Abstract
Continual human immunodeficiency virus type 1 (HIV-1) evolution and expansion within the human population have led to unequal distribution of HIV-1 group M subtypes. In particular, recent outgrowth of subtype C in southern Africa, India, and China has fueled speculation that subtype C isolates may be more fit in vivo. In this study, nine subtype B and six subtype C HIV-1 isolates were added to peripheral blood mononuclear cell cultures for a complete pairwise competition experiment. All subtype C HIV-1 isolates were less fit than subtype B isolates (P < 0.0001), but intrasubtype variations in HIV-1 fitness were not significant. Increased fitness of subtype B over subtype C was also observed in primary CD4+ T cells and macrophages from different human donors but not in skin-derived human Langerhans cells. Detailed analysis of the retroviral life cycle during several B and C virus competitions indicated that the efficiency of host cell entry may have a significant impact on relative fitness. Furthermore, phyletic analyses of fitness differences suggested that, for a recombined subtype B/C HIV-1 isolate, higher fitness mapped to the subtype B env gene rather than the subtype C gag and pol genes. These results suggest that subtype B and C HIV-1 may be transmitted with equal efficiency (Langerhans cell data) but that subtype C isolates may be less fit following initial infection (T-cell and macrophage data) and may lead to slower disease progression.
Rapid evolution of human immunodeficiency virus type 1 (HIV-1) within individuals and within the worldwide population stems from high mutation frequencies and rapid viral turnover in host cell infections (28, 40, 80). Divergent HIV-1 evolution coupled with continual introductions into susceptible human populations have now led to extreme viral diversification from original zoonotic jumps in central Africa (24, 39). HIV-1 has disseminated to and established an epidemic in nearly every geographic region in the world. At least two to three separate introductions of HIV-1 into the human population led to disproportionate spread of groups M (main), O (outlier), and N (non-M/non-O) (reviewed in references 22, 24, and 39). Although increased viral fitness may play a key role in the predominance and extreme variation of HIV-1 group M over group N or O isolates (reviewed in reference 61), few studies have investigated fitness differences between any primary HIV-1 isolates (reviewed in reference 43).
HIV-1 group M can be further subdivided into ten different subtypes or clades (A to J) based on envelope gene (env) diversity (40, 68). Molecular epidemiologic and evolutionary clock studies suggest that these subtypes may have evolved from different founder viruses in different African pandemics (39, 41). Expansion and overlap of these subtype-defined regions during the past two decades is associated with dual-subtype infections (9, 81) and detection of intersubtype HIV-1 recombinants (reviewed in references 60 and 54). Full genome sequencing has now identified 14 circulating recombinant forms of HIV-1, as well as many less stable forms of recombinants with ill-defined intersubtype breakpoints (54).
The proportions of subtypes in defined populations are not stable but are in constant flux due to new introductions of HIV-1 clades, changes in human behavior, therapeutic intervention, mode of transmission, and, possibly, subtype fitness (40, 52, 61). Preliminary in vitro studies and epidemiologic data suggest that the presence of some HIV-1 subtypes in various regions may be related to the mode of transmission (47, 48, 65). For example, the rapid spread of subtype B in the developed world, as opposed to other subtypes, may be due to specific introductions into male homosexual and intravenous drug user populations (29). By contrast, subtypes A, C, D, and E are found in heterosexual African populations, whereas subtype B is now nearly extinct (40).
However, one trend in subtype distribution throughout the world is certain: subtype C infections have risen in prevalence during the past 10 years and are now the predominant subtype worldwide (reviewed in reference 22). For example, subtype A had previously dominated in sub-Saharan Africa during the 1980s and early 1990s but has recently been overrun by subtype C infections (due in part to the raging epidemic in southern Africa). Although subtype B likely preceded subtype C as a founder clade in India and China, most new infections in these countries are attributable to subtype C isolates or intersubtype B/C recombinants (10, 22, 42, 57). Similar trends have been observed in Uganda, Kenya, Tanzania, and South America (e.g., Brazil and Argentina) (8, 15, 48, 65). However, it should be noted that many of these temporal changes in subtype distribution might be due simply to founder events in different susceptible human populations. For example, subtype A, associated with heterosexual transmission in Africa, has recently emerged in Russia among intravenous drug users (7). This population is typically infected by subtype B in most of the developed world.
To date, research on biological differences between HIV-1 subtypes have been limited, but there have been hundreds of reports on genotypic differences. The phyletic arrangement of nucleotide substitutions separating subtypes appears to have little consequence on HIV-1 replication kinetics (40). However, there is evidence that specific sequence alterations may affect virus replication. For example, most subtype C isolates appear to have an extra or third NF-κB element in the long terminal repeat (LTR), whereas subtype F isolates contain only one (33, 49). More NF-κB sites augment HIV-1 transcription from the LTR in the presence or absence of HIV-1 Tat protein as well as increase virus replication (33, 46). However, recent studies suggest that this extra NF-κB site in subtype C may be biologically inactive (69).
Additional studies suggest that subtype C HIV-1 isolates may also encode a more active protease (79). Increased activity of these individual functions may still be insufficient to overcome the decreased replicative capacity of the CCR5-tropic (R5) non-syncytium-inducing phenotype (4, 5, 75). Unlike other subtypes, subtype C isolates fail to switch from an R5, non-syncytium-inducing to a CXCR4-tropic (X4), syncytium-inducing phenotype, typically found in late HIV disease (11, 55, 56).
Most studies focusing on phenotypic differences between HIV-1 isolates of the same or different subtypes have only analyzed cloned fragments of the viral genome (33, 49, 79) and have not directly assessed the replication of primary HIV-1 isolates (reviewed in reference 43). In monoinfections, we observed no significant differences in replication kinetics of HIV-1 isolates of different subtypes. However, only gross differences in HIV-1 replication efficiencies (e.g., nef deletion [31]) are apparent in monoinfections due to inherent variations between infected cultures. Comparing the replication of one HIV-1 isolate with that of another in dual infections provides an internal control and permits accurate assessment of smaller differences in replicative capacity (21, 61, 62).
Here, we have compared the ex vivo fitness (or relative replication efficiencies) of nine subtype B and six C HIV-1 isolates by performing head-to-head pairwise competitions in different primary human cells. Subtype C HIV-1 isolates were outcompeted by subtype B HIV-1 isolates in primary blood mononuclear cells (PBMC), CD4+ T lymphocytes, and blood-derived macrophages. Detailed analyses of each step of the retroviral life cycle suggested that the efficiency of host cell entry could predict the winner of HIV-1 competitions. However, other steps in the retroviral life cycle likely contribute to differences in replication efficiency. Interestingly a subtype C and subtype B HIV-1 isolate were equally fit in epidermal Langerhans cells. Since epidermal Langerhans cells provide a model for primary HIV-1 infection in the genital tract, it is conceivable that subtype C HIV-1 isolates may be efficiently transmitted but less fit during disease progression. Although we developed a model for subtype distribution in the worldwide epidemic, the relevance of these ex vivo fitness results to disease progression, transmission, and spread must be investigated in longitudinal studies involving participants infected with different subtypes.
MATERIALS AND METHODS
Cells and viruses.
PBMC from HIV-seronegative blood donors were obtained by Ficoll-Hypaque density gradient centrifugation of heparin-treated venous blood. Prior to HIV-1 infection, the cells were stimulated with 2 μg of phytohemagglutinin (Gibco-BRL) per ml for 3 to 4 days and maintained in RPMI 1640 medium with 2 mM l-glutamine (Cellgro) supplemented with 10% fetal bovine serum (Cellgro), 10 mM HEPES buffer (Cellgro), 1 ng of interleukin-2 (Gibco-BRL) per ml, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (both from Cellgro).
Ten subtype B and subtype C HIV-1 isolates were obtained from the AIDS Research and Reference Reagent Program. Eight of nine non-syncytium-inducing/R5 subtype B and six of nine non-syncytium-inducing/R5 subtype C isolates could be propagated and expanded in phytohemagglutinin-stimulated, interleukin-2-treated PBMC (see Table 1). The stocks of subtype C and B isolates from the AIDS Research and Reference Reagent Program that could not be propagated have very low levels of reverse transcriptase activity and were not infectious for several PBMC cultures. All but one of the 15 isolates were non-syncytium-inducing HIV-1 isolates, as determined by the MT2 assay (75).
TABLE 1.
Characteristics and ex vivo fitness of primary HIV-1 isolates
| Laboratory reference name | Country of origin | Virus | Subtype in HIV-1 regiona
|
Phenotype/ coreceptor usageb | Mean intrasubtype fitness valuec ± SD | Mean intersubtype fitness value ± SD | Total relative fitness valued | |
|---|---|---|---|---|---|---|---|---|
| gag | env | |||||||
| B2 | Brazil | 92BR017 | B | B | NSI/R5 | 0.708 ± 0.243 | 1.68 ± 0.198 | 15.8 |
| B3 | USA | 91US005 | B | B | NSI/R5 | 0.453 ± 0.155 | 1.37 ± 0.270 | 11.8 |
| B4 | USA | 93US076 | B | B | SI/X4 | 1.482 ± 0.180 | 1.99 ± 0.005 | 23.8 |
| B5 | USA | 91US056 | B | B | NSI/R5 | 0.807 ± 0.156 | 1.65 ± 0.159 | 16.3 |
| B6 | USA | 91US714 | B | B | NSI/R5 | 1.28 ± 0.167 | 1.90 ± 0.070 | 21.6 |
| B7 | Brazil | 92BR023 | C | B | NSI/R5 | 0.979 ± 0.132 | 1.67 ± 0.181 | 17.7 |
| B8 | Brazil | 92BR028 | B | B | NSI/R5 | 1.56 ± 0.111 | 1.98 ± 0.019 | 24.3 |
| B9 | Brazil | 92BR018 | B | B | NSI/R5 | 1.17 ± 0.120 | 1.84 ± 0.104 | 20.5 |
| B10 | Brazil | 92BR003 | B | B | NSI/R5 | 0.561 ± 0.101 | 1.06 ± 0.135 | 10.8 |
| All B isolates | 1.00 ± 0.066 | 1.68 ± 0.061 | ||||||
| C2 | Nigeria | 97USNG30 | C | C | NSI/R5 | 1.31 ± 0.242 | 0.143 ± 0.104 | 7.83 |
| C3 | S. Africa | 97ZA012 | C | C | NSI/R5 | 1.55 ± 0.144 | 0.669 ± 0.194 | 13.8 |
| C5 | S. Africa | 97ZA003 | C | C | NSI/R5 | 1.00 ± 0.318 | 0.385 ± 0.176 | 8.45 |
| C6 | India | 98IN022 | C | C | NSI/R5 | 1.45 ± 0.216 | 0.539 ± 0.125 | 12.1 |
| C8 | Nigeria | 96USNG58 | C | C | NSI/R5 | 0.524 ± 0.160 | 0.108 ± 0.108 | 3.59 |
| C9 | Malawi | 93MW959 | C | C | NSI/R5 | 0.174 ± 0.174 | 0.0728 ± 0.045 | 1.53 |
| All C isolates | 1.00 ± 0.123 | 0.319 ± 0.061 | ||||||
MA and CA coding regions of gag were sequenced and analyzed as described in the text. C2 and C3 domains of the env gene were sequenced and analyzed as described in the text.
Biological phenotype and coreceptor usage were predicted by the charge of the amino acid residues at position 306 and 322. NSI and SI, non- and syncytium-inducing, respectively.
Relative fitness between two isolates of the same or different subtypes was calculated as the fraction of virus produced (f0) divided by the proportion of that virus in the initial inoculum (i0). Mean fitness value (w) is the average fitness of all intra- or intersubtype competitions plus or minus the standard deviation between each competition.
Total relative fitness value is the sum of all relative fitness values of each HIV-1 isolate in 14 competition experiments.
For all strains listed in Table 1, the letter before the dash indicates the subtype of the viral envelope and precedes the year of isolation, country of origin, and strain number (e.g., B2-92BR017 refers to a subtype B HIV-1 strain isolated in Brazil in 1992). Although we achieved greater success in propagating subtype B isolates, it appears that the inability to propagate the three subtype C stocks and one subtype B isolate was due to a lack of infectious virus in the stock. This problem has been observed frequently with several HIV-1 stocks and has been reported to the repository. However, in recent clinical studies in Uganda and Zimbabwe, it does appear that subtype C isolates are more difficult to propagate from patient PBMC, but this has not been correlated with viral load or CD4 cell count (K. Demers and E. J. Arts, unpublished results).
