. 2B and C). Thus, HIV-2 gp120 binding to CXCR4 as measured in this assay was highly specific and inhibitable by anti-CXCR4 MAbs to ECL2 as well as specific agonists and antagonists of CXCR4.
FIG. 2.
Inhibition of VCP gp120 binding to CXCR4 with anti-CXCR4 MAbs, SDF-1α, and AMD3100. Control pcDNA-transfected or CXCR4 and CD4 receptor-expressing QT6 cells were generated and preincubated with the indicated concentration of inhibitor for 15 min at 37°C. Purified VCP gp120 was then added at a concentration of 1 μg/ml, and cells were further incubated for 1 h at 37°C. Cells were washed and lysed, and lysates were analyzed for bound gp120 by SDS-PAGE and Western blotting. (A) MAbs were evaluated on pcDNA3 control cells or CXCR4-expressing cells. 4G10 is an anti-CXCR4 MAb directed against the N terminus. 12G5 and 44717.111 are MAbs directed against the CXCR4 ECL2. #19 is an anti-CD4 MAb. (B and C) SDF-1α (B) or AMD3100 (C) was evaluated on pcDNA3 control cells or on CXCR4- or CD4-expressing cells.
Evaluation of HIV-2 gp120 binding and Env fusion using CXCR4-CXCR2 chimeras.
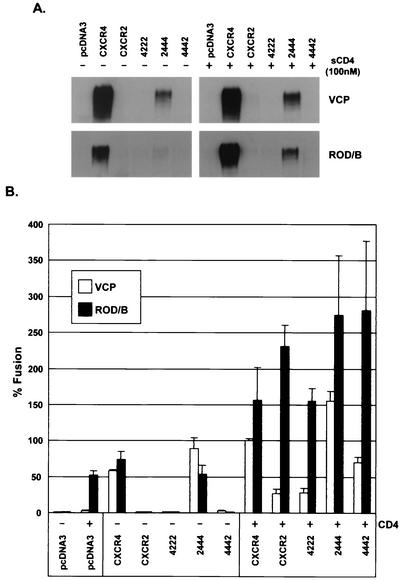
The ability of both VCP and ROD/B to efficiently bind to CXCR4 made it possible to investigate coreceptor determinants required for gp120 binding and to correlate this information with the ability of receptors to support membrane fusion. We analyzed several receptor chimeras containing specific domains from CXCR4 and CXCR2. Chimeras 2444 (containing the N-terminal domain from CXCR2 and the CXCR4 ECL1, -2, and -3 domains), 4442, and 4222 were evaluated because these were expressed on the cell surfaces to levels equivalent to 50 to 75% of the wild-type level (28; data not shown). We found that VCP gp120 could directly bind chimera 2444 but not CXCR2, 4222, or 4442 (Fig. 3A), consistent with the failure of an N-terminal CXCR4 MAb to prevent binding (Fig. 2A). Preincubation with sCD4 did not significantly enhance binding to any receptor. While ROD/B bound very weakly to chimera 2444 in the absence of sCD4, it bound nearly as well as VCP gp120 when sCD4 was present (Fig. 3A). Although these results suggest that the amino-terminal domain of CXCR4 is not required for CXCR4 binding, it likely plays some role in efficient use of CXCR4, since the binding of both gp120s to 2444 was reduced relative to binding to wild-type CXCR4. The fact that chimera 4442 was unable to bind VCP gp120 despite being well expressed also suggests a role for ECL3 (Fig. 3A). Unfortunately, a 2224 chimera was not well expressed on the cell surface (60) and therefore could not be used to assess the contribution of ECL3.
FIG. 3.
HIV-2 gp120 binding and Env fusion with CXCR4-CXCR2 chimeras. (A) HIV-2 VCP or ROD/B gp120 produced in 293T cells was bound to QT6 cells expressing either CXCR4, CXCR2, CXCR4-CXCR2 chimeras, or no receptor (pcDNA3) in the presence or absence of 100 nM sCD4 as indicated. Cells were washed and lysed, and lysates were analyzed for bound gp120 by SDS-PAGE and Western blotting. (B) HIV-2 env clones of VCP or ROD/B were evaluated in a cell-cell fusion assay on QT6 target cells expressing the receptors listed above in the presence or absence of CD4. Nomenclature of chimeras shows the first number representing the N terminus and the second, third, and fourth numbers representing the second, third, and fourth extracellular loops, respectively; “4” refers to CXCR4, and “2” refers to CXCR2. Percent fusion was calculated by using luciferase activity normalized to VCP fusion on CXCR4 with CD4. Values are means plus standard errors of the means.
CXCR4-CXCR2 chimeras were next evaluated in cell-cell fusion assays with VCP and ROD/B Env's. In the absence of CD4, both VCP and ROD/B could fuse with 2444 but not with 4222, 4442, or CXCR2 (Fig. 3B). However, with CD4, VCP and ROD/B could utilize all receptors, although a greater degree of fusion was observed for ROD/B than for VCP. Thus, the abilities of receptors to bind to the HIV-2 gp120s correlated with their abilities to function independently of CD4 in fusion assays. However, because these receptors were able to function to varying levels in the presence of CD4, gp120 binding was a less sensitive indicator for receptor utilization than cell-cell fusion.
Evaluation of the CXCR4 amino terminus in gp120 binding and Env fusion by using N-terminal deletion mutants.
