Nine neurodegenerative diseases are caused by expanding CAG repeats coding for polyglutamine (polyGln) (1–4). These include Huntington's disease, dentatorubral and pallidoluysian atrophy, several forms of spino-cerebellar ataxia, and spinal and bulbar muscular atrophy. Within the central nervous system, each disease has a distinctive pattern of degeneration, with considerable overlap among the diseases (5, 6). The genes containing CAG repeats show no homology to each other outside of the glutamine repeats, and most are genes of unknown function. Thus, speculation concerning pathogenesis has focused on the polyGln expansion itself.
For all of these diseases, there is a threshold of repeat length that causes disease. This threshold varies somewhat among the different diseases, but is generally in the range of 35–45 consecutive glutamines. In all polyGln diseases, the age of disease onset is strongly correlated with polyGln length, so that above the threshold, a longer repeat results in an earlier age of onset.
A pathological hallmark of these diseases is the aggregation of mutant polyGln protein, resulting in the formation of intranuclear inclusion bodies. In some of the diseases, inclusions have been observed in the cytoplasm, dendrites, and axonal processes. The inclusions are generally seen in affected areas of the brain (7, 8), though not limited to those neurons most likely to degenerate (9). Thus, whether inclusions are responsible for neurotoxicity has been controversial. Some studies have indicated a correlation between polyGln-containing inclusions and disease progression (10). However, in other studies, inclusion formation was dissociated from cytotoxicity (11, 12). In fact, inclusion formation may be, in part, a reflection of cellular protective mechanisms (13). Nevertheless, the inclusions are a useful marker for pathology and may provide clues to pathogenesis.
Aggregation of mutant polyGln proteins can be observed biochemically using a filter trap assay (14, 15). Aggregation in vitro proceeds by means of a nucleation-dependent process and results in the accumulation of β-sheet rich fibrillar structures detected by electron microscopy. Thus, the polyGln aggregation pathway appears to resemble that of Aβ protein in Alzheimer's disease and α-synuclein in Parkinson's disease, as well as other amyloidogenic proteins (16–18). Even if polyGln inclusions are not the major toxic species, the aggregation process appears linked to pathogenesis. Therefore, it is critical to understand the structure of both normal and mutant polyGln stretches.
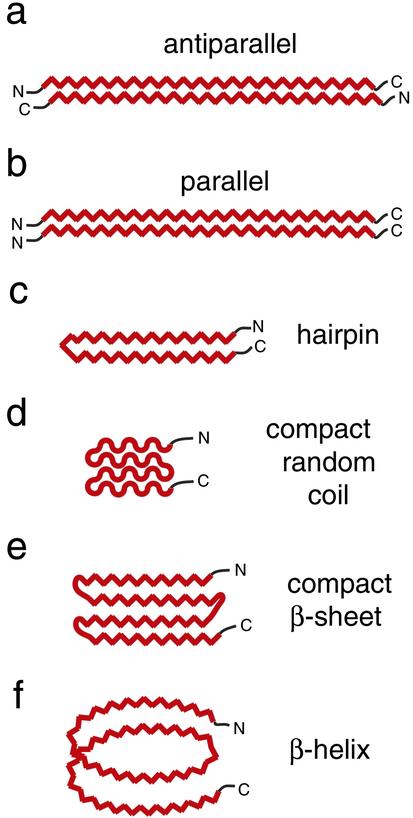
Detailed structural information on polyGln has been difficult to obtain, because both long and short stretches of synthetic polyGln peptides are quite insoluble. Nearly a decade ago, Max Perutz attempted to address this issue by using a Q15 peptide flanked by basic residues to improve its solubility. He found this peptide to adopt β-structure, and constructed an atomic model of poly(l-glutamine) consisting of antiparallel β-sheets held together by hydrogen bonds between main-chain and side-chain amides (Fig. 1a). This structure, described as a “polar zipper,” has been influential for studies of polyGln aggregation (14, 19, 20). Computer modeling studies have generated additional possible structures for expanded polyGln, such as parallel β-sheets (21), β-hairpins, and highly compact random coil (22) or β-sheet structures (Fig. 1 b–e). Based on x-ray diffraction and electron microscopy data, Perutz and his group (23, 24) have recently suggested a polyGln β-helix model with 20 residues per turn (Fig. 1f).