PBMC from three donors were purified from whole blood by density gradient centrifugation as described above. It is important to note that three donors were of two different racial groups (black and Caucasian) and from different ethnic backgrounds. Since no differences in HIV-1 fitness were observed for these cells from different donors, we have not reported ethnic and racial profiles. CD4+ T lymphocytes were then purified from the buffy coat with the MACS CD4+ T-cell isolation kit. Non-T cells, i.e., B cells, monocytes, NK cells, cytotoxic T cells, dendritic cells, early erythroid cells, platelets, and basophils, are magnetically depleted with a cocktail of anti-CD8, -CD11b, -CD16, -CD19, -CD36, and -CD56 antibodies. Isolation of highly pure CD4+ T cells was then obtained on MidiMACS columns (Miltenyi Biotech).
Macrophages were also purified from PBMC from the same three donors. Briefly, 10 × 106 unstimulated PBMC were added to 25-mm3 flask for 3 days. Three washes with phosphate-buffered saline (PBS) removed nonadherent cells. Ninety-five percent of the adherent cells were esterase and peroxidase positive, expressing CD14 (described in reference 67). Finally, Langerhans cells were obtained from suction blister roofs (i.e., epidermal sheets) induced on normal-appearing skin of healthy volunteers. These epidermal sheets are devoid of blood and contaminating fibroblasts; 2 to 3% of the cells within the sheets are Langerhans cells, whereas the remainder are epithelial cells (i.e., keratinocytes) (35).
U87.CD4 (human glioma) cells expressing CCR5 or CXCR4 were obtained through D. Littman and the AIDS Reagent Project. U87.CD4-CCR5 and U87.CD4-CXCR4 cells were grown in Dulbecco's modified Eagle's medium (complete medium) containing 1 mg of geneticin G418 (Life Technologies, Inc.) per ml to maintain CD4 expression (19).
Quantifying viral stocks and monitoring virus production.
Tissue culture dose for 50% tissue culture infectious dose (TCID50) was determined by serially diluting supernatants of each stock of virus and performing quadruplicate infections of phytohemagglutinin- and interleukin-2-treated PBMC. Reverse transcriptase activity in culture supernatants on day 10 of the infections was used to calculate TCID50 values with the Reed and Muench method (64). Titers were expressed as infectious units (IU) per milliliter. It is important to note that the TCID50 assay and subsequent dual virus competitions were performed with PBMC from the same donor and blood draw.
The assay to measure reverse transcriptase activity has been described previously (77). Viral RNA load was measured by the reverse transcription (RT)-PCR amplification technique as described before (3). Viral RNA was purified from pelleted virus particles (cell-free supernatants centrifuged at 32,000 × g for 40 min) with the Qiagen RNeasy kit and Qiashredder spin columns (Qiagen). Genomic HIV-1 RNA (specifically the LTR region) was reverse transcribed with the AG4 primer (3) and Moloney murine leukemia virus reverse transcriptase (Gibco-BRL), then PCR amplified with unlabeled S1 primer and 5′-end γ-32P-labeled A13 primer as described previously (3). As a positive control, 10-fold dilutions (10 to 108 copies) of HIV-1 pbs RNA (3) were RT-PCR amplified in parallel with the samples. All RT-PCR samples were then run on denaturing 8% polyacrylamide gels, which were dried, exposed to X-ray film (Kodak), and analyzed with the Bio-Rad Phosphor-imager. Viral RNA loads quantified by this method were equal to those obtained with Roche Amplicor assay (E. J. Arts, unpublished data).
Coreceptor expression and polymorphisms.
Unstimulated PBMC were left untreated for 12 h. Following this incubation with drugs, the cells were centrifuged at 800 × g for 10 min and incubated on ice for 15 min with 5% bovine serum albumin (Sigma, St. Louis, Mo.) in PBS (BioWhittaker, Walkersville, Md.). Cells were again centrifuged at 800 × g for 10 min and resuspended in 50 μl of PBS. Then 5 μl of fluorescein isothiocyanate-conjugated anti-human CD4 antibody (Becton Dickinson Immunocytochemistry Systems, San Jose, Calif.), 20 μl of phycoerythrin-conjugated anti-human CCR5 antibody, or 5 μl of phycoerythrin-conjugated mouse immunoglobulin G2a (IgG2a) κ isotype standard (PharmMingen, San Diego, Calif.) was added to the suspension and incubated in the dark on ice for 30 min (44). Cells were then washed with 5% bovine serum albumin-PBS and 500 μl of PBS. After the final wash, the cells were fixed with 300 μl of 1% paraformaldehyde and analyzed with a FacsScan flow cytometer and Lysis II software (Becton Dickinson, Bedford, Mass.).
DNA preparation and CCR5 genotyping.
One hundred thousand PBMC were obtained from each donor. DNA was purified with the QiaAmp blood kit as described above. PCR amplification was performed in a solution (25 ml) containing 2.5 pmol of the appropriate positive-strand and negative-strand primers, 67 mM Tris-HCl (pH 8.8), 6.7 mM MgSO4, 16.6 mM (NH4)2SO4, 10 mM 2-mercaptoethanol, 100 μM deoxynucleoside triphosphates, 2.5 U of thermostable DNA polymerase (PE Applied Biosystems, Foster City, Calif.), and 10 to 50 ng of purified human genomic DNA. Oligonucleotide primers used to direct amplification of CCR5 open reading frame products (wild-type, 312 bp; Δ32, 280 bp) were 62000+ and 62311− (70). Amplicons for the CCR5 open reading frame were visualized on 2% agarose gels following electrophoresis in Tris-borate-EDTA, stained with a 1:10,000 dilution of SYBR Gold (Molecular Probes, Eugene, Oreg.), and detected with a Storm 860 scanner (Molecular Dynamics, Sunnyvale, Calif.).
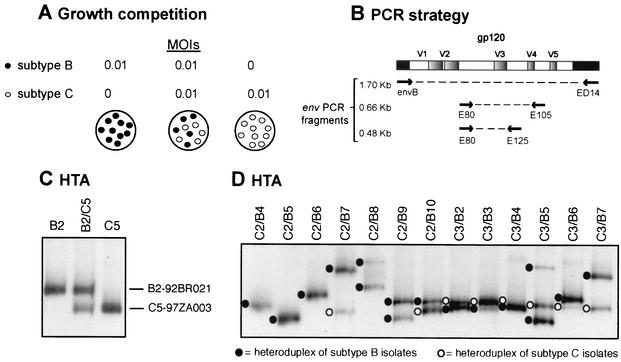
HIV-1 infections and growth competition assays.
All HIV-1 isolates listed above were used in mono- and dual infection studies of PBMC. Virus was added alone or in pairs to phytohemagglutinin- and interleukin-2-treated PBMC at a multiplicity of infection of 0.01 IU/cell in a 24-well plate (500 IU of virus and 5 × 105 cells). Following an 8-h incubation at 37°C with 5% CO2, cells were washed three times with 1× phosphate-buffered saline (PBS) and then resuspended in complete medium (106/ml). All monoinfection and dual-infection/competition experiments were performed in PBMC from one donor and in duplicate. As described for Fig. 2A, the dual-infection/competition assay involved the addition of two HIV-1 isolates (multiplicity of infection of 0.01 IU/cell) and were performed alongside the monoinfections. Uninfected cultures were used as HIV-negative control. Cell-free supernatants were assayed for reverse transcriptase activity at days 1, 2, 5, 10, and 15 postinfection. Growth curves were then plotted for each HIV-1 monoinfection (Fig. 1C). Two aliquots of cells were removed at day 10 in the 210 dual infections performed for the full pairwise competition study. Supernatants and two aliquots of cells were stored at −80°C for subsequent analysis.
FIG. 2.
Strategy for HIV-1 competition experiments and heteroduplex tracking method for dual virus detection and quantification. (A) Virus was added alone or in pairs to phytohemagglutinin- and interleukin-2-treated PBMC at a multiplicity of infection (MOI) of 0.01. Cells were washed after 8 h to remove virus. Cells and virus supernatant was harvested at day 10 and lysed. (B) Extracted DNA or RNA from dual infections was PCR or RT-PCR amplified with conserved HIV-1 env primers. PCR-amplified env products were denatured, annealed to a radiolabeled env probe, and then run on an 8% nondenaturing polyacrylamide gel (C and D). Heteroduplexes generated from monoinfections (C) were then used to identify those isolates found in the heteroduplex tracking analysis (HTA) of each dual infection (D). Phosphor-imaging analysis of each heteroduplex was used to quantify the production of each virus in a dual infection. Multiple heteroduplexes can track to a single HIV-1 isolate because these isolates represent the propagated quasispecies and not a single clone.
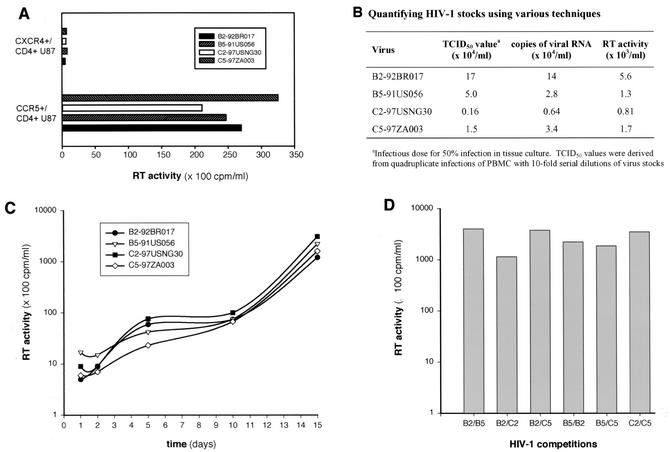
FIG. 1.
Measuring titers of HIV-1 stocks and replication kinetics. Four primary HIV-1 isolates, two subtype C and two subtype B strains (B2, B5, C2, and C5), were employed in monoinfection studies and are representative of the 15 isolates examined in this study. (A) Quantifying viral load in each stock by infection of PBMC with serially diluted virus (Reed-muench technique), by RT-PCR-amplifying HIV-1 RNA (3), and by measuring endogenous reverse transcriptase (RT) activity in virus stocks (77). Details of each assay are provided in the Materials and Methods section. (B) Coreceptor usage of each HIV-1 isolate. U87 cells expressing CD4 and CCR5 or CXCR4 were exposed to equal IU of each HIV-1 isolate. Virus production was measured by reverse transcriptase activity in the cell-free supernatant. All isolates except B4 were predicted to be R5/non-syncytium-inducing isolates, based on a more neutral V3 loop and neutral or negatively charged amino acids at position 306 and 322. (C) Virus production from PBMC monoinfected with 0.01 IU of B2, B5, C2, and C5 HIV-1 isolates. Reverse transcriptase activity in cell-free supernatant was measured during the 15-day monoinfection. (D) Virus production was measured by reverse transcriptase activity following a 15-day PBMC dual infection with B2, B5, C2, and C5. Two hundred and ten dual infections were analyzed with the same approach.
The same dual and monoinfection protocol was employed for experiments involving CD4+ T lymphocytes and macrophages. Langerhans cells within tissue were infected with HIV-1 as previously described (35). Briefly, skin was draped over 50-μl droplets containing 0.01 IU of B5 and/or C5 virus (250 IU of each virus for approximately 25,000 Langerhans cells within each explant) for 2 h. Excess virus was then washed from skin in PBS, and explants were floated on complete medium in six-well plates and cultured at 37°C in 5% CO2 for 3 to 4 days postinfection. HIV-1-infected Langerhans cells that had emigrated (i.e., crawled out) from explants were then harvested for DNA.
PCR and RT-PCR of HIV-1 env gene.
For all competition experiments, proviral DNA was extracted from lysed PBMC, T cells, macrophages, or Langerhans cells with the QIAamp DNA blood kit (Qiagen). Viral RNA was purified from pelleted virus particles (cell-free supernatants centrifuged at 32,000 × g for 40 min) with the Qiagen RNeasy kit and Qiashredder spin columns (Qiagen). Viral DNA isolated from infected cells or reverse transcribed from viral RNA, i.e., with Moloney murine leukemia virus reverse transcriptase and the ED14 primer (18), was (i) PCR amplified with a set of external primers, envB (25) and ED14 (gp120-coding region of env, ∼1.7 kb), followed by (ii) nested amplification with the E80-E125 primer pair (71) (C2-V3 env region, 0.48 kb) (Fig. 2B). Both external and nested PCRs were carried out in a 100-μl reaction mixture with defined cycling conditions (62). PCR-amplified products were isolated in agarose gels and then purified with the Qiaquick PCR purification kit (Qiagen).
In the dual infection/time course experiments, DNAs extracted from the five time points were PCR or RT-PCR amplified with conserved HIV-1 primer pairs specific for early minus-strand DNA, late minus-strand DNA, integrated HIV-1 proviral DNA, unspliced HIV-1 genomic RNA, and multiply spliced mRNA. Defined cycling conditions and annealing temperatures for each primer pair are available upon request. Quantitative assessment of each step in the retroviral life cycle involved a single round of PCR amplification with radiolabeled sense primer and unlabeled antisense primer. Linearized HXB2 DNA was PCR amplified with the same sets of primers and used as an amplification control (3). The amount of PCR-amplified product generated from 102 to 108 copies was plotted. For R2 values greater than 0.95, the equation of a line for each set of primers was used to calculate the copy number in the original samples used in the PCR analyses.