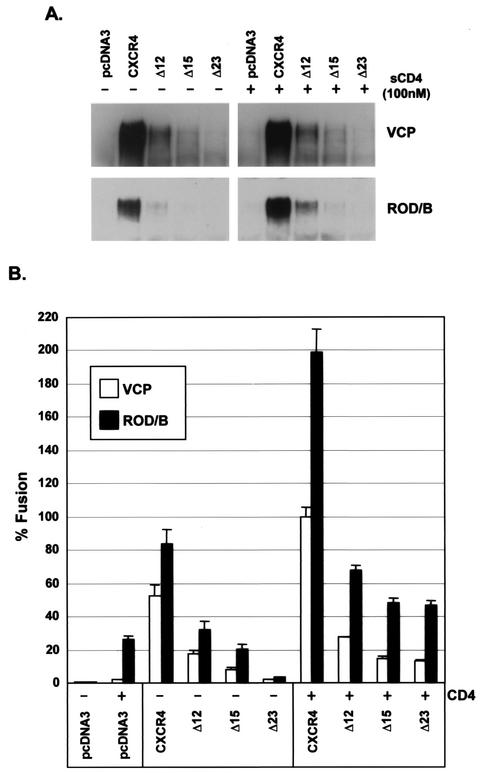
The role of the CXCR4 N terminus in gp120 binding was further assessed using a panel of mutant receptors containing deletions of the first 12, 15 or 23 amino acids (designated Δ12, Δ15, or Δ23, respectively). All mutants were expressed on the cell surface at levels between 20 and 30% that of wild-type CXCR4 (data not shown). VCP and ROD/B gp120 binding to these mutants was markedly reduced in the presence or absence of sCD4 and declined progressively with increasing length of the deletion (Fig. 4A). These findings correspond to the results of cell-cell fusion assays, where fusion with or without sCD4 was similarly reduced (Fig. 4B). Thus, these results provide additional support for the role of the CXCR4 N terminus in efficient gp120 binding as well as in CD4-independent and CD4-dependent fusion.
FIG. 4.
HIV-2 gp120 binding and Env fusion with CXCR4 N-terminal deletions. (A) HIV-2 VCP or ROD/B gp120 produced in 293T cells was bound to QT6 cells expressing CXCR4, CXCR4 with N-terminal deletions, or no receptor (pcDNA3) in the presence or absence of 100 nM sCD4 as indicated. Cells were washed and lysed, and lysates were analyzed for bound gp120 by SDS-PAGE and Western blotting. N-terminal CXCR4 deletions consisted of deletions of the first 12, 15, or 23 amino acids (Δ12, Δ15, or Δ23). (B) HIV-2 env clones of VCP or ROD/B were evaluated in a cell-cell fusion assay on QT6 target cells expressing the receptors listed above in the presence or absence of CD4. Percent fusion was calculated by using luciferase activity normalized to VCP fusion on CXCR4 with CD4. Values are means plus standard errors of the means.
Human-rat CXCR4 chimeras implicate ECL2 of CXCR4 in HIV-2 gp120 binding.
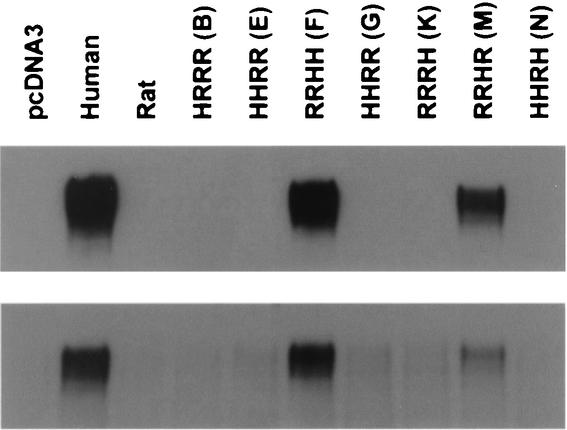
We also evaluated gp120 binding to a series of human-rat CXCR4 chimeras described previously (10) that allowed the contribution of individual extracellular domains to be assessed in a different context. Neither VCP nor ROD/B gp120 bound to rat CXCR4 (Fig. 5). However, for both gp120s, binding was detectable to RRHH and RRHR, but not to HRRR, HHRR, RRRH, or HHRH chimeras. These results suggest that the human ECL2 has elements that allow efficient gp120 binding, and they support infection results obtained with ROD/B and these chimeras in identifying ECL2 as important in viral entry (67). Thus, while the CXCR4-CXCR2 chimeras and N-terminal deletion mutants implicated determinants for gp120 binding in the N terminus and ECL3, the human-rat chimeras suggested that ECL2 was also a contributing factor.
FIG. 5.
HIV-2 gp120 binding to human-rat CXCR4 chimeras. HIV-2 VCP or ROD/B gp120 produced in 293T cells was bound to QT6 cells expressing human CXCR4, rat CXCR4, human-rat CXCR4 chimeras, or no receptor (pcDNA3). Cells were washed and lysed, and lysates were analyzed for bound gp120 by SDS-PAGE and Western blotting. VCP gp120 was evaluated without sCD4, while ROD/B gp120 was evaluated in the presence of 100 nM sCD4 (Fig. 1B). Nomenclature of chimeras shows the first letter representing the N terminus and the second, third, and fourth letters representing the second, third, and fourth extracellular loops, respectively. “H” refers to human CXCR4, and “R” refers to rat CXCR4. Previously used single-letter codes for human-rat CXCR4 chimeras are also provided (10). Construct G differs from construct E in that construct G contains a human cytoplasmic tail.
VCP and ROD/B gp120 binding to CXCR4 alanine mutants.
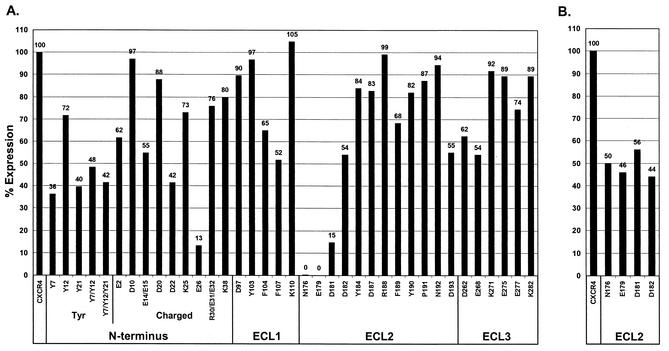
To identify individual residues important for gp120 binding, VCP and ROD/B gp120 binding was assessed on an extensive panel of CXCR4 mutants containing alanine substitutions in each extracellular domain. Given the importance of charged amino acids and tyrosines in CXCR4 fusion for HIV-1 (9, 14, 50), the mutations initially chosen were those replacing these residues either singly or in combination. Selected phenylalanines were also targeted. Using anti-CXCR4 MAbs to ECL2 or the N terminus, we found that all CXCR4 mutants were expressed at levels ranging from 36 to 105% that of wild-type CXCR4 except for E26A and D181A (13 and 15%, respectively, of the wild-type CXCR4 level) (Fig. 6).
FIG. 6.