Figure 1.
Schematic representation of proposed structural models for aggregated mutant polyGln. β-Sheet is shown as a zig-zag. Expanded polyGln as an extended antiparallel β-sheet, first described by Max Perutz as a “polar zipper” (a), or as a parallel β-sheet (b). (c) An antiparallel β-hairpin comprised of two β-strands and a single β-turn. A highly compact structure, consisting of four antiparallel random coil (d) or β-strand (e) elements. (f) A parallel β-helix with 20 residues per turn. For simplicity, two polyGln molecules are shown each for a and b, whereas a single polyGln molecule is depicted in c–f.
Biophysical analysis of synthetic or recombinant polyGln peptides with stretches containing 5–44 consecutive glutamines have demonstrated that monomeric polyGln is unstructured (25–27). In contrast, expanded polyGln aggregates derived from isolated polyGln peptides or from recombinant polyGln-containing proteins adopt β-sheet structure, as shown by x-ray fiber diffraction studies, circular dichroism, Fourier transform infrared spectroscopy, and other methods (20, 25, 28, 29). Thus, it is likely that expanded polyGln sequences in the aggregated state are β-sheets, though detailed structural information is currently not available.
In a recent issue of PNAS, Thakur and Wetzel (30) provide a mutational analysis to address the question of polyGln aggregate structure. Previously, the Wetzel laboratory demonstrated that polyGln peptides have aggregation properties similar to exon-1 huntingtin (25). In the current study, they used an ingenious strategy of inducing β-turns in synthetic peptides with Pro–Gly pairs at different intervals within a long polyGln stretch. To enhance the solubility of their peptides, they incorporated charged residues at both ends of the polyGln peptides, as introduced by Max Perutz and colleagues. Thakur and Wetzel investigated the influence of Pro–Gly mutations on formation of fibers, as detected by aggregation assays and electron microscopy, and found that peptides (ranging in total length from 46 to 50 aa) consisting of four Q9 or Q10 elements interspersed with three Pro–Gly elements (termed PGQ9 or PGQ10) undergo spontaneous aggregation nearly as efficiently as Q45. PGQ7 and PGQ8 peptides aggregate much less readily. Furthermore, a sequence containing d-prolines (PDGQ9), which have a stronger preference for β-turns, aggregates more efficiently than the same peptide with l-prolines. By contrast, addition of a P residue in the center of a Q9 element blocked the peptide's ability to aggregate.
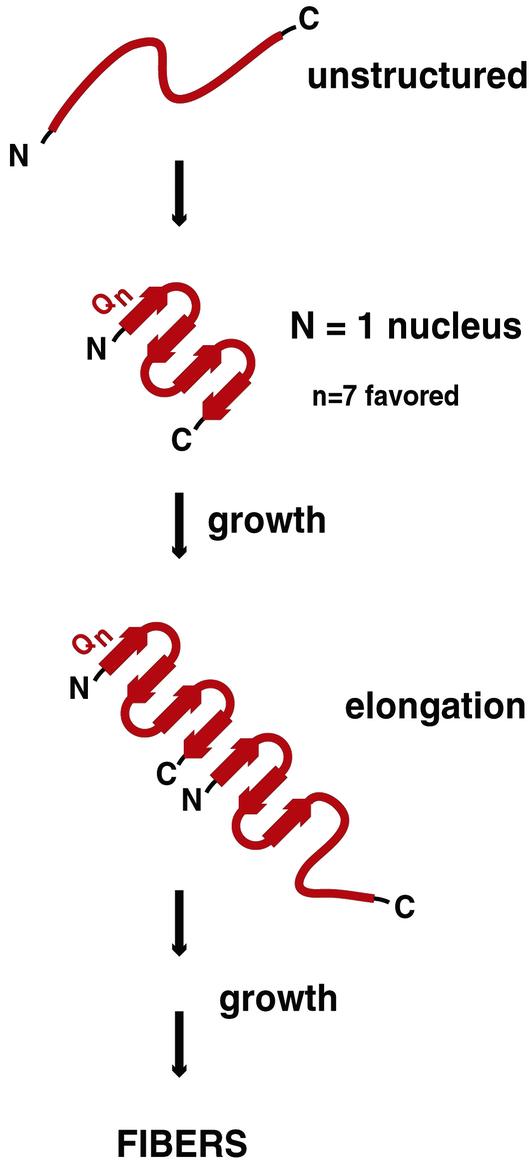
These data are suggestive of a model comprised of alternating β-strand and β-turn elements, with an optimum of seven consecutive glutamines within each β-strand (shown in Fig. 2). Based on electron microscopy data, the morphology of fibers generated with the PGQ9 peptide could be described as similar (though perhaps not identical) to that for the Q45 peptide. In addition, both the PGQ9 and the Q45 peptide were recognized by an anti-polyGln antibody selective for aggregates. Further, both peptides supported heterologous seeding (incorporation of normal length polyGln peptides into the expanded polyGln aggregate) in a similar manner. Thus, this study provides the first experimental data on the structure of aggregated polyGln, and is compatible with a model comprised of alternating β-strands and turns.