Relative production of each virus at the various steps in the retroviral life cycle involved external and nested PCR amplifications with unlabeled primer pairs. Five microliters of sample DNA was added to the external PCR, and 5 μl of the external PCR amplification product was transferred to the nested amplification. An early product of HIV-1 minus-strand DNA, incorporating the minus-strand strong stop and the region following the first template switch, was PCR amplified with the SU3-1 (nucleotide positions 432 to 456) in the HXB2 genome) and AU5-1 (nucleotides 600 to 581) external primer pairs and the SU3-1-AU5-2 (nucleotides 580 to 560) nested primer pair. The late minus-strand DNA product was PCR amplified with the SR-1 (nucleotides 467 to 490)-AUNS-8 (nucleotides 820 to 796) external primer pair and the SR-2 (nucleotides 521 to 544)-AUNS-7 (nucleotides 786 to 764) nested primer pair.
Unspliced and multiplyspliced HIV-1 RNAs were first reverse transcribed with Superscript II murine leukemia virus reverse transcriptase (Roche) and the AUNS-8 or E15 (nucleotides 8445 to 8424) primer, respectively. Unspliced cDNA was externally amplified with the SR-1-AUNS-8 primer pair and amplified with the SR-2-AUNS-7 nested primer pair, whereas multiply spliced cDNA was externally amplified with the SR-2-E15 primer pair and then PCR amplified with the SMS-7 (positions 5956 to 5979)-E15 nested primer pair.
External PCR amplification of the integrated HIV-1 was performed with the Alu and Alu-LTR primers. The Alu primer anneals to the highly redundant Alu sequence in the genome, while the Alu-LTR primer anneals to a sequence in the U3 region (14). The external PCR cycling conditions (35 cycles with a 72°C polymerization step for 2 min) were optimized for amplification of a 2-kbp fragment (i.e., HIV-1 DNA integrated within 2 kbp of an Alu sequence in the host cell genome). A nested PCR was then performed on the integrated HIV-1 DNA amplified in the external PCR. Three microliters of external amplification, unlabeled LTR-1 (nucleotides 59 to 79) primer and γ-32P-end-labeled AU3-1 (nucleotides 456 to 432) primer were added to the nested reaction mixture. To ensure that unintegrated DNA was not amplified in this PCR, 0.18 μl (or the equivalent amount of the original DNA sample carried over from the external to the nested amplification) was PCR amplified with the nested primer pair LTR-1 and γ-32P-labeled AU3-1) (44).
Heteroduplex tracking assay for detection of two HIV-1 env fragments.
Nested PCR products of the env gene were analyzed by heteroduplex tracking analysis (62). The same genomic regions (C2-V3) were PCR amplified from a subtype E HIV-1 env clone (E-pTH22) (18) for use as a DNA probe. For this amplification, the E80 primer was radiolabeled with T4 polynucleotide kinase and 2 μCi of [γ-32P]ATP. For the time course experiments, probe DNA was generated via PCR amplification of DNA from a subtype E-infected cell with the SU3-1-[32P]AU5-2 (early minus-strand DNA), SR-2-[32P]AUNS-7 (late minus-strand DNA), SR-2-[32P]AUNS-7 (unspliced RNA), and LTR-1-[32P]AU3-1 (integrated DNA) primer pairs.
Probe from the multiply spliced mRNA analysis was PCR amplified from the SVH6 Rev vector (containing a fusion of the two rev exons) with the SMS-7-[32P]E15 primer pair. This SVH6 Rev vector was kindly provided by A. Cochrane, University of Toronto (50). Radiolabeled PCR-amplified probes were separated on 1% agarose gels and purified with the Qiaquick gel extraction kit (Qiagen). Reaction mixtures contained DNA annealing buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.8], and 2 mM EDTA), 10 μl of unlabeled PCR-amplified DNA from the competition culture, and approximately 0.1 pmol of radioactive probe DNA (62). Reaction mixtures contained DNA annealing buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.8], and 2 mM EDTA), 10 μl of unlabeled PCR-amplified DNA from the competition culture, and approximately 0.1 pmol of radioactive probe DNA (62).
Reaction mixtures containing DNA amplified from the competition and probe were denatured at 95°C for 3 min and then rapidly annealed on wet ice. After 30 min on ice, the DNA heteroduplexes were resolved on Tris-borate-EDTA buffer on 5 to 8% nondenaturing polyacrylamide gels (30:0.8 acrylamide-bisacrylamide) for 2.5 h at 200 V. The percentage of polyacrylamide in the gel matrix was dependent on the size of the amplified product employed in the heteroduplex tracking analysis (see above). Gels were dried and exposed to X-ray film (Eastman Kodak Co., Rochester, N.Y.). Heteroduplexes representing production of each isolate in a dual infection were quantified with the Bio-Rad Phosphor-imager. Figures 2C and D show two representative heteroduplex tracking analyses involving competitions between subtype B and C isolates.
Estimation of viral fitness.
Classic models for estimating the replicative capacity or fitness of a virus are based on single competition experiments with equal multiplicities of infection of each virus. In our HIV-1 competition experiments, the final ratio of the two viruses produced from a dual infection was determined by heteroduplex tracking analysis and compared to production in the monoinfections. Production of individual HIV-1 isolates in a dual infection (f0) was divided by the initial proportion in the inoculum (io) and is referred to as relative fitness (w = f0/i0) (62). The ratio of the relative fitness values of each HIV-1 variant in the competition is a measure of the fitness difference (WD) between the two HIV-1 strains (WD = wM/wL), where wM and wL correspond to the relative fitness of the more and less fit virus, respectively (62).
Nucleotide sequence analysis.
A segment of the HIV-1 env gene (C2-C3; 336 bp) was PCR amplified with the E80-E125 primer pair. This PCR product was then sequenced in the sense and antisense directions with the E80 and E125 primers, respectively. env sequences were obtained for B4 (accession number AY090772), B5 (U79719), B6 (AY090773), B9 (AY090774), C3 (af286227), C5 (AY090775), and C6 (AF286232) with the ABI Prism BigDye terminator cycle sequencing ready reaction kit (Perkin-Elmer). The University of California-Davis sequencing facility performed all sequencing reactions. Nucleotide sequences of the env gene from the other HIV-1 isolates had been determined previously and submitted to GenBank under the following accession numbers: B2 (U08687), B3 (U27434), B7 (U08779), B8 (U16218), B10 (U08670), C2 (AF096349), C8 (AF096329), and C9 (U08453).
Two segments of the gag gene were also PCR amplified and sequenced for almost all of the subtype B and C isolates used in this study. The MA-CA segment was PCR amplified with the GS6 (nucleotide positions 794 to 815 in the HXB2 genome) and GA9 (nucleotides 1625 to 1605) primer pair and sequenced with the same primers, whereas the CA-NC fragment was PCR amplified and sequenced with the GS7 (nucleotides 890 to 911) and GA8 (nucleotides 1427 to 1404) primer pair. Only the B7 sequence of the MA-CA gag fragment was obtained from GenBank (U86559). Accession numbers of the nucleotide sequences encoding the MA-CA region of gag are as follows: B2 (AY090741), B3 (AY090746), B4 (AY090739), B5 (AY090740), B6 (AY090744), B8 (AY090743), B9 (AY090742), B10 (AY090745), C2 (AY090752), C3 (AY090751), C5 (AY090749), C6 (AY090747), C8 (AY090748), and C9 (AY090750).
Accession numbers of the nucleotide sequences encoding the CA-NC coding region of gag are as follows: B2 (AY090762), B3 (AY090757), B4 (AY090759), B5 (AY090756), B6 (need sequence), B7 (AY090758), B8 (AY090763), B9 (AY090761), B10 (AY090760), C2 (AY090765), C3 (AY090764), C5 (AY090753), C6 (af286232), C8 (AY090754), and C9 (AY090755). Nucleotide sequences were edited and translated with the BioEdit software and then aligned with the Clustal X version 1.63b program.
Phylogenetic analyses.
The gag and env sequences of the HIV-1 isolates used in this study and the set of reference strains were aligned with the Clustal X version 1.63b program. Pairwise DNA distance matrices were also generated with the Kimura two-parameter model (37). Phylogenetic analyses were performed with the MEGA version 1.02 program. Tree topologies were confirmed by the neighbor-joining method with the Kimura two-parameter distance matrices (37). Bootstrap resampling (1,000 data sets) of the multiple alignment tested the statistical robustness of the trees. Finally, phylogenetic trees based on nucleotide distance were schematically represented with the TreeView program (53).
Pairwise matrices were also constructed with the log10 value of the fitness differences in the pairwise competition experiments (Fig. 3 and 4). First, fitness differences were always calculated as wM/wL as described above. Virus pairs of equal fitness would have WD values equal to 1 in competition, which is comparable to HIV-1 isolates with identical nucleotide sequences, i.e., a genetic distance of zero. A log base 10 value would equate a WD of 1 to 0. Phylogenetic trees based on fitness difference were then determined with the Neighbor.exe program in the PHYLIP package (66). The TreeView program was used to draw the phylogenetic tree.
FIG. 3.
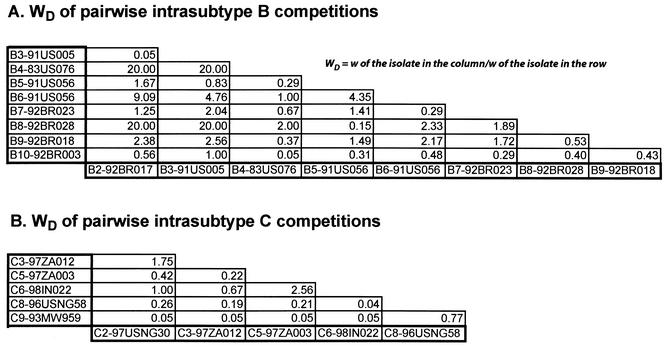
Pairwise competitions in PBMC to compare intrasubtype HIV-1 fitness differences. Nine subtype B and six subtype C HIV-1 isolates were employed in pairwise intrasubtype competitions. Fitness differences were plotted for the paired competitions between subtype B (panel A) and C (panel C) isolates. Fitness difference of each competition pair is presented as the fitness value of the isolate in the column divided by the fitness value of the isolate in the row. Average relative fitness values from these intrasubtype competitions in PBMC are provided in Table 1 and plotted with a phyletic analysis in Fig. 6.
FIG. 4.
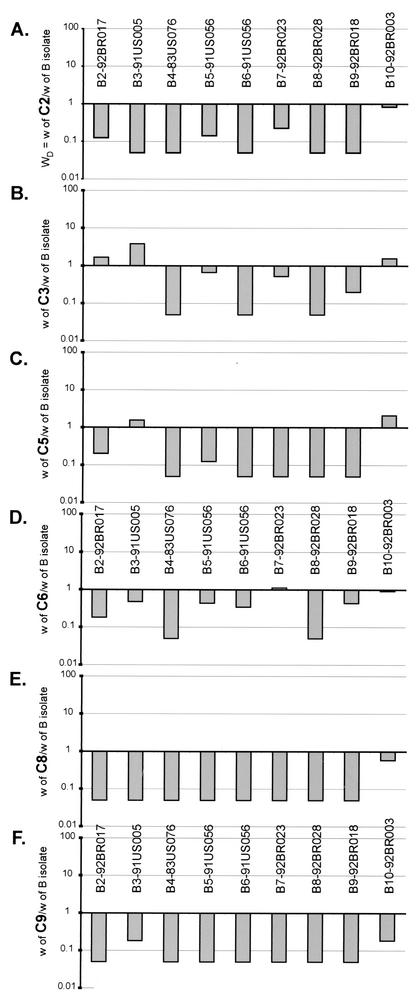
Comparing the relative fitness of subtype B and C HIV-1 isolates in direct competition. Subtype C isolates C2 (A), C3 (B), C5 (C), C6 (D), C8 (E), and C9 (F) were used to compete against each of the B isolates (B2, B3, B4, B5, B6, B8, B9, and B10) with equal multiplicities of infection for each isolate and PBMC cultures.
RESULTS
Characterization of HIV-1 isolates.
Several primary HIV-1 isolates were propagated and counted on phytohemagglutinin-stimulated, interleukin-2-treated PBMC isolated from the same donor and blood draw. Table 1 lists the subtype B and C HIV-1 isolates employed in the pairwise competition experiments. With the exception of HIV-1 B4, all isolates were R5 and non-syncytium inducing. Coreceptor utilization was predicted from the amino acid sequence in the V3 region of env, i.e., positively charged amino acids at positions 306 and 322 in the loop. Coreceptor utilization was confirmed for the B2, B4, B5, C2, and C5 isolates with CD4-positive U87 cells expressing CXCR4 or CCR5 (Fig. 1B). All R5/non-syncytium-inducing HIV-1 isolates were unable to infect CXCR4+/CD4+ U87 cells (Fig. 1A). By contrast, only the CXCR4 coreceptor mediated entry and replication of the B4 isolate (data not shown). To control for biological phenotype, this study focused on the ex vivo fitness of R5/non-syncytium-inducing HIV-1 isolates. Of note, the X4/syncytium-inducing phenotype is rare among subtype C isolates in the world (76).