Cell surface expression of CXCR4 alanine mutants. QT6 quail cells were infected with vaccinia virus vTF1.1 and transfected with CXCR4 or CXCR4 with alanine substitutions by the standard calcium phosphate method. After overnight expression, cells were detached and analyzed by FACS using MAbs to CXCR4, either 44717.111 (A) or 6H8 (B). The mean channel fluorescence intensity (MCF) for each construct is expressed relative to that of wild-type CXCR4 after subtraction of the MCF of cells transfected with vector alone. Because residues N176, E179, and D181 in ECL2 have been shown to be part of the epitope for MAb 44717.111 (6), MAb 6H8, which binds the N terminus of CXCR4 (62), was used to assess expression of these constructs.
CXCR4 N terminus mutants.
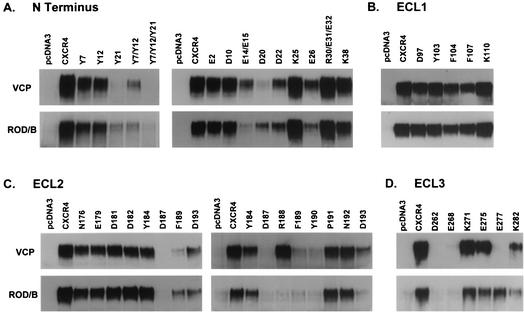
Because sCD4 was found to enhance ROD/B but not VCP gp120 binding to CXCR4, sCD4 was added to ROD/B gp120 while VCP gp120 binding was performed without sCD4. We found that alanines at Y7 and Y12 did not by themselves abolish VCP binding to CXCR4, although in combination they reduced binding (Fig. 7A). However, an alanine at Y21 severely affected VCP binding, and no binding was detected to a construct lacking tyrosines at all three positions. Similar results were found for the ROD/B gp120 (Fig. 7A).
FIG. 7.
HIV-2 gp120 binding to CXCR4 with alanine substitutions in the N terminus, ECL1, ECL2, or ECL3. HIV-2 VCP or ROD/B gp120 produced in 293T cells was bound to QT6 cells expressing CXCR4, CXCR4 with alanine substitutions, or no receptor (pcDNA3). Cells were washed and lysed, and lysates were analyzed for bound gp120 by SDS-PAGE and Western blotting. VCP gp120 was evaluated without sCD4, while ROD/B gp120 was evaluated in the presence of 100 nM sCD4. The original amino acids replaced by alanine and their numerical positions are shown for the N terminus (A), ECL1 (B), ECL2 (C), and ECL3 (D).
gp120 binding to mutants with alanines replacing N-terminal charged amino acids identified additional residues, and results with VCP and ROD/B were largely similar. For VCP, an alanine at D20 severely reduced binding (Fig. 7A) while mutations at E14/E15 and D22 produced more-modest reductions. Similar results were found for ROD/B, although the E14/E15 mutant produced a greater decrease. Reductions in both VCP and ROD/B binding were seen with the E26 mutant, although this may have been due to its reduced surface expression (Fig. 6A). Thus, in the CXCR4 N terminus, tyrosine residues (particularly Y21) and acidic residues, particularly E14, E15, D20, and D22, play important roles in mediating efficient gp120 binding. Subtle differences were observed between the two different gp120 proteins tested, with VCP depending more on the 20-21 region of the CXCR4 N terminus and ROD/B depending more on the 14-15 region.
CXCR4 extracellular loop mutants.
In ECL1, none of the five alanine mutations introduced at positions D97, Y103, F104, F107, and K110 affected either VCP or ROD/B gp120 binding (Fig. 7B), consistent with findings with CXCR4-CXCR2 and human-rat CXCR4 chimeras, which failed to support a role for this domain. In ECL2, alanines at D187 and F189 severely impaired gp120 binding (Fig. 7C), while an alanine at D193 reduced binding but to a more limited extent. We further assessed the region between D187 and D193, since Y190 has been shown to be involved in SDF-1α signaling (86). We found that an alanine at Y190 reduced gp120 binding, while alanines at P191 and N192 had no effect (Fig. 7C, right panels). In addition, an alanine at R188 reduced ROD/B gp120 binding but had no effect on VCP gp120 binding.
Mutations at charged amino acids in ECL3 also yielded some that affected gp120 binding. Alanines at D262 and E268 markedly reduced binding for both VCP and ROD/B proteins, while an alanine at K282 had a greater effect on ROD/B binding and an alanine at E277 had a greater effect on VCP binding (Fig. 7D). Interestingly, D262 has previously been shown to be important for HIV-1 entry (9), while E268 has been shown to be important for SDF-1α binding (86).
Effects of alanine mutations in CXCR4 on VCP and ROD/B Env fusion.
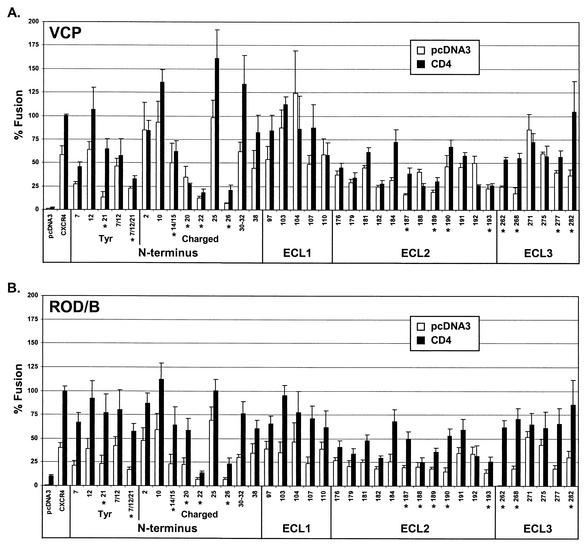
The above studies identified several alanine substitutions in the CXCR4 extracellular domains that affected VCP and/or ROD/B gp120 binding. In order to determine the impact of these mutations on CXCR4 function, we evaluated VCP and ROD/B Env's in cell-cell fusion assays with each of the CXCR4 alanine mutants with or without CD4 coexpression. In general, mutations that produced a marked reduction in gp120 binding (Fig. 7) produced a reduction in CD4-independent fusion by at least 50%, and in most cases by 75%, from that for wild-type CXCR4 (Fig. 8). Fusion was typically increased for these constructs when they were coexpressed with CD4, again indicating that binding of monomeric gp120 to cell surface CXCR4 is a less sensitive indicator of coreceptor activity than functional assays, particularly in the presence of CD4. Even constructs that exhibited no gp120 binding for either VCP or ROD/B (e.g., Y7/Y12/Y21, D187, D262, and E268) supported cell fusion with CD4 at levels >50% that of wild-type CXCR4. In one case, a mutation at D262 totally abolished CD4-independent fusion by ROD/B Env, but it was largely restored when CD4 was coexpressed.