Figure 2.
Model of polyGln aggregate initiation and elongation as proposed by Thakur and Wetzel (30). Before the conformational change that initiates disease pathogenesis, mutant polyGln lacks secondary structure. A polyGln monomer undergoes a structural transition to a four-stranded antiparallel β-sheet, with an optimum of seven glutamine residues per β-strand (extended chain). This structured monomer serves as a nucleus for binding of a second unstructured monomer. Binding of the disordered monomer to the ordered nucleus results in acquisition of β-structure in the newly added monomer, providing a new elongation site, and is referred to as template-assisted or “dock-and-lock” elongation. Adapted from Chen et al. (28).
To further investigate the pathway of fiber assembly, Wetzel and colleagues recently conducted a kinetic analysis of polyGln aggregation at different concentrations of synthetic peptide (28). Their results, along with results of the present study, are summarized in Fig. 2. From their kinetic data, they conclude that aggregation of polyGln is initiated by a monomer that functions as the critical nucleus. They suggest that fibril formation proceeds via linear additions of single polyGln molecules, after nucleation events consisting of a random coil to β-sheet transition within an individual monomer. However, this analysis is indirect. An alternative pathway involving oligomeric intermediates has been suggested by biochemical and morphological studies of polyGln aggregation in the context of huntingtin exon-1 protein (29).
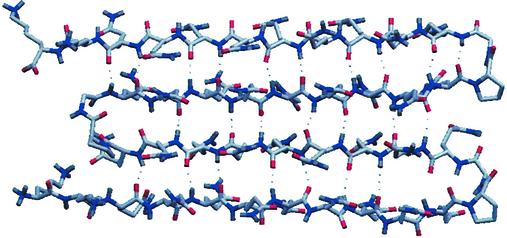
The present study provides the first experimental interventional data on the structure of polyGln within aggregates. It strongly supports a compact β-structure, rather than an extended strand model. Importantly, this structure is compatible with the polar zipper model, as hydrogen bonds are between main chains atoms and do not involve the side chains. A computer-generated drawing of the compact β-structure, with Pro–Gly residues at each β-turn, is depicted in Fig. 3. The strategy of using prolines for the introduction of β-turns is likely to be useful in future studies. This model provides testable hypotheses that can be addressed using other biophysical techniques, such as atomic force microscopy, NMR spectroscopy, electron paramagnetic resonance, and Raman spectroscopy. However, one limitation of the present study is that all data were derived from synthetic polyGln. Similar studies will need to be carried out with exon-1 huntingtin and constructs encoding fragments or full-length versions of other polyGln proteins.
Figure 3.
Computer-generated drawing of PGQ9 as a four-stranded antiparallel β-sheet. Only main-chain–main-chain hydrogen bonds were built into the model. The stretches containing nine glutamine residues were built as antiparallel β-strands and the Pro–Gly pairs were built as turns. Atoms are colored as follows: carbons, red; nitrogens, blue; oxygens, red.