Accurate TCID50 values (infectious dose of virus for 50% infection of PBMC) were derived from quadruplicate PBMC infections with serially diluted virus stocks (Fig. 1B). These TCID50 values, representing the infectious virus titer (IU per milliliter), were compared to assays that do not distinguish between infectious and inactive virus particles. Differences in the reverse transcriptase activity or viral RNA load of four virus stocks roughly corresponded to the differences observed in infectious titers (i.e., TCID50 values) (Fig. 1B). Accurate quantification of virus rules out the possibility that outgrowth of one isolate in a dual infection was due to major differences in virus inocula. The amount of HIV-1 p24 in virus stocks did not provide an accurate estimate of infectious titers (data not shown).
Nine subtype B and six subtype C HIV-1 isolates (0.01 multiplicity of infection) were used in mono- or dual infections of PBMC treated with phytohemagglutinin and interleukin-2. Duplicate pairwise competition experiments with all 15 viruses required 210 dual infection comparisons. Virus production in culture supernatants was monitored 1, 2, 5, and 10 days postinfection with a radioactively labeled reverse transcriptase assay. Differences in B2, B5, C2, and C5 HIV-1 production were not observed in PBMC monoinfections (Fig. 1C) or in dual infection with this particular combination of HIV-1 isolates (Fig. 1D). Similar results were obtained with the other 11 monoinfections and with the 210 dual infections (175,000 ± 4,500 cpm/ml at day 10).
Ex vivo fitness analyses.
Conventional assays involving reverse transcriptase activity or p24 antigen capture do not distinguish between two HIV-1 isolates in a dual infection (Fig. 1C and D). We have previously modified the heteroduplex tracking assay to monitor and quantify dual virus production in these competitions (62). This technique is schematically outlined in Fig. 2A and B. HIV-1 env DNA is PCR amplified, then denatured, and annealed to a radiolabeled env DNA probe. DNA heteroduplexes representing the two HIV-1 isolates in each dual infection migrate to different positions in a nondenaturing polyacrylamide gel (Fig. 2C).
Figure 2D represents a heteroduplex tracking analysis of a small subset of the pairwise competitions performed with the subtype B and C HIV-1 isolates. Heteroduplexes corresponding to the subtype B or C HIV-1 isolate in the dual infection were matched with the heteroduplex from the respective monoinfection (Fig. 2C). It is important to note that the entire HIV-1 quasispecies, propagated from individual patient samples and not from single HIV-1 clones, was used in these dual infections. Thus, more than one heteroduplex band was observed with several primary HIV-1 isolates (e.g., B5 and B8, Fig. 2D).
Relative production of each isolate in a dual infection assessed by heteroduplex tracking analysis provides a measure of ex vivo fitness for that virus (i.e., relative fitness, or w) as well as fitness difference of the two isolates in the dual infection (typically WD = wM/wL) (62). Mean relative fitness values for each isolate in pairwise inter- and intrasubtype competitions are shown in Table 1. Recombination between two isolates in a competition can result in a heteroduplex's migrating between the two parental heteroduplexes. Due in part to a low multiplicity of infection, recombination within this 400-nucleotide region is a low-frequency event (<0.01% of parental virus production) and is below the limit of detection by heteroduplex tracking analysis (62).
Intrasubtype B competitions revealed that not a single subtype B HIV-1 isolate outcompeted all other isolates of the same subtype (Fig. 3A). As an example, HIV-1 B2 had greater relative fitness in competition with B3, B5, and B10, but was less fit than B4, B6, B7, B8, and B9 (Fig. 3A). In the pairwise subtype B competitions, B10 and B3 were the least fit, whereas B4 (X4/syncytium inducing) and B8 were the most fit (Fig. 3A and Table 1). Although the X4/syncytium-inducing phenotype is generally associated with increased replication efficiency, the B4 isolate was outcompeted by the R5/non-syncytium-inducing B8 isolate in PBMC (WD = 2; Fig. 3A). Similar exceptions to X4 dominance have been described previously (62). Fitness differences between B isolates in competitions with B4 (P = 0.76, paired t test value = 0.31), B6 (P = 0.96, t = 0.058), or any other subtype B isolate were not significant (Table 1), as reflected by mean relative fitness values (w) of ∼1 for each subtype B competition.
Similar findings were obtained with the intrasubtype C competitions (Fig. 3B and Table 1). Mean w between subtype C isolates in intrasubtype competitions were similar to those between subtype B isolates (Table 1). The C9 isolate was the least fit, whereas C2 and C3 were the most fit (Table 1 and Fig. 3B). With the exception of C9, fitness differences among different subtype C isolates were not significant (C's versus C2, P = 0.38; C's versus C6, P = 0.156).
In contrast to the intrasubtype competitions, significant differences in fitness were observed when subtype B isolates were placed in competition with subtype C isolates (Fig. 4; Table 1). Almost all subtype B isolates outcompeted subtype C isolates in phytohemagglutinin- and interleukin-2-treated PBMC cultures (Fig. 4). Although there appears to be a dichotomy between ex vivo and in vivo subtype C fitness, reduced replicative capacity and pathogenesis may actually increase spread in the human population (23).
The mean w for all subtype C isolates in intersubtype B versus C competitions (w = 0.319 ± 0.0610; Table 1) was less than the mean w of the subtype B or C isolates in the pairwise intrasubtype competitions (w = 1.00 ± 0.0664 for intrasubtype B and 1.00 ± 0.123 for intrasubtype C competitions; Table 1) (P < 0.0001; Student's t test). In the intersubtype competitions, C9 remained the least fit of all HIV-1 isolates (Fig. 4F) whereas the X4/syncytium-inducing B4 isolate outcompeted every R5/non-syncytium-inducing subtype C isolate (Fig. 4). C3 and C5 were the only subtype C isolates that could compete with subtype B isolates (Fig. 4B and C), but most of the “wins” were against the B10 isolate, again the least fit isolate in intrasubtype B competitions (Fig. 3 and 4). Interestingly, the R5/non-syncytium-inducing B8 isolate had a slightly greater fitness than the X4/syncytium-inducing B4 isolate in both the intra- and intersubtype competitions (Table 1). In addition, B8 was slightly more fit than B4 in head-on competitions in PBMC (WD = 2; Fig. 3A).
Effect of primary human cell type on dual HIV-1 competitions.
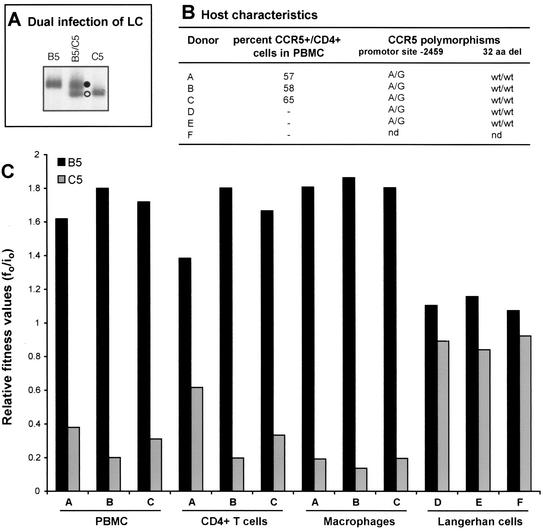
Although the results in Table 1 and Fig. 4 suggest that subtype C isolates are less fit than the subtype B isolates, these assays were performed in duplicate with PBMC from a single donor and blood draw. We have, however, observed limited variability in the relative production of two HIV-1 isolates (WD) when PBMC from different donors were used in dual-infection/competition assays (62). To test for possible host-specific variations in HIV-1 fitness, the fitness of B5 and C5 HIV-1 isolates was compared in PBMC from three different donors. In addition, we examined the fitness of these two isolates in establishing systemic infections (e.g., blood-derived macrophages and CD4+ T lymphocytes) as well as in Langerhans cells, cells thought to be the initial target during sexual transmission of HIV-1 (30, 35, 74, 82). Dual infection between B5 and C5 was selected because of moderate competition in PBMC (WD of B5/C5 = 8; Fig. 4C).
There was no significant difference in the B5 and C5 relative fitness values derived from competitions in the PBMC of three different donors (Fig. 5C). Relative fitness values were unaffected by variable CCR5 expression on the CD4+ cell population in phytohemagglutinin- and interleukin-2-treated PBMC (57 to 65%; Fig. 5B). All three donors were wild-type homozygotes for the CCR5 allele (73), i.e., they lacked the 32-amino-acid CCR5 deletion (Fig. 5B). In addition, each donor was heterozygous for the −2459 promoter polymorphism (A/G), which is one genetic factor responsible for various levels of surface CCR5 expression (45).
FIG. 5.
Comparing the relative fitness values of B5 and C5 HIV-1 isolates derived from competitions in CD4+ T cells, macrophages, and skin-derived Langerhans cells from different donors. (A) An example of B5 and C5 monoinfections and dual infection of skin-derived Langerhans cells (LC) from donor D. Skin explants were obtained from suction blister roofs isolated from the thighs of healthy donors (35) and exposed to a multiplicity of infection of 0.01 of the virus (250 IU added to approximately 25,000 Langerhans cells embedded within each skin explant). Langerhans cells were then allowed to emigrate from explants and were harvested on day 3 or 4 postinfection. DNA extracted from lysed cells was subjected to HIV-specific PCR amplifications and heteroduplex tracking analysis. Purification and subsequent HIV infections of macrophages and CD4+ T cells were done as described in Materials and Methods. Similar heteroduplex tracking analyses were performed on these samples. (B) Genetic characteristics and coreceptor expression on host PBMC cells. T cells and macrophages were purified from PBMC from three donors. Fluorescence-activated cell sorting analysis was performed on the PBMC populations prior to cell isolation to measure CCR5 and CD4 expression. PBMC labeled with fluorescein isothiocyanate-labeled anti-CD4 antibody and phycoerythrin-labeled anti-CCR5 antibodies were analyzed with the FACScan flow cytometer and Lysis II software (44). Percentages represent the fraction of CCR5-positive cells in the CD4-positive-gated lymphocyte population. Genetic polymorphisms at position −2459 in CCR5 promoter and the presence or absence of a 32-codon deletion (32 aa del) in the CCR5 open reading frame were also determined. wt, wild type; nd, not determined. (C) Relative fitness of the B5 and C5 HIV-1 isolates derived from dual infection of PBMC, CD4+ T cells, macrophages, and skin-derived Langerhans cells from different donors. Relative fitness value was measured as the proportion of each virus produced from the dual infection (f0) divided by the initial fraction of that virus added to the culture (i0) (62).
Increased CCR5 surface expression may mediate increased entry of R5/non-syncytium-inducing HIV-1 isolates and may augment HIV-1 production in monoinfections (58, 78). If, however, both viruses are restricted to enter through the same coreceptor, individual host cell differences in surface coreceptor expression should have a similar effect on both viruses in a given dual-infection assay. Use of different PBMC in these competitions did affect overall but not relative production of these viruses.
The B5-versus-C5 competition was also performed in CD4+ T cells and in macrophages from the same three donors. There were slight but insignificant differences in the relative fitness values in the CD4+ T cells, macrophages, and PBMC (Fig. 5C). This, however, does not imply that these cells supported the same level of HIV-1 replication. Total virus production in macrophage infections was at least 10-fold less than that obtained in CD4+ T-cell infections. Finally, the B5-versus-C5 HIV-1 competition was also performed in skin-derived Langerhans cells from three different donors. In these experiments, epidermal sheets were obtained from suction blister roofs induced on the thighs of healthy donors and then exposed to virus. Four days following exposure to virus, PCR-amplified env DNA was then used in heteroduplex tracking analysis as described above (Fig. 5A). In these competitions, C5 replicated relatively well in skin-derived Langerhans cells, as reflected by a lower WD value (wB5/wC5 ≈ 1) compared to other primary cell types (PBMC, CD4+ T cells, and macrophages; Fig. 5C). The possibility that subtype C isolates may have a slightly greater fitness in Langerhans cells than in blood-derived CD4+ cells will be the subject of future studies.
Where in the replication scheme do HIV-1 isolates compete?