FIG. 8.
HIV-2 Env fusion with CXCR4 with alanine substitutions in the N terminus, ECL1, ECL2, or ECL3. HIV-2 env clones of VCP (A) or ROD/B (B) were evaluated in a cell-cell fusion assay on QT6 target cells expressing CXCR4, CXCR4 with alanine substitutions, or no receptor (pcDNA3) in the presence or absence of CD4. Percent fusion was calculated by using luciferase activity normalized either to VCP fusion on CXCR4 with CD4 (A) or to ROD/B fusion on CXCR4 with CD4 (B). Values are means plus standard errors of the means. Asterisks indicate mutations that produced a marked reduction in gp120 binding (Fig. 7).
An intriguing finding was that some mutations showed no effects or only minor effects on gp120 binding but markedly reduced both CD4-independent and CD4-dependent fusion. These included mutations of N176, E179, D181, and D182 in the amino-terminal region of ECL2. It is possible that these mutations affect a function after gp120 binding or that binding to Env in its native trimeric conformation is more severely affected than is binding to monomeric gp120. Finally, although fusion with the E26 mutant was markedly reduced with or without CD4, this effect was most likely due to reduced surface expression, as noted above.
DISCUSSION
For primary HIV-1 strains, CD4 binding induces structural alterations in gp120 that enable it to interact with CCR5 and/or CXCR4 (77, 83). For R5-tropic viruses, studies using gp120 binding, cell-cell fusion, and viral entry assays have implicated the N terminus and multiple residues in the extracellular loops of CCR5 in mediating gp120 binding and viral entry (reviewed in reference 30). In contrast, analysis of Env-CXCR4 interactions has been hampered because the few HIV-1 strains evaluated to date bind to CXCR4 with low affinity (4, 45). Results with cell-cell fusion and viral entry assays have supported the view that the CXCR4 N terminus and ECL2 are important for Env function (9, 28, 50, 60, 67). However, these assays have not provided insight into exactly how gp120 binds CXCR4 and which residues are most important.
Since enhanced exposure and binding efficiency for the chemokine receptor can be characteristics that confer CD4 independence for certain Env's (34, 58, 61), we hypothesized that some CD4-independent isolates might exhibit higher binding affinities for CXCR4, and we evaluated gp120s from two well-characterized HIV-2 strains that have been shown to utilize CXCR4 in the absence of CD4 (38, 69). In contrast to HIV-1 gp120s, binding of VCP and ROD/B gp120s to CXCR4 was readily detectable. This binding was highly specific and inhibitable by anti-CXCR4 MAbs and by AMD3100 and SDF-1α, supporting the view that these gp120 proteins bind to CXCR4 with a higher affinity than that of HIV-1 gp120 proteins. However, we note that not all gp120s from CD4-independent isolates necessarily exhibit high-affinity binding to coreceptors. The HIV-1 strain 8x can infect CD4-negative cells in a CXCR4-dependent manner, even though its gp120 still binds poorly to CXCR4 (Kd, 500 nM) (45). Apparently, simply exposing the chemokine receptor binding site can itself be sufficient for CD4 independence. Of note, in contrast to HXB gp120, we did detect minimal binding of 8x gp120 to CXCR4 in this study (Fig. 1A), suggesting that its affinity, though low, is higher than that of HXB gp120.
Using CXCR4-CXCR2 and human-rat CXCR4 chimeras and an extensive panel of alanine mutants, we showed that the N terminus, ECL2, and ECL3 all contribute to gp120 binding, and we identified specific residues in each domain that are involved. For VCP and/or ROD/B, mutations at E14/E15, D20, Y21, and D22 (in the N terminus), D187, R188, F189, Y190, and D193 (in ECL2), and D262, E268, E277, and K282 (in ECL3) all reduced gp120-CXCR4 binding (summarized in Fig. 9). Previous studies using cell-cell fusion and viral entry assays have shown that most of these residues play some role in CXCR4's ability to function as an HIV coreceptor and have supported the view that Env-CXCR4 interactions involve tyrosines and negatively charged residues in the N terminus, ECL2, and ECL3 (9, 14, 50). Our results indicate that the mechanism for this effect likely occurs at the level of binding. Moreover, tyrosines in the N termini of CCR5 (40) and CXCR4 (39) have been shown to be sulfated, again supporting the view that negatively charged regions on the coreceptor contribute substantially to gp120 interactions. In the present study we also show that both Tyr190 and Phe189 in the CXCR4 ECL2 contribute to gp120 binding, suggesting that aromatic or uncharged amino acids in this region are required.
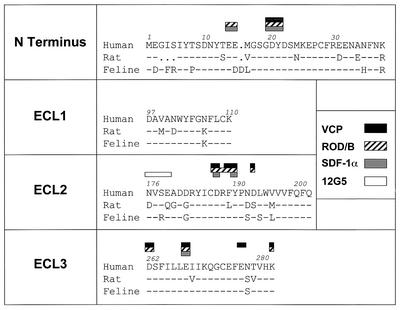
FIG. 9.
Summary of binding determinants on CXCR4 for VCP and ROD/B gp120s. A sequence alignment of extracellular regions in human, rat, and feline CXCR4 is shown. The first, second, and third extracellular loops are referred to as ECL1, ECL2, and ECL3, respectively. Binding sites identified in this study are shown for VCP gp120 (solid rectangles) and ROD/B gp120 (hatched rectangles). Shaded rectanges, residues important for SDF-1α binding and signaling (9, 86); open rectangle, epitope for 12G5 (6, 10).