At present, the mechanism of polyGln-mediated toxicity is not well understood. Mutant polyGln may assume an abnormal conformation, allowing it to associate with short polyGln stretches in other critical cellular proteins, rendering them inactive. Biochemical data from the Wetzel laboratory support this possibility (25). The transcription factor CREB binding protein has been proposed as a target (31–33), as has the normal allele of huntingtin itself (34). Alternatively, abnormal interactions with transcription factors could occur by mechanisms not directly involving the expanded polyGln region (35–38). Another interesting possible mechanism of toxicity is inhibition of the proteasome by the aggregated form of mutant polyGln protein (39, 40).
A key issue is the relationship between polyGln aggregation and cellular toxicity. As described above, the role of aggregation in toxicity has been controversial. Recent studies from the Wetzel laboratory have indicated that delivery of aggregated polyGln into cell nuclei is toxic (41). However, the exact nature of the material in the cells is uncertain. For delivery into cells, the aggregates were first sonicated and filtered. Thus, the toxic material could potentially be soluble monomer, an intermediate in the aggregation pathway, such as a monomeric nucleus as proposed by Thakur and Wetzel (30), or an oligomeric intermediate (29), as well as a small aggregate.
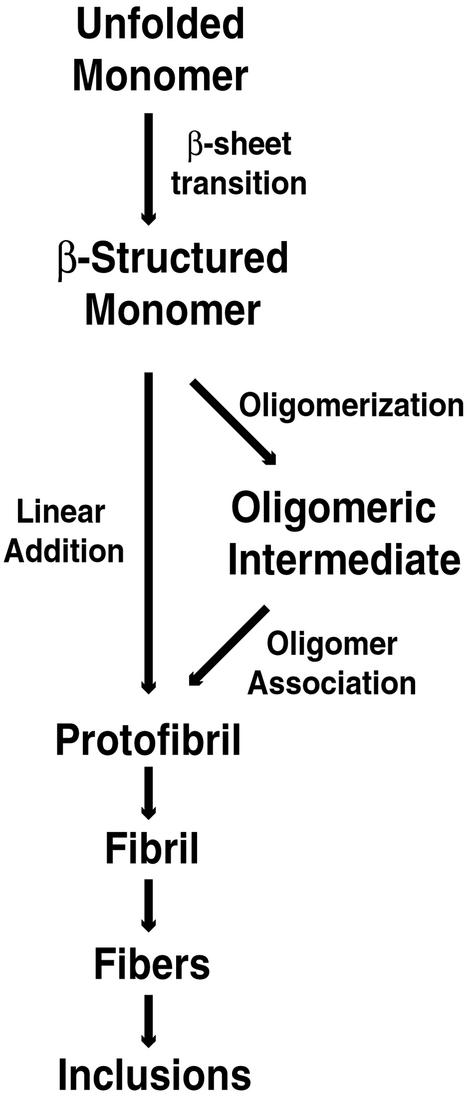
The pathway of polyGln-mediated aggregation is beginning to emerge, but is still poorly understood (Fig. 4). It is unclear whether aggregation proceeds via linear addition of single molecules or whether there are oligomeric intermediates. In addition, the stage in the aggregation pathway that causes cell death is unknown. Whatever the mechanism, a critical first step to understanding the pathogenesis associated with polyGln-based neurodegenerative diseases is elucidating the structural basis of polyGln aggregation. Mutational analysis can now be used to answer some of these questions. PolyGln aggregation has already been used as a target for therapeutic development (42–47). A better structural understanding aggregation of polyGln, as well as aggregation of other proteins, may lead to development of rational therapeutics for polyGln diseases and for other neurodegenerative and protein aggregation diseases (48).
Figure 4.
Hypothetical pathway of polyGln-mediated aggregation and inclusion formation. Unstructured polyGln monomer undergoes a structural conversion to β-sheet, resulting in the formation of protofibrillar intermediates. This step may proceed through a linear growth mechanism or through assembly of oligomeric intermediates. Protofibril assembly is followed by fibril formation, resulting in the characteristic inclusions observed in polyGln diseases and other amyloid-like diseases.