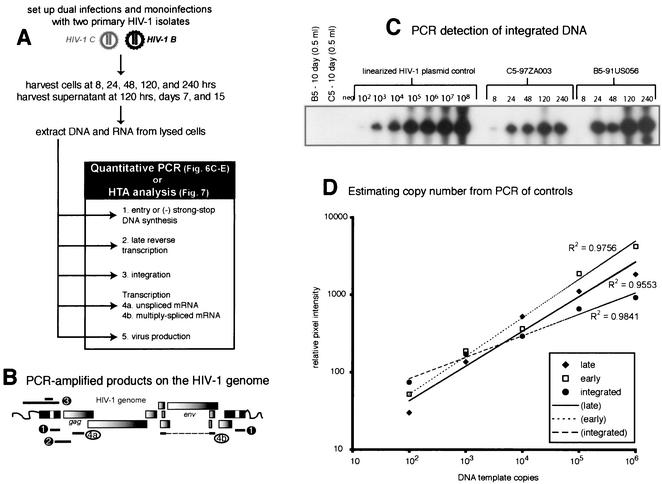
We have developed a method to determine which steps in the retroviral life cycle have the greatest impact on the replication efficiency and outcomes of these competition experiments. A schematic outline of this approach is shown in Fig. 6A. PBMC cultures were monoinfected and coinfected with 0.01 IU of the C2, C5, B2, and B5 HIV-1 isolates. All of these isolates had different fitness values in the full pairwise competition experiments (Table 1 and Fig. 3 and 4) but showed similar production in monoinfections (Fig. 1C). Various HIV-1 fragments representing different steps in the HIV-1 life cycle were PCR or RT-PCR amplified from each sample of the time course competition (8, 24, 48, 110, and 240 h; Fig. 6A).
FIG. 6.
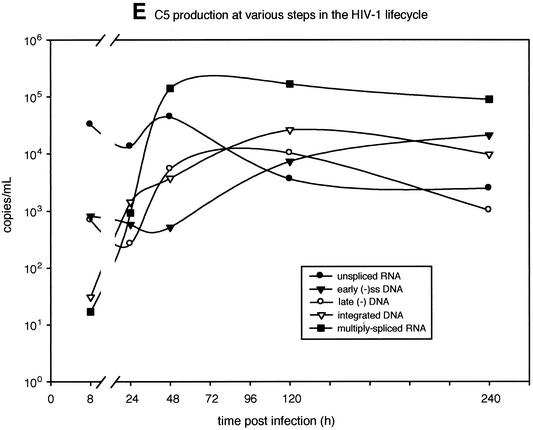
Time course competitions in PBMC with subtype B and C HIV-1 isolates and detection of HIV-1 products at different steps in the retroviral life cycle. (A) Schematic representation of pairwise dual infections with B2, B5, C2, and C5. PBMC were exposed to virus for 8 h, washed extensively, and reincubated at 37°C. Cells and supernatant were harvested at 8, 24, and 48 h and at 5 and 10 days postinfection. (B) Viral RNA or DNA extracted from infected cells was RT-PCR or PCR amplified with primer sets specific for early reverse transcripts, late reverse transcripts, integrated DNA, unspliced mRNA, and multiply spliced mRNA. Approximate positions of the primers are indicated. Radiolabeled primer pairs were used to detect and quantify products in both mono- and dual infections (C, D, and E), whereas an external-nested PCR amplification was necessary to generate sufficient product for heteroduplex tracking analysis (HTA) (Fig. 7). (C) Quantifying the level of HIV-1 integration during monoinfections with C5 and B5 HIV-1 isolates. HIV-1 DNA integrated within 4 kb of an Alu sequence in the human genome was PCR amplified with Alu and HIV-1 Alu-LTR primers (14). The primer pair LTR-1 and γ-32P-end-labeled AU3-1 was used for nested amplification of the previous PCR products. Time course samples for the C5 and B5 monoinfections were subjected to this external-nested PCR. To control for carryover of unintegrated HIV-1 DNA from the sample into the external and then nested PCR amplifications, 0.5 ml of the 240-h sample was added straight to the nested amplification (44). (D) Tenfold dilutions of linearized HXB2 plasmid (108 to 102) were PCR amplified with the nested primer pair as an amplification control and to estimate copy number for each specific product. Similar amplifications were performed on the different RNA and DNA HIV-1 products (A and B). However, in vitro-transcribed HIV-1 RNA was quantified, diluted, and used as template for control RT-PCR amplifications (data not shown). Copy number of sample RNA/DNA was derived from amplifications of known copies of control template. Copy number of control DNA and resulting PCR-amplified early, late, or integrated DNA product was plotted to obtain an equation of the line. All power regressions had R2 values of greater than 0.95 with a template copy range of 102 to 106. (E) Approximate copy numbers of early and late reverse-transcribed DNA, integrated DNA, unspliced mRNA, and multiply spliced mRNA from the C5 monoinfection were plotted against time. Similar results were observed in the B5 monoinfection as well as in the B5-versus-C5 dual infection (data not shown). (-), minus strand; (-)ss, minus-strand strong stop.
For these studies, we designed several sets of conserved HIV-1 primers with sequence identity to all known HIV-1 clades (Fig. 6B; the positions of these primer sets are outlined in Materials and Methods). Quantitative PCR with these radiolabeled primer pairs was used to estimate the amounts of each HIV-specific (B5 or C5) product during the virus life cycle (Fig. 6A). In contrast, heteroduplex tracking analysis performed with unlabeled PCR products provided a relative measure of dual virus production during each step in the life cycle. Copy numbers of each PCR-amplified HIV-1 DNA product were calculated as described previously (see Materials and Methods). The amount of PCR-amplified product was plotted against the copy number of the control template (102 to 108 copies; Fig. 6D). All regressions had an R2 value of greater than 0.95 within the range of 102 to 105 copies but reached an amplification plateau at 106 to 107 copies. As described below, all of the HIV-1-specific products amplified from the B5 and C5 monoinfections fit within the linear range.
The timing of various events in the HIV-1 C5 life cycle (Fig. 6D) was similar to that characterized previously (36) and representative of other mono- and dual infections. For example, entry of viral RNA and reverse transcription occurred within 8 h of C5 virus exposure, whereas HIV-1 integration and mRNA transcription were only detected by quantitative PCR at 24 to 48 h postinfection (Fig. 6C and D). It is important to note that virus titers were carefully calculated in TCID50 experiments and confirmed by comparison with viral RNA loads and reverse transcriptase activity in the same stocks (Fig. 1B). However, it was surprising to observe a decrease in unspliced HIV-1 RNA in cells following initial entry and a single round of infection (24 to 48 h). This decrease may be attributable to the rapid processing of HIV-1 RNA to multiply spliced messages and RNase H degradation of the RNA genome during reverse transcription.
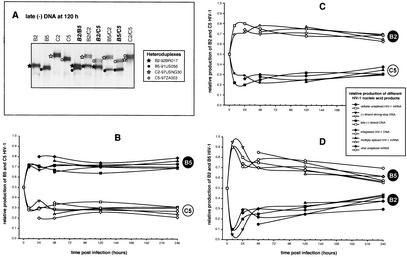
To measure dual virus production throughout the retroviral life cycle, the same sets of primers were used to amplify DNA from samples at different time points as well as from reference HIV-1 genomes. These two PCR products were mixed, denatured, and annealed as described previously. Figure 7A provides an example of a heteroduplex tracking analysis performed on late minus-strand DNA amplified samples of four mono- and six dual infections at the 120-h time point. Relative production of each nucleic acid-mediated step (entry, early and late reverse transcription, integration, and HIV-1 transcription, i.e., unspliced and multiply spliced mRNA) was then plotted against time (Fig. 6B, C, and D).
FIG. 7.
Measuring isolate-specific products that represent different steps in the retroviral life cycle during a dual-infection/competition experiment. Relative production of two HIV-1 isolates was measured during different steps in the HIV-1 life cycle, i.e., early and late reverse transcription, integration, and unspliced and multiply spliced mRNA transcription. Panel A provides an example of one heteroduplex tracking analysis performed on the late product of HIV-1 reverse transcription that was PCR amplified at 120 h postinfection. (-), minus strand. The products in the lanes identified with bold and italic labels were quantified and plotted in panels A, B, and C. These panels show the results of three time course dual infections involving B5 versus C5, B2 versus C5, and B5 versus B2, respectively.
Unspliced and multiply spliced mRNA was not analyzed by heteroduplex tracking analysis for the B2 versus C2, B5 versus B2, or C2 versus C5 dual infections. For these competitions, relative production of the other steps in the retroviral life cycle was determined (data not shown). In each of these competitions, WD values calculated from heteroduplex tracking analysis of PCR-amplified proviral DNA (Fig. 3 and 4) were nearly identical to the WD calculated from heteroduplex tracking analysis of RT-PCR-amplified viral RNA from cell-free supernatant (Fig. 7). As described previously (62), relative virus production in a dual infection can be measured 10 to 15 days postinfection by using proviral DNA within infected cells or viral RNA in HIV-1 particles released into the medium.
Reverse transcription of minus-strand strong-stop DNA and the first template switch are the first replication steps following host cell entry and core dissolution. Early minus-strand DNA was detected by PCR as early as 8 h postinfection in the B5 and C5 monoinfections (Fig. 6D). After only 8 h of competition, heteroduplex tracking analysis revealed an increase in the production of B5 early minus-strand DNA over that of C5 or B2 in the competition (Fig. 7B and D). This finding suggests an increase in host cell entry by the B5 HIV-1 isolate over C5 or B2. The WD or ratio of early minus-strand DNA production was maintained throughout the dual infection (10 days). Although B2 minus-strand DNA rebounded during competition, the relative amount of B5 early DNA still remained greater than that of B2 (Fig. 7D). Early minus-strand DNA synthesis could not be analyzed in the B2-versus-C5 competition (Fig. 7C).
The U5 of the long terminal repeat and noncoding region, i.e., a product of late reverse transcription, was also PCR amplified (Fig. 6B) and subjected to the same heteroduplex tracking analysis (Fig. 7B). Again, amounts of the late reverse-transcribed B5 DNA were greater than those of the B2 or C5 product in the dual infections (Fig. 7B and D). Increased fitness of the B2 isolate over C5 (Fig. 4) was reflected in a fourfold increase in late minus-strand DNA synthesis as early as 8 h postinfection (Fig. 6C). In all dual infections shown in Fig. 7, relative production of early and late reverse-transcribed DNA was nearly identical, suggesting that entry, and not efficiency of reverse transcription, was responsible for the fitness difference.
Integrated DNA was PCR amplified as described previously (14, 44) and in Materials and Methods, quantified (Fig. 6C and D), and then used in heteroduplex tracking analysis (Fig. 7). Proviral DNA integration is the step that follows reverse transcription in the retroviral life cycle. Consistent with the estimated 12 to 24 h required for this event (36), significant levels of integrated HIV-1 DNA were only detected at 24 h postinfection with B5 (data not shown) or C5 (Fig. 6D). Again, the ratio or WD of B5 to C5 integrated DNA was similar to the WD of B5 to C5 early or late reverse-transcribed DNA (Fig. 7B). Similar results were observed in the other dual infections (Fig. 7C and D).
RNA was extracted from cells at all time points and subjected to RT-PCR specific for unspliced HIV-1 mRNA and multiply spliced message. Relative amounts of unspliced HIV-1 RNA in the cell can be used as a measure of host cell entry because encapsidated HIV-1 RNA is released into the cell immediately following membrane fusion (Fig. 6D). Consistent with an early entry effect, the relative amount of unspliced B5 RNA was greater than that of unspliced C5 or B2 RNA in the dual infections (Fig. 7B and D). Increased HIV-1 RNA entry also appeared to predict the “winner” of the B2-versus-C5 competition (Fig. 7C). In contrast to unspliced RNA, multiply spliced RNA was only detected 48 h postinfection and represented active mRNA transcription from the HIV-1 LTR. Unlike entry, relative transcription from each proviral template (measured by heteroduplex tracking analysis) appeared to have little effect on the outcome of the competition (Fig. 7).
It is likely that an early event in the replication cycle (e.g., entry) is controlling these HIV-1 competitions (B2 versus B5, B2 versus C5, and B5 versus C5), whereas subsequent events in retroviral replication (e.g., reverse transcription, integration, and transcription) have minimal affect on the outcome. However, the initial impact of entry was not always maintained throughout the dual infection. For example, increases in HIV-1 B5 over B2 entry into host cells were diminished over the time course. After a 15-day dual infection, the WD of B2 versus B5 was approximately 1.8. The effect of entry on predicting the winner of a competition was most evident in the B5-versus-C9 dual infection (data not shown). The inability of C9 to compete with B5 (WD > 20; Fig. 4) was evident early in the infection; a 15-fold increase in B5 over C9 minus-strand DNA was observed just 8 h after infection. This difference was maintained throughout subsequent steps in the retroviral life cycle.
Variations in the efficiency of translation, posttranslational modifications, or assembly could not be analyzed without specific monoclonal antibodies or other markers that accurately differentiate between viral proteins of two isolates. Lack of continual outgrowth of the fitter variant is likely due to depletion of susceptible CCR5+/CD4+ cells in the culture (2) as well as receptor downregulation, which would prevent superinfection (17, 26). Competition during entry may also be due to viral interference on host cell receptors, i.e., subtype B isolates may bind more efficiently to CD4/CCR5 and exclude subtype C isolates from binding. These hypotheses will be tested in subsequent studies with HIV-1 clones pseudotyped with subtype B or C env genes.