Differences were noted in the effects of CXCR4 mutations on VCP versus ROD/B gp120 binding. Alanines at D20 and E277 reduced VCP binding more than ROD/B binding, while an alanine at R188 reduced ROD/B binding more than VCP binding. We also observed that VCP gp120, but not ROD/B gp120, could bind to feline CXCR4 in the absence of CD4 (data not shown). Differential effects of CXCR4 mutations on HIV-1 isolates have been noted in functional assays, supporting the view that X4 viruses vary in their structural requirements for CXCR4 utilization (9, 14). Similar findings for CCR5 utilization have been noted among R5 isolates (reviewed in reference 30). Our findings indicate that for HIV-2, these strain-specific differences result at least in part from differences in gp120 binding to CXCR4. Of note, these differences may account for the different sensitivities exhibited by different virus strains to coreceptor agonists and antagonists (25, 69a, 78) as well as for differences in their sensitivities to neutralizing antibodies (11, 74, 85).
It is intriguing that the CXCR4 residues implicated in VCP and ROD/B gp120 binding overlap substantially with those involved in SDF-1α binding and/or signaling. E14, E15, D20, Y21, D187, and E268 were each found to be important for SDF-1α binding, while Y190 and D187 were important for signaling (9, 86) (Fig. 9). Apparently, gp120 has evolved a mechanism of interacting with CXCR4 that closely mimics that of the natural ligand. It will be of interest to determine if gp120s that exhibit high-affinity binding to CXCR4 are also able to induce signaling through this receptor. A variety of biological effects have been attributed to gp120 interactions with CXCR4 in the presence or absence of CD4 (21, 23, 49, 59). While signaling is clearly not required for HIV entry (3, 9, 28), high-affinity interactions with CXCR4 could be relevant to pathogenesis, as has recently been suggested for neurovirulent HIV-1 isolates that exhibit high-affinity CCR5 interactions and induce neuronal apoptosis in vitro (44).
Despite the identification of residues important for efficient gp120 binding to CXCR4, mutation of these residues did not abolish CD4-independent fusion and in most cases only modestly reduced fusion levels. Moreover, fusion could often be enhanced or restored to near-wild-type levels in the presence of CD4. Discrepancies between the ability of a coreceptor mutant to support detectable gp120 binding in the context of an equilibrium binding assay and its ability to support virus infection or cell-cell fusion have been noted previously (5, 27, 28). If a coreceptor mutation abolishes detectable gp120 binding without eliminating membrane fusion activity, we interpret this to mean that the mutation reduces gp120-coreceptor affinity to a level where it cannot be easily detected by standard binding assays. Indeed, both VCP and ROD/B (58) are capable of using CCR5 for fusion, although binding of their gp120s to CCR5 was not detectable in our assay (Fig. 1B). There is likely to be a range of affinities that are compatible with membrane fusion, and a threshold Kd above which membrane fusion does not occur. Indeed, a broad range of Env-coreceptor affinities that are compatible with virus infection have been measured; they range from 5 to at least 500 nM. However, it is possible that the affinity of monomeric gp120 binding to coreceptors measured in these assays may not reflect that of gp120 expressed on the native Env trimer.
Env-coreceptor avidity and utilization likely reflect a complex interplay of gp120 affinity and interactions with CD4 and other cellular molecules. In the context of virus infection, binding of Env trimers to cell surface attachment factors such as DC-SIGN (43, 57) or DC-SIGNR (66, 76) or to CD4 likely constrains the virus to the two-dimensional plane of the cellular membrane, making it more likely that the coreceptor will be encountered. Also, being trimeric, Env can engage multiple CD4 molecules, likely providing a marked increase in total avidity for the fusion reaction. At first glance, the cooperative nature of this entry mechanism illustrates potential difficulties in inhibiting the fusion process through strategies aimed at blocking gp120 binding to chemokine receptors with small-molecule inhibitors alone. However, since multiple receptor binding events are needed to trigger Env trimers (53), and several trimers are likely needed to form a fusion pore, inhibition of a single binding event or Env trimer could effectively eliminate the activities of other subunits and trimers (C. M. McManus et al., submitted for publication). This may help explain the effectiveness of entry inhibitors such as T20, which have been shown to have significant antiviral activity both in vitro and in vivo (51, 81).
CXCR4 and CCR5 are the major viral coreceptors, even though they share relatively low amino acid identity in their extracellular domains. However, they share a conserved disulfide bond structure, and each contains sulfated tyrosines and/or negatively charged residues in the amino-terminal domain that are needed for efficient coreceptor activity. That there is structural similarity between the major coreceptors is evidenced by the fact that some viral Env proteins can interact with both coreceptors, and also that relatively subtle mutations in CXCR4 can make it possible for some R5 viruses to use this receptor (12, 13, 79). As with CCR5, the N terminus and ECL2 of CXCR4 are important for efficient coreceptor activity. It has been proposed that the N terminus of CCR5 engages the C4 region and the V3 base of gp120 (20, 41) while the CCR5 extracellular loops engage the V3 crown (20). Indeed, sulfated peptides from the CCR5 N terminus have been shown to bind to gp120 and to inhibit entry of R5-tropic HIV-1 isolates (19, 20, 41). We do not yet know if the CXCR4 N terminus and/or sulfated peptides from this domain also engage the C4 domain and the V3 base of gp120, or if the CXCR4 extracellular loops bind the gp120 V3 loop. However, the Env proteins studied here that bind CXCR4 with an apparently increased affinity can be used to more fully address which coreceptor domains and residues interact with the V3 loop, bridging sheet, and other regions in Env needed for successful interactions.
In summary, dissecting the requirements of HIV entry is pivotal in understanding HIV entry mechanisms and pathogenesis. The CD4-independent isolates of HIV-2 that we have described, which exhibit an apparently increased affinity for CXCR4, should be highly useful in identifying determinants on Env and the receptor that contribute to coreceptor affinity and in addressing the biological consequences of mutations that alter this interaction. In addition, there are multiple compounds currently being evaluated that inhibit various stages of HIV entry including Env-CD4 binding, Env-coreceptor binding, and formation of other Env fusion intermediates (reviewed in reference 55). Since HIV usage of CXCR4 has been correlated with rapid progression to AIDS, a small-molecule inhibitor that inhibits this interaction in conjunction with other antiviral strategies may be useful in preventing the development of AIDS. Thus, a better understanding of how HIV Env engages CXCR4 for entry is needed to design molecules effective in preventing this interaction.