Footnotes
See companion article on page 17014 in issue 26 of volume 99.
References
- 1.Fischbeck K H. Brain Res Bull. 2001;56:161–163. doi: 10.1016/s0361-9230(01)00577-9. [DOI] [PubMed] [Google Scholar]
- 2.Huang C C, Faber P W, Persichetti F, Mittal V, Vonsattel J P, MacDonald M E, Gusella J F. Somatic Cell Mol Genet. 1998;24:217–233. doi: 10.1023/b:scam.0000007124.19463.e5. [DOI] [PubMed] [Google Scholar]
- 3.Zoghbi H Y, Orr H T. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 4.Ross C. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 5.Ross C A. Neuron. 1995;15:493–496. doi: 10.1016/0896-6273(95)90138-8. [DOI] [PubMed] [Google Scholar]
- 6.Orr H T. Genes Dev. 2001;15:925–932. doi: 10.1101/gad.888401. [DOI] [PubMed] [Google Scholar]
- 7.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 8.Becher M W, Kotzuk J A, Sharp A H, Davies S W, Bates G P, Price D L, Ross C A. Neurobiol Dis. 1998;4:387–397. doi: 10.1006/nbdi.1998.0168. [DOI] [PubMed] [Google Scholar]
- 9.Kuemmerle S, Gutekunst C A, Klein A M, Li X J, Li S H, Beal M F, Hersch S M, Ferrante R J. Ann Neurol. 1999;46:842–849. [PubMed] [Google Scholar]
- 10.Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 11.Saudou F, Finkbeiner S, Devys D, Greenberg M E. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 12.Cummings C J, Reinstein E, Sun Y, Antalffy B, Jiang Y, Ciechanover A, Orr H T, Beaudet A L, Zoghbi H Y. Neuron. 1999;24:879–892. doi: 10.1016/s0896-6273(00)81035-1. [DOI] [PubMed] [Google Scholar]
- 13.Kopito R R. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 14.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates G P, Davies S W, Lerach H, Wanker E E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 15.Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates G P, Lehrach H, Wanker E E. Proc Natl Acad Sci USA. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serpell L C, Berriman J, Jakes R, Goedert M, Crowther R A. Proc Natl Acad Sci USA. 2000;97:4897–4902. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rochet J C, Lansbury P T., Jr Curr Opin Struct Biol. 2000;10:60–68. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 18.Serio T R, Cashikar A G, Kowal A S, Sawicki G J, Moslehi J J, Serpell L, Arnsdorf M F, Lindquist S L. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 19.Perutz M. Protein Science. London: Cambridge Univ. Press; 1994. pp. 1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perutz M F. Brain Res Bull. 1999;50:467. doi: 10.1016/s0361-9230(99)00137-9. [DOI] [PubMed] [Google Scholar]
- 21.Lathrop R H, Casale M, Tobias D J, Marsh J L, Thompson L M. Proc Int Conf Intell Syst Mol Biol. 1998;6:105–114. [PubMed] [Google Scholar]
- 22.Starikov E B, Lehrach H, Wanker E E. J Biomol Struct Dyn. 1999;17:409–427. doi: 10.1080/07391102.1999.10508374. [DOI] [PubMed] [Google Scholar]
- 23.Perutz M F, Pope B J, Owen D, Wanker E E, Scherzinger E. Proc Natl Acad Sci USA. 2002;99:5596–5600. doi: 10.1073/pnas.042681599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perutz M F, Finch J T, Berriman J, Lesk A. Proc Natl Acad Sci USA. 2002;99:5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Berthelier V, Yang W, Wetzel R. J Mol Biol. 2001;311:173–182. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- 26.Masino L, Kelly G, Leonard K, Trottier Y, Pastore A. FEBS Lett. 2002;513:267–272. doi: 10.1016/s0014-5793(02)02335-9. [DOI] [PubMed] [Google Scholar]