Applying phyletic analyses to fitness differences.
Considering that HIV-1 subtypes are subdivided based on env sequence diversity, we also used a phyletic approach to compare fitness differences to specific regions in the HIV-1 genome. Pairwise comparisons of the genetic distances between the gag (matrix-capsid coding regions) and env (C2-C3 domains) genes of these subtype B and C isolates were used to construct phylogenetic trees (Fig. 8B and C). We also employed this neighbor-joining method to construct phyletic trees based on fitness differences rather than genetic distances. Equal fitness or no competition between a pair of HIV-1 isolates would result in a fitness difference of 1 (WD = wM/wL), which would be analogous to two HIV-1 isolates' having identical sequences. Thus, nondirectional measures of fitness difference in the pairwise inter- and intrasubtype competitions (Fig. 3 and 4) could be derived from log10 values of WD.
FIG. 8.
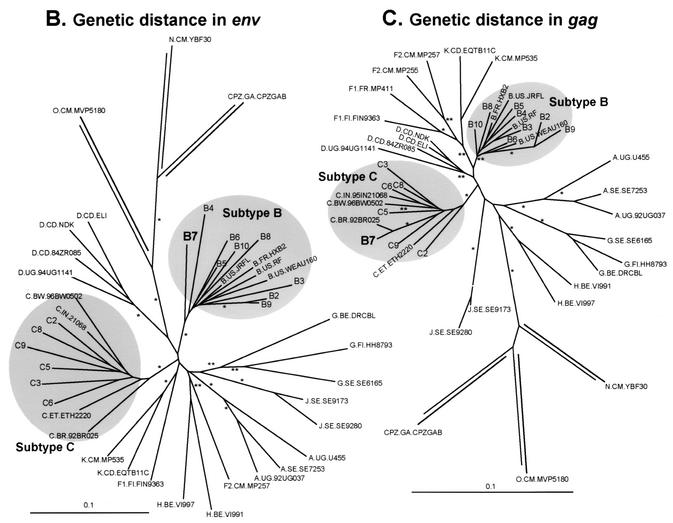
Analysis of phenotypic and genotypic relationships of HIV-1 isolates with the neighbor-joining method. Phyletic neighbor-joining trees were constructed from matrices of pairwise log10 fitness differences as well as from nucleotide distances in the env and gag genes. (A) The neighbor.exe program in the PHYLIP package was used to draw a phyletic tree based on the log10 values of the fitness difference in the pairwise dual-infection/competition experiments. Equal relative fitness values result in a fitness difference of 1. Thus, log10 values of wM/wL provide a unidirectional measure of fitness difference for comparison to genotypic distances. (B and C) Phylogenetic analyses of the gag and env sequences, respectively, of 15 subtype B and C isolates and of several reference HIV-1 isolates (see Materials and Methods for accession numbers). A 400-bp region of the env gene C2-C3 region (B) and a 500-bp region of the gag gene (C) were used to construct phylogenetic trees by the neighbor-joining method. env and gag subtypes are indicated. Bootstrap resampling values of 70 to 90% and >90% are indicated by ** and *, respectively. The branch lengths are drawn to scale. The scale bar represents 0.1 substitution per nucleotide.
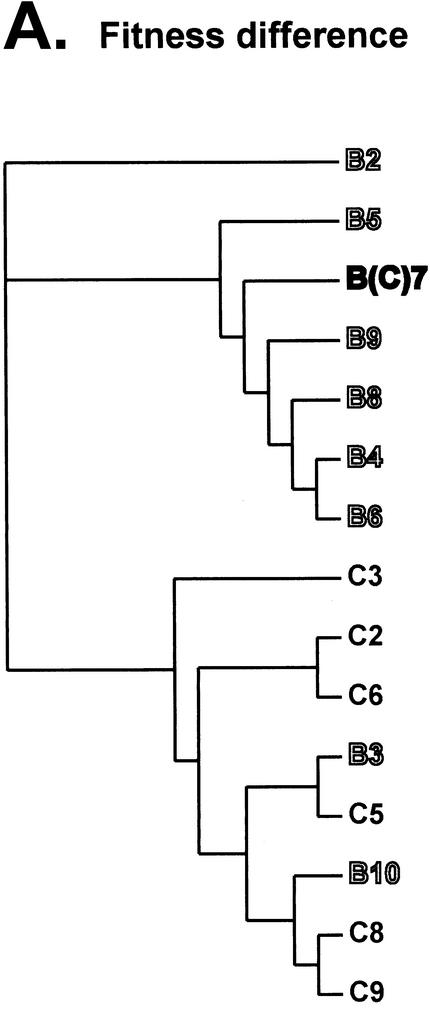
Although the phyletic fitness trees were not rooted with reference strains (e.g., HIV-1 group O isolates or simian immunodeficiency virus strain cpz [SIVcpz]), subtype B isolates still formed a cluster at the bottom of the fitness tree (Fig. 8A), which corresponded to a similar cluster of subtype B env or gag sequences in the genetic distance tree (Fig. 8B and C). Subtype C isolates also grouped together in the fitness tress as well as in the gag and env phylogenetic trees (Fig. 8). In the fitness tree, the presence of the B3 and B10 isolates in the subtype C cluster reflects the poor replicative capacity of these subtype B strains in the pairwise competitions (Table 1; Fig. 3 and 4). In addition to losing competitions with nearly every subtype B isolate (Fig. 3A), the B3 and B10 isolates were also less fit than some subtype C isolates, hence their position on the fitness tree.
Analyses involving the B7-92BR023 isolate provided the only notable exception to this similarity between the tress based on fitness and genetic distance. This isolate is closely related to other subtype B isolates when comparing fitness and env sequences. By contrast, the gag sequences (region encoding the matrix and part of the capsid) of B7 were not closely related to other subtype B gag sequences, but instead clustered with the gag sequences of subtype C isolates. Previous reports have characterized the B7-92BR023 isolate as an intersubtype B/C HIV-1 recombinant with subtype B env and C gag (16). Sequence analyses from this study suggest a subtype C pol gene and a possible breakpoint in the env gene (data not shown). Thus, the fitness (or replication efficiency) of at least the B7 isolate maps to the env gene and not the gag (Fig. 8C) or pol gene (protease-reverse transcriptase coding regions; data not shown).
DISCUSSION
During the past 15 years, the mechanics and specifics of HIV-1 replication have been resolved by sophisticated molecular and biochemical experiments. However, the vast majority of these studies have utilized a handful of subtype B laboratory strains to define the steps of HIV-1 replication. Although general mechanisms may be conserved, the extreme HIV-1 heterogeneity found in the current epidemic may also predict phenotypic and catalytic variations in virus replication. To date, there are few examples of natural polymorphisms in HIV-1 associated with altered replication kinetics (33, 38, 46, 49). Even fewer studies have compared the fitness or replication efficiency of heterogeneous primary HIV-1 isolates (6, 43, 62). Phenotypic differences between HIV-1 subtypes have typically focused on specific genomic segments or sequences without comparing the replication efficiency of the complete virus (33, 38, 46, 49). Hence, this study has investigated the ex vivo fitness differences between primary HIV-1 isolates of subtypes B and C. It is also part of a series of investigations comparing ex vivo HIV-1 fitness to disease progression (62) and intersubtype recombination (2, 63).
Fitness is an evolutionary term used to define an organism's replicative capacity in a given environment (21). Although the most “fit” isolate may win a given competition in the confines of the tissue culture microenvironment, ex vivo fitness may be unrelated to the ability of the same virus to survive in a host or to be transmitted within a given population (61). However, our results suggest a possible relationship between ex vivo and in vivo fitness of subtype C that may be used to model HIV-1 subtype C spread in the human population.
In the competition experiments, all of the R5/non-syncytium-inducing subtype B isolates outcompeted the R5/non-syncytium-inducing subtype C isolates, whereas differences in intrasubtype HIV-1 fitness were not significant. Dominance of HIV-1 subtype B over C was not due to genetic variability in the host PBMC population or differences in CCR5 expression. Increased fitness of the B5 over the C5 isolate was also observed in CD4+ T-cell and macrophage cultures. By contrast, C5 efficiently competed with the B5 isolate in skin-derived Langerhans cells. Phyletic analyses of fitness differences and genetic distances suggest that the efficiency of HIV-1 replication may map to the env gene, as opposed to either the gag or pol gene. Although this hypothesis is based on one isolate (B7), preliminary data suggest that the efficiency of host cell entry may control fitness (A. J. Marozsan and E. J. Arts, unpublished data).
Average relative fitness values were not significantly different in intrasubtype competitions. This does not imply that all subtype B isolates have equal fitness. In fact, several subtype B and C strains were fitter than some but not all isolates of the same subtype. In intrasubtype competitions, a lower intrinsic fitness (e.g., replication efficiency) of subtype C isolates may imply that all HIV-1 group M subtypes are segregated based on both genetic distance and a phenotypic attribute (such as replication efficiency). It is important to note that the standard classification of HIV-1 subtypes is based solely on sequence diversity in specific genomic regions (40), and not on phenotype or even serotype (12). Preliminary results suggest that HIV-1 isolates of subtypes A, B, D, and E have similar relative fitness values in pairwise competitions. We used neighbor joining to construct and compare phyletic trees based on genetic and fitness differences. Only subtype C isolates appeared to form a distinct cluster in the phyletic trees based on fitness differences. By contrast, the vast majority of subtype A, B, and D isolates have similar replication efficiencies and group together in these fitness tress.
Subtype C infections have increased in prevalence during the past 10 years of the AIDS epidemic (22). Increased spread over other subtypes is due in part to founder effects and raging pandemics in southern Africa and increasing subtype C infections in South America, India, China, and Southeast Asia (8, 10, 22, 42, 48, 57, 65). Introductions of subtype C into these regions may be a founder event, but other subtypes did preexist in some populations (e.g., A and B in southern Africa and India) (40). In regions such as the Yunnan Province of China, subtype C is responsible for the recent pandemic, even though this province is part of the Golden Triangle of intravenous drug use and surrounded by areas with a high prevalence of subtype B and CRF01 (subtype E) viruses (L. Zhang, Y. Cao, J. Yu, T. He, Z. Chen, N. Yin, S. Mei, Z. Zhou, Y. He, W. Lu, Z. Chen, and D. D. Ho, 9th Conference on Retroviruses and Opportunistic Infections, abstr., p. 58, 2002).
The rapid insurgence of subtype C throughout the world suggests that this clade may be fitter within the population. It is also quite conceivable that a subtype such as C may be introduced into human subpopulations with different social and sexual mixing patterns than populations infected by another subtype(s) (32, 51, 72). Thus, subtype C may not be in direct competition with other subtypes in these geographical regions. However, subtype C has obviously expanded beyond these possible subpopulations and appears to be spreading more rapidly than other subtypes. Based on these findings, we have devised a preliminary model of subtype prevalence in the HIV-1 epidemic.
First, subtype C isolates may replicate less efficiently than subtype B and possibly other group M subtypes in PBMC. Second, we and others have shown that ex vivo fitness (or replication efficiency) is a strong correlate of the rate of HIV-1 disease progression, that is, individuals infected with a less-fit HIV-1 isolate progress to AIDS at a slower rate and vice versa (6, 62). However, we may be overextending our observations (62; this study) to suggest that HIV-1 subtype C-infected individuals may progress to AIDS more slowly. Cross-sectional studies have actually described higher viral loads in patients infected with subtype C than in patients infected with other group M subtypes (e.g., A and D) (48). In addition, subtype C-infected individuals definitely progress to AIDS, as do most HIV-1-infected patients not receiving antiretroviral therapy.
Unfortunately, there are only a few prospective longitudinal studies analyzing disease progression in cohorts infected with different subtypes. A preliminary study observed similar rates of disease progression among individuals infected with different subtypes (A through D) (1). By contrast, Kaleebu et al. (34) observed faster rates of disease progression among a large cohort of Ugandans infected with subtype D compared to subtype A. They presuppose that subtype D HIV-1 isolates may replicate more efficiently than subtype A, a hypothesis similar to that developed from these ex vivo fitness studies on subtype C. To place these findings in the context of the model, it is possible that individuals infected with subtype C could survive longer but still maintain viral loads for efficient transmission. Infection of macaques with SIV strains pseudotyped with HIV-1 subtype B or C env genes (SHIV) may be an appropriate animal model to compare subtype-specific differences in disease progression (13, 27, 47). Longitudinal studies of defined cohorts following acute or early infections with various subtypes should also provide direct evidence of differential disease progression.