Acknowledgments
We thank Marc Alizon, Anne Brelot, Mark Biscone, Trevor L. Hoffman, and Jacqueline D. Reeves for plasmids; Tim Hart for sCD4; Chris Broder for MAb 4G10; Fernando Arenzana-Seisdedos for MAb 6H8; Mark Marsh for SDF-1α; Anormed, Inc., for AMD3100; and Navid Ahmad for technical assistance. We also thank A. Paul Godillot and the Structural Biology Core at the University of Pennsylvania Center for AIDS Research for providing purified HIV-2 VCP gp120.
G.L. was supported by the National Institutes of Health (NIH) Medical Scientist Training Program. R.W.D. was supported by NIH grants R01 40880 and R01 35383, by a Burroughs Wellcome Fund translational research award, and by an Elizabeth Glaser Scientist award from the Pediatric AIDS Foundation. J.A.H. was supported by NIH grant R01 AI45378. Support was also provided by the Penn Center for AIDS Research, NIH grant P30 AI45008.
REFERENCES
- 1.Alexander, W. A., B. Moss, and T. R. Fuerst. 1992. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J. Virol. 66:2934-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 3.Alkhatib, G., M. Locati, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1997. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology 234:340-348. [DOI] [PubMed] [Google Scholar]
- 4.Babcock, G. J., T. Mirzabekov, W. Wojtowicz, and J. Sodroski. 2001. Ligand binding characteristics of CXCR4 incorporated into paramagnetic proteoliposomes. J. Biol. Chem. 276:38433-38440. [DOI] [PubMed] [Google Scholar]
- 5.Baik, S. S., R. W. Doms, and B. J. Doranz. 1999. HIV and SIV gp120 binding does not predict coreceptor function. Virology 259:267-273. [DOI] [PubMed] [Google Scholar]
- 6.Baribaud, F., T. G. Edwards, M. Sharron, A. Brelot, N. Heveker, K. Price, F. Mortari, M. Alizon, M. Tsang, and R. W. Doms. 2001. Antigenically distinct conformations of CXCR4. J. Virol. 75:8957-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, E. A. 1998. HIV entry and tropism. When one receptor is not enough. Adv. Exp. Med. Biol. 452:151-157. [PubMed] [Google Scholar]
- 8.Bleul, C. C., M. Farzan, H. Choe, C. Parolin, I. Clark-Lewis, J. Sodroski, and T. A. Springer. 1996. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382:829-833. [DOI] [PubMed] [Google Scholar]
- 9.Brelot, A., N. Heveker, M. Montes, and M. Alizon. 2000. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J. Biol. Chem. 275:23736-23744. [DOI] [PubMed] [Google Scholar]
- 10.Brelot, A., N. Heveker, O. Pleskoff, N. Sol, and M. Alizon. 1997. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J. Virol. 71:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burton, D. R., and D. C. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11:S87-S98. [PubMed] [Google Scholar]
- 12.Chabot, D. J., and C. C. Broder. 2000. Substitutions in a homologous region of extracellular loop 2 of CXCR4 and CCR5 alter coreceptor activities for HIV-1 membrane fusion and virus entry. J. Biol. Chem. 275:23774-23782. [DOI] [PubMed] [Google Scholar]
- 13.Chabot, D. J., H. Chen, D. S. Dimitrov, and C. C. Broder. 2000. N-linked glycosylation of CXCR4 masks coreceptor function for CCR5-dependent human immunodeficiency virus type 1 isolates. J. Virol. 74:4404-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chabot, D. J., P. F. Zhang, G. V. Quinnan, and C. C. Broder. 1999. Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. J. Virol. 73:6598-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakrabarti, S., J. R. Sisler, and B. Moss. 1997. Compact, synthetic, vaccinia virus early/late promoter for protein expression. BioTechniques 23:1094-1097. [DOI] [PubMed] [Google Scholar]
- 16.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 17.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 18.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cormier, E. G., M. Persuh, D. A. Thompson, S. W. Lin, T. P. Sakmar, W. C. Olson, and T. Dragic. 2000. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 97:5762-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cormier, E. G., D. N. Tran, L. Yukhayeva, W. C. Olson, and T. Dragic. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 75:5541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis, C. B., I. Dikic, D. Unutmaz, C. M. Hill, J. Arthos, M. A. Siani, D. A. Thompson, J. Schlessinger, and D. R. Littman. 1997. Signal transduction due to HIV-1 envelope interactions with chemokine receptors CXCR4 or CCR5. J. Exp. Med. 186:1793-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, S. J. O'Brien, et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 23.Del Corno, M., Q. H. Liu, D. Schols, E. de Clercq, S. Gessani, B. D. Freedman, and R. G. Collman. 2001. HIV-1 gp120 and chemokine activation of Pyk2 and mitogen-activated protein kinases in primary macrophages mediated by calcium-dependent, pertussis toxin-insensitive chemokine receptor signaling. Blood 98:2909-2916. [DOI] [PubMed] [Google Scholar]
- 24.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 25.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, Z. Zhang, W. A. O'Brien, L. Ratner, G. M. Shaw, and E. Hunter. 2001. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J. Virol. 75:8605-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donzella, G. A., D. Schols, S. W. Lin, J. A. Este, K. A. Nagashima, P. J. Maddon, G. P. Allaway, T. P. Sakmar, G. Henson, E. De Clercq, and J. P. Moore. 1998. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 4:72-77. [DOI] [PubMed] [Google Scholar]
- 27.Doranz, B. J., S. S. Baik, and R. W. Doms. 1999. Use of a gp120 binding assay to dissect the requirements and kinetics of human immunodeficiency virus fusion events. J. Virol. 73:10346-10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doranz, B. J., M. J. Orsini, J. D. Turner, T. L. Hoffman, J. F. Berson, J. A. Hoxie, S. C. Peiper, L. F. Brass, and R. W. Doms. 1999. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J. Virol. 73:2752-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 30.Dragic, T. 2001. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J. Gen. Virol. 82:1807-1814. [DOI] [PubMed] [Google Scholar]