- 27.Altschuler E L, Hud N V, Mazrimas J A, Rupp B. J Pept Res. 1997;50:73–75. doi: 10.1111/j.1399-3011.1997.tb00622.x. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Ferrone F A, Wetzel R. Proc Natl Acad Sci USA. 2002;99:11884–11889. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirier M A, Li H, Macosko J, Cai S, Amzel M, Ross C A. J Biol Chem. 2002;277:41032–41037. doi: 10.1074/jbc.M205809200. [DOI] [PubMed] [Google Scholar]
- 30.Thakur A K, Wetzel R. Proc Natl Acad Sci USA. 2002;99:17014–17019. doi: 10.1073/pnas.252523899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nucifora F C, Jr, Sasaki M, Peters M F, Huang H, Cooper J K, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson V L, et al. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 32.McCampbell A, Taylor J P, Taye A A, Robitschek J, Li M, Walcott J, Merry D, Chai Y, Paulson H, Sobue G, et al. Hum Mol Genet. 2000;9:2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- 33.McCampbell A, Taye A A, Whitty L, Penney E, Steffan J S, Fischbeck K H. Proc Natl Acad Sci USA. 2001;98:15179–15184. doi: 10.1073/pnas.261400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuccato C, Ciammola A, Rigamonti D, Leavitt B R, Goffredo D, Conti L, MacDonald M E, Friedlander R M, Silani V, Hayden M R, et al. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 35.Steffan J S, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu Y Z, Gohler H, Wanker E E, Bates G P, Housman D E, Thompson L M. Proc Natl Acad Sci USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steffan J S, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol B L, Kazantsev A, Schmidt E, Zhu Y Z, Greenwald M, et al. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 37.Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, Naruse S, Kimura T, Koide R, Nozaki K, Sano Y, et al. Nat Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 38.Dunah A W, Jeong H, Griffin A, Kim Y M, Standaert D G, Hersch S M, Mouradian M M, Young A B, Tanese N, Krainc D. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 39.Bence N F, Sampat R M, Kopito R R. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 40.Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker E E. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang W, Dunlap J R, Andrews R B, Wetzel R. Hum Mol Genet. 2002;11:2905–2917. doi: 10.1093/hmg/11.23.2905. [DOI] [PubMed] [Google Scholar]
- 42.Heiser V, Scherzinger E, Boeddrich A, Nordhoff E, Lurz R, Schugardt N, Lehrach H, Wanker E E. Proc Natl Acad Sci USA. 2000;97:6739–6744. doi: 10.1073/pnas.110138997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heiser V, Engemann S, Bröcker W, Dunkel I, Boeddrich A, Waelter S, Nordhoff E, Lurz R, Schugardt N, Rautenberg S, et al. Proc Natl Acad Sci USA. 2002;99,(Suppl. 4):16400–16046. doi: 10.1073/pnas.182426599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kazantsev A, Walker H A, Slepko N, Bear J E, Preisinger E, Steffan J S, Zhu Y Z, Gertler F B, Housman D E, Marsh J L, et al. Nat Genet. 2002;30:367–376. doi: 10.1038/ng864. [DOI] [PubMed] [Google Scholar]
- 45.Khoshnan A, Ko J, Patterson P H. Proc Natl Acad Sci USA. 2002;99:1002–1007. doi: 10.1073/pnas.022631799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marsh J L, Walker H, Theisen H, Zhu Y Z, Fielder T, Purcell J, Thompson L M. Hum Mol Genet. 2000;9:13–25. doi: 10.1093/hmg/9.1.13. [DOI] [PubMed] [Google Scholar]
- 47.Sacchettini J C, Kelly J W. Nat Rev Drug Discovery. 2002;1:267–275. doi: 10.1038/nrd769. [DOI] [PubMed] [Google Scholar]
- 48.Bennett M J, Huey-Tubman K E, Herr A B, West A P, Jr, Ross S A, Bjorkman P J. Proc Natl Acad Sci USA. 2002;99:11634–11639. doi: 10.1073/pnas.182393899. [DOI] [PMC free article] [PubMed] [Google Scholar]