Maintenance of efficient subtype C transmission in lieu of reduced replicative capacity in blood cells may be supported by the competition experiments in Langerhans cells. A subtype C isolate could compete efficiently with a subtype B strain in skin-derived Langerhans cells but not in macrophages or T cells. Importantly, we exposed Langerhans cells to HIV-1 subtypes B and/or C in a relatively physiological state (i.e., while they remained “immature” within epithelial sheets) (35), as opposed to exposing these cells to HIV-1 after they had been allowed to crawl out of tissue and become activated, or “mature” (20, 59). Although it is controversial, we and others have suggested that Langerhans cells are the initial target cells infected following sexual exposure to HIV (29, 35, 74, 82). Our current finding that subtype C HIV-1 is able to compete with subtype B HIV-1 in Langerhans cells may be an important factor in the efficient transmission and spread of subtype C HIV-1 within a given population.
In summary, increased survival time of subtype C-infected individuals and maintenance of transmission efficiency in Langerhans cells (but not PBMC) may favor subtype C spread in geographical regions harboring several subtypes. Computer simulations are in development to test this epidemiological model.
HIV-1 competition experiments can be used to determine which steps in the HIV-1 life cycle have the greatest impact on relative fitness. Ultimately, knowing how primary HIV-1 isolates compete may reduce the necessity for these laborious competition experiments as well as identify HIV-1 genes or proteins controlling fitness. Time course competition experiments suggest that the fitness of wild-type HIV-1 is controlled by the efficiency of host cell entry and not by reverse transcription, integration, or transcription from the LTR. The impact of entry on replication efficiency is not surprising considering the following points: (i) the HIV-1 env gene displays the greatest heterogeneity among HIV-1 isolates, whereas the gag and pol genes are more conserved (40), and (ii) increases in heterogeneity likely augment differences related to the function or activity of that region or gene. Comparisons with neighbor-joining analyses revealed that fitness differences were more closely related to the genetic distances in env than in gag or pol. Specifically, one subtype B isolate, B7-92BR023, was a B/C recombinant containing a subtype B env gene but subtype C gag and pol genes (16).
Based on these time course experiments and phylogenetic analyses, it is somewhat surprising that the fitter isolate did not continue to outgrow the less-fit isolate after initial entry. At a multiplicity of infection of 0.01, all susceptible cells in a PBMC culture are infected after three rounds of HIV-1 replication (E. J. Arts et al., submitted for publication). Thus, intracellular steps would have a greater effect on competitions with longer incubation times (>120 h). We have preliminary data that constant addition of new cells would enhance the effect of HIV-1 entry on these competitions. Other steps in the retroviral life cycle must contribute to replication efficiency but do not override the entry effect. It is quite possible that an extra NF-κB site in the LTR (33, 49) and/or a more active protease (79) in subtype C isolates may have evolved to compensate for poor entry efficiency. The impact of the HIV-1 env gene on the efficiency of entry and relative fitness can only be confirmed with HIV-1 clones pseudotyped with these exogenous env genes. These experiments are currently under way in our laboratory.
In conclusion, subtype C HIV-1 isolates appear to be less fit than subtype B isolates in PBMC but equally fit in skin-derived Langerhans cell cultures. Fitness difference is not due to differential coreceptor usage, but may be due to the efficiency of host cell entry mediated by CD4 and CCR5. Although the relationship between ex vivo replication efficiency and subtype fitness in the human population is tenuous, these observations will be used to build epidemiological disease models for host-virus interactions and to add insight into the disproportionate spread of a given HIV-1 subtype within the human population.
Acknowledgments
The first four authors contributed equally to the data presented herein.
Research for this study was performed at Case Western Reserve University. E.J.A. was supported by research grants from the National Institute of Child Health and Development, NIH (NO1-HD-0-3310-502-02), and from the National Institute of Allergy and Infectious Diseases, NIH (AI49170). M.E.Q.-M. was supported by research grants from the National Heart, Lung, and Blood Institute, NIH (HL67610-02). All virus work was performed in the biosafety level 2 and 3 facilities of the CWRU Center for AIDS Research (AI36219).
We thank Korey Demers, Case Western Reserve University, Cleveland, Ohio, and Debra L. Borris, National Cancer Institute, Bethesda, Md., for technical assistance.
REFERENCES
- 1.Alaeus, A., K. Lidman, A. Bjorkman, J. Giesecke, and J. Albert. 1999. Similar rate of disease progression among individuals infected with HIV-1 genetic subtypes A-D. AIDS 13:901-907. [DOI] [PubMed] [Google Scholar]
- 2. Reference deleted.
- 3.Arts, E. J., J. Mak, L. Kleiman, and M. A. Wainberg. 1994. DNA found in human immunodeficiency virus type 1 particles may not be required for infectivity. J. Gen. Virol. 75:1605-1613. [DOI] [PubMed] [Google Scholar]
- 4.Asjo, B., L. Morfeldt-Manson, J. Albert, G. Biberfeld, A. Karlsson, K. Lidman, and E. M. Fenyo. 1986. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet 2:660-662. [PubMed] [Google Scholar]
- 5.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaak, H., M. Brouwer, L. J. Ran, F. de Wolf, and H. Schuitemaker. 1998. In vitro replication kinetics of human immunodeficiency virus type 1 (HIV-1) variants in relation to virus load in long-term survivors of HIV-1 infection. J. Infect. Dis. 177:600-610. [DOI] [PubMed] [Google Scholar]
- 7.Bobkov, A., E. Kazennova, T. Khanina, M. Bobkova, L. Selimova, A. Kravchenko, V. Pokrovsky, and J. Weber. 2001. An HIV type 1 subtype A strain of low genetic diversity continues to spread among injecting drug users in Russia: study of the new local outbreaks in Moscow and Irkutsk. AIDS Res. Hum. Retrovir. 17:257-261. [DOI] [PubMed] [Google Scholar]
- 8.Bongertz, V., D. C. Bou-Habib, L. F. Brigido, M. Caseiro, P. J. Chequer, J. C. Couto-Fernandez, P. C. Ferreira, B. Galvao-Castro, D. Greco, M. L. Guimaraes, M. I. Linhares de Carvalho, M. G. Morgado, C. A. Oliveira, S. Osmanov, C. A. Ramos, M. Rossini, E. Sabino, A. Tanuri, and M. Ueda. 2000. HIV-1 diversity in Brazil: genetic, biologic, and immunologic characterization of HIV-1 strains in three potential HIV vaccine evaluation sites. Brazilian Network for HIV Isolation and Characterization. J. Acquir. Immune Defic. Syndr. 23:184-193. [DOI] [PubMed] [Google Scholar]
- 9.Butto, S., C. Argentini, A. M. Mazzella, M. P. Iannotti, P. Leone, P. Leone, A. Nicolosi, and G. Rezza. 1997. Dual infection with different strains of the same HIV-1 subtype. AIDS 11:694-696. [PubMed] [Google Scholar]
- 10.Cassol, S., B. G. Weniger, P. G. Babu, M. O. Salminen, X. Zheng, M. T. Htoon, A. Delaney, M. O'Shaughnessy, and C. Y. Ou. 1996. Detection of HIV type 1 env subtypes A, B, C, and E in Asia with dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res. Hum. Retrovir. 12:1435-1441. [DOI] [PubMed] [Google Scholar]
- 11.Cecilia, D., S. S. Kulkarni, S. P. Tripathy, R. R. Gangakhedkar, R. S. Paranjape, and D. A. Gadkari. 2000. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology 271:253-258. [DOI] [PubMed] [Google Scholar]
- 12.Cheingsong-Popov, R., S. Osmanov, C. P. Pau, G. Schochetman, F. Barin, H. Holmes, G. Francis, H. Ruppach, U. Dietrich, S. Lister, and J. Weber. 1998. Serotyping of HIV type 1 infections: definition, relationship to viral genetic subtypes, and assay evaluation. UNAIDS Network for HIV-1 Isolation and Characterization. AIDS Res. Hum. Retrovir. 14:311-318. [DOI] [PubMed] [Google Scholar]
- 13.Chen, Z., Y. Huang, X. Zhao, E. Skulsky, D. Lin, J. Ip, A. Gettie, and D. D. Ho. 2000. Enhanced infectivity of an R5-tropic simian/human immunodeficiency virus carrying human immunodeficiency virus type 1 subtype C envelope after serial passages in pig-tailed macaques (Macaca nemestrina). J. Virol. 74:6501-6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins, K. R., M. E. Quinones-Mateu, M. Wu, H. Luzze, J. L. Johnson, C. Hirsch, Z. Toossi, and E. J. Arts. 2002. Human immunodeficiency virus type 1 (HIV-1) quasispecies at the sites of Mycobacterium tuberculosis infection contribute to systemic HIV-1 heterogeneity. J. Virol. 76:1697-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornelissen, M., B. R. van den, F. Zorgdrager, V. Lukashov, and J. Goudsmit. 1997. pol gene diversity of five human immunodeficiency virus type 1 subtypes: evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J. Virol. 71:6348-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crise, B., and J. K. Rose. 1992. Human immunodeficiency virus type 1 glycoprotein precursor retains a CD4-p56lck complex in the endoplasmic reticulum. J. Virol. 66:2296-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 19.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 20.Dittmar, M. T., G. Simmons, S. Hibbitts, M. O'Hare, S. Louisirirotchanakul, S. Beddows, J. Weber, P. R. Clapham, and R. A. Weiss. 1997. Langerhans cell tropism of human immunodeficiency virus type 1 subtype A through F isolates derived from different transmission groups. J. Virol. 71:8008-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 22.Essex, M. 1999. Human immunodeficiency viruses in the developing world. Adv. Virus Res. 53:71-88. [DOI] [PubMed] [Google Scholar]
- 23.Ewald, P. W. 1994. Evolution of infectious disease. Oxford University Press, Oxford, United Kingdom.