- 31.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 32.Dumonceaux, J., S. Nisole, C. Chanel, L. Quivet, A. Amara, F. Baleux, P. Briand, and U. Hazan. 1998. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J. Virol. 72:512-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edinger, A. L., M. Ahuja, T. Sung, K. C. Baxter, B. Haggarty, R. W. Doms, and J. A. Hoxie. 2000. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J. Virol. 74:7922-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edinger, A. L., C. Blanpain, K. J. Kunstman, S. M. Wolinsky, M. Parmentier, and R. W. Doms. 1999. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J. Virol. 73:4062-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edinger, A. L., and R. W. Doms. 1999. A cell-cell fusion assay to monitor HIV-1 Env interactions with chemokine receptors. Methods Mol. Med. 17:41-49. [DOI] [PubMed] [Google Scholar]
- 36.Edinger, A. L., J. L. Mankowski, B. J. Doranz, B. J. Margulies, B. Lee, J. Rucker, M. Sharron, T. L. Hoffman, J. F. Berson, M. C. Zink, V. M. Hirsch, J. E. Clements, and R. W. Doms. 1997. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc. Natl. Acad. Sci. USA 94:14742-14747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards, T. G., T. L. Hoffman, F. Baribaud, S. Wyss, C. C. LaBranche, J. Romano, J. Adkinson, M. Sharron, J. A. Hoxie, and R. W. Doms. 2001. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J. Virol. 75:5230-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Endres, M. J., P. R. Clapham, M. Marsh, M. Ahuja, J. D. Turner, A. McKnight, J. F. Thomas, B. Stoebenau-Haggarty, S. Choe, P. J. Vance, T. N. Wells, C. A. Power, S. S. Sutterwala, R. W. Doms, N. R. Landau, and J. A. Hoxie. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87:745-756. [DOI] [PubMed] [Google Scholar]
- 39.Farzan, M., G. J. Babcock, N. Vasilieva, P. L. Wright, E. Kiprilov, T. Mirzabekov, and H. Choe. 2002. The role of post-translational modifications of the CXCR4 amino terminus in stromal-derived factor 1α association and HIV-1 entry. J. Biol. Chem. 277:29484-29489. [DOI] [PubMed] [Google Scholar]
- 40.Farzan, M., T. Mirzabekov, P. Kolchinsky, R. Wyatt, M. Cayabyab, N. P. Gerard, C. Gerard, J. Sodroski, and H. Choe. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96:667-676. [DOI] [PubMed] [Google Scholar]
- 41.Farzan, M., N. Vasilieva, C. E. Schnitzler, S. Chung, J. Robinson, N. P. Gerard, C. Gerard, H. Choe, and J. Sodroski. 2000. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J. Biol. Chem. 275:33516-33521. [DOI] [PubMed] [Google Scholar]
- 42.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 43.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 44.Gorry, P. R., J. Taylor, G. H. Holm, A. Mehle, T. Morgan, M. Cayabyab, M. Farzan, H. Wang, J. E. Bell, K. Kunstman, J. P. Moore, S. M. Wolinsky, and D. Gabuzda. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 76:6277-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffman, T. L., G. Canziani, L. Jia, J. Rucker, and R. W. Doms. 2000. A biosensor assay for studying ligand-membrane receptor interactions: binding of antibodies and HIV-1 env to chemokine receptors. Proc. Natl. Acad. Sci. USA 97:11215-11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffman, T. L., and R. W. Doms. 1998. Chemokines and coreceptors in HIV/SIV-host interactions. AIDS 12:S17-S26. [PubMed] [Google Scholar]
- 47.Hoffman, T. L., C. C. LaBranche, W. Zhang, G. Canziani, J. Robinson, I. Chaiken, J. A. Hoxie, and R. W. Doms. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. USA 96:6359-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang, Y., W. A. Paxton, S. M. Wolinsky, A. U. Neumann, L. Zhang, T. He, S. Kang, D. Ceradini, Z. Jin, K. Yazdanbakhsh, K. Kunstman, D. Erickson, E. Dragon, N. R. Landau, J. Phair, D. D. Ho, and R. A. Koup. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 2:1240-1243. [DOI] [PubMed] [Google Scholar]
- 49.Iyengar, S., D. H. Schwartz, and J. E. Hildreth. 1999. T cell-tropic HIV gp120 mediates CD4 and CD8 cell chemotaxis through CXCR4 independent of CD4: implications for HIV pathogenesis. J. Immunol. 162:6263-6267. [PubMed] [Google Scholar]
- 50.Kajumo, F., D. A. Thompson, Y. Guo, and T. Dragic. 2000. Entry of R5X4 and X4 human immunodeficiency virus type 1 strains is mediated by negatively charged and tyrosine residues in the amino-terminal domain and the second extracellular loop of CXCR4. Virology 271:240-247. [DOI] [PubMed] [Google Scholar]
- 51.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 52.Kolchinsky, P., T. Mirzabekov, M. Farzan, E. Kiprilov, M. Cayabyab, L. J. Mooney, H. Choe, and J. Sodroski. 1999. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J. Virol. 73:8120-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuhmann, S. E., E. J. Platt, S. L. Kozak, and D. Kabat. 2000. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol. 74:7005-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.LaBranche, C. C., G. Galasso, J. P. Moore, D. P. Bolognesi, M. S. Hirsch, and S. M. Hammer. 2001. HIV fusion and its inhibition. Antivir. Res. 50:95-115. [DOI] [PubMed] [Google Scholar]
- 56.LaBranche, C. C., T. L. Hoffman, J. Romano, B. S. Haggarty, T. G. Edwards, T. J. Matthews, R. W. Doms, and J. A. Hoxie. 1999. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J. Virol. 73:10310-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee, B., G. Leslie, E. Soilleux, U. O'Doherty, S. Baik, E. Levroney, K. Flummerfelt, W. Swiggard, N. Coleman, M. Malim, and R. W. Doms. 2001. cis expression of DC-SIGN allows for more efficient entry of human and simian immunodeficiency viruses via CD4 and a coreceptor. J. Virol. 75:12028-12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin, G., B. Lee, B. S. Haggarty, R. W. Doms, and J. A. Hoxie. 2001. CD4-independent use of rhesus CCR5 by human immunodeficiency virus type 2 implicates an electrostatic interaction between the CCR5 N terminus and the gp120 C4 domain. J. Virol. 75:10766-10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu, Q. H., D. A. Williams, C. McManus, F. Baribaud, R. W. Doms, D. Schols, E. De Clercq, M. I. Kotlikoff, R. G. Collman, and B. D. Freedman. 2000. HIV-1 gp120 and chemokines activate ion channels in primary macrophages through CCR5 and CXCR4 stimulation. Proc. Natl. Acad. Sci. USA 97:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu, Z., J. F. Berson, Y. Chen, J. D. Turner, T. Zhang, M. Sharron, M. H. Jenks, Z. Wang, J. Kim, J. Rucker, J. A. Hoxie, S. C. Peiper, and R. W. Doms. 1997. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc. Natl. Acad. Sci. USA 94:6426-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin, K. A., R. Wyatt, M. Farzan, H. Choe, L. Marcon, E. Desjardins, J. Robinson, J. Sodroski, C. Gerard, and N. P. Gerard. 1997. CD4-independent binding of SIV gp120 to rhesus CCR5. Science 278:1470-1473. [DOI] [PubMed] [Google Scholar]