- 24.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 25.Gao, F., L. Yue, S. Craig, C. L. Thornton, D. L. Robertson, F. E. McCutchan, J. A. Bradac, P. M. Sharp, and B. H. Hahn. 1994. Genetic variation of HIV type 1 in four World Health Organization-sponsored vaccine evaluation sites: generation of functional envelope (glycoprotein 160) clones representative of sequence subtypes A, B, C, and E. WHO Network for HIV Isolation and Characterization. AIDS Res. Hum. Retrovir. 10:1359-1368. [DOI] [PubMed] [Google Scholar]
- 26.Garcia, J. V., J. Alfano, and A. D. Miller. 1993. The negative effect of human immunodeficiency virus type 1 Nef on cell surface CD4 expression is not species specific and requires the cytoplasmic domain of CD4. J. Virol. 67:1511-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harouse, J. M., A. Gettie, R. C. Tan, J. Blanchard, and C. Cheng-Mayer. 1999. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science 284:816-819. [DOI] [PubMed] [Google Scholar]
- 28.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 29.Hu, D. J., A. Buve, J. Baggs, G. G. van der, and T. J. Dondero. 1999. What role does HIV-1 subtype play in transmission and pathogenesis? An epidemiological perspective. AIDS 13:873-881. [DOI] [PubMed] [Google Scholar]
- 30.Hu, J., M. B. Gardner, and C. J. Miller. 2000. Simian immunodeficiency virus rapidly penetrates the cervicovaginal mucosa after intravaginal inoculation and infects intraepithelial dendritic cells. J. Virol. 74:6087-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, Y., L. Zhang, and D. D. Ho. 1995. Biological characterization of nef in long-term survivors of human immunodeficiency virus type 1 infection. J. Virol. 69:8142-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudgens, M. G., I. M. Longini, Jr., S. Vanichseni, D. J. Hu, D. Kitayaporn, P. A. Mock, M. E. Halloran, G. A. Satten, K. Choopanya, and T. D. Mastro. 2002. Subtype-specific transmission probabilities for human immunodeficiency virus type 1 among injecting drug users in Bangkok, Thailand. Am. J. Epidemiol. 155:159-168. [DOI] [PubMed] [Google Scholar]
- 33.Jeeninga, R. E., M. Hoogenkamp, M. Armand-Ugon, M. de Baar, K. Verhoef, and B. Berkhout. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 74:3740-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaleebu, P., N. French, C. Mahe, D. Yirrell, C. Watera, F. Lyagoba, J. Nakiyingi, A. Rutebemberwa, D. Morgan, J. Weber, C. Gilks, and J. Whitworth. 2002. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J. Infect. Dis. 185:1244-1250. [DOI] [PubMed] [Google Scholar]
- 35.Kawamura, T., S. S. Cohen, D. L. Borris, E. A. Aquilino, S. Glushakova, L. B. Margolis, J. M. Orenstein, R. E. Offord, A. R. Neurath, and A. Blauvelt. 2000. Candidate microbicides block HIV-1 infection of human immature Langerhans cells within epithelial tissue explants. J. Exp. Med. 192:1491-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, S. Y., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. E vol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 38.Kirchhoff, F., T. C. Greenough, D. B. Brettler, J. L. Sullivan, and R. C. Desrosiers. 1995. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 332:228-232. [DOI] [PubMed] [Google Scholar]
- 39.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 40.Kuiken, C., B. Foley, B. H. Hahn, P. Marx, F. E. McCutchan, J. Mellors, J. Mullins, J. Sodroski, S. Wolinsky, and B. Korber. 2002. HIV-1 sequence compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 41.Leitner, T., and J. Albert. 1999. The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc. Natl. Acad. Sci. USA 96:10752-10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo, C. C., C. Tian, D. J. Hu, M. Kai, T. Dondero, and X. Zheng. 1995. HIV-1 subtype C in China. Lancet 345:1051-1052. [DOI] [PubMed] [Google Scholar]
- 43.Malim, M. H., and M. Emerman. 2001. HIV-1 sequence variation: drift, shift, and attenuation. Cell 104:469-472. [DOI] [PubMed] [Google Scholar]
- 44.Marozsan, A. J., V. S. Torre, M. Johnson, S. C. Ball, J. V. Cross, D. J. Templeton, M. E. Quinones-Mateu, R. E. Offord, and E. J. Arts. 2001. Mechanisms involved in stimulation of human immunodeficiency virus type 1 replication by aminooxypentane RANTES. J. Virol. 75:8624-8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDermott, D. H., P. A. Zimmerman, F. Guignard, C. A. Kleeberger, S. F. Leitman, and P. M. Murphy. 1998. CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS Cohort Study. Lancet 352:866-870. [DOI] [PubMed] [Google Scholar]
- 46.Montano, M. A., C. P. Nixon, T. Ndung'u, H. Bussmann, V. A. Novitsky, D. Dickman, and M. Essex. 2000. Elevated tumor necrosis factor-alpha activation of human immunodeficiency virus type 1 subtype C in Southern Africa is associated with an NF-κB enhancer gain-of-function. J. Infect. Dis. 181:76-81. [DOI] [PubMed] [Google Scholar]
- 47.Ndung'u, T., Y. Lu, B. Renjifo, N. Touzjian, N. Kushner, V. Pena-Cruz, V. A. Novitsky, T. H. Lee, and M. Essex. 2001. Infectious simian/human immunodeficiency virus with human immunodeficiency virus type 1 subtype C from an African isolate: rhesus macaque model. J. Virol. 75:11417-11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neilson, J. R., G. C. John, J. K. Carr, P. Lewis, J. K. Kreiss, S. Jackson, R. W. Nduati, D. Mbori-Ngacha, D. D. Panteleeff, S. Bodrug, C. Giachetti, M. A. Bott, B. A. Richardson, J. Bwayo, J. Ndinya-Achola, and J. Overbaugh. 1999. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J. Virol. 73:4393-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novitsky, V. A., M. A. Montano, M. F. McLane, B. Renjifo, F. Vannberg, B. T. Foley, T. P. Ndung'u, M. Rahman, M. J. Makhema, R. Marlink, and M. Essex. 1999. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J. Virol. 73:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olsen, H. S., A. W. Cochrane, P. J. Dillon, C. M. Nalin, and C. A. Rosen. 1990. Interaction of the human immunodeficiency virus type 1 Rev protein with a structured region in env mRNA is dependent on multimer formation mediated through a basic stretch of amino acids. Genes Dev. 4:1357-1364. [DOI] [PubMed] [Google Scholar]
- 51.Op de Coul, E. L., R. A. Coutinho, S. A. van der, G. J. van Doornum, V. V. Lukashov, J. Goudsmit, and M. Cornelissen. 2001. The impact of immigration on env HIV-1 subtype distribution among heterosexuals in the Netherlands: influx of subtype B and non-B strains. AIDS 2277-2286. [DOI] [PubMed]
- 52.Osmanov, S., C. Pattou, N. Walker, B. Schwardlander, and J. Esparza. 2002. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J. Acquir. Immune Defic. Syndr. 29:184-190. [DOI] [PubMed] [Google Scholar]
- 53.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 54.Peeters, M. 2000. Recombinant HIV sequences: their role in the global epidemic, p. 54-72. In C. Kuiken, B. Foley, B. H. Hahn, P. Marx, F. E. McCutchan, J. Mellors, J. Mullins, J. Sodroski, S. Wolinsky, and B. Korber (ed.), HIV-1 sequence compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 55.Peeters, M., R. Vincent, J. L. Perret, M. Lasky, D. Patrel, F. Liegeois, V. Courgnaud, R. Seng, T. Matton, S. Molinier, and E. Delaporte. 1999. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:115-121. [DOI] [PubMed] [Google Scholar]
- 56.Ping, L. H., J. A. Nelson, I. F. Hoffman, J. Schock, S. L. Lamers, M. Goodman, P. Vernazza, P. Kazembe, M. Maida, D. Zimba, M. M. Goodenow, J. J. Eron, Jr., S. A. Fiscus, M. S. Cohen, and R. Swanstrom. 1999. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J. Virol. 73:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piyasirisilp, S., F. E. McCutchan, J. K. Carr, E. Sanders-Buell, W. Liu, J. Chen, R. Wagner, H. Wolf, Y. Shao, S. Lai, C. Beyrer, and X. F. Yu. 2000. A recent outbreak of human immunodeficiency virus type 1 infection in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. J. Virol. 74:11286-11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage tropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pope, M., S. S. Frankel, J. R. Mascola, A. Trkola, F. Isdell, D. L. Birx, D. S. Burke, D. D. Ho, and J. P. Moore. 1997. Human immunodeficiency virus type 1 strains of subtypes B and E replicate in cutaneous dendritic cell-T-cell mixtures without displaying subtype-specific tropism. J. Virol. 71:8001-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinones-Mateu, M. E., and E. J. Arts. 1998. Recombination in human immunodeficiency virus type-1 (HIV-1): update and implications. AIDS Rev. 2:89-100. [Google Scholar]
- 61.Quinones-Mateu, M. E., and E. J. Arts. 2002. HIV-1 fitness: implications for drug resistance, disease progression, and global epidemic evolution, p. 134-170. In C. Kuiken, B. Foley, B. H. Hahn, P. Marx, F. E. McCutchan, J. Mellors, J. Mullins, J. Sodroski, S. Wolinsky, and B. Korber (ed.), HIV-1 sequence compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 62.Quinones-Mateu, M. E., S. C. Ball, A. J. Marozsan, V. S. Torre, J. L. Albright, G. Vanham, G. G. van der Groen, R. L. Colebunders, and E. J. Arts. 2000. A dual infection-competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 74:9222-9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quinones-Mateu, M. E., Y. Gao, S. C. Ball, A. J. Marozsan, A. Abraha, and E. J. Arts. 2002. In vitro intersubtype recombinants of human immunodeficiency virus type 1: comparison to recent and circulating in vivo recombinant forms. J. Virol. 76:9600-9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 65.Renjifo, B., B. Chaplin, D. Mwakagile, P. Shah, F. Vannberg, G. Msamanga, D. Hunter, W. Fawzi, and M. Essex. 1998. Epidemic expansion of HIV type 1 subtype C and recombinant genotypes in Tanzania. AIDS Res. Hum. Retrovir. 14:635-638. [DOI] [PubMed] [Google Scholar]
- 66.Retief, J. D. 2000. Phylogenetic analysis with PHYLIP. Methods Mol. Biol. 132:243-258. [DOI] [PubMed] [Google Scholar]
- 67.Rich, E. A., C. A. Elmets, H. Fujiwara, R. S. Wallis, and J. J. Ellner. 1987. Deleterious effect of ultraviolet-B radiation on accessory function of human blood adherent mononuclear cells. Clin. Exp. Immunol. 70:116-126. [PMC free article] [PubMed] [Google Scholar]
- 68.Robertson, D. L., J. P. Anderson, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, C. Kuiken, G. H. Learn, T. Leitner, F. E. McCutchan, S. Osmanov, M. Peeters, M. L. Kalish, M. O. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 1999. HIV-1 nomenclature proposal, p. 492-505. In C. Kuiken, B. Foley, B. H. Hahn, P. Marx, F. E. McCutchan, J. Mellors, J. Mullins, J. Sodroski, S. Wolinsky, and B. Korber (ed.), HIV-1 sequence compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 69.Roof, P., M. Ricci, P. Genin, M. A. Montano, M. Essex, M. A. Wainberg, A. Gatignol, and J. Hiscott. 2002. Differential regulation of HIV-1 clade-specific B, C, and E long terminal repeats by NF-κB and the Tat transactivator. Virology 296:77-83. [DOI] [PubMed] [Google Scholar]
- 70.Salkowitz, J. R., S. F. Purvis, H. Meyerson, P. Zimmerman, T. R. O'Brien, L. Aledort, M. E. Eyster, M. Hilgartner, C. Kessler, B. A. Konkle, G. C. White, J. J. Goedert, and M. M. Lederman. 2001. Characterization of high-risk HIV-1 seronegative hemophiliacs. Clin. Immunol. 98:200-211. [DOI] [PubMed] [Google Scholar]
- 71.Sanders-Buell, E., M. O. Salminen, and F. E. McCutchan. 1995. Sequencing primers for HIV-1, p. III-15-III-21. In B. Korber, B. D. Walker, J. P. Moore, G. Myers, C. Brander, R. A. Koup, and B. Haynes (ed.), HIV-1 sequence compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 72.Santiago, M. L., E. G. Santiago, J. C. Hafalla, M. A. Manalo, L. Orantia, M. N. Cajimat, C. Martin, C. Cuaresma, C. E. Dominguez, M. E. Borromeo, A. S. De Groot, T. P. Flanigan, C. C. Carpenter, K. H. Mayer, and B. L. Ramirez. 1998. Molecular epidemiology of HIV-1 infection in the Philippines, 1985 to 1997: transmission of subtypes B and E and potential emergence of subtypes C and F. J. Acquir. Immune Defic. Syndr. 18:260-269. [DOI] [PubMed] [Google Scholar]
- 73.Smith, M. W., M. Dean, M. Carrington, C. Winkler, G. A. Huttley, D. A. Lomb, J. J. Goedert, T. R. O'Brien, L. P. Jacobson, R. Kaslow, S. Buchbinder, E. Vittinghoff, D. Vlahov, K. Hoots, M. W. Hilgartner, and S. J. O'Brien. 1997. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 277:959-965. [DOI] [PubMed] [Google Scholar]
- 74.Soto-Ramirez, L. E., B. Renjifo, M. F. McLane, R. Marlink, C. O'Hara, R. Sutthent, C. Wasi, P. Vithayasai, V. Vithayasai, C. Apichartpiyakul, P. Auewarakul, C. Pena, V., D. S. Chui, R. Osathanondh, K. Mayer, T. H. Lee, and M. Essex. 1996. HIV-1 Langerhans' cell tropism associated with heterosexual transmission of HIV. Science 271:1291-1293. [DOI] [PubMed] [Google Scholar]
- 75.Tersmette, M., R. E. de Goede, B. J. Al, I. N. Winkel, R. A. Gruters, H. T. Cuypers, H. G. Huisman, and F. Miedema. 1988. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J. Virol. 62:2026-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tien, P. C., T. Chiu, A. Latif, S. Ray, M. Batra, C. H. Contag, L. Zejena, M. Mbizvo, E. L. Delwart, J. I. Mullins, and D. A. Katzenstein. 1999. Primary subtype C HIV-1 infection in Harare, Zimbabwe. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:147-153. [DOI] [PubMed] [Google Scholar]
- 77.Torre, V. S., A. J. Marozsan, J. L. Albright, K. R. Collins, O. Hartley, R. E. Offord, M. E. Quinones-Mateu, and E. J. Arts. 2000. Variable sensitivity of CCR5-tropic human immunodeficiency virus type 1 isolates to inhibition by RANTES analogs. J. Virol. 74:4868-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tuttle, D. L., J. K. Harrison, C. Anders, J. W. Sleasman, and M. M. Goodenow. 1998. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J. Virol. 72:4962-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Velazquez-Campoy, A., M. J. Todd, S. Vega, and E. Freire. 2001. Catalytic efficiency and vitality of HIV-1 proteases from African viral subtypes. Proc. Natl. Acad. Sci. USA 98:6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, and B. H. Hahn. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 81.Xin, K. Q., X. H. Ma, K. A. Crandall, H. Bukawa, Y. Ishigatsubo, S. Kawamoto, and K. Okuda. 1995. Dual infection with HIV-1 Thai subtype B and E. Lancet 346:1372-1373. [DOI] [PubMed] [Google Scholar]
- 82.Zaitseva, M., A. Blauvelt, S. Lee, C. K. Lapham, V. Klaus-Kovtun, H. Mostowski, J. Manischewitz, and H. Golding. 1997. Expression and function of CCR5 and CXCR4 on human Langerhans cells and macrophages: implications for HIV primary infection. Nat. Med. 3:1369-1375. [DOI] [PubMed] [Google Scholar]