- 62.Mondor, I., M. Moulard, S. Ugolini, P. J. Klasse, J. Hoxie, A. Amara, T. Delaunay, R. Wyatt, J. Sodroski, and Q. J. Sattentau. 1998. Interactions among HIV gp120, CD4, and CXCR4: dependence on CD4 expression level, gp120 viral origin, conservation of the gp120 COOH- and NH2-termini and V1/V2 and V3 loops, and sensitivity to neutralizing antibodies. Virology 248:394-405. [DOI] [PubMed] [Google Scholar]
- 63.Moore, J. P., Q. J. Sattentau, R. Wyatt, and J. Sodroski. 1994. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J. Virol. 68:469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oberlin, E., A. Amara, F. Bachelerie, C. Bessia, J. L. Virelizier, F. Arenzana-Seisdedos, O. Schwartz, J. M. Heard, I. Clark-Lewis, D. F. Legler, M. Loetscher, M. Baggiolini, and B. Moser. 1996. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 382:833-835. [DOI] [PubMed] [Google Scholar]
- 66.Pohlmann, S., E. J. Soilleux, F. Baribaud, G. J. Leslie, L. S. Morris, J. Trowsdale, B. Lee, N. Coleman, and R. W. Doms. 2001. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc. Natl. Acad. Sci. USA 98:2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reeves, J. D., N. Heveker, A. Brelot, M. Alizon, P. R. Clapham, and L. Picard. 1998. The second extracellular loop of CXCR4 is involved in CD4-independent entry of human immunodeficiency virus type 2. J. Gen. Virol. 79:1793-1799. [DOI] [PubMed] [Google Scholar]
- 68.Reeves, J. D., S. Hibbitts, G. Simmons, A. McKnight, J. M. Azevedo-Pereira, J. Moniz-Pereira, and P. R. Clapham. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates infect CD4-negative cells via CCR5 and CXCR4: comparison with HIV-1 and simian immunodeficiency virus and relevance to cell tropism in vivo. J. Virol. 73:7795-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reeves, J. D., and T. F. Schulz. 1997. The CD4-independent tropism of human immunodeficiency virus type 2 involves several regions of the envelope protein and correlates with a reduced activation threshold for envelope-mediated fusion. J. Virol. 71:1453-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69a.Reeves, J. D., S. A. Gallo, N. Ahmad, J. Miamidian, P. Harvey, M. Sharron, S. Pöhlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. Sensitivity of HIV-1 to entry inhibitors correlates with envelope: coreceptor affinity, receptor density and fusion kinetics. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 70.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 71.Rucker, J., B. J. Doranz, A. L. Edinger, D. Long, J. F. Berson, and R. W. Doms. 1997. Cell-cell fusion assay to study role of chemokine receptors in human immunodeficiency virus type 1 entry. Methods Enzymol. 288:118-133. [DOI] [PubMed] [Google Scholar]
- 72.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 73.Schols, D., S. Struyf, J. Van Damme, J. A. Este, G. Henson, and E. De Clercq. 1997. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J. Exp. Med. 186:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Si, Z., M. Cayabyab, and J. Sodroski. 2001. Envelope glycoprotein determinants of neutralization resistance in a simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) derived by passage in monkeys. J. Virol. 75:4208-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh, A., and R. G. Collman. 2000. Heterogeneous spectrum of coreceptor usage among variants within a dualtropic human immunodeficiency virus type 1 primary-isolate quasispecies. J. Virol. 74:10229-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Soilleux, E. J., R. Barten, and J. Trowsdale. 2000. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J. Immunol. 165:2937-2942. [DOI] [PubMed] [Google Scholar]
- 77.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 78.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor, does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang, Z. X., J. F. Berson, T. Y. Zhang, Y. H. Cen, Y. Sun, M. Sharron, Z. H. Lu, and S. C. Peiper. 1998. CXCR4 sequences involved in coreceptor determination of human immunodeficiency virus type-1 tropism. Unmasking of activity with M-tropic Env glycoproteins. J. Biol. Chem. 273:15007-15015. [DOI] [PubMed] [Google Scholar]
- 80.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 81.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Willett, B. J., K. Adema, N. Heveker, A. Brelot, L. Picard, M. Alizon, J. D. Turner, J. A. Hoxie, S. Peiper, J. C. Neil, and M. J. Hosie. 1998. The second extracellular loop of CXCR4 determines its function as a receptor for feline immunodeficiency virus. J. Virol. 72:6475-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 84.Xiao, X., D. Norwood, Y. R. Feng, M. Moriuchi, A. Jones-Trower, T. S. Stantchev, H. Moriuchi, C. C. Broder, and D. S. Dimitrov. 2000. Inefficient formation of a complex among CXCR4, CD4 and gp120 in U937 clones resistant to X4 gp120-gp41-mediated fusion. Exp. Mol. Pathol. 68:139-146. [DOI] [PubMed] [Google Scholar]
- 85.Ye, Y., Z. H. Si, J. P. Moore, and J. Sodroski. 2000. Association of structural changes in the V2 and V3 loops of the gp120 envelope glycoprotein with acquisition of neutralization resistance in a simian-human immunodeficiency virus passaged in vivo. J. Virol. 74:11955-11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou, N., Z. Luo, J. Luo, D. Liu, J. W. Hall, R. J. Pomerantz, and Z. Huang. 2001. Structural and functional characterization of human CXCR4 as a chemokine receptor and HIV-1 co-receptor by mutagenesis and molecular modeling studies. J. Biol. Chem. 276:42826-42833. [DOI] [PubMed] [Google Scholar]